Mice expressing a version of Bub1 that lacks kinase activity have increased chromosome segregation errors and aneuploidy but not increased susceptibility to tumors.
Abstract
The mitotic checkpoint protein Bub1 is essential for embryogenesis and survival of proliferating cells, and bidirectional deviations from its normal level of expression cause chromosome missegregation, aneuploidy, and cancer predisposition in mice. To provide insight into the physiological significance of this critical mitotic regulator at a modular level, we generated Bub1 mutant mice that lack kinase activity using a knockin gene-targeting approach that preserves normal protein abundance. In this paper, we uncover that Bub1 kinase activity integrates attachment error correction and mitotic checkpoint signaling by controlling the localization and activity of Aurora B kinase through phosphorylation of histone H2A at threonine 121. Strikingly, despite substantial chromosome segregation errors and aneuploidization, mice deficient for Bub1 kinase activity do not exhibit increased susceptibility to spontaneous or carcinogen-induced tumorigenesis. These findings provide a unique example of a modular mitotic activity orchestrating two distinct networks that safeguard against whole chromosome instability and reveal the differential importance of distinct aneuploidy-causing Bub1 defects in tumor suppression.
Introduction
Aneuploidy, an abnormal number of individual chromosomes, is a common cause of infertility, early embryonic lethality, and congenital birth defects in humans as well as a hallmark of cancer (Torres et al., 2008). Two intricate molecular networks, the attachment error correction machinery and the mitotic checkpoint, evolved across eukaryotic species to promote accurate chromosome segregation during mitotic and meiotic divisions and protect against aneuploidy (Pinsky and Biggins, 2005; Peters, 2006; Ruchaud et al., 2007; Holland and Cleveland, 2009). The mitotic checkpoint inhibits separation of sister chromatids until all kinetochores are attached to spindle microtubules, and the attachment error correction machinery ensures that each of the two sister chromatids are properly biorientated. Although tremendous progress has been made in elucidating the molecular framework of the mitotic checkpoint and error correction machinery, insights into the interplay and coordination between these two machineries have remained largely elusive.
One component that is key to high fidelity chromosome segregation is the mitotic kinase Bub1. At the onset of mitosis, Bub1 accumulates at unattached kinetochores, where it facilitates recruitment of other core mitotic checkpoint components, including Mad1, Mad2, BubR1, Bub3, and Cenp-E, to create inhibitors of Cdc20, one of two activating subunits of the anaphase-promoting complex/cyclosome (APC/C; Sharp-Baker and Chen, 2001; Taylor et al., 2001; Johnson et al., 2004; Wong and Fang, 2006; Jeganathan et al., 2007). Attachment of the last kinetochore to the mitotic spindle triggers the release of Cdc20 from the inhibitory kinetochore-derived complex of Mad2, BubR1, and Bub3 (Sudakin et al., 2001; Kulukian et al., 2009), thereby relinquishing APC/C inhibition and triggering the ubiquitin-mediated degradation of securin and cyclin B. Their clearance results in the activation of separase, a protease that initiates anaphase by opening the cohesin ring structures that hold sister chromosomes together. In addition to its role as a scaffolding component at unattached kinetochores, Bub1 has been proposed to contribute to APC/C inhibition directly by phosphorylation of Cdc20 (Chung and Chen, 2003; Tang et al., 2004a; Kang et al., 2008). However, the role of Bub1 kinase activity in checkpoint signaling remains unclear, as some studies have demonstrated an intact checkpoint with Bub1 kinase mutants (Sharp-Baker and Chen, 2001; Perera and Taylor, 2010), whereas others have observed that a robust checkpoint response requires Bub1 catalysis (Chen, 2004; Kang et al., 2008; Klebig et al., 2009; McGuinness et al., 2009).
Besides contributing to accurate chromosome segregation through mitotic checkpoint activation, Bub1 has been proposed to have other mitotic roles. For example, it was recently reported that Bub1 mediates the recruitment of Sgo1 to inner centromeres through phosphorylation of histone H2A at residue T121 (Kawashima et al., 2010). Sgo1 has been proposed to protect centromere cohesin from premature separation through its association with PP2A phosphatase, which counteracts kinases targeting cohesin (Tang et al., 2004b, 2006; Kitajima et al., 2006; Lee et al., 2008; Xu et al., 2009). Thus, Bub1-mediated Sgo1 recruitment could represent a checkpoint-independent function for Bub1 in chromosome segregation. Studies in budding yeast and mice have suggested, however, that mitotic Sgo1 is dispensable for protecting centromeric cohesin and that Bub1 protects cohesin in a checkpoint-dependent manner (Katis et al., 2004; Indjeian et al., 2005; Perera et al., 2007; Perera and Taylor, 2010).
In addition to protecting cohesin, centromeric Sgo1 has been proposed to serve as an adaptor to facilitate Aurora B inner centromeric accumulation (Kawashima et al., 2007; Tsukahara et al., 2010; Yamagishi et al., 2010). Aurora B kinase performs a crucial task in attachment error correction by destabilizing erroneous kinetochore–microtubule attachments (Ruchaud et al., 2007). Localization of Aurora B to inner centromeres not only requires Sgo1 but also histone H3 modification at threonine 3 (H3T3) by Haspin kinase (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). Whether Sgo1 facilitates Aurora B localization remains to be further investigated, as depletion of Sgo1 does not interfere with Aurora B accumulation at inner centromeres (McGuinness et al., 2005; Dai et al., 2006; Kawashima et al., 2007).
Like many other mitotic regulators, Bub1 is essential for embryogenesis and survival of proliferating cells (Perera et al., 2007). Much of what we know about the physiological relevance of Bub1 has come from mouse models that express abnormal levels of Bub1 protein (Jeganathan et al., 2007; Ricke et al., 2011). A key finding was that high fidelity chromosome segregation is exceedingly dependent on proper level of Bub1 expression, with Bub1 insufficiency or overabundance causing chromosome segregation errors and driving tumorigenesis. Given that Bub1 has tumor-suppressive and oncogenic properties, a key question that emerges is how the various modular functions of Bub1 drive pathology. Whereas in vitro structure–function studies using Bub1 depletion have provided insight into the roles of specific Bub1 domains in the scaffolding functions at kinetochores, this approach has been problematic for analysis of the kinase domain because of residual enzyme and incomplete replacement of endogenous Bub1 with exogenously produced protein (Qi and Yu, 2007; Klebig et al., 2009; Perera and Taylor, 2010).
To determine the phenotypic relevance of Bub1 kinase activity at the cellular and organismal levels, we mutated a single residue in the catalytic loop of the endogenous Bub1 gene in mice. Using this approach, we obtain a clean genetic system that meets the criteria of complete inactivation of catalytic activity without any alterations to protein stability, to pinpoint the mechanistic contribution of this enzyme. We find that the major defect is a direct consequence of deregulated Aurora B activity. Unexpectedly, although mice deficient for Bub1 kinase activity harbor significant aneuploidy, they do not show increased susceptibility to spontaneous or carcinogen-induced tumorigenesis. This is surprising for two reasons. First, aneuploidy is a putative cancer-promoting event, and second, Bub1 hypomorphism causes near-diploid aneuploidy and tumors (Jeganathan et al., 2007; Schliekelman et al., 2009). Thus, we present here the novel concept of a multifunctional mitotic regulator that causes aneuploidy through different mechanisms, not all of which are tumor suppressive.
Results
Generation of mice deficient in Bub1 catalytic activity
To understand the in vivo role of Bub1 kinase activity, we used gene targeting in murine embryonic stem (ES) cells to mutate the endogenous Bub1 sequence such that the catalytic residue D892 (Kang et al., 2008) was changed to N (Fig. 1 A). We obtained correctly targeted ES clones (Fig. 1 B) and used these to create mice carrying the kinase-dead Bub1 allele, hereafter referred to as Bub1KD. Intercrosses of Bub1+/KD mice yielded Bub1KD/KD offspring at the expected Mendelian frequency. Bub1KD/KD mice were overtly indistinguishable from Bub1+/KD and wild-type littermates.
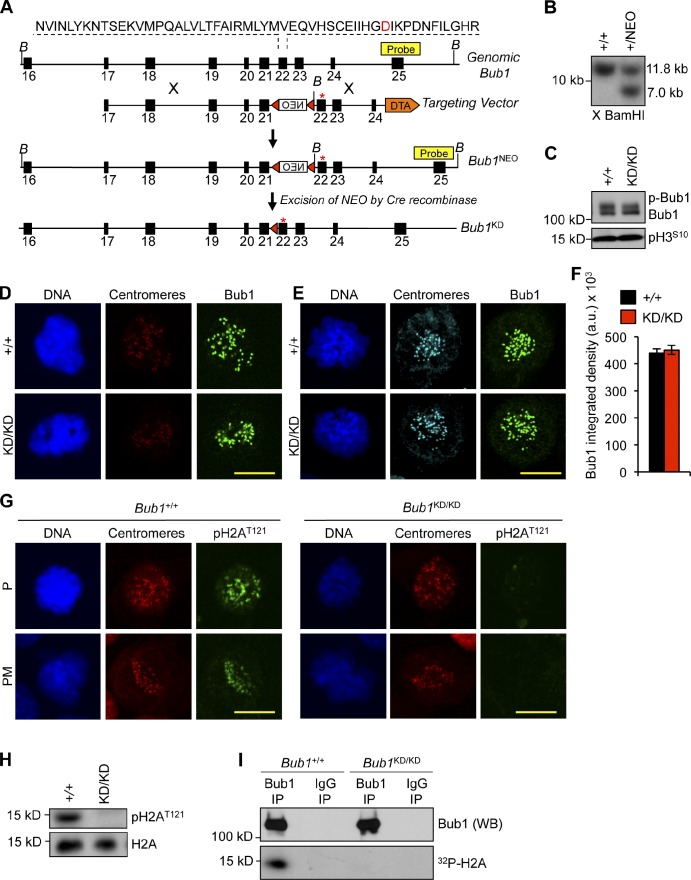
Figure 1.
Generation of Bub1 kinase–deficient mice. (A) Bub1 targeting strategy. Bub1 locus (+), D892N targeting vector, targeted allele (Bub1NEO), the targeted allele after Cre recombination (Bub1KD), BamHI (B) restriction sites, loxP sites (red triangles), and the Southern probe are indicated. DTA, diphtheria toxin A. Asterisks mark the D892N point mutation. (B) Southern blot of targeted ES clones digested with BamHI. X, digested. (C) Western blot of lysates from nocodazole-treated shake-off MEFs probed for Bub1 and pH3S10. (D) Immunostaining of Bub1+/+ and Bub1KD/KD MEFs. (E) Immunostaining of monastrol-treated Bub1+/+ and Bub1KD/KD MEFs. (F) Quantification of Bub1 staining shown in E from three independent MEF lines. Error bars indicate SEM. (G) Immunostaining of Bub1+/+ and Bub1KD/KD MEFs. (H) Western blot of lysates from taxol blocked MEFs probed for pH2AT121 and histone H2A. (I) Bub1 was immunoprecipitated from taxol-blocked Bub1+/+ and Bub1KD/KD immortalized MEFs and incubated with histone H2A in the presence of γ-[32P]ATP. a.u., arbitrary unit; NEO, neomycin; KD, kinase dead; IP, immunoprecipitation; WB, Western blot. Bars, 10 µm.
Western blot analysis of mitotic extracts prepared from Bub1KD/KD and wild-type mouse embryonic fibroblasts (MEFs) confirmed that the Bub1KD and Bub1 proteins were expressed at similar levels (Fig. 1 C). Kinetochore-associated Bub1 protein levels were the same in Bub1KD/KD and wild-type MEFs (Fig. 1, D–F). To confirm that Bub1KD encoded a catalytically deficient protein, we monitored for phosphorylation of a known Bub1 substrate, histone H2A at Thr121 (Boyarchuk et al., 2007; Kawashima et al., 2010). Although wild-type MEFs showed abundant H2AThr121 phosphorylation in mitosis, no such modification was detectable in Bub1KD/KD cells (Fig. 1, G and H). Moreover, Bub1 protein immunoprecipitated from Bub1KD/KD MEFs was unable to phosphorylate H2A in vitro (Fig. 1 I). Based on these findings, we conclude that the Bub1KD allele encodes a Bub1 kinase-dead protein with comparable stability as its wild-type counterpart.
Loss of Bub1 kinase activity causes chromosome misalignment and aneuploidy
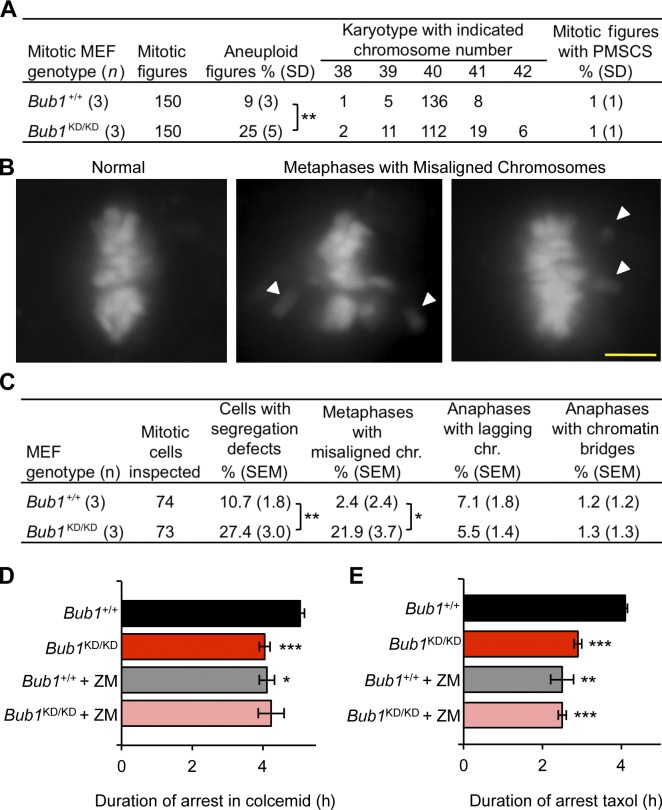
To assess whether Bub1 kinase activity affects karyotypic stability, we performed counts on chromosome spreads from Bub1KD/KD and wild-type MEFs. Consistent with earlier data, aneuploidy was observed in 9% of wild-type spreads. In contrast, aneuploidy rates were substantially higher in Bub1KD/KD MEFs, with 25% showing near-diploid aneuploidy (Fig. 2 A). Metaphase spreads from Bub1KD/KD MEFs showed no evidence of increased premature sister chromatid separation (PMSCS; Fig. 2 A). To determine the nature of the chromosome segregation errors that drive aneuploidization in the absence of Bub1 catalytic activity, we transduced Bub1KD/KD and wild-type MEFs with a lentivirus encoding mRFP-H2B to permit visualization of chromosomes and monitored chromosome segregation by live-cell imaging. Rates of anaphase onset with misaligned chromosomes were markedly increased in the absence of catalytic activity, with 21.9% of Bub1KD/KD MEFs showing this defect compared with 2.4% of wild-type MEFs (Fig. 2, B and C). Other mitotic defects that can cause aneuploidy, such as lagging chromosomes and chromatin bridges, were not increased. The time from prophase to anaphase onset was normal in Bub1KD/KD MEFs, even in those cells experiencing chromosome misalignment (Fig. S1 A).
Figure 2.
Bub1 kinase activity is required for chromosome alignment and maintenance of chromosomal stability. (A) Karyotype analysis of passage 5 MEFs. Statistics indicate comparison with wild type. (B) Representative images for chromosome segregation defects by live-cell imaging. White arrowheads shows misaligned chromosomes. Bar, 10 µm. (C) Live-cell imaging of chromosome segregation defects in primary MEFs. Statistics indicate comparison with wild type. (D) Analysis of mitotic duration in Bub1+/+ and Bub1KD/KD MEFs after colcemid addition (n = 3 or more lines per genotype). Error bars indicate SEM. The time at which 50% of the cells exited mitosis was compared. Statistics indicate comparison with wild type. (E) Same as D but with 0.5 µM taxol. chr., chromosome. *, P < 0.5; **, P < 0.1; ***, P < 0.001.
Aurora B–dependent mitotic checkpoint activity is impaired in Bub1 kinase-dead cells
Inability to delay anaphase onset in the presence of misaligned chromosomes might be caused by mitotic checkpoint failure. To examine mitotic checkpoint robustness in the absence of Bub1 kinase activity, we performed colcemid- and taxol-challenge assays on Bub1KD/KD and wild-type MEFs that were transduced with lentivirus encoding mRFP-H2B. Cells undergoing nuclear envelope breakdown (NEBD) were marked and monitored at 30-min intervals for chromatin decondensation that signals mitotic exit without cytokinesis. The time of arrest of Bub1KD/KD MEFs was 20 and 30% reduced in colcemid and taxol, respectively (Fig. 2, D and E). This reduction of mitotic checkpoint activity is moderate compared with BubR1 hypomorphic MEFs, which consistent with earlier studies (Baker et al., 2004, 2006), show a 73% reduction in time of arrest in colcemid (Fig. S1 B). Kinetochores of Bub1KD/KD MEFs harbored normal levels of BubR1, Mad2, Mad1, and Cenp-E (Fig. S1 C), indicating that the observed impairment in checkpoint activity is not caused by mislocalization of Bub1-dependent mitotic regulators (Jeganathan et al., 2007). Consistently, Bub1KD/KD MEFs challenged with spindle poisons had the same amount of kinetochore-associated Bub1 as wild-type MEFs (Fig. S1 D).
It has been previously shown that Aurora B inhibition sensitizes cells to taxol more exquisitely than microtubule depolymerization (Ditchfield et al., 2003; Hauf et al., 2003). This prompted us to examine the possible role of this mitotic kinase in the impairment of mitotic checkpoint activity in Bub1KD/KD MEFs. Strikingly, wild-type MEFs showed an 18% decline in mitotic checkpoint activity in colcemid when Aurora B kinase was inhibited with the drug ZM447439 (ZM), which is similar to the level of impairment we observed in Bub1KD/KD MEFs (Fig. 2 D). ZM treatment of Bub1KD/KD MEFs did not exaggerate the mitotic checkpoint defect, suggesting that loss of Bub1 kinase activity impairs checkpoint activity through an Aurora B–dependent mechanism. Similar results were obtained when the aforementioned experiments were repeated with taxol instead of colcemid (Fig. 2 E).
Bub1 kinase activity is required for centromeric accumulation of shugoshins and Aurora B
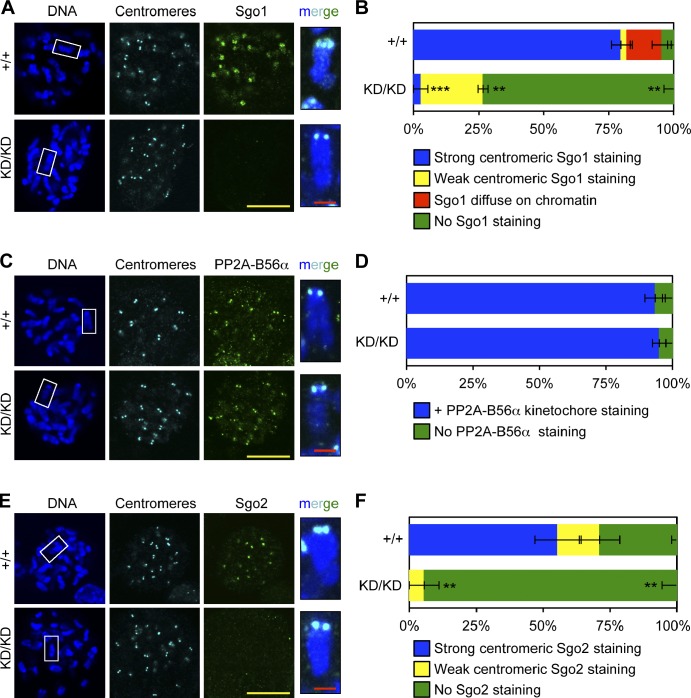
Next, we sought to determine how Bub1 kinase activity might regulate Aurora B activity. It has been proposed that Bub1-mediated phosphorylation of H2AT121 at inner centromeres provides a histone mark for recruitment of Sgo1, which serves as an adaptor for binding of Aurora B (Kawashima et al., 2010; Tsukahara et al., 2010; Yamagishi et al., 2010). Consistent with previous studies, we found that Bub1KD/KD MEFs failed to accumulate Sgo1 at mitotic centromeres (Figs. 3, A and B; and S2, A and B; Kitajima et al., 2005; Fernius and Hardwick, 2007). Surprisingly, phosphatase PP2A-B56α, which is thought to be recruited to inner centromeres by Sgo1 to counteract phosphorylation-mediated release of cohesion and prevent PMSCS (McGuinness et al., 2005; Riedel et al., 2006; Tang et al., 2006; Xu et al., 2009), accumulated normally at mitotic centromeres of Bub1KD/KD MEFs (Fig. 3, C and D). In keeping with this, metaphase spreads of Bub1KD/KD MEFs showed no evidence of PMSCS (Fig. 2 A). In addition to Sgo1, Bub1 has been implicated in the centromeric recruitment of Sgo2 (Tsukahara et al., 2010). Bub1KD/KD MEFs failed to accumulate Sgo2 at inner centromeres of mitotic chromosomes (Figs. 3, E and F; and S2, C and D), revealing that this localization process is dependent on Bub1 kinase activity.
Figure 3.
Optimal loading of shugoshins at inner centromeres requires Bub1 kinase activity. (A) IF of spreads from colcemid-blocked MEFs. Insets show enlarged chromosomes from boxed region (merged). (B) Quantification of Sgo1 localization (n = 3 lines per genotype). Error bars indicate SEM. Statistics indicate comparison with wild type. (C) Same as A except with PP2A-B56α. (D) Same as B except with PP2A-B56α. (E) Same as A except with Sgo2 (green). (F) Same as B except with Sgo2. **, P < 0.1; ***, P < 0.001. KD, kinase dead. Bars: (yellow) 10 µm; (red) 1 µm.
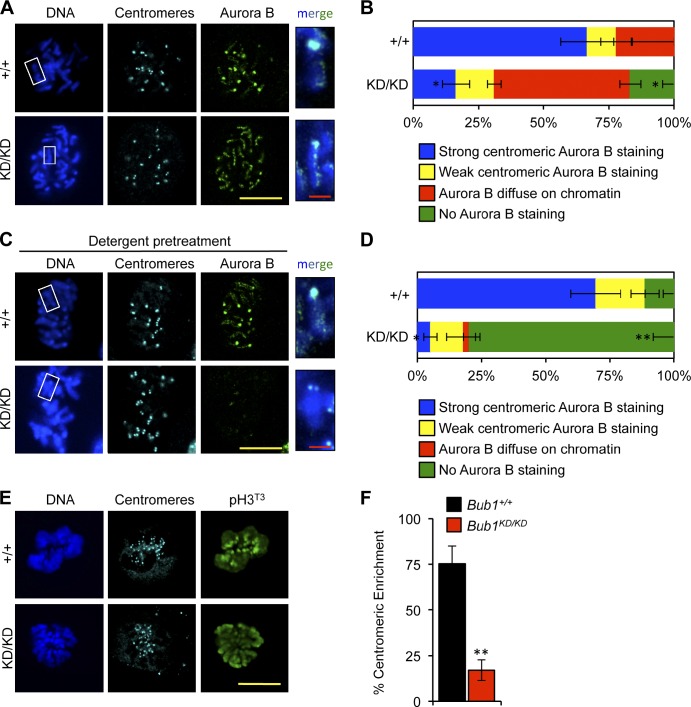
To examine whether loss of Bub1 kinase activity might alter Aurora B localization, we immunostained chromosome spreads from Bub1KD/KD and wild-type MEFs for Aurora B and centromeres. Although Aurora B concentrated at inner centromeres of mitotic chromosomes of wild-type MEFs, it localized along chromosome arms in Bub1KD/KD spreads (Figs. 4, A and B; and S2, E–H). When chromosomes were pretreated with detergent to dissociate low-affinity interactions (Nozawa et al., 2010), wild-type spreads retained Aurora B staining at inner centromeres, whereas Bub1KD/KD spreads lost staining along arms and showed only very weak staining at inner centromeres (Figs. 4, C and D; and S2, E–H). Similarly, Aurora B localized to chromosome arms when intact cells were stained (Fig. S3, A and B). Importantly, total protein levels of Aurora B were unaffected by loss of Bub1 kinase activity (Fig. S3 C). We conclude that Aurora B associates weakly with chromosome arms in the absence of Bub1 kinase activity and fails to achieve stable inner centromeric loading.
Figure 4.
Bub1 kinase activity contributes to Aurora B accumulation at inner centromeres. (A) IF of spreads from unperturbed MEFs. Insets shows enlarged chromosomes from the boxed region (merged). Statistics indicate comparison with wild type. (B) Quantification of Aurora B localization (n = 3 MEF cell lines per genotype). Error bars indicate SEM. Statistics indicate comparison with wild type. (C) Same as A except with detergent pretreatment. (D) Same as B except with detergent pretreatment. (E) IF of mitotic MEFs. (F) Quantification of pH3T3 centromeric enrichment (n = 3 lines per genotype). Error bars indicate SEM. Statistics indicate comparison with wild type. *, P < 0.5; **, P < 0.1. KD, kinase dead. Bars: (yellow) 10 µm; (red) 1 µm.
Several recent studies have indicated that Aurora B localization to inner centromeres is mediated by histone H3T3 phosphorylation (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). Haspin, the kinase responsible for phosphorylation of H3T3, is a substrate of Aurora B, which led to the idea of a positive feedback loop between Haspin and Aurora B (Wang et al., 2011). Notably, pH3T3 dispersed along chromosomes in Bub1KD/KD MEFs rather than concentrating at inner centromeres (Fig. 4, E and F). Total pH3T3 levels and the temporal pattern of pH3T3 were unchanged in Bub1KD/KD cells (Fig. S3 D and not depicted). Aurora B localization was unchanged by Haspin overexpression in Bub1+/+ and Bub1KD/KD MEFs despite increased H3T3 phosphorylation (Fig. S3 E and not depicted), suggesting that pH3T3 density alone is insufficient to drive Aurora B accumulation.
Survivin has been proposed to function as the pH3T3-dependent inner centromeric receptor for Aurora B (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010), which prompted us to test whether survivin, like Aurora B, is unable to accumulate at inner centromeres in MEFs lacking Bub1 kinase activity. Endogenous survivin in Bub1+/+ and Bub1KD/KD MEFs showed accumulation at inner centromeres (Fig. S3 F). However, the staining efficiency was rather low irrespective of genotype, thus indicating the need for an alternative approach to detect survivin. To this end, we expressed HA-tagged survivin in Bub1+/+ and Bub1KD/KD MEFs. Nearly all wild-type MEFs showed accumulation of HA-survivin at inner centromeres of mitotic chromosomes (Fig. S3 F). An identical staining pattern was observed in about half of the Bub1KD/KD MEFs (Fig. S3 F). In contrast to survivin, Aurora B failed to accumulate at inner centromes, displaying its typical redistribution to chromosome arms. These findings indicate that Aurora B and survivin can exist in distinct mitotic protein complexes and support the idea that Aurora B delocalization in Bub1KD/KD cells is survivin independent. The other half of Bub1KD/KD cells showed diffuse staining of HA-survivin and Aurora B along chromosome arms, implying that the affinity of survivin at centromeric chromosomes weakened in the absence of Bub1 kinase activity (Fig. S3 H). Collectively, we conclude that Bub1 kinase activity is required for accumulation of Aurora B at inner centromeric regions.
Loss of Bub1 catalysis diminishes centromeric Aurora B activity
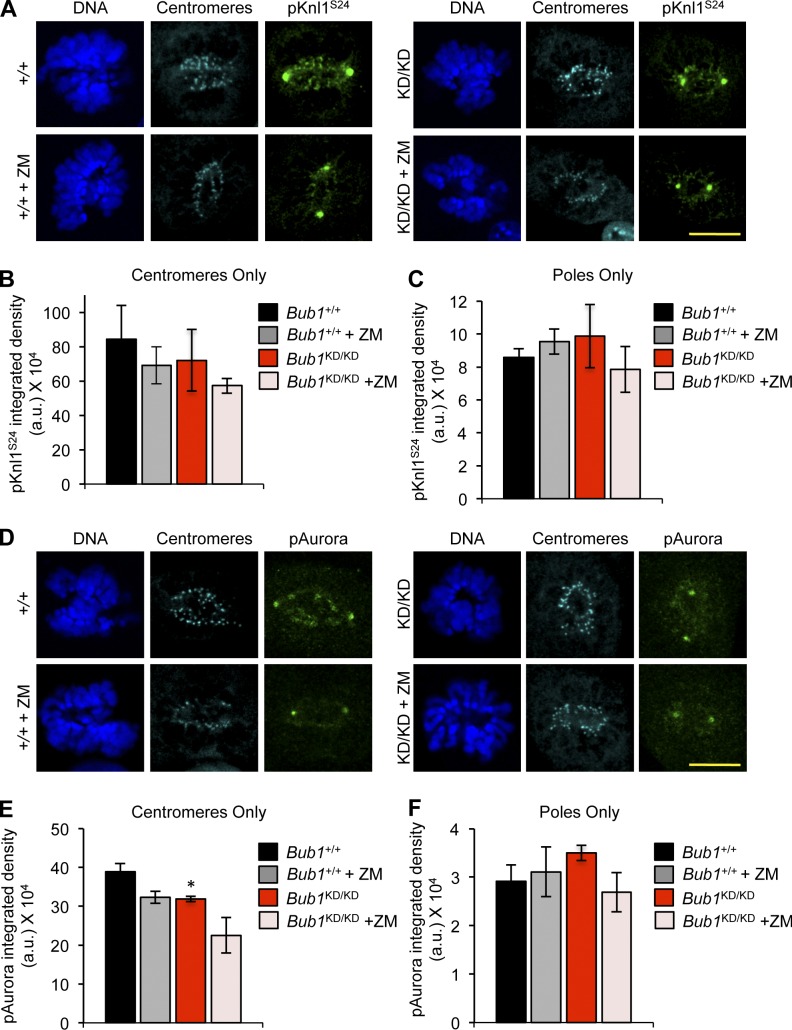
To assess whether loss of Bub1 kinase activity resulted in a decline of Aurora B kinase activity at inner centromeres, we stained Bub1KD/KD and wild-type MEFs for phosphorylated Knl1, a key kinetochore-associated substrate of Aurora B (Welburn et al., 2010). Although phosphorylated Knl1S24 was detectable at mitotic inner centromeres of Bub1KD/KD MEFs, its level was reduced compared with wild type (Fig. 5, A–C). Autophosphorylation of Aurora BT232 was diminished in Bub1KD/KD MEFs during prometaphase (Fig. 5, D–F; Yasui et al., 2004).
Figure 5.
Loss of Bub1 catalysis reduces inner centromeric Aurora B activity. (A) IF of mitotic MEFs after treatment with 1 µM ZM for 3 h. (B) Quantification of centromeric region (n = 3 MEF lines per genotype). Error bars indicate SEM. (C) Same as B except signal at spindle poles is quantified. (D) Same as A except with p-Aurora. Note that p-Aurora is a pan-Aurora antibody, and signal at poles is p-Aurora A, whereas signal at inner centromeres is p-Aurora B. (E) Same as B except with p-Aurora. Statistics indicate comparison with wild type without ZM. (F) Same as C but with p-Aurora. a.u., arbitrary unit; KD, kinase dead. *, P < 0.5. Bars, 10 µm.
The extent to which Knl1 and Aurora B phosphorylation was inhibited in Bub1KD/KD MEFs suggested that there might be residual Aurora B activity at inner centromeres. Consistently, treatment of Bub1KD/KD MEFs with 1 µM ZM further reduced Knl1 and Aurora B phosphorylation (Fig. 5, A–F). ZM also inhibited substrate phosphorylation in wild-type MEFs but not very strongly. To examine the efficacy of ZM in MEFs, we measured the level of reduction of another Aurora B substrate, histone H3S10, and compared it to that in HeLa cells, in which ZM is known to inhibit Aurora B robustly (Ditchfield et al., 2003; Girdler et al., 2008). Western blotting demonstrated that ZM dramatically reduced H3S10 phosphorylation in HeLa cells, but not in MEFs (Fig. S3, I and J), indicating that ZM is not nearly as effective in inhibiting Aurora B substrate phosphorylation in MEFs as in HeLa cells.
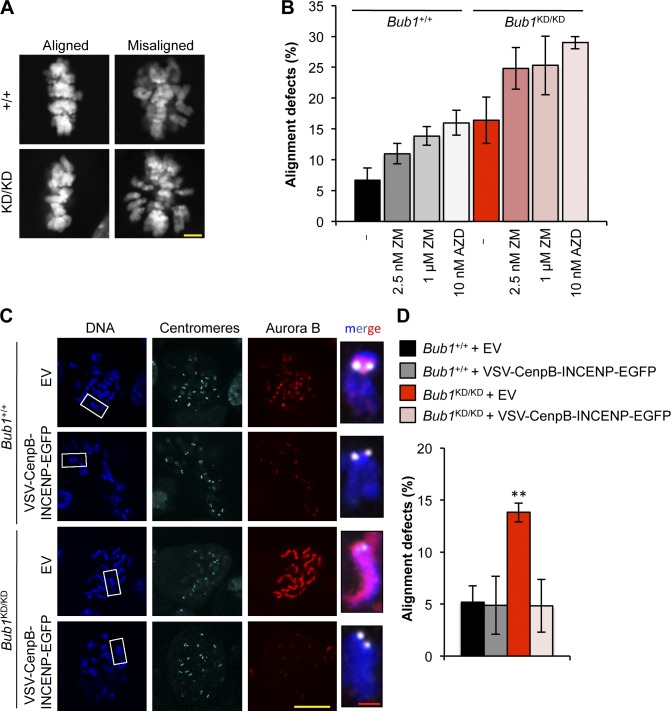
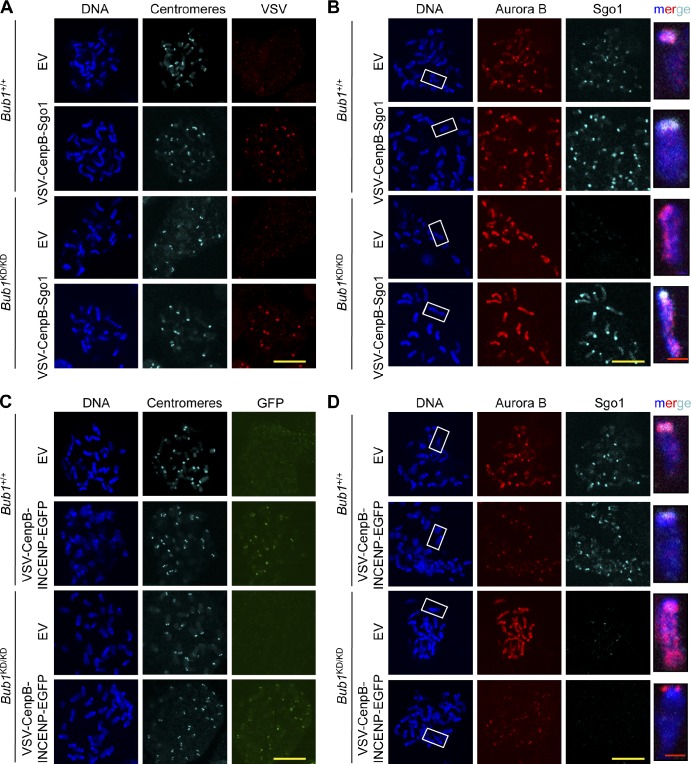
Next, we examined whether the reduction in Aurora B activity affects the efficiency of attachment error correction by performing a monastrol washout assay on Bub1KD/KD and wild-type MEFs (Lampson and Kapoor, 2005). In this assay, cells are arrested in monastrol, which causes monopolar spindles with reversible syntelic attachments. After monastrol removal, a bipolar spindle forms, and duplicated chromosomes align in the metaphase plate upon Aurora B–dependent correction of any syntelic attachments. Bub1KD/KD MEFs subjected to this assay had an increased incidence of chromosome alignment defects compared with wild-type MEFs (Fig. 6, A and B). Importantly, chemical inhibition of Aurora B with ZM or AZD1152-HQPA (hereafter AZD) escalated the alignment defects of Bub1KD/KD MEFs (Fig. 6 B), suggesting that Bub1 kinase–deficient MEFs have residual Aurora B–mediated error correction activity. To confirm that impaired targeting of Aurora B to inner centromeres causes chromosome misalignment in Bub1KD/KD cells, we forced Aurora B accumulation at centromeres by ectopically expressing vesicular stomatitis virus (VSV)–CenpB–inner centromere protein (INCENP)–EGFP (CenpB-INCENP hereafter) as previously reported (Liu et al., 2009). Expression of CenpB-INCENP in wild-type and Bub1KD/KD MEFs mediated Aurora B accumulation at centromeres (Fig. 6 C). Importantly, the chromosome misalignment defect of Bub1KD/KD MEFs observed in monastrol washout experiments was fully corrected by ectopic expression of CenpB-INCENP (Fig. 6 D). Together, the aforementioned data suggest that reduced Aurora B activity is responsible for the misalignment defects observed in Bub1KD/KD MEFs.
Figure 6.
Deregulated Aurora B activity promotes chromosome alignment defects in Bub1 kinase–deficient cells. (A) Representative images for cells with aligned or misaligned chromosomes after monastrol washout. (B) Analysis of chromosome alignment in cells that were treated with monastrol for 1 h and then with monastrol and MG132 for 1 h and released for 90 min into MG132 alone, with ZM or AZD (n = 3 independent MEF lines with ≥75 cells per group). Error bars indicate SEM. (C) IF of chromosome spreads from the indicated cells. Insets show enlarged chromosome from the boxed regions. (D) Analysis of chromosome alignment in cells released from monastrol as in B. Statistics indicate comparison with wild type with empty vector. EV, empty vector; KD, kinase dead. **, P < 0.1. Bars: (yellow) 10 µm; (red) 1 µm.
The Mad3 homology domain of Bub1 is required for robust Bub1 kinase activity
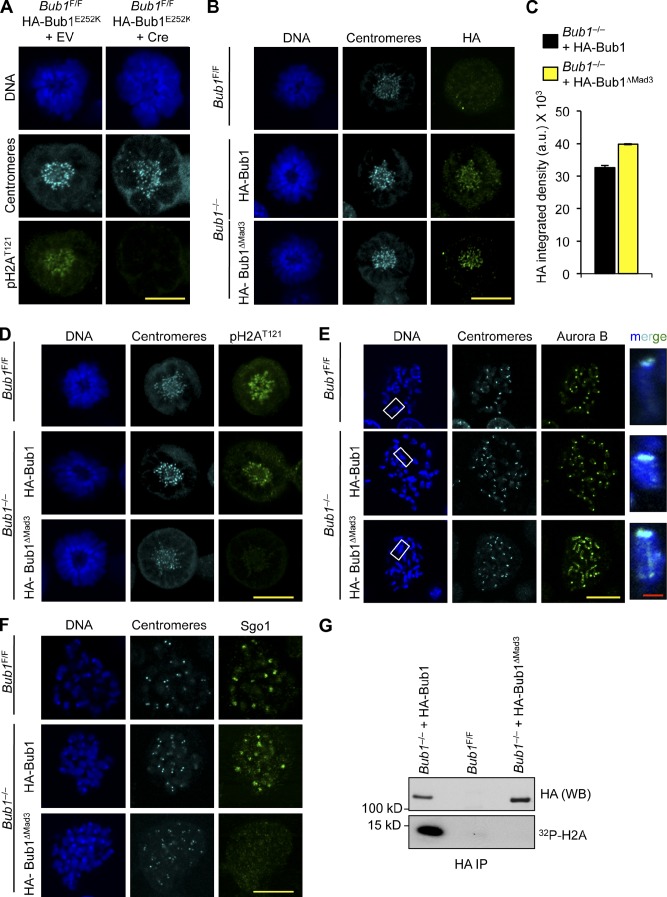
To determine whether kinetochore localization of Bub1 is a requirement for its enzymatic functions, we sought to (a) stably express Bub1 mutants that are unable to accumulate at kinetochores in Bub1 conditional knockout (Bub1F/F) MEFs (Fig. S4, A and B), (b) inactivate endogenous Bub1 with Cre-expressing lentivirus, and (c) monitor these cells for H2AT121 phosphorylation. The Gle2-binding sequence–like motif of Bub1, which binds Bub3, is known to be required for kinetochore localization (Taylor et al., 1998; Wang et al., 2001). We inactivated this motif by mutating the Bub1 cDNA sequence such that E252 was substituted to K (Bub1-E252K), thus preventing kinetochore accumulation (Fig. S4, C and D; Larsen et al., 2007; Krenn et al., 2012). The N-terminal Mad3 homology domain of Bub1, comprising the first 126 amino acids, has been implicated in kinetochore association through Knl1 binding (Kiyomitsu et al., 2007, 2011; Bolanos-Garcia et al., 2009; Klebig et al., 2009). We inactivated this domain by removing the Bub1 cDNA sequence encoding for the Mad3 homology domain (Bub1-ΔMad3). Consistent with a previous study (Perera et al., 2007), treatment of Bub1F/F MEFs with Cre-expressing lentivirus resulted in cell death within 5–6 d (unpublished data). Stable expression of full-length Bub1 cDNA with an N-terminal HA tag coding sequence before Cre expression fully rescued cell growth and survival, but the Bub1-E252K mutant did not (unpublished data). However, before cell death, cells undergoing mitosis in the absence of Bub1 lack phosphorylated histone H2A at kinetochore regions (Fig. S4 E). Similarly, cells expressing Bub1-E252K in the absence of endogenous Bub1 lack phosphorylated histone H2A at centromeres (Fig. 7 A), demonstrating that Bub1 kinetochore association is required for histone modification at this locale. On the other hand, expression of Bub1-ΔMad3 fully supported cell growth in the absence of endogenous Bub1 (unpublished data). In contrast to earlier studies (Kiyomitsu et al., 2007; Klebig et al., 2009), Bub1-ΔMad3, which binds Bub3, accumulates normally at kinetochores (Figs. 7, B and C; and S4 F). Another surprising observation was that, despite an intact kinase domain, H2AT121 phosphorylation was markedly diminished (Figs. 7 D and S4 G). Consistently, Aurora B distributed weakly along chromosome arms in Bub1−/− MEFs expressing Bub1-ΔMad3 (Figs. 7 E and S4 H), and Sgo1 kinetochore association was lost (Fig. 7 F). The same defects were observed in Bub1−/− MEFs expressing Bub1 lacking catalytic activity (Bub1-D892N) or the entire kinase domain (unpublished data).
Figure 7.
The N terminus of Bub1 is required for histone H2A phosphorylation. (A) IF of monastrol-blocked MEFs 4 d after infection with indicated viruses. (B) IF of monastrol-blocked MEFs. (C) Quantification of the HA signal, normalized from three independent experiments, is shown. Error bars indicate SEM. (D) IF of monastrol-blocked MEFs. (E) IF of spreads from colcemid-blocked MEFs. Staining was performed on spreads directly fixed in paraformaldehyde. Insets show enlarged chromosomes from the boxed regions. (F) IF of spreads from colcemid-blocked MEFs. (G) In vitro kinase assay for HA-Bub1 and HA-Bub1-ΔMad3. a.u., arbitrary unit; EV, empty vector; IP, immunoprecipitation; WB, Western blot. Bars: (yellow) 10 µm; (red) 1 µm.
Because deletion of the N terminus of Bub1 profoundly reduced H2A phosphorylation in vivo and because the kinase domain is intact, we monitored the in vitro kinase activity of HA-Bub1-ΔMad3. Bub1 was immunoprecipitated with the HA tag from Bub1−/− cells expressing HA-Bub1-ΔMad3 or HA-Bub1 and Bub1F/F cells as a control. Immunoprecipitates were incubated with recombinant histone H2A in the presence of γ-[32P]ATP. Whereas wild-type Bub1 was sufficient to phosphorylate H2A in vitro, there was little detectable incorporation of 32P into H2A with HA-Bub1-ΔMad3 (Fig. 7 G), indicating the N terminus is required for H2A phosphorylation in vitro. Collectively, these data uncover a novel requirement for Mad3 homology domain in activating Bub1 kinase activity. Finally, we note that consistent with in vitro kinase assays on Bub1 protein precipitated from wild-type and Bub1KD/KD MEFs (Fig. 1 I), HA-Bub1D892N precipitated from Bub1−/− MEFs with anti-HA antibody failed to phosphorylate histone H2A substrate in vitro (Fig. S4 I), providing additional evidence that Bub1D892N is catalytically deficient.
Phosphorylated histone H2A and H3 stably bind Aurora B but not Sgo1
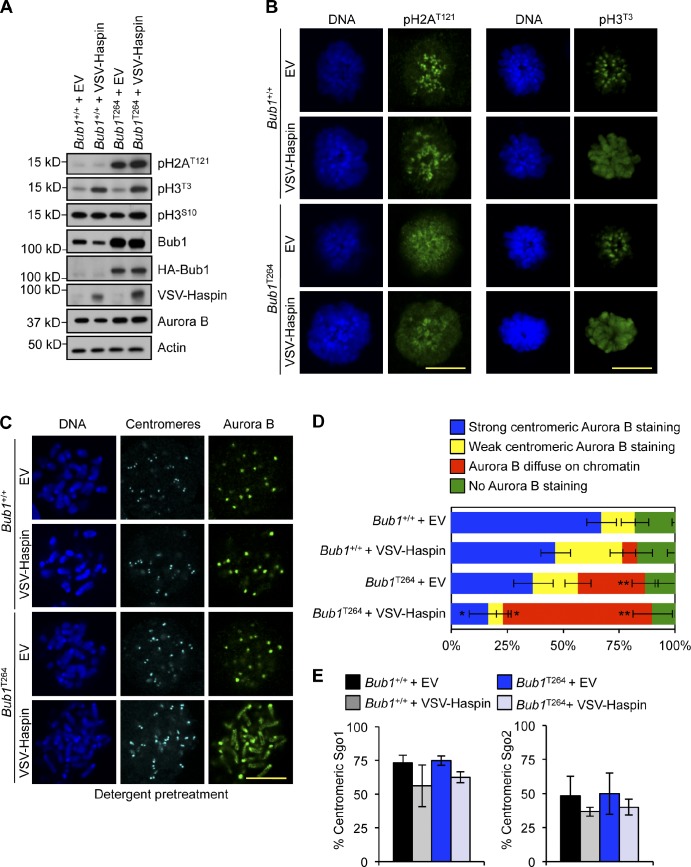
It has been proposed that H2A phosphorylation at T121 by Bub1 creates a mark for inner centromeric localization of Sgo1, in which one of its proposed functions is to load the chromosomal passenger complex (Kawashima et al., 2007; Yamagishi et al., 2010). We recently reported that phosphorylation of H2AT121 is not restricted to centromeres and spreads to chromosome arms in MEFs derived from Bub1 transgenic (Bub1T264) mice that overexpress wild-type Bub1 (Ricke et al., 2011). Interestingly, we observed that phosphorylated H2AT121 at chromosome arms of Bub1T264 MEFs is not sufficient to load Sgo1 or Aurora B. Several studies have pointed to the importance of H3T3 phosphorylation in inner centromeric Aurora B localization (Kelly et al., 2010; Wang et al., 2010, 2011; Yamagishi et al., 2010), which prompted us to examine whether phosphorylation of H2AT121 and H3T3 together might serve as a binding site for Sgo1, Sgo2, or Aurora B. To this end, we transduced wild-type and Bub1T264 MEFs with a lentivirus encoding doxycycline (DOX)-inducible Haspin kinase. Overexpression of Haspin in wild-type MEFs resulted in robust H3T3 phosphorylation along the entire chromosome but was insufficient to delocalize Sgo1, Sgo2, or Aurora B (Fig. 8). In contrast, overexpression of Haspin in Bub1T264 MEFs localized Aurora B in a detergent-resistant manner along chromosomes (Fig. 8 D), indicative of binding comparable with inner centromeric Aurora B. Strikingly, neither Sgo1 nor Sgo2 loaded onto chromosome arms (Fig. 8 E), suggesting that neither shugoshin serves as an adaptor for Aurora B. Collectively, these data indicate that phosphorylated H2AT121 and H3T3 act in concert to create a high-affinity binding site for shugoshin-independent loading of Aurora B at inner centromeres.
Figure 8.
Phosphorylated histone H3T3 and histone H2AT121 together serve as the minimal binding element for stable Aurora B association. (A) Western blot of lysates from MEF extracts. Actin was used as a loading control. (B) IF of monastrol-blocked MEFs. (C) IF of spreads from colcemid-blocked MEFs. Staining was performed on spreads that were first treated with detergent before fixation. (D) Quantification of Aurora B (n = 3 lines per group). Staining was performed on spreads with detergent before fixation. Statistics indicate comparison with wild type with empty vector. (E) Quantification of cells with centromeric Sgo1 or Sgo2. Error bars indicate SEM. EV, empty vector. *, P < 0.5; **, P < 0.1. Bars, 10 µm.
To obtain additional evidence for this hypothesis, we artificially targeted Sgo1 to centromeres by transducing wild-type and Bub1KD/KD MEFs with a lentivirus encoding DOX-inducible CenpB-Sgo1 fusion protein. As expected, Aurora B remained weakly associated with chromosome arms and failed to accumulate at centromeres of Bub1KD/KD MEFs despite centromeric localization of ectopically expressed CenpB-Sgo1 (Fig. 9, A and B). Furthermore, ectopically expressed CenpB-INCENP localized properly at centromeres and promoted Aurora B targeting to centromeres in Bub1KD/KD MEFs (Fig. 9, C and D) but did not restore Sgo1 localization (Fig. 9 D), underscoring that Sgo1 recruitment is Aurora B independent, at least in the absence of Bub1 kinase activity.
Figure 9.
Inner centomeric recruitment of Aurora B and Sgo1 lacks interdependence in the absence of Bub1 kinase activity. (A) IF of spreads from colcemid-blocked MEFs. (B) IF of spreads from colcemid-blocked MEFs. Chromosomes were stained for Aurora B (red), Sgo1 (cyan), and DNA (blue). Staining was performed on spreads directly fixed in paraformaldehyde. Insets shows enlarged chromosomes from boxed regions (merged). (C) IF of spreads from colcemid blocked MEFs. (D) IF of spreads from colcemid blocked MEFs. Staining was performed on spreads directly fixed in paraformaldehyde. Insets show enlarged chromosomes from boxed regions (merged). EV, empty vector. Bars: (yellow) 10 µm; (red) 1 µm.
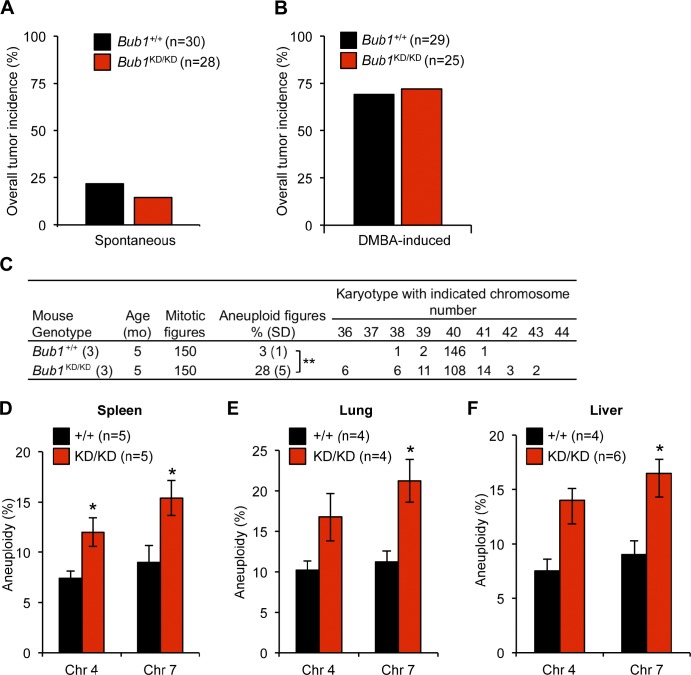
Bub1 kinase-dead mice harbor significant aneuploidy but are not tumor prone
Murine strains engineered with defects in chromosomal instability genes generally demonstrate increased susceptibility to tumor development (Ricke et al., 2008; Holland and Cleveland, 2009). More specifically, deregulating Bub1 expression promotes increased susceptibility to spontaneous tumor development, concomitant with increased aneuploidization (Jeganathan et al., 2007; Schliekelman et al., 2009; Ricke et al., 2011). To determine the extent to which Bub1 kinase-dead mice are prone to cancer, cohorts of Bub1+/+ and Bub1KD/KD mice were aged to 14–15 mo and screened for tumors. However, there was no difference in the incidence or tumor type of Bub1 kinase-dead mice compared with wild type (Fig. 10 A and not depicted). To further examine whether Bub1 kinase deficiency contributes to tumorigenesis, we challenged Bub1+/+ and Bub1KD/KD mice with a low dose of the carcinogen 7, 12-dimethylbenz(a)anthracene (DMBA) at postnatal day 3–5. Again, there was no difference in tumor incidence or burden (Fig. 10 B and not depicted). Chromosome counts on splenocytes from 5-mo-old animals confirmed that aneuploidy results in vivo as Bub1 kinase–deficient mice harbored 28% aneuploidy. Similarly, aneuploidy rates in spleen, lung, and liver of 14–15-mo-old Bub1KD/KD mice were significantly higher than those in wild type (Fig. 10, D–F). Collectively, this indicates, rather surprisingly, that despite substantial aneuploidy, Bub1 kinase–deficient mice are not prone to spontaneous or carcinogen-induced tumorigenesis.
Figure 10.
Bub1 kinase activity is required for protection from aneuploidization but not tumor suppression. (A) Spontaneous tumor incidence of 14-mo-old mice. (B) Overall tumor incidence of DMBA-treated mice. (C) Karyotype analysis of splenocytes from 5-mo-old mice. (D–F) Quantification of chromosome (Chr) 4 and 7 copies by interphase FISH. Statistics indicate comparison with wild type. KD, kinase dead. Error bars indicate SEM. *, P < 0.5; **, P < 0.1.
Bub1 kinase-dead males are subfertile
While intercrossing Bub1KD/KD mice, we noticed that litter sizes were frequently small, which prompted us to measure male and female fertility. Bub1KD/KD males and females were bred to wild-type females and males, respectively, and the number of litters and pups produced was recorded over 90 d. Although there was no difference in the mean litter size of Bub1KD/KD females compared with wild type, Bub1KD/KD males produced significantly fewer pups per litter (Fig. S5, A and B). Testis size and weight of Bub1KD/KD males was markedly reduced (Fig. S5 C), and Bub1KD/KD males contained on average 3.5-fold less spermatozoa in the cauda epididymis than wild type (Fig. S5 D). Histological analysis of testis showed that the diameter of seminiferous tubules in Bub1KD/KD mice was markedly reduced (Fig. S5 E). Spermatogenesis involves a mitotic proliferative phase that precedes meiosis to maintain spermatogonia stem cells and to amplify the spermatogonia progenitor cells that precede meiosis. Whereas E-cadherin, a spermatogonia stem cell marker, was unchanged in testis from Bub1 kinase–deficient mice, c-kit, a marker unique to spermatocyte progenitors, was markedly reduced (unpublished data), suggesting that the fertility defect is partially independent of meiosis. In addition, chromosome counts on primary and secondary spermatocytes showed substantial aneuploidy (Fig. S5, F and G). We propose that chromosome missegregation resulting from loss of Bub1 kinase activity affects male fertility by reducing sperm cell number and karyotypic integrity. Together, this indicates that whereas aneuploidization caused by Bub1 kinase deficiency in germ cells causes phenotypic abnormalities, aneuploidization in somatic cells does not.
Discussion
Tremendous progress has been made in elucidating the components and overall mode of action of the two key surveillance mechanisms that ensure proper chromosome segregation during mitosis, the mitotic checkpoint and the attachment error correction machinery. How individual components of these networks function at the modular level and how both networks operate in an integrated fashion remain poorly understood. Here, we studied the role of the cancer-critical mitotic regulator Bub1 at a modular level in primary cells by introducing a point mutation in the endogenous Bub1 locus that completely ablates Bub1 catalytic activity. Of critical importance is that the mutation did not influence Bub1 steady-state levels, as bidirectional deviations from normal Bub1 levels drive chromosome missegregation, aneuploidy, and tumorigenesis. Several key observations from the analysis of these mice and MEFs indicate that Bub1 kinase activity integrates attachment error correction and mitotic checkpoint signaling by controlling the localization and activity of Aurora B kinase through phosphorylation of histone H2A at threonine 121. First, cells lacking Bub1 kinase activity fail to concentrate Aurora B at inner centromeres. Second, phosphorylation of inner centromeric Aurora B substrates, such as Knl1 and Aurora B itself, is reduced when Bub1 kinase activity is lacking. Third, the error correction activity of Aurora B at inner centromeres is compromised in Bub1KD/KD cells and is corrected by artificial reconstitution of Aurora B at centromeres. Fourth, male mice deficient for Bub1 kinase activity are subfertile, similar to transgenic mice overexpressing a kinase-inactive Aurora B from a testis-specific promoter and Aurora B+/− mice (Kimmins et al., 2007; Fernández-Miranda et al., 2011).
How might Bub1’s actions promote Aurora B positioning at inner centromeres? We propose that phosphorylation of histones H2AT121 and H3T3 creates a high-affinity binding site for Aurora B. This model is strongly supported by our finding that ectopic phosphorylation of pH2AT121 and pH3T3 along chromosome arms by Bub1 and Haspin overexpression is sufficient for detergent-resistant Aurora B binding in the absence of Sgo1 or Sgo2. Although little is understood about Sgo2, several studies have implicated Sgo2 in Aurora B attachment error correction activities (Huang et al., 2007; Lee et al., 2008; Tsukahara et al., 2010). On the other hand, an earlier study has suggested that accumulation of Aurora B at inner centromeres is dependent on Sgo1’s recruitment to phosphorylated H2AT121 by Bub1 (Yamagishi et al., 2010). However, artificial reconstitution of centromeric Sgo1 in Bub1KD/KD MEFs failed to accumulate Aurora B at inner centromeres, supporting the view that Aurora B recruitment to inner centromeres is pH2AT121 dependent but Sgo1 independent. Our experiments reveal that accumulation of Sgo1 at centromeres is not required for centromeric accumulation of phosphatase PP2A or Aurora B. Previously, we reported that Bub1-mediated phosphorylation of H2A is necessary but not sufficient for Sgo1 recruitment (Ricke et al., 2011), indicating the involvement of another factor. An attractive possibility is that Bub1 mediates Sgo1 recruitment through its enzymatic and scaffold functions, although the role of Bub1-dependent mitotic Sgo1 is unclear.
The catalytic functions of both Bub1 and Haspin are sensitive to deregulation, as overexpression of either protein leads to aberrant phosphorylation of chromosomes. However, unlike ectopically expressed Haspin (Wang et al., 2011), overexpressed Bub1 does not decorate chromosome arms (Ricke et al., 2011), indicating that accumulation on chromosomes is not a strict requirement for aberrant histone phosphorylation. Importantly, the defects in chromosome alignment and Aurora B substrate phosphorylation that characterize Bub1 kinase–deficient MEFs were exacerbated upon treatment with Aurora inhibitors, indicating that loss of Bub1 kinase activity causes a partial rather than a complete loss of Aurora B error correction ability. The use of Aurora B inhibitors in our study further revealed that inhibitory potential of ZM is significantly lower in primary MEFs than in HeLa cells. The basis for this difference is currently unclear.
Here, we demonstrate that kinetochore localization of Bub1 is pivotal for H2AT121 phosphorylation at inner centromeres. Dependency of Bub1 on Bub3 for kinetochore targeting is well established (Taylor et al., 1998; Wang et al., 2001). However, recent studies have suggested a role for the N-terminal tetratricopeptide repeat (TPR) motif in kinetochore association of Bub1 through Knl1 (Kiyomitsu et al., 2007; Klebig et al., 2009). Unexpectedly, we found that a Bub1 truncation mutant lacking this domain concentrates normally at kinetochores in cells lacking endogenous Bub1, suggesting that Bub1 kinetochore accumulation is solely Bub3 dependent. This is consistent with a study that the Knl1–Bub1 interaction is not required for Bub1 kinetochore targeting (Krenn et al., 2012). Similarly, BubR1 also localizes properly at kinetochores when its TPR motif is mutated (Elowe et al., 2010; Lara-Gonzalez et al., 2011; Krenn et al., 2012).
Our discovery that the Bub1 N terminus is required for H2A phosphorylation raises the possibility that Bub1’s enzymatic activity is subject to regulation. Biochemical characterization identified two functions for the N-terminal TPR motif of Bub1, self-association and Knl1 interaction (Bolanos-Garcia et al., 2009). Whereas biochemically purified Knl1 does not impact Bub1 kinase activity in vitro (Krenn et al., 2012), Bub1 self-association through the N-terminal TPR domain could contribute to kinase activation in a manner similar to Mps1. Mps1 harbors a structurally similar TPR domain that drives dimerization, which is a prerequisite for robust Mps1 catalysis (Lee et al., 2012). Alternatively, Bub1 may undergo a conformational change before enzymatic activation in vivo, and the N terminus is necessary for this step. It is also possible that the N terminus is necessary to expose residues whose phosphorylation is a prerequisite for Bub1 activation (Chen, 2004).
Our analysis at the organismal level demonstrates that loss of Bub1 kinase activity causes significant aneuploidy but is dispensable for tumorigenesis. The observation that Bub1 kinase-dead mice were neither prone to spontaneous nor carcinogen-induced tumors is very surprising in light of the increased tumor susceptibility of Bub1 hypomorphic mice and the high degree of aneuploidization in Bub1KD/KD mice, which exceeds other chromosomal instability models prone to spontaneous tumorigenesis (van Ree et al., 2010; Ricke et al., 2011). Compared with Bub1 kinase deficiency, the distinguishing feature of Bub1 hypomorphism is defects in kinetochore assembly. Therefore, the novel observation of a multifunctional mitotic regulator that can cause aneuploidy through divergent mechanisms, not all of which are tumor suppressive, provides a mechanistic basis for the complexity of the aneuploidy paradox. Conversely, loss of Bub1 kinase activity causes subfertility selectively in males. In this manner, Bub1 kinase deficiency phenocopies mice with deregulated Aurora B activity (Kimmins et al., 2007; Fernández-Miranda et al., 2011). Because perturbation of Bub1 kinase activity is not overtly affecting viability and health in mice, this raises the possibility that loss of Bub1 kinase activity is a potential cause of reduced fertility in men.
In summary, by using a knockin approach, we made several novel and insightful findings on the role of Bub1 and its enzymatic activity. We find that histone H2A phosphorylation by Bub1 provides an essential mark for Aurora B inner centromeric recruitment, which is necessary for attachment error correction and robust mitotic checkpoint activity. These findings underscore the significance of studying mitotic regulators with a modular structure. Although variation in expression level has been studied in many mitotic regulators, precise mutants that disrupt a particular function are few (Huang et al., 2008; Li et al., 2009). This highlights the importance of understanding whether a particular chromosomal instability gene causes aneuploidy when defective and the mechanism underlying the aneuploidization.
Materials and methods
Mouse strains
The Bub1-D892N construct was generated by recombineering as previously described (Malureanu, 2011). The bacterial artificial chromosome was obtained from Thermo Fisher Scientific. The Bub1-flox construct was generated by genetic modification of the Bub1 hypomorphic targeting construct (Jeganathan et al., 2007). Mice were housed in a pathogen-free barrier environment. All mice were maintained on a mixed 129SV/E × C57BL/6 genetic background. The Bub1N allele was converted to the Bub1KD allele by crossing with protamine-Cre mice (O’Gorman et al., 1997). Males heterozygous for Bub1NEO and hemizygous for protamine-Cre were crossed with wild-type female mice to obtain Bub1+/KD mice. Mice heterozygous for Bub1KD were intercrossed to obtain Bub1KD/KD mice. The Bub1FNEO allele was converted to the Bub1F allele by crossing with mice harboring homozygous insertion of Flp recombinase at the Rosa26 locus (Raymond and Soriano, 2010). Mouse protocols were reviewed and approved by the Mayo Clinic institutional animal care and use committee. Mice at 14–15 mo of age were sacrificed, and their major organs were screened for overt tumors. For carcinogen-induced tumorigenesis, mice were given a single application of 50 µl of 0.5% DMBA to the dorsal surface on postnatal day 3–5 and sacrificed after 4 mo. Single-cell suspensions of various tissues were hybridized with probes to chromosomes 4 and 7 for interphase FISH (Baker et al., 2009). In brief, each tissue was minced using a dissociator (gentleMACS; Miltenyi Biotec) and enzymatically digested with liberase (Roche). At least 200 cells were analyzed per sample.
Fertility and histology
Male fertility was measured by breeding 2-mo-old Bub1+/+ and Bub1KD/KD male mice to 2-mo-old wild-type females for 3 mo. Female fertility was measured by breeding 2-mo-old Bub1+/+ and Bub1KD/KD female mice to 2-mo-old wild-type males for 3 mo. For histology, testes were fixed for 48 h in modified Davidson’s fluid, rinsed, and stored in PBS. For chromosome counts on spermatocytes, testes were minced, swelled with 75 mM KCl, fixed, and stained with Giemsa (Jin et al., 2010).
Generation and culture of MEFs
MEFs were generated at embryonic day 13.5 and cultured as previously described (Babu et al., 2003). At least three independently generated MEF lines per genotype were used. Mitotic MEFs were harvested after either preincubation with nocodazole or taxol. MEFs grown in the presence of 100 ng/ml nocodazole (Sigma-Aldrich) for 4 h were collected after shake off. Alternatively, cells were starved from serum (0.1% FBS) for 12 h. 12 h after release into 10% FBS containing DMEM, 0.5 µM taxol was added, and cells were harvested 6 h later. All experiments with Bub1KD/KD MEFs were performed with primary MEFs from passages 3–4. All experiments with Bub1F/F cells were performed with MEFs that were immortalized by expression of SV40 large T antigen.
Statistical analyses
Prism software (GraphPad Software) was used for the generation of all analysis. Mann-Whitney tests were used for pairwise significance analysis in Figs. 10 (D–F) and S5 A. An unpaired t test was used for comparisons in the following figures: Figs. 2 (A, C, D, and E), 3 (B and F), 4 (B, D, and F), 5 E, 6 D, 8 (D and E), S2 (B, E, and F), S3 G, and S5 (C, D, and F). All graphs are indicated with the significance score of *, P < 0.5; **, P < 0.1; and ***, P < 0.001.
Indirect immunofluorescence (IF), chromosome spreads, and Western blotting
For IF on chromosome spreads, cells were swelled in 75 mM KCl and cytospun onto coverslips. In all cases, DNA was visualized with Hoechst. For Aurora B staining, cells that were detergent extracted were treated with 0.1% Triton X-100/PBS for 2 min, fixed with 4% PFA/PBS for 12 min, and treated with 0.1% Triton X-100/PBS for 10 min. For Aurora B and Sgo1 staining, cells were fixed with 4% PFA/PBS for 12 min and extracted with 0.1% Triton X-100/PBS for 10 min. A laser-scanning microscope (LSM 510 or LSM 780; Carl Zeiss) with an inverted microscope (Axiovert 100M; Carl Zeiss) with a C-Apochromat 100× oil immersion objective was used to analyze immunostained cells and capture images. ImageJ (National Institutes of Health) was used for quantification by first converting to an 8-bit grayscale and then tracing the cell edges using the freehand tool and calculating the mean pixel intensity within the marked area. The integrated density measures signal strength (mean pixel intensity multiplied by area). Monastrol-blocked cells were treated for 90 min at 100 µM and fixed using the kinetochore protocol. Standard fixations for kinetochore stainings were 1% paraformaldehyde for 5 min at room temperature. For the monastrol washout experiment, 100 µM monastrol was added for 60 min, after which, 10 µM MG132 was added for 60 min. Cells were then released for 90 min into 10 µM MG132 alone. For chromosome spreads, cells were either unperturbed or 0.5 µg/ml colcemid, 100 ng/ml nocodazole, or 0.5 µM taxol treated for 4 h before swelling as noted. For quantification, a minimum of 30 cells per group was analyzed.
Primary antibodies for IF were as follows: Aurora B (BD), pThr232-Aurora (Cell Signaling Technology), Bub1 (rabbit; Jeganathan et al., 2007); BubR1 (BD), centromeres (Antibodies Inc.), Cdc20 (Santa Cruz Biotechnology, Inc.), Cenp-E (rabbit; D. Cleveland, Ludwig Institute for Cancer Research, La Jolla, CA); HA (16B12 [Covance] or 3F10 [Roche]), pH2AT121 (Active Motif), pH3T3 (EMD Millipore), pKnl1S24 (rabbit; I. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, MA), Mad1 (rabbit; J.M. van Deursen); Mad2 (rabbit; D. Cleveland), Sgo1 (rabbit; S. Taylor, University of Manchester, Manchester, England, UK), Sgo2 (rabbit; Y. Watanabe, University of Tokyo, Tokyo, Japan), and survivin (Novus Biologicals). Primary antibodies for Western blotting were as follows: actin (Sigma-Aldrich), Aurora B (BD), Bub1 (rabbit; Jeganathan et al., 2007), BubR1 (BD), pH2AT121 (rabbit; Y. Watanabe), pH3T3 (EMD Millipore), pH3S10 (EMD Millipore), H2A (Abcam), HA (Covance), INCENP (Abcam), Sgo1 (Abcam), and VSV (Sigma-Aldrich).
Live-cell imaging microscopy
For chromosome segregation analysis, MEFs positive for H2B-mRFP were followed through an unchallenged mitosis at interframe intervals of 3 min as previously described (Jeganathan et al., 2007). Primary MEFs positive for H2B-mRFP were seeded onto 35-mm glass-bottomed dishes. All experiments were performed using a laser-scanning system (Axio Observer.Z1; Carl Zeiss) with CO2 Module S, TempModule S, Heating Unit XL S, Plan Apochromat 63×, NA 1.4 oil differential interference contrast III objective, a camera (AxioCam MRm; Carl Zeiss), and AxioVision 10.6 software (Carl Zeiss). The imaging medium was kept at 37°C. Colcemid and taxol challenge assays were performed as previously described (van Ree et al., 2010). In brief, cells undergoing NEBD were marked and monitored at 30-min intervals. The duration of arrest is defined by the interval between NEBD, and chromatin decondensation was then calculated. The time at which 50% of the cells had exited mitosis was compared. Where noted, 1 µM ZM was added at the same time as spindle poisons. At least three independent lines per genotype were used.
In vitro kinase assay
Bub1 was immunoprecipitated using the HA epitope in NP-40 lysis buffer (0.1% NP-40 and 10% glycerol in PBS with protease and phosphatase inhibitor pellets). Beads were serially washed with kinase buffer, and 1 µg histone H2A (New England Biolabs, Inc.) and γ-[32P]ATP were added. The reactions were incubated at 30°C for 30 min, quenched with SDS Laemmli buffer, and separated by SDS-PAGE. Kinase buffer was 50 mM Tris-HCl, pH 7.7, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM ATP, and 10 mM β-glycerophosphate.
Plasmid constructs
Stable expression of HA-tagged wild-type murine Bub1 cDNA was achieved by Tol2-based transposition using pKTol2C-hygromycin and pKC-Tol2 as previously described (Hamada et al., 2011). All Bub1 mutants were cloned into the retroviral vector pMSCV-puromycin with a HA epitope, and stable cell lines were generated as previously described (Hamada et al., 2011). Epitope-tagged survivin expression was attained by cloning a HA-tagged human survivin (cDNA obtained from J. Higgins, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) into TSIN-puro2 (van Ree et al., 2010). DOX-inducible Haspin expression was attained by cloning a VSV-tagged human Haspin (cDNA obtained from J. Higgins) into pTRIPZ3 (van Ree et al., 2010). DOX-inducible expression of VSV-CenpB-Sgo1 was attained by cloning into pTRIPZ3 (Sgo1 cDNA obtained from W. Dai, New York University School of Medicine, Tuxedo, NY; CenpB cDNA obtained from S. Lens, University Medical Center Utrecht, Utrecht, Netherlands). DOX-inducible expression of VSV-CenpB-INCENP-EGFP was attained by cloning the construct into pTRIPZ3 (construct obtained from S. Lens; Liu et al., 2009).
Online supplemental material
Fig. S1 shows the analysis of mitotic checkpoint functions in wild-type and Bub1 kinase–deficient MEFs. Fig. S2 shows the localization of Sgo1, Sgo2, and Aurora B in wild-type and Bub1 kinase–deficient MEFs. Fig. S3 shows the localization of Aurora B in wild-type and Bub1 kinase–deficient MEFs after Haspin overexpression. Fig. S4 describes the generation of cell lines that express Bub1 mutants and harbor mutant endogenous Bub1 alleles with loxP sites. Fig. S5 shows how Bub1 kinase deficiency influences fertility. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205115/DC1.
Acknowledgments
We gratefully acknowledge reagents from D. Cleveland, I. Cheeseman, W. Dai, S. Lens, J. Higgins, S. Taylor, and Y. Watanabe. We thank W. Zhou and M. Li from the gene knockout facility for help with mouse generation, D. Baker and P. Galardy for critical reading of the manuscript, and M. Hamada for assistance with cloning. We acknowledge the Microscopy and Cell Analysis Core Facility for microscope usage.
This work was supported by the National Institutes of Health (CA126828 and CA96985 to J.M. van Deursen).
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DMBA
- 12-dimethylbenz(a)anthracene
- DOX
- doxycycline
- ES
- embryonic stem
- IF
- immunofluorescence
- INCENP
- inner centromere protein
- MEF
- mouse embryonic fibroblast
- NEBD
- nuclear envelope breakdown
- PMSCS
- premature sister chromatid separation
- TPR
- tetratricopeptide repeat
- VSV
- vesicular stomatitis virus
- ZM
- ZM447439
References
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 10.1083/jcb.200211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Malureanu L., Perez-Terzic C., Terzic A., van Deursen J.M. 2006. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172:529–540 10.1083/jcb.200507081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jin F., Jeganathan K.B., van Deursen J.M. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 16:475–486 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Kiyomitsu T., D’Arcy S., Chirgadze D.Y., Grossmann J.G., Matak-Vinkovic D., Venkitaraman A.R., Yanagida M., Robinson C.V., Blundell T.L. 2009. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure. 17:105–116 10.1016/j.str.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y., Salic A., Dasso M., Arnaoutov A. 2007. Bub1 is essential for assembly of the functional inner centromere. J. Cell Biol. 176:919–928 10.1083/jcb.200609044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H. 2004. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint. EMBO J. 23:3113–3121 10.1038/sj.emboj.7600308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E., Chen R.H. 2003. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 5:748–753 10.1038/ncb1022 [DOI] [PubMed] [Google Scholar]
- Dai J., Sullivan B.A., Higgins J.M. 2006. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 11:741–750 10.1016/j.devcel.2006.09.018 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., Nigg E.A. 2010. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell Sci. 123:84–94 10.1242/jcs.056507 [DOI] [PubMed] [Google Scholar]
- Fernández-Miranda G., Trakala M., Martín J., Escobar B., González A., Ghyselinck N.B., Ortega S., Cañamero M., Pérez de Castro I., Malumbres M. 2011. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 138:2661–2672 10.1242/dev.066381 [DOI] [PubMed] [Google Scholar]
- Fernius J., Hardwick K.G. 2007. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 3:e213 10.1371/journal.pgen.0030213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdler F., Sessa F., Patercoli S., Villa F., Musacchio A., Taylor S. 2008. Molecular basis of drug resistance in aurora kinases. Chem. Biol. 15:552–562 10.1016/j.chembiol.2008.04.013 [DOI] [PubMed] [Google Scholar]
- Hamada M., Haeger A., Jeganathan K.B., van Ree J.H., Malureanu L., Wälde S., Joseph J., Kehlenbach R.H., van Deursen J.M. 2011. Ran-dependent docking of importin-β to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 194:597–612 10.1083/jcb.201102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10:478–487 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J.B., Liu S.T., Kao G.D., Muschel R., Chan G.K., Yen T.J. 2007. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 177:413–424 10.1083/jcb.200701122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Andreu-Vieyra C.V., York J.P., Hatcher R., Lu T., Matzuk M.M., Zhang P. 2008. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol. 6:e15 10.1371/journal.pbio.0060015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V.B., Stern B.M., Murray A.W. 2005. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 307:130–133 10.1126/science.1101366 [DOI] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Hamada M., Malureanu L., Jeganathan K.B., Zhou W., Morbeck D.E., van Deursen J.M. 2010. Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet. 6:e1001147 10.1371/journal.pgen.1001147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- Kang J., Yang M., Li B., Qi W., Zhang C., Shokat K.M., Tomchick D.R., Machius M., Yu H. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell. 32:394–405 10.1016/j.molcel.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis V.L., Galova M., Rabitsch K.P., Gregan J., Nasmyth K. 2004. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 14:560–572 10.1016/j.cub.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Tsukahara T., Langegger M., Hauf S., Kitajima T.S., Watanabe Y. 2007. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 21:420–435 10.1101/gad.1497307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S., Crosio C., Kotaja N., Hirayama J., Monaco L., Höög C., van Duin M., Gossen J.A., Sassone-Corsi P. 2007. Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol. Endocrinol. 21:726–739 10.1210/me.2006-0332 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15:353–359 10.1016/j.cub.2004.12.044 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 441:46–52 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., Yanagida M. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011 10.1128/MCB.00815-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1–kinetochore subunit Knl1 interaction. J. Cell Biol. 196:451–467 10.1083/jcb.201110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98 10.1038/ncb1208 [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Scott M.I., Diez M., Sen O., Taylor S.S. 2011. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 124:4332–4345 10.1242/jcs.094763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N.A., Al-Bassam J., Wei R.R., Harrison S.C. 2007. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc. Natl. Acad. Sci. USA. 104:1201–1206 10.1073/pnas.0610358104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kitajima T.S., Tanno Y., Yoshida K., Morita T., Miyano T., Miyake M., Watanabe Y. 2008. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 10:42–52 10.1038/ncb1667 [DOI] [PubMed] [Google Scholar]
- Lee S., Thebault P., Freschi L., Beaufils S., Blundell T.L., Landry C.R., Bolanos-Garcia V.M., Elowe S. 2012. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J. Biol. Chem. 287:5988–6001 10.1074/jbc.M111.307355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang X., Wei Z., York J.P., Zhang P. 2009. Loss of spindle assembly checkpoint–mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 185:983–994 10.1083/jcb.200904020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A. 2011. Targeting vector construction through recombineering. Methods Mol. Biol. 693:181–203 10.1007/978-1-60761-974-1_11 [DOI] [PubMed] [Google Scholar]
- McGuinness B.E., Hirota T., Kudo N.R., Peters J.M., Nasmyth K. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3:e86 10.1371/journal.pbio.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness B.E., Anger M., Kouznetsova A., Gil-Bernabé A.M., Helmhart W., Kudo N.R., Wuensche A., Taylor S., Hoog C., Novak B., Nasmyth K. 2009. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr. Biol. 19:369–380 10.1016/j.cub.2009.01.064 [DOI] [PubMed] [Google Scholar]
- Nozawa R.S., Nagao K., Masuda H.T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. 2010. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12:719–727 10.1038/ncb2075 [DOI] [PubMed] [Google Scholar]
- O’Gorman S., Dagenais N.A., Qian M., Marchuk Y. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 94:14602–14607 10.1073/pnas.94.26.14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D., Taylor S.S. 2010. Sgo1 establishes the centromeric cohesion protection mechanism in G2 before subsequent Bub1-dependent recruitment in mitosis. J. Cell Sci. 123:653–659 10.1242/jcs.059501 [DOI] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 13:566–579 10.1016/j.devcel.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493 10.1016/j.tcb.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Qi W., Yu H. 2007. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 282:3672–3679 10.1074/jbc.M609376200 [DOI] [PubMed] [Google Scholar]
- Raymond C.S., Soriano P. 2010. ROSA26Flpo deleter mice promote efficient inversion of conditional gene traps in vivo. Genesis. 48:603–606 10.1002/dvg.20659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., van Ree J.H., van Deursen J.M. 2008. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 24:457–466 10.1016/j.tig.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., Jeganathan K.B., van Deursen J.M. 2011. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J. Cell Biol. 193:1049–1064 10.1083/jcb.201012035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Gálová M., Petronczki M., Gregan J., Cetin B., et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441:53–61 10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Schliekelman M., Cowley D.O., O’Quinn R., Oliver T.G., Lu L., Salmon E.D., Van Dyke T. 2009. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 69:45–54 10.1158/0008-5472.CAN-07-6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R.H. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250 10.1083/jcb.153.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. 2004a. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 16:387–397 10.1016/j.molcel.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Tang Z., Sun Y., Harley S.E., Zou H., Yu H. 2004b. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 101:18012–18017 10.1073/pnas.0408600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H. 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 10:575–585 10.1016/j.devcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1–11 10.1083/jcb.142.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395 [DOI] [PubMed] [Google Scholar]
- Torres E.M., Williams B.R., Amon A. 2008. Aneuploidy: cells losing their balance. Genetics. 179:737–746 10.1534/genetics.108.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- van Ree J.H., Jeganathan K.B., Malureanu L., van Deursen J.M. 2010. Overexpression of the E2 ubiquitin–conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 188:83–100 10.1083/jcb.200906147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M., Higgins J.M. 2011. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21:1061–1069 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Babu J.R., Harden J.M., Jablonski S.A., Gazi M.H., Lingle W.L., de Groen P.C., Yen T.J., van Deursen J.M. 2001. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J. Biol. Chem. 276:26559–26567 10.1074/jbc.M101083200 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong O.K., Fang G. 2006. Loading of the 3F3/2 antigen onto kinetochores is dependent on the ordered assembly of the spindle checkpoint proteins. Mol. Biol. Cell. 17:4390–4399 10.1091/mbc.E06-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Cetin B., Anger M., Cho U.S., Helmhart W., Nasmyth K., Xu W. 2009. Structure and function of the PP2A-shugoshin interaction. Mol. Cell. 35:426–441 10.1016/j.molcel.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. 2004. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 279:12997–13003 10.1074/jbc.M311128200 [DOI] [PubMed] [Google Scholar]