Abstract
Synaptic architecture and its adaptive changes require numerous molecular events that are both highly ordered and complex. A majority of neuropsychiatric illnesses are complex trait disorders, in which multiple etiologic factors converge at the synapse via many signaling pathways. Investigating the protein composition of synaptic microdomains from human patient brain tissues will yield valuable insights into the interactions of risk genes in many disorders. These types of studies in postmortem tissues have been limited by the lack of proper study paradigms. Thus, it is necessary not only to develop strategies to quantify protein and post-translational modifications at the synapse, but also to rigorously validate them for use in postmortem human brain tissues. In this study we describe the development of a liquid chromatography-selected reaction monitoring method, using a stable isotope-labeled neuronal proteome standard prepared from the brain tissue of a stable isotope-labeled mouse, for the multiplexed quantification of target synaptic proteins in mammalian samples. Additionally, we report the use of this method to validate a biochemical approach for the preparation of synaptic microdomain enrichments from human postmortem prefrontal cortex. Our data demonstrate that a targeted mass spectrometry approach with a true neuronal proteome standard facilitates accurate and precise quantification of over 100 synaptic proteins in mammalian samples, with the potential to quantify over 1000 proteins. Using this method, we found that protein enrichments in subcellular fractions prepared from human postmortem brain tissue were strikingly similar to those prepared from fresh mouse brain tissue. These findings demonstrate that biochemical fractionation methods paired with targeted proteomic strategies can be used in human brain tissues, with important implications for the study of neuropsychiatric disease.
Synaptic architecture and its adaptive changes require numerous molecular events that are both highly ordered and complex (1, 2). Molecular and cellular research into these processes has, until recently, been limited to technologies that can examine one molecule to the next within a cascade. In the last decade, mass spectrometry (MS)-based proteomic methodologies, paired with biochemical fractionation techniques, have enabled us to begin cataloging the proteomes and signaling of mammalian synaptic microdomains (1, 3–7) and microdomain specific protein complexes (8–10). These studies have revealed that synaptic architecture and plasticity involve numerous interactions between multiple signaling mechanisms with robust crosstalk and protein interactions (8, 9, 11, 12). These findings also dovetail with decades of genetic studies into neuropsychiatric disease, which have identified dozens of risk genes spread across multiple pathways at the synapse (13, 14). Historically, signaling cascades and molecular disease models have been depicted as a string of molecular events that are connected in tandem. Such views were shaped in part by the methodologies that permitted the measure of a handful of protein events at a time. MS-based proteomic methodologies can monitor numerous proteins and post-translational modifications simultaneously, permitting us to examine signaling pathways in the context of many other intracellular molecular events. The next challenge in the field of neuroscience will be to establish methodologies for quantitative assessment of proteomes in specific microdomains of neural tissues, to monitor numerous molecular events simultaneously, and to understand them within the context of nonlinear intracellular trafficking, protein interactions and post-translational modifications.
Postmortem brain studies are a critical component of neuropsychiatric research as brain tissues of patients may harbor pathophysiologic information of the illnesses. A majority of neuropsychiatric illnesses are complex trait disorders in which multiple etiologic factors converge at the synapse via many signaling pathways (13, 14). The application of advanced molecular and cellular technologies to access synaptic microdomains, however, has been limited in postmortem brains by confounds such as therapeutic intervention, agonal state, and postmortem interval. Thus, there is a pressing need to validate and focus these biochemical fractionation and MS proteomic methods for use with human postmortem brain tissue. The vast majority of previous neuroproteomic studies have employed two dimensional separation/tandem MS approaches to perform qualitative analyses of mammalian synaptic preparations (3, 7, 9). Protein quantification has been accomplished using either synthetic stable isotope-labeled peptide standards (5) or isobaric peptide labeling methods, such as Isobaric tags for relative and absolute quantitation (12, 15). Sample workup can complicate the application of isobaric tagging in an efficient and reproducible manner, (16) whereas use of synthetic peptide standards is costly and can lead to difficulty controlling for confounds in sample preparation, most notably protein digestion (17). Stable isotope labeling by amino acids in cell culture (SILAC)1 can produce only a partially labeled neural proteome (18) and we have found that full expression levels and synaptic architecture of brain tissue are difficult to reproduce in fully labeled synaptic cultures. The recent availability of stable isotope labeling in mammals (SILAM) mouse tissue allows for the generation of a stable isotope labeled neuroproteome within the context of native tissues to serve as an internal standard for the quantification of a vast number of proteins (19). Walther et al. recently used SILAM tissue to quantify over 4000 proteins in mouse hippocampus and cortex (20). Considering the high level of homology between mammalian neuronal transcripts, we investigated the suitability of this stable isotope-labeled proteome to serve as a protein standard for human tissue as well as additional model organisms.
In this study we first describe the development and validation of liquid chromatography-selected reaction monitoring (LC-SRM)/MS-based methodology for studies of mammalian synapses. Quantification of 189 neuronal proteins was conducted using a [13C6]lysine-labeled brain proteome internal standard ([13C6]-brain ISTD) prepared from [13C6]lysine SILAM mouse brain tissue. We also report the application of this method to further validate biochemical fractionation of human postmortem brain tissue for the study of synaptic biology in human disease. We observed highly reproducible enrichment of proteins into synaptic microdomains in a function- and family-specific manner. These protein enrichments were remarkably comparable between fresh-frozen mouse and human postmortem tissues. Additionally, bioinformatic analyses indicate that this method has the capability to quantify thousands of additional neuronal proteins in many model systems. These experiments demonstrate that the biochemical fractionation coupled with LC-SRM/MS and [13C6]-brain ISTDs provides a powerful tool for investigating the protein composition of synaptic microdomains in mammalian tissues, with broad applications to neuropathology and neurological studies in humans and model systems.
EXPERIMENTAL PROCEDURES
Brain Tissue
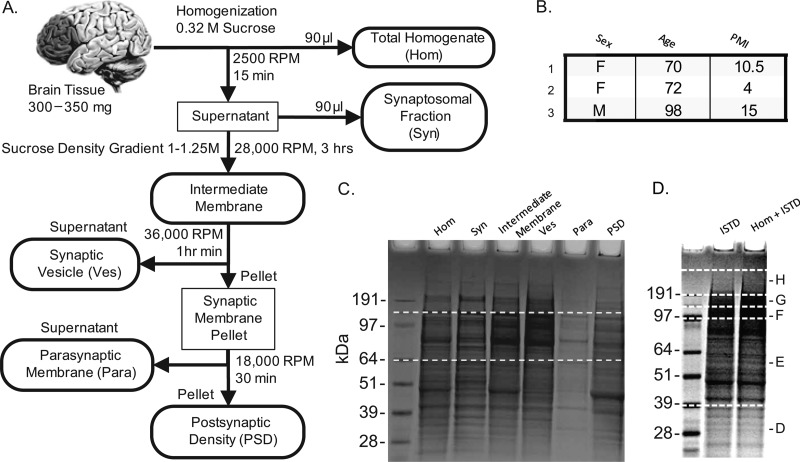
Prefrontal cortex (Fig. 1B) tissue slices from three cognitively and neurpathologically normal subjects were obtained from the University of Pennsylvania (UPenn) Brain Bank. Consent for tissue collection was granted prospectively by patients. Autopsy consent was granted by the next-of-kin at time of death. Postmortem enrollment, brain autopsy, and neuropathological assessment were conducted under UPenn Conte Core A, IRB numbers 703835 and 188200. After death, the cases were stored at 2–4 °C until transport to UPenn, where all autopsies were performed. Upon sagittal bisection, a hemisphere from each case was cut into coronal slabs, which were frozen overnight at −80 °C and then sealed in plastic bags for long-term storage at −80 °C. Tissue samples from the other hemisphere were fixed and prepared for microscopic analysis. The cases were examined by trained neuropathologists for gross and microscopic abnormalities diagnostic of diverse neurodegenerative dementias. We selected fresh frozen tissue from the prefrontal cortex because its large volume allowed repeated sampling without depleting resources available for other studies on precious normal human brain tissue. Within this restriction, we selected tissues with a wide range in age, sex, and postmortem interval (PMI) (Fig. 1B). Mouse brain tissues consisted of the forebrain plus midbrain olfactory bulb, were dissected from three male C57/BL6 mice 8–12 months old.
Fig. 1.
Biochemical fractionation and sample preparation. A, 300–350 mg of white matter was dissected from three human postmortem prefrontal cortexes. Three mouse brains, minus olfactory bulbs and cerebellum, were obtained. These tissues were subject to biochemical fractionation to obtain total homogenate (Hom), synaptosomal (Syn) and intermediate membrane fractions as well as preparations enriched for vesicular (Ves), Parasynaptic (Para), and postsynaptic density (PSD) membranes. A crude membrane fraction was prepared from [13C6] lysine labeled (>97%) mouse brain tissue for use as a stable isotope labeled internal standard proteome ([13C6]brain ISTD). B, human subject demographic information. C, two proteomes were prepared for LC-MS/MS: 1) 30 μg of the [13C6]brain ISTD and 2) 30 μg of fraction H, from human subject 1, mixed with 15 μg of the [13C6]brain ISTD. The proteomes were incubated at 95 °C for 20 min with 1× LDS loading buffer, separated on a 1.5 mm 4–12% Bis-Tris Gel (Invitrogen), cut into five fractions, diced into 2 mm cubes, reduced alkylated, digested with trypsin. Peptides were recovered from the gel cubes in 50/50 H2O/ACN with 3% formic acid, desalted and filtered. D, the biochemical fractions were mixed with the [13C6]brain ISTD at a 2:1 ratio by mass and prepared for LC-SRM/MS. Specifically, 30 μg of the total homogenate, synaptosomal, intermediate membrane and postsynaptic density preparations were prepared with 15 μg ISTD and 6 μg of the parasynaptic membrane preparation was prepared with 3 μg [13C6]brain ISTD .
Biochemical Fractionation
Synaptic fractions were obtained using a biochemical method we previously validated for use in human postmortem brain tissue (Fig. 1A).(3) 300–350 mg gray matter was homogenized in 1.5 ml solution A (0.32 m sucrose, 1 mm MgCl2, and 0.1 mm CaCl2) with a Teflon pestle. Approximately 100 μl of the homogenate was saved, solubilized with 1% SDS, and clarified by centrifugation. The remaining homogenate was centrifuged at 2500 rotations per minute (RPM) for 15 min. The pellet was discarded and 100 μl of the supernatant agitated on a rocker at 4 °C with a final concentration of 0.5% digitonin, 0.2% sodium cholate, and 0.5% Nonidet P-40 for 1 h, centrifuged at 14,000 RPM for 20 min, and the supernatant was saved as the synaptosomal fraction. The remainder of the supernatant prepared from the 2500 RPM spin was adjusted to 1.25 m sucrose with 2 m sucrose and 1 mm CaCl2 to a final volume of 5 ml. Five ml of 1 m sucrose was overlaid and the gradient ultracentrifuged at 28,000 rpm for 3 h in a SW 40 Ti rotor using a Beckmann L7 Ultracentrifuge. The band at the interface was collected, diluted 1:10 in 0.1 mm CaCl2 and centrifuged at 12,000 RPM for 20 min. The supernatant was discarded and the pellet, the intermediate membrane fraction, was dissolved in 1 ml 20 mm Tris pH 6.0 and sonicated with three 10 s pulses. One ml of 20 mm Tris pH 6.0 with 2% Triton X-100 (Sigma) was added to the solution and agitated on a rocker at 4 °C for 30 min, followed by centrifugation at 18,000 RPM for 30 min. The supernatant, the Vesicular fraction (V), was precipitated in acetone at –20°C overnight. The pellet was air dried, dissolved in 1 ml 20 mm Tris pH 8.0, sonicated with three 10-s pulses, mixed with 1 ml of 20 mm Tris pH 8.0 with 2% Triton X-100, agitated on a rocker at 4 °C for 45min, and centrifuged at 36,000 RPM for 1 h. The resulting supernatant, the parasynaptic membrane fraction, was precipitated in acetone at –20°C overnight. The pellet, the postsynaptic density (PSD) fraction, was air dried, washed three times with 20 mm Tris pH 8.0 and dissolved in 300 μl 20 mm Tris pH 7.4 with 1% SDS. Following acetone precipitation, the vesicular and parasynaptic fractions were centrifuged at 3000 RPM for 30 min and dissolved in 100 μl 20 mm Tris pH 7.4 with 1% SDS.
To prepare the [13C6]brain ISTD, 400 mg labeled MouseExpress brain tissue (Cambridge Isotopes, Cambridge, MA) was homogenized in 4 ml buffer A as described above, centrifuged at 1000 × g for 10 min to clarify, agitated on a rocker at 4 °C for 30 min, and centrifuged at 10,000 × g for 20 min. The pellet, a crude membrane fraction, was dissolved in 2 ml 20 mm Tris pH 7.4 with 1% SDS. Total protein in all preparations was quantified with the micro BCA assay (Pierce, Waltham, MA) and all solutions were supplemented with protease and phosphate inhibitor cocktails (Sigma) as well as 1 mm sodium fluoride and sodium orthovanadate (Sigma).
Sample Preparation for LC-SRM/MS and LC-tandem MS (MS/MS)
Synaptic preparations were mixed with the [13C6]brain ISTD (1 μg/μl) at a ratio of 2:1(μg/μg) and processed for MS analysis as described in Cheng et al. 2006. Briefly, 40 μl preparations were heated with lithium dodecyl sulfate (LDS) (Invitrogen, Carlsbad, CA) buffer at 95 °C for 20 min, separated on a 1.5 mm 4–12% Bis-Tris Gel (Invitrogen), cut into three fractions for LC-SRM/MS and five for LC-MS/MS (Figs. 1C and 1D), chopped into ∼2 mm cubes, washed in 200 μl 50% acetonitrile (ACN) containing 25 mm NH4HCO3, reduced in 300 μl 10 mm dithiothreitol, alkylated in 300 μl 55 mm iodoacetamide (Sigma), and digested with 80–120 μl trypsin (.025 μg/μl) (Promega) overnight at 37 °C, so that the amount of trypsin was 1:5 of the total protein to be digested by mass. Peptides were recovered from gel cubes into 200 μl 50/50 H2O/ACN with 3% formic acid by vortex-mixing and sonication for 20 min each, twice. Samples were then evaporated to a 100 μl volume, brought to 1 ml in H20 with 0.1% formic acid, desalted with Oasis® HLB cartridges (Waters, Milford, MA), evaporated almost to completion and suspended in ∼45 μl H2O with 0.1% formic acid, and filtered with 0.22 μm Ultra free-MC filter cartridges (Millipore).
Dilution Curve Preparation
To confirm that light/heavy peptide SRM measures were within linear range, a dilution curve was prepared by varying the ratio of a human total homogenate fraction/[13C6]brain ISTD from 0.05 to 19. Five mixtures were prepared, (μg homogenate/μg [13C6]brain ISTD): 1/19, 3/17. 10/10, 17/3 and 19/1. These preparations were analyzed by LC-SRM/MS in duplicate.
MS Methods
For LC-SRM/MS analyses, all peptide preparations were run on a TSQ Vantage triple stage quadrupole mass spectrometer (ThermoFisher Scientific) with an Eksigent 2Dnano LC (Eksigent) and a CaptiveSpray source (Michrom). Five μl (∼2.5 μg protein) sample was loaded on to a 3μ 200A 1 × 150 mm Magic C18 column (Michrom) at 1 μl/min for 12 min, and eluted at 750 nl/min over a 25 min gradient from 3–35% mobile phase B (ACN containing 0.1% formic acid). SRM transitions were timed using 1–1.5 min retention windows, depending on the number of SRMs to be assayed. Transitions were monitored, allowing for a cycle time of 1 s, resulting in a dynamic dwell time, never falling below 10 msec. The MS instrument parameters were as follows: capillary temperature 275οC, spray voltage 1100 V, and a collision gas of 1.4 mTorr (argon). The resolving power of the instrument was set to 0.7 Da full width at half maximum (FWHM) for Q1 and Q3. Data were acquired using a chrom filter peak width of 4.0 s.
LC-data dependant -MS/MS analyses were conducted on a Q Exactive (ThermoFisher Scientific) quadrupole orbitrap hybrid instrument with an Easy nLC-II (ThermoFisher Scientific) nano-pump/autosampler. Three μl peptide sample (∼1 ug) was loaded and resolved on a 20 cm × 75 μm proteopepII C18 packed tip column over a 90 min gradient at a flow rate of 350 nl/min, 0–35% mobile phase B (ACN, 0.1 formic acid). A data-dependent top 10 method was used to acquire a 70,000 resolution full scan to trigger ten 17,500 resolution HCD scans. Ions were isolated for MS/MS analysis with a 2.0 Da window on the quadrupole. Average cycle times were 1.2 s. Each sample was analyzed in triplicate.
LC-data Dependant MS/MS Data Analysis
Raw files from triplicate injections were searched together within Proteome Discoverer 1.3 (ThermoFisher Scientific) using SEQUEST and the human refseq. database (release 47, ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot/) with 34,340 entries. Search parameters allowed trypsin to cleave after Lysine and Arginine and have two missed cleavages. Precursor ion mass tolerance was set to 15 ppm and fragment ion mass tolerance was 20 mmu. The dynamic modification of methionine (oxidation = 15.995 Da) and lysine (13C6 = 6.020 Da), in addition to the static modification of cysteine (carbamidomethylation = 57.021 Da) was accepted on up to four residues per peptide. Within the Proteome Discoverer Software, SILAC pairs were identified using the “2-plex” workflow node, and all peptides were rescored using the Percolator (21) algorithm node. Finally, peptides were filtered at 1% false discovery rate (FDR).
SRM Transition Design and Validation
Method development began with the selection of ∼300 synaptic proteins of interest from published multidimensional MS/MS analyses of mouse and human brain tissue biochemical fractions (27, 12) as well as the LC-data dependant MS/MS analysis described in this study. RAW data from the two analysis of human tissue were searched in Proteome Discoverer™ 1.1 (ThermoFisher Scientific) using SEQUEST. Proteome Discoverer™ files were loaded into ProteinCenter™ (ThermoFisher Scientific) and sorted by function and subcellular location using Gene Ontology (GO) terms. Targets for inclusion in the LC-SRM/MS assay were selected with a bias toward well annotated synaptic proteins, such as glutamate receptors, kinases, phosphatases, vesicular fusion, amino acid metabolism, protein trafficking and scaffolding. Peptides for proteins of interest were then filtered based on the following criteria: 1) presence of lysine, 2) nonredundant to a selected protein or protein group (determined by BLAST search) and 3) 100% homology across mouse and human sequences (determined by BLAST search). Acceptable peptide sequences, along with MS2 spectra, were imported into Pinpoint™ (Thermo-Scientific). Initially, five mass transitions were selected for each target peptide and its “heavy” counter-part. Validation experiments were performed in 1:1 mixtures of human intermediate membrane fraction and the [13C6]brain ISTD. Pinpoint™ was used to visually and statistically validate SRM transitions.
Modeling of Postmortem Interval in Mice
Ten male 8–12 month old C57/B6 mice were sacrificed by CO2 and cervical dislocation, per protocol, and placed at 4 °C. The brains were removed from the crania at 0, 4, 8, 12, and 16 h postsacrifice, separated from the cerebellum and olfactory bulbs, frozen on dry ice, stored at −80 °C and subject to biochemical fractionation. The PSD fractions were analyzed by LC-SRM/MS as described above.
LC-SRM/MS Data and Statistical Analysis
Raw files generated by LC-SRM/MS analysis were loaded into Pinpoint files containing target proteins/peptides/transitions. All individual SRM transitions and integration areas were inspected both manually and by Pinpoint.
Pinpoint first identifies the peak based on co-elution of transitions, and then calculates the peak start and peak stop. This helps calculate the area under the curve for that transition. Summing this area for all transitions from the light gives the total area for the light peptides, and taking the ratio against the total area of the heavy peptide calculates the relative intensity with respect to the heavy internal standard. After computing the area under the peak for each transition for both the light and the heavy peptides, Pinpoint computes three scores, each of which help the user evaluate the quantitation fidelity. The first score calculates the overlap in the apex for each transition, and is meant to identify transitions that might be contaminated by an isobaric interference with a slightly different chromatographic peak shape. The threshold for acceptance is defined as 1/4th of the peak width at FWHM. The second score calculates the overlap in the peak shape of all the transitions so as to identify transitions that might have a shoulder on the leading or the tailing edge indicating an underlying isobaric interference with slightly different retention time. For this, Pinpoint calculates the time difference between the right half and the left half of the peak and compares it to the average difference across all transitions for that peptide. The threshold is also defined as 1/4th of the peak width at FWHM. The third score compares the ratios of transitions from the light and heavy peptides. For both the light and heavy SRM spectra the transitions are normalized to the total ion current. Pinpoint then reports the maximum difference between these normalized intensities of corresponding transitions from the light and heavy peptides, a contaminating co-eluting species should alter the ratio of either the light or heavy peptide so as to not be in agreement. The threshold for this score is 25%. Any transition that scores above the threshold gets flagged and will carry this flag to the peptide level, resulting in the peptide being scrutinized by manual inspection.
Protein enrichment in the synaptosomal, vesicular, parasynaptic, and PSD fractions were calculated by normalizing protein measures in each fraction to measures in the total homogenate fraction. The coefficient of variation (CV), expressed as a percent, was calculated to assess precision of peptide quantification across injections, accuracy of protein quantification from multiple peptides, and reproducibility of fractionation.
Unsupervised hierarchical clustering was used to construct self-organized heat maps of protein and fraction by enrichment values with Cluster 3.0 and Treeview (22, 23). Enrichment values were log2 transformed, median centered by enrichment and fraction, then normalized by protein and fraction. Uncentered Pearson correlation was used as the similarity metric and the clustering method was centroid linkage. Pearson correlation coefficients were calculated in Cluster 3.0. ANOVA and Student's t test were used to calculate significance for the effects of species and fraction on enrichment CVs, species and protein group on enrichment values, and postmortem interval (PMI) and protein group on protein measure.
Comparison of Human and Mouse Protein Sequences
Human and mouse protein sequence databases were assembled from the orthologous protein accessions listed in the HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene, release 65). The Entrez efetch utility was used to retrieve the mouse and human fasta format sequences from the Entrez protein database. Proteins were digested in-silico with trypsin while retaining peptides ending with lysine and 6 ≤ length ≤ 25. Human peptides were additionally filtered to retain only proteotypic peptides, yielding a set comprising 246,630 peptides from 16,271 proteins. For each human protein, the number of peptides having an identical amino acid sequence to any mouse peptide was tabulated. A separate analysis was limited to the 1400 proteins identified in a proteomic study of mouse (Bl6) synaptosomes (Seeholzer and Kalb, unpublished).
RESULTS
LC-data Dependant MS/MS Analysis of Postmortem Human Brain Total Homogenate Spiked with [13C6]Brain ISTD
The combined analysis of all five gel bands, each analyzed in triplicate, resulted in the identification of 3025 protein groups, 2556 by two or more unique peptide sequences. In total, 230,841 peptide spectral matches were attained, allowing for 27,650 unique peptide sequence identifications. A total of 3025 labeled lysine containing peptides unique to a single protein group were identified by MS/MS (supplemental Table S1). The results of these searches as well as the other data sets (24), used to generate SRMs can be viewed at the National Institute of Standards and Technology Libraries of Peptide Tandem Mass Spectra and at PRoteomics IDEntifications database (PRIDE) (accession: PXD000004). Supplemental Fig. S1A shows the ratio distribution of all observed unique peptide pairs in all triplicate analysis, across all fractions. These unique labeled peptide pairs corresponded to 1305 proteins, 755 of which were represented by two or more unique pairs. Supplemental Fig. S1B shows the protein distribution by the number of unique peptide pairs.
Fidelity of Peptide SRMs
To ensure that the desired peptide was assayed, rigorous selection criteria were employed for the inclusion of peptide LC-SRM/MS transitions. After a peptide met the three initial criteria described above: nonredundancy, presence of lysine, and 100% sequence homology between mouse and human tissues, multiple validation experiments were conducted to confirm peptide SRM accuracy. Candidate “light”/“heavy” peptide SRMs were evaluated manually during method development for A) retention time, B) similar y-ion ratios (within 25%), C) presence in the correct gel fraction, and D) a signal to noise ratio greater than 3. Peptide SRM pairs for which more than one identical peak was observed were omitted. Out of the ∼300 initial proteins selected, 2–4 SRMs for 1–4 peptides from 189 proteins met these criteria (supplemental Table S2). Of these 189, 110 were measured by one peptide, 53 by two, and 25 by three or more.
Precision of Protein Quantification
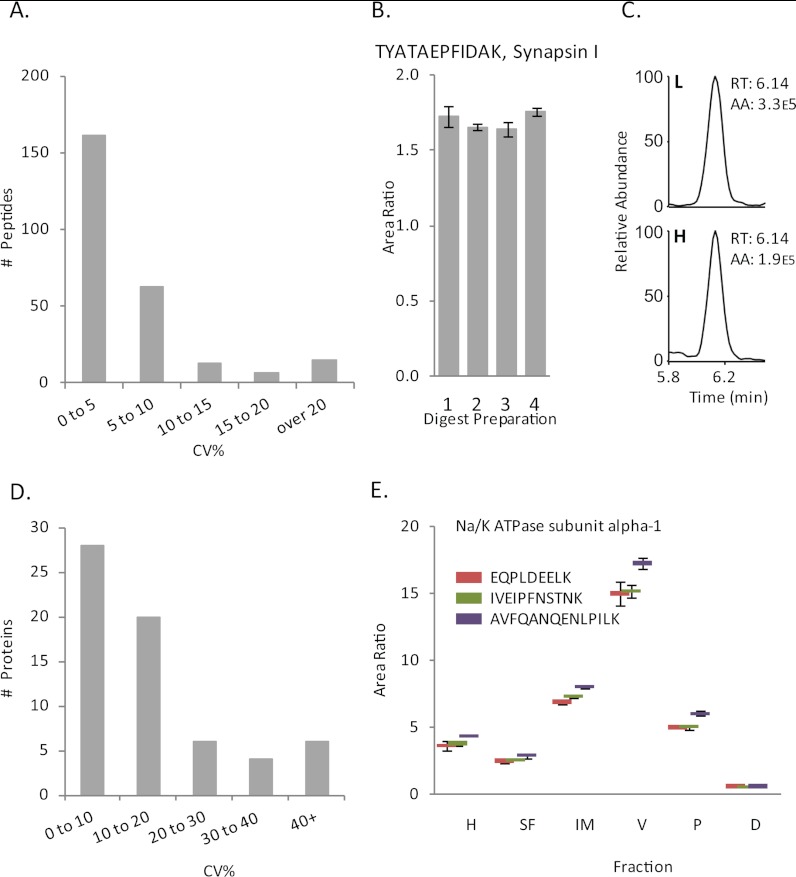
To assess LC-SRM/MS technical precision, four 30 μg aliquots of a human intermediate membrane fraction were individually mixed with 15 μg [13C6]brain ISTD, prepared for LC-SRM/MS analysis and injected in triplicate over several days. A subset of 118 proteins was quantified in this experiment. First, analytical precision of peptide quantification over repeated injections was assessed. The median CV for peptide measures across three repeated injections was 4.3% and CV correlated with the signal intensity of the light peptide SRM (R2 = .7) (supplemental Fig. S2A). Variability from sample preparation was also assessed. The median CV% across 4 repeated preparations was 3.8%, with the vast majority under 10% (Figs. 2A and 2B). The variability from sample preparation with and without the [13C6]-brain ISTD was compared as well. We observed both an increase in median CV% (from 3.8% to 7.5%) as well as a broadening of the CV distribution (supplemental Fig. S2B). Finally, we assessed precision of protein quantification by calculating variability between the peptides that were quantified for the same protein. The median CV for proteins quantified with two or more peptides was 12%, with the majority of CVs being under 20% (Fig. 2D). This indicates that LC-SRM/MS with a [13C6]brain ISTD imparts excellent precision and highly reproducible sample preparation. These Pinpoint files are available at Proteome Commons (proteomecommons.org/dataset.jsp?i=77704) and a free Pinpoint viewer is available at http://portal.thermo-brims.com/.
Fig. 2.
Precision of peptide quantification. To assess variability in sample preparation, four 30 μg aliquots of a human intermediate membrane fraction were individually mixed with 15 μg [13C6]brain ISTD and analyzed by LC-SRM/MS with triplicate injections. The coefficient of variance expressed as percentage (CV%) was calculated for each peptide measured across the four fractions. A, Depicts the distribution of the peptide CV%s, showing the vast majority under 10%. B, Shows light/heavy area ratios for the TYATAEPFIDAK peptide, which is unique to Synapsin I. Error bars are standard deviation of the triplicate injections. C, Chromatographic peaks from LC-SRM/MS analysis of TYATAEPFIDAK. CV% of peptide measures for the same proteins were calculated to assess accuracy of protein measures derived from multiple peptide SRMs. D, Shows the distribution of these CV%s. E, shows the consistency of peptide measures for the same protein (Na/K ATPase subunit alpha-1) across six fractions prepared from human subject 1.
The light/heavy peptide ratios were found to be linear over the range of the dilution curve for most peptides, with some deviating from linearity at the first and/or last point. The average R2 value for 195 peptides assayed in this experiment was 0.96. Fig. 5 in the discussion shows representative dilution curves of four proteins: NR1, CDK5, PSD-95, and CAMKIIα.
Fig. 5.
Comparison of human and mouse vesicular and postsynaptic density fractions. A and B, depict representative chromatographic peaks of light and heavy peptides from four peptides with different expression values. Note that the y axis is log10. This figure highlights the similarity in relative protein expression between the standard and analyte proteomes. C–F, Human total homogenate fraction was mixed with the [13C6]brain ISTD in the following ratios (μg): 1/19, 3/17. 10/10, 17/3, and 19/1. These preparations were analyzed by LC-SRM/MS in duplicate and the prepared L/H ratio plotted against the observed average L/H peptide ratio (peptide measure) for each protein. This figure show's representative curves for CaMKIIα, CDK5, NR1, and PSD95. The 19/1 point is not shown to highlight the midrange of the curve, but does not deviate from the slope reported here.
Comparison of LC-SRM/MS and LC-data Dependant MS/MS Quantification
The 1305 proteins identified by labeled pairs of homologous heavy and light peptides returned 1418 gene symbols in DAVID. Of these 131 were monitored in the experiment described above (supplemental Fig. S3A). We compared the light/heavy ratios of 84 peptides that were monitored in both experiments and were included for quantification by manual inspection. In general we observed good agreement between the two methods (supplemental Fig. S3B).
Quantitative Analysis of Subcellular Fractions
We analyzed the relative protein composition of five neuronal biochemical fractions: Total homogenate, synaptosomal, vesicular, parasynaptic, and PSD, prepared from three human postmortem prefrontal cortex sections and three mouse brains (Fig. 1). Enrichment was confirmed by Western blot for protein markers of vesicular, parasynaptic, and PSD fractions (data not shown). The enrichment of all fractions except for parasynaptic from mouse 3 was confirmed. This preparation was analyzed with the other samples and included in hierarchical clustering, but was dropped from average enrichment and enrichment variability calculations. The fractions were mixed, 2:1 by mass, with [13C6]brain ISTD, separated into three fractions by SDS-PAGE (Fig. 1C) and analyzed by LC-SRM/MS with duplicate injections (supplemental Table S2). To ensure a continued high level of precision in peptide and protein measurement, peptide SRMs in this data set were re-evaluated for variability, intensity, elution time, L/H ratio, and signal-to-noise ratio using the criteria described above. To evaluate the flagging algorithms employed by Pinpoint, we plotted the distribution of the two populations of peptides (included for—and excluded from—quantification by manual analysis) by L/H score and apex score (supplemental Fig. S4). We observed that the population of peptides excluded from quantification by manual evaluation was composed mostly of flagged peptides (those with higher scores). Additionally, as post-translational modifications can confound protein measurements derived from peptide SRMs, peptide trends across different fractions from the same individual were manually inspected for consistency, as in Fig. 2E. Proteins lacking consistent peptide measures, defined as a difference of > 50%, were removed. Peptide measurements for only one protein met these criteria. It is important to note that even minute differences in peptide measurement for the same protein were consistent across all fractions. For example the AVFQANQENLPILK peptide, from Na/K ATPase subunit alpha-1, was slightly elevated in all fractions compared with the other two peptides (Fig. 2E). The 150 proteins that passed these criteria in both human and mouse preparations were used as the basis for comparing fractionation between the two species (supplemental Table S3). These Pinpoint files are available at Proteome Commons (Mouse: proteomecommons.org/dataset.jsp?i=77703 Human: proteomecommons.org/dataset.jsp?i=77702) and a free Pinpoint viewer is available at http://portal.thermo-brims.com/.
Protein enrichment values were calculated by normalizing protein measures in the synaptosomal, vesicular, parasynaptic, and PSD fractions to protein measures in the total homogenate fraction. Averages and CVs of protein enrichment values and protein measures in total homogenate were calculated for each species. Median CVs for enrichment values were under 30% for all preparations from both species (Fig. 4A). In all preparations, except the vesicular, the average CV was significantly lower in mice but a two-way ANOVA did not reveal a significant interaction between species and fraction (p = 0.2). A substantial number of proteins had CVs above 30% (supplemental Fig. S5). At first, we suspected that proteins with high CVs in a fraction would have low average enrichment values, whereas more abundant “bona fide” proteins would have more consistent values. Regression analysis, however, found no correlation between average enrichment value and CV in any of the fractions (data not shown), necessitating the sorting of proteins by both enrichment value and variability.
Fig. 4.
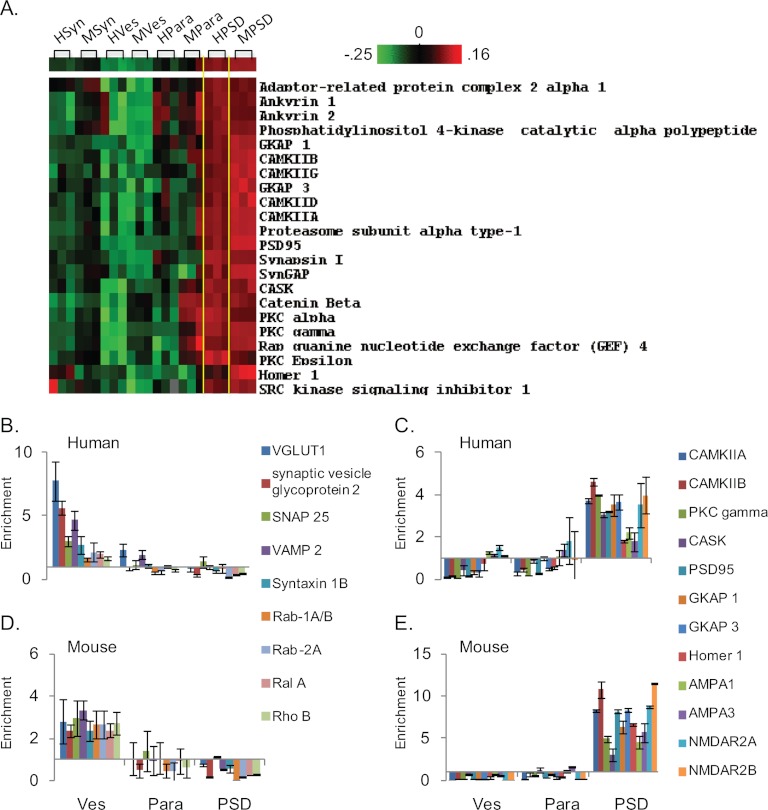
Comparison of human and mouse vesicular and postsynaptic density fractions. A, rotated and expanded view of Fig. 3B showing protein enrichment in PSD fractions prepared from human and mouse tissues. The Pearson's Correlation Coefficient for this grouping was 0.72. To more closely contrast human and mouse fractions we compared enrichment values for a subset of the proteins grouped in the clustering analysis. B and D, Show the enrichment of vesicular proteins in vesicular (Ves), parasynaptic (Para), and PSD fractions from human and mouse tissues. C and E, Show enrichment of kinases, scaffolding proteins, and glutamate receptors in the same fractions from humans and mice. Error bars are standard deviation of three individuals/animals. Note that in C and E, protein enrichment is higher in the fractions prepared from mouse tissue.
To gain this more comprehensive view of the data, unsupervised hierarchical clustering was used to group proteins and fraction by protein enrichment values (Fig. 3B, supplemental Figs. S6, S7, and S8). In both species we observed segregation of preparations into discreet groups by fraction identity (supplemental Figs. S7 and S8). Clustering analysis of human and mouse preparations together revealed that biochemical fractions within species had the tightest grouping, followed by fraction identity, regardless of species (Fig. 3B, supplemental Fig. S8). Many protein groups clustered by high enrichment values were both similar across species and in agreement with previously published analyses of these preparations. Kinases, scaffolding proteins, and glutamate receptors were enriched in PSD fractions (Fig. 4A, supplemental Fig. S8A), Soluble NSF attachment protein receptor complex proteins and vesicular amino acid transporters were enriched in vesicular fractions (supplemental Fig. S8B), and guanine nucleotide binding protein subunits and voltage-dependent ion channels were enriched in parasynaptic fractions (supplemental Fig. S8C). Synaptosomal fractions were rich in cytosolic enzymes (supplemental Fig. S8D). Clustering analysis of human and mouse fractions together also highlighted two striking differences between mouse and human preparations. First, it identified a subset of “contaminant” proteins that were enriched in human PSD fractions, but not in mouse fractions (supplemental Fig. S8E). Second, all septin proteins measured were enriched in mouse PSD and parasynaptic fractions, but not in human fractions (supplemental Fig. S8F).
Fig. 3.
Overview of protein enrichment. Protein enrichment values were calculated for each protein in the synaptosomal (Syn), vesicular (Ves), parasynaptic (Para), and PSD fractions by normalizing measures in each fraction to protein measures in the total homogenate (Hom). The averages of these values for each fraction were then taken for the three human and three mouse samples and CV%s calculated. A, Reports the median CV% of protein enrichment values for each fraction. To sort out protein groups and fractions by enrichment value unsupervised hierarchical clustering, with Pearson's Correlation as the similarity metric, was used. B, Shows this clustering analysis of the mouse and human fractions as a heat map. synaptosomal (SF), vesicular (V), parasynaptic (P), and PSD (D).
The enrichment values were heavily processed before hierarchical clustering. Therefore, to more closely investigate the similarities and differences between human and mouse preparations, we compared the average enrichment values for the protein groups identified by the clustering analysis in vesicular and PSD fractions. In the vesicular fraction, enrichment values for vesicular proteins were more similar to each other in mice, as compared with humans (Figs. 4B and 4D). In PSD fractions, enrichment values for kinases, scaffolding proteins and glutamate receptors were roughly twice as high in mouse (Figs. 4C and 4E). A two-way ANOVA confirmed a significant interaction between species and the protein group for average enrichment value (p < 0.0001). Enrichment values for the “contaminant” proteins were, on average, 12-fold higher in the human PSD compared with mouse (p = 0.022), the most extreme of which was creatine kinase brain isoform with a 72-fold higher enrichment value (supplemental Fig. S9A). The septins had, on average, threefold higher enrichment values in the mouse PSD compared with human PSD (p = 0.017) (supplemental Fig. S10A and S10B). Interestingly, we observed some reciprocal enrichment between the PSD and synaptosomal fractions for both of these protein families (supplemental Figs. S9 and S10).
To test if PMI contributed to these differences, we analyzed PSD fractions prepared from mouse tissues with PMIs ranging from 0 to 16 h. Measures of “contaminant” proteins in PSD increased with postmortem interval, whereas measures for glutamate receptors and scaffolding proteins decreased (supplemental Fig. S9C and S9D). A two-way ANOVA confirmed a significant interaction between the two protein groups, PSD proteins versus contaminants, over PMI (p = .007). The protein measures for the septins in fraction D, however, increased steadily to a PMI of 12 h, before decreasing after 16 h (supplemental Fig. S10C).
DISCUSSION
Deregulated synaptic activity is implicated in a host of neurological and psychiatric diseases, but strategies to address synaptic protein function in human postmortem brain tissues are limited. Many neuropsychiatric diseases, such as autism, depression, and schizophrenia are believed to arise from complex interactions between many synaptic signaling cascades in specific subcellular microdomains (10, 13, 14, 25, 26), the remnants of which are present in the postmortem brain tissues of patients. Therefore, the ability to assess protein expression and trafficking events in postmortem brain tissue with high precision and accuracy is essential. Various MS-based proteomics strategies have been applied in rodent and nonhuman primate tissue with limited application in postmortem human tissue (5, 7, 12, 27). Here we report, for the first time, the targeted and precise relative quantification of over 150 synaptic proteins in five distinct subcellular preparations from human postmortem brain tissues. This study demonstrates that the core protein components of synaptic microdomains are consistently enriched in biochemical fractions prepared from human tissue. Additionally, we identified proteins whose partitioning through fractionation is affected by the PMI and other postmortem confounds. This validated fractionation - stable isotope dilution LC-SRM/MS methodology will further the investigation of synaptic function in a variety of neuropsychiatric diseases.
LC-SRM/MS with a [13C6]brain ISTD allowed the simultaneous quantification of over 150 proteins with precision and reproducibility. We observed low variability in peptide measures over repeated injections and preparations (Fig. 2). Although the analytic precision correlated with intensity of the analyte signal (R2 = 0.7), the CV for the vast majority of peptides was < 20% (supplemental Fig. S2). The [13C6]brain ISTD is prepared from [13C6]lysine SILAM mouse brain tissue, and thus contains labeled peptides at ratios close to those found in human brain tissues over the large dynamic range of protein expression (supplemental Fig. S1). Fig. 5A and 5B illustrates this point in four proteins with vastly different expression levels: βTubulin, VAMP2, PSD-95, and GAD67. The median CV for proteins quantified with two or more peptides was 12%, with the majority of CVs being under 20% (Fig. 2D). It is important to note that in cases where the peptide measures for the same protein differed, the relative enrichment differences between fractions were often consistent, for example in the case of the AVFQANQENLPILK peptide, from Na/K ATPase subunit alpha-1 (Fig. 2E). Several factors could contribute to this discrepancy; such as a difference in AA sequence around this peptide between species resulting in differential trypsin cleavage efficiency or the expression of different splice variants not cataloged in the NCBI database. Confounds such as this may complicate attempts to get at absolute protein quantities, but still allow for the assessment of relative differences between samples with a high level of precision.
LC-SRM/MS analysis using a [13C6]brain ISTD has several advantages over multidimensional separation tandem MS strategies for protein quantification. First, time requirements for both experiment and data analysis are greatly reduced. Sample preparation time is reduced, as compared with strong cation exchange, because gel fractionation can yield a minimal number of fractions, and allows for multiplexed sample preparations within a single gel. Indeed, it is likely that for relatively “simple” proteomes, the first dimension of separation could be omitted. Gradients for SRM/MS are much shorter than tandem MS analyses in which full MS/MS data for peptide identification must be collected. Software such as Pinpoint and Skyline has streamlined peptide SRM design, validation, and analysis. We validated 189 proteins for LC-SRM/MS quantification with the [13C6]brain ISTD. This number is dwarfed by multidimensional separation tandem MS strategies that use labeled ISTDs or isobaric labeling. However, our bioinformatic analysis indicates that this strategy could theoretically be applied to ∼60% of the whole proteome and >90% of the proteins in the representative synaptic proteome (Seeholzer and Kalb, unpublished) (Fig. 6), the vast majority of which have peptides (proteotypic, ending in lysine, between 6 and 25 AA long) that have been recorded in the National Institute of Standards and Technology peptide database. As these libraries grow from incorporation of our own and others discovery data, so will the number of proteins that can be assayed. Additionally, this methodology allows researchers to target specific protein families and/or pathways. While the number of peptides deemed useful for quantification will be diminished by those containing methionyl residues (∼20%) or beginning with glutamine (∼1%), this number will also be augmented by allowing for the conservative isobaric substitution of leucine for isoleucine and visa versa.
Fig. 6.
Comparison of human and protein sequences. Protein sequences were retrieved for human and mouse homologenes from the total proteome and a representative synaptic proteome. Lysine containing peptides, between 6 and 25 amino acids in length were compared between the two species. This analysis shows that the [13C6]brain ISTD can theoretically be used to quantify 80% of the human synaptic proteins identified by more than two peptides. Additionally, the vast majority of these synaptic proteins have peptides in the NIST Libraries of Peptide Tandem Mass Spectra.
The study of addiction and neuropsychiatric illnesses often focuses on the activity, expression and trafficking of specific proteins identified by genetic or pharmacological studies such as disrupted in schizophrenia 1 (DISC1) and NMDA receptor subunits. Thus the ability to target these and other implicated proteins is highly desirable. Several of these proteins such as PSD95, CDK5, CAMKIIα, and several NMDA receptor subunits were quantified in the experiments described here. We were able to reproducibly quantify these proteins over a large dynamic range (Figs. 5C–5F). Others, most notably DISC1, ErbB4, nNOS, and PLCγ were not included in the assay for two main reasons. For some proteins (e.g. ErbB4 and nNOS) peptides suitable for quantification have not been observed either our or any other MS/MS analyses of synaptic preparations to which we had access. Other proteins, such as DISC1 and PLCγ, have been the subject of targeted MS/MS studies and will be included in future iterations of the assay described here. Indeed, we are confident that as the sophistication of proteomics methods improves with time the vast majority of synaptic proteins of interest to the field of neuroscience will be assayable by this or a similar approach.
Previously, we have reported that postsynaptic densities can be fractionated from the majority of human postmortem brain tissues (24). In the present study, we used the accuracy and precision of LC-SRM/MS with a [13C6]brain ISTD to further examine the reproducibility of subcellular fractionation in human postmortem brain tissue. We compared the relative enrichment of over 150 proteins in four fractions prepared from the prefrontal cortexes of three human subjects and the brain tissues of three mice (Fig. 1).
Mouse brains flash frozen on dry ice immediately after dissection, are mostly free of the postmortem confounds as well as individual variability inherent to human brains. In these tissues the median variability of protein enrichment was under 30% for all fractions (Fig. 3A, supplemental Fig. S5). Unsupervised hierarchical clustering of proteins and fractions by enrichment values revealed tight grouping of fraction types (supplemental Fig. S6). The clustering analysis also returned enriched protein groups in line with current views of synaptic architecture (see “Results”). Despite the fact that the parasynaptic fraction from mouse 3 was identified as an outlier, it still clustered with the parasynaptic fractions from the other two mice (supplemental Fig. 6A), demonstrating the power gained from comparing multiple protein measures.
The unsupervised hierarchical clustering performed may also be viewed as a protein significance analysis. Within the analysis, peptide SRM quality is first judged based on the agreement of the light and heavy peptide retention time apex, symmetry, and transition percent relative abundance. Interferences in the SRM signal will cause one or more of these three criteria to flag the measurement, indicating that the peptide signal is being distorted by underlying noise. This analysis was automated within the Pinpoint program whereas other SRM data processing software are available offering similar tools (21, 28, 29). When multiple peptides per protein are being monitored, the relative ratio between light and heavy forms should remain consistent, giving some statistical confidence to the protein measurement (20). Although a single peptide can be used to measure a specific protein, it is very difficult to judge the specificity of the measurement without performing a rigorous controlled analytical analysis (30, 31). In these experiments, single peptide measurements were included which were unique to a single protein and that passed the quality assessment described above. Furthermore, these single peptide measurements were enriched in their appropriate subcellular fraction based on the unsupervised hierarchical clustering. The assessment of protein significance in SRM analysis is beginning to be addressed by researchers, but will be dependent on the experimental goals (29). In this preliminary experiment, we considered these single peptide measurements to meet the experimental fitness of purpose if they were correctly tracked with subcellular fraction between mouse and human brain and showed consistency across multiple preparations.
Human and mouse preparations were compared by enrichment value variability, hierarchical clustering, and relative enrichment levels. Variance in protein expression and enrichment values tended to be smaller in preparations from mouse tissue (Fig. 3A, supplemental Fig. S5). Considering the individual variability in the humans from which tissue was derived, most notably in age (70, 72, and 98), sex (2F, 1 m), and PMI (4, 10, and 15 h), protein enrichment in human fractions should be considered rather consistent. Hierarchal clustering also revealed a striking similarity in enrichment patterns between the same fractions prepared from different species, while highlighting some key differences (Fig. 3B). The enrichments that drove this clustering were composed of well-documented synaptic molecules (supplemental Fig. S8). Subcellular fractions prepared from human tissue are consistently enriched for key synaptic proteins. Indeed, the relative ratios of these proteins, compared with each other, is similar between mouse and human tissues, suggesting that the core protein complexes composing these microdomains are largely intact in human postmortem brain despite biochemical procedures for fractionation and postmortem confounds (Fig. 4). Indeed, we have previously shown that protein-protein interactions with PSD95, a central protein in postsynaptic protein complexes, are preserved in human brain tissue and survive biochemical fractionation (24). Thus, it should be possible to investigate alterations to the relative amounts of PSD proteins in neuropsychiatric diseases.
Despite this high level of similarity, we did observe two distinct differences between flash frozen mouse and postmortem human brain tissues. First, although PSD proteins were enriched in PSD fractions from both species, the enrichment values were twofold higher in fractions prepared from mice (Fig. 4). We also identified a subset of “contaminant” proteins enriched in PSD fractions prepared from humans but not from mice (supplemental Figs. S4 and S8). Interestingly, some of these proteins were enriched in synaptosomal fractions from mice, but none were enriched in human synaptosomal fractions (supplemental Figs. S8 and S9). Experiments with PSD fractions prepared from mouse brain tissues with simulated PMIs support this assertion. We observed increased amounts of these “contaminant” proteins with decreased amounts of PSD proteins as PMI increased (supplemental Fig. S9C and 9D). Finally, septin proteins were enriched in the PSD and parasynaptic fractions from mice, but not from humans (supplemental Figs. S8 and S10). Septins are conserved eukaryotic proteins that form cytoskeletal components that provide membrane rigidity and create barriers between subcellular domains in many cell types, including neurons (32). Additionally, more specialized roles for individual septin family members have been identified on both sides of the synapse. For example, Septin 5 has been shown to inhibit vesicle exocytosis by interacting directly with the Soluble NSF attachment protein receptor complex, (33, 34) whereas Septin 11 plays a functional role in dendritic arborization of GABAergic synapses (35). PMI could not explain the differences in enrichment as septin proteins increased with PMI, in contrast to the observations in human tissue (supplemental Fig. S10C). Although individual septins have known roles at the synapse, the septin cytoskeletons have not been observed to associate tightly with postsynaptic protein complexes (8–10). In addition to PMI, another difference between mouse and human tissue is the ratio of white to gray matter. Human cortex samples were cut to maximize the amount of gray matter in the tissue sample, whereas it is not possible to separate the two in mouse brain. We suspect this composition of human tissue yields a more pure PSD fraction in terms of septin contamination. Further experiments, ideally in gray matter dissected from fresh-frozen nonhuman primate tissue, are required to confirm this hypothesis.
Neuropsychiatric illnesses are associated with alterations in synaptic biology, which involve a multitude of proteins and signaling mechanisms in a microdomain-specific manner. The ability to assay the proteomes of synaptic microdomains, in which disease factors may converge, is essential to advancing our understanding of these illnesses. Here, we have validated a fractionation-stable isotope dilution LC-MS methodology for the precise and targeted quantitation of over 150 synaptic proteins in subcellular fractions prepared from human postmortem brain tissue. Additionally, we have demonstrated that this method has the potential to be expanded to >1000 synaptic proteins. Thus, this study represents an important first step in bringing targeted proteomic strategies to bear on the study of neuropsychiatric disease in human brain tissue.
Supplementary Material
Footnotes
* This work was supported by NIMH grants RO1MH075916, P50MH096891 and F31 MH087106 and a grant from the Stanley Medical Research Institute and from the National Alliance for Research on Schizophrenia and Depression.
 This article contains supplemental Figs. S1 to S10 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S10 and Tables S1 to S4.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- [13C6]brain ISTD
- [13C6]lysine-labeled brain proteome internal standard ([13C6]brain ISTD)
- ANOVA
- analysis of variance
- CV
- coefficient of variation
- FWHM
- full width at half maximum
- LC-SRM/MS
- liquid chromatography-selected reaction monitoring/mass spectrometry
- PMI
- postmortem interval
- PSD
- postsynaptic density
- RPM
- rotations per minute
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Coba M. P., Pocklington A. J., Collins M. O., Kopanitsa M. V., Uren R. T., Swamy S., Croning M. D., Choudhary J. S., Grant S. G. (2009) Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci. Signal. 2, ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bard L., Groc L. (2011) Glutamate receptor dynamics and protein interaction: lessons from the NMDA receptor. Mol. Cell. Neurosci. 48, 298–307 [DOI] [PubMed] [Google Scholar]
- 3. Hahn C. G. (2010) A road less travelled: Unpacking the complex traits of schizophrenia. Brain Res. Bull. 83, 85. [DOI] [PubMed] [Google Scholar]
- 4. Bayés A., van de Lagemaat L. N., Collins M. O., Croning M. D., Whittle I. R., Choudhary J. S., Grant S. G. (2011) Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J. (2006) Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 6. English J. A., Dicker P., Föcking M., Dunn M. J., Cotter D. R. (2009) 2-D DIGE analysis implicates cytoskeletal abnormalities in psychiatric disease. Proteomics 9, 3368–3382 [DOI] [PubMed] [Google Scholar]
- 7. Peng J., Kim M. J., Cheng D., Duong D. M., Gygi S. P., Sheng M. (2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011 [DOI] [PubMed] [Google Scholar]
- 8. Husi H., Ward M. A., Choudhary J. S., Blackstock W. P., Grant S. G. (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669 [DOI] [PubMed] [Google Scholar]
- 9. Dosemeci A., Makusky A. J., Jankowska-Stephens E., Yang X., Slotta D. J., Markey S. P. (2007) Composition of the synaptic PSD-95 complex. Mol. Cell. Proteomics 6, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernández E., Collins M. O., Uren R. T., Kopanitsa M. V., Komiyama N. H., Croning M. D., Zografos L., Armstrong J. D., Choudhary J. S., Grant S. G. (2009) Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grant S. G. (2010) Targeted TAP tags, phosphoproteomes and the biology of thought. Expert Rev. Proteomics 7, 169–171 [DOI] [PubMed] [Google Scholar]
- 12. Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 13. Keller M. A., Gwinn K., Nash J., Horsford J., Zhang R., Rich S. S., Corriveau R. A. (2007) Whole genome association studies of neuropsychiatric disease: An emerging era of collaborative genetic discovery. Neuropsychiatr. Dis. Treat. 3, 613–618 [PMC free article] [PubMed] [Google Scholar]
- 14. Uhl G. R., Grow R. W. (2004) The burden of complex genetics in brain disorders. Arch. Gen. Psychiatry 61, 223–229 [DOI] [PubMed] [Google Scholar]
- 15. Li K., Hornshaw M. P., van Minnen J., Smalla K. H., Gundelfinger E. D., Smit A. B. (2005) Organelle proteomics of rat synaptic proteins: Correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res. 4, 725–733 [DOI] [PubMed] [Google Scholar]
- 16. Ishihama Y., Sato T., Tabata T., Miyamoto N., Sagane K., Nagasu T., Oda Y. (2005) Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat. Biotechnol. 23, 617–621 [DOI] [PubMed] [Google Scholar]
- 17. Brun V., Dupuis A., Adrait A., Marcellin M., Thomas D., Court M., Vandenesch F., Garin J. (2007) Isotope-labeled protein standards: Toward absolute quantitative proteomics. Mol. Cell. Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 18. Spellman D. S., Deinhardt K., Darie C. C., Chao M. V., Neubert T. A. (2008) Stable isotopic labeling by amino acids in cultured primary neurons: Application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol. Cell. Proteomics 7, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zanivan S., Krueger M., Mann M. (2012) In vivo quantitative proteomics: The SILAC mouse. Methods Mol. Biol. 757, 435–450 [DOI] [PubMed] [Google Scholar]
- 20. Walther D. M., Mann M. (2011) Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics 10, M110.004523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 22. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Page R. D. (1996) TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 24. Hahn C. G., Banerjee A., Macdonald M. L., Cho D. S., Kamins J., Nie Z., Borgmann-Winter K. E., Grosser T., Pizarro A., Ciccimaro E., Arnold S. E., Wang H. Y., Blair I. A. (2009) The post-synaptic density of human postmortem brain tissues: An experimental study paradigm for neuropsychiatric illnesses. PLoS One 4, e5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrison P. J., Weinberger D. R. (2005) Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol. Psychiatry 10, 40–68; image 5 [DOI] [PubMed] [Google Scholar]
- 26. Balu D. T., Coyle J. T. (2011) Neuroplasticity signaling pathways linked to the pathophysiology of schizophrenia. Neurosci. Biobehav. Rev. 35, 848–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. English J. A., Pennington K., Dunn M. J., Cotter D. R. (2011) The neuroproteomics of schizophrenia. Biol. Psychiatry 69, 163–172 [DOI] [PubMed] [Google Scholar]
- 28. Abbatiello S. E., Mani D. R., Keshishian H., Carr S. A. (2010) Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem. 56, 291–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 30. Yang Z., Hayes M., Fang X., Daley M. P., Ettenberg S., Tse F. L. (2007) LC-MS/MS approach for quantification of therapeutic proteins in plasma using a protein internal standard and 2D-solid-phase extraction cleanup. Anal. Chem. 79, 9294–9301 [DOI] [PubMed] [Google Scholar]
- 31. Berna M. J., Zhen Y., Watson D. E., Hale J. E., Ackermann B. L. (2007) Strategic use of immunoprecipitation and LC/MS/MS for trace-level protein quantification: Myosin light chain 1, a biomarker of cardiac necrosis. Anal. Chem. 79, 4199–4205 [DOI] [PubMed] [Google Scholar]
- 32. Estey M. P., Kim M. S., Trimble W. S. (2011) Septins. Curr. Biol. 21, R384–7 [DOI] [PubMed] [Google Scholar]
- 33. Beites C. L., Campbell K. A., Trimble W. S. (2005) The septin Sept5/CDCrel-1 competes with alpha-SNAP for binding to the SNARE complex. Biochem. J. 385, 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amin N. D., Zheng Y. L., Kesavapany S., Kanungo J., Guszczynski T., Sihag R. K., Rudrabhatla P., Albers W., Grant P., Pant H. C. (2008) Cyclin-dependent kinase 5 phosphorylation of human septin SEPT5 (hCDCrel-1) modulates exocytosis. J. Neurosci. 28, 3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X., Serwanski D. R., Miralles C. P., Nagata K., De Blas A. L. (2009) Septin 11 is present in GABAergic synapses and plays a functional role in the cytoarchitecture of neurons and GABAergic synaptic connectivity. J. Biol. Chem. 284, 17253–17265 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.