Abstract
Lipid rafts are highly ordered membrane domains rich in cholesterol and sphingolipids that provide a scaffold for signal transduction proteins; altered raft structure has also been implicated in cancer progression. We have shown that 25 μm 10-(octyloxy) decyl-2-(trimethylammonium) ethyl phosphate (ODPC), an alkylphospholipid, targets high cholesterol domains in model membranes and induces apoptosis in leukemia cells but spares normal hematopoietic and epithelial cells under the same conditions. We performed a quantitative (SILAC) proteomic screening of ODPC targets in a lipid-raft-enriched fraction of leukemic cells to identify early events prior to the initiation of apoptosis. Six proteins, three with demonstrated palmitoylation sites, were reduced in abundance. One, the linker for activation of T-cell family member 2 (LAT2), is an adaptor protein associated with lipid rafts in its palmitoylated form and is specifically expressed in B lymphocytes and myeloid cells. Interestingly, LAT2 is not expressed in K562, a cell line more resistant to ODPC-induced apoptosis. There was an early loss of LAT2 in the lipid-raft-enriched fraction of NB4 cells within 3 h following treatment with 25 μm ODPC. Subsequent degradation of LAT2 by proteasomes was observed. Twenty-five μm ODPC inhibited AKT activation via myeloid growth factors, and LAT2 knockdown in NB4 cells by shRNA reproduced this effect. LAT2 knockdown in NB4 cells also decreased cell proliferation and increased cell sensitivity to ODPC (7.5×), perifosine (3×), and arsenic trioxide (8.5×). Taken together, these data indicate that LAT2 is an early mediator of the anti-leukemic activity of alkylphospholipids and arsenic trioxide. Thus, LAT2 may be used as a target for the design of drugs for cancer therapy.
The development of resistance to drugs that inhibit signaling pathways in cancer cells has emerged as a major limitation of targeted therapy. While the major mechanism of acquired resistance is the emergence of additional mutations or growth factor receptor overexpression (1), recent studies have shown an interesting mechanism of constitutional resistance to epidermal growth factor receptor inhibitors in breast cancer cells, which involves structural alterations in lipid rafts and is independent of the kinase itself (2).
Lipid rafts or membrane rafts are highly ordered membrane domains that are rich in cholesterol and sphingolipids which function by compartmentalizing diverse cellular processes (3, 4), including signal transduction (5–7). Emerging evidence associates altered raft structure with cancer progression (8–10). Therefore, the development of therapeutic strategies for disrupting raft-based cell signaling in cancer represents a potentially useful approach. We and others have presented evidence that alkylphospholipid (APL)1 drugs target raft structure in leukemia (11) and lymphoma cells (12). One such APL, perifosine, is currently in clinical trials as an anti-cancer therapeutic agent (13).
We demonstrated that 10-(octyloxy) decyl-2-(trimethylammonium) ethyl phosphate (ODPC) targets high cholesterol raft-like domains in model membranes and induces apoptosis in leukemia cells, with an effective dose of 25 μm after 24 h in NB4 cells, but has no effect on normal hematopoietic and epithelial cells under the same conditions (11).
Here we present evidence based on quantitative proteomics (14) that the APL ODPC targets proteins recovered in a lipid raft-enriched fraction of leukemic cells. Proteins with predicted palmitoylation sites located in lipid rafts are reduced in abundance after treatment with ODPC. We provide evidence that an adaptor protein for cell signaling, linker for activation of T-cells-2 (LAT2)/non-T-cell activation linker (NTAL)/linker for activation of B-cells (LAB) (15), is involved in early events of ODPC anti-leukemic activity. Additionally, we show that LAT2 knockdown cells obtained with shRNA have suppressed AKT activation, decreased cell proliferation, and increased cell sensitivity to drugs such as ODPC, perifosine, and arsenic trioxide (ATO), indicating that LAT2 is a potential target for the design of drugs for cancer therapy.
EXPERIMENTAL PROCEDURES
Cell Culture and Viability Measurements
The human cell lines NB4 (acute promyelocytic leukemia) (16), U937 (histiocytic lymphoma with myeloid markers) (17), and K562 (chronic myeloid leukemia in blast crisis) (18) were cultured at 37 °C with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cell viability was determined via trypan blue assay, and only cultures with ≥95% viability were used. ODPC was synthesized as described elsewhere (11, 19) and tested at 25 μm concentration for 3, 6, 12, and 24 h with phosphate buffered saline (PBS) as the vehicle control. Apoptotic events were detected via annexin-V and propidium iodide assays using flow cytometry (11). All cell lines were purchased from the American Tissue Culture Collection (Rockville, MD). Perifosine was purchased from Selleck Chemicals (Houston, TX) and dissolved in PBS. The effective dose (ED50) of perifosine for the inhibition of 50% of the proliferation of leukemic cells was determined by means of median dose effect analysis using commercially available software (Calcusyn) (Biosoft, Ferguson, MO) (20).
Effect of ODPC on the PI3K/AKT Pathway
NB4 cells were maintained serum-free overnight (18 h). ODPC (25 μm), Wortmannin (1 μm) (positive control for inhibition of phosphoinositide 3-kinase (PI3K)), or PBS (negative control) was then added for 15 min, and cells were stimulated with a mixture of myeloid growth factors (10 ng/ml each of hr-IL-3, hr-GM-CSF, hr-FLT3-L, and hr-SCF) (PeproTech, Mexico City, Mexico). Aliquots were removed 5, 15, and 30 min after stimulation, and phosphorylation of Ser 473 of AKT was measured via Western blotting.
Effect of the Inhibition of Palmitoyl Transferase and Cholesterol Depletion on LAT2
Palmitoyl transferases were inhibited via the incubation of NB4 cells with 100 μm 2-bromopalmitic acid (2-BrPA) (Sigma, St. Louis, MO), or its vehicle (0.1% v/v dimethyl sulfoxide (DMSO)) as a negative control, for 1 h to deplete palmitoyl residues in raft proteins (21). The cells were collected after 3, 6, 12, and 24 h. For cholesterol depletion, 2.5 × 105 cells were treated with 2.5 mg/ml methyl-β-cyclodextrin (MCD) (product number C4555, lot number 054K01461V; Sigma) for 30 min in serum-free medium (22). After incubation the cells were washed three times with PBS and suspended in complete culture medium containing 25 μm ODPC or PBS as a vehicle control.
Effect of Proteasome Inhibition on APL Induction of LAT2 Degradation
NB4 cells were treated with 10 μm MG132 (Sigma) or 0.1% DMSO (v/v) as a vehicle control for 3 h. Cells were then treated with 25 μm ODPC or PBS as a vehicle control and harvested at 3, 6, 12, and 24 h for LAT2 analysis via Western blotting. Alternatively, NB4 cells were treated with 5 μm MLN9708 (Selleck Chemicals, Houston, TX) for 30 min or 0.1% DMSO (v/v) as a vehicle control and then exposed to 25 μm ODPC or 25 μm perifosine or PBS as a vehicle control and harvested at 3, 6, 12, and 24 h for LAT2 analysis via Western blotting.
Caspase-3 Activity Assay
The tetrapeptide Asp-Glu-Val-Asp (DEVD), modified by N-acetylation of the N terminus and with p-nitroanilide (pNA) to form an alpha peptide bond with the C-terminal aspartic acid residue (Sigma), was used as a substrate. DEVD-dependent protease activity was determined via spectrophotometric measurement of pNA at 405 nm released from the substrate. Caspase activity is reported as nmol substrate hydrolyzed/min. Briefly, following the induction of apoptosis, between 1 × 106 and 2 × 106 cells were rinsed twice with PBS, resuspended in lysis buffer (50 mm HEPES, 100 mm NaCl, 0.1% Chaps, 1 mm DTT, 100 μm EDTA, pH 7.4), incubated on ice for 5 min, and centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatants were denoted total cell lysates (held on ice until use). The protein concentration was determined according to the Bradford method (Bio-Rad, Hercules, CA), using bovine serum albumin as a standard. Thirty micrograms of proteins were incubated with 200 μm substrate for 1 h at 37 °C in reaction buffer (50 mm HEPES, 100 mm NaCl, 0.1% Chaps, 10 mm DTT, 100 μm EDTA, and 10% glycerol, pH 7.4).
SILAC Labeling
NB4 cells were cultured with SILAC, RPMI 1640 medium Kit (Life Tech, Carlsbad, CA), containing light lysine (natural l-Lys) or heavy lysine ([U13C-l-Lys) and supplemented with 10% (v/v) dialyzed fetal bovine serum plus 0.01% (w/v) penicillin/streptomycin as described elsewhere (14). Cells underwent at least seven duplication cycles. The “heavy” labeled NB4 cells were treated with 25 μm ODPC for 3 h, and the “light” labeled NB4 cells were treated with PBS as a control. For each treatment, 2.0 × 108 cells were used.
Cell Fractionation
For total cell extracts, equal amounts of light (control) or heavy (ODPC-treated) NB4 cells were mixed, washed twice with cold PBS, and resuspended in buffer (0.1 ml 25 mm Tris-HCl pH 8.5, 2% SDS plus a protease inhibitor mixture (product number P8340; Sigma)). A d-130 tissue homogenizer (Biosystem, São José dos Pinhais, PR, Brazil) was used at 15,000 rpm for 2 min to lyse the cells. Lysates were centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatants were denoted total cell lysates. Lipid raft-enriched fractions, designated as detergent-resistant membranes (DRMs), were isolated using a successive detergent extraction method exactly as described elsewhere (23), except for the use of a 25-gauge needle instead of a Dounce homogenizer to disrupt the cells. Briefly, NB4 cells were resuspended in buffer M (50 mm HEPES, pH 7.4, 10 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA plus a protease inhibitor mixture, 1 mm Na3VO4, 1 mm NaF, and 1 mm Na4P2O7.10 dH2O) and broken by being passed through a 25-gauge needle 20 times and centrifuged at 500 × g for 10 min at 4 °C to pellet nuclei and intact cells. The supernatant was centrifuged at 16,000 × g for 20 min at 4 °C to pellet membranes. The pellets were resuspended in buffer A (25 mm MES (2-(N-morpholino)-ethanesulfonic acid), 150 mm NaCl, pH 6.5) and samples combined with an equal volume of buffer A containing 2% Triton X-100 and protease inhibitor. Samples were incubated on ice for 60 min and centrifuged at 16,000 × g for 20 min at 4 °C, and the supernatant (the Triton-soluble material) was designated as DSM. Pellets were rinsed briefly with buffer A and resuspended in buffer B (10 mm Tris-Cl, pH 7.6, 150 mm NaCl, 60 mm β-octyl glucoside and phosphatase and protease inhibitor). Samples were incubated on ice for 30 min and centrifuged at 16,000 × g for 20 min at 4 °C, and supernatants were collected as the lipid raft-enriched fraction that was designated as DRM. Western blot analysis was performed to determine the level of efficacy of enrichment with the lipid raft marker Lyn, the non-raft marker Ergic-53, and the nucleus marker histone H4.
Sucrose Density Gradient Centrifugation
NB4 cells (4 × 108) were washed twice with cold PBS and lysed in 2 ml of 25 mm MES buffer (pH 6.5), 150 mm NaCl containing a protease inhibitor mixture (product number P8340; Sigma), and 1% Triton X-100 for 30 min at 4 °C. Cells were disrupted by being passed through a 25-gauge needle 20 times followed by use of a d-130 tissue homogenizer (Biosystem) (15,000 rpm) for 2 min. The suspension of disrupted cells was then centrifuged at 500 × g for 10 min at 4 °C to pellet nuclei and intact cells. About 1 ml of supernatant was layered onto a continuous sucrose density gradient (0%–63% (w/v), 13 ml) in 25 mm MES buffer (pH 6.5) containing 150 mm NaCl, and centrifugation was carried out for 4 h at 180,000 × g using a Hitachi vertical rotor (P65VT3) at 4 °C. Fractions of 1 ml were collected and assayed for protein content and refractive index. The protein concentration was determined according to the Bradford method (Bio-Rad) using bovine serum albumin as standard. Five micrograms of protein from each fraction were subjected to SDS-polyacrylamide gel electrophoresis and Western blot analysis.
LC-MS/MS Analysis
The proteins present in the total protein extract or DRM fraction were partially separated by 12.5% SDS-PAGE. Each lane was cut into 6 or 10 pieces, washed, and digested with trypsin as described elsewhere (24). Tryptic peptides were successively extracted with 0.1% formic acid and 50% acetonitrile and then 70% acetonitrile and dried with a SpeedVac apparatus (Thermo Scientific, Marietta, OH). Peptide mixtures were dissolved in 45 μl 4% acetonitrile, 0.1% formic acid, and 0.5 m urea and analyzed via LC-MS/MS in a nanoflow reversed-phase HPLC system connected to an LTQ Orbitrap mass spectrometer (Thermo Scientific). Chromatography was carried out with an in-house packed 75-μm inner diameter (New Objectives) × 25-cm long C18 column packed with Magic C18 resin at 250 nl/min with 90 min linear gradients from 5% to 40% acetonitrile in 0.1% formic acid. MS/MS scans of the five most abundant doubly or triply charged peaks in the FT-MS scan were recorded in a data-dependent mode in the linear ion trap (25). Two additional verification replicate runs were performed using the same amount of DRM extract mixtures. The data for these replicates were collected as described above. Peptides and proteins were identified with the Computational Proteomics Analysis System (26) using the X!Tandem search engine (January 2007 release) (27) and Peptide Prophet (28) and Protein Prophet (29) algorithms for the statistical validation of data and protein grouping. MS data were searched in the human International Protein Index (IPI version 3.52; 73,950 entries). Search parameters for tryptic peptides included up to two missed cleavages, mass allowances of 0.5 Da for fragment ions, fixed cysteine modification with carbamidomethylation (+57.02146), variable methionine oxidation (+15.99491), and variable lysine modification (+6.020129) to account for both heavy and light SILAC labels. Only peptides with a Peptide Prophet score above 0.90 and precursor ions with a delta mass less than 20 ppm were considered for protein identification and quantification. The list of proteins was generated with a Protein Prophet cut-off value of 0.9, representing an overall protein false discovery rate of ∼2% based on the Protein Prophet estimate and including proteins identified based on single peptide hits. Proteins were quantitated as previously described, using the Q3 algorithm to measure SILAC peak intensities (30, 31).
Western Blotting
NB4, U937, or K562 cells were washed twice in cold PBS; lysed with lysis buffer, 50 mm Tris-HCl, pH 8.5, 2% SDS, 1 mm Na3VO4 containing the protease inhibitor mixture (product number P8340; Sigma); and homogenized in a d-130 tissue homogenizer (15,000 rpm) (Biosystems, Sao Jose dos Pinhais, PR, Brazil) for 2 min on ice. Lysates were centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatants were designated as total cell lysates. The protein concentration was determined according to the Bradford method (Bio-Rad). Proteins were submitted to SDS-PAGE and transferred to polyvinylidene fluoride membranes (GE Lifesciences, Pittsburgh, PA). Membranes were blocked with 5% nonfat milk in 0.05% Tween-TBS and incubated with the specific antibodies. Mouse anti-β-actin (sc-81178), goat anti-Lyn(44)-G (sc-15G), goat anti-ERGIC-53 (sc-32442), rabbit anti-histone H4 (H-97) (sc-10810), rabbit anti-AKT 1(5G12) (sc-81435), rabbit anti-phospho-AKT 1/2/3 (Ser-473) (sc-101629), rabbit anti-PTEN (C-20)-R (sc-6817-R), and horseradish peroxidase-conjugated secondary donkey anti-goat IgG (sc-2033) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-phospho-PTEN (Ser-380/Thr-382/383) (#9549), rabbit anti-caspase-3 (#9662), rabbit anti-cleaved caspase-3 (#9661), rabbit anti-PARP (#9542), rabbit anti-cleaved PARP (#9541), rabbit anti-caspase-7 (#9492), rabbit anti-cleaved caspase-7 (#9491), rabbit anti-caspase-9 (#9502), rabbit anti-cleaved caspase-9 (#9501), rabbit anti-cleaved caspase-8 (#9496), rabbit anti-NTAL/LAB (#9533), rabbit anti-phospho-S6 ribosomal protein (Ser-235/236) (#4858), rabbit anti-AKT (#9272), rabbit anti-phospho-AKT (Ser-473)(#4058), and horseradish peroxidase-conjugated secondary antibody goat anti-rabbit IgG were purchased from Cell Signaling (Beverly, MA). Goat anti-mouse IgG (NA931VS) was purchased from GE Healthcare Life Sciences. The apparent molecular weights reported in the figures were obtained via comparison with a biotinylated protein ladder (#7727 and #7075) (Cell Signaling, Beverly, MA). The antibody–protein complex was detected using ECL Western blotting detection reagents (GE Healthcare Life Sciences). Western blotting experiments were quantified with image analysis software (ImageQuant TL) (GE Healthcare Life Sciences).
Lentiviral Transduction for Knockdown of LAT2
Stable NB4 cell line knockdown of LAT2 was obtained using MISSION lentiviral shRNA transduction particles (catalogue number SHCLNV-NM_014146; Sigma) according to the manufacturer's protocol. The shRNA sequence against LAT2 used was CCGGGAAGATGAGGAATCTGAGGATCTCGAGATCCTCAGATTCCTCATCTTCTTTTTTG(TRCN0000129029). MISSION TurboGFP™ Control Transduction Particles (Sigma) were used as a control for transduction efficiency. Empty vector virus (pLKO.1, catalogue number SHC003V; Sigma) was used as a negative control for LAT2 knockdown. RPMI 1640 medium was used for viral transduction of NB4 cells in the presence of polybrene (8 μg/ml) (Sigma). NB4 cells were transduced at a multiplicity of infection of 0.25. After 16 to 18 h, cells were cultured in fresh medium, and 1 week after transduction cells were selected with puromycin (3 μg/ml) (Sigma) for 1 week to obtain a stable cell line. Reduced basal LAT2 expression was detected via Western blotting after puromycin selection. NB4 cells transfected with an empty vector maintained the same behavior as the parental nontransfected NB4 cell line and were used as NB4 wild type cells (NB4 WT) in all comparisons with the NB4 LAT2 knockdown (NB4 LAT2KD) cell line.
Effect of LAT2 on AKT Activation
NB4 WT and NB4 LAT2KD cells were maintained serum-free overnight (18 h). Cells were assayed via trypan blue exclusion assay, and only cultures with viability > 95% were used. Cells were stimulated with a mixture of myeloid growth factors (10 ng/ml each of hr-IL-3, hr-GM-CSF, hr-FLT3-L, and hr-SCF) (PeproTech, Mexico City, Mexico). Aliquots were obtained at 5 and 15 min after stimulation and assayed for AKT phosphorylation at Ser 473 (product number 4058; Cell Signaling) via Western blotting.
Population Doubling Assay of NB4 WT and NB4 LAT2KD Cells
On day 0, the cell concentrations of exponentially growing NB4 WT and NB4 LAT2KD cells were determined in a Coulter counter (T890 automatic cell counter) (Beckman Coulter, Brea, CA), and cells were then plated in 25 cm2 culture flasks in triplicate at a cell concentration of 5 × 105 cells/ml with a final volume of 5 ml, where they were cultured for a total of 10 days. At 48-h intervals, the cells were counted and re-plated in fresh flasks under the same conditions. Nonlinear regression analysis was used to determine the doubling time (DT) and the growth rate (GR) of both cell lines. DT is defined as the time in hours needed for one population doubling, and GR is the number of doublings in 1 day. DT values were determined using Graph Pad Prism 5 software (GraphPad Software, Inc., La Jolla, CA) to carry out nonlinear regression using the least squares method and the exponential growth equation as a model. The comparison of the two growth rates was made via the extra sum-of-squares F test.
The values of the cell concentrations determined each 48 h were used as inputs for y, and y0 was fixed at 5 × 105 cells/ml. The constant k was equal to the GR that was reported as an inverse of time and was calculated by the equation: y = yo.exp(kx), where x is the time. The value of DT was a transformation of the growth rate:
Dose-effect Analysis with Arsenic Trioxide, ODPC, or Perifosine on NB4 WT or NB4 LAT2KD Cells
For the measurements of cytotoxicity we used the Annexin V apoptosis detection kit APC (e-Biosciences, San Diego, CA) as recommended by the manufacturer. We acquired the samples using a FACSCalibur flow cytometer (BD Biosciences, Heidelberg, Germany) and analyzed the data using FlowJo software (TreeStar Inc., San Carlos, CA). We considered a toxic effect to be indicated by the externalization of phosphatidylserine (annexin-positive cells) and/or the loss of plasma membrane integrity (propidium iodide-positive cells). The double negative fraction of cells was considered not affected. NB4 WT or NB4 LAT2KD in exponential growth were plated in 24-well plates at a density of 5 × 105 cell/ml with a final volume of 1 ml per well. Cells were treated for 24 h with ATO at final concentrations of 0.0625, 0.125, 0.25, 0.5, 1, 2, and 4 μm; ODPC at 6.25, 12.5, 25, 50, and 100 μm; and perifosine at 2.5, 5, 10, 20, and 40 μm. Dose-effect analysis is based on the median effect equation of Chou and Talalay (32). We used the Calcusyn software (Biosoft Inc., Cambridge, UK) to calculate the data and create a dose-effect table and graphics for each drug in each cell line. The effective-dose 50% (ED50), the 95% confidence interval (95% CI), and the regression coefficient (r) are reported.
RESULTS
Induction of Apoptosis in NB4 and U937 Leukemia Cells by ODPC
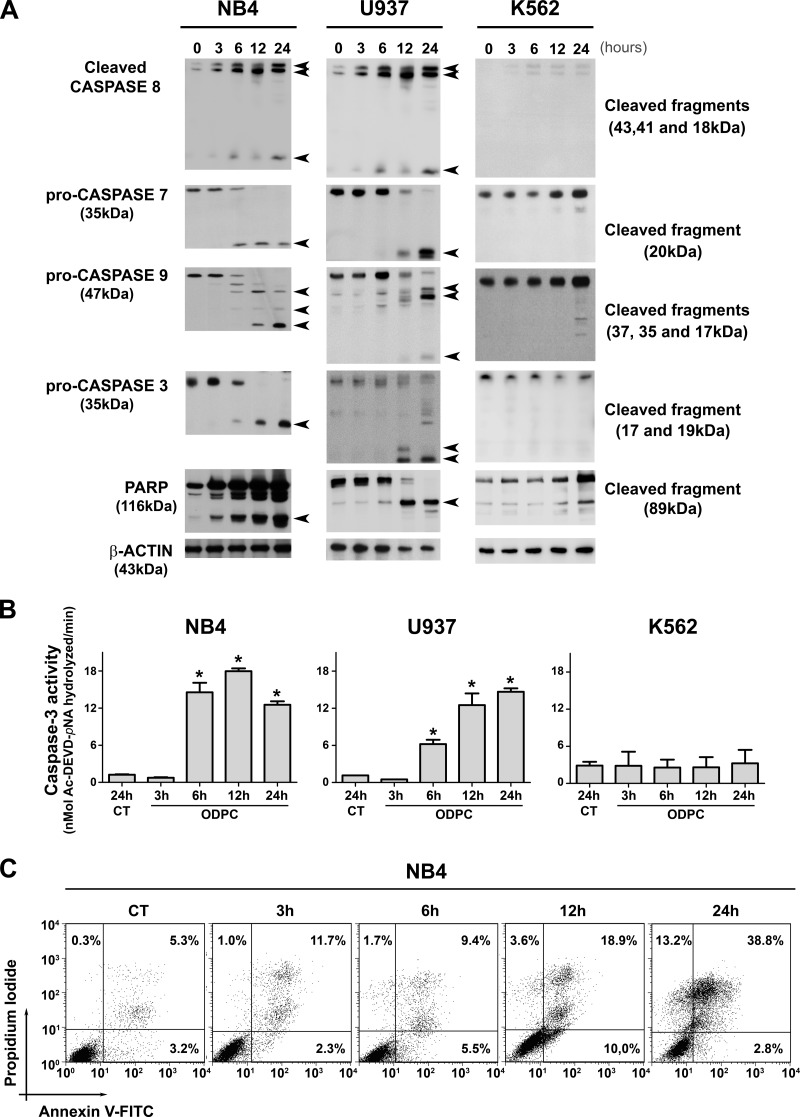
In order to identify the early events of ODPC treatment of acute myeloid leukemia (AML) cells, we determined the time of appearance of some of the proteins that participate in apoptosis after treatment with 25 μm ODPC. The concentration of ODPC used was the ED50 for these cells for 24 h (11). In a time course study, NB4 (acute promyelocytic leukemia cells) and U937 (myelomonocytic leukemic cells) were treated with 25 μm ODPC, initiating apoptosis after 6 h, based on caspase activation (caspase-3, -7, -8, and -9) as indicated by Western blotting (Fig. 1A) and caspase-3 enzyme activity (Fig. 1B). These results were also confirmed by measuring annexin-V and propidium iodide staining using flow cytometry in NB4 cells (Fig. 1C). In contrast, there was no evidence of apoptosis following treatment of the K562 chronic myeloid leukemia cell line with 25 μm ODPC for up to 24 h (Figs. 1A and 1B). To identify changes in protein abundance induced by ODPC, we carried out mass spectrometric analysis of stable isotope-labeled proteins of a lipid raft-enriched membrane fraction from NB4 cells.
Fig. 1.
Kinetics of apoptosis induced in leukemia cells by treatment with ODPC. A, apoptosis was initiated within 6 h of exposure to 25 μm ODPC in AML cell lines. Antibodies were used to detect activated caspases, pro-caspase, or cleaved fragments. B, colorimetric caspase-3 activity assay using the specific peptide substrate (AcDEVD-pNA) for activated caspase-3. Measurements were carried out using 30 μg total protein; p < 0.05 compared with control (analysis of variance). C, annexin V and propidium iodide staining was measured via flow cytometry of NB4 cells. The K562 cell line showed a pattern of resistance compared with the U937 and NB4 cell lines (A and B). On the basis of these results, we used NB4 cells treated with 25 μm ODPC for 3 h to perform the proteomic screening. Arrows indicate the cleaved fragments.
Isolation of the Lipid Raft-enriched Detergent-resistant Membrane Fraction from NB4 Cells
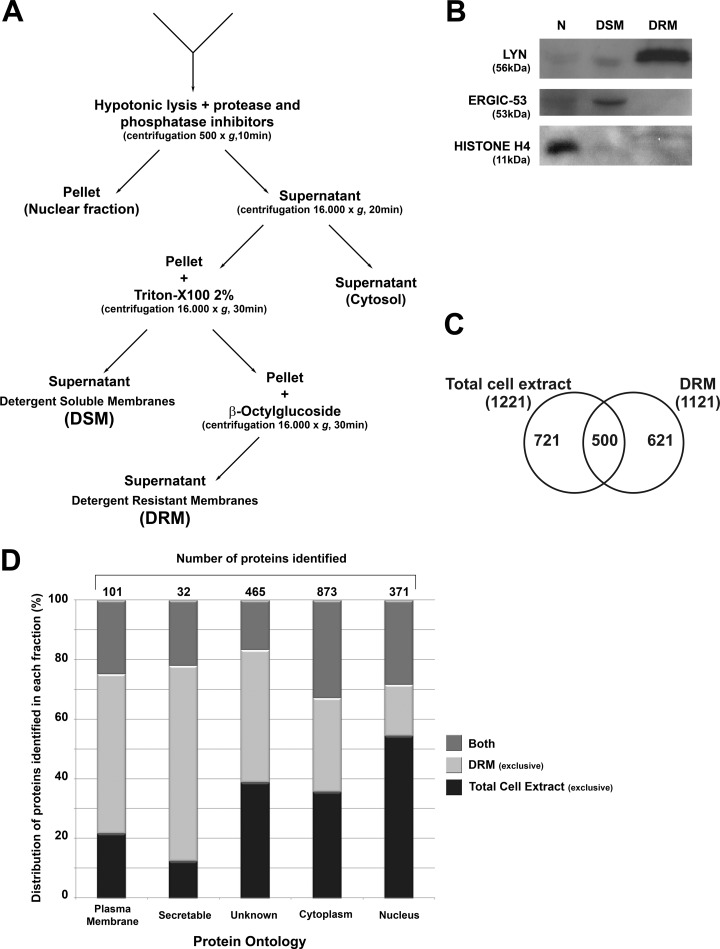
DRMs were isolated on the basis of their differential detergent solubility (23) (see Fig. 2A). The selectivity of the method is demonstrated in Fig. 2B by the low level of cross-contamination in blots prepared with antibodies to LYN, a specific marker of lipid rafts (33), to ERGIC-53, a marker of endoplasmic reticulum Golgi intermediate compartment (34), and to the nuclear marker histone H4 (35).
Fig. 2.
Isolation of detergent-resistant membrane (DRM) and detergent-soluble membrane (DSM) fractions from NB4 cells and the identification of proteins by proteomics. A, successive detergent extraction method to isolate Triton X-100-resistant membranes (DRM) that are rich in lipid rafts. B, quality control Western blots of specific markers for lipid rafts, non-raft membranes, and nuclear proteins. Lyn, a raft-associated kinase, appears only in the DRM fraction; ERGIC-53, an endoplasmic reticulum-Golgi network protein, appears only in the non-raft membranes; and histone-H4 is present only in the nuclear fraction. C, number of proteins identified exclusively in the total cell extract, the DRM fraction, or both fractions. D, cellular compartment ontology identified proteins exclusively in the total cell extract, exclusively in the DRM fraction, or in both fractions. Annotation was obtained with Ingenuity Pathway Analysis software.
Quantitative Proteomics of Identified ODPC Targets in Detergent-resistant Membranes
Cultured cells were grown with SILAC reagents (with complete incorporation of labeled amino acids) before ODPC treatment. Mass spectrometry was used to identify the proteins whose abundance was modified by ODPC treatment. A 3-h exposure to 25 μm ODPC was used because apoptosis had not yet been initiated at that early time (Figs. 1A and 1B). Both the whole cell lysate and the DRM fraction were analyzed via GEL-LC-MS/MS, and 1842 proteins were identified with less than a 2% false discovery rate based on Protein Prophet Algorithm estimations (supplemental Table S1). Fig. 2C shows that a total of 621 proteins were identified exclusively in the DRM fraction extract (i.e. not in the total cell extract) and that 721 proteins were identified exclusively in the total cell extract. The fact that only 500 proteins were detected in both extracts was somewhat surprising, given that the DRM fraction was prepared from the total cell lysate. Cellular compartment ontology of proteins was used to identify proteins exclusively in the total cell extract, exclusively in the DRM fraction, or in both fractions. Annotation was obtained with Ingenuity Pathway Analysis software. The DRM fraction sub-proteome was, as expected, relatively enriched with membrane and membrane-associated proteins relative to the total cell lysate (Fig. 2D).
For quantitative analysis based on a discovery SILAC experiment, proteins were considered to be regulated only if at least two lysine-containing tryptic peptides were identified with peptide prophet scores > 0.90 and the ratio of ODPC-treated versus non-treated NB4 cells was greater than 2-fold. Based on these criteria, ODPC treatment increased the abundance of only one protein in the total cell extract, small nuclear ribonucleoprotein. In contrast, six proteins were significantly decreased in abundance in the DRM fraction after ODPC treatment, as shown in Table I. Two additional replicates were performed for verification purposes. The replicates confirmed a significant decrease in abundance (≥1.3-fold) of all six proteins initially selected in the discovery SILAC experiment. The proteins reduced in abundance were two ribosomal proteins (RPL23 and RPL38), one ATP-dependent protease (CLPP), one membrane chlorine channel (CLIC1), one adaptor protein for signal transduction named LAT2, and one Splicing factor 3A subunit 1 (SF3A1). A complete summary of our quantitative data analysis based on SILAC is presented in supplemental Table S2.
Table I. Lipid raft proteins reduced in abundance by 3 h of treatment of NB4 cells with 25 μm ODPC.
| IPI | Protein | Gene | Experimentally validated palmitoylationa | Ratiob (ODPC treated/control) | Standard deviationb | Total quant. eventsb |
|---|---|---|---|---|---|---|
| IPI00395993 | Linker for activation of T-cell family member 2 | LAT2 | Yes (36–38) | 0.58 | 0.23 | 10 |
| IPI00003870 | Clp protease proteolytic subunit, mitochondrial precursor | CLPP | No | 0.59 | 0.24 | 5 |
| IPI00010896 | Chloride intracellular channel 1 | CLIC1 | Yes (38) | 0.68 | 0.15 | 23 |
| IPI00010153 | 60S ribosomal protein L23 | RPL23 | Yes (37) | 0.74 | 0.35 | 35 |
| IPI00017451 | Splicing factor 3 subunit 1 | SF3A1 | No | 0.75 | 0.26 | 10 |
| IPI00215790 | 60S ribosomal protein L38 | RPL38 | No | 0.75 | 0.10 | 12 |
a Experimentally validated palmitoylation is reported in the references cited within parentheses.
b Values obtained from three replicate proteomic experimental experiments.
Of these six downregulated proteins, only LAT2 has been identified as a lipid raft protein (36). LAT2, RPL23, and CLIC1 contain sites of palmitoylation (37, 38), a post-translational modification that is believed to target proteins to lipid rafts. Furthermore, when these six proteins were evaluated with respect to tissue expression using BioGPS (39), LAT2 was shown to be specifically expressed in hematopoietic tissues. In these analyses, the normal counterparts of B-acute lymphoblastic leukemia, AML, and myelomonocytic leukemia presented LAT2 mRNA expression levels almost 100 times greater than the basal gene expression (supplemental Fig. S1). LAT2 was selected for in-depth study as a target of ODPC on the basis of these considerations.
LAT2 as a Target of APLs
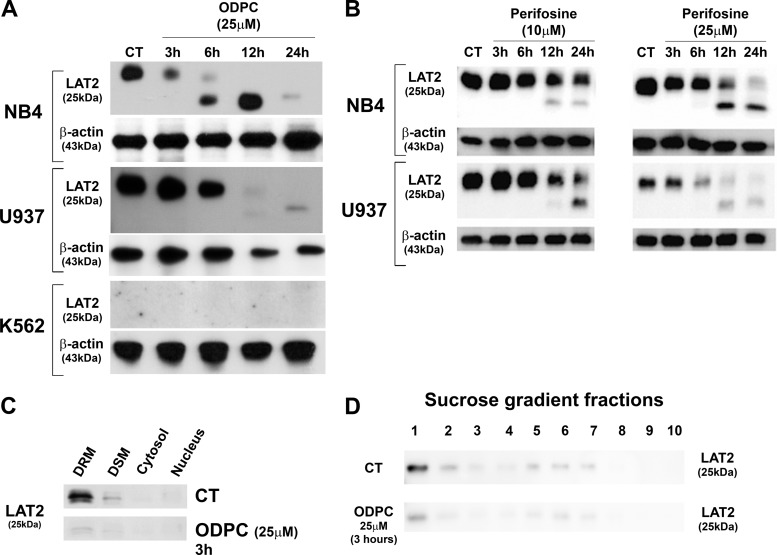
LAT2, an adaptor protein for signal transduction in lipid rafts of mast cells (40), was reduced in abundance by treatment of NB4 cells with ODPC (Table I) and in U937 cells, based on Western blotting. However, LAT2 was not detected in K562 cells (Fig. 3A), which were relatively more resistant to ODPC treatment than NB4 and U937 cells (Figs. 1A and 1B). Perifosine, an APL currently in clinical trials for cancer treatment, also induced a reduction of LAT2 abundance in NB4 and U937 cells (Fig. 3B). Interestingly, ODPC induced a reduction of LAT2 in the DRM fraction of NB4 cells after only 3 h of treatment (Fig. 3C). A similar reduction of LAT2 was obtained in lipid rafts isolated with a sucrose gradient after 3 h of treatment with ODPC (Fig. 3D) (41).
Fig. 3.
Reduction of the abundance of LAT2 by alkylphospholipids. A, Western blot validation (total cell extract) of the effect of ODPC on LAT2 in NB4 and U937 cells. K562 does not express LAT2. B, Western blots (total cell extract) show the reduction of LAT2 after treatment with perifosine (10 and 25 μm) in NB4 and U937 cells. C, D, Western blotting of LAT2 in cell fractions of control and ODPC-treated NB4 cells (25 μm for 3 h). The successive detergent fractionation method was used in C, and the sucrose density gradient centrifugation method was used in D. DSM, detergent soluble membranes; DRM, detergent resistant membranes.
Chemically Induced Perturbations of the Structure of Lipid Rafts Mimic or Potentiate the Effect of ODPC on LAT2
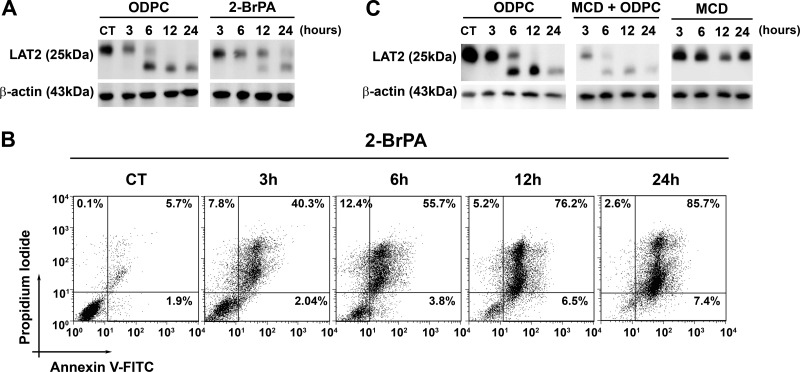
Palmitoylation of cysteine residues is a post-translational modification that targets proteins to lipid rafts (42) and is involved in mechanisms of regulation of signal transduction (43). There is strong experimental evidence that LAT2 is palmitoylated in vivo (36, 44). We have demonstrated that ODPC disrupts model lipid bilayers (11). We tested whether other substances that target lipid rafts would have the same effects on LAT2 as observed for ODPC in NB4 cells. Indeed, the depletion of palmitoylated residues in proteins via the inhibition of palmitoyl protein transferases using 2-BrPA had an effect very similar to that of ODPC treatment on NB4 cells, including a reduction of LAT2 and the induction of apoptosis (Figs. 4A and 4B). Moreover, cholesterol depletion by MCD significantly increased the effect of ODPC on LAT2 in NB4 cells (Fig. 4C), but MCD alone had no effect on LAT2 levels and did not induce apoptosis (data not shown).
Fig. 4.
Chemically induced perturbations of lipid raft structure mimic or potentiate the effect of ODPC on LAT2. Inhibition of protein palmitoylation by 100 μm 2-bromopalmitic acid (2-BrPA) has an effect on LAT2 abundance (A) and apoptosis induction (B) that is very similar to that of 25 μm ODPC treatment. NB4 cells were exposed to 2-BrPA. LAT2 abundance was evaluated via Western blot, and apoptosis induction via annexin-V and propidium iodide staining by flow cytometry at the indicated times. C, cholesterol depletion by methyl-β-cyclodextrin (MCD) increases the effect of ODPC on LAT2 abundance; however, MCD has a very limited effect when used alone. NB4 cells were exposed to MCD (2.5 mg/ml) in serum-free medium for 30 min, and subsequently MCD was washed out and complete medium was restored. LAT2 abundance was evaluated via Western blot at the indicated times.
LAT2 Is Degraded by Proteasomes Following Treatment with APLs
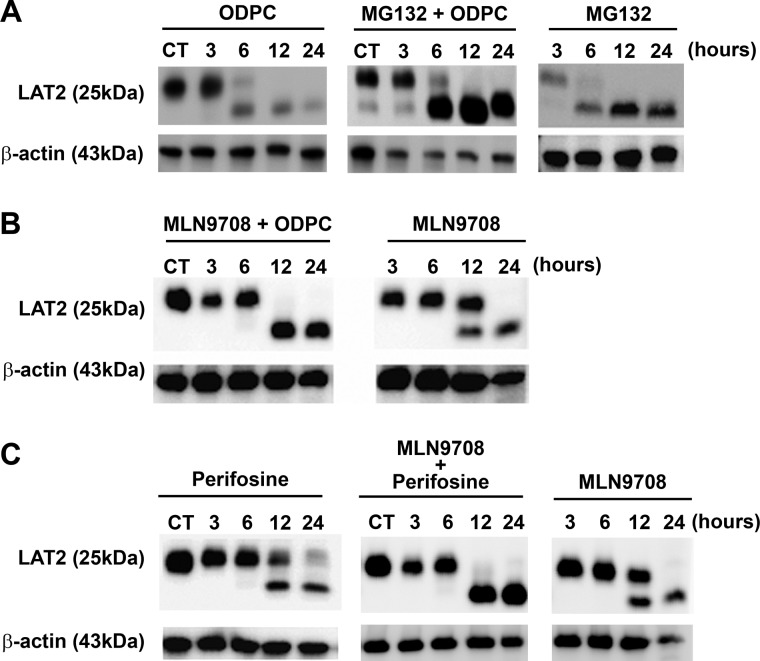
The regulation of LAT2 through degradation by proteasomes has been reported (36). To determine whether proteasome-mediated degradation follows LAT2 displacement from lipid rafts by ODPC treatment, we exposed NB4 cells to MG132, a potent and specific cell-permeable proteasome inhibitor, followed by treatment with 25 μm ODPC. LAT2 was accumulated in ODPC-treated cells in the presence of the proteasome inhibitor MG132, suggesting that ODPC treatment makes LAT2 susceptible to proteasomal degradation (Fig. 5A). To further evaluate the role of the proteasome in LAT2 degradation following APL exposure, we used a second proteasome inhibitor (MLN9708) on ODPC- or perifosine-treated cells. In these experiments, the presence of MLN9708 caused an accumulation of LAT2 after ODPC (Fig. 5B) or perifosine (Fig. 5C) treatment, confirming that LAT2 is degraded by proteasomes after treatment with APL.
Fig. 5.
LAT2 is degraded by proteasomes after treatment with alkylphospholipids. A, 3-h treatment of NB4 cells before exposure to the proteasome inhibitor MG132 (10 μm) prevented the reduction of LAT2 induced by 25 μm ODPC. B, C, a similar effect was observed after exposure (30 min) of NB4 cells to the proteasome inhibitor MLN9708 (5 μm) followed by treatment with 25 μm ODPC (B) or 25 μm perifosine (C).
Reduction of the Abundance of LAT2 by ODPC Is Associated with Inhibition of the AKT Signaling Pathway
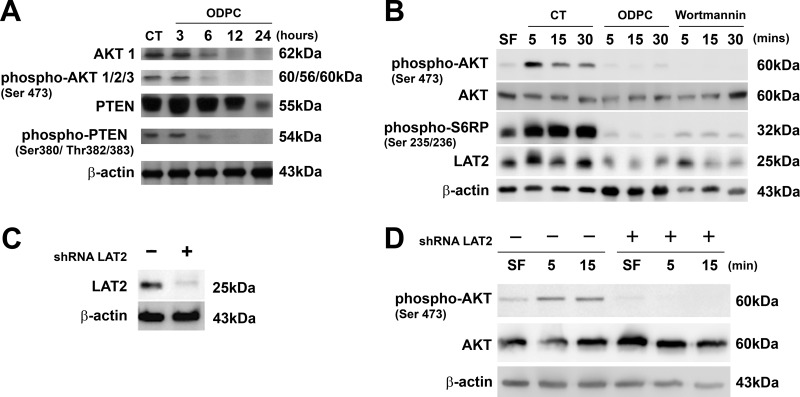
There is evidence that LAT2 functions as an adaptor molecule that positively regulates mast cell survival by inhibiting the recruitment of protein phosphatases to lipid rafts, leading to AKT hyperphosphorylation and activation (45). In addition, perifosine has been shown to be an inhibitor of AKT signaling (46). On the basis of these considerations, we determined whether ODPC treatment had an effect on the AKT signaling pathway. Treatment with 25 μm ODPC reduced AKT and phospho-AKT levels after 3 h of treatment (Fig. 6A). There was no effect on PTEN, the main negative regulator of the AKT pathway, except at 24 h of treatment, and this effect occurred after the onset of apoptosis. Thus there was no evidence for the reduction of PTEN at earlier times. We also observed that the C-terminal region of PTEN was dephosphorylated after 3 h in the presence of ODPC (Fig. 6A). C-terminal dephosphorylation has been considered as an indicator of PTEN phosphatase activity (47) that negatively regulates the AKT pathway.
Fig. 6.
Reduction of the abundance of LAT2 by ODPC or LAT2 knockdown by shRNA (shRNA LAT2) is associated with lack of activation of the AKT pathway. A, ODPC (25 μm) reduced AKT and phospho-AKT levels after 3 h of treatment; however, PTEN was reduced only at 24 h, when apoptosis was fully installed. ODPC treatment induced a state of hypophosphorylation at the C terminus of PTEN, which has been reported to be required in order to initiate phosphatase activity (41). B, ODPC inhibited AKT activation by myeloid growth factors (MGFs) in a manner similar to Wortmannin, a specific PI3K inhibitor. ODPC inhibited the up-regulation of LAT2 induced by MGFs under the same conditions and inhibited ribosomal S6P phosphorylation induced by MGFs. NB4 cells were maintained serum-free (SF) overnight, treated with PBS as control with 25 μm ODPC or 1 μm Wortmannin for 15 min, stimulated with an MGF mixture (10 ng/ml IL-3, GM-CSF, l-FLT-3, and SCF), and harvested at the indicated times. C, LAT2 stable knockdown was obtained from NB4 cells via lentiviral transduction of shRNA that targets LAT2. D, AKT activation was suppressed in LAT2 stable knockdown NB4 cells. Both cell lines were maintained SF overnight and subsequently stimulated with an MGF mixture (same as described above) and harvested at the indicated times.
Given prior evidence linking LAT2 to signaling in myeloid cells (15, 48), we next determined whether ODPC could impair AKT activation and alter LAT2 levels as a result of treatment with myeloid growth factors (MGFs). In order to maintain a state of hypophosphorylation of AKT, NB4 cells were maintained in serum-free medium overnight and were subsequently treated with 25 μm ODPC or its vehicle control (PBS) for 15 min. The functionality of the pathway was tested by the addition of a growth factor mixture containing 10 ng/ml each of GM-CSF, IL-3, FLT3-L, and SCF. Analysis of AKT phosphorylation at Ser 473, a cancer cell survival signal (49), showed that ODPC inhibited AKT activation (phosphorylation at Ser 473) in the first minutes of the test (Fig. 6B) and to the same extent as treatment with Wortmannin, a classical irreversible inhibitor of the upstream kinase PI3K (50). The absence of AKT activation in ODPC- and Wortmannin-treated cells was supported by hypophosphorylation of one of its downstream targets, S6-ribosomal protein (Fig. 6B). The phosphorylated state of S6-ribosomal protein is essential for proper translation of proteins by the ribosome (51). In our experiments, LAT2 was rapidly synthesized after MGF addition to control-treated cells but suppressed in ODPC- and Wortmannin-treated cells, suggesting that LAT2 mRNA translation is a highly regulated process that depends on the AKT pathway.
The reduction of the abundance of LAT2 and the inhibition of AKT signaling observed upon treatment with ODPC (Figs. 3A, 3B, and 6B) or perifosine (52) suggest that LAT2 contributes to the cytotoxic effect of these drugs. To further test this hypothesis, we created a LAT2 knockdown cell line derived from NB4 by the stable expression of shRNA that targets LAT2 expression. Fig. 6C shows a representative Western blot that confirms the reduction of LAT2 protein by more than 70% in the NB4 LAT2KD.
The Knockdown of LAT2 in NB4 Cells Impairs AKT Activation by MGF
To further determine whether LAT2 is essential for AKT activation, we compared the response of NB4 WT with NB4 LAT2KD to AKT activation triggered by MGF. We observed an absence of AKT phosphorylation at Set 473 in NB4 LAT2KD after the MGF stimulus (Fig. 6D), supporting the importance of this adaptor protein for AKT activation (45).
The Knockdown of LAT2 in the NB4 Cell Line Results in a Reduction of Cell Proliferation
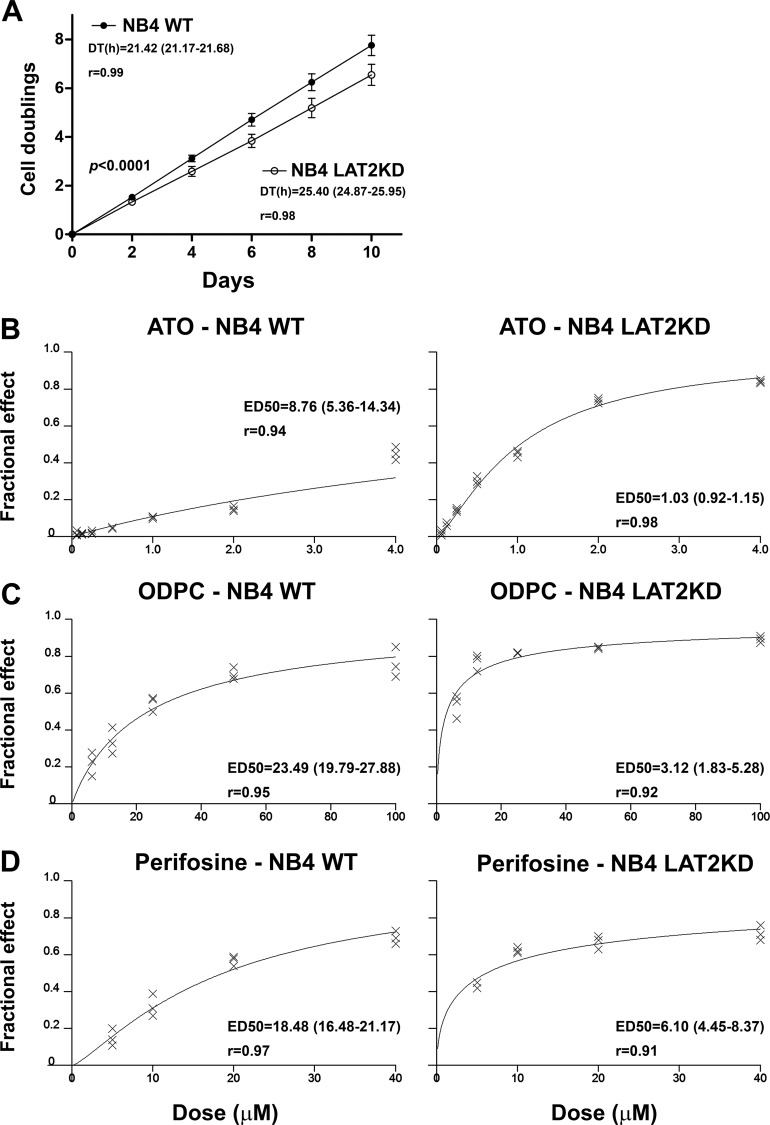
We were able to maintain NB4 cells as viable despite the LAT2 knockdown. However, NB4 LAT2KD presented a lower rate of cell proliferation as determined by a cell population doubling assay. The mean and 95% CI for the GR, reported as the number of cell duplications per day, were 0.776 (0.767–0.786) for NB4 WT and 0.655 (0.641–0.669) for NB4 LAT2KD. The DT was lower in the parental NB4 cell line (NB4 WT) than in NB4 LAT2KD (Fig. 7A). This result demonstrates that the knockdown of LAT2 reduces the proliferation potential of NB4 cells, probably by inducing AKT suppression (53, 54).
Fig. 7.
LAT2 knockdown NB4 cells (NB4 LAT2KD) have a lower proliferation potential and a higher sensitivity to APLs or ATO. A, NB4 LAT2KD cells have a lower proliferation potential than the parental NB4 cells, as determined by the doubling time, which is higher in NB4 LAT2KD. B–D, NB4 LAT2KD showed greater sensitivity to apoptosis induction by ATO (B), ODPC (C), and perifosine (D). Values in parentheses indicate the 95% CI.
The Knockdown of LAT2 in NB4 Cells Results in Increased Sensitivity to the Action of APLs and ATO
Because AKT is a conserved pro-survival pathway (55), we hypothesized that the down-regulation of LAT2 and consequent AKT pathway impairment might induce a state of increased sensitivity to drug action, irrespective of the mechanism of action of the agent. We tested this hypothesis by comparing NB4 WT and NB4 LAT2KD cell lines with respect to sensitivity to ATO. Additionally, we compared the sensitivity of the cell lines to the APLs ODPC and perifosine. Figs. 7B–7D show dose-response curves for the three agents, based on the dose-effect equation described by Chou and Talalay (32), and show that NB4 LAT2KD presented greater sensitivity to all three treatments. Values of ED50 and the respective increased sensitivity to the treatment are presented in Table II. Note that the sensitivity of NB4 LAT2KD cells to these drugs is 3- to 8-fold greater than that of the NB4 WT cells.
Table II. Effective dose 50% (ED50) of ATO, ODPC, or perifosine for the inhibition of the proliferation of NB4 wild type (NB4 WT) and LAT2 knockdown NB4 (NB4 LAT2KD) cells.
| NB4 WT |
NB4 LAT2KD |
Relative drug susceptibilitya |
|||||
|---|---|---|---|---|---|---|---|
| ED50 (μm) | 95% CIb | r | ED50 (μm) | 95% CIb | r | NB4 WT/NB4 LAT2KD | |
| ATO | 8.76 | 5.36 to 14.34 | 0.94 | 1.03 | 0.92 to 1.15 | 0.98 | 8.50 |
| ODPC | 23.49 | 19.79 to 27.88 | 0.95 | 3.12 | 1.83 to 5.28 | 0.92 | 7.53 |
| Perifosine | 18.67 | 16.48 to 21.17 | 0.97 | 6.10 | 4.45 to 8.37 | 0.91 | 3.06 |
a Ratio calculated by averaging ED50.
b 95% CI is the 95% confidence interval for ED50, expressed in μm.
DISCUSSION
ODPC induces apoptosis in the AML cells NB4 and U937, but K562 leukemia cells are relatively more resistant. Normal cells are also resistant to ODPC at the same concentrations that initiate apoptosis in leukemia cells (11). We have demonstrated here via a quantitative proteomic analysis that some lipid raft proteins are reduced in abundance in the DRM fraction by ODPC treatment. This result illustrates the importance of the use of subcellular fractionation to enrich and identify raft proteins. For example, we identified 621 proteins in the DRM fraction that were not demonstrable in the total cell extract from which DRM was prepared. The simplest explanation for this is that the preparation of the DRM fraction enriched the proteins sufficiently for them to be detected, but they were too dilute to be detected in the total cell extract. Quantitative mass spectrometric analysis of the DRM fraction indicated that LAT2 and five other proteins are potential targets of ODPC in leukemia cells. In addition, it is important to emphasize the efficacy of the method recently described by Adam et al. (25) for obtaining the DRM fraction enriched with lipid raft proteins.
After the proteomic discovery replication experiments, LAT2 was selected for extensive study based on its significant regulation and its specificity for hematopoietic tissues, as shown in supplemental Fig. S1. LAT2 is a 25- to 30-kDa transmembrane adaptor protein associated with lipid rafts. It has been experimentally demonstrated that the location of LAT2 in the lipid raft is due to a double Cys-palmitoylation at a C25-V-R-C28 site (36, 44). In myeloid cells and B-cells, extracellular signals bind to transmembrane receptors, which trigger LAT2 phosphorylation that recruits signaling molecules such as Grb2, Gab2, and GADS into receptor-signaling complexes (15, 48). LAT2 function has been studied in detail in mast cells, in which this protein functions as a negative regulator of LAT-induced degranulation signals (56). LAT was the first protein of this family to be described (57). However, the role of LAT2 in the survival signaling of cancer cells has not been reported previously. Here, we demonstrated the role of LAT2 in the early events that induce apoptosis in leukemia cells by means of functional experiments. We have previously presented preliminary evidence that ODPC induces a state of disorganization in the membranes of model systems. Our experiments with mimetic membrane models indicated that ODPC has a primary action on the lipid bilayer, and these effects were more pronounced in liposomes containing cholesterol, a model of lipid raft-enriched membranes (11). Similarly, 2-BrPA and MCD were used as tools to induce similar perturbations of lipid rafts in order to characterize the behavior of LAT2. We showed that 2-BrPA, an inhibitor of palmitoyl transferase, caused an effect similar to that of ODPC by reducing the abundance of LAT2 and triggering apoptosis. MCD, which disrupts lipid rafts by scavenging cholesterol, potentiated the effect of ODPC on LAT2 abundance. These results emphasize the importance of the post-translational modification palmitoylation for stabilizing adaptor proteins in lipid rafts, as previously reported (58, 59). Our data suggest that the main effect of ODPC on membranes appears to be interference with the interaction of lipid rafts and palmitoylated proteins, which leads to an inefficient scaffold for maintaining signal transduction proteins.
We demonstrated that LAT2 is degraded by proteasomes after experimentally induced perturbations in lipid raft structure. Proteasome regulation of protein stability is expected in the case of a highly regulated protein involved in cell signaling. Indeed, Brdicka et al. (36) have reported that B-cell receptor activation in a B-cell line induces LAT2 phosphorylation and ubiquitination and that the E3-ubiquitin ligase c-Cbl can interact with phosphorylated LAT2 after stimulation of THP-1 cells. Moreover, we consistently detected the appearance of a lower apparent molecular weight band of LAT2 in ODPC- and 2-BrPA-treated cells before the complete degradation of LAT2. This same band also increased in the presence of MG132 or MLN9708, two proteasome inhibitors. This suggests that LAT2 might be first cleaved by a non-proteasome mechanism that generates a protein with a mass almost 3 KDa lower. Whether this cleaved product has lost its N-terminal portion, including its palmitoylation sites, or represents another cleaved product was not determined. However, the complete loss of LAT2 in the DRM fraction or lighter membrane fraction from the sucrose gradient after only 3 h of exposure to ODPC (Figs. 3C and 3D) suggests that the most probable peptide lost in this LAT2 processing step is the N-terminal portion that is essential for lipid raft association.
Although we have confirmed the involvement of LAT2 and other proteins of the AKT pathway in the process of apoptosis triggered by ODPC, we cannot conclude that LAT2 is the only target of ODPC or perifosine, because APLs have a broad range of actions on diverse cancer subtypes (60) and LAT2 is an adaptor protein expressed specifically in hematopoietic cells (15, 48). However, we demonstrated that the down-regulation of LAT2 by shRNA in NB4 cells mimics the action of ODPC, impairing AKT activation by MGFs (Fig. 6D). Because AKT is a well-described pro-survival pathway in cancer cells (55), we hypothesized that LAT2 down-regulation and consequent AKT impairment might induce cell death or cause reduction in cell proliferation. We could maintain NB4 cells as viable even after a stable reduction of more than 70% of LAT2 protein levels. In fact, NB4 LAT2 knockdown cells exhibited a lower level of cell proliferation (Fig. 7A) and an increased sensitivity to APLs and ATO (Figs. 7B–7D). This finding highlights LAT2 and possible other adaptor proteins as potential cancer therapeutic targets.
Most studies of the mechanisms of action of APLs, especially those using edelfosine, have focused on altered phosphatidylcholine biosynthesis as the major mechanism of action of the drug (61, 62). In the case of perifosine, the main focus was on the consequences of AKT signaling inhibition (63–65). However, our results indicate that cell signaling disruption by APLs is probably due to a primary action on the organization of lipids in rafts and that protein function is consequently altered (Fig. 8). ODPC inhibited AKT activation in acute leukemia cells by MGFs after a few minutes of incubation in a way similar to that of the specific PI3K inhibitor Wortmannin. However, there is no evidence that APLs function as direct kinase inhibitors, although there is evidence that proper lipid raft assembly is essential for the highly constitutive AKT activity of cancer cells (66). Importantly, we demonstrated that a lack of LAT2 impairs AKT activation by MGFs in leukemia cells. This indicates that some adaptor proteins might be essential for signal transduction in cancer cells and suggests that ODPC and probably other APLs have their primary action on cell signaling pathways by inhibiting raft assembly. A proposed model of the mechanism of action of APLs is shown in Fig. 8.
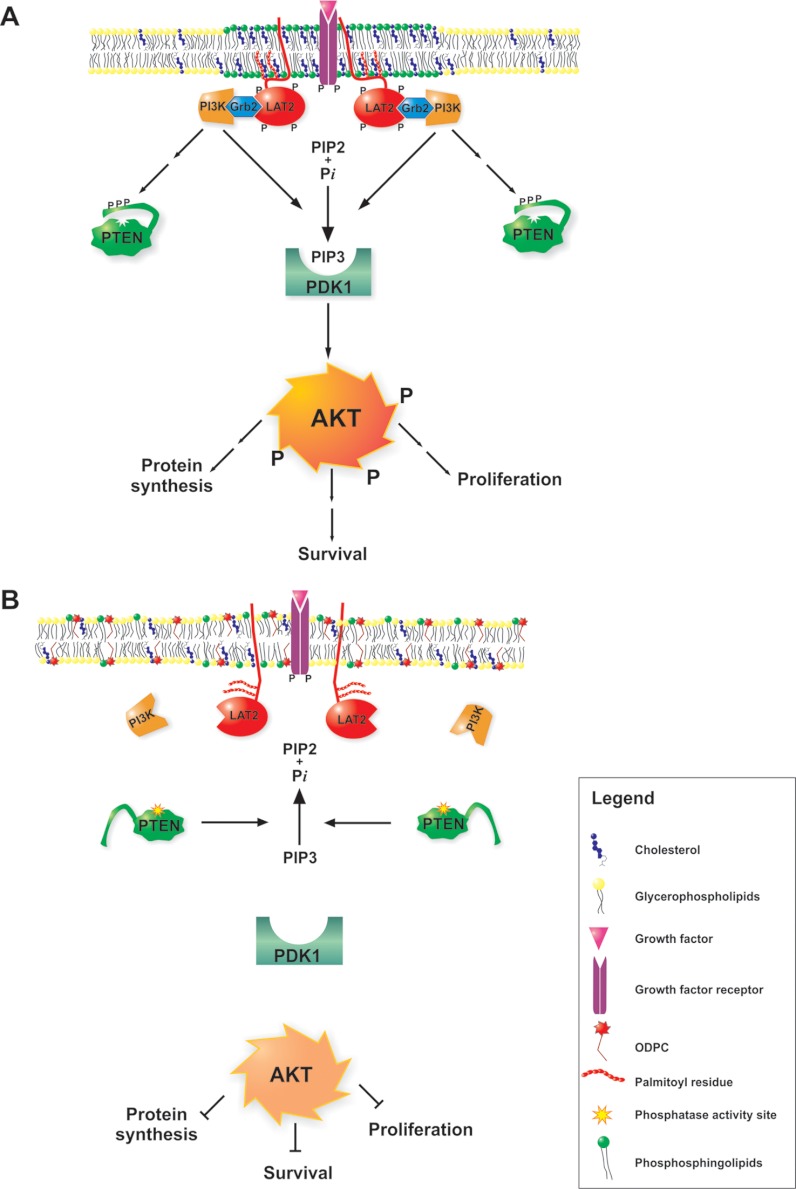
Fig. 8.
Proposed mechanism of alkylphospholipid (ODPC)-induced raft-based inhibition of cell signaling. A, lipid rafts are membrane domains enriched in shingolipids (green) and cholesterol (blue) that serve as a scaffold for signaling through growth factor receptors (purple). The correct assembly of lipid rafts containing adaptor proteins such as LAT2 and Grb2 is essential for proper signaling of the AKT survival pathway (shown in A). Alkylphospholipids (ODPC) disrupt the assembly of lipid rafts, displacing raft-associated adaptor proteins. As a result, signal transduction is inhibited (shown in B). ODPC, 10-(octyloxy) decyl-2-(trimethylammonium) ethyl phosphate; PI3K, phosphatidylinositol 3-kinases; Grb2, growth factor receptor-bound protein 2; LAT2, linker for activation of T-cell family member 2; PTEN, phosphatase and tensin homolog; PDK1, phosphoinositide-dependent protein kinase 1); AKT, v-AKT murine thymoma viral oncogene homolog 1; PIP2, phosphatidylinositol (3,4)-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate.
Supplementing serum-free culture medium with MGF induced a rapid up-regulation of the expression of LAT2 in vehicle control-treated NB4 cells. This effect was partially reduced by ODPC treatment and was suppressed by Wortmannin. This rapid up-regulation of LAT2 after exposure to MGF and its reduction by a PI3K inhibitor suggest that LAT2 mRNA translation depends on a functional AKT pathway. Indeed, phosphorylation of 4EBP-1 and ribosomal S6 kinase (RS6K) and up-regulation of cap-dependent translation in the ribosome is downstream of PI3K/AKT/mTOR (51, 67, 68). To support this hypothesis, we showed that S6RP, the main target of RS6K, was hyperphosphorylated by the addition of MGF to control cells but was hypophosphorylated in the presence of ODPC or Wortmannin. Additionally, the up-regulation of LAT2 induced by MGF was blocked by two classical inhibitors of translation (cycloheximide and rapamycin) but not by a classical transcription inhibitor, actinomycin D (data not shown). These results suggest that ODPC might have a direct or indirect inhibitory effect on the translation apparatus. Interestingly, we have shown here that the ribosomal proteins (RP) L23 and L38 are reduced in abundance in the lipid raft fraction of ODPC-treated cells. This finding confirms a previous report stating that a raft disruption treatment with MCD causes down-regulation of several RP (69). Moreover, Yang et al. (38), using a specific chemical approach to evaluate palmitoylated proteins, have reported that palmitoylation might target ribosome proteins to lipid rafts. They have demonstrated via immunofluorescence that the ribosomal protein L10A partially co-localizes with lipid rafts and that 2-BrPA treatment reverses this association. Whether this represents only a mechanism that mediates the association between RP and kinases/phosphatases of lipid rafts, in order to regulate the function of RP before the final ribosomal assembly, or the physical association between ribosomes and lipid rafts is not known. This inhibition of translation by ODPC might be caused by the inhibition of AKT/mTOR activation or by direct down-regulation of RP in lipid rafts, or by both mechanisms. This might be associated with the specificity of ODPC for malignant cells, as previous reports have linked an altered translational control to the malignant phenotype (70, 71).
The induction of apoptosis by APLs has been reported to be predominantly related to Fas/CD95 death receptor activation in lipid rafts with secondary activation of the extrinsic caspase pathway (72–74). In the present study we showed that both intrinsic and extrinsic caspase pathways are activated after ODPC treatment. This result is consistent with the report of Chiarini et al. (75) that the proapoptotic action of perifosine is only partially dependent on Fas/CD95 and is also mediated by the mitochondrial intrinsic pathway.
Our results are relevant to the mechanisms of action of APLs. We emphasize the importance of palmitoylation for cell signaling in lipid rafts and provide evidence that the APLs, ODPC and perifosine, create a state of disorganization of these microdomains that has a profound impact on cell function. Moreover, lipid raft resident adaptor proteins that function in signal transduction, here exemplified by LAT2, emerge as potential therapeutic targets in human cancer.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Stefano Servi, Dipartimento di Chimica, Materiali, Ingegneria Chimica “Giullio Natta,” Politecnico di Milano, for providing a sample of the original compound that was used as a standard for ODPC synthesis. We are grateful to Dr. Elettra Greene for assistance in revising the manuscript.
Footnotes
* This research was supported by FAPESP, FINEP, and CNPq. C.H.T., G.A.S., and P.S.S. received fellowships from FAPESP Proc. No. 07/58649-1, 2011/07387-2, and 2011/09718-6, respectively. G.A.F. received a fellowship from CAPES.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- AML
- acute myeloid leukemia
- APL
- alkylphospholipid
- ATO
- arsenic trioxide
- DMSO
- dimethyl sulfoxide
- DRM
- detergent-resistant membrane
- DT
- doubling time
- GR
- growth rate
- LAB
- linker for activation of B-cells
- LAT2
- linker for activation of T cells-2
- MCD
- methyl-β-cyclodextrin
- MGF
- myeloid growth factor
- NTAL
- non-T-cell activation linker
- ODPC
- 10-(octyloxy) decyl-2-(trimethylammonium) ethyl phosphate
- RP
- ribosomal proteins.
REFERENCES
- 1. Kosaka T., Yamaki E., Mogi A., Kuwano H. (2011) Mechanisms of resistance to EGFR TKIs and development of a new generation of drugs in non-small-cell lung cancer. J. Biomed. Biotechnol. 2011, 165214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Irwin M. E., Mueller K. L., Bohin N., Ge Y., Boerner J. L. (2011) Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell. Physiol. 226, 2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pike L. J. (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47, 1597–1598 [DOI] [PubMed] [Google Scholar]
- 4. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 5. Seminario M. C., Bunnell S. C. (2008) Signal initiation in T-cell receptor microclusters. Immunol. Rev. 221, 90–106 [DOI] [PubMed] [Google Scholar]
- 6. Gupta N., DeFranco A. L. (2007) Lipid rafts and B cell signaling. Semin. Cell Dev. Biol. 18, 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jury E. C., Flores-Borja F., Kabouridis P. S. (2007) Lipid rafts in T cell signalling and disease. Semin. Cell Dev. Biol. 18, 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jahn K. A., Su Y., Braet F. (2011) Multifaceted nature of membrane microdomains in colorectal cancer. World J. Gastroenterol. 17, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patra S. K. (2008) Dissecting lipid raft facilitated cell signaling pathways in cancer. Biochim. Biophys. Acta 1785, 182–206 [DOI] [PubMed] [Google Scholar]
- 10. Hitosugi T., Sato M., Sasaki K., Umezawa Y. (2007) Lipid raft specific knockdown of SRC family kinase activity inhibits cell adhesion and cell cycle progression of breast cancer cells. Cancer Res. 67, 8139–8148 [DOI] [PubMed] [Google Scholar]
- 11. dos Santos G. A., Thomé C. H., Ferreira G. A., Yoneda J. S., Nobre T. M., Daghastanli K. R., Scheucher P. S., Gimenes-Teixeira H. L., Constantino M. G., de Oliveira K. T., Faça V. M., Falcão R. P., Greene L. J., Rego E. M., Ciancaglini P. (2010) Interaction of 10-(octyloxy) decyl-2-(trimethylammonium) ethyl phosphate with mimetic membranes and cytotoxic effect on leukemic cells. Biochim. Biophys. Acta 1798, 1714–1723 [DOI] [PubMed] [Google Scholar]
- 12. van der Luit A. H., Vink S. R., Klarenbeek J. B., Perrissoud D., Solary E., Verheij M., van Blitterswijk W. J. (2007) A new class of anticancer alkylphospholipids uses lipid rafts as membrane gateways to induce apoptosis in lymphoma cells. Mol. Cancer Ther. 6, 2337–2345 [DOI] [PubMed] [Google Scholar]
- 13. Pinton G., Manente A. G., Angeli G., Mutti L., Moro L. (2012) Perifosine as a potential novel anti-cancer agent inhibits EGFR/MET-AKT axis in malignant pleural mesothelioma. PLoS ONE 7, e36856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 15. Iwaki S., Jensen B. M., Gilfillan A. M. (2007) Ntal/Lab/Lat2. Int. J. Biochem. Cell Biol. 39, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Berger R. (1991) NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 77, 1080–1086 [PubMed] [Google Scholar]
- 17. Strefford J. C., Foot N. J., Chaplin T., Neat M. J., Oliver R. T., Young B. D., Jones L. K. (2001) The characterisation of the lymphoma cell line U937, using comparative genomic hybridisation and multi-plex FISH. Cytogenet. Cell Genet. 94, 9–14 [DOI] [PubMed] [Google Scholar]
- 18. Naumann S., Reutzel D., Speicher M., Decker H. J. (2001) Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk. Res. 25, 313–322 [DOI] [PubMed] [Google Scholar]
- 19. Agresta M., D'Arrigo P., Fasoli E., Losi D., Pedrocchi-Fantoni G., Riva S., Servi S., Tessaro D. (2003) Synthesis and antiproliferative activity of alkylphosphocholines. Chem. Phys. Lipids 126, 201–210 [DOI] [PubMed] [Google Scholar]
- 20. Chou T. C. (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681 [DOI] [PubMed] [Google Scholar]
- 21. Resh M. D. (2006) Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 40, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vink S. R., Luit A. H. v. d., Klarenbeek J. B., Verheij M., Blitterswijk W. J. v. (2007) Lipid rafts and metabolic energy differentially determine uptake of anti-cancer alkylphospholipids in lymphoma versus carcinoma cells. Biochem. Pharmacol. 74, 1456–1465 [DOI] [PubMed] [Google Scholar]
- 23. Adam R. M., Yang W., Di Vizio D., Mukhopadhyay N. K., Steen H. (2008) Rapid preparation of nuclei-depleted detergent-resistant membrane fractions suitable for proteomics analysis. BMC Cell Biol. 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pereira S. R., Faça V. M., Gomes G. G., Chammas R., Fontes A. M., Covas D. T., Greene L. J. (2005) Changes in the proteomic profile during differentiation and maturation of human monocyte-derived dendritic cells stimulated with granulocyte macrophage colony stimulating factor/interleukin-4 and lipopolysaccharide. Proteomics 5, 1186–1198 [DOI] [PubMed] [Google Scholar]
- 25. Faça V. M., Ventura A. P., Fitzgibbon M. P., Pereira-Faça S. R., Pitteri S. J., Green A. E., Ireton R. C., Zhang Q., Wang H., O'Briant K. C., Drescher C. W., Schummer M., McIntosh M. W., Knudsen B. S., Hanash S. M. (2008) Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PLoS One 3, e2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rauch A., Bellew M., Eng J., Fitzgibbon M., Holzman T., Hussey P., Igra M., Maclean B., Lin C. W., Detter A., Fang R., Faca V., Gafken P., Zhang H., Whiteaker J., Whitaker J., States D., Hanash S., Paulovich A., McIntosh M. W. (2006) Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J. Proteome Res. 5, 112–121 [DOI] [PubMed] [Google Scholar]
- 27. MacLean B., Eng J. K., Beavis R. C., McIntosh M. (2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 28. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 29. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 30. Faca V., Coram M., Phanstiel D., Glukhova V., Zhang Q., Fitzgibbon M., McIntosh M., Hanash S. (2006) Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J. Proteome Res. 5, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 31. Faca V. M., Song K. S., Wang H., Zhang Q., Krasnoselsky A. L., Newcomb L. F., Plentz R. R., Gurumurthy S., Redston M. S., Pitteri S. J., Pereira-Faca S. R., Ireton R. C., Katayama H., Glukhova V., Phanstiel D., Brenner D. E., Anderson M. A., Misek D., Scholler N., Urban N. D., Barnett M. J., Edelstein C., Goodman G. E., Thornquist M. D., McIntosh M. W., DePinho R. A., Bardeesy N., Hanash S. M. (2008) A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 5, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chou T. C., Talaly P., Chou T. C., Talaly P. (1977) A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. Journal of Biological Chemistry 252, 6438–6442 [PubMed] [Google Scholar]
- 33. Suzuki K. G., Fujiwara T. K., Sanematsu F., Iino R., Edidin M., Kusumi A. (2007) GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Appenzeller-Herzog C., Hauri H. P. (2006) The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 119, 2173–2183 [DOI] [PubMed] [Google Scholar]
- 35. Burgess R. J., Zhang Z. (2010) Histones, histone chaperones and nucleosome assembly. Protein Cell 1, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brdicka T., Imrich M., Angelisová P., Brdicková N., Horváth O., Spicka J., Hilgert I., Lusková P., Dráber P., Novák P., Engels N., Wienands J., Simeoni L., Osterreicher J., Aguado E., Malissen M., Schraven B., Horejsí V. (2002) Non-T cell activation linker (NTAL): a transmembrane adaptor protein involved in immunoreceptor signaling. J. Exp. Med. 196, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson J. P., Raghavan A. S., Yang Y. Y., Charron G., Hang H. C. (2011) Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteomics 10, M110.001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tkaczyk C., Horejsi V., Iwaki S., Draber P., Samelson L. E., Satterthwaite A. B., Nahm D. H., Metcalfe D. D., Gilfillan A. M. (2004) NTAL phosphorylation is a pivotal link between the signaling cascades leading to human mast cell degranulation following Kit activation and Fc epsilon RI aggregation. Blood 104, 207–214 [DOI] [PubMed] [Google Scholar]
- 41. Delmas D., Rébé C., Lacour S., Filomenko R., Athias A., Gambert P., Cherkaoui-Malki M., Jannin B., Dubrez-Daloz L., Latruffe N., Solary E. (2003) Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells. J. Biol. Chem. 278, 41482–41490 [DOI] [PubMed] [Google Scholar]
- 42. Salaun C., Greaves J., Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sorek N., Bloch D., Yalovsky S. (2009) Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12, 714–720 [DOI] [PubMed] [Google Scholar]
- 44. Janssen E., Zhu M., Zhang W., Koonpaew S. (2003) LAB: a new membrane-associated adaptor molecule in B cell activation. Nat. Immunol. 4, 117–123 [DOI] [PubMed] [Google Scholar]
- 45. Roget K., Malissen M., Malbec O., Malissen B., Daëron M. (2008) Non-T cell activation linker promotes mast cell survival by dampening the recruitment of SHIP1 by linker for activation of T cells. J. Immunol. 180, 3689–3698 [DOI] [PubMed] [Google Scholar]
- 46. Elrod H. A., Lin Y. D., Yue P., Wang X., Lonial S., Khuri F. R., Sun S. Y. (2007) The alkylphospholipid perifosine induces apoptosis of human lung cancer cells requiring inhibition of Akt and activation of the extrinsic apoptotic pathway. Mol. Cancer Ther. 6, 2029–2038 [DOI] [PubMed] [Google Scholar]
- 47. Vazquez F., Ramaswamy S., Nakamura N., Sellers W. R. (2000) Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 20, 5010–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orr S. J., McVicar D. W. (2011) LAB/NTAL/Lat2: a force to be reckoned with in all leukocytes? J. Leukoc. Biol. 89, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carracedo A., Pandolfi P. P. (2008) The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene 27, 5527–5541 [DOI] [PubMed] [Google Scholar]
- 50. Knight Z. A. (2010) Small molecule inhibitors of the PI3-kinase family. Curr. Top. Microbiol. Immunol. 347, 263–278 [DOI] [PubMed] [Google Scholar]
- 51. Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. (2006) mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 52. Bhatt A. P., Bhende P. M., Sin S.-H., Roy D., Dittmer D. P., Damania B., Bhatt A. P., Bhende P. M., Sin S.-H., Roy D., Dittmer D. P., Damania B. (2010) Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood 115, 4455–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R., Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R. (1999) Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fei H. R., Chen G., Wang J. M., Wang F. Z. (2010) Perifosine induces cell cycle arrest and apoptosis in human hepatocellular carcinoma cell lines by blockade of Akt phosphorylation. Cytotechnology 62, 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vivanco I., Sawyers C. L. (2002) The phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat. Rev. Cancer 2, 489–501 [DOI] [PubMed] [Google Scholar]
- 56. Volná P., Lebduska P., Dráberová L., Símová S., Heneberg P., Boubelík M., Bugajev V., Malissen B., Wilson B. S., Horejsí V., Malissen M., Dráber P. (2004) Negative regulation of mast cell signaling and function by the adaptor LAB/NTAL. J. Exp. Med. 200, 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang W., Sloan-Lancaster J., Kitchen J., Trible R. P., Samelson L. E. (1998) LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell 92, 83–92 [DOI] [PubMed] [Google Scholar]
- 58. Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 59. Pechlivanis M., Kuhlmann J. (2006) Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids—more than just membrane anchoring. Biochim. Biophys. Acta 1764, 1914–1931 [DOI] [PubMed] [Google Scholar]
- 60. van Blitterswijk W. J., Verheij M. (2008) Anticancer alkylphospholipids: mechanisms of action, cellular sensitivity and resistance, and clinical prospects. Curr. Pharm. Des. 14, 2061–2074 [DOI] [PubMed] [Google Scholar]
- 61. Boggs K. P., Rock C. O., Jackowski S. (1995) Lysophosphatidylcholine and 1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine inhibit the CDP-choline pathway of phosphatidylcholine synthesis at the CTP:phosphocholine cytidylyltransferase step. J. Biol. Chem. 270, 7757–7764 [DOI] [PubMed] [Google Scholar]
- 62. Caskey C. T. (1975) Identifiying inherited disease through the family history. Am. Fam. Physician 11, 118–127 [PubMed] [Google Scholar]
- 63. Li Z., Tan F., Liewehr D. J., Steinberg S. M., Thiele C. J. (2010) In vitro and in vivo inhibition of neuroblastoma tumor cell growth by AKT inhibitor perifosine. J. Natl. Cancer Inst. 102, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Engel J. B., Schönhals T., Häusler S., Krockenberger M., Schmidt M., Horn E., Köster F., Dietl J., Wischhusen J., Honig A. (2011) Induction of programmed cell death by inhibition of AKT with the alkylphosphocholine perifosine in in vitro models of platinum sensitive and resistant ovarian cancers. Arch. Gynecol. Obstet. 283, 603–610 [DOI] [PubMed] [Google Scholar]
- 65. Cirstea D., Hideshima T., Rodig S., Santo L., Pozzi S., Vallet S., Ikeda H., Perrone G., Gorgun G., Patel K., Desai N., Sportelli P., Kapoor S., Vali S., Mukherjee S., Munshi N. C., Anderson K. C., Raje N. (2010) Dual inhibition of akt/mammalian target of rapamycin pathway by nanoparticle albumin-bound-rapamycin and perifosine induces antitumor activity in multiple myeloma. Mol. Cancer Ther. 9, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Elhyany S., Assa-Kunik E., Tsory S., Muller T., Fedida S., Segal S., Fishman D. (2004) The integrity of cholesterol-enriched microdomains is essential for the constitutive high activity of protein kinase B in tumour cells. Biochem. Soc. Trans. 32, 837–839 [DOI] [PubMed] [Google Scholar]
- 67. She Q. B., Halilovic E., Ye Q., Zhen W., Shirasawa S., Sasazuki T., Solit D. B., Rosen N. (2010) 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 18, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jastrzebski K., Hannan K. M., Tchoubrieva E. B., Hannan R. D., Pearson R. B. (2007) Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25, 209–226 [DOI] [PubMed] [Google Scholar]
- 69. Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barna M., Pusic A., Zollo O., Costa M., Kondrashov N., Rego E., Rao P. H., Ruggero D. (2008) Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hsieh A. C., Costa M., Zollo O., Davis C., Feldman M. E., Testa J. R., Meyuhas O., Shokat K. M., Ruggero D. (2010) Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 17, 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mollinedo F., de la Iglesia-Vicente J., Gajate C., Estella-Hermoso de Mendoza A., Villa-Pulgarin J. A., de Frias M., Roué G., Gil J., Colomer D., Campanero M. A., Blanco-Prieto M. J. (2010) In vitro and in vivo selective antitumor activity of edelfosine against mantle cell lymphoma and chronic lymphocytic leukemia involving lipid rafts. Clin. Cancer Res. 16, 2046–2054 [DOI] [PubMed] [Google Scholar]
- 73. Gajate C., Gonzalez-Camacho F., Mollinedo F. (2009) Involvement of raft aggregates enriched in Fas/CD95 death-inducing signaling complex in the antileukemic action of edelfosine in Jurkat cells. PLoS One 4, e5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gajate C., Mollinedo F. (2007) Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood 109, 711–719 [DOI] [PubMed] [Google Scholar]
- 75. Chiarini F., Del Sole M., Mongiorgi S., Gaboardi G. C., Cappellini A., Mantovani I., Follo M. Y., McCubrey J. A., Martelli A. M. (2008) The novel Akt inhibitor, perifosine, induces caspase-dependent apoptosis and downregulates P-glycoprotein expression in multidrug-resistant human T-acute leukemia cells by a JNK-dependent mechanism. Leukemia 22, 1106–1116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.