The kinesin KIF14 associates with the PDZ domain of Radil and negatively regulates Rap1-mediated inside-out integrin activation by tethering Radil on microtubules.
Abstract
The small GTPase Rap1 regulates inside-out integrin activation and thereby influences cell adhesion, migration, and polarity. Several Rap1 effectors have been described to mediate the cellular effects of Rap1 in a context-dependent manner. Radil is emerging as an important Rap effector implicated in cell spreading and migration, but the molecular mechanisms underlying its functions are unclear. We report here that the kinesin KIF14 associates with the PDZ domain of Radil and negatively regulates Rap1-mediated inside-out integrin activation by tethering Radil on microtubules. The depletion of KIF14 led to increased cell spreading, altered focal adhesion dynamics, and inhibition of cell migration and invasion. We also show that Radil is important for breast cancer cell proliferation and for metastasis in mice. Our findings provide evidence that the concurrent up-regulation of Rap1 activity and increased KIF14 levels in several cancers is needed to reach optimal levels of Rap1–Radil signaling, integrin activation, and cell–matrix adhesiveness required for tumor progression.
Introduction
The small G-protein Rap1 is an important mediator of integrin inside-out signaling, which plays a pivotal role in adhesion, spreading, and migration of cells (Bos et al., 2003; Arthur et al., 2004; Kinashi and Katagiri, 2004, 2005; Bos, 2005). Rap1 acts as a molecular switch that cycles between active GTP-bound and inactive GDP-bound states. Rap1 activity is regulated by guanine nucleotide exchange factors (GEFs) such as Epac1 (de Rooij et al., 1998) and GTPase activating proteins (GAPs) such as Rap1GAP (Rubinfeld et al., 1991). Upon activation, Rap1 has the ability to increase the affinity of integrins for their extracellular matrix (ECM) ligands and to promote their clustering (Sebzda et al., 2002; Lafuente et al., 2004; Han et al., 2006; Kim et al., 2011). In recent years the identification and characterization of downstream Rap effector proteins such as RIAM (Lafuente et al., 2004), RapL (Katagiri et al., 2003), Krit1 (Glading et al., 2007), AF-6 (Boettner et al., 2000), and Radil (Smolen et al., 2007) have shed light on the molecular mechanisms underlying the cellular effects mediated by Rap1. We previously identified the Rap1 effector Radil as a protein associating with Gβγ subunits of heterotrimeric G-proteins (Ahmed et al., 2010). Radil was found to be required for the Rap1a-mediated inside-out activation of integrins, adhesion, and spreading of human fibrosarcoma cells (Ahmed et al., 2010). Radil is also known to have important functions in Epac1-mediated spreading of lung carcinoma cells (Ross et al., 2011) and to be indispensable for the migration of neural crest cells during zebrafish development (Smolen et al., 2007).
The control of cell–matrix adhesion plays a fundamental role in controlling cancer cell migration during metastasis (McLean et al., 2005; Desgrosellier and Cheresh, 2010; Arjonen et al., 2011). The implication of Rap1 signaling in the modulation of integrin activity has thus provided a framework to study its implication in tumor progression. Both hyper-activation as well as decreased Rap1 activity is known to affect the migration of breast, melanoma, and prostate cancer cells (Bailey et al., 2009; Zheng et al., 2009; Kim et al., 2012). This highlights that precise control of cellular adhesion by Rap1 and its effectors is required for efficient cell movements. This requirement for the fine-tuning of Rap1-mediated inside-out signaling for optimal control of cell–matrix adhesion implies the existence of positive and negative mechanisms of regulation. Although how Rap1 leads to integrin inside-out activation is becoming better defined, the identification of mechanisms buffering or negatively impinging this process is not well understood. Such regulators may be especially relevant in the context of aggressive cancer cells to optimally adjust Rap1 activity where it is known to be elevated (Lorenowicz et al., 2008; Lyle et al., 2008; Bailey et al., 2009; Zheng et al., 2009; Freeman et al., 2010; Huang et al., 2012).
Kinesins are molecular motors associated with intracellular transport (Hirokawa et al., 2009; Verhey and Hammond, 2009). Kinesin superfamily proteins (KIFs) are important molecular motors that transport various cargoes along microtubules tracks. Several kinesins have been implicated in cancer progression due to their role in mitotic cell division (Huszar et al., 2009). Recently, kinesins were uncovered as playing important regulatory roles in adhesion and migration of cells (Uchiyama et al., 2010; Zhang et al., 2010). Blocking kinesin-1 activity using inactivating antibodies was also shown to lead to increase in the size and number of substrate adhesions (Kaverina et al., 1997; Krylyshkina et al., 2002). Although the precise mechanisms are unclear, kinesins were suggested to control the delivery of factors at adhesion sites to retard their growth or promote their disassembly. KIF14 was initially characterized as a protein involved in cytokinesis by interacting with protein-regulating cytokinesis-1 (PRC1) and Citron kinase (Gruneberg et al., 2006). KIF14 was also demonstrated to be highly up-regulated in several cancers including retinoblastomas, breast cancers, lung cancers, and ovarian cancers and its high expression levels has been clinically correlated with increased breast cancer invasiveness and mortality (Corson et al., 2005, 2007; Corson and Gallie, 2006; Thériault et al., 2012).
We previously established that the C-terminal PDZ domain of Radil was critical for its function, but the identity of the protein(s) binding to the Radil PDZ domain and how it contributed to Rap1–Radil signaling was, however, not addressed. PDZ domains are present in many scaffolding proteins and are involved in the organization of multi-protein complexes important for numerous biological processes including cell adhesion, spreading, migration, and polarization (Hall et al., 1998; Fanning and Anderson, 1999; Harris and Lim, 2001; Sheng and Sala, 2001; Humbert et al., 2003; Kim and Sheng, 2004; Ludford-Menting et al., 2005; Meerschaert et al., 2007; Radziwill et al., 2007; Baumgartner et al., 2008). In this study, we use an integrative proteomic approach based on phage display and mass spectrometry to identify KIF14 as a novel PDZ-binding protein that interacts with Radil. Our results describe KIF14 as a negative regulator of Rap1–Radil-mediated signaling and uncover an essential role for Radil in breast cancer progression.
Results
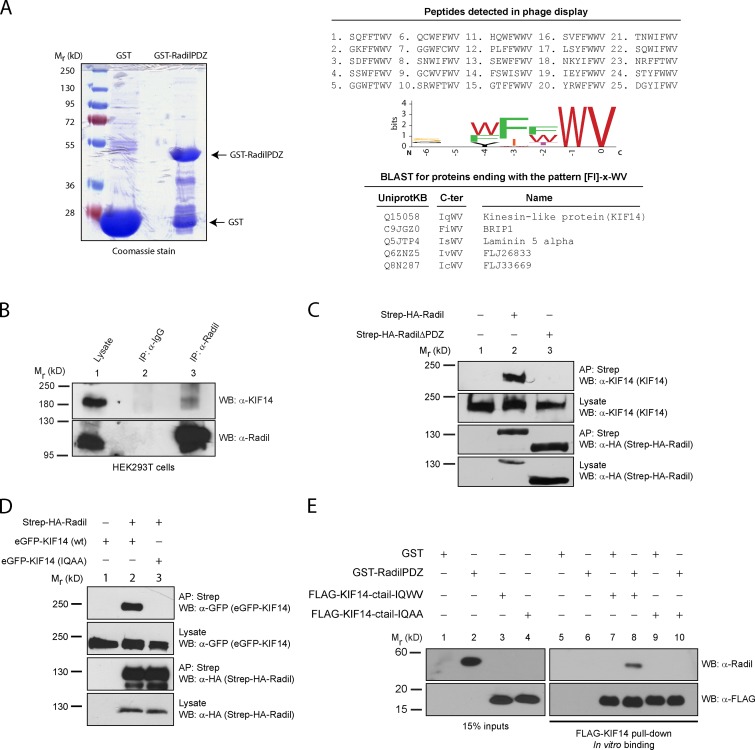
Radil associates with kinesin family protein 14 (KIF14)
To gain further insights into Radil function and regulation, we set out to identify proteins interacting with its C-terminal PDZ domain. We first used a phage display approach to define the preferred C-terminal peptides recognized by the Radil PDZ domain. We panned a phage-displayed peptide library with a recombinant Radil PDZ domain fused to GST (Fig. 1 A) and repeated multiple rounds of binding selection. Binding phages were isolated and the sequences of the displayed peptides were determined (Fig. 1 A). The analysis of the 25 best binding sequences revealed that the Radil PDZ domain recognizes hydrophobic C-terminal ligands with a strong consensus to the sequence [FI]-[FWT]-WV for the last four amino acids as represented by the motif enrichment diagram in Fig. 1 A. Most PDZ domains prefer ligands containing either S/T (class I) or hydrophobic amino acids (class II) at the −2 position (third residue from the end; Tonikian et al., 2008). Radil appears to contain an atypical PDZ domain because it can bind to both ligands classes. Because the Radil PDZ domain can accommodate both hydrophobic and hydrophilic residues at the −2 position, we performed a BLAST search against the Swiss-Prot human proteome database to assemble a list of natural proteins with C termini matching the consensus sequence [FI]-x-WV. We compared this list of putative Radil PDZ domain ligands to a list of proteins that we identified in FLAG-Radil complexes using immunoprecipitation and mass spectrometry (Fig. 1 A; Table S1), and we found that KIF14 was present in both lists. Consistent with a physical association between these proteins, the immunoprecipitation of Radil from HEK293T cells led to the coprecipitation of KIF14 (Fig. 1 B). The phage display results suggest that KIF14 binds to the PDZ domain of Radil. To confirm this result in a cellular context, lysates from control HEK293T or from cells stably expressing Strep-HA-Radil or Strep-HA-RadilΔPDZ were subjected to affinity purification using Sepharose-streptavidin followed by Western blotting using antibodies specific to KIF14. Whereas endogenous KIF14 copurifies with full-length Radil, it is absent in control or RadilΔPDZ affinity-purified samples (Fig. 1 C, compare lane 2 with lanes 1 and 3). To validate that KIF14 interacts with Radil via its atypical PDZ binding motif, we coexpressed Strep-HA-Radil with wild-type KIF14 or a mutant KIF14-IQAA in which the C-terminal tryptophan and valine were substituted with alanine residues, and we assessed their interaction by affinity purification. Although wild-type KIF14 associates with Radil, KIF14-IQAA does not (Fig. 1 D, compare lanes 2 and 3). To also test for the specificity of the Radil-KIF14 interaction we performed affinity purification of Radil and blotted for the unrelated kinesin family protein KIF7 and showed that these proteins do not interact (Fig. S1 A). We also showed that KIF14 does not interact with AF-6, which is closely related to Radil in terms of domain architecture and also possesses a C-terminal PDZ domain whose sequence is 33% identical and 53% homologous to the PDZ domain of Radil (Fig. S1, B and C). To determine if the interaction between KIF14 and Radil is direct, we performed in vitro binding experiments using recombinant proteins. Although the recombinant C-terminal domain of wild-type KIF14 binds to the PDZ domain of Radil, the IQAA mutant did not (Fig. 1 E, compare lane 8 and 10). From these experiments we conclude that Radil and KIF14 directly associate through a PDZ domain–ligand interaction.
Figure 1.
KIF14 interacts with the Radil PDZ domain. (A) Recombinant GST-RadilPDZ domain was used as bait in phage display selections using a library of random heptapeptides. 25 unique binding peptides were recovered from the screen and analyzed using a sequence logo generator to identify the preferred binding sequence. The resulting C-terminal binding motif [FI]-x-WV was searched against UniProtKB/Swiss-Prot human database; and the C termini of five proteins were found to match this sequence. (B) KIF14 coimmunoprecipitates with Radil. Lysates from HEK293T cells were immunoprecipitated (IP) using α-Radil or control IgG antibodies, followed by Western blotting (WB) with α-Radil or α-KIF14 antibodies. (C) Radil interacts with KIF14 via its PDZ domain. Lysates from HEK293T cells stably expressing Strep-HA-Radil or Strep-HA-RadilΔPDZ were affinity purified (AP) using streptavidin Sepharose followed by WB using α-HA or α-KIF14 antibodies. (D) The KIF14 PDZ ligand is required to bind to Radil. HEK293T cells stably expressing Strep-HA-Radil were transiently transfected with eGFP-KIF14 wild-type (wt) or eGFP-KIF14-IQAA (WV→AA mutant) constructs. Lysates were subjected to AP with streptavidin beads followed by Western blotting with α-KIF14 and α-HA antibodies. (E) Radil binds to KIF14 directly. Recombinant GST-RadilPDZ was incubated with purified FLAG-tagged KIF14-ctail (wild-type or IQAA mutant) that were immobilized on FLAG-M2 beads. After 1 h of binding, beads were washed and protein binding determined by Western blotting using α-Radil and α-FLAG (rabbit) antibodies.
Radil localizes with KIF14 on the microtubule network
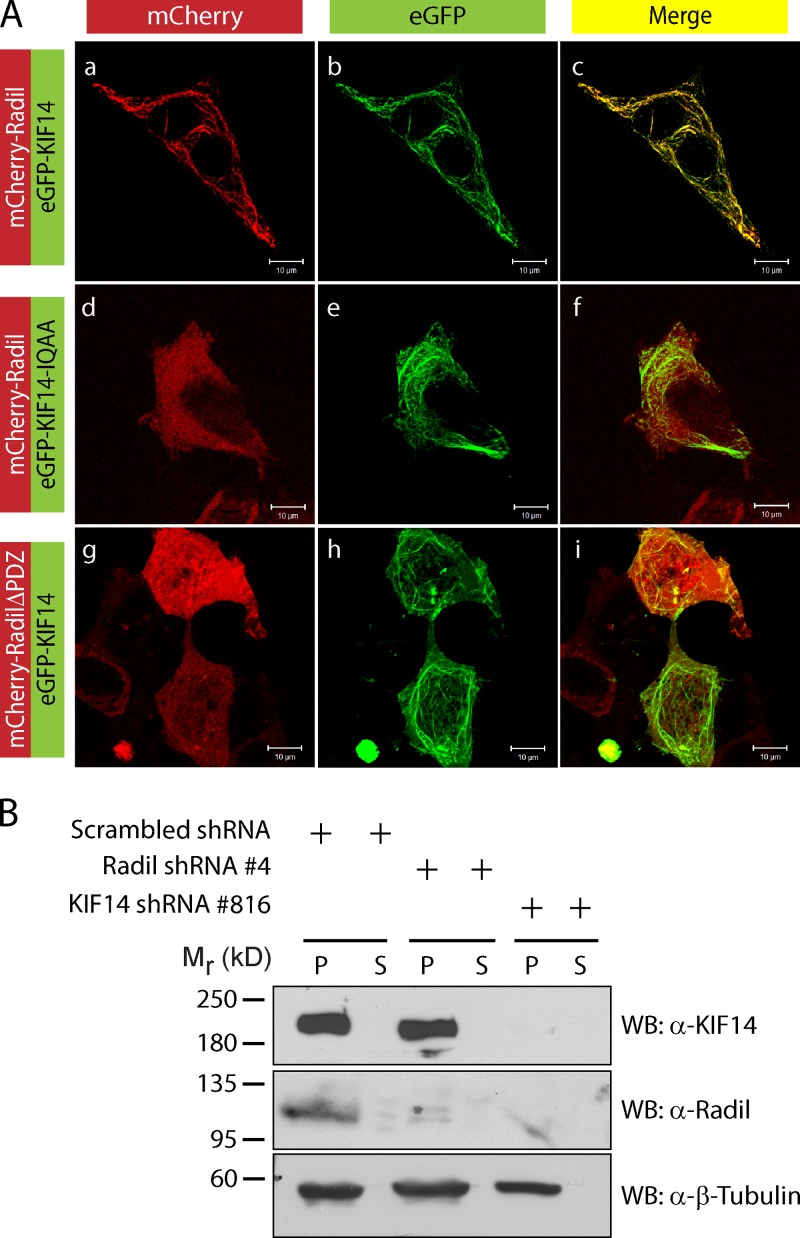
Kinesins are ATP-consuming motor proteins that travel along the microtubule network to deliver proteins or organelles as cargo to different cellular locations (Hirokawa et al., 2009; Verhey and Hammond, 2009). We hypothesized that Radil could associate with microtubules through its binding to KIF14. To address this we used confocal microscopy to study the subcellular localizations of Radil and KIF14 in interphase cells. The strong colocalization of KIF14 with β-tubulin and acetyl-tubulin validates the presence of KIF14 on microtubules (Fig. S1 D). When coexpressed in HEK293T cells, mCherry-Radil and eGFP-KIF14 colocalize on microtubules (Fig. 2 A, a–c). The mutant KIF14-IQAA also localizes on microtubules (Fig. 2 A, e) but fails to recruit Radil (Fig. 2 A, d–f). Similarly, RadilΔPDZ does not colocalize with KIF14 on microtubules (Fig. 2 A, g–i). To support these findings, we performed microtubule sedimentation experiments from MDA-MB-231 cells to determine the subcellular localization relationship of endogenous Radil and KIF14. In cells expressing scrambled shRNAs, KIF14 and Radil cosediment with microtubules (Fig. 2 B). Sedimentation of KIF14 with microtubules is not affected when Radil’s levels are reduced using shRNA (Fig. 2 B and Fig. S1 E), but knockdown of KIF14 inhibits the sedimentation of Radil in microtubule fractions (Fig. 2 B and Fig. S1 F). We conclude that KIF14 recruits Radil to the microtubule network through a physical association.
Figure 2.
KIF14 recruits Radil on microtubules. (A) Shown is localization of mCherry-Radil with eGFP-KIF14 (a–c), mCherry-Radil with eGFP-KIF14-IQAA (d–f), and mCherry-RadilΔPDZ with eGFP-KIF14 (g–i). HEK293T cells were cotransfected with low amounts of Radil expression plasmids together with KIF14 cDNA constructs. (B) Localization of endogenous Radil on microtubules is KIF14 dependent. MDA-MB-231 cells expressing scrambled- Radil or KIF14 shRNA were lysed in BRB80 buffer and cytosolic tubulin was polymerized by addition of 2 mM GTP and 20 µM paclitaxel. Rigor binding of motor proteins was allowed by addition of 2 mM AMP-PNP and the microtubule–motor protein mixture pelleted by centrifugation. The fractionated proteins were detected by Western blotting. P, pellet fraction; S, supernatant fraction.
KIF14 negatively regulates Radil function in cell spreading
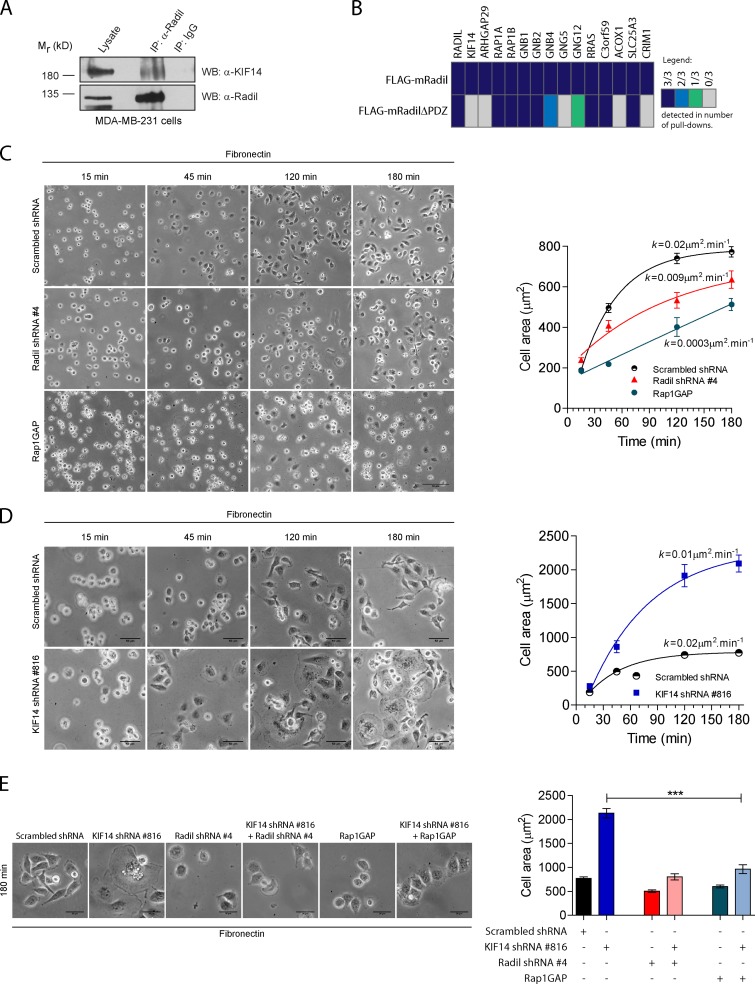
KIF14 was first identified as an oncogene in retinoblastoma and is overexpressed in several cancers. Interestingly, KIF14 is highly overexpressed in breast tumors and its expression correlates with a worst outcome of the disease (Corson et al., 2005; Corson and Gallie, 2006). We extended these data by measuring KIF14 levels using qPCR in 103 individual primary breast tumors and show that KIF14 expression is increased on average 12.32-fold (Fig. S2 A). In contrast, Radil mRNA expression remains unchanged among different tumor grades (unpublished data). We could not evaluate KIF14 and Radil levels during breast cancer progression because we did not have access to the metastatic tissues, but elevated KIF14 expression in primary breast tumors is known to correlate with the severity and invasiveness of cancer (Corson and Gallie, 2006). High levels of KIF14 expression in many primary breast tumors and its correlation with breast cancer severity prompted us to measure its abundance in three different breast cancer cell lines. KIF14 expression was 1.6-, 5-, and 14-fold higher in the transformed MCF7, MDA-MB-468, and MDA-MB-231 breast cancer cell lines, respectively, compared with normal breast tissues (Fig. S2 B). Interestingly, KIF14 expression in these cell lines appears to correlate with their migratory and invasive properties because MCF7 and MDA-MB-468 cells are poorly invasive compared with MDA-MB-231 (Meng et al., 2000; Wang et al., 2007; Naik et al., 2008). We established that the Radil–KIF14 complex exists in MDA-MB-231 cells using coimmunoprecipitation (Fig. 3 A). We also analyzed Radil and RadilΔPDZ protein complexes isolated from MDA-MB-231 cells using mass spectrometry. As expected, several peptides corresponding to Rap1 and Gβγ subunits were identified in both wild-type Radil and RadilΔPDZ, but KIF14 peptides were only identified in Radil complexes. (Fig. 3 B and Table S2). These results confirm that KIF14 interacts with the PDZ domain of Radil and suggest that the RadilΔPDZ mutant is not grossly misfolded because it still interacts with most interactors but selectively lost the ability to bind KIF14. These observations led us to examine a potential functional role for Radil in breast cancer and test the possibility that KIF14 overexpression modulates Radil functions during tumorigenesis.
Figure 3.
KIF14 negatively regulates Radil during MDA-MB-231 cell spreading. (A) The KIF14–Radil complex is present in MDA-MB-231 cells. Immunoprecipitation was performed using α-Radil or control IgG antibodies followed by Western blotting (WB) with α-Radil or α-KIF14 antibodies. (B) Shown is a protein–protein interaction heat map for FLAG-mRadil and FLAG-mRadilΔPDZ. Lysates from cells stably expressing FLAG-mRadil or FLAG-mRadilΔPDZ were subjected to pull-downs with α-FLAG M2 beads and the purified protein complexes were subsequently analyzed by LC-MS/MS. Colors indicate the number of experiments in which the identified proteins were detected as depicted in the legend (right). (C) Rap1–Radil signaling is required for MDA-MB-231 cell spreading. Cells expressing control shRNA, Radil shRNA#4, or overexpressing Rap1GAP cDNA were plated on fibronectin for the indicated times and cell spreading was quantified by measuring the cell area (right). (D) KIF14 negatively regulates cell spreading. Cells expressing KIF14 shRNA# 816 were plated on fibronectin-coated plates and cell spreading was monitored over the time points indicated by quantifying cell surface areas. Images were captured from five random fields for each time point. The curves were fitted by one phase-association model (see Materials and methods). (E) KIF14 negatively regulates Rap1a–Radil signaling. The increased cell spreading observed in cells expressing KIF14 shRNA is rescued by coexpressing Radil shRNA or Rap1GAP. Cells were plated on a fibronectin-coated surface (0.5 µg/ml) and allowed to adhere and spread for 180 min. At the end of the experiment cells were imaged (left) and their spreading quantified (right). Also see Fig. S2. Bars, 50 µm. Error bars, ±SEM.
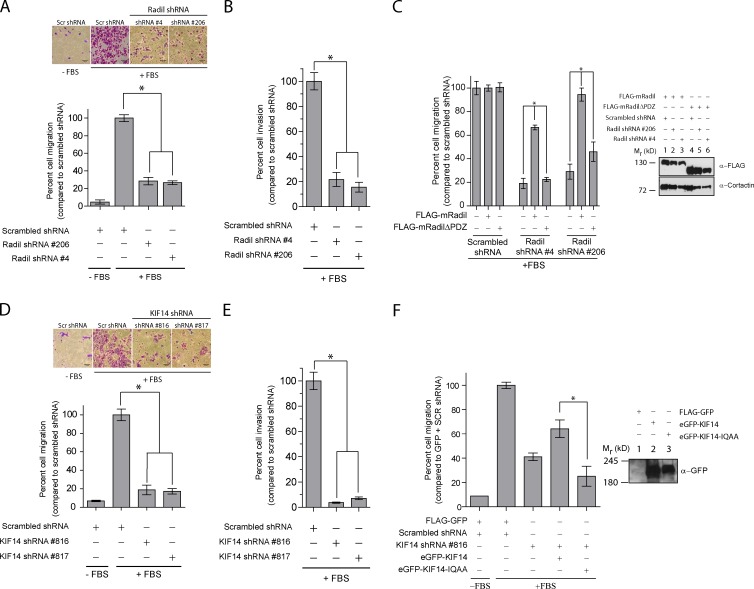
Given the established role of Rap1a–Radil signaling in cell adhesion and spreading in other contexts (Smolen et al., 2007; Ahmed et al., 2010; Ross et al., 2011), we next tested its requirement for MDA-MB-231 cell spreading. We validated two shRNAs (#4 and #206) that knock down Radil mRNA levels by more than 75% and that efficiently decrease Radil protein levels (Fig. S1 E). The depletion of Radil led to a marked impairment in the ability of cells to spread on fibronectin-coated coverslips (Fig. 3 C and Fig. S2 C), a phenotype similar to cells expressing Rap1GAP and therefore have low Rap1a activity (Fig. 3 C). Notably, 120 min after plating, most of the scrambled-shRNA–expressing cells are fully spread, whereas Radil-depleted cells and Rap1GAP-expressing cells still exhibit a rounded morphology (Fig. 3 C, quantification of the rate of spreading in right panel). The Radil shRNA is on target because expressing a knockdown-resistant mouse Radil cDNA rescued the spreading defects induced by both shRNAs (Fig. S2 D). However, expression of mouse RadilΔPDZ could not rescue the spreading defects (Fig. S2 D). We conclude that Radil is required for ECM- and Rap1a-mediated spreading in breast cancer cells.
Because KIF14 interacts physically with Radil we were interested to test whether it was important for cell spreading. After knockdown of KIF14 using shRNAs (Fig. 3 D and Fig. S1 F), MDA-MB-231 cells behave opposite to cells in which Radil was depleted, exhibiting increased spreading efficiency and accelerated kinetics (Fig. 3 D, compare cell spreading at 45 min). Indeed, KIF14-depleted cells are on average three times the area of control shRNA-expressing cells (Fig. 3 D, compare cells at 180 min and quantification on right panel). To explore the functional relationship between KIF14, Rap1a, and Radil, we asked if blocking Rap1a activity or decreasing Radil expression could modify the enhanced cell spreading phenotype manifest upon KIF14 depletion. Overexpression of Rap1GAP and depletion of Radil with shRNA both reversed the enhanced spreading phenotype of KIF14-depleted cells (Fig. 3 E, quantification in right panel). These findings indicate that KIF14 negatively regulates cell spreading by controlling Rap1a–Radil activity.
KIF14 negatively regulates Rap1a- and Radil-dependent inside-out signaling
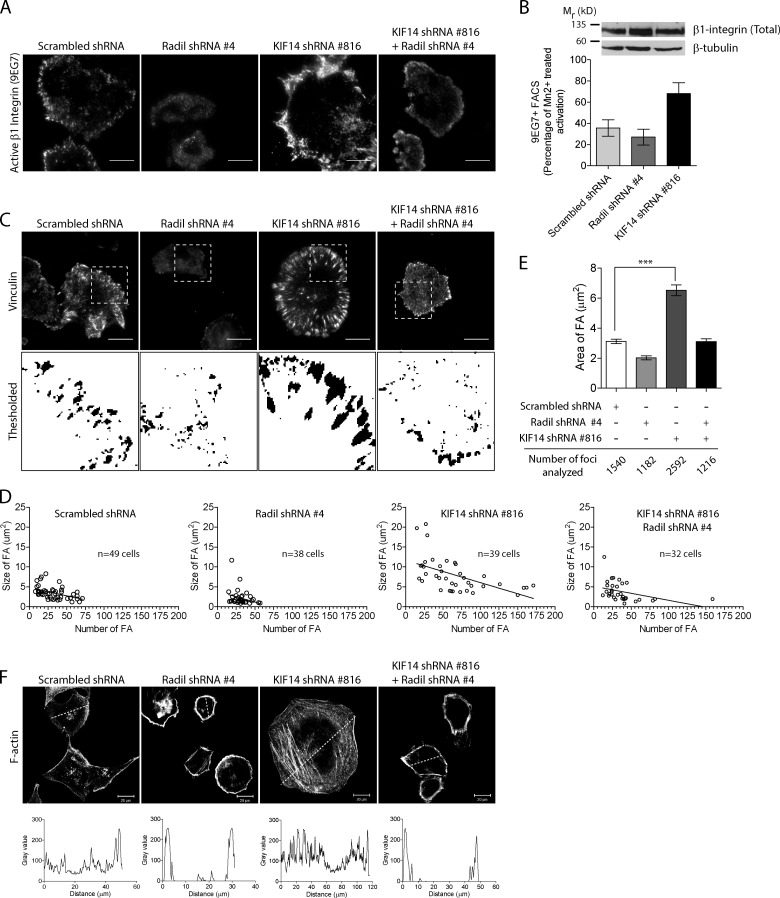
Through the engagement of different effectors such as Radil, Rap1a regulates cell spreading by modulating inside-out signaling influencing integrin activation. Using the 9EG7 antibody that specifically recognizes activated β1-integrin, we show that Radil knockdown impairs integrin activation during spreading in MDA-MB-231 cells (Fig. 4 A, quantification in B). We note that this is likely an underestimation because cells with efficient Radil knockdown have impaired adhesion and were washed off during the experiment. Consistent with the enhanced spreading phenotype, depletion of KIF14 leads to hyperactivation of β1-integrin when MDA-MB-231 cells are plated on fibronectin (Fig. 4, A and B). This results from increased Rap1a-dependent inside-out signaling because integrin activation returns to normal when Radil expression is reduced within KIF14 shRNA-expressing cells (Fig. 4 A). We confirmed that the effects are due to change in integrins activation rather than expression because β1-integrin levels did not vary (Fig. 4 B).
Figure 4.
Radil and KIF14 differentially regulate integrin activation and focal adhesion dynamics. (A) Shown are representative TIRF images of active β1 integrin (9EG7) staining in MDA-MB-231 cells expressing scrambled shRNA, Radil shRNA #4, KIF14 shRNA #816, or Radil shRNA #4 + KIF14 shRNA #816. Cells were plated on fibronectin-coated coverslips (0.5 µg/ml) for 120 min, fixed, and stained with 9EG7 antibody. (B) FACS analysis showing increase in activated β1 integrin (9EG7) upon depletion of KIF14. Total integrin expression levels were determined by Western blot using a β1 integrin antibody that recognizes the cytoplasmic tail. β-Tubulin was used as loading control. (C) Focal adhesion sites were imaged using TIRF microscopy and vinculin staining in cells treated as above. Threshold images were used to quantify FA areas and numbers. (D) Graphical representation of FA area versus number in individual cells taken from three independent experiments. (E) Average FA area for all cells quantified in D. (F) Confocal images of F-actin in cells treated as indicated and following spreading for 120 min on fibronectin coated coverslips (n = 3). A line was drawn across cells and the intensity profile plotted (below) to show the relative distribution of actin stress fibers in cells. Also see Fig. S3. Bars, 20 µm. Error bars, ±SEM.
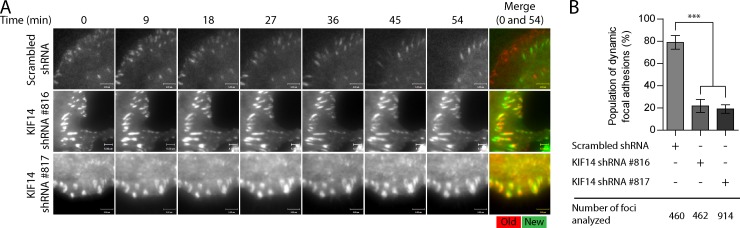
Integrin engagement by ECM proteins leads to their clustering and to the recruitment and activation of several proteins implicated in the ontogeny and maturation of focal contacts. Hence, we predicted that Radil- and KIF14-depleted cells would have altered focal contact properties given their defects in spreading and integrin activation. We thus visualized focal adhesions by imaging vinculin in cells expressing control, Radil, or KIF14 shRNAs (Fig. 4 C; Fig. S3, A and B), quantified their abundance and size in individual cells (Fig. 4 D), and calculated the average focal adhesion sizes in all the cells examined (Fig. 4 E). Knockdown of Radil results in cells with fewer and smaller focal contacts, whereas KIF14 depletion dramatically increases focal contact numbers and results in enlarged focal adhesions that are on average approximately twice the size of control cells (Fig. 4, C and E). After the stabilization of adhesion sites, F-actin is known to increase in density leading to formation of compact bundles of stress fiber, which in turn leads to focal adhesion maturation (Chrzanowska-Wodnicka and Burridge, 1996; Amano et al., 1997; Geiger et al., 2009; Gardel et al., 2010; Parsons et al., 2010; Oakes et al., 2012). Predictably, hyperactivated integrin signaling and the presence of large mature focal adhesions observed in KIF14-depleted cells correlate with increased stress fiber formation (Fig. 4 F; Fig. S3, A and B). In all three staining conditions we also noted that Radil- as well as KIF14-depleted cells have unpolarized morphology when compared with control shRNA-infected cells. Presence of large focal adhesions is indicative of mature adhesion and suggests defective adhesion disassembly. To test this hypothesis we performed time-lapse imaging experiments to look at the dynamics of the focal adhesion marker Paxillin fused to Venus (Fig. 5 A). Approximately 80% of adhesion foci in control cells displayed rapid and dynamic turnover of Venus-Paxillin (Fig. 5, A and B; Video 1). On the other hand, KIF14-shRNA–expressing cells exhibited stable and large focal adhesions that seldom disassembled over a period of approximately 1 h (Fig. 5, A and B; Videos 2 and 3). We conclude that KIF14 is a negative regulator of inside-out integrin signaling regulated by Rap1 and its effector Radil.
Figure 5.
KIF14 knockdown alters focal adhesion dynamics. (A) Venus-Paxillin dynamics was assessed by time lapse TIRF microscopy in MDA-MB-231 cells expressing scrambled or two different KIF14 shRNAs. Cells were plated on fibronectin (0.2 µg/ml)-coated glass slides. Each cell was observed over 54 min and a picture was taken every 3 min. Merge shows overlay of images taken at time 0 and 54. Red = 0 min; green = 54 min. (B) Quantification of Venus-Paxillin dynamics shown in A. Bars, 5 µM. Error bars, ±SEM.
KIF14 spatially restricts Radil from interacting with activated Rap1a
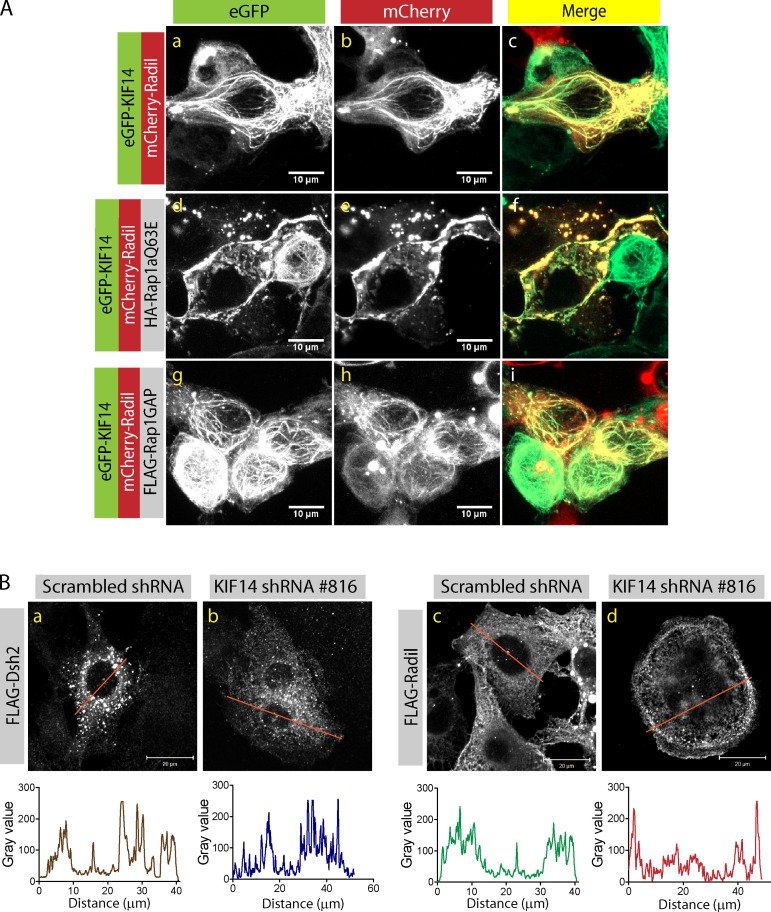
Expression of the constitutively active Rap1a-Q63E mutant leads to plasma membrane recruitment of Radil (Ahmed et al., 2010). Having established that KIF14 negatively regulates inside-out signaling and recruits Radil on microtubules, we reasoned that Rap1a activity could modulate the recruitment of Radil on microtubules. Supporting this prediction, expressing the Rap1a-Q63E mutant leads to the dislodging of Radil from the microtubules and to its recruitment at the plasma membrane (Fig. 6 A). The release of Radil from microtubules in KIF14-shRNA–expressing cells could thus explain the increased inside-out integrin signaling observed in these cells. We therefore monitored the localization of Radil in MDA-MB-231 cells while varying the levels of KIF14. Because they express high levels of KIF14, Radil is predominantly localized on microtubules in MDA-MB-231 (Fig. 6 B, c). Upon knockdown of KIF14, Radil is redistributed at the cell cortex, presumably at sites of Rap1a activation (Bivona et al., 2004; Li et al., 2009), whereas the localization of the control protein Dishevelled2 is not affected (Fig. 6 B, compare b and d). Likewise, disrupting microtubule networks with nocodazole results in Radil redistribution at the cell periphery (Fig. S3 C). We also tested if Rap1 activity is affected in the knockdown conditions. Rap1-GTP pull-down assays did not reveal any change in Rap1 activity in KIF14-depleted cells compared with control cells but depletion of Radil leads to a slight decrease in Rap1 activity (Fig. S3 D). These observations suggest that KIF14 controls inside-out integrin activation by tethering the Rap1a effector Radil on microtubules and thereby spatially controlling Rap1 signaling.
Figure 6.
Rap1a activation releases Radil and KIF14 from microtubules. (A) Confocal images of HEK293T cells expressing mCherry-Radil and eGFP-KIF14 with or without HA-Rap1aQ63E or FLAG-Rap1GAP. Expression of constitutively active Rap1a leads to dislocation of Radil and KIF14 from microtubules and recruitment of Radil to the plasma membrane. Bars, 10 µm. (B) Confocal images of MDA-MB-231 cells expressing FLAG-Dishevelled2 (FLAG-Dsh2) or FLAG-Radil transduced with scrambled shRNA or KIF14 shRNA #816. A line was drawn across each cell and the profile of protein distribution depicted below each image (The data shown are from a single representative experiment out of three repeats.). Bars, 20 µm.
The tuning of Rap1a–Radil-mediated inside-out signaling is required for efficient breast cancer cell migration and invasion
Both decreased or hyperactivated Rap1a or integrin signaling inhibit cell migration and invasion (Tsygankova et al., 2007; Lyle et al., 2008; Bailey et al., 2009; Freeman et al., 2010). With this in mind, together with the known role of Radil in neural crest cell migration (Smolen et al., 2007), we tested the requirement of Radil for the migration of MDA-MB-231 cells. Knockdown of Radil with two different shRNAs (#4 and #206) severely impairs the migratory (Fig. 7 A) and invasive (Fig. 7 B) properties of these cells. Interestingly, depletion of KIF14 using two independent shRNAs (#816 and #817), which we show leads to hyperactivated Rap1–Radil signaling, also leads to decreased MDA-MB-231 cell migration and matrigel invasion (Fig. 7, D and E). Several lines of evidence support the specificity of the phenotypes that we observed. First we obtained similar results with two independent Radil and KIF14 shRNAs. Second, we were able to rescue the migration defects by expressing the RNAi-resistant mouse Radil cDNA or the eGFP-KIF14 coding sequence because the KIF14 shRNAs target the 3′ UTR region (Fig. 7, C and F). In contrast, expression of mouse RadilΔPDZ and eGFP-KIF14-IQAA mutants fails to rescue the cell migration defects associated with Radil and KIF14 depletion (Fig. 7, C and F). From these findings we conclude that the optimal balance of Rap1a–Radil signaling controlled by KIF14 is required to sustain the efficient migration and invasion of breast cancer cells.
Figure 7.
Radil and KIF14 are required for MDA-MB-231 cell migration and invasion. (A) MDA-MB-231 cells were transduced with the indicated shRNAs. 72 h after transduction 5 × 104 cells were seeded on the upper chamber of Transwells and 20% fetal bovine serum (FBS) in DMEM was applied to the lower chamber. Cells were allowed to migrate for 10–12 h and migrated cells counted from pictures of 4–5 random fields. (B) Cell invasion was assessed as above except Transwells coated with 1 µg/ml of matrigel were used. Cells were allowed to invade through the matrigel for 20 h. (C) MDA-MB-231 cells stably expressing the shRNA-resistant murine full-length mRadil or mRadilΔPDZ were transduced with scrambled, or two different Radil shRNAs. The migratory potential of these cells was assessed as described above. Expression levels of FLAG-mRadil and FLAG-mRadilΔPDZ are shown on the right. Cortactin used as loading control. (D) Transwell cell migration and (E) invasion assays with cells expressing KIF14 shRNAs. Cells were processed as above. (F) MDA-MB-231 cells were transduced with scrambled shRNA or KIF14 shRNA #816 in the presence of FLAG-GFP, eGFP-KIF14, or eGFP-KIF14-IQAA. The different cells were then subjected to Transwell assays. Expression levels of eGFP-KIF14 and eGFP-KIF14-IQAA are shown on the right. KIF14 shRNA #816 targets the 3′ UTR and was thus used for rescue experiments. Bars, 50 µm. Error bars, mean ± SEM.
Knockdown of Radil blocks breast cancer progression
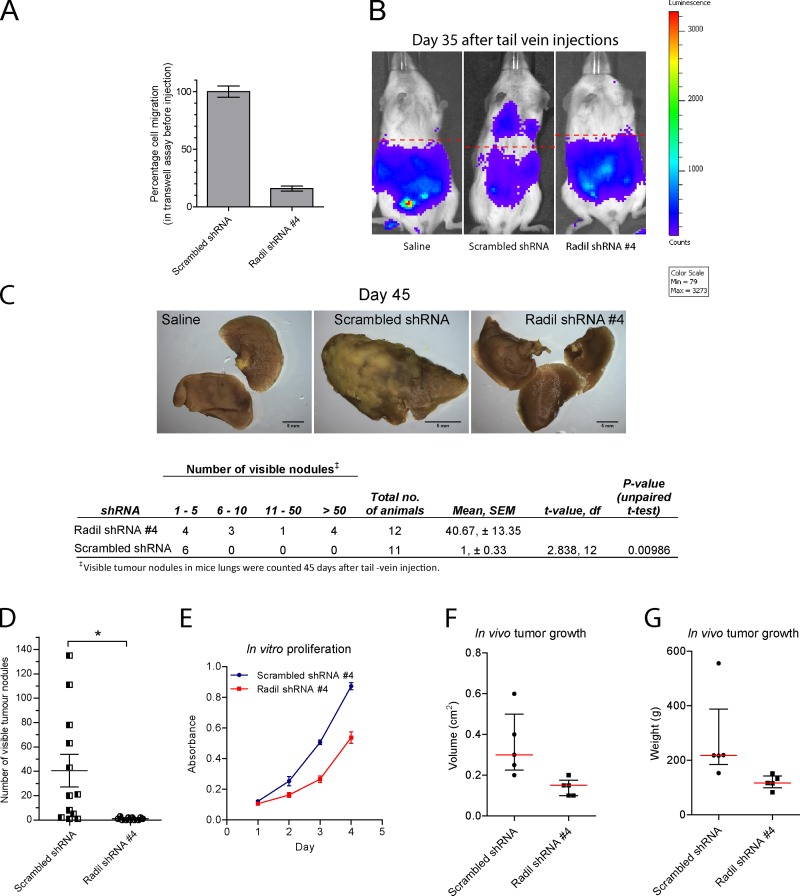
Because knockdown of Radil efficiently blocks cell migration and invasion of breast cancer cells in vitro, we further studied the requirement of Radil during breast cancer cell metastasis using a mouse model. MDA-MB-231 cells constitutively expressing Renilla luciferase (MDA-MB-231-Rluc) were derived to assess metastatic activity to the lungs using bioluminescence imaging. Control or Radil-shRNA–expressing cells were injected in the tail veins of immunodeficient mice. Before injections, we validated the functional knockdown of Radil by measuring the in vitro migratory properties of the cells using Transwell assays (Fig. 8 A). After injections, we monitored lung metastasis by bioluminescence imaging on days 10, 20, and 35 and began to detect bioluminescence signals on day 35 only in control shRNA-treated animals (Fig. 8 B). By day 45 some of the control shRNA-treated animals exhibited signs of respiratory distress and lethargy, at which point the experiment was terminated. Gross anatomical review confirmed massive tumor growths in the lungs of several control shRNA-treated mice, accompanied by extensive vascularization and increase in organ sizes (Fig. 8, C and D). In contrast, all the animals injected with Radil shRNA-expressing cells appeared healthy on day 45 and only a few metastatic nodules were detected on the lungs of 6 of the 11 animals (Fig. 8, C and D).
Figure 8.
Radil is required for breast cancer cell metastasis. (A) MDA-MB-231 cells stably expressing Renilla luciferase were transduced with scrambled shRNA or Radil shRNA #4 and their migratory potential was tested using Transwell assays before injection in mice. (B) MDA-MB-231 cells were injected via tail vein in NOD-SCID mice. Metastasis and homing of cells to the lungs was monitored over time using in vivo bioluminescence imaging. Shown are images from day 35. (C) Mice were sacrificed 45 d after inoculations. Lungs from mice injected with saline or with scrambled shRNA or Radil shRNA–expressing cells were collected, fixed, and stained with Bouin’s solution. Shown are representative images from each condition (top). Quantification of the visible tumor nodules is provided in the table (below). Scrambled shRNA, n = 12; Radil shRNA #4, n = 11; statistics, Student’s t test. (D) Total number of nodules counted on the lungs of each animal. Error bars, ±SEM. (E) In vitro proliferation of MDA-MB-231 cells treated with scrambled or Radil shRNA #4. (F) Quantification of tumor end volume (in cm3) on day 31. Scrambled shRNA, n = 5; Radil shRNA #4, n = 5. Red line denotes median value. Bars, interquartile range. (G) Quantification of tumor end weight (in grams) on day 31.
It is well established that integrins (Kuwada and Li, 2000; Cruet-Hennequart et al., 2003) and Rap1 (Dao et al., 2009) as well as KIF14 (Gruneberg et al., 2006; Thériault et al., 2012) are also important during cell proliferation. We were therefore interested to determine if Radil controls the proliferation of breast cancer cells. Cells expressing Radil shRNA show 42.5% inhibition of cell proliferation when cultured in vitro for 4 d (Fig. 8 E). We also assessed their growth in vivo by injecting cells expressing control or Radil shRNAs subcutaneously in nude mice. 31 d after injection, subcutaneous tumors from all mice were retrieved and their weights and volumes were measured. Radil shRNA-expressing tumors were on average 44% lighter than control shRNA-expressing tumors (Fig. 8, F and G; Fig. S4, A and B). We conclude that Radil is required during breast cancer progression by controlling cell proliferation and cell dissemination to the lungs.
Discussion
Our results identify the oncogene KIF14 as a negative regulator of Rap1a–Radil signaling. We provide evidence that KIF14 tethers Radil on microtubules and thereby controls inside-out integrin activation by tuning the availability of this Rap effector for binding Rap1-GTP at the plasma membrane. Because KIF14 is frequently up-regulated in several human cancers, this led us to study the role of Radil in cancer progression. Our data show that depletion of Radil and KIF14 using shRNAs affects the migratory and invasive properties of breast cancer cells in vitro. Using xenograft models, we further demonstrated the requirement of Radil for tumor cell proliferation and for the efficient dissemination of breast cancer cells to lungs.
The regulation of Rap1–Radil signaling by KIF14 highlights the necessity for the fine regulation of integrin activation and cell–matrix adhesion during cancer cell migration. Indeed, it is known that reduced and hyperactivated Rap1 signaling similarly lead to impaired cell motility (Lorenowicz et al., 2008; Lyle et al., 2008; Bailey et al., 2009; Zheng et al., 2009; Freeman et al., 2010; Huang et al., 2012). Accordingly, we show that Radil- and KIF14-depleted cells that respectively exhibit impaired and hyperactivated integrin signaling do not migrate efficiently. In addition to increased spreading and reduced migration, KIF14-depleted cells have lost the polarized morphology that characterizes highly migratory cancer cells. We propose that the spatial control of Radil availability, controlled by microtubules and KIF14, enables rapid changes in localized Rap1–Radil signaling and integrin activation that are known to be critical for the regulation of cell–matrix adhesion and the establishment of cell polarity (Arthur et al., 2004; Gérard et al., 2007; Itoh et al., 2007; Jeon et al., 2007; Freeman et al., 2010). The modulation of Rap1 signaling through the availability of Radil thus represents another regulatory step for the inside-out activation of integrins during breast cancer cells migration (Fig. 9, model). Although mechanisms differ, other Rap1 effectors localize and are regulated by microtubules. For example, the Rap1 effectors RapL (Fujita et al., 2005) and Krit1 (Béraud-Dufour et al., 2007) directly interact with microtubules from where they are dislodged and recruited to the plasma membrane when Rap1 is activated. Although a kinesin protein may not be needed in these cases, this suggests that the control of Rap1 effectors by microtubules may be a general mechanism to govern spatiotemporal Rap1 signaling in various contexts.
Figure 9.
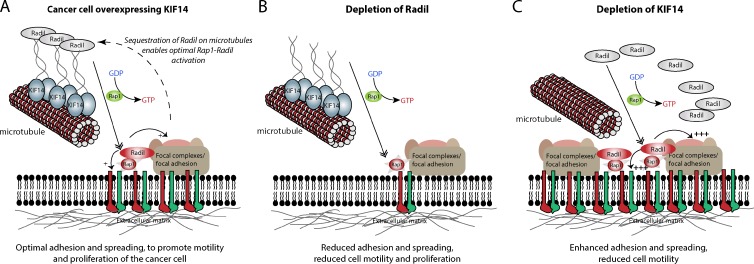
Proposed model describing the roles of Radil and KIF14 during cancer cell migration. (A) In highly motile cancer cells, KIF14 is up-regulated and sequesters Radil on microtubules. This enables optimal Radil–Rap1 signaling, inside-out integrin signaling, and cell–matrix adhesive properties required for efficient cell migration. (B) Depletion of Radil inhibits integrin activation, reduces cell–matrix adhesion, and causes loss of traction and cell motility. (C) Depletion of KIF14 leads to the release of Radil from microtubules, thereby increasing the pool of Radil available to associate with activated Rap1 at the plasma membrane. This leads to hyperactivated integrins, increases focal adhesion formation, and decreases motility.
At present, the mechanism by which Radil leads to inside-out activation of integrins is not completely clear. Although other Rap1 effectors such as RIAM have been shown to mediate the formation of an integrin–activation complex containing Rap1 and Talin (Lee et al., 2009) that can directly associate with integrins, our mass spectrometry interrogation of the Radil complex did not reveal the presence of Talin or other integrin-associated proteins. However, we previously identified Gβγ subunits of heterotrimeric G-proteins as Radil interactors and demonstrated that Gβγ overexpression leads to Rap1a-dependent inside-out activation of integrins and cell spreading (Ahmed et al., 2010). Interestingly, Gγ subunits were previously shown to localize to the tip of stress fibers and focal adhesions (Hansen et al., 1994). After G-protein–coupled receptor activation, the release of Gβγ could thus direct the assembly of a Gβγ–Rap1a–Radil signaling complex, leading to spatiotemporal activation of integrins. Especially relevant to our findings showing that the depletion of Radil inhibits the metastasis of MDA-MB-231 cells to lungs, activation of the CXCR4 receptor (Müller et al., 2001) and Gβγ subunits (Kirui et al., 2010; Tang et al., 2011) were previously shown to be required for this process. Additionally, given that loss of function for the ERM protein ezrin was recently demonstrated to be required for the Rap1-induced spreading to similar extent than Radil (Ross et al., 2011), studying the functional relationship between these proteins may reveal further clues about their mechanisms. As the expression of Rap1GAP and knockdown of Radil similarly lead to decreased activation of integrins, impaired focal adhesion formation and defects in spreading of breast cancer cells, our results strongly implicate a requirement for Rap1–Radil signaling in the context of cancer cell migration.
We previously established that the PDZ domain of Radil was important for its activity in cell adhesion (Ahmed et al., 2010). The integration of the phage-display screen to identify the preferred PDZ ligand for the Radil PDZ domain and the mass spectrometry analysis of Radil-containing protein complex enabled the identification of KIF14 as a protein interacting with the Radil PDZ domain. The C-terminal of KIF14 contains the atypical hydrophobic [F/I]-x-W-V motif revealed by phage display and was found to be critical for its binding to Radil and for recruitment of the latter on microtubules. Further studies will be needed to understand how the binding of Radil to KIF14 is modulated. One possibility is that the recruitment of Rap1-GTP to the N-terminal RA domain of Radil could modulate the availability of the C-terminal PDZ ligand for binding to KIF14.
KIF14 is frequently overexpressed in several human cancers. Its overexpression in breast cancer was previously shown to be a predictor of poor survival, as it correlates with tumor grade and invasiveness (Corson et al., 2005; Corson and Gallie, 2006). KIF14 in a complex with citron kinase and PRC1 was identified as a mitotic kinesin with a role during cytokinesis because its knockdown led to binucleated HeLa cells (Carleton et al., 2006; Gruneberg et al., 2006). Interestingly, Rap1 activity and integrin signaling need to be optimally regulated for normal cell division (Dao et al., 2009). Indeed, at the onset of mitosis, most adherent cells undergo cell retraction characterized by the disassembly of focal adhesions and actin stress fibers, processes that are controlled by Rap1 and integrins. Given the importance of Radil as a Rap1 effector and its regulation by KIF14, we show that Radil, perhaps not surprisingly, controls cell proliferation and tumor growth. Our results thus position the KIF14–Rap1–Radil signaling axis as an important mediator of tumor progression, as it affects both cell proliferation and migration (Bailey et al., 2009; Freeman et al., 2010; Huang et al., 2012).
The ability of cells to dynamically change their adhesion to the ECM during cell migration is a property optimized by cancer cells to support metastasis and cancer progression (Felding-Habermann et al., 2001; Felding-Habermann, 2003; Desgrosellier and Cheresh, 2010). In the light of our findings, KIF14 overexpression detected in several human cancers may reflect this optimization process required to tune the level of Rap1–Radil signaling and resulting integrin activation needed for efficient cell motility and tumor progression. We validated that KIF14 is highly up-regulated in a cohort of primary breast tumors and showed that KIF14 overexpression in breast cancer cell lines correlates with their invasive properties. In contrast, Radil expression is not significantly changed in breast tumors or in cell lines but Rap1 activity is known to be highly up-regulated in cancer cells (Sjöblom et al., 2006; Zheng et al., 2009; Banerjee et al., 2011). Interestingly, similar to KIF14, Sharpin was recently identified as an inhibitor of β1-integrins by preventing the interaction with Talin and Kindlin, thereby inhibiting the dynamic switching between active and inactive conformations (Rantala et al., 2011). Sharpin levels are also up-regulated in different cancers and this was found to be important for cell migration.
The creation and regulation of traction forces in response to interactions of the ECM with different regions of the cell are intrinsic to cell migration and proliferation (Kaverina et al., 2002; Dao et al., 2009). Regulated cycles of assembly and disassembly of focal adhesions and focal complexes at adhering and retracting sides of the cells produce the tractions required for cells to move. It is becoming increasingly clear that the precise control of focal adhesion dynamics is important for efficient cell migration (Gardel et al., 2008, 2010; Nagano et al., 2012). As such, the presence of fewer focal complexes at the periphery and large focal adhesions are signs of reduced adhesion dynamics and cell movement. The depletion of Radil in MDA-MB-231 impairs integrin activation and leads to reduced focal adhesions, whereas KIF14 depletion leads to dramatically enlarged focal adhesions and increased stress fibers (Fig. 4). Consistent with this notion, both Radil- and KIF14-depleted cells exhibit reduced cell motility (Fig. 7).
In summary this study identifies the oncogene KIF14 as a PDZ ligand–containing protein interacting with Radil. KIF14 negatively regulates Rap1a signaling and inside-out integrin activation by tethering Radil on microtubules. Our results describe the role of Radil in cancer progression, and suggest that it is an important nexus integrating GPCR, Gβγ, and Rap1 signaling in this context. Given that KIF14 is frequently overexpressed in several cancers, further studies examining the requirement of Radil for the progression of other cancers will be important. Furthermore, guided by other studies showing the targeting of PDZ-mediated interactions with small molecules (Houslay, 2009; Thorsen et al., 2010; Patra et al., 2012) and synthetic peptides (Tonikian et al., 2008; Zhang et al., 2009), inhibiting KIF14–Radil interaction could provide a novel strategy for the treatment of cancer.
Materials and methods
Plasmid constructs and reagents
Human Radil and amino acids 1–869 of Radil (RadilΔPDZ) were cloned into the pIRESpuro-GLUE plasmid. The Radil and Radil-ΔPDZ coding sequences were also cloned into pLentiGlue-HA-mCherry lentiviral vector. Mouse Radil (mRadil) and mRadil-ΔPDZ coding sequences were amplified by PCR from MGC mouse I.M.A.G.E. clone #5696312 and inserted into the PSL9-FLAG lentiviral vector. Paxillin was also cloned downstream of Venus in the PSL9 lentiviral vector. The eGFP-KIF14 expression plasmid was a kind gift from Dr. Francis Barr (Max Planck Institute of Biochemistry, Martinsreid, Germany; Gruneberg et al., 2006). The eGFP-KIF14-IQAA mutant was generated by QuikChange PCR mutagenesis to convert the last two amino acids of eGFP-KIF14 from WV to AA. pGEX-Radil-PDZ encodes for the last 206 amino acid residues of Radil that include the PDZ domain fused to GST. Flag-KIF14-ctail-IQWV and Flag-KIF14-ctail–IQAA that encodes for the last 166 amino acids of human KIF14 were cloned into the pIRES-puro vector downstream of a Flag tag. All cDNA constructs were verified by sequencing and detailed maps are available on the laboratory website (http://phm.utoronto.ca/angers).
Antibodies were purchased from the following vendors: mouse anti-HA.11 clone 16B12 (Covance); mouse anti-FLAG, rabbit anti-FLAG, mouse anti-vinculin clone hVin-1, mouse anti–β-tubulin clone TUB 2.1, and mouse anti-acetylated-tubulin clone 6-11B-1 (Sigma-Aldrich); rabbit anti-GFP (University of Alberta, Edmonton, Alberta, Canada); mouse anti-cortactin clone 4F11 (EMD Millipore); rabbit anti–β1-integrin C-terminal antibody (EMD Millipore); rabbit anti-KIF14 (Bethyl Laboratories); rat anti-CD29 clone 9EG7 (active β1-integrin antibody; BD); and rabbit anti-CDC25C (Santa Cruz Biotechnology, Inc.). The Radil polyclonal antibody was raised in rabbit using the entire human Radil protein as antigen and was described previously (Ahmed et al., 2010). Rabbit anti-KIF7 antibodies were a gift from C.C. Hui (SickKids, Toronto, Canada). Secondary antibodies conjugated to Alexa Fluor 488 and 594 were purchased from Life Technologies. Phalloidin conjugated to CF647 dye was purchased from Biotium, Inc. Secondary antibodies conjugated to horseradish peroxidase were purchased from Jackson ImmunoResearch Laboratories, Inc. Fibronectin from bovine plasma was obtained from Sigma-Aldrich. Matrigel growth factor reduced basement membrane matrix was purchased from BD.
KIF14 MISSION shRNA clones in the lentiviral plasmid pLKO.1-puro were acquired from Sigma-Aldrich. TRCN0000113816 and TRCN0000113817 labeled as KIF14 shRNAs #816 and #817 respectively throughout the paper, were the most efficient according to our validation using Western blot analysis (Fig. S1 F). Radil MISSION shRNA clone TRCN0000155169, which led to effective knockdown (Radil shRNA#4), was also purchased from Sigma-Aldrich. Another shRNA targeting human Radil (Radil shRNA#206) was designed in pLKO.1-puro with the following sequence: 5′-CCGGGCCCACCAAGAGCAAACTGAACTCGAGTTCAGTTTGCTCTTGGTGGGCTTTTTG-3′. A scrambled nontargeting control shRNA was also created in the pLKO.1-puro vector with the sequence: 5′-CCGGTCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG-3′.
Phage display analysis
Recombinant Radil PDZ domain was purified from Escherichia coli as a GST-fusion protein and used as the bait in phage display selections (Tonikian et al., 2008). In brief, phage-displayed C-terminal peptide libraries containing >1010 unique, random heptapeptides were used to isolate ligands for the Radil PDZ domain. To obtain a concise representation of the preferred binding motif sequence of the Radil PDZ domain, amino acid sequence enrichment analysis was performed on 25 unique binding peptide sequences using a web-based sequence logo generator WebLogo (http://weblogo.berkeley.edu; Schneider and Stephens, 1990; Crooks et al., 2004). The resulting sequence logo was used to find proteins in the human proteome ending with the given sequence pattern using PHI-BLAST algorithm to search the UniProt knowledgebase/Swiss-Prot database using Prosite’s ScanProsite tool.
Tissue culture, transfections, and lentiviral transductions
HEK293T and MDA-MB-231 cells were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich), glutamine, and penicillin/streptomycin. HEK293T cells were transfected using calcium phosphate precipitation (Jordan et al., 1996; Ahmed et al., 2011). MDA-MB-231 cells were transiently transfected with Lipofectamine LTX supplemented with PLUS reagent according to manufacturer guidelines (Life Technologies). All lentiviral particles were produced in HEK293T cells by cotransfection of VSV-G (3 µg), psPAX2 (6 µg), and lentiviral plasmids (8 µg) in 30–40% confluent monolayer cell culture grown in 10-cm plates. Media was changed 12–16 h after transfection and virus subsequently collected after 24 and 48 h. MDA-MB-231 cells were subsequently transduced in the presence of 10 µg/ml polybrene (Sigma-Aldrich). 24 h after infection the viral media was replaced with fresh DMEM/10% FBS media. Where appropriate cells were selected with puromycin (2 µg/ml) 48 h after viral infection and typically used for our experiments within 4–5 d after transduction.
RT-PCR and quantitative real-time PCR
Frozen sections were obtained from surgical samples of 99 breast tumors and 10 reduction mammoplasties from the Manitoba Breast Tissue Bank (MBTB; Watson et al., 1996; Liu et al., 2010). All tumors were from female patients who had not received any therapy at the time of resection. The University Health Network Research Ethics Board and the MBTB Access Review Committee approved this study, and all subjects provided informed consent to the MBTB.
Total RNA was extracted from breast tumors, and normal breast tissue samples using TRIzol (Life Technologies) according to the manufacturer’s instructions. The concentration and quality of total RNA were determined using a spectrophotometer (Nanodrop-1000; Thermo Fisher Scientific). For cDNA synthesis, 1 µg of total RNA was reverse transcribed using random primers (Invitrogen) and Superscript II reverse transcription (Life Technologies).
Real-time PCR was performed in a 7900HT Fast Real-Time PCR system using 1.5 µl of the synthesized cDNA product plus the Universal PCR Master Mix in a final volume of 20 µl (Applied Biosystems). Taqman gene expression assays (Applied Biosystems) were used to measure the mRNA expression of Radil (Hs01020348_m1), KIF14 (Hs_00978216_m1), and GAPDH (Hs99999905_m1) in triplicate. Mean relative gene expression was determined using the ΔΔCt method built into the SDS 2.2 software (Applied Biosystems). GAPDH was used as the endogenous control gene.
Immunofluorescence microscopy and image acquisition
Cells grown on coverslips were fixed with 4% paraformaldehyde in 5% sucrose-PBS, pH 7.4, for 15 min. Cells were permeabilized with 0.5% Triton X-100 followed by blocking in 2% BSA for 30 min. β1-Integrin staining was performed in nonpermeabilized cells. For microtubule disruption experiments cells were plated on fibronectin-coated coverslips (0.5 µg/ml) overnight and treated with nocodazole (10 µM) or DMSO for 20 min. Cells were subsequently washed with PBS gently and processed for immunostaining as described above. Slides were mounted on coverslips using Vectashield mounting media (Vector Laboratories). For total internal reflection fluorescence microscopy (TIRF), cells were imaged directly on 18-mm circular no. 1.5 coverglasses (Thermo Fisher Scientific).
Laser scanning confocal images were acquired using a Plan-Apochromat 63×/1.4 NA oil immersion objective on a confocal microscope (LSM510 Meta; Carl Zeiss) operated by LSM510 software. eGFP/Alexa Fluor 488 and mCherry/Alexa Fluor 594 fluorophores were excited individually with 488- and 543-nm lasers, respectively, with appropriate filter sets. Uncompressed images were processed with Zeiss LSM Image Browser version 4.2 and ImageJ software version 1.44p (National Institutes of Health). Where indicated, images were taken as z-stacks and rendered as 3D projection for detailed visualization of the microtubule networks. To measure the distribution of fluorescence in cells, a line was drawn from end to end on a single confocal slice and measured using the Plot Profile function in ImageJ.
Images of focal adhesions were obtained by TIRF microscopy using a microscope (cellTIRF/Lambert FLIM; Olympus). Images were acquired with a 60×/1.49 NA Apon oil immersion objective and an inverted microscope (model IX81; Olympus) equipped with a back-thinned EM-CCD camera (C9100-13; Hamamatsu Photonics) operated by Volocity version 4 software. For fixed cell TIRF imaging cells were kept in PBS. Evanescent field depth for TIRF microscopy was kept at ∼200 nm. Focal adhesion size and abundance were quantified using the particle analysis function of ImageJ from thresholded images using a fixed region of interest (27.5 × 27.5 µm2) from two different areas at the cell periphery. For live-cell imaging, cells were kept in DMEM supplemented with 10% FBS and 10 mM Hepes, pH 7.4. Temperature was maintained at 37°C and CO2 at 5% using a live-cell chamber (Chamlide). Images were captured every 3 min for 54 min. Percent dynamics was calculated by overlaying images from time 0 min and 54 min and counting the percentage of focal adhesions that disappeared within 54 min as described previously (Matsumoto et al., 2010). Image contrasts to generate figures were adjusted using ImageJ or Volocity version 6 softwares.
Affinity purification, immunoprecipitation, and Western blot analysis
Cells were lysed and protein complexes affinity purified on streptavidin Sepharose column, or immunoprecipitated with the indicated antibodies as described previously (Ahmed et al., 2011). In brief, cells were solubilized in buffer containing 0.5% Igepal CA630, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 10 mM NaF, 0.25 mM NaOVO3, 100 mM β-glycerophosphate, and protease inhibitor cocktail (Sigma-Aldrich) for 1 h at 4°C. Streptavidin affinity purification was performed on streptavidin Sepharose resin (GE Healthcare) for 16 h, followed by extensive washing of the beads in lysis buffer. For immunoprecipitations the lysates were incubated with the antibodies and protein A–Agarose beads (Sigma-Aldrich) for 16 h. α-FLAG pull-downs were performed using α-FLAG-M2 Agarose beads (Sigma-Aldrich). Co-purified proteins were eluted from the beads at 95°C for 5 min using 2× Laemmli buffer containing β-mercaptoethanol (Sigma-Aldrich), resolved by SDS-PAGE, and transferred onto nitrocellulose or PVDF membranes (Pall) for Western blot analysis.
In vitro binding assay
Flag-KIF14-ctail-IQWV and Flag-KIF14-ctail–IQAA were transiently transfected and expressed in HEK293T cells. Cells were lysed in buffer containing 0.5% Igepal CA630, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and protease inhibitor cocktail and immunopurified using Flag-M2 beads (Sigma-Aldrich) for 2 h. Purified proteins bound to Flag beads were rinsed twice in the above buffer also containing 0.1% SDS and 0.5% sodium deoxycholate to remove any noncovalent interactors bound to the bait proteins. For in vitro binding, equimolar amounts of GST-Radil-PDZ and Flag-tagged KIF14 c-tails immobilized on Flag-M2 beads were mixed in the same buffer and incubated with gentle rocking at room temperature for 1 h. Beads were subsequently washed with lysis buffer three times and binding analyzed by Western blotting.
Microtubule sedimentation assay
Approximately 6 × 106 MDA-MB-231 cells were lysed in 500 µl of BRB80 buffer (80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, pH 6.8, with KOH and protease inhibitors) followed by sonication for 15 s. Lysates were then centrifuged at 100,000 g for 30 min at 4°C using an ultracentrifuge (Optima; Beckman Coulter) with an MLA-130 rotor. All the following steps were performed at room temperature. Tubulin in the supernatant was polymerized by adding 2 mM GTP (Sigma-Aldrich), 2 mM MgCl2, and stepwise addition of paclitaxel (Sigma-Aldrich) to a final concentration of 20 µM. Kinesins were then allowed to bind by adding 2 mM AMP-PNP and letting the reaction incubate at 37°C for 30 min. Tubulin–kinesin complex was pelleted by centrifugation at 165,000 g for 1 h and the gelatinous precipitate was resuspended in SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
LC-MS/MS analysis of protein complexes
HEK293T cells or MDA-MB-231 cells expressing FLAG-mRadil or FLAG-mRadilΔPDZ were lysed in TAP lysis buffer (0.1% Igepal CA630, 10% glycerol, 50 mM Hepes-NaOH, pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM dithiothreitol, 10 mM NaF, 0.25 mM NaOVO3, 100 mM β-glycerophosphate, and protease inhibitor cocktail) and immunoprecipitated with α-FLAG-M2 beads overnight at 4°C. Purified proteins were eluted from the beads using 500 mM ammonium hydroxide at pH 11.0. The proteins in the complex were reduced in 25 mM dithiothreitol and alkylated using 100 mM iodoacetimide (Sigma-Aldrich), and brought to 1 mM CaCl2. The proteins were then directly digested with sequenced-grade trypsin (Promega). Tryptic peptides were analyzed by LC-MS/MS using a linear ion trap mass spectrometer (model LTQ-XL, Thermo Fisher Scientific; Ahmed et al., 2010). The tandem mass spectra generated were searched against the human NCBI protein sequences using SEQUEST (Eng et al., 1994) running on the Sorcerer platform (Sage-N Research). The peptides identified by SEQUEST were validated using PeptideProphet and assigned a protein identification using ProteinProphet (Keller et al., 2002). The identified proteins were further filtered against other unrelated FLAG pull-down experiments to subtract background proteins using ProHits (Liu et al., 2010).
Cell spreading
Plates or coverslips were coated with 0.5 µg/ml fibronectin at 4°C overnight, and rinsed with PBS twice the following day and dried before performing the experiments. Cells were incubated at 37°C and allowed to adhere and spread for different times. Several phase contrast images of cells were taken from random fields for each condition with a 10×/0.25 NA Ph1 objective using an inverted microscope (Eclipse TS100; Nikon). The cell areas were measured by outlining the perimeters of each cell using ImageJ software version 1.44p. Curve was fitted and the rate constant estimated by nonlinear regression analysis using the one-phase association equation: Y = Y0 + (Plateau − Y0)(1−e−KX), where Y is the cell area and X is time, plateau is the size of the cell when fully spread. Quantified results are reported as ±SEM. Pictures of cell spreading shown in figures were processed using ImageJ (for scale bars) and Adobe Photoshop CS4 (for size and resolution).
Integrin activation assay and flow cytometry
MDA-MB-231 cells were detached from plates using PBS containing 4 mM EDTA followed by centrifugation at 500 g. Cells were rinsed 2× in PBS, resuspended in DMEM containing 0.2% BSA and 20 mM Hepes, pH 7.2, and were allowed to recover for 1 h with gentle agitation on a rocker. Cells from each experimental condition were then incubated at 4°C for 40 min with α-CD29 (9EG7) antibody in the presence or absence of 10 mM MnCl2. Cells were then washed in PBS three times followed by fixation in 4% paraformaldehyde containing 5% sucrose in PBS for 10 min and followed by three more washes in PBS before labeling with Alexa Fluor 647 conjugated goat α-rat IgG (Life Technologies) for 30 min. Flow cytometry analysis was performed with a flow cytometer (FACSCanto II; BD) running on FACSDiva software v6.1.3 (BD). Post-acquisition data analysis was performed using FlowJo version 7.6.5 (Tree Star).
Cell migration and invasion assays
For migration assays, 5 × 104 cells were seeded on the top chamber of Falcon cell culture Transwell inserts with 8-µm pores (BD). For Matrigel invasion assays, 50 µg of growth factor–reduced Matrigel (50 µl; BD) was polymerized on the top chamber of the Transwell inserts for 2 h at 37°C and used immediately as described for migration assays. Cells were allowed to settle on the Transwell inserts with serum-free media in the bottom chambers for 1 h. After 1 h, the media in the bottom chamber was supplemented with 20% FBS and cells were incubated at 37°C for 10–12 h. At the end of this time point cells at the bottom of the Transwell inserts were fixed with 2% glutaraldehyde (Sigma-Aldrich) for 15 min followed by staining with 0.1% crystal violet in 200 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Sigma-Aldrich) for 1 h. The cells that did not invade or migrate were carefully wiped off the top of the membrane with a cotton swab. Cell migration and invasion were quantified by averaging cell counts from pictures taken from 4–5 random areas on the membranes for each condition using an inverted microscope (Eclipse TS100; Nikon) with a 10× objective. Quantified results are presented as ±SEM.
Rap activity assay
MDA-MB-231 cells expressing scrambled, Radil-, or KIF14-specific shRNAs were lysed in RBD lysis buffer (0.5% Igepal CA630, 150 mM NaCl, 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 50 mM NaF, 2 mM sodium vanadate, and protease inhibitor cocktails) and solubilized. 600 µg of protein from each condition was then incubated with GST-RalGDS-RBD coupled to glutathione-Sepharose 4B beads for 1 h at 4°C and GTP bound Rap1 levels assessed by Western blotting.
Mouse xenograft experiments
For in vivo metastasis experiments, 6–10-wk-old NOD-SCID mice were injected via the tail vein with MDA-MB-231-RLuc cells treated with the indicated shRNAs at a density of 106 per mice. Cell metastasis and homing to the lung was monitored by injecting mice intraperitoneally with XenoLight RediJect Coelenterazine h (Caliper) and measured by in vivo bioluminescence imaging using an IVIS Spectrum Bioluminescence system (Xenogen). Animals were sacrificed 45 d after inoculation, and lung tissues were collected and fixed in 3.7% paraformaldehyde followed by 70% ethanol. Lungs were stained with Bouin’s stain (Sigma-Aldrich) overnight. Visible tumor nodules were counted to assess the extent of tumor metastasis. To assess effects on tumor growth 106 MDA-MB-231 cells treated with scrambled shRNA or Radil shRNA #4 were implanted subcutaneously into 6-wk-old NOD-SCID mice. After letting tumors grow for 31 d the mice were sacrificed, and the tumors were dissected and photographed using a fluorescence stereomicroscope. Each tumor was weighed and their volume measured by liquid displacement. All animals were studied using protocols approved by the Animal Care Committee of the Ontario Cancer Institute, and are in accordance with Canadian Council on Animal Care guidelines.
In vitro cell proliferation assay
In vitro cell proliferation was measured using MTT assay described previously (Alley et al., 1988). In brief, on day 0 cells were counted and plated on 24 well plates. Subsequently one plate was taken out each day over the time course, and media replaced with 600 µl of fresh media containing 200 µg/ml of methylthiazolyldiphenyl-tetrazolium bromide (Sigma-Aldrich). Absorbance was measured at 570 nm with background subtraction at 630 nm using a spectrophotometer (DU 730 UV/Vis; Beckman Coulter).
Statistical analysis
Data are generally reported as ±SEM and analyzed by analysis of variance (ANOVA) followed by Tukey’s post-hoc multiple comparison tests (GraphPad Prism 5 software), unless otherwise stated. Data in Fig. S2 A are reported as ±SD. All statistical analysis was considered significant at P < 0.05. Student’s t test was performed to analyze the data represented in Fig. 8, C and D.
Online supplemental material
Fig. S1 shows the specificity of interaction between Radil and KIF14. Radil does not interact with the unrelated KIF7, and KIF14 does not interact with AF-6. The figure also illustrates sequence homology between Radil and AF-6 PDZ domains. Microscopy images show KIF14 localization on microtubules. Fig. S2 shows expression levels of KIF14 in primary human breast tumors and in human breast cancer cell lines. Radil but not RadilΔPDZ cDNA rescues cell spreading defects of Radil shRNA-expressing cells. Cells expressing two different KIF14 shRNAs exhibit increased spreading phenotypes. Fig. S3 shows F-actin and vinculin staining of cells expressing different Radil and KIF14 shRNAs than the ones used in the main figures. Also shown is localization of FLAG-Radil upon nocodazole treatment and Rap1 activation status in cells expressing Radil- or KIF14-specific shRNAs. Fig. S4 shows knockdown efficiency of Radil in cells before subcutaneous injections in mice. Tumors were dissected 31 d after injection and GFP expression confirms the continuous expression of the shRNA containing plasmids. Videos 1–3 accompany Fig. 5 and show the effect of KIF14 depletion on focal adhesion dynamics. Table S1 shows FLAG-Radil pull-downs in HEK293T cells. Table S2 shows FLAG-mRadil and FLAG-mRadilΔPDZ pull-downs in MDA-MB-231 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206051/DC1.
Acknowledgments
The authors are thankful to the members of the Angers laboratory for important discussions and to Rabindra Shivnaraine for his insights with statistical analysis. S. Angers is a Canada Research Chair in Architecture of Signal Transduction. Animal experiments were carried out at the Princess Margaret Hospital/Ontario Cancer Institute animal facility. Microscopy images were taken using the Imaging Facility of The Hospital for Sick Children, Toronto, Ontario, Canada.
The project was supported by a Canadian Breast Cancer Foundation-Ontario grant to S. Angers. S.M. Ahmed was supported by the Alexander Graham Bell Canada Graduate Scholarship from Natural Sciences and Engineering Research Council of Canada.
Footnotes
Abbreviation used in this paper:
- GAP
- GTPase-activating protein
References
- Ahmed S.M., Daulat A.M., Meunier A., Angers S. 2010. G protein betagamma subunits regulate cell adhesion through Rap1a and its effector Radil. J. Biol. Chem. 285:6538–6551 10.1074/jbc.M109.069948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.M., Daulat A.M., Angers S. 2011. Tandem affinity purification and identification of heterotrimeric g protein-associated proteins. Methods Mol. Biol. 756:357–370 10.1007/978-1-61779-160-4_22 [DOI] [PubMed] [Google Scholar]
- Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R. 1988. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 48:589–601 [PubMed] [Google Scholar]
- Amano M., Chihara K., Kimura K., Fukata Y., Nakamura N., Matsuura Y., Kaibuchi K. 1997. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 275:1308–1311 10.1126/science.275.5304.1308 [DOI] [PubMed] [Google Scholar]
- Arjonen A., Kaukonen R., Ivaska J. 2011. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 5:421–430 10.4161/cam.5.5.17723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur W.T., Quilliam L.A., Cooper J.A. 2004. Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J. Cell Biol. 167:111–122 10.1083/jcb.200404068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.L., Kelly P., Casey P.J. 2009. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 69:4962–4968 10.1158/0008-5472.CAN-08-4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R., Mani R.S., Russo N., Scanlon C.S., Tsodikov A., Jing X., Cao Q., Palanisamy N., Metwally T., Inglehart R.C., et al. 2011. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 30:4339–4349 10.1038/onc.2011.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner M., Radziwill G., Lorger M., Weiss A., Moelling K. 2008. c-Src-mediated epithelial cell migration and invasion regulated by PDZ binding site. Mol. Cell. Biol. 28:642–655 10.1128/MCB.01024-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béraud-Dufour S., Gautier R., Albiges-Rizo C., Chardin P., Faurobert E. 2007. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. FEBS J. 274:5518–5532 10.1111/j.1742-4658.2007.06068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona T.G., Wiener H.H., Ahearn I.M., Silletti J., Chiu V.K., Philips M.R. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164:461–470 10.1083/jcb.200311093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B., Govek E.E., Cross J., Van Aelst L. 2000. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc. Natl. Acad. Sci. USA. 97:9064–9069 10.1073/pnas.97.16.9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L. 2005. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17:123–128 10.1016/j.ceb.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Bos J.L., de Bruyn K., Enserink J., Kuiperij B., Rangarajan S., Rehmann H., Riedl J., de Rooij J., van Mansfeld F., Zwartkruis F. 2003. The role of Rap1 in integrin-mediated cell adhesion. Biochem. Soc. Trans. 31:83–86 10.1042/BST0310083 [DOI] [PubMed] [Google Scholar]
- Carleton M., Mao M., Biery M., Warrener P., Kim S., Buser C., Marshall C.G., Fernandes C., Annis J., Linsley P.S. 2006. RNA interference-mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol. Cell. Biol. 26:3853–3863 10.1128/MCB.26.10.3853-3863.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson T.W., Gallie B.L. 2006. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int. J. Cancer. 119:1088–1094 10.1002/ijc.21954 [DOI] [PubMed] [Google Scholar]
- Corson T.W., Huang A., Tsao M.S., Gallie B.L. 2005. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 24:4741–4753 10.1038/sj.onc.1208641 [DOI] [PubMed] [Google Scholar]
- Corson T.W., Zhu C.Q., Lau S.K., Shepherd F.A., Tsao M.S., Gallie B.L. 2007. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin. Cancer Res. 13:3229–3234 10.1158/1078-0432.CCR-07-0393 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruet-Hennequart S., Maubant S., Luis J., Gauduchon P., Staedel C., Dedhar S. 2003. alpha(v) integrins regulate cell proliferation through integrin-linked kinase (ILK) in ovarian cancer cells. Oncogene. 22:1688–1702 10.1038/sj.onc.1206347 [DOI] [PubMed] [Google Scholar]
- Dao V.T., Dupuy A.G., Gavet O., Caron E., de Gunzburg J. 2009. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J. Cell Sci. 122:2996–3004 10.1242/jcs.041301 [DOI] [PubMed] [Google Scholar]
- de Rooij J., Zwartkruis F.J., Verheijen M.H., Cool R.H., Nijman S.M., Wittinghofer A., Bos J.L. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 396:474–477 10.1038/24884 [DOI] [PubMed] [Google Scholar]
- Desgrosellier J.S., Cheresh D.A. 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 10:9–22 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J., McCormack A., Yates J. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Fanning A.S., Anderson J.M. 1999. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Invest. 103:767–772 10.1172/JCI6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felding-Habermann B. 2003. Integrin adhesion receptors in tumor metastasis. Clin. Exp. Metastasis. 20:203–213 10.1023/A:1022983000355 [DOI] [PubMed] [Google Scholar]
- Felding-Habermann B., O’Toole T.E., Smith J.W., Fransvea E., Ruggeri Z.M., Ginsberg M.H., Hughes P.E., Pampori N., Shattil S.J., Saven A., Mueller B.M. 2001. Integrin activation controls metastasis in human breast cancer. Proc. Natl. Acad. Sci. USA. 98:1853–1858 10.1073/pnas.98.4.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S.A., McLeod S.J., Dukowski J., Austin P., Lee C.C., Millen-Martin B., Kubes P., McCafferty D.M., Gold M.R., Roskelley C.D. 2010. Preventing the activation or cycling of the Rap1 GTPase alters adhesion and cytoskeletal dynamics and blocks metastatic melanoma cell extravasation into the lungs. Cancer Res. 70:4590–4601 10.1158/0008-5472.CAN-09-3414 [DOI] [PubMed] [Google Scholar]
- Fujita H., Fukuhara S., Sakurai A., Yamagishi A., Kamioka Y., Nakaoka Y., Masuda M., Mochizuki N. 2005. Local activation of Rap1 contributes to directional vascular endothelial cell migration accompanied by extension of microtubules on which RAPL, a Rap1-associating molecule, localizes. J. Biol. Chem. 280:5022–5031 10.1074/jbc.M409701200 [DOI] [PubMed] [Google Scholar]
- Gardel M.L., Sabass B., Ji L., Danuser G., Schwarz U.S., Waterman C.M. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183:999–1005 10.1083/jcb.200810060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel M.L., Schneider I.C., Aratyn-Schaus Y., Waterman C.M. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Spatz J.P., Bershadsky A.D. 2009. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10:21–33 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Gérard A., Mertens A.E., van der Kammen R.A., Collard J.G. 2007. The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. J. Cell Biol. 176:863–875 10.1083/jcb.200608161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A., Han J., Stockton R.A., Ginsberg M.H. 2007. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J. Cell Biol. 179:247–254 10.1083/jcb.200705175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., Chan E.H., Chalamalasetty R.B., Nigg E.A., Barr F.A. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172:363–372 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Premont R.T., Chow C.W., Blitzer J.T., Pitcher J.A., Claing A., Stoffel R.H., Barak L.S., Shenolikar S., Weinman E.J., et al. 1998. The beta2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature. 392:626–630 10.1038/33458 [DOI] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., Ginsberg M.H. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796–1806 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Hansen C.A., Schroering A.G., Carey D.J., Robishaw J.D. 1994. Localization of a heterotrimeric G protein gamma subunit to focal adhesions and associated stress fibers. J. Cell Biol. 126:811–819 10.1083/jcb.126.3.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B.Z., Lim W.A. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114:3219–3231 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Houslay M.D. 2009. Disrupting specific PDZ domain-mediated interactions for therapeutic benefit. Br. J. Pharmacol. 158:483–485 10.1111/j.1476-5381.2009.00359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Anand S., Murphy E.A., Desgrosellier J.S., Stupack D.G., Shattil S.J., Schlaepfer D.D., Cheresh D.A. 2012. EGFR-dependent pancreatic carcinoma cell metastasis through Rap1 activation. Oncogene. 31:2783–2793 10.1038/onc.2011.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert P., Russell S., Richardson H. 2003. Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays. 25:542–553 10.1002/bies.10286 [DOI] [PubMed] [Google Scholar]
- Huszar D., Theoclitou M.E., Skolnik J., Herbst R. 2009. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 28:197–208 10.1007/s10555-009-9185-8 [DOI] [PubMed] [Google Scholar]
- Itoh M., Nelson C.M., Myers C.A., Bissell M.J. 2007. Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 67:4759–4766 10.1158/0008-5472.CAN-06-4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon T.J., Lee D.J., Merlot S., Weeks G., Firtel R.A. 2007. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 176:1021–1033 10.1083/jcb.200607072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M., Schallhorn A., Wurm F.M. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596–601 10.1093/nar/24.4.596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Maeda A., Shimonaka M., Kinashi T. 2003. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4:741–748 10.1038/ni950 [DOI] [PubMed] [Google Scholar]
- Kaverina I.N., Minin A.A., Gyoeva F.K., Vasiliev J.M. 1997. Kinesin-associated transport is involved in the regulation of cell adhesion. Cell Biol. Int. 21:229–236 10.1006/cbir.1997.0136 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J.V. 2002. Regulation of substrate adhesion dynamics during cell motility. Int. J. Biochem. Cell Biol. 34:746–761 10.1016/S1357-2725(01)00171-6 [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kim C., Ye F., Ginsberg M.H. 2011. Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27:321–345 10.1146/annurev-cellbio-100109-104104 [DOI] [PubMed] [Google Scholar]
- Kim E., Sheng M. 2004. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5:771–781 10.1038/nrn1517 [DOI] [PubMed] [Google Scholar]
- Kim W.J., Gersey Z., Daaka Y. 2012. Rap1GAP regulates renal cell carcinoma invasion. Cancer Lett. 320:65–71 10.1016/j.canlet.2012.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi T., Katagiri K. 2004. Regulation of lymphocyte adhesion and migration by the small GTPase Rap1 and its effector molecule, RAPL. Immunol. Lett. 93:1–5 10.1016/j.imlet.2004.02.008 [DOI] [PubMed] [Google Scholar]
- Kinashi T., Katagiri K. 2005. Regulation of immune cell adhesion and migration by regulator of adhesion and cell polarization enriched in lymphoid tissues. Immunology. 116:164–171 10.1111/j.1365-2567.2005.02214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirui J.K., Xie Y., Wolff D.W., Jiang H., Abel P.W., Tu Y. 2010. Gbetagamma signaling promotes breast cancer cell migration and invasion. J. Pharmacol. Exp. Ther. 333:393–403 10.1124/jpet.109.164814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylyshkina O., Kaverina I., Kranewitter W., Steffen W., Alonso M.C., Cross R.A., Small J.V. 2002. Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. J. Cell Biol. 156:349–359 10.1083/jcb.200105051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwada S.K., Li X. 2000. Integrin alpha5/beta1 mediates fibronectin-dependent epithelial cell proliferation through epidermal growth factor receptor activation. Mol. Biol. Cell. 11:2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., Boussiotis V.A. 2004. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 7:585–595 10.1016/j.devcel.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Lee H.S., Lim C.J., Puzon-McLaughlin W., Shattil S.J., Ginsberg M.H. 2009. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284:5119–5127 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.T., Alemayehu M., Aziziyeh A.I., Pape C., Pampillo M., Postovit L.M., Mills G.B., Babwah A.V., Bhattacharya M. 2009. Beta-arrestin/Ral signaling regulates lysophosphatidic acid-mediated migration and invasion of human breast tumor cells. Mol. Cancer Res. 7:1064–1077 10.1158/1541-7786.MCR-08-0578 [DOI] [PubMed] [Google Scholar]
- Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z.Y., Breitkreutz B.J., Ding Y., Colwill K., Pasculescu A., et al. 2010. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28:1015–1017 10.1038/nbt1010-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenowicz M.J., Fernandez-Borja M., Kooistra M.R., Bos J.L., Hordijk P.L. 2008. PKA and Epac1 regulate endothelial integrity and migration through parallel and independent pathways. Eur. J. Cell Biol. 87:779–792 10.1016/j.ejcb.2008.05.004 [DOI] [PubMed] [Google Scholar]
- Ludford-Menting M.J., Oliaro J., Sacirbegovic F., Cheah E.T., Pedersen N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., et al. 2005. A network of PDZ-containing proteins regulates T cell polarity and morphology during migration and immunological synapse formation. Immunity. 22:737–748 10.1016/j.immuni.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Lyle K.S., Raaijmakers J.H., Bruinsma W., Bos J.L., de Rooij J. 2008. cAMP-induced Epac-Rap activation inhibits epithelial cell migration by modulating focal adhesion and leading edge dynamics. Cell. Signal. 20:1104–1116 10.1016/j.cellsig.2008.01.018 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Fumoto K., Okamoto T., Kaibuchi K., Kikuchi A. 2010. Binding of APC and dishevelled mediates Wnt5a-regulated focal adhesion dynamics in migrating cells. EMBO J. 29:1192–1204 10.1038/emboj.2010.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean G.W., Carragher N.O., Avizienyte E., Evans J., Brunton V.G., Frame M.C. 2005. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat. Rev. Cancer. 5:505–515 10.1038/nrc1647 [DOI] [PubMed] [Google Scholar]
- Meerschaert K., Bruyneel E., De Wever O., Vanloo B., Boucherie C., Bracke M., Vandekerckhove J., Gettemans J. 2007. The tandem PDZ domains of syntenin promote cell invasion. Exp. Cell Res. 313:1790–1804 10.1016/j.yexcr.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Meng Q., Qi M., Chen D.Z., Yuan R., Goldberg I.D., Rosen E.M., Auborn K., Fan S. 2000. Suppression of breast cancer invasion and migration by indole-3-carbinol: associated with up-regulation of BRCA1 and E-cadherin/catenin complexes. J. Mol. Med. 78:155–165 10.1007/s001090000088 [DOI] [PubMed] [Google Scholar]
- Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature. 410:50–56 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- Nagano M., Hoshino D., Koshikawa N., Akizawa T., Seiki M. 2012. Turnover of focal adhesions and cancer cell migration. Int. J. Cell Biol. 2012:310616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik M.U., Naik T.U., Suckow A.T., Duncan M.K., Naik U.P. 2008. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res. 68:2194–2203 10.1158/0008-5472.CAN-07-3057 [DOI] [PubMed] [Google Scholar]
- Oakes P.W., Beckham Y., Stricker J., Gardel M.L. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.T., Horwitz A.R., Schwartz M.A. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra C.R., Rupasinghe C.N., Dutta S.K., Bhattacharya S., Wang E., Spaller M.R., Mukhopadhyay D. 2012. Chemically modified peptides targeting the PDZ domain of GIPC as a therapeutic approach for cancer. ACS Chem. Biol. 7:770–779 10.1021/cb200536r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill G., Weiss A., Heinrich J., Baumgartner M., Boisguerin P., Owada K., Moelling K. 2007. Regulation of c-Src by binding to the PDZ domain of AF-6. EMBO J. 26:2633–2644 10.1038/sj.emboj.7601706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala J.K., Pouwels J., Pellinen T., Veltel S., Laasola P., Mattila E., Potter C.S., Duffy T., Sundberg J.P., Kallioniemi O., et al. 2011. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat. Cell Biol. 13:1315–1324 10.1038/ncb2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.H., Post A., Raaijmakers J.H., Verlaan I., Gloerich M., Bos J.L. 2011. Ezrin is required for efficient Rap1-induced cell spreading. J. Cell Sci. 124:1808–1818 10.1242/jcs.079830 [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Munemitsu S., Clark R., Conroy L., Watt K., Crosier W.J., McCormick F., Polakis P. 1991. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 65:1033–1042 10.1016/0092-8674(91)90555-D [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebzda E., Bracke M., Tugal T., Hogg N., Cantrell D.A. 2002. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3:251–258 10.1038/ni765 [DOI] [PubMed] [Google Scholar]
- Sheng M., Sala C. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1–29 10.1146/annurev.neuro.24.1.1 [DOI] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Smolen G.A., Schott B.J., Stewart R.A., Diederichs S., Muir B., Provencher H.L., Look A.T., Sgroi D.C., Peterson R.T., Haber D.A. 2007. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes Dev. 21:2131–2136 10.1101/gad.1561507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Sun Z., Runne C., Madsen J., Domann F., Henry M., Lin F., Chen S. 2011. A critical role of Gbetagamma in tumorigenesis and metastasis of breast cancer. J. Biol. Chem. 286:13244–13254 10.1074/jbc.M110.206615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault B.L., Pajovic S., Bernardini M.Q., Shaw P.A., Gallie B.L. 2012. Kinesin family member 14: an independent prognostic marker and potential therapeutic target for ovarian cancer. Int. J. Cancer. 130:1844–1854 10.1002/ijc.26189 [DOI] [PubMed] [Google Scholar]
- Thorsen T.S., Madsen K.L., Rebola N., Rathje M., Anggono V., Bach A., Moreira I.S., Stuhr-Hansen N., Dyhring T., Peters D., et al. 2010. Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc. Natl. Acad. Sci. USA. 107:413–418 10.1073/pnas.0902225107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R., Zhang Y., Sazinsky S.L., Currell B., Yeh J.H., Reva B., Held H.A., Appleton B.A., Evangelista M., Wu Y., et al. 2008. A specificity map for the PDZ domain family. PLoS Biol. 6:e239 10.1371/journal.pbio.0060239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsygankova O.M., Prendergast G.V., Puttaswamy K., Wang Y., Feldman M.D., Wang H., Brose M.S., Meinkoth J.L. 2007. Downregulation of Rap1GAP contributes to Ras transformation. Mol. Cell. Biol. 27:6647–6658 10.1128/MCB.00155-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y., Sakaguchi M., Terabayashi T., Inenaga T., Inoue S., Kobayashi C., Oshima N., Kiyonari H., Nakagata N., Sato Y., et al. 2010. Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc. Natl. Acad. Sci. USA. 107:9240–9245 10.1073/pnas.0913748107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wang C., Navab R., Iakovlev V., Leng Y., Zhang J., Tsao M.S., Siminovitch K., McCready D.R., Done S.J. 2007. Abelson interactor protein-1 positively regulates breast cancer cell proliferation, migration, and invasion. Mol. Cancer Res. 5:1031–1039 10.1158/1541-7786.MCR-06-0391 [DOI] [PubMed] [Google Scholar]
- Watson P.H., Snell L., Parisien M. 1996. The NCIC-Manitoba Breast Tumor Bank: a resource for applied cancer research. CMAJ. 155:281–283 [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhu C., Chen H., Li L., Guo L., Jiang W., Lu S.H. 2010. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 31:1676–1684 10.1093/carcin/bgq134 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Appleton B.A., Wiesmann C., Lau T., Costa M., Hannoush R.N., Sidhu S.S. 2009. Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5:217–219 10.1038/nchembio.152 [DOI] [PubMed] [Google Scholar]
- Zheng H., Gao L., Feng Y., Yuan L., Zhao H., Cornelius L.A. 2009. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. 69:449–457 10.1158/0008-5472.CAN-08-2399 [DOI] [PubMed] [Google Scholar]