Abstract
The non-receptor tyrosine kinase SRC is frequently deregulated in human colorectal cancer (CRC), and SRC increased activity has been associated with poor clinical outcomes. In nude mice engrafted with human CRC cells, SRC over-expression favors tumor growth and is accompanied by a robust increase in tyrosine phosphorylation in tumor cells. How SRC contributes to this tumorigenic process is largely unknown. We analyzed SRC oncogenic signaling in these tumors by means of a novel quantitative proteomic analysis. This method is based on stable isotope labeling with amino acids of xenograft tumors by the addition of [13C6]-lysine into mouse food. An incorporation level greater than 88% was obtained in xenograft tumors after 30 days of the heavy lysine diet. Quantitative phosphoproteomic analysis of these tumors allowed the identification of 61 proteins that exhibited a significant increase in tyrosine phosphorylation and/or association with tyrosine phosphorylated proteins upon SRC expression. These mainly included molecules implicated in vesicular trafficking and signaling and RNA binding proteins. Most of these proteins were specific targets of SRC signaling in vivo, as they were not identified by analysis via stable isotope labeling by amino acids in cell culture (SILAC) of the same CRC cells in culture. This suggests that oncogenic signaling induced by SRC in tumors significantly differs from that induced by SRC in cell culture. We next confirmed this notion experimentally with the example of the vesicular trafficking protein and SRC substrate TOM1L1. We found that whereas TOM1L1 depletion only slightly affected SRC-induced proliferation of CRC cells in vitro, it drastically decreased tumor growth in xenografted nude mice. We thus concluded that this vesicular trafficking protein plays an important role in SRC-induced tumor growth. Overall, these data show that SILAC analysis in mouse xenografts is a valuable approach for deciphering tyrosine kinase oncogenic signaling in vivo.
The non-receptor tyrosine kinase (TK)1 SRC mediates cellular signaling induced by growth factors and integrins (1) leading to cell growth, survival, and migration. It also has oncogenic activity when deregulated, a role originally described for the constitutively active v-SRC (2) that has since been observed in a large variety of human cancers (3). Remarkably, elevated SRC kinase activity has been found in more than 80% of colorectal cancers (CRCs) to levels (5- to 10-fold) consistent with oncogenic properties (4). Moreover, SRC deregulation has been associated with poor clinical outcomes (3), suggesting an additional function of SRC during late tumorigenesis. SRC deregulation largely occurs in the absence of mutations in the SRC gene. Instead, it primarily involves protein over-expression (2) and inhibition of SRC negative regulators, such as the transmembrane protein Cbp/PAG (5, 6). A large body of evidence indicates that SRC deregulation is an important event in colon tumorigenesis (3, 6). Indeed, SRC controls growth, survival, and invasion of some CRC cell lines in vitro (4). Moreover, it contributes to tumor growth, angiogenesis, and metastasis formation in mouse colon tumor xenograft models (7–11). However, our knowledge of the SRC-dependent oncogenic signaling pathway in CRC is largely incomplete, mostly because the majority of data have been obtained in two-dimensional cell culture models. Moreover, the standard culture conditions of CRC cells do not allow the recapitulation of all the SRC-dependent signaling cascades that are activated during tumorigenesis to promote tumor growth, angiogenesis, and interactions with the microenvironment.
MS-based quantitative phosphoproteomic technology has been a valuable tool for deciphering signaling pathways initiated by a given TK (12). Particularly, the method of stable isotope labeling with amino acids in cell culture (SILAC) has been employed for the characterization of oncogenic TK signaling pathways in cell culture, including HER2 (13) and BCR-ABL (14). We recently used this powerful approach to investigate SRC-dependent oncogenic signaling in CRC cells (15) and identified the first SRC-dependent tyrosine “phosphoproteome” in these cells. Additionally, we found that SRC phosphorylated a small cluster of TKs that mediate its oncogenic activity, thus uncovering a TK network that is important for the induction of CRC cell growth (15). Whether these signaling processes also operate in vivo is, however, currently unknown.
SRC oncogenic signaling could be investigated in vivo using similar MS-based quantitative phosphoproteomic approaches in animal models or tumor biopsies. However, the application of the SILAC method in vivo has been challenging until recently because it requires efficient protein labeling in different tissues, which is conditioned by the rate of de novo protein synthesis. Recently, Mann et al. described the successful development of a SILAC approach for labeling mice that is based on the addition of [13C6]-lysine to their food (16). They reported complete labeling from the F2 generation. Similar SILAC approaches were then described for additional multicellular organisms, such as worms (17), flies (18), and zebrafish (19). Here, we report a similar SILAC approach in which we labeled tumors in nude mice xenografted with human CRC cells. We reasoned that the high rate of de novo protein synthesis occurring in tumors should allow efficient tumor labeling in a short period of time. Indeed, we obtained consistent (>88%) labeling of the tumor proteome by feeding xenografted mice with the SILAC mouse diet for only 30 days. We then used this approach to compare the tyrosine phosphoproteome of SRC over-expressing tumors (labeled with heavy amino acids) and of control tumors (labeled with light amino acids) and report the first SRC-dependent tyrosine phosphoproteome of CRC in vivo. Finally, comparison of the in vivo and in vitro SRC-dependent tyrosine phosphoproteomes showed that some of the SRC substrates were specifically activated only in CRC xenograft tumors, and not in cultured CRC cells.
EXPERIMENTAL PROCEDURES
Reagents
Human SRC and mouse Tom1L1 were sub-cloned in, respectively, pMX-ps-CESAR and pBABE-puro (20). The sequences used for the generation of the shRNA constructs (RNAi-Ready pSIREN-RetroQ, Clontech) were GACACTCGGTAGTCTATAC (negative control) and GATGAGTTATTAGCAGAAG (human TOM1L1). 13C615N4-arginine and 13C615N2-lysine were from EURISO-TOP, Saint Aubin, France, respectively; the anti-pTyr columns were obtained from Upstate Inc. (Charlottesville, NC). Antibodies used in this study were 4G10 (a gift from P. Mangeat), pY100 (CST, Danvers, MA), anti-CBL (sc-170, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Ezrin (21), anti-HRS (A-5, Alexis Biochemicals San Diego, CA), anti-HnRNPA1 (clone 4B10, Sigma Aldrich), anti-HuR (sc-5261, Santa Cruz Biotechnology, Santa Cruz, CA), anti-TOM1L1 (22), anti-Tensin 3 (sc-134908; Santa Cruz Biotechnology, Santa Cruz, CA), anti-pY118-Paxillin (BIOSOURCE), anti-Cortactin p80/85 (clone 4F11; Upstate Inc.), anti-Paxillin (clone 349, a gift from A. Blangy), anti-pY410-p130 CAS (Cell Signaling Technology Danvers, MA), anti-p130 CAS (BD Biosciences), anti-HIF1-α (H1alpha67; Novus Biologicals, Cambridge, UK), anti-Ki67 (Sp6; Thermo Scientific), anti-CD34 (sc-18917; Santa Cruz Biotechnology), anti-cleaved Caspase-3 (5AE1, Cell Signaling Technology), anti-SRC family kinases (23), anti-pY418-SRC (BIOSOURCE), and anti-Tubulin (a gift from N. Morin).
Cell Infections and Growth
SW620 cells (ATCC, Rockville, MD) were grown, infected, and selected as described in Ref. 24 to generate stable expressors in a polyclonal background. SRC expressors were isolated via FACS (Montpellier RIO Imaging facility, IGMM Cytometry) and shRNA expressing cells by selection with 1 μg/ml puromycin.
Mouse Xenografts, [13C6]-lysine Tumor Labeling, and Protein Extraction
In vivo experiments were performed in compliance with the French guidelines for experimental animal studies (Direction des Services Vétérinaires, Ministère de l'Agriculture, Agreement No. B 34–172-27) and fulfilled the UK Coordinating Committee on Cancer Research guidelines for the welfare of animals in experimental neoplasia. Swiss nu/nu (nude) mice (Charles River, L'Arbresle, France) were injected subcutaneously with 2 × 106 cells in the flank. Tumor growth was assessed by calculating the tumor volume on the indicated days. For stable isotope labeling in mouse xenografts, animals were fed with a heavy [13C6]-lysine diet for 18 or 30 days using the CIL's Mouse Feed Labeling Kit (EURISO-TOP, Saint Aubin, France). Animals were then sacrificed, and tumors were dissected, flash frozen in liquid nitrogen, and kept at −80 °C. Protein extraction from frozen tumors was performed at 4 °C in lysis buffer (20 mm Hepes, 150 mm NaCl, 0.5% Triton, 6 mm β-octylglucoside, 100 μm orthovanadate, 100 μm aprotinin, 100 mm DTT, 100 mm NAF) using a size 21 Duall Glass Tissue Grinder.
Mass Spectrometry Analysis
Phosphotyrosine immunoaffinity purification (using a mixture of 4G10 and pY100 antibodies) and tryptic digestion were essentially performed as described in Ref. 25. Purified proteins were separated on SDS-PAGE gels. Lysine C-digested samples (1 μl) obtained from 15 gel slices were analyzed on-line using nanoflow HPLC-nano-electrospray ionization on an LTQ-Orbitrap XL mass spectrometer (ThermoScientific, Waltham, MA) coupled with an Ultimate 3000 HPLC apparatus (Dionex, Amsterdam, The Netherlands). Desalting and pre-concentration of the samples were performed on-line on a Pepmap® precolumn (0.3 mm × 10 mm). A gradient consisting of 0%–40% B for 60 min and 80% B for 15 min (A = 0.1% formic acid, 2% acetonitrile in water; B = 0.1% formic acid in acetonitrile) at 300 nl/min was used to elute peptides from the capillary (0.075 mm × 150 mm) reverse-phase column (Pepmap®, Dionex) fitted with an uncoated silica PicoTip Emitter (NewObjective, Woburn, MA). Spectra were recorded using Xcalibur software (version 2.0.7; ThermoScientific). Spectra were acquired with the instrument operating in the information-dependent acquisition mode throughout the HPLC gradient. Survey scans were acquired in the Orbitrap system with the resolution set at a value of 60,000. Up to five of the most intense ions per cycle were fragmented and analyzed in the linear trap. Peptide fragmentation was performed using nitrogen gas on the most abundant and at least doubly charged ions detected in the initial MS scan with an active exclusion time of 1 min. Ion selection was set at 5.000 counts.
Analysis was performed using MaxQuant software (version 1.1.1.36). All MS/MS spectra were searched using Andromeda against a database consisting of a combination of Homo sapiens and Mus musculus CPS databases (97,681 entries; released June 2011) and 250 classical contaminants, containing forward and reverse entities. Search parameters were the default parameters when using MaxQuant with a slight modification: the search precursor mass tolerance was set at 20 ppm, and the main search (after recalibration) at 6 ppm. A maximum of two miscleavages were allowed. The search was performed allowing the following variable modifications: Oxidation (M) and Phospho (STY). FDR was set at 0.01 for peptides and proteins, and the minimal peptide length was six. Quantification was also performed using standard parameters; we considered only proteins with at least two identified/quantified peptides for further analysis, after the elimination of reverse and contaminant entries. The statistical validity of the results and the determination of over-represented proteins were assessed using significance B, as defined using Perseus (version 1.1.1.36, standard parameters) on the logarithmized normalized ratio (base 2). Identified proteins and peptides are included in supplemental Tables S1 and S4. To estimate the proportion of proteins specific to mice or humans or common to both species, we classified those proteins based on the identification of peptides specific to one of the two species. In the absence of any specific peptide, proteins were classified as “common to both.” Comparison of our in vivo labeling methodology with label-free analysis was performed as described in Ref. 18 and was based on data obtained from MaxQuant from analysis obtained from two subsequent LC-MS/MS runs using the same sample (two technical replicates). We then compared the ratio of the ratio to evaluate the precision of the calculated ratio using a labeling strategy and the ratio of the peptide intensity to evaluate the label-free precision. SRC targets were assigned as “novel” based on the absence of published data supporting any interaction with or phosphorylation by SRC. Interrogation of the literature was performed with the use of different names for the same protein available on the PhosphoSitePlus Web site.
SILAC Analysis
SILAC (13C615N4-Arg and 13C615N2-Lys as heavy amino acids), phosphotyrosine immunoaffinity purification, and tryptic digestion were performed essentially as described in Ref. 15, except that cell (2 × 108) treatment with orthovanadate was omitted. Purified proteins were separated on SDS-PAGE gels, and trypsin-digested samples (1 μl) obtained from 15 cut gel slices were analyzed using an LTQ-Orbitrap XL apparatus as described for the SILAC analysis in mouse xenografts. MaxQuant and Perseus analyses were performed as described in the mouse SILAC section, but spectra were searched against only human and contaminant entries of the database (46,713 entries, June 2011 release) with trypsin as an enzyme. Identified proteins and peptides are included in supplemental Tables S2 and S5.
Biochemistry
Immunoprecipitation and Western blotting were performed as described in Ref. 24. Immunoprecipitates or 20 to 50 μg of whole cell lysates/lane were loaded on 9% SDS-PAGE gels and then transferred onto Immobilon membranes (Millipore Molsheim, France). Detection was performed using the ECL System (Amersham Biosciences). Optimal exposure times of membranes were used, and protein expression and phosphorylation were quantified using ImageQuant TL software (GE Healthcare) and adjusted for background noise and protein loading.
Immunohistochemistry
Formalin-fixed and paraffin-embedded xenograft tissues were cut into 4-μm sections. After deparaffination and rehydration, tumor sections were subjected to 0.3% H2O2 for 20 min to quench endogenous peroxidase activity, treated with 20% normal horse serum in PBS for 45 min and Avidin/Biotin Kit (Vector Laboratories Burlingame, CA), and incubated at 4 °C overnight with the primary antibodies. Secondary biotinylated antibodies (Vector Laboratories) (1:400 dilution) were incubated for 1 h at room temperature. Detection was performed with Vectastain ABC kit (Vector Laboratories) using the peroxidase substrate kit (Vector Laboratories). Counterstaining with hematoxylin was done for 1 min. Sections were scanned using Nanozoomer Slide Scanner (Hamamatsu City, Japan), and quantifications were performed using Aperio's ImageScope by calculating the staining density as the number of positive pixels out of the total number of pixels.
In situ Hybridization
Tissue sections were incubated in 7 mm citrate buffer (pH 6) at 100 °C for 20 min, followed by pepsin digestion for 15 min. After dehydration in graduated ethanol series (70%, 85%, 100%), sections were hybridized with a mouse COT1 DNA probe (Invitrogen, Carlsbad, CA) that had been labeled using a FISHBright 550 Red/Orange labeling kit (Kreatech, Amsterdam, The Netherlands). Briefly, pretreated tissue sections were co-denatured with 100 ng of mouse COT1 DNA labeled probe at 80 °C for 10 min, followed by hybridization at 37 °C for 16 h. Slides were next washed, stained with DAPI, and mounted according to the FISH Digestion Kit protocol (Kreatech, Amsterdam, The Netherlands). Hybridization was observed using a Leica fluorescence microscope, and 10 images were captured for each slide using a charge-coupled device camera driven by Isis software (Metasystems, Altlussheim, Germany).
RESULTS
SRC Increases Tumor Growth and Protein pTyr Content in Human CRC Xenografts
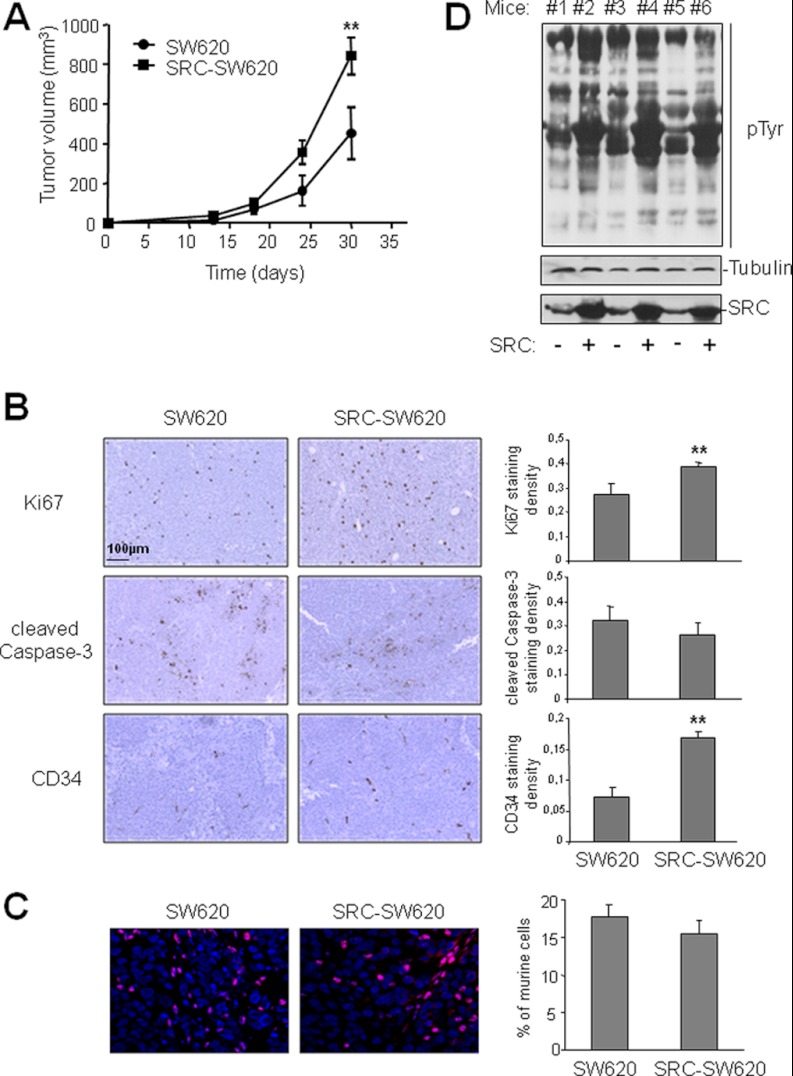
We first transduced wild type SRC retrovirally in SW620 cells, a human metastatic CRC cell line that exhibits a moderate level of endogenous SRC relative to other metastatic CRC cell lines (26). We previously reported that the expression of wild type SRC in these cells (SRC-SW620) is highly deregulated because of the absence of the SRC negative regulator Cbp/PAG (6). Indeed, a >5-fold increase in SRC protein level in SRC-SW620 cells was sufficient to strongly increase the pTyr content and significantly promote cell growth in both standard and soft agar culture conditions (15). Similarly, the growth of xenograft tumors induced by subcutaneous injection of SRC-SW620 cells in nude mice was >2-fold faster than that of tumors derived from parental SW620 cells (Fig. 1A). Although immuno-histochemical analysis of tumor sections showed no significant reduction of cell apoptosis upon SRC expression, 30% and 240% increases in cell proliferation and angiogenesis, respectively, were obtained in SRC-expressing tumors (Fig. 1B). Therefore, in vivo SRC oncogenic activity may rely on its capacity to promote cell proliferation and angiogenesis as previously reported (4). In situ hybridization of a mouse-specific probe indicated that between 16% and 18% of the cellular content of xenograft tumors was of mouse origin (fibroblasts, endothelial cells, pericytes, and leukocytes), and this proportion was not modified by SRC expression (Fig. 1C). This suggests that SRC overexpression does not affect stromal cell infiltration in the tumor. Similarly, SRC expression had no effect on tumor necrosis (10% to 12% of the whole surface of tumor sections analyzed was scored as necrotic; unpublished data). Interestingly, these SRC oncogenic effects were associated with a strong increase of the pTyr content in xenograft tumors in which SRC was over-expressed (Fig. 1D). Moreover, the SRC-dependent increase in pTyr was comparable in xenograft tumors, indicating that the level of SRC activity in the different tumors was similar. These data suggest that the increase in pTyr content upon SRC over-expression is directly linked to the enhanced growth of the xenograft tumors.
Fig. 1.
SRC increases tumor growth and pTyr content in CRC xenograft models. A, SRC promotes growth of xenograft tumors in nude mice. The mean tumor volume (in mm3) ± S.E. (n = 5) obtained after injection of parental SW620 cells (control, circle) or SW620 cells that over-express SRC (SRC-SW620, square) is presented over time. B, SRC promotes cell proliferation and angiogenesis in xenograft tumors. Representative sections (left-hand panels) and quantification (right-hand panels) of immunohistochemical analysis (left) showing cell proliferation (anti-ki67), apoptosis (anti-cleaved Caspase 3), and angiogenesis (anti-CD34) in xenograft tumors derived from SW620 and SRC-SW620 cells as described under “Experimental Procedures.” C, SRC expression does not affect stromal cell infiltration in tumors. A representative example (left) and quantification (right) of the percentage of mouse cells included in xenograft tumors derived from SW620 and SRC-SW620 cells. Tumor sections were labeled via in situ hybridization with a mouse-specific probe as described under “Experimental Procedures.” The merge between mouse labeled nuclei (red) and DAPI-stained nuclei (blue) is shown. D, SRC increased pTyr content in tumors. Western blotting of total tumor lysates from mice injected with SW620 (−) or SRC-SW620 (+) cells using the anti-phosphotyrosine 4G10 antibody. The levels of Tubulin and SRC are also shown. *p < 0.05 and **p < 0.01 using Student's t test.
Stable Isotope Labeling with Amino Acids in Mouse Xenografts
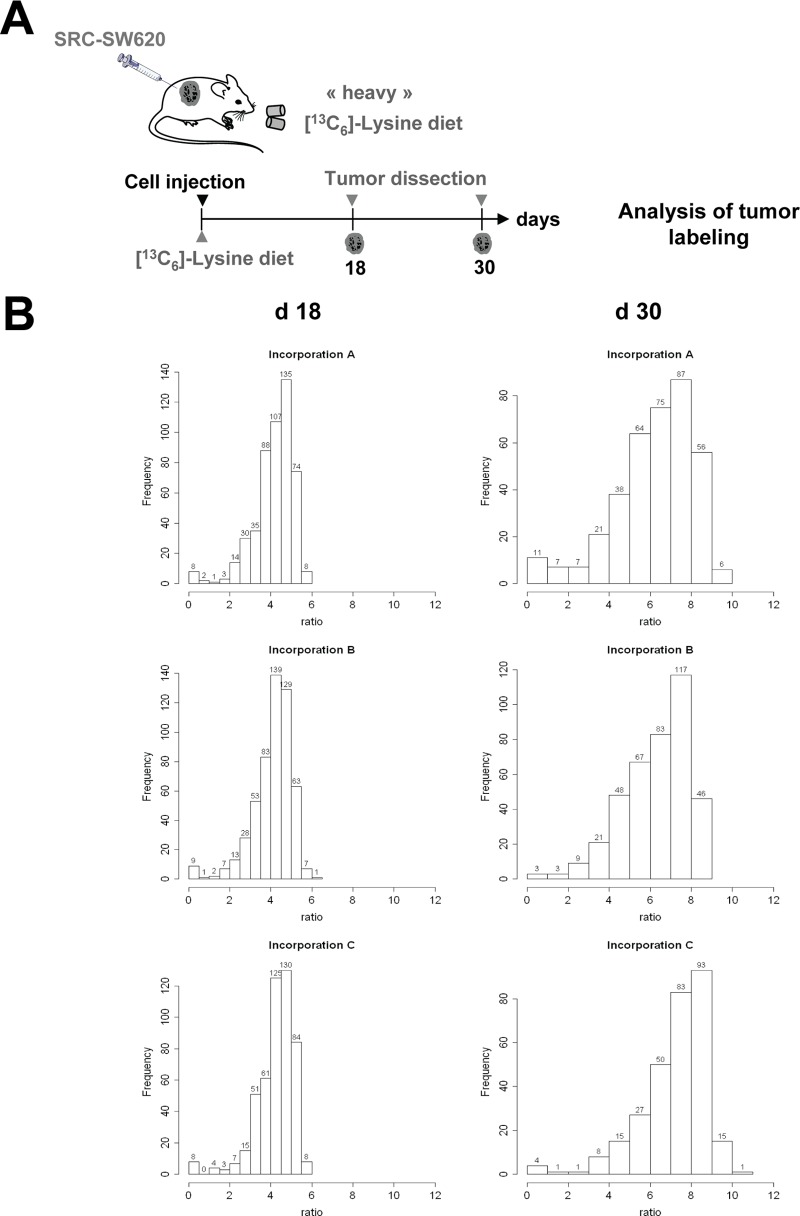
We then applied an MS-based quantitative phosphoproteomic method based on stable isotope labeling with amino acids in mouse xenografts to thoroughly characterize the SRC-dependent oncogenic signaling pathway in xenograft tumors. Mice were first subcutaneously injected with 2 × 106 SRC-SW620 cells and then fed with the heavy [13C6]-lysine diet, as done to obtain the SILAC mouse, but only during the time required for tumors to reach volumes of about 100 mm3 (18 days) and 900 mm3 (30 days). Indeed, we hypothesized that a short period of SILAC diet should be sufficient to achieve a good ratio of [13C6]-lysine incorporation in tumors, given their high de novo protein synthesis. Tumor proteins were then solubilized from isolated tumors and separated on one-dimensional SDS-PAGE gels. Three tumor samples for each time point (from a total of six animals) were then in-gel digested with the endoproteinase Lys-C and analyzed via LC-MS/MS. Digested peptides were then quantified based on the relative Lys intensities (see Fig. 2A for a schematic of the protocol). We observed a median SILAC ratio of 1:3.2 to 1:3.3 at day 18, indicating that at least 77% of the tumor protein had been newly synthesized during this period (Fig. 2B). This median ratio reached 1:7.4 at day 30, which corresponded to >88% of tumor protein labeling (Fig. 2B). These ratios were very consistent over time and in tumors from different animals, further validating our in vivo SILAC approach. In contrast, the median SILAC ratio of nontransformed surrounding tissue (i.e. muscle) reached 1.97, which corresponded to 66% of protein labeling (supplemental Fig. S1). Together, these results indicate that although it is insufficient for labeling nontransformed tissues of the host mice, a 30-day SILAC mouse diet is sufficient to label xenograft tumors to a level that is adequate for quantitative proteomic analysis.
Fig. 2.
Time course of [13C6]-lysine incorporation in xenograft tumors. A, schematic of the analysis of heavy [13C6]-lysine incorporation in mouse xenograft tumors. After subcutaneous injection of SRC-SW620 cells into the flanks of nude mice, animals were subjected to a heavy SILAC diet containing [13C6]-lysine for 18 or 30 days (three animals for each time point). B, histogram showing the distribution of the incorporation ratios in tumor proteins after 18 days (left) and 30 days (right) in six different animals.
Because between 16% and 18% of xenograft tumors are composed of mouse stromal cells, we also evaluated the SILAC ratio of the mouse-specific proteome present in xenograft tumor lysates. First, we observed that 16% to 21% of the tumoral proteome was composed of mouse-specific proteins, a proportion that was not significantly modified upon SRC expression (not shown). These data tightly correlate with the percentage of mouse cells observed in xenograft tumors. We next observed a median SILAC ratio of the mouse-specific proteome of 1:6 to 1:6.9 at day 30, indicating that at least 85% of the mouse tumor proteins had been newly synthesized during this period (not shown). We thus concluded that there was an 89.2% incorporation level of human cancer cells in xenograft tumors.
Finally, we aimed at comparing the precision of quantification of our in vivo labeling methodology with label-free analysis. To this end, we used the same strategy described by Sury et al. (18) in which the same sample is analyzed in both runs and the ratio of protein abundance in replicate 1 to that in replicate 2 should theoretically be 1 for all proteins. The spread of the experimental ratios allows the assessment of the precision of quantification. In the case of label-free analysis, we obtained ratio fold changes of between −2.96 and 2.44 following a normal distribution with a standard deviation of 0.62, and in the case of metabolic labeling, ratio fold changes varied from −1.65 to 0.78 following a normal distribution with a standard deviation of 0.15 (supplemental Fig. S2). Therefore, when using MaxQuant on xenograft samples, quantification with metabolic labeling exhibits higher precision than label-free quantification.
Quantitative Phosphoproteomics in Xenograft Tumors
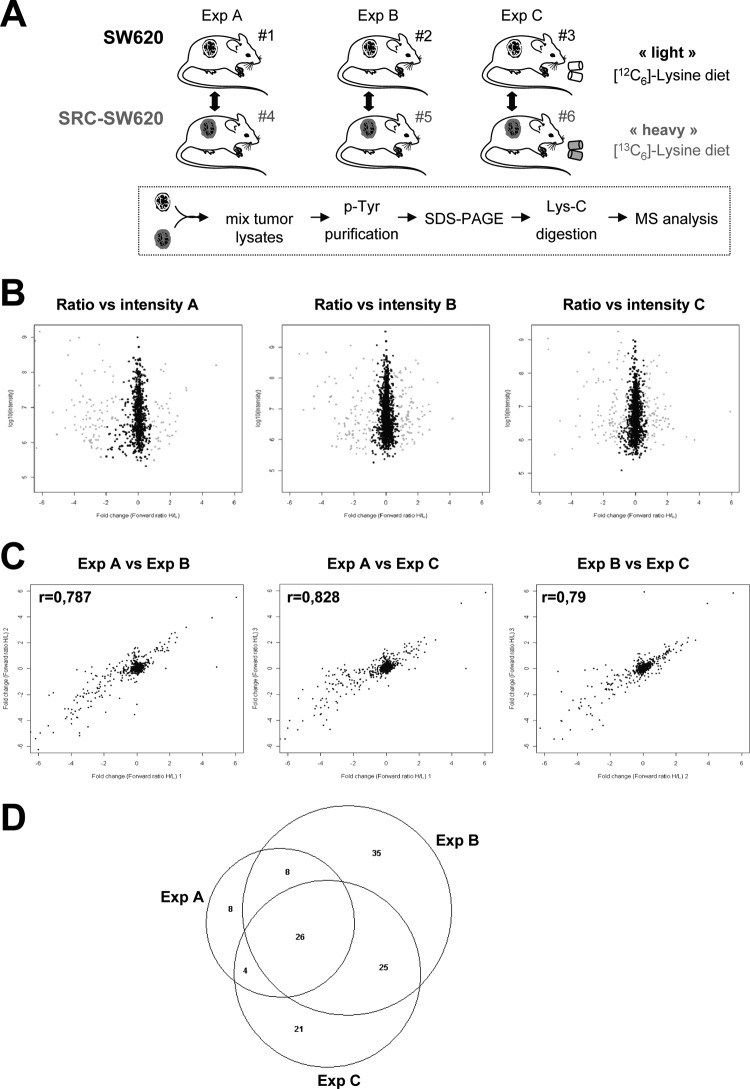
We next utilized this mouse SILAC approach to investigate the SRC-dependent oncogenic signaling pathway in xenografted tumors (Fig. 3A). SRC-SW620 cells were injected in three different animals that were fed with the “heavy” [13C6]-Lys-containing diet. As a control, parental SW620 cells were injected in three other mice that were fed with a “light” [12C6]-Lys-containing diet. After 30 days of this regimen, xenograft tumors were isolated and lysed, and three pairs of lysates were prepared by mixing (1:1) one SRC-SW620 xenograft tumor lysate with one control tumor lysate (experiments A, B, and C). pTyr proteins were then purified using anti-pTyr antibodies and analyzed via MS (25). Although this approach is not optimal for the identification of SRC phosphorylation sites, it allows the identification of SRC substrates and/or effectors (referred to as “SRC targets”), which provides unique and valuable information about the nature of SRC signaling operating in these tumors (27).
Fig. 3.
Analysis of SRC oncogenic signaling in xenograft tumors by SILAC mouse. A, schematic overview of the SILAC experimental procedure applied to mouse xenografts. B, SILAC in mouse xenografts after 30 days of heavy mouse diet allowed the quantification of tyrosine phosphorylation in three independent experiments. The heavy/light ratio for each protein is represented as a function of the signal intensity. Proteins are colored in gray when scored as significant by MaxQuant. C, scatter plot showing the correlation between pairs of the three independent experiments. Pearson correlation coefficient (r) calculated by Perseus is shown. D, Venn diagram showing the number of common and specific SRC targets identified in the three independent mouse SILAC experiments.
Quantitative phosphoproteomic analysis indicated that there were 46 proteins with a ratio significantly higher than 1 in experiment A, 94 in experiment B, and 72 in experiment C (Fig. 3B and supplemental Table S1). A pair-by-pair comparison of the MS data showed a good correlation (r = 0.8) (Fig. 3C). Some variability could be noticed for proteins with a ratio less than 1. This discrepancy could be explained at least in part by the incomplete labeling of purified mouse proteins with a ratio of <1 (supplemental Fig. S3). Additionally, it might also reflect the tyrosine phosphoproteome variability among grafted animals. When we focused on only the proteins with a ratio of >1, 26 were found to have increased pTyr content in all three experiments, and 61 had an increased in at least two experiments (Fig. 3D and Table I). We conclude that in SRC-SW620 xenograft tumors, the tyrosine phosphorylation of these 61 proteins or their association with pTyr proteins was increased by SRC over-expression. A list of these SRC targets is shown in Table I; 26 out of the 61 have been previously described as SRC targets using genetic (28, 29), proteomic (15, 30–35), or candidate strategies (36–38). Well-known substrates involved in SRC oncogenic signaling were identified in this analysis, including p130 CAS, FAK, and Tensin-3 (30, 39). Moreover, 35 of the identified proteins have never been described as potential SRC substrates, confirming the interest of such an approach. These include the seven transmembrane receptors coupled to heterotrimeric G-protein GPR155, vacuolar protein sorting-associated protein 37B, and (cytosine-5)-methyltransferase 1. We also attempted to evaluate the proportion of SRC targets that belonged to the host animal. Although at least 17% of all purified proteins contained mouse-specific peptides, only two mouse proteins (Clathrin Heavy Chain and Alix) had a significant ratio greater than 1 (supplemental Fig. S3). These two proteins are well-known SRC substrates that regulate endocytosis of growth factors and adhesive receptors (40, 41), a process that might contribute to the regulation of the interaction of the tumor with the micro-environment or the induction of tumor angiogenesis. This finding suggests that although most of the SRC targets identified via mouse SILAC can be attributed to the human CRC cells, at least 4.7% of them belong to the host animal and contribute to tumorigenesis.
Table I. SRC targets identified by SILAC mouse in xenograft tumors.
| Gene symbol | Protein name | Mean ratio | SRC target | |
|---|---|---|---|---|
| Significant in 3/3 | SRC | SRC | 7.24 | |
| TMEM192 | Transmembrane protein 192 | 5.63 | Novel | |
| PXN | Paxillin | 4.97 | Described in Refs. 5, 30, 35 | |
| SYT1 | Synaptotagmin | 4.80 | Novel | |
| BCAR1 | P130 CAS | 4.14 | Described in Refs. 5, 28, 30, 31, 35 | |
| GPR155 | G protein-coupled receptor 155 | 4.06 | Novel | |
| CTPS | CTP synthase | 3.83 | Described in Ref. 34 | |
| STX3 | Syntaxin-3 | 3.83 | Novel | |
| TNS3 | Tensin-3 | 3.49 | Described in Refs. 15, 35 | |
| PTK2 | FAK | 3.22 | Described in Refs. 15, 31, 35 | |
| ATP6V0D1 | V-ATPase subunit d 1 | 3.17 | Novel | |
| TOM1L1 | TOM1-like protein 1 | 2.90 | Described in Ref. 15 | |
| VPS37B | Vacuolar protein sorting 37B | 2.88 | Novel | |
| ARAP2 | Centaurin-delta-1 | 2.87 | Novel | |
| PDCD6IP | Alix | 2.79 | Described in Ref. 15 | |
| PEAK1 | Sgk269 | 2.79 | Described in Refs. 15, 30 | |
| STXBP2 | Syntaxin binding protein 2 | 2.78 | Novel | |
| MYOF | Myoferlin | 2.73 | Novel | |
| TRAPPC9 | Trafficking protein particle complex subunit 9/NIK-binding protein | 2.54 | Novel | |
| ROBO1 | Roundabout homolog 1 | 2.52 | Described in Ref. 15 | |
| PACSIN2 | Syndapin II | 2.51 | Novel | |
| TSG101 | Tumor susceptibility gene 101 | 2.45 | Novel | |
| TRAPPC10 | TRAPP subunit TMEM1 | 2.42 | Novel | |
| AGL | Glycogen debranching enzyme | 2.30 | Novel | |
| HGS | HRS | 1.96 | Described in Refs. 15, 30 | |
| Significant in 2/3 | DNMT1 | DNA methyltransferase 1 | 5.72 | Novel |
| HIST3H2A | Histone H2A type 3 | 4.96 | Novel | |
| RRAGD | Ras-related GTP binding D | 4.40 | Novel | |
| MGEA5 | Bifunctional protein NCOAT | 3.84 | Novel | |
| ANKS1A | Odin | 3.34 | Described in Ref. 15 | |
| CPPED1 | Calcineurin-like phosphoesterase domain-containing protein 1 | 3.32 | Novel | |
| DOK1 | Docking protein 1 | 3.20 | Described in Refs. 15, 30, 33, 34 | |
| ARAP3 | Centaurin-delta-3 | 3.13 | Described in Ref. 15 | |
| NEDD9 | Enhancer of filamentation 1 | 2.62 | Described in Ref. 36 | |
| LIMD1 | LIM domain-containing protein 1 | 2.60 | Described in Ref. 30 | |
| CBL | E3 ubiquitin-protein ligase CBL | 2.52 | Described in Refs. 5, 30, 35 | |
| CTPS2 | CTP synthase II | 2.51 | Novel | |
| SDCBP | Syntenin-1 | 2.45 | Described in Ref. 37 | |
| TRAPPC4 | Hematopoietic stem/progenitor cell protein 172 | 2.26 | Novel | |
| RALY | RNA-binding protein Raly | 2.26 | Novel | |
| HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 2.21 | Novel | |
| PROM1 | Prominin-1/CD133 | 2.06 | Described in Ref. 38 | |
| HNRNPUL2 | Heterogeneous nuclear ribonucleoprotein U-like 2 | 2.03 | Novel | |
| HNRPDL | Heterogeneous nuclear ribonucleoprotein d-like | 1.89 | Described in Ref. 32 | |
| MATR3 | Matrin-3 | 1.81 | Novel | |
| INPP5D | SHIP1 | 1.78 | Described in Ref. 15 | |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 1.73 | Described in Ref. 32 | |
| CLTA | Clathrin light chain A | 1.69 | Novel | |
| PCBP2 | Heterogenous nuclear ribonucleoprotein E2 | 1.64 | Novel | |
| HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 1.64 | Described in Ref. 32 | |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L | 1.61 | Novel | |
| EPHB2 | Ephrin type-B receptor 2 | 1.60 | Described in Ref. 15 | |
| HNRNPR | Heterogeneous nuclear ribonucleoprotein R | 1.57 | Novel | |
| RBMX | Heterogeneous nuclear ribonucleoprotein G | 1.56 | Novel | |
| ELAVL1 | HuR | 1.55 | Novel | |
| HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | 1.52 | Described in Ref. 32 | |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 1.50 | Novel | |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 1.50 | Novel | |
| CLTC | Clathrin heavy chain 1 CHC | 1.49 | Described in Refs. 15, 30, 34, 35 | |
| HNRNPA0 | Heterogeneous nuclear ribonucleoprotein A0 | 1.48 | Novel | |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 | 1.35 | Described in Ref. 32 | |
| DHX9 | ATP-dependent RNA helicase A2 | 1.33 | Novel |
Comparative Analysis of SRC Signaling In vivo and In vitro
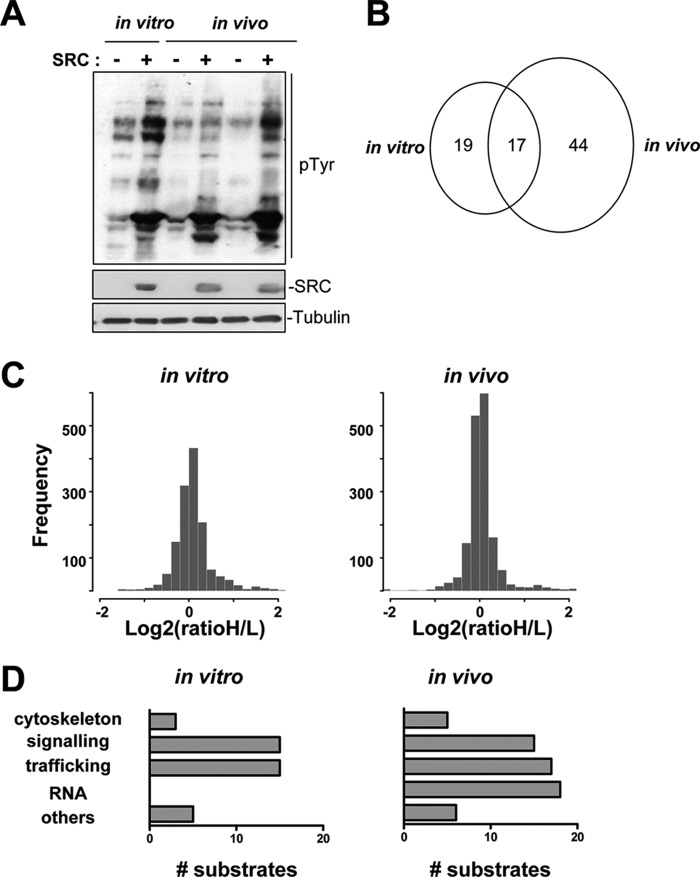
We then carried out a clustering analysis based on functional assignments described in the literature and found that 19 of the identified SRC substrates play a role in the vesicular trafficking activity involved in endo-/exocytosis (e.g. HRS, TSG101, TOM1L1, Alix, Syntaxin), and 15 have signaling functions that are important for the regulation of cell growth and adhesion (e.g. ARAP2 and 3, RRAGD, CBL, SHIP2) (Fig. 4D and supplemental Table S2). This finding predicts an important role for SRC in the cellular signaling and vesicular trafficking that mediate tumor growth. We also obtained nine transmembrane proteins as potential SRC targets, four of which (Robo, GPR155, EPHB2, and CD133) mediate signal transduction induced by extracellular stimuli. Therefore, membrane receptors might also contribute to SRC oncogenic activity in vivo. We also validated our previous in vitro finding (15) concerning the phosphorylation by SRC of a network of TKs that include EPHB2, FAK, and the atypical TK SgK269/PEAK1 and which could play an important role in cell transformation (15). Finally, we identified an additional cluster of 17 potential SRC substrates in CRC tumors that bind to mRNA (including HuR, hnRNPA1, C, D, L, R, and U), although only four proteins had cytoskeletal function (Tensin-3, p130 CAS, NEDD9, and Paxillin) (Fig. 4D and supplemental Table S3). These data suggest that SRC over-expression might have a profound effect on mRNA splicing and stability in vivo.
Fig. 4.
Comparison of in vivo and in vitro SRC signaling recorded via MS analysis. A, Western blot analysis using the 4G10 antibody of SRC-induced tyrosine phosphorylation in total protein lysates from parental SW620 and SRC-SW620 cells (in vitro) and from xenograft tumors derived from SW620 and SRC-SW620 cells (in vivo). The levels of tubulin and SRC are also shown. B, Venn diagram showing the number of common and specific SRC targets identified by mouse SILAC. C, histograms showing the distribution of in vitro and in vivo SILAC ratios. D, comparative clustering analysis of SRC targets identified via SILAC in vivo and in vitro. The number of proteins identified in each category is shown.
We then compared SRC oncogenic signaling in vivo and in vitro. Western blot analysis showed that the pTyr pattern in cultured SRC-SW620 cells was similar to the one observed in xenograft tumors derived from these cells, suggesting that SRC oncogenic signaling might not drastically vary between in vitro and in vivo conditions (Fig. 4A). Accordingly, no clear hypoxic activity has been detected in tumors (supplemental Fig. S4), indicating that oxygen concentration in vivo might not significantly affect SRC oncogenic signaling in xenograft tumors. We then investigated SRC oncogenic signaling in SRC-SW620 cells (three different experiments) via quantitative phosphoproteomics coupled to SILAC. Analysis of the proteomic data indicated that a group of 36 proteins had a significant ratio greater than 1 in at least two of the three experiments (Table II). The rather small number of proteins with a significant increased ratio may be attributed in part to the fact that we decided to retain only proteins with a significant ratio of >1 in at least two experiments. Accordingly, most of the SRC targets previously identified in slightly different conditions (15) were also detected in at least one of the three experiments, but they were not retained by the current selection procedure (supplemental Table S3). We then compared the in vitro and in vivo SILAC data. First, some differences in the ratio distribution were observed (Fig. 4C). Specifically, the ratio increase was more pronounced in the SILAC analysis, possibly because of a higher proficiency of SRC to phosphorylate its substrates in cell culture than in vivo. Second, a comparison of the in vivo and in vitro SRC targets showed that only 17 of the 36 proteins that had been identified in vitro exhibited an increased ratio in tumors as well (Fig. 4B). Most of these proteins have signaling activity, suggesting that the capacity of SRC to phosphorylate/activate signaling proteins is rather well conserved in both situations (Fig. 4D and supplemental Table S3). This analysis also highlighted important differences in the in vivo and in vitro SRC oncogenic signaling activities, as 18 proteins had a ratio of >1 only in vitro, and 45 had such a ratio only in vivo. This discrepancy was further illustrated by the fact that 8 of the 19 vesicular trafficking proteins and all 18 mRNA binding proteins identified in tumors as SRC substrates were not detected in SW620 cells in vitro. This finding indicates that SRC oncogenic signaling significantly differs in in vitro and in vivo conditions, and it validates our in vivo method as a valuable approach to decipher SRC oncogenic signaling during tumorigenesis.
Table II. SRC targets identified by SILAC.
| Gene symbol | Protein name | Mean ratio | |
|---|---|---|---|
| Significant in 3/3 | SRC | SRC | 15.26 |
| TMEM106B | Transmembrane protein 106B | 14.72 | |
| TMEM192 | Transmembrane protein 192 | 10.39 | |
| TNS3 | Tensin-3 | 5.28 | |
| ATP6V0D1 | V-ATPase subunit d 1 | 4.38 | |
| SYNGR2 | Synaptogyrin-2 | 3.95 | |
| SDCBP | Syntenin-1 | 3.49 | |
| DSP | Desmoplakin | 3.08 | |
| ANKS1A | Odin | 3.07 | |
| CTPS | CTP synthase | 3.06 | |
| Significant in 2/3 | KIDINS220 | Kinase d-interacting substrate, 220kDa | 6.71 |
| VTI1B | v-SNARE | 5.39 | |
| STX8 | Syntaxin-8 | 5.06 | |
| VPS18 | Vacuolar protein sorting protein 18 | 5.04 | |
| STX3 | Syntaxin-3 | 4.98 | |
| TTYH3 | Protein tweety homolog 3 | 4.49 | |
| VPS16 | Vacuolar protein sorting-associated protein 16 | 4.21 | |
| CEACAM1 | Antigen CD66 | 3.94 | |
| SYT1 | Synaptotagmin | 3.82 | |
| GPRC5C | Retinoic acid responsive gene protein | 3.44 | |
| MYOF | Myoferlin | 3.25 | |
| TRAPPC4 | Hematopoietic stem/progenitor cell protein 172 | 3.21 | |
| ARAP3 | Centaurin-delta-3 | 2.90 | |
| PXN | Paxillin | 2.89 | |
| PEAK1 | Sgk269 | 2.81 | |
| INPPL1 | SHIP2 | 2.65 | |
| INPP5D | SHIP1 | 2.52 | |
| PTK2 | FAK | 2.29 | |
| TOLLIP | Toll-interacting protein | 2.24 | |
| TFRC | Transferrin receptor (p90, CD71) | 2.09 | |
| GPRC5A | Retinoic acid-induced protein 3 | 2.06 | |
| BCAR3 | Breast cancer anti-estrogen resistance protein 3 | 2.03 | |
| PDCD6IP | Alix | 1.96 | |
| HGS | HRS | 1.89 | |
| PHB | Prohibitin | 1.89 | |
| CDKN1C | Cyclin-dependent kinase inhibitor p57 | 1.77 | |
| MLLT4 | Afadin | 1.27 |
The SRC Substrate TOM1L1 Specifically Regulates SRC-induced CRC Growth
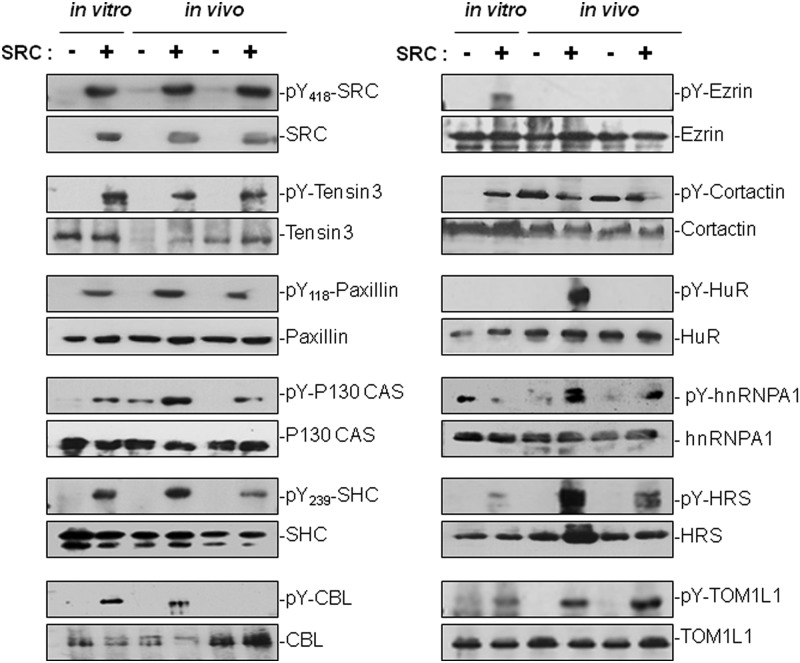
We then validated biochemically the MS data of a selected group of SRC targets (Fig. 5). These experiments confirmed most of the MS findings: (i) the pTyr levels of SRC, Paxillin, p130 CAS, Tensin-3, and CBL were increased upon SRC expression both in vitro and in vivo; (ii) SRC induced pTyr of the cytoskeletal-associated proteins Ezrin and Cortactin only in cell culture conditions; (iii) SRC induced pTyr of the mRNA-binding proteins HuR and hnRNPA1 only in tumors; and (iv) the pTyr levels of the vesicular proteins TOM1L1 and HRS were much more strongly increased in tumors derived from than in cultured SRC-SW620 cells. It should be noted that Cortactin, p130 CAS, CBL, and TOM1L1 belong to the group of in vitro SRC targets that had a ratio significantly greater than 1 in only one of the three experiments. Thus, although they were not scored as significant “SRC targets” in the MS analysis, biochemical experiments confirmed that SRC phosphorylates these proteins in cell culture. Similarly, while Cortactin had an in vivo SILAC ratio of 1, it showed elevated tyrosine phosphorylation in control tumors as well. This suggests that SRC over-expression was not needed to increase Cortactin phosphorylation in vivo. This might explain, at least partially, the low number of cytoskeletal proteins with a ratio greater than 1 in the SILAC mouse analysis.
Fig. 5.
Biochemical validation of selected SRC targets. Protein level and tyrosine phosphorylation of some selected SRC targets identified via SILAC in vitro and in vivo. Western blots showing the tyrosine phosphorylation level of the indicated proteins using 4G10 antibody following immunoprecipitation from SRC-SW620 cell lysates (in vitro) or from xenograft tumors lysates (in vivo) using the indicated antibodies.
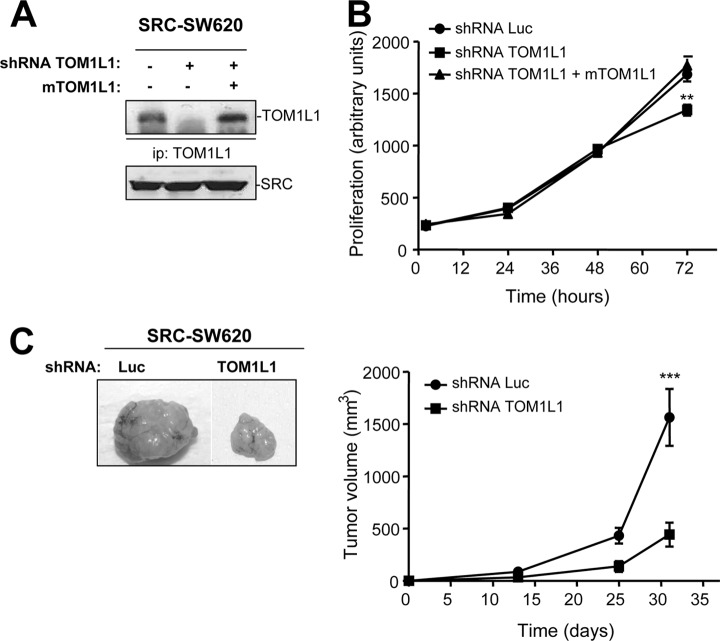
Finally, we wished to confirm the SRC oncogenic signaling specificity in vivo at the functional level for TOM1L1, one of the identified substrates. TOM1L1 is a novel adaptor protein with dual vesicular trafficking and intracellular signaling function (22, 42, 43). It contains vesicular-trafficking VHS and GAT homology domains at the N terminus and interaction motifs for signaling proteins at the C terminus. We previously identified TOM1L1 as a novel SRC substrate that negatively regulates its mitogenic (22) and transforming activity in fibroblasts (20). As TOM1L1 is heavily phosphorylated on Tyr in SRC-positive tumors, we wondered whether it had a specific function in SRC-induced CRC tumorigenesis. TOM1L1 depletion by shRNA only slightly affected SRC-induced proliferation of SRC-SW620 cells in culture (Figs. 6A and 6B). The specificity of the shRNA effect was shown by the capacity of mouse TOM1L1 to restore cell proliferation. Conversely, TOM1L1 depletion strongly affected the growth of SRC-SW620 xenograft tumors in nude mice (Fig. 6C). We thus conclude that TOM1L1 has a specific function in tumor growth of SRC-positive CRC.
Fig. 6.
A novel function of TOM1L1 in SRC-induced tumor growth. A, Western blot showing the level of TOM1L1 in SRC-SW620 cells that were transduced with the indicated shRNAs and following infection (or not) with the mouse TOM1L1 construct. B, TOM1L1 depletion has a minor effect on the growth of SRC-SW620 cells in vitro. The time course of growth (arbitrary units) of SRC-SW620 cells infected with the indicated constructs in standard culture conditions. **p < 0.01 using Student's t test. C, TOM1L1 depletion strongly reduces SRC-SW620 tumor growth in xenografted nude mice. Left-hand panel: A representative example of tumors obtained following subcutaneous injection of SRC-SW620 cells transduced with the indicated shRNAs. Right-hand panel: growth of tumors derived from SRC-SW620 cells depleted or not of TOM1L1 in nude mice. The mean tumor volume in mm3 ± S.E. (n = 8) is shown. ***p < 0.001 using Student's t test.
DISCUSSION
Here we describe a novel SILAC approach used to investigate oncogenic TK signaling in vivo in mouse xenografts. This method is based on the efficient labeling of tumor proteins by feeding xenografted mice with the mouse SILAC diet for a limited period of time (30 days), which is possible thanks to the high rate of de novo protein synthesis in tumors. Indeed, we could successfully label xenograft tumors derived from human CRC cells that are characterized by a much slower in vitro growth rate than human leukemic cells. Therefore, we think that this approach might be suitable for most human cancer cells that induce significant tumor growth in nude mice. We also predict that our mouse SILAC approach will have a large number of applications, including the analysis of the dynamic signaling of oncogenic TK during tumor progression from early tumorigenesis to metastasis formation, as well as evaluations of the activity of TK inhibitors on the tumor phosphoproteome over time. In this case, this methodology could be particularly useful for determining the molecular cause(s) of innate or acquired resistance to such inhibitors. Mann et al. recently developed the Super-SILAC approach, an alternative method of quantifying the tumor proteome of human biopsies (44, 45) that could be applied to mouse xenografts as well. The principle of this approach is to generate a mixture of several SILAC-labeled cell lines to be used as internal standards for comparison with human carcinoma tissues. However, our findings with regard to SRC-SW620 xenograft tumors and cells highlight significant differences in SRC signaling in vitro and in vivo and suggest that SILAC-labeled cell lines might not be the best internal standard. Therefore, our in vivo SILAC analysis applied to mouse xenografts should be considered as a complementary approach for deciphering TK oncogenic signaling in vivo.
Our mouse SILAC analysis allowed us to uncover new features of SRC oncogenic signaling in CRC tumors. Although we cannot formally rule out that some of the identified targets are due to SRC overexpression, the level of ectopic SRC in SW620 cells is consistent with the endogenous SRC level in metastatic CRC cells exhibiting high SRC activity. We thus believe that most of the identified candidates might be truly SRC targets in CRC, as previously suggested in vitro (15). We first identified a conserved signature of SRC oncogenic activity in tumors. Accordingly, several of the identified targets that regulate cell adhesion, growth, and survival were identified as important SRC substrates in tumors. These include FAK, p130 CAS, Paxillin (46), Tensin-3(39), and SgK269 (47). The role of the other SRC substrates in this transforming process has not been established yet. Second, we found that SRC might interact with a number of transmembrane receptors, suggesting that one of its functions is to amplify oncogenic signaling initiated by extracellular stimuli secreted by the tumor or the stroma. Alternatively, these receptors could mediate SRC oncogenic activity through a reverse signaling process, as observed in vitro (15). Whatever the mechanism, these data indicate that such transmembrane receptors might also contribute to SRC oncogenic activity during CRC tumorigenesis. Third, we found important differences between in vitro and in vivo SRC signaling. This is illustrated by the low percentage of identity between the in vivo and in vitro SILAC data, and particularly by the unexpectedly large cluster of mRNA binding proteins that were identified as SRC substrates in CRC tumors only. We thus hypothesize that SRC might have an additional role in mRNA splicing/stability in vivo, a process known to play important roles during tumorigenesis (48). In agreement with this idea, a similar function has been ascribed to SRC-like proteins in the promotion of normal and cancer cell growth (49–51).
Finally, our analysis identified a large number of vesicular trafficking proteins that are targeted by SRC in tumors, suggesting that one important function of SRC in CRC is to regulate vesicular trafficking processes related to endocytic and/or secretory pathways. The exact role of these proteins in CRC tumorigenesis is still obscure. Nevertheless, we started to address this issue by investigating the role of TOM1L1 in CRC. This vesicular trafficking protein was originally identified as a negative regulator of cell proliferation and an inducer of cell differentiation (22, 52). TOM1L1 interacts with clathrin heavy chain (CHC) and promotes endocytosis of growth factor receptors leading to signaling termination (43, 53); by interacting with SRC, the TOM1L1-CHC complex also modifies SRC membrane partitioning and hinders its proliferative and transforming activity (20). TOM1L1 might also have a tumor suppressor function, as shown by its capacity to block skin carcinogenesis in animal models (54). Surprisingly, our SILAC analysis in mouse xenografts uncovered a completely distinct function of TOM1L1 in CRC where it promotes SRC-induced tumor growth. How TOM1L1 regulates SRC oncogenic activity is completely unknown, but it could be through endocytosis of specific receptors important for oncogenic signaling. The receptor TK MET is a likely candidate, as it is an important regulator of SRC oncogenic signaling in CRC (15, 55), and two recent reports showed a direct role of MET endocytosis in tumorigenesis (56, 57). Based on these preliminary findings on TOM1L1, we hypothesize that other vesicular trafficking proteins might have pro-oncogenic activity in this cancer.
Supplementary Material
Acknowledgments
We thank Valérie Simon for generating constructs, William Ourliac and Imade Ait Arsa for technical assistance, Dr. Cédric Leroy for helpful discussions, and Dr. Christine Bénistant and Dr. Simon Descamp for reviewing the manuscript. We also thank Montpellier RIO Imaging (MRI), the RHEM Histology facilities, and the animal facilities of the IRCM of Montpellier. A.S. is supported by INCa and “la Fondation de France,” and S.R. is an INSERM investigator.
Footnotes
* This work was supported by the Ligue Nationale contre le Cancer (équipe labélisée) and INCa.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TK
- tyrosine kinase
- pTyr
- tyrosine phosphorylation
- SILAC
- stable isotope labeling with amino acids in cell culture
- CRC
- colorectal cancer
- MS
- mass spectrometry.
REFERENCES
- 1. Thomas S. M., Brugge J. S. (1997) Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 2. Yeatman T. J. (2004) A renaissance for SRC. Nat. Rev. Cancer 4, 470–480 [DOI] [PubMed] [Google Scholar]
- 3. Summy J. M., Gallick G. E. (2003) Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 22, 337–358 [DOI] [PubMed] [Google Scholar]
- 4. Sirvent A., Benistant C., Roche S. (2012) Oncogenic signaling by tyrosine kinases of the SRC family in advanced colorectal cancer. Am. J. Cancer Res. 2, 357–371 [PMC free article] [PubMed] [Google Scholar]
- 5. Oneyama C., Hikita T., Enya K., Dobenecker M. W., Saito K., Nada S., Tarakhovsky A., Okada M. (2008) The lipid raft-anchored adaptor protein cbp controls the oncogenic potential of c-Src. Mol. Cell 30, 426–436 [DOI] [PubMed] [Google Scholar]
- 6. Sirvent A., Benistant C., Pannequin J., Veracini L., Simon V., Bourgaux J. F., Hollande F., Cruzalegui F., Roche S. (2010) Src family tyrosine kinases-driven colon cancer cell invasion is induced by Csk membrane delocalization. Oncogene 29, 1303–1315 [DOI] [PubMed] [Google Scholar]
- 7. Nam J. S., Ino Y., Sakamoto M., Hirohashi S. (2002) Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. Clin. Cancer Res. 8, 2430–2436 [PubMed] [Google Scholar]
- 8. Weis S., Cui J., Barnes L., Cheresh D. (2004) Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J. Cell Biol. 167, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Golas J. M., Lucas J., Etienne C., Golas J., Discafani C., Sridharan L., Boghaert E., Arndt K., Ye F., Boschelli D. H., Li F., Titsch C., Huselton C., Chaudhary I., Boschelli F. (2005) SKI-606, a Src/Abl inhibitor with in vivo activity in colon tumor xenograft models. Cancer Res. 65, 5358–5364 [DOI] [PubMed] [Google Scholar]
- 10. Nautiyal J., Banerjee S., Kanwar S. S., Yu Y., Patel B. B., Sarkar F. H., Majumdar A. P. (2011) Curcumin enhances dasatinib-induced inhibition of growth and transformation of colon cancer cells. Int. J. Cancer 128, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopetz S., Lesslie D. P., Dallas N. A., Park S. I., Johnson M., Parikh N. U., Kim M. P., Abbruzzese J. L., Ellis L. M., Chandra J., Gallick G. E. (2009) Synergistic activity of the SRC family kinase inhibitor dasatinib and oxaliplatin in colon carcinoma cells is mediated by oxidative stress. Cancer Res. 69, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmelzle K., White F. M. (2006) Phosphoproteomic approaches to elucidate cellular signaling networks. Curr. Opin. Biotechnol. 17, 406–414 [DOI] [PubMed] [Google Scholar]
- 13. Bose R., Molina H., Patterson A. S., Bitok J. K., Periaswamy B., Bader J. S., Pandey A., Cole P. A. (2006) Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl. Acad. Sci. U.S.A. 103, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang X., Hajivandi M., Veach D., Wisniewski D., Clarkson B., Resh M. D., Pope R. M. (2006) Quantification of change in phosphorylation of BCR-ABL kinase and its substrates in response to Imatinib treatment in human chronic myelogenous leukemia cells. Proteomics 6, 4554–4564 [DOI] [PubMed] [Google Scholar]
- 15. Leroy C., Fialin C., Sirvent A., Simon V., Urbach S., Poncet J., Robert B., Jouin P., Roche S. (2009) Quantitative phosphoproteomics reveals a cluster of tyrosine kinases that mediates SRC invasive activity in advanced colon carcinoma cells. Cancer Res. 69, 2279–2286 [DOI] [PubMed] [Google Scholar]
- 16. Kruger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fassler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 17. Larance M., Bailly A. P., Pourkarimi E., Hay R. T., Buchanan G., Coulthurst S., Xirodimas D. P., Gartner A., Lamond A. I. (2011) Stable-isotope labeling with amino acids in nematodes. Nat. Methods 8, 849–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sury M. D., Chen J. X., Selbach M. (2010) The SILAC fly allows for accurate protein quantification in vivo. Mol. Cell. Proteomics 9, 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westman-Brinkmalm A., Abramsson A., Pannee J., Gang C., Gustavsson M. K., von Otter M., Blennow K., Brinkmalm G., Heumann H., Zetterberg H. (2011) SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J. Proteomics 75, 425–434 [DOI] [PubMed] [Google Scholar]
- 20. Collin G., Franco M., Simon V., Benistant C., Roche S. (2007) The Tom1L1-clathrin heavy chain complex regulates membrane partitioning of the tyrosine kinase Src required for mitogenic and transforming activities. Mol. Cell. Biol. 27, 7631–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andreoli C., Martin M., Le Borgne R., Reggio H., Mangeat P. (1994) Ezrin has properties to self-associate at the plasma membrane. J. Cell Sci. 107, 2509–2521 [DOI] [PubMed] [Google Scholar]
- 22. Franco M., Furstoss O., Simon V., Benistant C., Hong W. J., Roche S. (2006) The adaptor protien Tom1L1 is a negative regulator of Src mitogenic signaling induced by growth factors. Mol. Biol. Cell 26, 1932–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roche S., Koegl M., Barone M. V., Roussel M. F., Courtneidge S. A. (1995) DNA synthesis induced by some but not all growth factors requires Src family protein tyrosine kinases. Mol. Cell. Biol. 15, 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sirvent A., Boureux A., Simon V., Leroy C., Roche S. (2007) The tyrosine kinase Abl is required for Src-transforming activity in mouse fibroblasts and human breast cancer cells. Oncogene 26, 7313–7323 [DOI] [PubMed] [Google Scholar]
- 25. Amanchy R., Kalume D. E., Iwahori A., Zhong J., Pandey A. (2005) Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC). J. Proteome Res. 4, 1661–1671 [DOI] [PubMed] [Google Scholar]
- 26. Dehm S., Senger M. A., Bonham K. (2001) SRC transcriptional activation in a subset of human colon cancer cell lines. FEBS Lett. 487, 367–371 [DOI] [PubMed] [Google Scholar]
- 27. Zhang G., Neubert T. A. (2011) Comparison of three quantitative phosphoproteomic strategies to study receptor tyrosine kinase signaling. J. Proteome Res. 10, 5454–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lock P., Abram C. L., Gibson T., Courtneidge S. A. (1998) A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 17, 4346–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Courtneidge S. A. (2003) Isolation of novel Src substrates. Biochem. Soc. Trans. 31, 25–28 [DOI] [PubMed] [Google Scholar]
- 30. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 31. Qiao Y., Molina H., Pandey A., Zhang J., Cole P. A. (2006) Chemical rescue of a mutant enzyme in living cells. Science 311, 1293–1297 [DOI] [PubMed] [Google Scholar]
- 32. Amanchy R., Zhong J., Molina H., Chaerkady R., Iwahori A., Kalume D. E., Gronborg M., Joore J., Cope L., Pandey A. (2008) Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J. Proteome Res. 7, 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo W., Slebos R. J., Hill S., Li M., Brabek J., Amanchy R., Chaerkady R., Pandey A., Ham A. J., Hanks S. K. (2008) Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res. 7, 3447–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amanchy R., Zhong J., Hong R., Kim J. H., Gucek M., Cole R. N., Molina H., Pandey A. (2009) Identification of c-Src tyrosine kinase substrates in platelet-derived growth factor receptor signaling. Mol. Oncol. 3, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hochgrafe F., Zhang L., O'Toole S. A., Browne B. C., Pinese M., Porta Cubas A., Lehrbach G. M., Croucher D. R., Rickwood D., Boulghourjian A., Shearer R., Nair R., Swarbrick A., Faratian D., Mullen P., Harrison D. J., Biankin A. V., Sutherland R. L., Raftery M. J., Daly R. J. (2010) Tyrosine phosphorylation profiling reveals the signaling network characteristics of basal breast cancer cells. Cancer Res. 70, 9391–9401 [DOI] [PubMed] [Google Scholar]
- 36. Manie S. N., Beck A. R., Astier A., Law S. F., Canty T., Hirai H., Druker B. J., Avraham H., Haghayeghi N., Sattler M., Salgia R., Griffin J. D., Golemis E. A., Freedman A. S. (1997) Involvement of p130(Cas) and p105(HEF1), a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J. Biol. Chem. 272, 4230–4236 [DOI] [PubMed] [Google Scholar]
- 37. Boukerche H., Su Z. Z., Prevot C., Sarkar D., Fisher P. B. (2008) mda-9/Syntenin promotes metastasis in human melanoma cells by activating c-Src. Proc. Natl. Acad. Sci. U.S.A. 105, 15914–15919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boivin D., Labbe D., Fontaine N., Lamy S., Beaulieu E., Gingras D., Beliveau R. (2009) The stem cell marker CD133 (prominin-1) is phosphorylated on cytoplasmic tyrosine-828 and tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry 48, 3998–4007 [DOI] [PubMed] [Google Scholar]
- 39. Qian X., Li G., Vass W. C., Papageorge A., Walker R. C., Asnaghi L., Steinbach P. J., Tosato G., Hunter K., Lowy D. R. (2009) The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell 16, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le Roy C., Wrana J. L. (2005) Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6, 112–126 [DOI] [PubMed] [Google Scholar]
- 41. Schmidt M. H., Dikic I., Bogler O. (2005) Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 280, 3414–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seykora J. T., Mei L., Dotto G. P., Stein P. L. (2002) 'Srcasm: a novel Src activating and signaling molecule. J. Biol. Chem. 277, 2812–2822 [DOI] [PubMed] [Google Scholar]
- 43. Puertollano R. (2005) Interactions of TOM1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 280, 9258–9264 [DOI] [PubMed] [Google Scholar]
- 44. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 45. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frame M. C. (2004) Newest findings on the oldest oncogene; how activated src does it. J. Cell Sci. 117, 989–998 [DOI] [PubMed] [Google Scholar]
- 47. Wang Y., Kelber J. A., Tran Cao H. S., Cantin G. T., Lin R., Wang W., Kaushal S., Bristow J. M., Edgington T. S., Hoffman R. M., Bouvet M., Yates J. R., 3rd, Klemke R. L. (2010) Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 10920–10925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. David C. J., Manley J. L. (2010) Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bromann P. A., Korkaya H., Webb C. P., Miller J., Calvin T. L., Courtneidge S. A. (2005) Platelet-derived growth factor stimulates Src-dependent mRNA stabilization of specific early genes in fibroblasts. J. Biol. Chem. 280, 10253–10263 [DOI] [PubMed] [Google Scholar]
- 50. Paronetto M. P., Achsel T., Massiello A., Chalfant C. E., Sette C. (2007) The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 176, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brignatz C., Paronetto M. P., Opi S., Cappellari M., Audebert S., Feuillet V., Bismuth G., Roche S., Arold S. T., Sette C., Collette Y. (2009) Alternative splicing modulates autoinhibition and SH3 accessibility in the Src kinase Fyn. Mol. Cell. Biol. 29, 6438–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li W., Marshall C., Mei L., Dzubow L., Schmults C., Dans M., Seykora J. (2005) Srcasm modulates EGF and Src-kinase signaling in keratinocytes. J. Biol. Chem. 280, 6036–6046 [DOI] [PubMed] [Google Scholar]
- 53. Liu N. S., Loo L. S., Loh E., Seet L. F., Hong W. (2009) Participation of Tom1L1 in EGF-stimulated endocytosis of EGF receptor. EMBO J. 28, 3485–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao L., Li W., Marshall C., Griffin T., Hanson M., Hick R., Dentchev T., Williams E., Werth A., Miller C., Bashir H., Pear W., Seykora J. T. (2009) Srcasm inhibits Fyn-induced cutaneous carcinogenesis with modulation of Notch1 and p53. Cancer Res. 69, 9439–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Emaduddin M., Bicknell D. C., Bodmer W. F., Feller S. M. (2008) Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parachoniak C. A., Luo Y., Abella J. V., Keen J. H., Park M. (2011) GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev. Cell 20, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Joffre C., Barrow R., Menard L., Calleja V., Hart I. R., Kermorgant S. (2011) A direct role for Met endocytosis in tumorigenesis. Nat. Cell Biol. 13, 827–837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.