Postsynaptic Farp1 helps to assemble a synaptic protein complex that promotes F-actin assembly and spine formation, and modulates active zones.
Abstract
Synaptic adhesion organizes synapses, yet the signaling pathways that drive and integrate synapse development remain incompletely understood. We screened for regulators of these processes by proteomically analyzing synaptic membranes lacking the synaptogenic adhesion molecule SynCAM 1. This identified FERM, Rho/ArhGEF, and Pleckstrin domain protein 1 (Farp1) as strongly reduced in SynCAM 1 knockout mice. Farp1 regulates dendritic filopodial dynamics in immature neurons, indicating roles in synapse formation. Later in development, Farp1 is postsynaptic and its 4.1 protein/ezrin/radixin/moesin (FERM) domain binds SynCAM 1, assembling a synaptic complex. Farp1 increases synapse number and modulates spine morphology, and SynCAM 1 requires Farp1 for promoting spines. In turn, SynCAM 1 loss reduces the ability of Farp1 to elevate spine density. Mechanistically, Farp1 activates the GTPase Rac1 in spines downstream of SynCAM 1 clustering, and promotes F-actin assembly. Farp1 furthermore triggers a retrograde signal regulating active zone composition via SynCAM 1. These results reveal a postsynaptic signaling pathway that engages transsynaptic interactions to coordinate synapse development.
Introduction
Synapse formation in the brain involves concerted steps. Axons and dendrites of developing neurons interact through exploratory filopodia (Ziv and Smith, 1996; Fiala et al., 1998), and contact triggers cytoskeletal rearrangements, resulting in shorter and wider filopodia as stable synapses form (Hotulainen and Hoogenraad, 2010). Adhesion molecules guide these stages, assembling into transsynaptic complexes to regulate synapse number and morphology (Missler et al., 2012). These parameters are critical for neuronal connectivity (Kasai et al., 2003; Chklovskii et al., 2004; Yuste, 2011).
The actin cytoskeleton is prominent in dendritic spines, the postsynaptic specializations of mature excitatory synapses, and shapes these protrusions, anchors receptors, and participates in signaling (Okamoto et al., 2004; Frost et al., 2010). Spine actin is highly dynamic (Fischer et al., 1998), and its reorganization contributes to the formation and structural plasticity of spines (Bonhoeffer and Yuste, 2002). Regulators of postsynaptic actin include members of the Rho GTPase family—RhoA, Rac1, and Cdc42—that have distinct functions in modulating spine turnover and morphology (Tashiro et al., 2000; Tada and Sheng, 2006). Cell surface interactions can activate theses GTPases, notably via Ephrin-B receptors that bind guanine nucleotide exchange factors (GEFs) and, additionally, promote kinase signaling (Penzes et al., 2003; Moeller et al., 2006; Tolias et al., 2007).
The understanding of synapse organization will benefit from additional insight into the signaling pathways underlying dendritic contact exploration and spine development. To identify novel regulators of synapse formation, we focused on synaptic cell adhesion molecule 1 (SynCAM 1)-mediated synaptogenesis. SynCAM 1 (also known as Cadm1 and nectin-like 2 protein) is well-suited to study synaptic signaling because it first promotes excitatory synapse numbers and then acts in the mature brain to maintain this increase (Biederer et al., 2002; Fogel et al., 2007; Robbins et al., 2010). Further, it has an intracellular motif predicted to interact with 4.1 protein/ezrin/radixin/moesin (FERM) domains present in cytoskeletal regulators (Biederer, 2006).
In an unbiased proteomic analysis of synaptic membranes from SynCAM 1 knockout (KO) mice, we have now identified FERM, Rho/ArhGEF, and Pleckstrin domain protein 1 (Farp1) as a novel synapse-organizing molecule that binds via its FERM domain to the cytosolic tail of SynCAM 1. Functional studies revealed that Farp1 promotes the structural dynamics of dendritic filopodia and their stability early in development. In mature neurons, Farp1 is enriched at postsynaptic sites and regulates the number of spines in dissociated neurons and organotypic slice culture. Notably, SynCAM 1 requires Farp1 to promote synapse formation, and the synaptogenic activity of Farp1 is reduced in absence of SynCAM 1. Biochemical assays and live imaging of an optical probe demonstrate that Farp1 specifically binds the GTPase Rac1 and activates it in postsynaptic protrusions. In turn, Farp1 increases F-actin polymerization in spine heads. Moreover, SynCAM 1 and postsynaptic Farp1 signal retrogradely across the synaptic cleft to modulate the composition of presynaptic active zones. These results identify a novel signaling pathway that coordinates synaptic adhesion and pre- and postsynaptic organization.
Results
Proteomic identification of Farp1
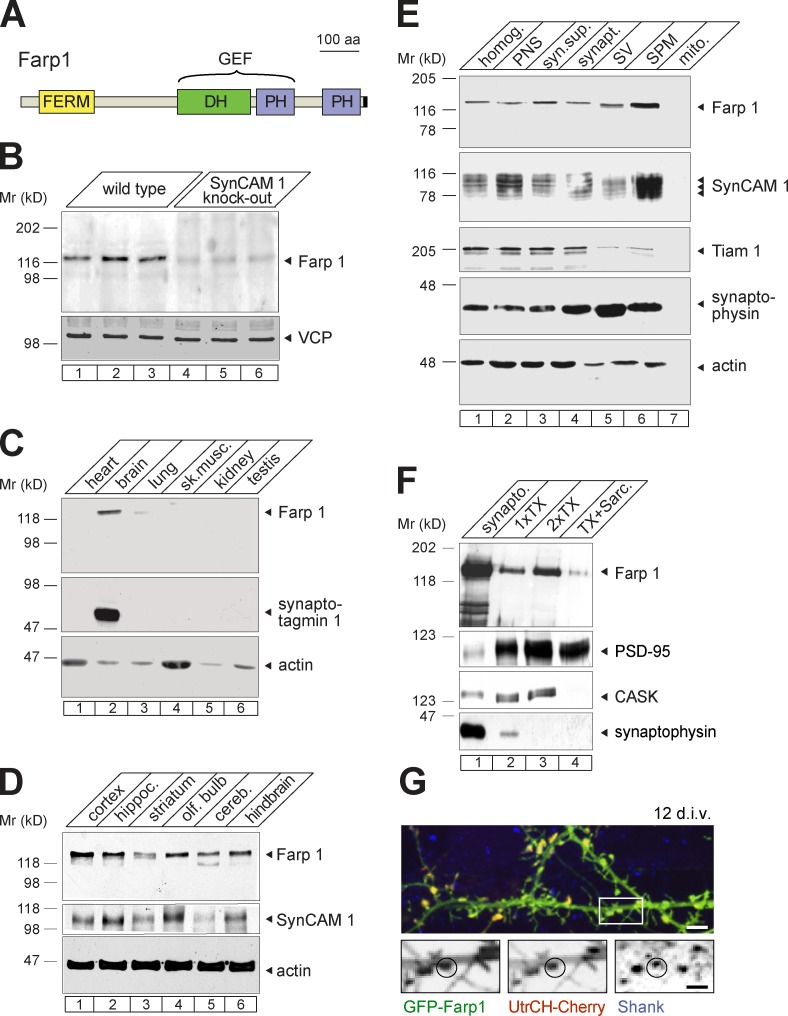
We performed a proteomic screen to compare the composition of synaptic membranes from forebrains of KO mice lacking SynCAM 1 (Robbins et al., 2010) versus wild-type (WT) littermates. This approach followed the rationale that intracellular synaptogenic signaling partners of SynCAM 1 may be recruited to or stabilized at synaptic membranes by this adhesion molecule, resulting in lower levels of such partners at synapses lacking SynCAM 1. Isobaric tagging for relative and absolute quantitation (iTRAQ) mass spectrometry identified 24 proteins that increased above a 1.3-fold cutoff in SynCAM 1 KO synaptic plasma membranes compared with WT. These hits included neurexin 1, neuroligin 2, and EphA4, synapse-organizing proteins that may be increased to compensate for the loss of SynCAM 1. Conversely, nine proteins were reduced below a 0.7-fold cutoff in SynCAM 1 KO synapses. Among them, Farp1 was selected for further analysis because of the high degree of reduction by 54% approximated by mass spectrometry, and its domain organization. Farp1 contains a FERM domain (Koyano et al., 1997) that is present in cytoskeletal regulators (Hoover and Bryant, 2000), together with Dbl oncogene homology (DH) and pleckstrin homology (PH) domains characteristic of GEFs that activate Rho family GTPases (Fig. 1 A; Hart et al., 1994). We generated an antibody that recognizes Farp1 in immunoblotting (Fig. S1). Quantitative analysis of hippocampal homogenates showed that Farp1 is reduced in SynCAM 1 KO mice by 79 ± 22% (P = 0.02; Fig. 1 B), validating its proteomic identification.
Figure 1.
Proteomic identification of Farp1, a synaptic protein reduced in SynCAM 1 KO mice. (A) Organization of Farp1 into FERM, DH, and PH domains. DH-PH domains are characteristic of GEFs. Black rectangle, epitope detected by the antibody. (B) Quantitative immunoblotting of SynCAM 1 KO hippocampi confirms Farp1 reduction compared with WT mice. Equal protein amounts were loaded. For quantification described in the text, signals were normalized to the loading control VCP. (C) Farp1 is enriched in brain determined by immunoblotting of equal protein amounts of adult rat tissues. Synaptotagmin 1 and actin were loading controls. (D) Farp1 expression throughout brain shown by immunoblotting of equal protein amounts. (E) Fractionation of P13 rat forebrain. Farp1 enriches in synaptic plasma membranes (SPM), similar to SynCAM 1. Farp1 in the crude synaptic vesicle (SV) preparation is presumably caused by nonvesicular content (Fogel et al., 2007). PNS, postnuclear supernatant; syn. sup., synaptosomal supernatant; synapt., synaptosomes. (F) Synaptosomal extracts prepared at P16 by sequential detergent extraction contain Farp1. A fraction is present in PSDs. PSD-95 served as a positive control and CASK and synaptophysin served as negative controls. (G) Farp1 localizes to dendritic protrusions. Confocal image of dissociated hippocampal neurons expressing GFP-Farp1 (green) at 12 div. Colocalization with the F-actin probe UtrCH-Cherry (red) and postsynaptic Shank (blue) is marked by circles. Single channels from the boxed area are enlarged below. Bars: (top) 5 µm; (insets) 2.5 µm.
Farp1 promotes dendrite outgrowth in motor neurons of the chick spinal cord (Zhuang et al., 2009). However, the expression, interactions, and functions of Farp1 protein in the mammalian central nervous system were unknown. Immunoblotting showed that Farp1 is most prominent in the brain (Fig. 1 C) and is expressed across brain regions (Fig. 1 D). At postnatal day 13 (P13), when synapse formation in rat hippocampus is rapid (Fiala et al., 1998), Farp1 was found in synaptosomes and enriched in synaptic plasma membranes, cofractionating with SynCAM 1 (Fig. 1 E, lanes 4 and 6). Farp1 was present in Triton X-100–resistant fractions from synaptosomes, which is indicative of a cytoskeletal association, and a small fraction of Farp1 cofractionated with postsynaptic densities (Fig. 1 F).
Our antibody detected Farp1 when it was overexpressed in neurons (Fig. S1), but was not suitable for staining endogenous protein. To localize Farp1, we generated a full-length construct tagged at its amino terminus with GFP. In dissociated cultures of rat hippocampal neurons at 15 d in vitro (div), GFP-Farp1 was present in dendrites but not axons (Fig. S2). GFP-Farp1 was enriched at 12 div in most dendritic protrusions positive for the excitatory postsynaptic protein Shank (Fig. 1 G). GFP-Farp1–containing protrusions were additionally marked by the F-actin probe UtrCH-Cherry (utrophin calponin homology domain fused to Cherry; Burkel et al., 2007), which we used as marker for spine heads, as they contain dense actin networks (Matus et al., 1982). These results show that Farp1 is a novel postsynaptic protein that is strongly reduced at synaptic membranes lacking SynCAM 1.
SynCAM 1–Farp1 form a synaptic complex through a FERM domain interaction
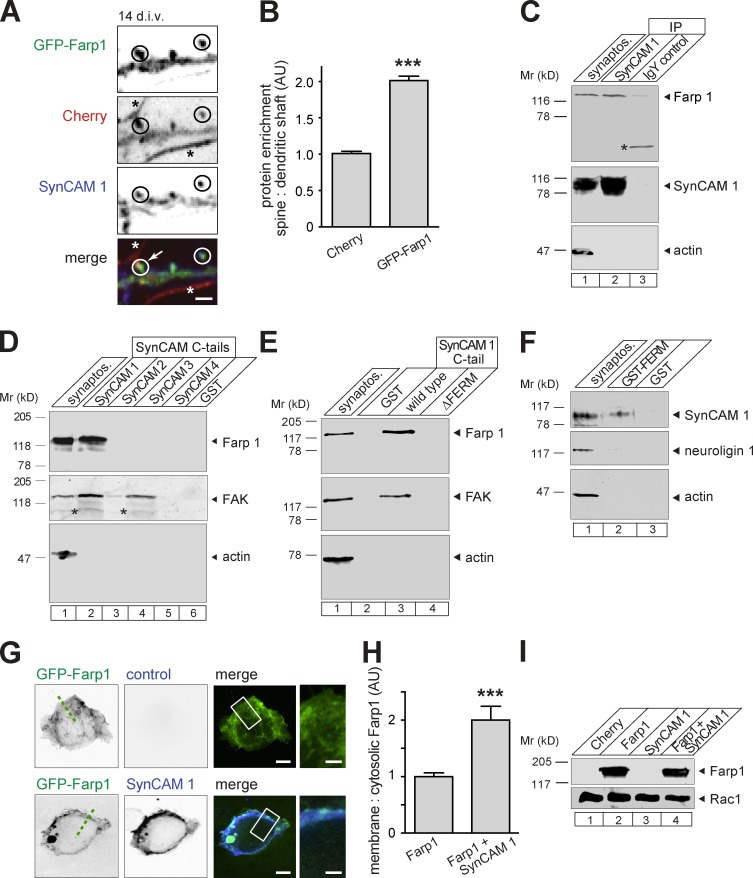
In hippocampal neurons, GFP-Farp1 was enriched in spine heads relative to the volume marker Cherry, and colocalized with endogenous SynCAM 1 (Fig. 2, A and B). In support of an interaction between SynCAM 1 and Farp1 at synapses and consistent with the rationale of our proteomic screen, Farp1 coimmunoprecipitated with SynCAM 1 from synaptosomes (Fig. 2 C). The high degree of sequence similarity of the cytosolic FERM domain–binding motifs of SynCAMs 1–4 (Biederer, 2006) raised the possibility that Farp1 may bind more than one family member. However, affinity chromatography of forebrain extracts on GST fusions of different SynCAM cytosolic tails showed that Farp1 only binds to SynCAM 1 (Fig. 2 D). This agreed with the selective reduction of Farp1 measured by mass spectrometry and immunoblotting in mice lacking SynCAM 1 alone. Farp1 is the only known specific intracellular partner for SynCAM 1, in contrast to FAK (Stagi et al., 2010), that binds both SynCAMs 1 and 3 (Fig. 2 D).
Figure 2.
SynCAM 1 forms a synaptic complex with Farp1 and recruits it to membranes. (A and B) GFP-Farp1 colocalizes with SynCAM 1 at dendritic protrusions. (A) Confocal image of dendrites from neurons coexpressing GFP-Farp1 (green) and Cherry (red). Neurons were stained at 14 div for endogenous, surface-expressed SynCAM 1 (blue). Circles mark colocalization at spine protrusions. Asterisks label Farp1-negative neurites that are probably axons based on narrowness and length. The arrow indicates a likely en passant synapse with a Farp1-positive spine. Bar, 1 µm. (B) GFP-Farp1 is enriched in spines by 100 ± 6.8% relative to Cherry measured from images as in A (99 spines/condition; three independent experiments; P < 0.0001; error bars indicate mean ± SEM). (C) Farp1 coimmunoprecipitates with SynCAM 1 from rat forebrain synaptosomes at P13. IgY was a control for antibody specificity, and actin was a negative control for binding. The asterisk marks an unidentified, cross-reactive protein. (D) Synaptosomal Farp1 is retained by a GST fusion of the SynCAM 1 cytosolic tail, but not other SynCAMs. FAK and actin served as positive and negative controls for binding. The asterisks mark unidentified bands. (E) Binding of Farp1 extracted from synaptosomes at P13 to the cytosolic tail of SynCAM 1 requires the SynCAM 1 FERM-binding motif. (F) A GST fusion of the Farp1 FERM domain retains SynCAM 1 solubilized from synaptosomes. Neuroligin 1 and actin were negative controls. (G–I) SynCAM 1 recruits GFP-Farp1 to plasma membranes. (G) Confocal images of HEK293 cells expressing GFP-Farp1 alone (green; top) or with SynCAM 1 (blue; bottom). GFP-Farp1 distribution was measured by line scans as indicated by the broken line. Boxed areas are enlarged on the right. Bars: (overview panels) 5 µm; (insets) 2 µm. (H) Quantification of data as in G normalized to cells expressing GFP-Farp1 alone (GFP-Farp1, 39 cells; GFP-Farp1+SynCAM 1, 51; three independent experiments; error bars indicate mean ± SEM; ***, P < 0.001). (I) Farp1 amount in transfected HEK293 cells is unaffected by SynCAM 1. Lysates were immunoblotted for Farp1. Rac1 served as loading control.
We mapped the interaction of SynCAM 1 and Farp1 using affinity chromatography of synaptosomal fractions. SynCAM 1/Farp1 binding was FERM domain–dependent, as a GST fusion of the cytosolic SynCAM 1 tail lacking five amino acids in the FERM motif did not bind Farp1 from brain (Fig. 2 E, lane 4). Reciprocally, a GST fusion of the Farp1 FERM domain was sufficient to retain synaptosomal SynCAM 1 (Fig. 2 F).
To assess whether SynCAM 1 can alter the subcellular localization of Farp1, we transfected HEK293 cells with GFP-Farp1 alone or with SynCAM 1. GFP-Farp1 expressed alone was present both in the cytosol and at the plasma membrane, to which it may be recruited through its PH domains (Fig. 2 G). Line scan analysis showed that the fraction of GFP-Farp1 at the membrane perimeter relative to cytosol doubled in the presence of SynCAM 1 (200 ± 25%, P < 0.0001; Fig. 2, G and H). Heterologous coexpression of SynCAM 1 did not alter the amount of overexpressed Farp1 in HEK293 cells (Fig. 2 I). Together, these data support the fact that SynCAM 1 interacts with Farp1 at plasma membranes, presumably directly.
Farp1 promotes filopodial dynamics and stability
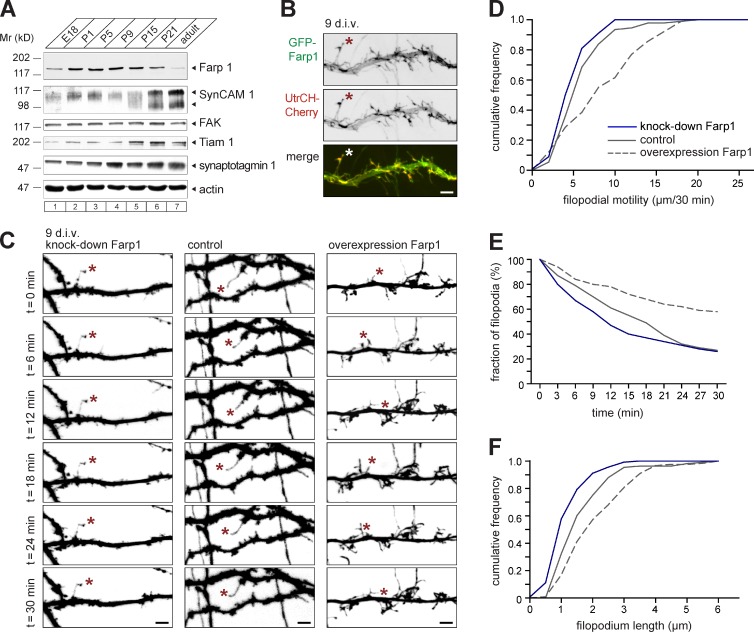
Farp1 is highly expressed in rat forebrain already at P1, i.e., before the peak of synaptogenesis, and remains elevated until P15, when SynCAM 1 increases synapse numbers (Fig. 3 A; Robbins et al., 2010). Farp1 continues to be expressed to a lower extent in adulthood. In developing neurons at 9 div, GFP-Farp1 was present in filopodia-like dendritic protrusions (Fig. 3 B) that can be precursors to dendritic spines. We imaged live filopodia at 9 div over 30-min periods and quantified their properties (Fig. 3 C and Video 1). This was performed blind to the condition, as were all other measurements in this study. We compared Farp1 overexpression and knockdown, for which we used a short-hairpin construct (shFarp1) that reduced the amounts of Farp1 overexpressed in heterologous cells by 75 ± 13% and in neurons by 68 ± 2% (Fig. S1). Farp1 knockdown reduced the motility of dendritic filopodia compared with neurons expressing an shScramble control construct (Fig. 3, C and D; shScramble, 6.3 ± 0.3 µm/30 min; shFarp1, 5.3 ± 0.2; P = 0.007). This effect was rescued by coexpression of knockdown-resistant Farp1sh res, and the increase in motility under this condition was presumably caused by Farp1 expression above endogenous levels (shFarp1 + Farp1sh res, 7.5 ± 0.4 µm/30 min; P = 0.03). Overexpressing Farp1 in the presence of endogenous protein further elevated filopodial motility (Farp1sh res, 8.9 ± 0.5 µm/30 min; P < 0.0001).
Figure 3.
Farp1 regulates the motility, turnover, and morphology of dendritic filopodia. (A) Profile of Farp1 in rat forebrain at indicated embryonic (E) and postnatal (P) days detected by immunoblotting of equal protein amounts. SynCAM 1 appears at multiple molecular weights due to N-glycan modifications (Fogel et al., 2007). (B) Confocal imaging of live hippocampal neurons at 9 div coexpressing GFP-Farp1 (green) with the F-actin probe UtrCH-Cherry (red) visualizes Farp1 in dendritic filopodia (asterisks). Grayscale, individual channels. Bar, 2.5 µm. (C) Confocal images of live hippocampal neurons at 9 div transfected with a shFarp1 knockdown vector (left), shScramble control (center), or shScramble plus Farp1sh res (right). Images were taken at 3-min intervals for 30 min. Asterisks mark filopodia tracked throughout the imaging period. Bars, 2.5 µm. (D) Filopodial tip motility imaged as in C. Knockdown of Farp1 reduces filopodial dynamics, whereas overexpressing Farp1 increases it (D’Agostino and Pearson test, distributions are significantly different; shScramble, 92 filopodia; shFarp1, 100; shScramble + Farp1sh res, 91; three independent experiments). n.s., not significant. (E) Filopodial turnover imaged as in C. 100 filopodia per condition were tracked from t0. Stability is plotted as a survival curve. Farp1 overexpression doubles the fraction of stable filopodia, defined as present for 30 min (three independent experiments). (F) Farp1 knockdown shortens filopodia (D’Agostino and Pearson test, distributions were significantly different; shScramble, 1.7 ± 0.2 µm length from n = 108 filopodia; shFarp1, 1.4 ± 0.1 µm from n = 135; shScramble + Farp1sh res, 2.5 ± 0.2 µm from n = 119; P = 0.002 and 0.005, respectively; three independent experiments).
Tracking dendritic filopodia, we found that Farp1 prolonged filopodial lifetimes (Fig. 3 E; shScramble, 16 ± 1.0 min; shScramble + Farp1sh res, 22 ± 1.0; P = 0.0003), whereas Farp1 knockdown did not significantly decrease filopodial stability (shFarp1, 14 ± 1.0 min; P = 0.14). In addition, Farp1 knockdown and overexpression shortened and lengthened filopodia, respectively (Fig. 3 F; shScramble, 1.83 ± 0.09 µm; shFarp1, 1.32 ± 0.05; shFarp1 + Farp1sh res, 2.18 ± 0.10; shScramble + Farp1sh res, 2.25 ± 0.10; P < 0.0001, < 0.0001, and < 0.002, respectively). Farp1 therefore contributes to the motility, stability, and morphology of dendritic filopodia.
Farp1 regulates excitatory synapse number and acts downstream of SynCAM 1
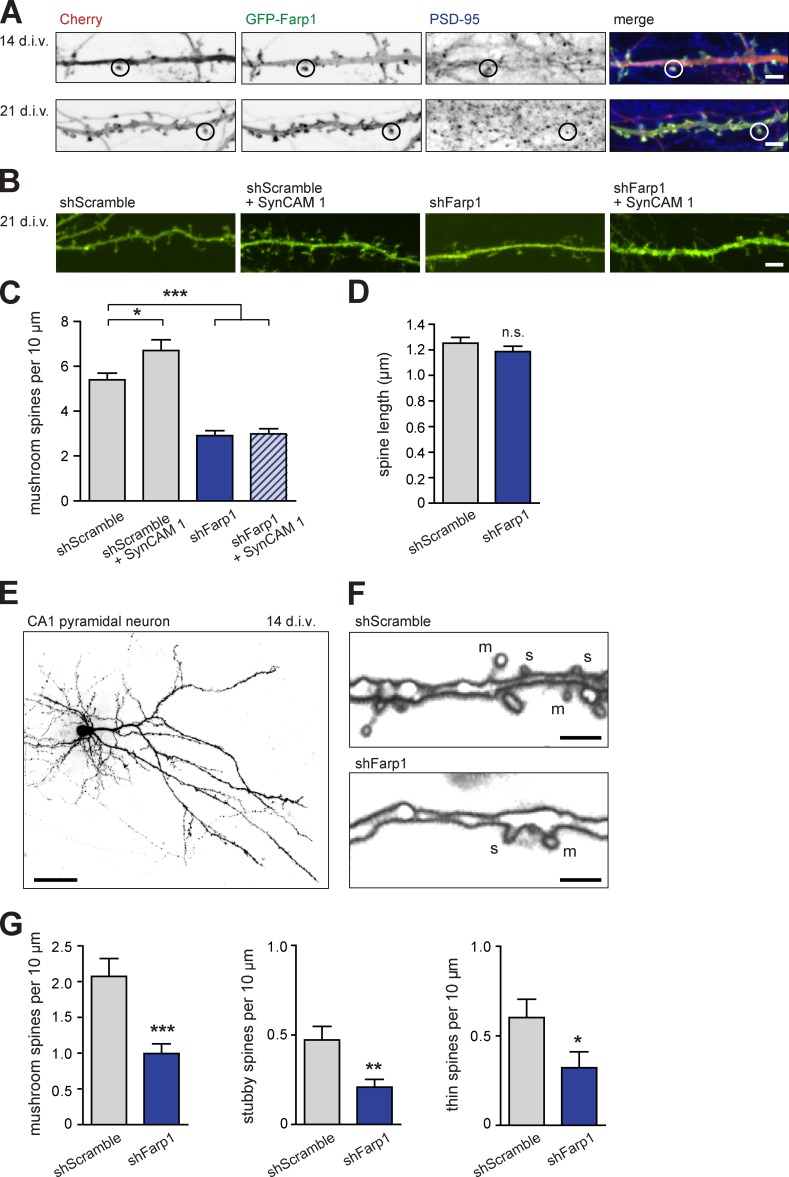
The regulation of filopodia in early development may affect the later formation of synapses. Consistent with synaptic functions, GFP-Farp1 is present in dendritic spine heads at 14 and 21 div (Fig. 4 A). Spines in these cultures are apposed to presynaptic sites with recycling vesicles (Fogel et al., 2007), allowing for the approximation of spines as sites of excitatory synapses. Knockdown of Farp1 in neurons significantly lowered mushroom spine density at 21 div compared with controls (Fig. 4, B and C; shScramble control, 5.4 ± 0.3 mushroom spines per 10 µm; shFarp1, 2.9 ± 0.2; P < 0.0001). This decrease was rescued by coexpression of Farp1sh resist (Fig. S3). Knockdown of Farp1 did not alter spine length (Fig. 4 D), and we observed no effect of reduced Farp1 on stubby or thin spine densities in dissociated neurons.
Figure 4.
Knockdown of Farp1 reduces spine numbers and abrogates SynCAM-mediated increases in spine density. (A) Farp1 is postsynaptic in developing and mature neurons. Confocal images show dissociated hippocampal neurons at 14 (top) and 21 div (bottom) coexpressing GFP-Farp1 (green) and Cherry. Postsynaptic PSD-95 was detected by immunostaining. Circles, dendritic protrusions where Farp1 and PSD-95 colocalize. Grayscale, individual channels. Bars, 2 µm. (B and C) Knockdown of Farp1 lowers spine density and precludes the synaptogenic effect of SynCAM 1. (B) Confocal images of dissociated hippocampal neurons at 21 div expressing shScramble control or shFarp1 vectors. GFP (green)-filled transfected neurons. SynCAM 1 was overexpressed where indicated. Bars, 2 µm. (C) Mushroom spine densities imaged as in B (shScramble, 550 spines; shScramble + SynCAM 1, 437; shFarp1, 275; shFarp1 + SynCAM 1, 274; three independent experiments). (D) No effect on mushroom spine length in dissociated neurons expressing shFarp1 (shScramble, 220 spines; shFarp1, 132; three independent experiments). n.s., not significant. (E–G) Farp1 is required for normal spine density in organotypic slice culture. (E) Confocal overview of a CA1 pyramidal neuron expressing the shScramble vector in slice culture at 14 div. Neurons were visualized by GFP expressed from the knockdown vector. Bar, 100 µm. (F) 3D renderings of dendrites of CA1 neurons expressing shScramble (top) or shFarp1 (bottom). m, mushroom; s, stubby; t, thin spines. Bars, 2 µm. (G) Spine densities imaged as in F (mushroom spines, shScramble 2.1 ± 0.25 per 10 µm dendrite, shFarp1 1.0 ± 0.13; stubby, shScramble 0.47 ± 0.08, shFarp1 0.21 ± 0.04; thin, shScramble 0.60 ± 0.10, shFarp1 0.32 ± 0.09; n = 22 neurons for shScramble, 20 for shFarp1; two independent experiments). Error bars indicate mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We hypothesized that SynCAM 1 promotes synapse formation through its partner Farp1, and tested this by coexpressing SynCAM 1 with shScramble or shFarp1. As expected, SynCAM 1 overexpression significantly increased mushroom spine density (Fig. 4, B and C; SynCAM 1 + shScramble, 6.7 ± 0.5 per 10 µm; P = 0.017). Importantly, neurons that overexpressed SynCAM 1 while Farp1 was knocked down showed the same low spine density as neurons expressing shFarp1 alone (SynCAM 1 + shFarp1, 3.0 ± 0.2 per 10 µm). Thus, endogenous Farp1 regulates spine number, and SynCAM 1 requires Farp1 to increase spine density.
Does Farp1 also regulate synapse numbers in an intact environment corresponding to the packing of neuropil in vivo? We investigated this in organotypic slice cultures, which maintain hippocampal connectivity while allowing the manipulation of protein expression in individual neurons. Slices were prepared from rat pups at P4/5 and biolistically transfected with shScramble or shFarp1 at 10 div. 4 d after transfection, we analyzed dendritic spines of pyramidal CA1 neurons (Fig. 4, E and F). Knockdown of Farp1 prominently decreased the density of mushroom spines by 52 ± 3.8% (P < 0.001), stubby spines by 56 ± 2.5% (P < 0.01), and thin spines by 47 ± 3.8% (P < 0.05; Fig. 4 G), resulting in 52 ± 5.0% fewer total spines (P = 0.0001). The density of the less abundant stubby and thin spines was lowered by Farp1 knockdown in slice cultures, but not in dissociated neurons. This may indicate that physical constraints in tissue affect synapse development, and that the loss of synaptogenic proteins such as Farp1 could render thin and stubby spines in slice cultures less resilient and hence more unstable than in dissociated neurons. Together, these results support the fact that Farp1 is required for normal synapse numbers in CA1 neurons.
Farp1 rescues synapse loss in SynCAM 1 KO neurons
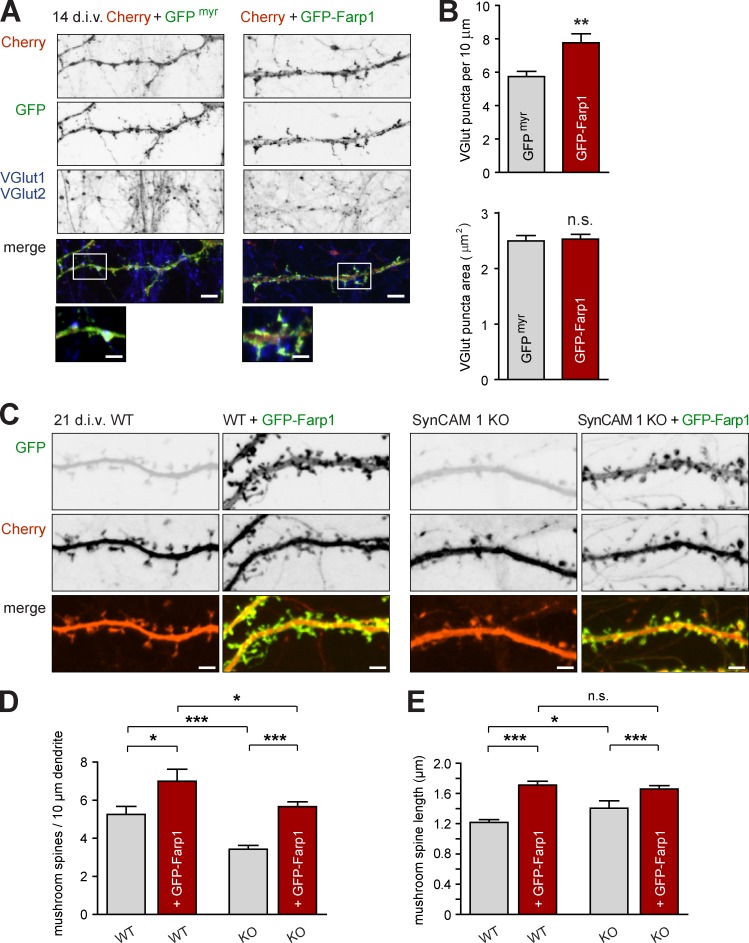
Is Farp1 sufficient to promote mature synapse numbers? Immunostaining for the excitatory presynaptic markers VGlut1/2 at 14 div showed that dendrites of dissociated rat neurons overexpressing GFP-Farp1 had 35 ± 11% more presynaptic sites compared with controls expressing myristoylated GFP (GFPmyr; Fig. 5, A and B; GFPmyr, 5.8 ± 0.30 VGlut puncta per 10 µm; GFP-Farp1, 7.8 ± 0.53; P = 0.0016). The VGlut puncta area was unaffected by Farp1 (Fig. 5 B). GFP-Farp1 also increased mushroom spine numbers later in development, as determined in Cherry-filled WT mouse neurons at 21 div (Fig. 5 C, left two panels; and Fig. 5 D; Cherry, 5.3 ± 0.4 mushroom spines per 10 µm; Cherry + GFP-Farp1, 7.0 ± 0.6; P = 0.02). This was associated with increases in the density of stubby (Cherry, 1.1 ± 0.1 per 10 µm; Cherry + GFP-Farp1, 1.6 ± 0.2; P = 0.02) and thin spines (Cherry, 0.27 ± 0.07; Cherry + GFP-Farp1, 0.95 ± 0.22; P = 0.005). Farp1 selectively promoted mushroom spine density in dissociated rat neurons, leaving stubby and thin spine numbers unaltered (Fig. S4).
Figure 5.
Farp1 increases excitatory synapse number. (A and B) Dendrites expressing GFP-Farp1 carry more excitatory presynaptic terminals than controls expressing myristoylated GFP (GFPmyr). (A) Confocal images of hippocampal neurons at 14 div coexpressing soluble Cherry (red) and GFPmyr (green; left column) or GFP-Farp1 (green; right). Cultures were immunostained for the excitatory presynaptic markers VGlut1/2 (blue). Boxed areas are enlarged below. Bars: (overview) 5 µm; (enlarged insets) 2 µm. (B) Quantification of images as in A. Farp1 increased VGlut puncta number (top) but size was unaffected (bottom). (438 and 456 puncta, respectively; three independent experiments.) (C–E) Lower spine density in SynCAM 1 KO neurons is rescued by Farp1. (C) Confocal images of WT and SynCAM 1 KO neurons at 21 div. Neurons expressed Cherry (red) as volume marker with or without GFP-Farp1 (green). Bars, 3 µm. (D) Quantification of mushroom spine density imaged as in C. Neurons were prepared from littermates. (WT, 430 spines; KO, 470; WT + GFP-Farp1, 422; KO + GFP-Farp1, 720; three independent experiments.) (E) Mushroom spine length imaged as in C (WT, 223 spines; KO, 98; WT + GFP-Farp1, 224; KO + GFP-Farp1, 305; three independent experiments). Error bars indicate mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant.
We next asked whether Farp1 rescues spine density in SynCAM 1 mouse KO neurons, consistent with a role downstream of SynCAM 1. Neurons were transfected with Cherry to count spines, and KO cultures were prepared from littermates in parallel to the WT cultures in Fig. 5 C. As expected, KO neurons had fewer mushroom spines than WT neurons (Fig. 5 C, right two panels; and Fig. 5 D; KO + Cherry, 3.4 ± 0.19 spines per 10 µm, P < 0.0001). Expression of GFP-Farp1 in SynCAM 1 KO neurons rescued this reduction (KO + GFP-Farp1, 5.7 ± 0.3 spines per 10 µm, P = 0.02). KO neurons overexpressing GFP-Farp1 formed fewer spines than WT neurons expressing GFP-Farp1 (P = 0.02), which indicates that SynCAM 1 contributes to the ability of Farp1 to promote spine numbers.
Morphological analysis showed that GFP-Farp1 increased spine length independently of SynCAM 1 (Fig. 5 E; WT, 1.22 ± 0.04 µm; WT + GFP-Farp1, 1.71 ± 0.05, P < 0.0001; KO + GFP-Farp1, 1.66 ± 0.04, P = 0.009). SynCAM 1 loss also moderately increased spine length (KO, 1.40 ± 0.10, P = 0.03). These results support the finding that SynCAM 1 and Farp1 share synaptogenic functions, yet have distinct roles in shaping spines.
Presynaptic active zones are organized by transsynaptic Farp1 signaling via SynCAM 1
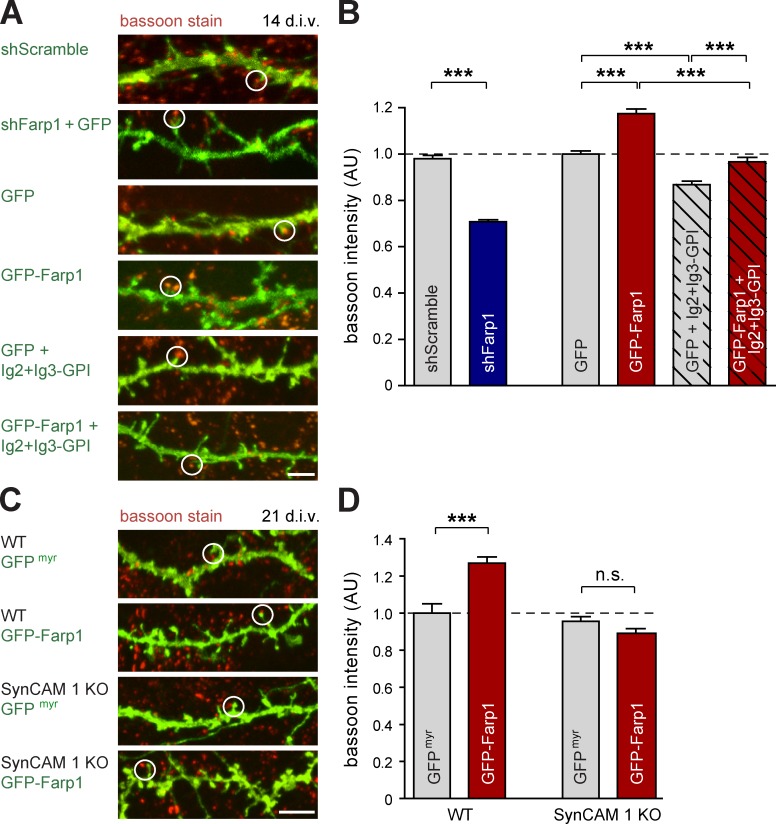
As postsynaptic Farp1 expression promotes presynaptic terminal number, we tested to what extent Farp1/SynCAM 1 signaling may act across the synaptic cleft to regulate presynaptic composition. We measured the effects of altering postsynaptic Farp1 expression on presynaptic terminals by staining hippocampal neurons for the active zone marker bassoon (Fig. 6 A). This confirmed the synaptogenic effect of Farp1, as its overexpression increased bassoon puncta number by 44 ± 11% (GFP, 3.5 ± 0.3 puncta per 10 µm; GFP-Farp1, 5.1 ± 0.3; P = 0.0002; n, see legend Fig. 6 B). Farp1 knockdown lowered the bassoon puncta number by 31 ± 13% (P = 0.03), further supporting its endogenous role in controlling synapse numbers.
Figure 6.
Postsynaptic Farp1 organizes presynaptic active zones via SynCAM 1. (A and B) Bassoon intensity is regulated by Farp1 across the synaptic cleft. (A) Confocal images of hippocampal neurons at 14 div. Green, GFP or GFP-Farp1. Red, bassoon detected by immunostaining. Circles mark representative spines apposed to bassoon. (B) Bassoon intensity imaged as in A normalized to staining atop GFP control dendrites (shScramble, n = 15 neurons; shFarp1, 17; GFP, 22; GFP-Farp1, 25; GFP + Ig2+Ig3-GPI, 14; GFP-Farp1 + Ig2+Ig3-GPI, 21; 167, 143, 296, 284, 182, and 286 puncta were analyzed, respectively). (C and D) Farp1 regulates bassoon intensity in dependence on SynCAM 1. (C) Hippocampal neurons from WT and SynCAM 1 KO littermate mice (three mice each) were transfected at 5–7 div to express GFPmyr or GFP-Farp1. Bassoon immunostaining at 21 div was imaged by confocal microscopy. Green, GFPmyr or GFP-Farp1. Red, bassoon. Circles, representative spines apposed to bassoon. (D) Bassoon puncta intensity imaged as in C, normalized to staining atop GFPmyr -expressing WT dendrites. (WT + GFPmyr, n = 173 puncta from 14 neurons; WT + GFP-Farp1, n = 338 from 18 neurons; KO + GFPmyr; n = 309 from 21 neurons; KO + GFP-Farp1, n = 293 from 26 neurons). The broken lines mark the control’s value of AU = 1.0, to which the other values were normalized. Error bars indicate mean ± SEM; ***, P < 0.001. n.s., not significant. Bars, 4 µm.
Knockdown of Farp1 in dendrites additionally decreased the immunostaining intensity of the remaining apposed bassoon puncta by 29 ± 2% compared with GFP controls (P < 0.0001; Fig. 6 B). In turn, overexpression of GFP-Farp1 increased the intensity of bassoon puncta by 18 ± 2% (P < 0.0001). To assess contributions of SynCAM 1, we used a construct of its Ig2+Ig3 domains linked to the membrane surface via a glycophosphatidylinositol (GPI) anchor that interferes with lateral SynCAM 1 assembly in cis and reduces transsynaptic adhesion (Fogel et al., 2011). It lacks the tail of SynCAM 1 and its effects are therefore independent of cytosolic partners. Notably, coexpression of Ig2+Ig3-GPI impaired the ability of elevated Farp1 to promote presynaptic bassoon staining intensity above controls (P < 0.0001). Farp1 still increased bassoon intensity in neurons expressing Ig2+Ig3-GPI, albeit moderately, which may have been caused by incomplete disruption of SynCAM assembly. We addressed this next in SynCAM 1 KO neurons.
We analyzed transsynaptic roles of Farp1 in hippocampal neurons cultured from SynCAM 1 WT and KO littermate mice (Fig. 6 C). Studying bassoon puncta density first, we observed that the postsynaptic overexpression of Farp1 in WT neurons increased the number of bassoon puncta by 55 ± 16%, as expected (WT + GFP, 3.70 ± 0.24 puncta per 10 µm; WT + GFP-Farp1, 5.72 ± 0.55; P = 0.0013). KO neurons had fewer bassoon puncta (KO + GFP, 3.03 ± 0.21 puncta per 10 µm, P < 0.05), and elevating Farp1 in SynCAM 1 KO neurons increased bassoon puncta density above control KO neurons by 71 ± 15% (KO + GFP-Farp1, 5.20 ± 0.39, P < 0.001). This effect indicates that overexpressed Farp1 can engage SynCAM 1–independent pathways to promote synapse numbers. Measuring the intensity of bassoon puncta next (Fig. 6 D), we determined that Farp1 overexpression in postsynaptic WT neurons promoted this parameter by 27 ± 6% (P < 0.0001), as expected. Importantly, Farp1 required SynCAM 1 to promote bassoon puncta intensity, as Farp1 overexpression in SynCAM 1 KO neurons failed to alter bassoon signals.
Together, these results support the finding that postsynaptic Farp1 can modulate the composition of presynaptic active zones. The lateral assembly of SynCAM 1 in dendritic membranes contributes to this transsynaptic signaling by Farp1. Notably, SynCAM 1 is required for the ability of postsynaptic Farp1 to increase presynaptic bassoon intensity across the cleft.
Farp1 activates the GTPase Rac1 at synapses
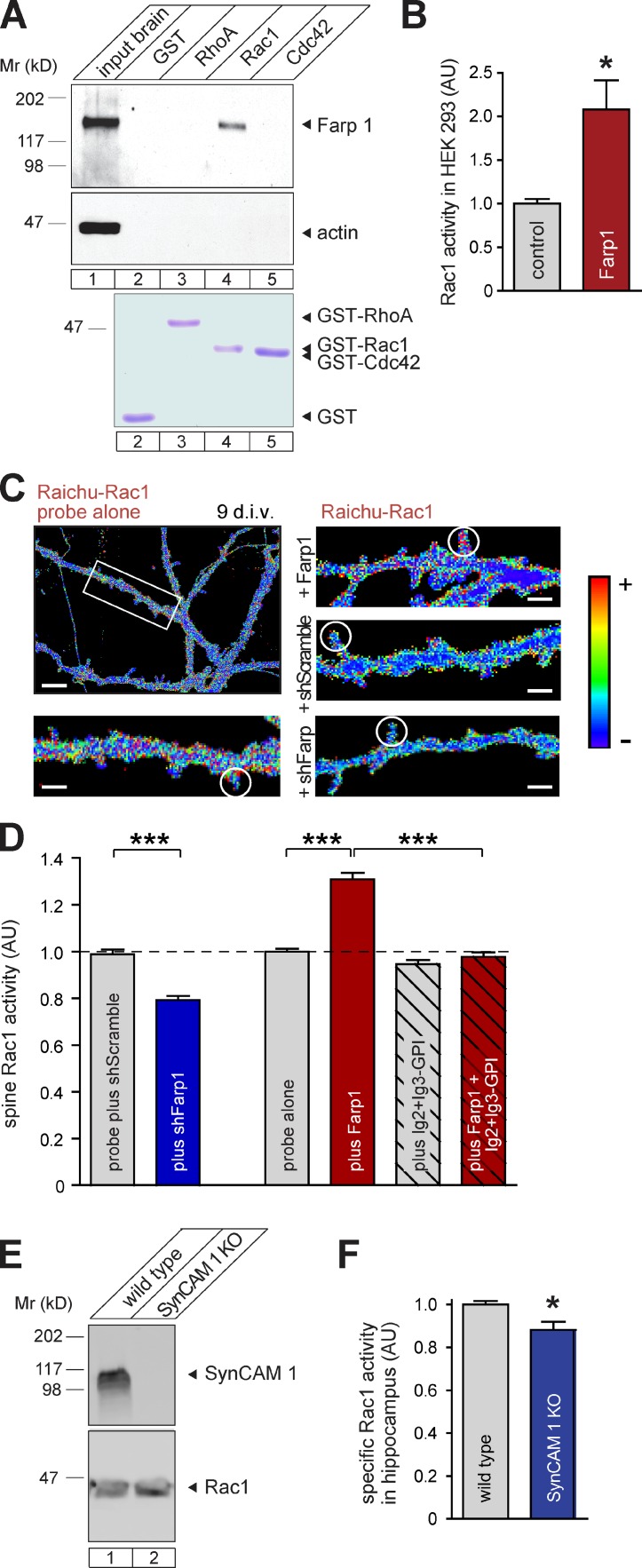
Aiming to define the mechanism of postsynaptic Farp1, we were guided by the presence of DH-PH domains that indicated a role as a GEF for a Rho family GTPase. To test whether Farp1 interacts with RhoA, Rac1, or Cdc42, the best-characterized GTPases in spines, we performed affinity chromatography on empty nucleotide mutants of these enzymes that retain activating GEFs more strongly (García-Mata et al., 2006). Heterologously expressed Farp1 was retained on Rac1-containing beads but not on RhoA or Cdc42, which indicates that Farp1 may selectively activate Rac1 (Fig. 7 A). This interaction is indeed functionally relevant, as HEK293 cells expressing Farp1 contained twice the amount of active GTP-Rac1 compared with control cells (200 ± 47%, P = 0.03; Fig. 7 B). Expression of Farp1 in these cells had no effect on total Rac1 levels (Fig. 2 I). Because Rac1 promotes spine numbers (Nakayama et al., 2000), the ability of Farp1 to activate Rac1 agrees with a shared role of both proteins in promoting spines.
Figure 7.
Farp1 activates postsynaptic Rac1. (A) Farp1 expressed in HEK293 cells binds the Rac1 G15A empty nucleotide mutant but not RhoA G17A or Cdc42 G15A measured by affinity chromatography on GST fusions. Actin was a negative control. Bottom row, comparable amounts of immobilized GST fusion proteins detected by Coomassie staining. (B) Farp1 activates Rac1. HEK293 cells expressing Farp1 contain twice as much active GTP-Rac1 compared with cells expressing Cherry (n = 3 independent experiments). (C and D) Farp1 overexpression increases whereas Farp1 knockdown decreases Rac1 activity in spines. (C) Rac1 activity imaged in live neurons at 9 div with the Raichu-Rac1 probe in dendritic protrusions vs. adjacent shaft areas. Ratiometric heat maps represent a higher YFP/CFP ratio, which is indicative of greater Rac1 activity, as warmer colors. The boxed area in the overview is enlarged below. Circles mark representative dendritic spines. Bars: (overview) 5 µm; (enlarged panel) 1 µm. (D) Quantification of images obtained as in C (probe plus shScramble, n = 149 spines; plus shFarp1, 189; probe alone, 269; plus Farp1, 223; plus Ig2+Ig3-GPI, 259; plus Farp1 + Ig2+Ig3-GPI, 234; three independent experiments). The broken lines mark the control’s value of AU = 1.0, to which the other values were normalized. (E) Same Rac1 levels in WT and SynCAM 1 KO hippocampi by quantitative immunoblotting. (F) Lower Rac1 activity in SynCAM 1 KO hippocampi at P13 than in WT littermates (n = 6 mice each). Specific Rac1 activity was measured by normalizing active GTP-Rac1 levels to Rac1 protein amounts determined by quantitative immunoblotting. Error bars indicate mean ± SEM. *, P < 0.05; ***, P < 0.001.
To determine whether Farp1 directly regulates local Rac1 activity at synapses, we imaged live neurons expressing the optical probe Raichu-Rac1 (Itoh et al., 2002). This FRET-based probe contains a Rac1 GTPase sequence proximal to a binding domain for Rac1-GTP flanked by YFP and CFP, allowing the measurement of Rac1 activity as the YFP/CFP ratio. We imaged Rac1 activity live at 9 div in dendritic protrusions. Signals were normalized for each protrusion to Rac1 activity along the adjacent dendritic shaft (Fig. 7 C). Farp1 knockdown decreased postsynaptic Rac1 activity by 20 ± 3% (P < 0.0001; Fig. 7 D). Other activators of Rac1 in spines therefore fail to fully compensate for a reduction in Farp1. In turn, Farp1 overexpression increased synaptic Rac1 activity significantly by 31 ± 3% (P < 0.0001; Fig. 7, C and D). Co-expression of SynCAM 1 Ig2+Ig3-GPI with Farp1 abrogated the ability of Farp1 to activate Rac1, which indicates that Farp1 requires interactions with SynCAM 1 clusters that are laterally assembled in dendritic membranes to activate Rac1.
As Farp1 can activate Rac1, the reduction of Farp1 in SynCAM 1 KO brains may result in lower levels of active Rac1. We biochemically measured this in hippocampal homogenates of SynCAM 1 KO and WT littermates. Because of loss of Rac1 activity over time, activity levels could only be analyzed in freshly prepared hippocampal homogenates, but not in subcellular fractions. Measurements were performed at P13, when Farp1 is abundant in WT controls (Fig. 3 A) and synaptogenesis is high. SynCAM 1 KO mice showed a modest but significant 12 ± 4% reduction in the specific activity of Rac1 compared with WT littermates (P = 0.010), whereas total Rac1 levels were unchanged (Fig. 7, E and F). Rac1 signaling is therefore impaired in the brains of SynCAM 1 KO mice, concurrent with a reduction of Farp1.
Spine actin is regulated by Farp1
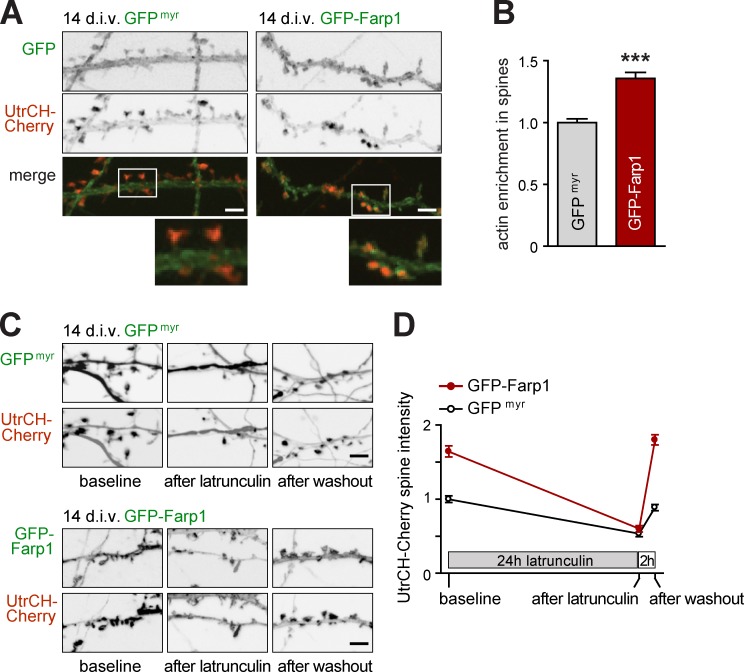
Rac1 promotes actin polymerization by regulating actin nucleation (Jaffe and Hall, 2005). To test whether Farp1 promotes F-actin in spines, consistent with signaling roles upstream of Rac1, we used the UtrCH-Cherry probe (Burkel et al., 2007). Neurons coexpressing UtrCH-Cherry with either GFPmyr or GFP-Farp1 were imaged live at 14 div, when spines have become more abundant. We quantified amounts of F-actin in spines by measuring UtrCH-Cherry fluorescence intensity within the center of spine heads and normalized each measurement to the probe signal along the adjacent dendritic shaft. Spines of neurons expressing GFP-Farp1 showed 36 ± 5% higher UtrCH-Cherry intensity (P < 0.0001; Fig. 8, A and B), which is consistent with an increase in F-actin by postsynaptic Farp1.
Figure 8.
Farp1 promotes F-actin assembly in spines. (A and B) GFP-Farp1 increases F-actin amounts in spine heads. (A) Confocal images of live neurons at 14 div coexpressing the F-actin probe UtrCH-Cherry (red) with GFPmyr (green, left column) or GFP-Farp1 (green, right). Enlarged panels show UtrCH-Cherry intensity in spine heads and along adjacent dendritic shafts, with measurement from the latter used to normalize each spine signal. Bars: (overview) 5 µm. (B) Quantification of images as in A (GFPmyr neurons, 381 spines; GFP-Farp1, 352; three independent experiments; error bars indicate mean ± SEM; ***, P < 0.001). (C and D) Farp1 increases spine actin polymerization. (C) Dendritic segments were imaged live at 14 div to detect UtrCH-Cherry. Neurons were then treated with Lat-A for 24 h to depolymerize actin, followed by recovery in Lat-A–free medium for 2 h. Panels show representative images. Bars, 5 µm. (D) Quantification of images as in C. GFP-Farp1 increases baseline UtrCH-Cherry signal in spine heads over control, and Lat-A decreases spine UtrCH-Cherry signal in both conditions. After Lat-A washout, spines expressing GFP-Farp1 exhibit a stronger increase in F-actin signal than controls (GFPmyr neurons vs. GFP-Farp1 before Lat-A, 248 and 316 spines; after Lat-A, 152 and 165; after washout, 169 and 203; three independent experiments).
To determine whether this may result from increased actin polymerization, we treated GFPmyr- or GFP-Farp1–expressing neurons for 24 h with 2.5 µM Latrunculin A (Lat-A), a drug that leads to actin depolymerization. Lat-A treatment decreased spine F-actin in both GFPmyr- and GFP-Farp1–expressing neurons, reducing UtrCH-Cherry intensity by 47 ± 6% and 63 ± 10%, respectively (Fig. 8, C and D). Lat-A reduced F-actin to the same low level in both conditions, which indicates that Farp1 does not alter actin depolymerization. 2 h after washout of Lat-A, GFP-Farp1–expressing neurons exhibited a strong 199 ± 8% increase of UtrCH-Cherry fluorescence in spines, completely recovering their high baseline level. Control neurons expressing GFPmyr showed only a 66 ± 6% increase in F-actin in the same period. This accelerated increase of spine F-actin supports the conclusion that Farp1 promotes F-actin polymerization in spines.
Discussion
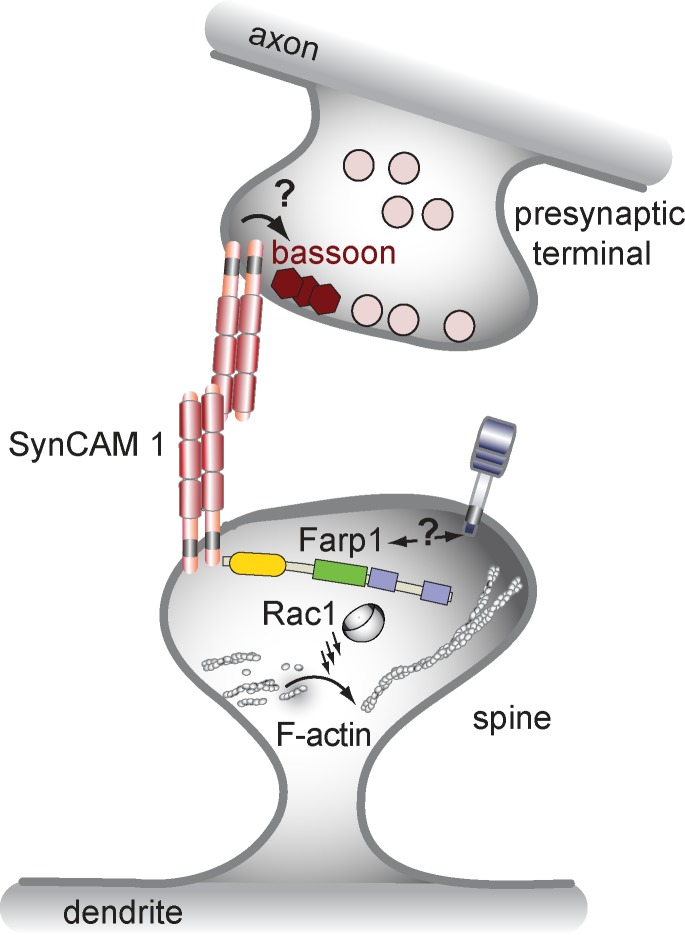
Our proteomic analysis of SynCAM 1 KO synapses led us to identify and characterize Farp1 as a novel postsynaptic protein that signals during concerted steps of excitatory synapse development (Fig. 9). In immature neurons, Farp1 regulates filopodial dynamics. At later stages, Farp1 localizes to dendritic spines, where it activates the GTPase Rac1 and promotes actin assembly. Moreover, postsynaptic Farp1 contributes to the control of excitatory synapse density and signals across the synaptic cleft to modulate active zone composition. Two effects of Farp1 may enable it to promote synapse number. On the one hand, the Farp1-mediated increase in filopodial dynamics and lifetime may make axo-dendritic contacts more frequent. This could culminate in a conversion of filopodia to spines (Knott et al., 2006; Kayser et al., 2008), although this model is a matter of debate (Yuste and Bonhoeffer, 2004). On the other hand, Farp1 elevates Rac1 activity and F-actin assembly in mature spines, molecular changes that can increase synapse numbers (Nakayama et al., 2000; Zhang and Benson, 2001; Zito et al., 2004).
Figure 9.
Model of transsynaptic organization by the SynCAM 1–Farp1 pathway.
Interplay of Farp1 with SynCAM 1 in vivo is indicated by our findings that Farp1 is strongly reduced at SynCAM 1 KO synapses and that both proteins form a synaptic complex. In agreement, SynCAM 1 requires Farp1 to drive synaptogenesis, which elucidates the first synaptic signaling mechanism of SynCAMs. Farp1 additionally rescues the lower synapse density in SynCAM 1 KO neurons, which is consistent with acting downstream of SynCAM 1. Moreover, the functions of Farp1 as GEF for spine Rac1 and in transsynaptic signaling involve the assembly of SynCAM complexes in dendritic membranes. The reduction in Farp1-driven spine formation in the absence of SynCAM 1 may point to roles of SynCAM 1 in activating Farp1, although molecular evidence for this is not yet available.
Although endogenous Farp1 is less abundant at SynCAM 1 KO synapses, and the extent to which Farp1 increases spine number is lower in the absence of SynCAM 1, additional proteins likely cooperate with Farp1 to promote synapse density, at least in the absence of SynCAM 1. One candidate binding partner is Plexin A4, with which Farp1 interacts in the chick spinal cord to promote dendritic outgrowth of motor neurons (Zhuang et al., 2009). Yet, the brains of mice lacking Plexins A3 or A4 have increased spine numbers and larger spines, supporting roles of Plexin signaling in restricting synapse number and size (Tran et al., 2009). As Farp1 has opposite functions, it appears unlikely to operate in a synapse-organizing Plexin A4 pathway. Additional evidence for SynCAM-independent functions of Farp1 is provided by our finding that elevating Farp1 and losing SynCAM 1 both result in increased spine length. Future studies can now identify additional upstream partners of Farp1 in mammalian neurons.
Farp1-regulated GTPase signaling may converge with other postsynaptic pathways. Notably, Ephrin B receptors signal through Tiam1 and Kalirin-7, GEFs for Rac1, to promote synapse development (Penzes et al., 2003; Tolias et al., 2007), and may act in concert with N-cadherins to regulate Rho family GTPases in spines (Elia et al., 2006; Xie et al., 2008). Likewise, the activity-dependent remodeling of spines is regulated by the GEF Epac2, which binds neuroligin and activates the Ras-like GTPase Rap1 (Woolfrey et al., 2009). Spine number and Rac1 activity are also modulated by the GEF α-PIX (Zhang et al., 2005). These findings support the idea that GTPases and Rac1 in particular play central roles in synaptic differentiation and can act downstream of synapse-organizing adhesion proteins. This agrees with aberrations of synaptic adhesion molecules and GTPase signaling in developmental and psychiatric disorders (Govek et al., 2005; Südhof, 2008; Penzes et al., 2011). Interestingly, a chromosomal microdeletion that includes the FARP1 gene is also linked to mental retardation (Amor et al., 2005).
Our data indicate that Farp1 plays a significant role in activating spine Rac1, as other GEFs do not fully compensate once Farp1 is knocked down. Our study also reveals an unexpected regulatory mechanism of a GEF: Farp1 depends on the lateral assembly of SynCAM 1 in postsynaptic membranes to activate Rac1. This agrees with the notion that the clustering of signaling molecules into modular domains can underlie their function (Groves and Kuriyan, 2010). Subspine domains where SynCAM 1 forms adhesion complexes may therefore be sites of preferential Farp1/Rac1 activation.
Because SynCAM 1 can induce synapses through its interactions across the synaptic cleft, we analyzed effects of Farp1 on presynaptic organization. Our results show that postsynaptic Farp1 promotes bassoon puncta intensity only when SynCAM 1 is present. Additionally, Farp1 depends on lateral SynCAM clustering in dendritic membranes to act across the cleft. This provides evidence that SynCAM 1 and Farp1 cooperate to signal retrogradely. SynCAM 1/Farp1 signaling across the synaptic cleft may contribute to the shortening of the presynaptic active zone that occurs in SynCAM 1 KO brains (Robbins et al., 2010). The transsynaptic role of postsynaptic Farp1 is reminiscent of the effects of PSD-95 and SAP-97 on presynaptic markers (El-Husseini et al., 2000; Regalado et al., 2006), with PSD-95 acting via neuroligin 1 to also regulate presynaptic plasticity (Futai et al., 2007). Postsynaptic neuroligin 1 further promotes the maturation of presynaptic vesicle release and active zone stability (Wittenmayer et al., 2009). These retrograde effects may complement the bidirectional signaling of ephrins and their Eph receptors across synapses (Klein, 2009) and the roles of glutamate receptors in presynaptic maturation and homeostasis (see, e.g., Lindskog et al., 2010; Tracy et al., 2011). The interaction of postsynaptic SynCAM 1 and Farp1 in organizing presynaptic terminals described here presents a novel function for a GTPase regulator.
Last, the molecular complexity of SynCAMs, with all four family members expressed in the brain (Thomas et al., 2008), raises the question of how this diversity may impact synaptic signaling. Only SynCAM 1 binds Farp1, and postsynaptic sites can contain both SynCAM 1 (this paper) and SynCAM 2 (Shu et al., 2011). Synaptic signaling could hence be regulated by differential SynCAM expression across postsynaptic sites, enabling those spines that contain SynCAM 1 to engage Farp1 for regulating their cytoskeleton and signaling across the cleft. With respect to nonsynaptic roles of Farp1, it can now also be tested to what extent SynCAM 1 contributes to Farp1 function in dendritic growth (Zhuang et al., 2009).
Together, this study reveals Farp1 as a postsynaptic signaling protein that binds SynCAM 1 to instruct and integrate excitatory synapse development.
Materials and methods
Antibodies
For immunoblotting, a polyclonal antibody was raised in rabbit against the peptide CSRAHILSHKESHLY corresponding to the extreme C-terminal sequence of rat Farp1 with an amino-terminal cysteine for coupling to beads (used in immunoblotting at 1:500; see also Fig. S1). An antibody raised against SynCAM 1 in chicken (clone 3E1; MBL Laboratories) was used in immunoblotting (1:500), immunoprecipitation (1:200), and immunofluorescence (1:200). Mouse anti-bassoon (Assay Designs VAM-PS003; 1:400; Enzo Life Sciences) was used for quantitative immunostaining. Mouse monoclonal antibodies used for immunoblotting detected actin (clone C4 69100; 1:2,000; MP Biomedicals), FAK (clone 4.47, 1:1,000; Millipore), neuroligin 1 (clone 4C12.1; 1:1,000; Synaptic Systems), synaptotagmin 1 (clone 41.1; 1:100; Synaptic Systems), Tiam 1 (C-16 sc-872; 1:100; Santa Cruz Biotechnology, Inc.), synaptophysin (clone C1604.4; 1:5,000; Synaptic Systems), PSD-95 (1:2,000; Synaptic Systems), CASK (clone K56A_50; 1:1,000; NeuroMab), and Rac1 (ARC03; 1:1,000; Cytoskeleton). Polyclonal antibodies for immunoblotting detected GFP (600-101-215; 1:2,000; Rockland Immunochemicals Inc.), and VCP (Sugita and Südhof, 2000), a gift from Thomas Südhof (Stanford University, Palo Alto, CA). Antibodies used in immunofluorescence detected Shank (clone N23B_49; 1:500; NeuroMab), MAP2 (AB364; 1:500; EMD Millipore), tau (A0024; 1:500; Dako), and VGlut 1+2 (raised in guinea pig against the peptides CGATHSTVQPPRPPPPVRDY [GP71, corresponding to EMD Millipore AB5905] and CVQESAQDAYSYKDRDDYS [GP73, corresponding to EMD Millipore AB5907], respectively [Tagliaferro and Morales, 2008]; 1:300 each).
Expression vectors
To generate a plasmid for the expression of Farp1, the full-length coding sequence was amplified from a rat brain P11 cDNA library. A 5′ NheI site was introduced during amplification with forward and reverse oligos LC0002, 5′-GAGCTAGCATGGGAGAAATAGAGCAGAAGC-3′, and LC0003, 5′-GCTCAGTACAGGTGAGATTCTTTGTGAC-3′. The amplified sequence was inserted into the pCR-BluntII-TOPO vector (Invitrogen), then subcloned into the pCAGGS-BGHpA vector (a gift from Y. Nakagawa, University of Minnesota, Minneapolis, MN) via BamHI and XhoI restriction sites. A sequence encoding GFP was subcloned into the pCAGGS-BGHpA-Farp1 vector via the 5′ NheI site. pCAGGS-SynCAM 1 encodes full-length mouse SynCAM 1 splice product 4 (Biederer, 2006), with the coding sequence subcloned via 5′ and 3′ EcoRI sites. The pCAGGS-myrGFP vector was generated by PCR amplifying myrEGFP and subcloning the insert after partial EcoRI digestion, and pCAGGS-Cherry was generated after amplification of the sequence encoding Cherry from pRSETB-Cherry (a gift from R. Tsien, University of California, San Diego, La Jolla, CA) and subcloning using EcoRI sites as described previously (Stagi et al., 2010). pCMV-SynCAM 1–Ig2+Ig3-GPI was cloned by subcloning the sequence encoding SynCAM 1 Ig2 and Ig3 with a 5′ SynCAM 1 signal peptide sequence via EcoRI–SalI into pCMV-GPI as described previously (Fogel et al., 2011). pCS2-UtrCH-Cherry (UtrCH-Cherry) encodes an F-actin–binding probe based on the calponin homology domain of utrophin (a gift from W. Bement, University of Wisconsin, Madison, WI; Burkel et al., 2007; Ferguson et al., 2009). The vector pRaichu-Rac1-1011x was a gift from M. Matsuda (Kyoto University, Kyoto, Japan; Itoh et al., 2002).
GST-Farp1FERM was generated by amplifying the first 1,152 nucleotides of the Farp1 coding sequence from the pTOPO-Farp1 vector using the oligo pair 5′-GAGCTAGCATGGGAGAAATAGAGCAGAAGC-3′ and 5′-CAGCTCGAGCTATTCTGAATTCGGTGCT-3′. The resulting Farp1FERM construct was subcloned into the pGEX-KG vector via SmaI and XhoI restriction sites. GST fusion constructs of the WT and ΔFERM SynCAM 1 cytosolic tail were generated as described previously (Biederer et al., 2002; Stagi et al., 2010) by amplifying the full cytosolic sequence of SynCAM 1 corresponding to amino acids 399–455 of mouse SynCAM 1 splice product 4 (Biederer, 2006) or amino acids 399–403/411–405 lacking seven amino acids in the FERM-binding motif, and subcloning into pGEX-KG via SmaI or SmaI–HindIII, respectively. GST fusion constructs of empty-nucleotide mutants of RhoA, Rac1, and Cdc42 that preferentially bind GEFs were a gift from K. Burridge (University of North Carolina at Chapel Hill, Chapel Hill, NC; García-Mata et al., 2006).
Short-hairpin constructs against Farp1 (shFarp1) and a scrambled control hairpin (shScramble) expressed from pGENECLIP-GFP vectors that target rat and mouse orthologues and coexpress soluble GFP from the vector backbone were developed with SABiosciences (QIAGEN). shFarp1 targets the sequence 5′-GGGGCGTCCTTCCGGTTTAGTGTG-3′ beginning at nucleotide 1,005 of Farp1, which is conserved in rat and mouse orthologues. shScramble targets the nonspecific sequence 5′-GGAATCTCATTCGATGCATAC-3′. A shFarp1-resistant Farp1 construct (Farp1sh resist) was generated through introducing seven silent mutations within the shFarp1 targeting sequence using the QuikChange mutagenesis kit (Agilent Technologies). This construct with the modified shFarp1 target sequence of 5′-GGATCTTCTTTTAGATTCAGTGGT-3′ was used for rescue experiments (see Figs. S1 and S3). To perform studies with the Raichu-Rac1 probe and avoid emission overlap, versions of knockdown vectors not expressing GFP were generated by cutting AgeI–SmaI restriction sites to remove the GFP sequence and religation.
Biochemical procedures
Brain homogenates were fractionated by the method of Jones and Matus (1974) with modifications (Biederer et al., 2002). For coimmunoprecipitation, synaptosomes were prepared from forebrain, washed by repeated centrifugation, and solubilized with 1% Triton X-100 in the presence of 10 mM Hepes-KOH, pH 7.4, 25 mM KAc, and 320 mM sucrose with protease inhibitors added. Synaptosomal extracts were precleared on anti–chicken IgY agarose beads (DAIgY-AGA-1; Gallus Immunotech) for 1 h at 4°C. Pre-cleared material was then incubated as described previously (Fogel et al., 2007) with either anti-SynCAM 1 antibody raised in chicken (clone 3E1, 1:200; MBL Laboratories) or IgY protein, and samples were rotated overnight at 4°C. The following morning, anti-IgY beads were added for 90 min at 4°C to collect antibody complexes, the supernatant was removed, beads were washed four times in homogenization buffer, and bound proteins were eluted from beads using 2% SDS and analyzed by immunoblotting.
Postsynaptic density (PSD) extraction was performed as described previously (Cho et al., 1992). In brief, synaptosomes were prepared from the forebrain as described in the preceding paragraph, collected after sucrose density gradient centrifugation at a 1,000/1,200 mM sucrose interphase, and extracted two times for 20 min with 0.5% Triton X-100 at 4°C, followed by a third extraction with 3.0% sarkosyl to obtain a PSD fraction in the final pellet.
For affinity chromatography, synaptosomes were prepared and solubilized as above, precleared with glutathione beads for 2 h at 4°C, and then incubated with GST fusion proteins overnight. Proteins were eluted from washed beads with 2% SDS. For profiling protein expression, frozen tissue samples were rapidly homogenized using microtip-aided sonication in 50 mM Hepes, pH 7.4, 8.0 M urea, and 0.5 mM PMSF. Quantitative immunoblotting was performed on an Odyssey Imaging System (LI-COR Biosciences).
Rac1 activity was measured using a G-LISA kit (Cytoskeleton) according to the manufacturer’s instructions. In brief, HEK293 cells were lysed, 1.3 mg/ml of protein per condition was incubated with GTP-Rac1 affinity plates, and GTP-Rac1 was detected through specific antibody binding and HRP development. For analysis of GTP-Rac1 in brain tissue, hippocampi and some surrounding cortex were rapidly dissected in PBS, then homogenized, and 1.0 mg/ml protein for each condition was analyzed.
Proteomic screen
Synaptic plasma membranes were purified from 6-wk-old SynCAM 1 KO and WT littermate mice as described previously (Biederer et al., 2002). In brief, synaptosomes were prepared from forebrain by differential centrifugation, hypotonically lysed and pelleted, adjusted to 1.1 M sucrose, and overlayed with layers of 855 mM and 320 mM sucrose for ultracentrifugation in a SW40 rotor; the synaptic plasma membrane fraction was collected at the interface of 855 mM/1.1 M sucrose. Quality of the fractionation was controlled by measuring the enrichment of synaptic markers by immunoblotting. Samples were subjected to 4-plex iTRAQ with technical replicates as described previously (Dávalos et al., 2010). Two WT and two KO synaptic plasma membrane samples were trypsin digested, labeled with iTRAQ 114/115 and 116/117 reagents, respectively, pooled, and purified using a strong cation exchange column (Applied Biosystems). For QSTAR Elite liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis, each cation exchange fraction was dried and resuspended for reverse phase LC, individual peptides were separated, and data collection was performed by electrospray ionization on a mass spectrometer (AB Sciex API QSTAR Elite; Applied Biosystems). iTRAQ quantitation and protein identification were performed using the Paragon search algorithm (Shilov et al., 2007) in ProteinPilot 3.0 software against the International Protein Index mouse database. The amounts of 24 proteins were increased in synaptic plasma membranes from SynCAM 1 KO mice above a 1.3-fold cutoff as determined by mass spectrometry, and 9 proteins were reduced, scoring those proteins that were identified with sufficient confidence by sequencing more than one peptide. All proteomic hits will require independent validation, as performed for Farp1.
Cell culture
HEK293 cells were obtained from the American Type Culture Collection and cultured using standard conditions. Cells were transfected using either Fugene (Roche) or calcium chloride transfection. For immunoblotting, cell lysates were homogenized and analyzed 48–72 h after transfection. For imaging, HEK293 cells were plated onto glass coverslips at 30,000 cells/ml and immunostained.
Primary hippocampal neurons were dissected at P0 from rats or SynCAM 1 KO mice and their WT littermates as described previously (Biederer and Scheiffele, 2007). In brief, animals were decapitated and their hippocampi dissected in Hank’s buffered salt solution (Invitrogen), then incubated in dissociation solution including 0.4 U/ml papain (LS003127; Worthington Biochemical Corporation), 1.5 mM calcium, 0.75 mM EDTA, and 0.2 mg/ml l-cysteine at 37°C for 30 min. Neurons were dissociated by manual trituration using a sterile glass pipette and plated on Matrigel (BD). At 5–7 div, neurons were transfected using Lipofectamine LTX and PLUS reagent (Invitrogen). For live imaging, neurons were plated on glass-bottom 35-mm dishes (MatTek Corporation). 2.5 µM Lat-A (Cayman Chemical) was added to the medium for 24 h before acquiring live imaging data. For select conditions indicated in Fig. 8 (C and D), Lat-A–containing medium was replaced with preconditioned neuronal medium and cells were allowed to recover for 2 h before live imaging.
Organotypic slice culture
Organotypic slice cultures were prepared as described previously (Stoppini et al., 1991). In brief, hippocampi were dissected from rat pups at P4 or P5, and 400-µm sagittal slices were obtained using a McIlwain tissue chopper (Mickle Laboratory Engineering). Slices were grown on Teflon membranes and cell culture inserts (both from EMD Millipore) for 4 d at 37°C, then moved to 33°C. Media containing 10% horse serum was changed every 4 d. At 10 div, slices were biolistically transfected with either shScramble or shFarp1 using a Helios Gene Gun (Bio-Rad Laboratories). At 13 div, slices were fixed in 4% PFA and mounted on slides with Aqua Poly/Mount (Polysciences, Inc.). Slices were imaged by confocal microscope (LSM710; Carl Zeiss). Pyramidal neurons in the CA1 region were identified based on their morphology, and secondary and tertiary dendrites were analyzed for spine densities.
Immunocytochemistry
Immunocytochemistry using the primary antibodies described above was performed on cultured hippocampal neurons at the indicated ages. Neurons were washed with PBS and fixed with 4% paraformaldehyde in PBS containing 4% sucrose for 15 min at room temperature. For SynCAM 1 surface staining, live neurons were stained for 10 min at room temperature in PBS with SynCAM 1 antibody (MBL clone 3E1, 1:200), then washed and fixed in PFA. Neurons were washed again and blocked and permeabilized in PBS containing 3% FBS and 0.1% Triton X-100. Neurons were then incubated with primary antibodies against proteins other than SynCAM 1 in blocking buffer overnight at 4°C and then secondary Alexa Fluor dye-conjugated antibodies (Invitrogen) for 90 min at 4°C, then washed and mounted on glass coverslips in Vectashield medium (Vector Laboratories).
Microscopy and analysis
Imaging of fixed and live samples was performed on a spinning disk microscope (UltraView VoX; Perkin-Elmer) equipped with a camera (C9100-50; Hamamatsu Photonics) and an autofocus system (Perfect Focus; Nikon) unless noted otherwise. Fluorochromes imaged include GFP (488 nm), mCherry (568 nm), and Alexa Fluor 647 (Invitrogen). Unless otherwise stated, images were acquired with a 60× oil Plan-Apochromat VC lens (Nikon) with 1.4 NA. Live imaging of neurons plated on glass-bottom dishes (MatTek Corporation) was performed in modified Tyrode buffer (Biederer and Scheiffele, 2007) on a stage heated to 37°C.
Imaging of live neurons expressing the Raichu-Rac1 probe were performed on a confocal microscope (LSM710; Carl Zeiss) equipped with an argon laser (Lasos; Lasertechnik GmbH) and acquired with Zen 2010 software (Carl Zeiss). Images were acquired using a 63× oil Plan-Apochromat lens with 1.4 NA (Carl Zeiss). Raichu-Rac1 imaging was performed as described previously (Itoh et al., 2002; Nakamura et al., 2005). In brief, CFP was excited at 458 nm, and z stacks were obtained alternately in both CFP (465–515 nm) and YFP (520–650 nm) channels, and collapsed into 2D images. Local Rac1 activity was measured as the ratio of the fluorescence intensity of YFP to the intensity of CFP, and regions of interest were selected in dendritic protrusions versus the adjacent shaft. Care was taken to minimize variation in exposure time, laser power, and gain between conditions and experiments. For imaging of fixed neuronal cultures, z stacks of neurons and dendrites were taken and collapsed into 2D frames for analysis.
Spine fluorescence intensities of UtrCH-Cherry were normalized to the average UtrCH-Cherry fluorescence intensity measured in three areas along the dendritic shaft adjacent to each analyzed spine. UtrCH-Cherry was imaged live throughout this study as the signal tended to fade after fixation.
Analyses of spine density and morphology, VGlut1 and -2 puncta density, and bassoon puncta density were performed using ImageJ. Analyses of bassoon puncta intensity, line scans of protein distribution in HEK293 cells, tracking of live dendritic filopodia over time, fluorescence intensity measurements of the UtrCH-Cherry probe, and the ratiometric analysis of the Raichu-Rac1 probe were performed using Volocity (PerkinElmer).
Data and statistical analysis
All imaging data acquisitions for quantitated analyses and their quantitation were performed with the researcher blind to the conditions. Data analysis was performed using GraphPad Prism 5 (Graph Pad Software) and MATLAB (MathWorks). Statistical analyses were performed as indicated using either a Student’s t test with errors corresponding to the standard error of the mean or the D’Agostino and Pearson normality test. *, t test P < 0.05; **, P < 0.01; ***, P < 0.001.
Animal procedures
All animal procedures undertaken in this study were approved by the Yale University Institutional Animal Care and Use Committee and were in compliance with National Institutes of Health (NIH) guidelines.
Online supplemental material
Fig. S1 demonstrates the efficiency of Farp1 knockdown by the shFarp1 versus the shScramble construct, and rescue of Farp1 knockdown by Farp1sh resist (in HEK293 cells, Fig. S1 A; in cultured neurons, Fig. S1, B and C). Fig. S2 shows the subcellular localization of Farp1 in dendrites stained with MAP2 and the absence of Farp1 in tau-stained axons. Fig. S3 complements Fig. 4 and confirms that overexpression of Farp1sh resist rescues spine loss after Farp1 knockdown in cultured neurons. Fig. S4 complements Fig. 5 by demonstrating that overexpression of GFP-Farp1 in rat hippocampal neurons increases dendritic mushroom spine number. Video 1 shows time lapse videos of live neurons expressing shScramble control vector, shFarp1, or shFarp1 + Farp1sh resist as analyzed in Fig. 3. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205041/DC1.
Acknowledgments
We thank Drs. K. Perez de Arce and A. Ribic for experimental advice and Y. Lei for technical assistance, Y. Lee for support, S. Warren for help with GTPase assays, and Drs. S. Chandra, C. Greer, T. Koleske, and T. Pollard for comments. We thank Dr. C. Colangelo for proteomic studies, and Dr. T. Momoi (National Institute of Neuroscience, Tokyo, Japan) for providing the SynCAM 1/RA175 KO mice. We thank the Cellular Neuroscience, Neurodegeneration and Repair program for access to its imaging core run by Dr. S. Wilson.
This work was supported by NIH grant R01 DA018928 (to T. Biederer), a NARSAD award (to T. Biederer), a Dana Foundation Brain and Immuno-Imaging Grant (to T. Biederer), a National Science Foundation Graduate Research Fellowship DGE-0644492 (to L. Cheadle), and NIH grant P30 DA018343 (to the Yale/NIDA Neuroproteomics Center).
Author contributions: L. Cheadle and T. Biederer conceived the approaches, L. Cheadle and T. Biederer performed experiments, and L. Cheadle and T. Biederer wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- DH
- Dbl oncogene homology
- div
- days in vitro
- Farp1
- FERM, Rho/ArhGEF, and Pleckstrin domain protein 1
- FERM
- 4.1 protein/ezrin/radixin/moesin
- GEF
- guanine nucleotide exchange factor
- GPI
- glycosylphosphatidylinositol
- KO
- knockout
- Lat-A
- Latrunculin A
- P
- postnatal day
- PH
- pleckstrin homology
- PSD
- postsynaptic density
- SynCAM
- synaptic cell adhesion molecule
- WT
- wild type
References
- Amor D.J., Voullaire L., Bentley K., Savarirayan R., Choo K.H. 2005. Mosaic monosomy of a neocentric ring chromosome maps brachyphalangy and growth hormone deficiency to 13q31.1-13q32.3. Am. J. Med. Genet. A. 133A:151–157 10.1002/ajmg.a.30527 [DOI] [PubMed] [Google Scholar]
- Biederer T. 2006. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 87:139–150 10.1016/j.ygeno.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Biederer T., Scheiffele P. 2007. Mixed-culture assays for analyzing neuronal synapse formation. Nat. Protoc. 2:670–676 10.1038/nprot.2007.92 [DOI] [PubMed] [Google Scholar]
- Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E.T., Südhof T.C. 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 297:1525–1531 10.1126/science.1072356 [DOI] [PubMed] [Google Scholar]
- Bonhoeffer T., Yuste R. 2002. Spine motility. Phenomenology, mechanisms, and function. Neuron. 35:1019–1027 10.1016/S0896-6273(02)00906-6 [DOI] [PubMed] [Google Scholar]
- Burkel B.M., von Dassow G., Bement W.M. 2007. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton. 64:822–832 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chklovskii D.B., Mel B.W., Svoboda K. 2004. Cortical rewiring and information storage. Nature. 431:782–788 10.1038/nature03012 [DOI] [PubMed] [Google Scholar]
- Cho K.O., Hunt C.A., Kennedy M.B. 1992. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 9:929–942 10.1016/0896-6273(92)90245-9 [DOI] [PubMed] [Google Scholar]
- Dávalos A., Fernández-Hernando C., Sowa G., Derakhshan B., Lin M.I., Lee J.Y., Zhao H., Luo R., Colangelo C., Sessa W.C. 2010. Quantitative proteomics of caveolin-1-regulated proteins: characterization of polymerase i and transcript release factor/CAVIN-1 IN endothelial cells. Mol. Cell. Proteomics. 9:2109–2124 10.1074/mcp.M110.001289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini A.E., Schnell E., Chetkovich D.M., Nicoll R.A., Bredt D.S. 2000. PSD-95 involvement in maturation of excitatory synapses. Science. 290:1364–1368 [PubMed] [Google Scholar]
- Elia L.P., Yamamoto M., Zang K., Reichardt L.F. 2006. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 51:43–56 10.1016/j.neuron.2006.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.M., Raimondi A., Paradise S., Shen H., Mesaki K., Ferguson A., Destaing O., Ko G., Takasaki J., Cremona O., et al. 2009. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev. Cell. 17:811–822 (published erratum appears in Dev. Cell. 2010. 18:332) 10.1016/j.devcel.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C., Feinberg M., Popov V., Harris K.M. 1998. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J. Neurosci. 18:8900–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Kaech S., Knutti D., Matus A. 1998. Rapid actin-based plasticity in dendritic spines. Neuron. 20:847–854 10.1016/S0896-6273(00)80467-5 [DOI] [PubMed] [Google Scholar]
- Fogel A.I., Akins M.R., Krupp A.J., Stagi M., Stein V., Biederer T. 2007. SynCAMs organize synapses through heterophilic adhesion. J. Neurosci. 27:12516–12530 10.1523/JNEUROSCI.2739-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel A.I., Stagi M., Perez de Arce K., Biederer T. 2011. Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. EMBO J. 30:4728–4738 10.1038/emboj.2011.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost N.A., Kerr J.M., Lu H.E., Blanpied T.A. 2010. A network of networks: cytoskeletal control of compartmentalized function within dendritic spines. Curr. Opin. Neurobiol. 20:578–587 10.1016/j.conb.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K., Kim M.J., Hashikawa T., Scheiffele P., Sheng M., Hayashi Y. 2007. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci. 10:186–195 10.1038/nn1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R., Wennerberg K., Arthur W.T., Noren N.K., Ellerbroek S.M., Burridge K. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406:425–437 10.1016/S0076-6879(06)06031-9 [DOI] [PubMed] [Google Scholar]
- Govek E.E., Newey S.E., Van Aelst L. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49 10.1101/gad.1256405 [DOI] [PubMed] [Google Scholar]
- Groves J.T., Kuriyan J. 2010. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 17:659–665 10.1038/nsmb.1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M.J., Eva A., Zangrilli D., Aaronson S.A., Evans T., Cerione R.A., Zheng Y. 1994. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J. Biol. Chem. 269:62–65 [PubMed] [Google Scholar]
- Hoover K.B., Bryant P.J. 2000. The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr. Opin. Cell Biol. 12:229–234 10.1016/S0955-0674(99)00080-0 [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Hoogenraad C.C. 2010. Actin in dendritic spines: connecting dynamics to function. J. Cell Biol. 189:619–629 10.1083/jcb.201003008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jones D.H., Matus A.I. 1974. Isolation of synaptic plasma membrane from brain by combined flotation-sedimentation density gradient centrifugation. Biochim. Biophys. Acta. 356:276–287 10.1016/0005-2736(74)90268-5 [DOI] [PubMed] [Google Scholar]
- Kasai H., Matsuzaki M., Noguchi J., Yasumatsu N., Nakahara H. 2003. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 26:360–368 10.1016/S0166-2236(03)00162-0 [DOI] [PubMed] [Google Scholar]
- Kayser M.S., Nolt M.J., Dalva M.B. 2008. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron. 59:56–69 10.1016/j.neuron.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. 2009. Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nat. Neurosci. 12:15–20 10.1038/nn.2231 [DOI] [PubMed] [Google Scholar]
- Knott G.W., Holtmaat A., Wilbrecht L., Welker E., Svoboda K. 2006. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat. Neurosci. 9:1117–1124 10.1038/nn1747 [DOI] [PubMed] [Google Scholar]
- Koyano Y., Kawamoto T., Shen M., Yan W., Noshiro M., Fujii K., Kato Y. 1997. Molecular cloning and characterization of CDEP, a novel human protein containing the ezrin-like domain of the band 4.1 superfamily and the Dbl homology domain of Rho guanine nucleotide exchange factors. Biochem. Biophys. Res. Commun. 241:369–375 10.1006/bbrc.1997.7826 [DOI] [PubMed] [Google Scholar]
- Lindskog M., Li L., Groth R.D., Poburko D., Thiagarajan T.C., Han X., Tsien R.W. 2010. Postsynaptic GluA1 enables acute retrograde enhancement of presynaptic function to coordinate adaptation to synaptic inactivity. Proc. Natl. Acad. Sci. USA. 107:21806–21811 10.1073/pnas.1016399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A., Ackermann M., Pehling G., Byers H.R., Fujiwara K. 1982. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc. Natl. Acad. Sci. USA. 79:7590–7594 10.1073/pnas.79.23.7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M., Südhof T.C., Biederer T. 2012. Synaptic cell adhesion. Cold Spring Harb. Perspect. Biol. 4:a005694 10.1101/cshperspect.a005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller M.L., Shi Y., Reichardt L.F., Ethell I.M. 2006. EphB receptors regulate dendritic spine morphogenesis through the recruitment/phosphorylation of focal adhesion kinase and RhoA activation. J. Biol. Chem. 281:1587–1598 10.1074/jbc.M511756200 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Aoki K., Matsuda M. 2005. Monitoring spatio-temporal regulation of Ras and Rho GTPase with GFP-based FRET probes. Methods. 37:146–153 10.1016/j.ymeth.2005.05.021 [DOI] [PubMed] [Google Scholar]
- Nakayama A.Y., Harms M.B., Luo L. 2000. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Nagai T., Miyawaki A., Hayashi Y. 2004. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7:1104–1112 10.1038/nn1311 [DOI] [PubMed] [Google Scholar]
- Penzes P., Beeser A., Chernoff J., Schiller M.R., Eipper B.A., Mains R.E., Huganir R.L. 2003. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 37:263–274 10.1016/S0896-6273(02)01168-6 [DOI] [PubMed] [Google Scholar]
- Penzes P., Cahill M.E., Jones K.A., VanLeeuwen J.E., Woolfrey K.M. 2011. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14:285–293 10.1038/nn.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado M.P., Terry-Lorenzo R.T., Waites C.L., Garner C.C., Malenka R.C. 2006. Transsynaptic signaling by postsynaptic synapse-associated protein 97. J. Neurosci. 26:2343–2357 10.1523/JNEUROSCI.5247-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E.M., Krupp A.J., Perez de Arce K., Ghosh A.K., Fogel A.I., Boucard A., Südhof T.C., Stein V., Biederer T. 2010. SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron. 68:894–906 10.1016/j.neuron.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., Schaeffer D.A. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics. 6:1638–1655 10.1074/mcp.T600050-MCP200 [DOI] [PubMed] [Google Scholar]
- Shu X., Lev-Ram V., Deerinck T.J., Qi Y., Ramko E.B., Davidson M.W., Jin Y., Ellisman M.H., Tsien R.Y. 2011. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9:e1001041 10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagi M., Fogel A.I., Biederer T. 2010. SynCAM 1 participates in axo-dendritic contact assembly and shapes neuronal growth cones. Proc. Natl. Acad. Sci. USA. 107:7568–7573 10.1073/pnas.0911798107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L., Buchs P.A., Muller D. 1991. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 37:173–182 10.1016/0165-0270(91)90128-M [DOI] [PubMed] [Google Scholar]
- Südhof T.C. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903–911 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S., Südhof T.C. 2000. Specificity of Ca2+-dependent protein interactions mediated by the C2A domains of synaptotagmins. Biochemistry. 39:2940–2949 10.1021/bi9920984 [DOI] [PubMed] [Google Scholar]
- Tada T., Sheng M. 2006. Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16:95–101 10.1016/j.conb.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Tagliaferro P., Morales M. 2008. Synapses between corticotropin-releasing factor-containing axon terminals and dopaminergic neurons in the ventral tegmental area are predominantly glutamatergic. J. Comp. Neurol. 506:616–626 10.1002/cne.21576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A., Minden A., Yuste R. 2000. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb. Cortex. 10:927–938 10.1093/cercor/10.10.927 [DOI] [PubMed] [Google Scholar]
- Thomas L.A., Akins M.R., Biederer T. 2008. Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J. Comp. Neurol. 510:47–67 10.1002/cne.21773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Bikoff J.B., Kane C.G., Tolias C.S., Hu L., Greenberg M.E. 2007. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc. Natl. Acad. Sci. USA. 104:7265–7270 10.1073/pnas.0702044104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy T.E., Yan J.J., Chen L. 2011. Acute knockdown of AMPA receptors reveals a trans-synaptic signal for presynaptic maturation. EMBO J. 30:1577–1592 10.1038/emboj.2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.S., Rubio M.E., Clem R.L., Johnson D., Case L., Tessier-Lavigne M., Huganir R.L., Ginty D.D., Kolodkin A.L. 2009. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 462:1065–1069 10.1038/nature08628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenmayer N., Körber C., Liu H., Kremer T., Varoqueaux F., Chapman E.R., Brose N., Kuner T., Dresbach T. 2009. Postsynaptic Neuroligin1 regulates presynaptic maturation. Proc. Natl. Acad. Sci. USA. 106:13564–13569 10.1073/pnas.0905819106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfrey K.M., Srivastava D.P., Photowala H., Yamashita M., Barbolina M.V., Cahill M.E., Xie Z., Jones K.A., Quilliam L.A., Prakriya M., Penzes P. 2009. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat. Neurosci. 12:1275–1284 10.1038/nn.2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Photowala H., Cahill M.E., Srivastava D.P., Woolfrey K.M., Shum C.Y., Huganir R.L., Penzes P. 2008. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J. Neurosci. 28:6079–6091 10.1523/JNEUROSCI.1170-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. 2011. Dendritic spines and distributed circuits. Neuron. 71:772–781 10.1016/j.neuron.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R., Bonhoeffer T. 2004. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5:24–34 10.1038/nrn1300 [DOI] [PubMed] [Google Scholar]
- Zhang W.D., Benson D.L. 2001. Stages of synapse development defined by dependence on F-actin. J. Neurosci. 21:5169–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Webb D.J., Asmussen H., Niu S., Horwitz A.F. 2005. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 25:3379–3388 10.1523/JNEUROSCI.3553-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang B., Su Y.S., Sockanathan S. 2009. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 61:359–372 10.1016/j.neuron.2008.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K., Knott G., Shepherd G.M., Shenolikar S., Svoboda K. 2004. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 44:321–334 10.1016/j.neuron.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Ziv N.E., Smith S.J. 1996. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 17:91–102 10.1016/S0896-6273(00)80283-4 [DOI] [PubMed] [Google Scholar]