Abstract
The cellular microenvironment comprises soluble factors, support cells, and components of the extracellular matrix (ECM) that combine to regulate cellular behavior. Pluripotent stem cells utilize interactions between support cells and soluble factors in the microenvironment to assist in the maintenance of self-renewal and the process of differentiation. However, the ECM also plays a significant role in shaping the behavior of human pluripotent stem cells, including embryonic stem cells (hESCs) and induced pluripotent stem cells. Moreover, it has recently been observed that deposited factors in a hESC-conditioned matrix have the potential to contribute to the reprogramming of metastatic melanoma cells. Therefore, the ECM component of the pluripotent stem cell microenvironment necessitates further analysis.
In this study we first compared the self-renewal and differentiation properties of hESCs grown on Matrigel™ pre-conditioned by hESCs to those on unconditioned Matrigel™. We determined that culture on conditioned Matrigel™ prevents differentiation when supportive growth factors are removed from the culture medium. To investigate and identify factors potentially responsible for this beneficial effect, we performed a defined SILAC MS-based proteomics screen of hESC-conditioned Matrigel™. From this proteomics screen, we identified over 80 extracellular proteins in matrix conditioned by hESCs and induced pluripotent stem cells. These included matrix-associated factors that participate in key stem cell pluripotency regulatory pathways, such as Nodal/Activin and canonical Wnt signaling. This work represents the first investigation of stem-cell-derived matrices from human pluripotent stem cells using a defined SILAC MS-based proteomics approach.
The two defining characteristics of human embryonic stem cells (hESCs),1 self-renewal and pluripotency, are maintained by a delicate balance of intracellular and extracellular signaling processes. Extracellular regulation is primarily the result of changes in the microenvironment surrounding the cells during growth in vitro or in vivo. HESCs interact with this “niche ” through support cells, extracellular matrix (ECM) components, and autocrine/paracrine signaling (reviewed in Refs. 1–3). Modulation of any of these supportive elements individually or in combination has been used extensively to alter hESC behavior (1–3).
The culture of hESCs, as well as that of human induced pluripotent stem cells (hiPSCs), is conventionally performed on a layer of irradiated mouse embryonic fibroblast cells (MEFs). These MEFs are believed to promote the maintenance of hESCs and hiPSCs through the secretion of beneficial support proteins and cytokines into the soluble microenvironment. A number of proteomic studies have been conducted that examine the secretome of feeder-cell layers in an attempt to elucidate proteins and pathways essential for hESC and hiPSC survival (4–7). Alternatively, hESCs and hiPSCs can be cultured in feeder-free conditions in the absence of support cells. In feeder-free conditions, hESCs and hiPSCs are most often grown on the basement membrane matrix Matrigel™ in medium that has been previously conditioned by MEFs (MEF-CM). Matrigel™ is a gelatinous mixture that is secreted by Engelbreth-Holm-Swarm mouse sarcoma cells (8). Although recent studies have proposed that a variety of defined matrices can support the growth of hESCs and hiPSCs, few of these can maintain a wide range of stem cell lines and therefore are typically not used in place of Matrigel™. The properties of Matrigel™ that make it such an effective matrix for hESC and hiPSC culture remain poorly understood. Because of the complexity of matrices like Matrigel™, the majority of proteomic studies that examine the hESC and hiPSC microenvironment have focused on contributions from support cells and soluble extracellular factors.
The ECM is typically a complex network of structural proteins and glycosaminoglycans that function to support cells through the regulation of processes such as adhesion and growth factor signaling (9). Thus, it is not surprising that the generation of a well-defined matrix capable of facilitating hESC and hiPSC self-renewal has remained difficult (10). Previous proteomic investigations of Matrigel™ and other matrices supportive of hESC maintenance in vitro have revealed the presence of numerous growth, binding, and signaling proteins (11, 12). Further examination of how hESCs and hiPSCs interact with these complex matrices would provide critical information about what role the ECM plays in the organization of processes involved in the regulation of self-renewal and pluripotency.
A recent study has established the ability of hESC-derived matrix microenvironments to alter tumorigenic properties through the reprogramming of metastatic melanoma cells (13). Importantly, this effect was found to be dependent on the exposure of metastatic cells to hESC-derived conditioned Matrigel™. Culture of metastatic melanoma cells in hESC-conditioned medium did not promote the reprogramming effect. These data suggest that the proteins responsible for this effect were integrated in the matrix. With the use of immunochemical techniques, it was later found that the left-right determination (Lefty) proteins A and B that were deposited in the matrix by hESCs during conditioning were at least in part responsible for the cellular change observed in metastatic cells (14). The Lefty A and B proteins are antagonists of transforming growth factor (TGF)-β signaling that act directly on Nodal protein, a critical regulator of the stem cell phenotype (15, 16). Subsequent studies of conditioned matrix utilizing mESCs implicated the bone morphogenic protein (BMP) 4 antagonist Gremlin as a primary regulator of the observed changes in metastatic cells (17). Collectively, these studies were all biased by a targeted analysis of potential effectors of metastatic cells. A comprehensive proteomic analysis of conditioned matrix could potentially reveal other factors involved in metastatic cell reprogramming. Furthermore, proteomic examination of hESC and hiPSC conditioned matrix could expose factors important in the regulation of self-renewal and pluripotency by the microenvironment in vitro.
To this end, we have analyzed both types of human pluripotent stem cells, hESCs and hiPSCs, via a mass spectrometry (MS)-based proteomics approach to identify proteins deposited during growth in feeder-free conditions in vitro on Matrigel™. To investigate the hESC- and hiPSC-derived matrix, the metabolic labeling technique known as stable isotope labeling with amino acids in cell culture (SILAC) was used (18). SILAC facilitates the identification of hESC- and hiPSC-derived proteins that would otherwise be confounded by the presence of mouse-derived protein background from Matrigel™. From the proteomic analysis of three cells lines, namely, the hESC lines H9 and CA1 and the hiPSC line BJ-1D, we identified a total of 621, 1355, and 1350 total unique proteins, respectively. This work represents the first analysis of a hESC- and hiPSC-derived conditioned matrix and resulted in the identification of at least one novel microenvironmental contributor responsible for the regulation of human pluripotent stem cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Harvest
H9 (passage 26) and CA1 (passage 20) hESCs and BJ-1D (passage 69) hiPSCs were maintained on CF-1 irradiated MEF feeder layers (GlobalStem, Rockville, MD) using media composed of knockout DMEM/F12, 20% knockout serum replacement, 1% non-essential amino acids, 2 mm glutamine (CellGro, Manassas, VA), 0.1 mm 2-mercaptoethanol (Fisher, Toronto, ON, Canada), and 4 ng/ml of basic fibroblast growth factor (bFGF). Irradiated MEF feeder layers were seeded at a density of 2 × 105 cells/well in a six-well dish. During MEF culture, hESCs and hiPSCs were passaged mechanically when they reached 70% confluency (∼6 days after plating). For feeder-free growth, standard hESC medium that was conditioned on a layer of irradiated MEF feeders (MEF-CM) was used with plates coated with Matrigel™ (BD Biosciences, Franklin Lake, NJ). For general feeder-free growth, culture dishes were coated with a 1:30 dilution of Matrigel™ in DMEM/F12. Prior to use, MEF-CM was supplemented with an additional 8 ng/ml of bFGF and filtered. During feeder-free culture, hESCs and hiPSCs were passaged mechanically when they reached 70% confluency (∼6 days after plating). H9 hESCs were obtained from WiCell (Madison, WI). The CA1 hESC line used in all experiments was obtained from Dr. Cheryle Seguin of the University of Western Ontario (19, 20). The hiPSC line BJ-1D was derived by reprogramming using retroviral transduction of BJ fibroblasts with the Yamanaka factors essentially as we have described elsewhere (21) and validated as pluripotent using in vitro and in vivo differentiation assays and expression profiling (unpublished results). Cells were fed every 24 h in both feeder-dependent and feeder-free conditions.
For SILAC experiments, hESC and hiPSC cell lines were grown in feeder-free conditions on Matrigel™ (BD Biosciences, Franklin Lake, NJ). Custom DMEM/F12-glutamax that contained no l-arginine or l-lysine was ordered from Invitrogen and used in all SILAC experiments. Culture dishes were coated with 1:30 dilutions of Matrigel™ in DMEM/F12 containing no l-arginine or l-lysine. Complete SILAC medium was prepared using the StemPro® hESC Media system (Invitrogen) according to the manufacturer's instructions, substituting the provided basal medium with the arginine- and lysine-free custom DMEM/F12. [13C6, 15N4]-l-arginine and [13C6, 15N2]-l-lysine (Cambridge Isotope Laboratories, Andover, MA) were supplemented into the SILAC medium at 90 mg/L and 92 mg/L, respectively. Unlabeled l-proline was added to the SILAC medium at a final concentration of 800 mg/L (22). During feeder-free maintenance in SILAC medium, cells were passaged mechanically when they reached 80% confluency (∼4 days after plating). Cells were fed every 24 h in feeder-free conditions with SILAC medium.
Embryoid bodies (EBs) were generated in all cases using Aggrewell 400Ex plates (StemCell Technologies, Vancouver, BC, Canada) according to the manufacturer's instructions. Briefly, ∼2 × 106 hESCs or hiPSCs were harvested enzymatically using Accutase (Invitrogen) from Matrigel™-coated plates. As Accutase will generate a single cell suspension, hESCs and hiPSCs were reconstituted in Aggrewell medium (StemCell Technologies) supplemented with ROCK inhibitor (Y-27632) (Sigma-Aldrich, St. Louis, MO) at a final concentration of 10 μm to promote aggregation. After plating in Aggrewell dishes, EBs were grown for 48 h at 37 °C prior to transfer to ultra-low adherence tissue culture dishes (Corning, Lowell, MA). EBs were fed every 2 to 3 days with fresh Aggrewell medium not supplemented with ROCK inhibitor. After 15 days of culture, EB RNA was harvested using Trizol extraction according to the manufacturer's instructions (Invitrogen). EBs were assayed for expression of CDX2, alpha-feto protein, GATA-6, GATA-4, Oct3/4, PAX-6, T-brachyury, Nanog, and Neurogenic differentiation 1 using TaqMan primer probe sets (Invitrogen). Details of these assays can be found in supplemental Table S7. All EB assays were performed in biological and technical duplicate for each cell line and each condition.
Generation and Harvest of Conditioned Matrix
To generate labeled populations of stem cells, H9, CA1, or BJ-1D cells were grown in feeder-free conditions on Matrigel™ in heavy-isotope ([13C6, 15N4]-l-arginine and [13C6, 15N2]-l-lysine) containing SILAC medium for 7 days (about four population doublings). Small colonies (∼5 to 10 cells based on visual inspection) generated using mechanical passaging were plated at low confluency (∼10%) to prevent overgrowth during the 7-day labeling phase. After the 7-day labeling period, hESC and hiPSC lines were harvested to determine isotopic label incorporation. Alternatively, hESCs or hiPSCs were mechanically passaged to Matrigel™-coated (1:10 dilution) dishes for the generation of the conditioned matrix. In this case, small (∼5 to 10 cells) hESC or hiPSC colonies were passaged to the Matrigel™-coated dishes to result in a confluency of ∼40% 24 h after plating. This was done to maximize the surface of the dish covered by hESCs and hiPSCs at harvest without compromising stem cell phenotype due to overconfluent dishes. Labeled stem cells were allowed to grow for a period of 5 days in Matrigel™-coated wells in heavy-isotope-containing SILAC medium. After this growth period, hESCs and hiPSCs were typically ∼90% confluent on the Matrigel™ layer.
Prior to harvest of the conditioned Matrigel™, wells were rinsed thoroughly with PBS to ensure removal of medium components. Cells were removed from the conditioned Matrigel™ layer using a commercial matrisperse mixture called Cell Recovery Solution (BD Biosciences) according to the manufacturer's instructions. The collected Cell Recovery Solution containing the hESCs or hiPSCs was spun for 10 min at 10,000g to pellet the cells. The stem cells in this pellet were examined through RT-PCR, flow cytometry, and EB assays. The remaining conditioned Matrigel™ layers were rinsed thoroughly with PBS to remove any residual cell debris. The conditioned Matrigel™ layers were examined manually to ensure complete cell removal. The remaining conditioned Matrigel™ layer was incubated in a solution containing 8 m urea, 100 mm ammonium bicarbonate, and 1 m NaCl for 24 h at 4 °C. This harvested conditioned Matrigel™ solution was concentrated using high-capacity C18 cartridges (Phenomenex, Torrance, CA). Eluted components were dried using a SpeedVac and reconstituted in 8 m urea with 100 mm ammonium bicarbonate prior to fractionation. Stem-cell-conditioned Matrigel™ was generated and analyzed in biological and technical duplicate or greater for each cell line (H9, n = 3; CA1, n = 3; BJ-1D, n = 2).
For examination of stem cell growth and pluripotency on hESC-conditioned Matrigel™, layers were prepared as above with minor alterations. Briefly, hESCs were grown in feeder-free conditions on Matrigel™ (1:30 dilution) in MEF-CM. hESCs were passaged mechanically as small colonies (∼5 to 10 cells) to Matrigel™-coated (1:10 dilution) dishes to generate a confluency of ∼30% 24 h after plating. hESCs were maintained on this Matrigel™ layer in MEF-CM for 5 days (∼70% confluent after this period) to produce the conditioned Matrigel™. Layers of conditioned Matrigel™ were rinsed and cells were removed as described above. Fresh hESCs grown in feeder-free conditions on Matrigel™ in MEF-CM were mechanically passaged to the conditioned Matrigel™ layers to produce a confluency of ∼30% 24 h after plating. During the first 24 h, hESCs were fed with MEF-CM. After this initial phase, the culture medium was changed to EB medium and fed daily. EB medium has the same composition as standard hESC media used for growth on MEF feeders as outlined above, minus bFGF. The absence of bFGF in this formulation will result in undirected hESC differentiation. After 4 days of growth in EB medium, hESCs were examined for pluripotency marker expression using RT-PCR, Western blotting (see “Methods” in the supplementary material), flow cytometry, and immunofluorescence (see “Methods” in the supplementary material).
MS Analysis
Samples of harvested conditioned Matrigel™ were subjected to fractionation with 1D-SDS-PAGE and subsequently digested using an in-gel protocol with trypsin (see “Methods” in the supplementary material). Alternatively, conditioned Matrigel™ samples were digested using an in-solution protocol with trypsin and subjected to fractionation with strong anion exchange or strong cation exchange columns packed in-house (see “Methods” in the supplementary material). Prepared fractions were injected and separated using a nanoAcquity system (Waters, Milford, MA) equipped with a 25 cm × 75 μm inner diameter C18 column. Fractions were separated using a 1% to 40% acetonitrile gradient over 150 min at a flow rate of 300 nL/min. MS analysis was done on a Q-ToF Ultima (Micromass/Waters) using data-dependent acquisition with selection of the four top precursor masses per survey scan. Survey scans were set at 1 s, and MS/MS acquisition was set at 1 s or 8000 cps TIC cut-off. Exclusion lists were generated using in-house software that automatically added 0.7 amu to the precursor masses selected in previous runs and output a file also containing retention time information. Each fraction was analyzed for four exclusion rounds (five injections total), with the injection volume adjusted depending on signal intensity in the MS survey scan.
Proteomic Data Analysis
Data analysis for all samples was performed with PEAKS 5.3 software (Bioinformatics Solutions Inc., Waterloo, ON, Canada) (23, 24). After being imported into PEAKS, MS/MS spectra from raw data files were refined using the following settings: merge spectra - true (100 ppm mass tolerance, 60 s retention time tolerance), correct precursor mass - true, determine precursor charge state - true (minimum charge +2, maximum charge +5), spectral quality filter - true (0.65 threshold), centroid, deisotope, and deconvolute - true. Resulting MS/MS spectra were then de novo sequenced using the following parameters: parent monoisotopic mass tolerance, 100 ppm; fragment monoisotopic mass tolerance, 0.15 Da; enzyme specificity, semi-Trypsin; fixed modifications, carbamidomethylation, +10 Da on Arginine and +8 Da on Lysine for SILAC; and variable modifications, N-terminal acetylation and oxidized methionine. After de novo analysis, data were searched against the UniProt sequence database (updated December 2011; Human taxonomy specified, 20,236 total entries, or Mouse taxonomy specified, 16,376 total entries) using the following parameters: parent monoisotopic mass tolerance, 100 ppm; fragment monoisotopic mass tolerance, 0.15 Da; enzyme specificity, semi-Trypsin (two missed cleavages permitted); fixed modifications, carbamidomethylation, +10 Da on arginine and +8 Da on lysine for SILAC; variable modifications, N-terminal acetylation and oxidized methionine; and estimate false-discovery rate - true.
The resultant proteins identified from the database search were further processed using PEAKS 5.3 software. Proteins were filtered by assigned score to give a <1.0% false-discovery rate for peptide matches based on estimation from a decoy-fusion method of database searching (24). Identified proteins were also required to be derived by at least two unique peptides. Proteins that contained overlapping peptides were grouped. Within each group, only the highest scoring member was reported when differentiation based on unique peptides could be made. When proteins could not be distinguished, they were all reported, as members of the same group. For all samples analyzed, keratin hits were manually removed from the final datasets. All raw peptide identifications can be found in supplemental Tables S8–S10. Gene ontology analysis was performed using STRAP software (CPC Tools) (25). The data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash: FB/HM1WSAUo4uZ/QX8FDnyxRqsq+xl7lFV5A4ri4Eik0px38w20CBv3E3WdmuIu1IF9ANsZGhpOdEHlcAxk+Q+MJr9gAAAAAAAAo/Q = = .
Real-time PCR
RNA was purified using Trizol reagent according to the manufacturer's instructions (Invitrogen) with the following modification: precipitated RNA pellets in isopropanol were centrifuged for 60 min at 12,000g at room temperature. After NanoDrop quantification, 1 μg of cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Invitrogen). Real-time PCR was performed using the TaqMan® Universal PCR Master Mix (Invitrogen). Samples were incubated at 50 °C for 2 min followed by 10 min at 95 °C. DNA was then amplified at 95 °C for 15 s followed by 1 min at 58 °C for 46 cycles. All samples were normalized to large ribosomal protein RPLPO. All primers were obtained from Invitrogen, and details of these assays can be found in supplemental Table S7. Each biological replicate was run in triplicate for every marker assayed. All reagents were used according to the manufacturer's instructions.
Flow Cytometry
All reagents and probes for flow cytometry were obtained from eBiosciences (San Diego, CA) unless otherwise noted. Cells were enzymatically harvested using Accutase (Invitrogen) to obtain a single cell suspension. After centrifugation, cell pellets were resuspended in 5% FBS in PBS. Alexa-488 conjugated SSEA-4 and Alexa-647 conjugated SSEA1 were diluted into the cell mixture according to the manufacturer's instructions. Negative control samples contained respective isotype controls for the primary antibodies used. Solutions were incubated for 2 h and analyzed using an Accuri C6 (BD Biosciences) flow cytometer after rinsing to remove unbound antibody. Populations were gated according to forward and side scatter patterns with filtering for viable cells using 7-aminoactinomycin D staining. Data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism software. Results are expressed as S.D. or S.E. as indicated in figure legends. Statistical significance was determined using one-way ANOVA with Tukey post hoc. Differences were reported as follows: * p < 0.05, ** p < 0.01, *** p < 0.001.
RESULTS
hESC-derived Matrix for the Maintenance of an Undifferentiated State
The ability of embryonic microenvironments to suppress the tumorigenic phenotype has been previously illustrated in a number of developmental and cellular systems (reviewed in Ref. 26). Analogous to metastatic cells, embryonic stem cells can direct cellular changes in response to cues from their microenvironment. Given the recent observation that a hESC-derived matrix microenvironment could modulate the behavior of metastatic cells (14), we sought to determine what effect exposure would have on the stem cell phenotype. To this end, we monitored changes in the gene and protein expression of hESCs grown on conditioned Matrigel™ in the absence of exogenous growth factor supplementation (Figs. 1A–1C). In this way, matrix-dependent changes in hESC behavior could be monitored without interference from soluble factors present in MEF-CM.
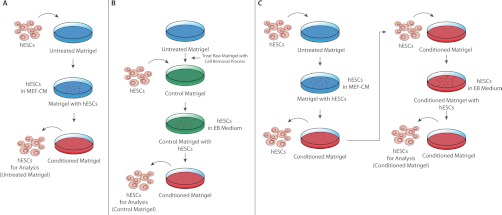
Fig. 1.
Generation of samples for hESC-conditioned Matrigel™ analysis. A, untreated Matrigel™ samples were generated by plating hESCs on a Matrigel™ layer and harvesting after 4 days of growth in MEF-CM. B, control Matrigel™ samples were generated by treating a Matrigel™ layer with cell recovery solution (see Experimental Procedures) and plating hESCs on this layer. hESCs were recovered from the treated layer after 4 days of growth in EB medium. C, conditioned Matrigel™ samples were produced by plating hESCs on a Matrigel™ layer and culturing for 4 to 5 days in MEF-CM. hESCs were subsequently removed from this layer using cell recovery solution to produce conditioned Matrigel™. Fresh hESCs were plated on the conditioned Matrigel™ layer and grown for 4 days in EB medium.
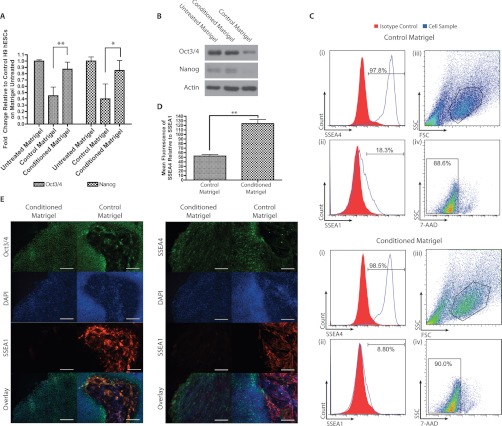
After 4 days of culture in medium that contained no exogenous bFGF (EB medium), a decrease in POU5F1 (herein referred to as Oct3/4) and Nanog gene and protein expression were observed in H9 hESCs grown on control or conditioned Matrigel™ relative to untreated cells. However, the observed reduction in Oct3/4 and Nanog gene and protein expression was significantly less in hESCs grown on conditioned Matrigel™ than in those grown on control Matrigel™ (Figs. 2A and 2B). A similar trend was observed when H9 hESCs were analyzed using flow cytometry for the expression of stage-specific embryonic antigen (SSEA) 4 and SSEA1, which are expressed by undifferentiated and differentiated pluripotent stem cells, respectively. H9 hESCs grown in EB medium on conditioned Matrigel™ exhibited an increase in SSEA4 mean fluorescence intensity relative to SSEA1 when compared with cells on control Matrigel™ (Figs. 2C and 2D). When cultures were examined using immunofluorescence, pockets of SSEA1-positive cells could be observed in colonies on control Matrigel™ layers, indicating differentiation (Fig. 2E). SSEA1-positive colonies were observed with a reduced frequency on conditioned Matrigel™. Based on visual microscopy inspection, we did not observe an increase in cell death or growth rate in the conditioned Matrigel™ cultures relative to the control. Subsequent analysis of a second hESC line, CA1, revealed similar trends (supplemental Figs. S1A–S1E). Taken together, these data indicate that conditioned Matrigel™ promotes the preservation of an undifferentiated state and that hESCs secrete factors into the matrix that are beneficial for their own maintenance.
Fig. 2.
hESC-conditioned Matrigel™ enhances the maintenance of an undifferentiated state in H9 cells relative to those grown on control Matrigel™. Untreated, control, and conditioned Matrigel™ hESC samples were generated as described in Fig. 1 and the Experimental Procedures section. A, RT-PCR data for H9 hESCs grown on the three separate matrix conditions. Values represent fold change of Oct3/4 or Nanog expression relative to untreated Matrigel™ samples. RPLPO was used as an internal control for all samples. Error bars represent S.E., n = 3. B, Western blot for Oct3/4 and Nanog in H9 hESCs grown on the different matrices. Actin is shown as a control. C, flow cytometry data for H9 hESCs grown on the different matrices. For each sample, plot (i) shows SSEA4 fluorescence (blue) relative to a matched isotype control (solid red), (ii) shows SSEA1 fluorescence relative to isotype control, (iii) is a scatter plot with the gated cell population shown, and (iv) is a scatter plot with the viable cell population gated. Percentages denote fractions of cells within the gates shown. D, results from flow cytometry analysis of H9 hESCs grown on the different matrices for the markers SSEA4 and SSEA1. Values represent mean fluorescence intensity of SSEA4 relative to SSEA1 for control Matrigel™ or conditioned Matrigel™ samples. Values are derived from the data in C. Error bars represent S.E., n = 3. E, immunocytochemistry for H9 hESCs grown on control Matrigel™ or conditioned Matrigel™. Cells were stained with Oct3/4, SSEA4, DAPI, and SSEA1. Scale bars represent 250 μm.
Proteomic Analysis of Pluripotent-Stem-Cell-Derived Conditioned MatrigelTM
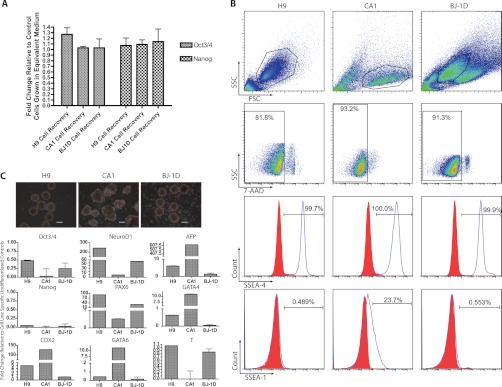
To determine which factors are deposited by hESCs and hiPSCs during growth on Matrigel™, we employed a global MS-based proteomics approach to analyze the corresponding conditioned Matrigel™. Previous studies that have illustrated the effects of conditioned Matrigel™ on metastatic cells have employed an NH4OH cellular lysis protocol to remove hESCs from Matrigel™. Lysis of cells on the matrix could potentially confound the proteomic analysis by releasing high abundance intracellular proteins. To limit the potential for intracellular protein contamination in the conditioned Matrigel™ resulting from lysis, we utilized a protocol based on Matrigel™ de-polymerization to remove intact cells from the matrix (see “Methods” in the supplementary material). Visual inspection via microscopy of the conditioned Matrigel™ layers revealed that no intact cells remained on the matrix after treatment with cell recovery solution. With this method, the vast majority of the hESCs and hiPSCs that were removed remained intact and viable, as indicated by 7-AAD staining in flow cytometry (Fig. 3B). Additionally, the removal of cell lysis from the protocol permitted analysis of the undifferentiated state and differentiation capacity of the cells that deposited the matrix (Figs. 3A and 3C).
Fig. 3.
Cell viability and pluripotency during generation and harvest of conditioned Matrigel™. H9 and CA1 hESCs and BJ-1D hiPSCs were used to generate conditioned Matrigel™. After harvesting using a cell recovery method that minimizes lysis, remaining cells were assayed for viability and markers of pluripotency. A, RT-PCR analysis of harvested cells for the markers Oct3/4 and Nanog. Values are relative to undifferentiated cells grown in feeder-free conditions not exposed to the cell recovery harvesting methods. Error bars represent S.D., n = 3. B, flow cytometry analysis of cells harvested using the cell recovery method. Cells were assayed for SSEA4 and SSEA1 surface marker expression. Viability was determining using 7-AAD staining. C, cells harvested using the cell recovery method were used to generate embryoid bodies. Following 15 days in culture, embryoid bodies were harvested and assayed for markers of pluripotency (Oct3/4, Nanog), trophoblast (CDX2), ectoderm (NeuroD1, PAX6), endoderm (GATA4, GATA6, alpha-fetoprotein), and mesoderm (T). Error bars represent S.D., n = 2. Values are fold change relative to undifferentiated cells. Phase contrast images of embryoid bodies are provided above. Scale bars represent 250 μm. In both A and C, RPLPO is used as a positive control.
After cell removal and concentration of the conditioned Matrigel™ solution, a combination of strong cation exchange, strong anion exchange, and 1D-SDS-Gel methods (see “Methods” in the supplementary material) were employed for fractionation of the complex ECM-like mixture with iterative exclusion MS analysis (12). MS analysis of these samples illustrated the limited ability of our assay to determine hESC-derived proteins in our list of identifications (supplemental Figs. S2A and S2B). This limitation is a direct result of performing the conditioned matrix experiments on Matrigel™. Because the mixture that is analyzed via MS represents a combination of hESC-derived and Matrigel™ proteins, identification specificity is reliant on nonhomology between human and mouse proteins. However, when we searched the dataset against a concatenated human and mouse sequence database, we observed that few proteins were identified as belonging to a single species, indicating significant interspecies sequence similarity between the identified peptides (supplemental Figs. S2A and S2B). Many of the identified proteins also were found in commercially prepared Matrigel™ in our previous proteomic analysis (supplemental Figs. S2A and S2B) (11). Taken together, these complications severely limited the specificity and sensitivity of the proteomic analysis.
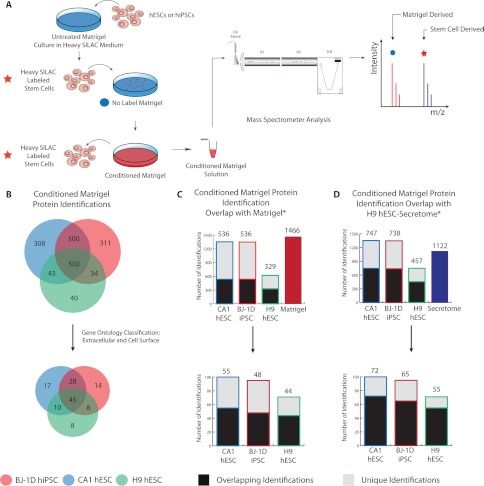
To overcome these challenges, we employed the metabolic labeling approach known as SILAC (18). We have adapted a commercially available, fully defined, serum-free medium formulation to perform SILAC in hESCs and hiPSCs (27) that builds on conditions previously optimized in our laboratory for the application of SILAC with hESCs (22). After 10 passages in SILAC medium (>60 days), H9, CA1, and BJ-1D stem cell lines maintained expression for Oct3/4, Nanog, and SSEA4 as indicated by RT-PCR, immunofluorescence, and flow cytometry (supplemental Figs. S3A–S3C). After the induction of differentiation in vitro using EB assays, all cell lines exhibited increased expression of markers characteristic of each of the three primary germ layers (supplemental Fig. S3D). Additionally, H9 and CA1 hESCs also retained the ability to differentiate in vivo as observed using a teratoma assay (supplemental Fig. S3E; also see “Methods” in the supplementary material). We observed an increase in growth rate in feeder-free culture in SILAC medium (doubling time reduced to ∼24 h in SILAC medium from ∼30 h in MEF-CM), resulting in >96% incorporation of the isotopic labels in just 7 days (supplemental Fig. S4A). Additionally, there was no observable conversion of arginine to proline when utilizing the exogenous addition of l-proline to the SILAC medium as previously reported (supplemental Figs. S4B and S4C) (22). Taken together, these data indicate that extended culture in StemPro®-based SILAC medium can maintain hESC and hiPSC lines in an undifferentiated and pluripotent state. In addition, this medium also represents one of the first fully defined culture media applicable to SILAC experimentation with hESCs and hiPSCs (28). In this SILAC approach, hESCs and hiPSCs would be fully labeled with “heavy ” arginine and lysine prior to being plated for the generation of conditioned Matrigel™ (Fig. 4A). In this way, proteins that are deposited during the generation of conditioned Matrigel™ carry a “heavy” label and can easily be distinguished via MS from the Matrigel™ proteins.
Fig. 4.
Proteomic analysis of SILAC hESC- and hiPSC-derived conditioned Matrigel™. A, H9 and CA1 hESCs, and BJ-1D hiPSCs were grown in defined SILAC medium containing [13C6, 15N4]-arginine and [13C6, 15N2]-lysine. After 7 days of culture, cells were passaged to culture dishes precoated with Matrigel™. After 4 to 5 days of culture on Matrigel™ in SILAC medium containing the heavy isotopic labels, cells were removed using a cell recovery method to minimize cell lysis. The remaining conditioned Matrigel™ layer was rinsed and harvested. Peptides derived from hESCs will carry an isotopic label and will display a resultant mass shift in the MS, whereas those previously present in commercial Matrigel™ will not have this difference. B, conditioned Matrigel™ from H9 and CA1 hESCs and from BJ-1D hiPSCs was analyzed in biological triplicate using an MS-based proteomics protocol. Numbers within the Venn diagrams represent the total number of protein identifications found in each conditioned Matrigel™ analysis for the specific overlap. Datasets were also filtered using gene ontology classification for cellular components based on the terms “extracellular” and “cell surface.” These values are represented in the lower Venn diagram. C, hESC and hiPSC conditioned Matrigel™ proteomic data were individually compared with those previously acquired for commercial Matrigel™ (11). Protein descriptions were used for comparison, as identifiers are species specific. D, hESC and hiPSC conditioned Matrigel™ identifications were further compared with a secretome dataset acquired through proteomic analysis of H9 hESC conditioned medium. Comparison was done manually based upon protein identifiers. In C and D, overlapping identifications are denoted by a black box, and unique identifications are in gray. The total number of overlapping identifications can be found at the top of the bar in text. The rightmost bars in the top graphs of C and D represent total identifications found in Matrigel™ (11) and the H9 hESC secretome (29).
With this protocol, we analyzed conditioned Matrigel™ derived from H9 and CA1 hESCs, as well as BJ-1D hiPSCs. MS analysis yielded a total of 621, 1355, and 1350 “heavy ” labeled proteins identified from the H9, CA1, and BJ-1D cell lines, respectively (Fig. 4B, supplemental Tables S1–S3). A comparison of proteins identified between the cell lines indicated a ∼30% overlap between the three datasets (Fig. 4B). The gain in the number of proteins in the CA1 and BJ-1D stem cell lines is due to their decreased stability in the StemPro® SILAC medium. We observed an increase in cell death and subsequent lysis during general growth of the CA1 and BJ-1D cell lines relative to H9 hESCs in StemPro® SILAC medium, resulting in the identification of additional intracellular proteins. However, we did not observe any significant differences in growth rate or cell morphology between the hESC and hiPSC cultures. Gene ontology analysis revealed that in the dataset corresponding to each cell line, proteins representative of several cellular components were present. This suggests that a variable amount of cell lysis is occurring during growth or harvest of the conditioned Matrigel™ for all the cell lines.
To limit the contaminants from cell lysis, the dataset was filtered using the “extracellular” and “cell surface” cellular component gene ontology terms. Filtering reduced the number of proteins identified in each dataset to 71, 100, and 95 labeled proteins for H9, CA1, and BJ-1D, respectively (Fig. 4B, supplemental Tables S4–S6). However, the data still contained numerous intracellular proteins (histones, mitochondrial, and cytoskeleton associated). This was primarily the result of the limited specificity of the gene ontology assignments. A comparison of the previously acquired commercial Matrigel™ dataset (11) with gene ontology filtered conditioned Matrigel™ datasets revealed that 62%, 55%, and 50% of proteins are identified in both matrices (Fig. 4C). These data illustrate the necessity of SILAC when performing this analysis because of the extensive overlap between the proteins in conditioned Matrigel™ and commercial Matrigel™. The overlap between the three individual conditioned Matrigel™ datasets is ∼35% based on protein identifiers after gene ontology filtering (Fig. 4B). However, coverage between biological replicates of the same cell line using the identical fractionation methods overlapped a minimum of 71% across the samples in the experiment. The consistencies between biological replicates from single stem cell lines illustrate the reproducibility of not only the proteomics method, but also the conditioned Matrigel™ assay itself.
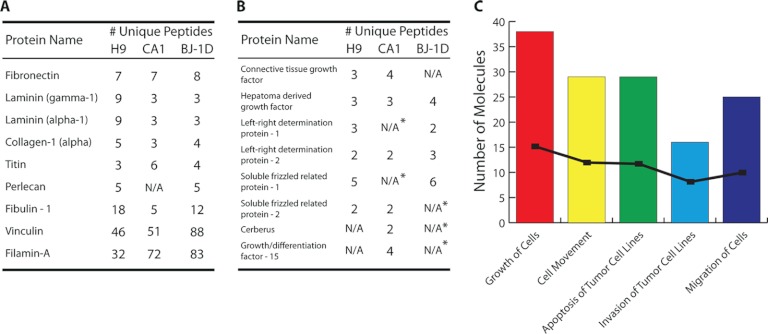
The proteomics data from analysis of conditioned Matrigel™ from hESCs and hiPSCs indicate that numerous cell-line-specific factors were deposited based on comparisons between the protein lists. In comparisons of core ECM proteins, isoforms of fibronectin and laminins and heparan sulfate proteoglycan linked proteins were present at similar abundance levels based on the number of unique peptides in conditioned Matrigel™ from H9 and BJ-1D cell lines. However, BJ-1D conditioned Matrigel™ was enriched by matrix remodeling factors, such as matrix metalloproteinase-14 and -15. Conditioned Matrigel™ derived from CA1 hESCs was found to contain multiple collagen and laminin isoforms not identified in the matrices from the other stem cell lines. Low abundance factors such as soluble frizzled related proteins (sFRPs) 1 and 2 and Lefty A and B were identified in conditioned Matrigel™ from all cell lines. Others, such as connective tissue growth factor (CTGF), were identified in conditioned Matrigel™ from both hESC lines, but not in that from BJ-1D hiPSCs. In addition, other low abundance factors such as Cerberus and fibroblast growth factor-15 were identified only in CA1 conditioned Matrigel™.
Secretome Analysis of the hESC and hiPSC Microenvironment
To further validate and enrich for candidates in the conditioned Matrigel™ data, a comparison was performed with a previously published proteomics screen of the H9 hESC-secretome obtained from MS analysis of hESC-conditioned medium (29). We performed a re-analysis of the H9 hESC-secretome raw dataset with the same software (PEAKS 5.3) used to process the conditioned Matrigel™ data to facilitate comparison and minimize variability from protein assignments or identifiers. Comparison with the H9 hESC-secretome protein identifiers revealed 73%, 55%, and 54% overlap between the unfiltered and 77%, 72%, and 68% between the filtered H9, CA1, and BJ-1D conditioned Matrigel™ datasets, respectively (Fig. 4D). The reduced similarity between the CA1 and BJ-1D conditioned Matrigel™ datasets and the H9 hESC-secretome further highlights the cell-line-specific differences between these hESCs and hiPSCs. However, the overall similarity with the hESC secretome highlights the extracellular nature of the conditioned Matrigel™ data set.
Further examination of the H9 hESC-secretome and conditioned Matrigel™ data revealed that many core ECM proteins, such as fibronectin, collagen, and laminin isoforms, were found in all of the datasets. However, most were significantly more abundant in the H9 hESC-secretome based on the numbers of unique peptides identified. Other factors, such as sFRP1 and CTGF, were also found in common between the conditioned Matrigel™ and H9 hESC-secretome datasets. However, these proteins were not found to have a higher abundance in the H9 hESC-secretome based on unique peptide identifications. Additionally, other factors, such as the Lefty A and B proteins, were not identified in our reanalyzed H9 hESC-secretome dataset. These differences illustrate that factors identified only in the conditioned Matrigel™ potentially have an ECM-specific role in the microenvironment. Additionally, similarities between the H9 hESC-secretome and conditioned Matrigel™ data suggest that overlapping factors might function in both soluble and insoluble portions of the microenvironment.
Stem Cell Factors Identified in Conditioned MatrigelTM
Proteomic investigation of the conditioned Matrigel™ dataset revealed the presence of numerous hESC and hiPSC regulatory proteins. In order to enrich for hESC and hiPSC derived proteins that facilitate the maintenance of an undifferentiated state on conditioned Matrigel™, we opted to focus on only those classified as extracellular or cell surface by gene ontology. Within this filtered dataset we found many essential core ECM proteins that are also present in commercial Matrigel™ preparations (Fig. 5A). Further investigation of the filtered conditioned Matrigel™ dataset revealed the presence of numerous factors with potential roles in hESC and hiPSC pluripotency (Fig. 5B). These included regulators of TGF-β/Nodal and Wnt signaling pathways, Lefty A/B, Cerberus, and sFRP1/2. Other identified factors, such as hepatoma-derived growth factor and CTGF, are of particular interest because of their relevance in the biology of cancer cells (30, 31). Interestingly, after pathway analysis of the filtered datasets, we observed a significant enrichment for molecules involved in cell motility, apoptosis, and invasion of cancer cells (Fig. 5C). Several of these have previously been found in proteomic screens of MEF and hESC-conditioned medium (29). However, their detection in conditioned Matrigel™ suggests that the ECM is playing a role in their activity.
Fig. 5.
Proteins and pathways identified in conditioned Matrigel™ from hESCs and hiPSCs. Conditioned Matrigel™ was analyzed using an iterative exclusion MS proteomics-based approach. Proteins were identified using PEAKS 5.3 software. A, subset of the core ECM proteins found in conditioned Matrigel™ that were also identified in a previous proteomic analysis of commercial Matrigel™ (11). In the case of collagen, the top isoform was chosen. B, growth factors and proteins known to regulate core hESC and hiPSC pluripotency pathways found in conditioned Matrigel™. Asterisks indicate when a protein was identified as a single peptide hit. Only proteins with two unique peptides are retained, such that these single peptide hits are not included in the final dataset. C, pathway analysis performed using Ingenuity Systems software. The top biological functions identified in all three cell lines were related to cellular movement, growth, and death or invasion of tumor cell lines. Number of molecules denotes the number of candidate proteins identified as being involved with that function found in our dataset. Black boxes represent the -log(p value) for the probability of identifying these pathways as being enriched in our dataset by chance.
DISCUSSION
In this study we have employed a SILAC MS-based proteomics approach to reveal, for the first time, the components of hESC- and hiPSC-derived conditioned Matrigel™. With this approach, we have identified numerous factors with recognized roles in hESC and hiPSC pluripotency, as well as several others not yet described. We observed that growth on stem-cell-derived conditioned Matrigel™ in the absence of bFGF promotes the maintenance of an undifferentiated state in hESCs. This indicates that hESCs are depositing factors in the matrix that can prevent stem cell differentiation and promote self-renewal. Although previous studies have identified candidate proteins in the hESC microenvironment through the analysis of conditioned medium, none have focused on proteins deposited in the ECM by pluripotent stem cells. Therefore, the methodology and the information within the dataset presented here are of substantial importance in promoting a comprehensive understanding of the hESC and hiPSC in vitro microenvironment.
The presence of core ECM components in conditioned Matrigel™, such as isoforms of fibronectin, collagen, and laminin, illustrates that hESCs and hiPSCs generate a self-supportive niche for attachment and proliferation. All three of these matrix proteins have been shown to sustain the growth of hESCs and hiPSCs when used as the sole growth support (10, 12, 32–34). These core proteins regulate critical cellular processes such as cell attachment through integrin binding. Integrin interactions can mediate signal transduction responses similar to those triggered by soluble ligands (35, 36). Therefore, hESCs and hiPSCs potentially utilize these interactions to regulate a diverse array of cellular functions. It is likely that the presence of multiple core ECM proteins in hESC- and hiPSC-supportive matrices such as Matrigel™ engages a complement of integrin-mediated events that collectively facilitate hESC and hiPSC growth (10, 37).
The identification of multiple growth factors and their binding partners in conditioned Matrigel™ highlights another potential function for these core ECM proteins in the matrix as a reservoir for mitogenic and morphogenic factors. The ECM has been proposed to contribute to the regulation of growth factor signaling in the microenvironment (reviewed in Ref. 9). Central to this function is the presence of specialized domains on fibronectin and laminin designed to bind growth factors such as epidermal growth factor, hepatocyte growth factor, and platelet derived growth factor. Fibronectin, for example, contributes to TGF-β signaling through interaction with latent-TGF-β binding proteins (38, 39). Binding between growth factors and these glycoproteins can also be mediated by interactions with heparan sulfate, a molecule that has been shown to be beneficial for the maintenance of hESCs and hiPSCs (9, 40). The result of these interactions implicates the ECM in the orchestration of cellular processes through the binding and regulation of growth factors.
Controlling the activity and concentration of growth factors present in the microenvironment is critical for maintenance of the hESC and hiPSC state. We identified proteins that regulate Wnt, TGF-β (through Activin/Nodal), and BMP signaling pathways in conditioned Matrigel™. The sFRP1 and sFRP2 proteins have active roles in numerous cell systems, primarily in the regulation of canonical Wnt signaling (reviewed in Ref. 41). Wnt signaling through the canonical pathway is important for the maintenance of mESC and hESC self-renewal and pluripotency (42–44). In a recent study comparing 59 different hESC lines, a positive correlation between Nanog and sFRP2 gene expression was observed (45). Additionally, sFRP2 was identified as potentially regulated by Nanog in ChIP analyses of hESCs (46). The C-terminal netrin domain on sFRP1 and sFRP2 is known to confer heparin-binding properties that potentially regulate the activity of the parent protein (47, 48). Lefty A and B regulate TGF-β signaling through antagonism of the activating protein Nodal (49). Activin/Nodal mediated signaling is one of the core pathways critical for the maintenance of self-renewal and pluripotency of hESCs and hiPSCs (15, 16, 50). Interestingly, Lefty was previously observed to mediate metastatic melanoma cellular reprogramming on conditioned Matrigel™ (14). The presence of Lefty and sFRP proteins in conditioned Matrigel™ indicates that the ECM might function to regulate these pathways.
Previous studies of ESC-derived matrix proteins have focused on the antagonism of the BMP pathway using Gremlin (17). The inhibition of BMP mediated signaling promotes the maintenance of hESC pluripotency (51). The activation of SMAD (SMA mothers against decapentalegic) 1/5/8 mediated gene expression following hESC exposure to BMP ligands has been implicated in the differentiation of hESCs (16, 50). Cerberus is an inhibitor of BMP mediated activation that acts through antagonism of BMP4 (52, 53). Interestingly, Cerberus has also been shown to inhibit Nodal and Wnt signaling pathways (53, 54). The microenvironment is considered to effect cellular change by providing a reservoir for growth factors, morphogens, and cytokines. Given such, it is likely that the core pathways we have identified here function together in conditioned Matrigel™ to generate a supportive environment. Indeed, multiple levels of cross-talk have been suggested among the FGF/TGF-β/Wnt pathways (55, 56).
Currently, there is significant interest within the scientific community in assessing the equivalency of hESCs and hiPSCs based on multiple genomic, proteomic, and developmental criteria (57, 58). However, these studies have revealed numerous differences between individual hESC and hiPSC lines (59). In this study, although the conditioned Matrigel™ datasets were found to contain numerous proteins unique to a single cell line, we did not observe a significant enrichment in factors contributing to any specific pathway or process when we focused on gene-ontology-filtered ECM proteins. These findings indicate that in the culture system utilized for this analysis, the stem-cell-deposited contribution to the microenvironment is highly similar in the individual hESC and hiPSC lines tested. However, as has recently been demonstrated in large-scale comparative analyses of hESC and hiPSCs, the ability to characterize differences between stem cell proteomes is dependent on the sampling of a sufficient number of cell lines (58). Expansion of the analysis performed here to include an increased number of cell lines could potentially facilitate comparative analysis of hESC and hiPSC conditioned Matrigel™. The inclusion of additional pluripotent stem cell lines would also aid in the validation of deposited proteins as opposed to those originating from cellular lysis during the generation of conditioned Matrigel™.
The contamination of conditioned Matrigel™ by intracellular proteins highlights the primary technical challenge of this work going forward. Comparisons with datasets of secreted proteins, such as the H9 hESC-secretome, give confidence that the proteins found in conditioned Matrigel™ are truly secreted. However, the limited specificity of gene ontology assignments renders bioinformatic confirmation challenging. In addition, the high abundance proteins present in Matrigel™ interfere with biological target validation of stem cell proteins using assays such as immunofluorescence because of off-target interactions with the matrix. Analysis of conditioned Matrigel™ in a defined growth system based on alternative matrices such as fibronectin, vitronectin, or synthetic growth surfaces could potentially facilitate the validation of identified candidates for true localization and function through the minimization of background interactions. The recent development of spotted matrix microarrays is an attractive alternative matrix support for use in combination with MS-based proteomics analysis of conditioned Matrigel™ (60). In addition to supporting high-throughput screening of identified proteins from MS, these methods permit colony-specific screening. Morphogen gradients have been observed within individual stem cell colonies that will be masked in population-level analysis (20). Direct analysis of colony-specific matrix interactions with MS-based proteomics can potentially provide valuable insight into the stem cell microenvironment.
In conclusion, we have developed a technique that allows the characterization of factors deposited by cells into complex ECMs such as Matrigel™. Using this innovative approach, we confirmed the presence of key pluripotency regulators such as Lefty in stem cell conditioned matrices. In addition, we identified other proteins, such as sFRP1/2, that could play important roles in the regulation of stem fate. The constituents of the ECM are largely unknown because of the complexity of this fraction, especially when matrices such as Matrigel™ are used as support. The new method presented in this work can be used to characterize other ECMs (e.g. cancer cells) so that key regulatory extracellular proteins can be identified.
Supplementary Material
Acknowledgments
C.H. is a recipient of an NSERC Canada Graduate Scholarship Doctoral Award. We are grateful to Courtney Brooks for MEF cell culture and conditioning.
Footnotes
* This work was supported by a grant from the NSERC Discovery program to G.L. and by grants from CIHR to W.L.S. (Grant No. MOP-89910), L.-M.P., and D.H.B.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- BMP
- bone morphogenic protein
- CM
- conditioned medium
- CTGF
- connective tissue growth factor
- DMEM
- Dulbeccos' modified Eagle's medium
- ECM
- extracellular matrix
- FBS
- fetal bovine serum
- FGF
- fibroblast growth factor
- hESC
- human embryonic stem cell
- hiPSC
- human induced pluripotent stem cell
- Lefty
- left-right determination factor
- MEF
- mouse embryonic fibroblast
- PBS
- phosphate buffered saline
- sFRP
- soluble frizzled related protein
- SILAC
- stable isotope labeling with amino acids in cell culture
- SMAD
- SMA mothers against decapentalegic
- SSEA
- stage-specific embryonic antigen
- TGF
- transforming growth factor.
REFERENCES
- 1. Li L., Xie T. (2005) Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21, 605–631 [DOI] [PubMed] [Google Scholar]
- 2. Spradling A., Drummond-Barbosa D., Kai T. (2001) Stem cells find their niche. Nature 414, 98–104 [DOI] [PubMed] [Google Scholar]
- 3. Watt F. M., Hogan B. L. M. (2000) Out of Eden: stem cells and their niches. Science 287, 1427–1430 [DOI] [PubMed] [Google Scholar]
- 4. Chin A. C. P., Fong W. J., Goh L. T., Philp R., Oh S. K. W., Choo A. B. H. (2007) Identification of proteins from feeder conditioned medium that support human embryonic stem cells. J. Biotechnol. 130, 320–328 [DOI] [PubMed] [Google Scholar]
- 5. Lim J. W. E., Bodnar A. (2002) Proteome analysis of conditioned medium from mouse embryonic fibroblast feeder layers which support the growth of human embryonic stem cells. Proteomics 2, 1187–1203 [DOI] [PubMed] [Google Scholar]
- 6. Prowse A. B. J., McQuade L. R., Bryant K. J., Marcal H., Gray P. P. (2007) Identification of potential pluripotency determinants for human embryonic stem cells following proteomic analysis of human and mouse fibroblast conditioned media. J. Proteome Res. 6, 3796–3807 [DOI] [PubMed] [Google Scholar]
- 7. Prowse A. B. J., McQuade L. R., Bryant K. J., Van Dyk D. D., Tuch B. E., Gray P. P. (2005) A proteome analysis of conditioned media from human neonatal fibroblasts used in the maintenance of human embryonic stem cells. Proteomics 5, 978–989 [DOI] [PubMed] [Google Scholar]
- 8. Kleinman H. K., McGarvey M. L., Liotta L. A., Robey P. G., Tryggvason K., Martin G. R. (1982) Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry 21, 6188–6193 [DOI] [PubMed] [Google Scholar]
- 9. Hynes R. O. (2009) The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hakala H., Rajala K., Ojala M., Panula S., Areva S., Kellomaki M., Suuronen R., Skottman H. (2009) Comparison of biomaterials and extracellular matrices as a culture platform for multiple, independently derived human embryonic stem cell lines. Tissue Eng. Part A 15, 1775–1785 [DOI] [PubMed] [Google Scholar]
- 11. Hughes C. S., Postovit L. M., Lajoie G. A. (2010) Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890 [DOI] [PubMed] [Google Scholar]
- 12. Hughes C. S., Radan L., Betts D., Postovit L. M., Lajoie G. A. (2011) Proteomic analysis of extracellular matrices used in stem cell culture. Proteomics 11, 3983–3991 [DOI] [PubMed] [Google Scholar]
- 13. Postovit L. M., Seftor E. A., Seftor R. E., Hendrix M. J. (2006) A three-dimensional model to study the epigenetic effects induced by the microenvironment of human embryonic stem cells. Stem Cells 24, 501–505 [DOI] [PubMed] [Google Scholar]
- 14. Postovit L. M., Margaryan N. V., Seftor E. A., Kirschmann D. A., Lipavsky A., Wheaton W. W., Abbott D. E., Seftor R. E., Hendrix M. J. (2008) Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 4329–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Besser D. (2004) Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J. Biol. Chem. 279, 45076–45084 [DOI] [PubMed] [Google Scholar]
- 16. Vallier L., Alexander M., Pedersen R. A. (2005) Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 [DOI] [PubMed] [Google Scholar]
- 17. Kim M. O., Kim S. H., Oi N., Lee M. H., Yu D. H., Kim D. J., Cho E. J., Bode A. M., Cho Y. Y., Bowden T. G., Dong Z. (2011) Embryonic stem-cell-preconditioned microenvironment induces loss of cancer cell properties in human melanoma cells. Pigment Cell Melanoma Res. 24, 922–931 [DOI] [PubMed] [Google Scholar]
- 18. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 19. Seguin C. A., Draper J. S., Nagy A., Rossant J. (2008) Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell 3, 182–195 [DOI] [PubMed] [Google Scholar]
- 20. Peerani R., Rao B. M., Bauwens C., Yin T., Wood G. A., Nagy A., Kumacheva E., Zandstra P. W. (2007) Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 26, 4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hotta A., Cheung A. Y., Farra N., Garcha K., Chang W. Y., Pasceri P., Stanford W. L., Ellis J. (2009) EOS lentiviral vector selection system for human induced pluripotent stem cells. Nat. Protoc. 4, 1828–1844 [DOI] [PubMed] [Google Scholar]
- 22. Bendall S. C., Hughes C., Stewart M. H., Doble B., Bhatia M., Lajoie G. A. (2008) Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 7, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma B., Zhang K. Z., Hendrie C., Liang C. Z., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 4, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhatia V. N., Perlman D. H., Costello C. E., McComb M. E. (2009) Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal. Chem. 81, 9819–9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hendrix M. J., Seftor E. A., Seftor R. E., Kasemeier-Kulesa J., Kulesa P. M., Postovit L. M. (2007) Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer 7, 246–255 [DOI] [PubMed] [Google Scholar]
- 27. Wang L., Schulz T. C., Sherrer E. S., Dauphin D. S., Shin S., Nelson A. M., Ware C. B., Zhan M., Song C. Z., Chen X., Brimble S. N., McLean A., Galeano M. J., Uhl E. W., D'Amour K. A., Chesnut J. D., Rao M. S., Blau C. A., Robins A. J. (2007) Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood 110, 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang S., Tian R., Li L., Figeys D., Wang L. (2011) An enhanced chemically defined SILAC culture system for quantitative proteomics study of human embryonic stem cells. Proteomics 11, 4040–4046 [DOI] [PubMed] [Google Scholar]
- 29. Bendall S. C., Hughes C., Campbell J. L., Stewart M. H., Pittock P., Liu S., Bonneil E., Thibault P., Bhatia M., Lajoie G. A. (2009) An enhanced mass spectrometry approach reveals human embryonic stem cell growth factors in culture. Mol. Cell. Proteomics 8, 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu C. Y., Chang C. C., Prakash E., Kuo M. L. (2008) Connective tissue growth factor (CTGF) and cancer progression. J. Biomed. Sci. 15, 675–685 [DOI] [PubMed] [Google Scholar]
- 31. Everett A. D., Bushweller J. (2003) Hepatoma derived growth factor is a nuclear targeted mitogen. Curr. Drug Targets 4, 367–371 [DOI] [PubMed] [Google Scholar]
- 32. Miyazaki T., Futaki S., Hasegawa K., Kawasaki M., Sanzen N., Hayashi M., Kawase E., Sekiguchi K., Nakatsuji N., Suemori H. (2008) Recombinant human laminin isoforms can support the undifferentiated growth of human embryonic stem cells. Biochem. Biophys. Res. Commun. 375, 27–32 [DOI] [PubMed] [Google Scholar]
- 33. Baxter M. A., Camarasa M. V., Bates N., Small F., Murray P., Edgar D., Kimber S. J. (2009) Analysis of the distinct functions of growth factors and tissue culture substrates necessary for the long-term self-renewal of human embryonic stem cell lines. Stem Cell Res. 1, 28–38 [DOI] [PubMed] [Google Scholar]
- 34. Furue M. K., Na J., Jackson J. P., Okamoto T., Jones M., Baker D., Hata R., Moore H. D., Sato J. D., Andrews P. W. (2008) Heparin promotes the growth of human embryonic stem cells in a defined serum-free medium. Proc. Natl. Acad. Sci. U.S.A. 105, 13409–13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berrier A. L., Yamada K. M. (2007) Cell-matrix adhesion. J. Cell. Physiol. 213, 565–573 [DOI] [PubMed] [Google Scholar]
- 36. Legate K. R., Wickstrom S. A., Fassler R. (2009) Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23, 397–418 [DOI] [PubMed] [Google Scholar]
- 37. Ludwig T. E., Levenstein M. E., Jones J. M., Berggren W. T., Mitchen E. R., Frane J. L., Crandall L. J., Daigh C. A., Conard K. R., Piekarczyk M. S., Llanas R. A., Thomson J. A. (2006) Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187 [DOI] [PubMed] [Google Scholar]
- 38. Munger J. S., Sheppard D. (2011) Cross talk among TGF-beta signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb. Perspect. Biol. 3, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez F., Rifkin D. B. (2009) Extracellular microfibrils: contextual platforms for TGFbeta and BMP signaling. Curr. Opin. Cell Biol. 21, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sasaki N., Okishio K., Ui-Tei K., Saigo K., Kinoshita-Toyoda A., Toyoda H., Nishimura T., Suda Y., Hayasaka M., Hanaoka K., Hitoshi S., Ikenaka K., Nishihara S. (2008) Heparan sulfate regulates self-renewal and pluripotency of embryonic stem cells. J. Biol. Chem. 283, 3594–3606 [DOI] [PubMed] [Google Scholar]
- 41. Esteve P., Bovolenta P. (2010) The advantages and disadvantages of sfrp1 and sfrp2 expression in pathological events. Tohoku J. Exp. Med. 221, 11–17 [DOI] [PubMed] [Google Scholar]
- 42. ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R. K., Nusse R. (2011) Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13, 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004) Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 [DOI] [PubMed] [Google Scholar]
- 44. Dravid G., Ye Z., Hammond H., Chen G., Pyle A., Donovan P., Yu X., Cheng L. (2005) Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells 23, 1489–1501 [DOI] [PubMed] [Google Scholar]
- 45. Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P. W., Beighton G., Bello P. A., Benvenisty N., Berry L. S., Bevan S., Blum B., Brooking J., Chen K. G., Choo A. B., Churchill G. A., Corbel M., Damjanov I., Draper J. S., Dvorak P., Emanuelsson K., Fleck R. A., Ford A., Gertow K., Gertsenstein M., Gokhale P. J., Hamilton R. S., Hampl A., Healy L. E., Hovatta O., Hyllner J., Imreh M. P., Itskovitz-Eldor J., Jackson J., Johnson J. L., Jones M., Kee K., King B. L., Knowles B. B., Lako M., Lebrin F., Mallon B. S., Manning D., Mayshar Y., McKay R. D., Michalska A. E., Mikkola M., Mileikovsky M., Minger S. L., Moore H. D., Mummery C. L., Nagy A., Nakatsuji N., O'Brien C. M., Oh S. K., Olsson C., Otonkoski T., Park K. Y., Passier R., Patel H., Patel M., Pedersen R., Pera M. F., Piekarczyk M. S., Pera R. A., Reubinoff B. E., Robins A. J., Rossant J., Rugg-Gunn P., Schulz T. C., Semb H., Sherrer E. S., Siemen H., Stacey G. N., Stojkovic M., Suemori H., Szatkiewicz J., Turetsky T., Tuuri T., van den Brink S., Vintersten K., Vuoristo S., Ward D., Weaver T. A., Young L. A., Zhang W. (2007) Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816 [DOI] [PubMed] [Google Scholar]
- 46. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uren A., Reichsman F., Anest V., Taylor W. G., Muraiso K., Bottaro D. P., Cumberledge S., Rubin J. S. (2000) Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 275, 4374–4382 [DOI] [PubMed] [Google Scholar]
- 48. Lee J. L., Lin C. T., Chueh L. L., Chang C. J. (2004) Autocrine/paracrine secreted frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis. J. Biol. Chem. 279, 14602–14609 [DOI] [PubMed] [Google Scholar]
- 49. Schier A. F. (2003) Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19, 589–621 [DOI] [PubMed] [Google Scholar]
- 50. James D., Levine A. J., Besser D., Hemmati-Brivanlou A. (2005) TGF beta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem. Development 132, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 51. Xu R. H., Peck R. M., Li D. S., Feng X., Ludwig T., Thomson J. A. (2005) Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods 2, 185–190 [DOI] [PubMed] [Google Scholar]
- 52. Belo J. A., Bachiller D., Agius E., Kemp C., Borges A. C., Marques S., Piccolo S., De Robertis E. M. (2000) Cerberus-like is a secreted BMP and nodal antagonist not essential for mouse development. Genesis 26, 265–270 [PubMed] [Google Scholar]
- 53. Piccolo S., Agius E., Leyns L., Bhattacharyya S., Grunz H., Bouwmeester T., De Robertis E. M. (1999) The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397, 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawano Y., Kypta R. (2003) Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 116, 2627–2634 [DOI] [PubMed] [Google Scholar]
- 55. Guo X., Wang X. F. (2009) Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 19, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Singh A. M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A. L., Sun Y., Menendez L., Kulik M., Dalton S. (2012) Signaling network crosstalk in human pluripotent cells: a SMAD2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Narsinh K. H., Plews J., Wu J. C. (2011) Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol. Ther. 19, 635–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phanstiel D. H., Brumbaugh J., Wenger C. D., Tian S., Probasco M. D., Bailey D. J., Swaney D. L., Tervo M. A., Bolin J. M., Ruotti V., Stewart R., Thomson J. A., Coon J. J. (2011) Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods 8, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Allegrucci C., Young L. E. (2007) Differences between human embryonic stem cell lines. Hum. Reprod. Update 13, 103–120 [DOI] [PubMed] [Google Scholar]
- 60. Gobaa S., Hoehnel S., Roccio M., Negro A., Kobel S., Lutolf M. P. (2011) Artificial niche microarrays for probing single stem cell fate in high throughput. Nat. Methods 8, 949–955 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.