Abstract
Cellular defects that impair the fidelity of mitosis promote chromosome missegregation and aneuploidy. Increasing evidence reveals that errors in mitosis can also promote the direct and indirect acquisition of DNA damage and chromosome breaks. Consequently, deregulated cell division can devastate the integrity of the normal genome and unleash a variety of oncogenic stimuli that may promote transformation. Recent work has shed light on the mechanisms that link abnormal mitosis with the development of DNA damage, how cells respond to such affronts, and the potential impact on tumorigenesis.
Introduction
Progression from a nontransformed normal cell to a malignant cancer cell requires multiple genetic changes that hyperactivate oncogenes while restraining tumor suppressors (Hanahan and Weinberg, 2011). This occurs via two distinct, but not mutually exclusive, mechanisms: the acquisition of genetic mutations, and gene copy number changes.
Genetic mutations arise as a consequence of cells failing to efficiently detect, repair, and/or respond to DNA damage, and may be subtle (e.g., single nucleotide changes) or more complex (e.g., amplifications, deletions, insertions, translocations; Negrini et al., 2010). Mutations can arise spontaneously, as a consequence of endogenous genotoxic stress, such as from stalled/collapsing replication forks generated during S phase or reactive oxygen species produced by normal metabolic activity (Spry et al., 2007; Hoeijmakers, 2009; Branzei and Foiani, 2010; Ciccia and Elledge, 2010). However, environmental and/or genetic perturbations that markedly increase DNA damage—and subsequent mutation rates—greatly facilitate oncogenesis. This is best illustrated by the significant predisposition to cancer in familial genetic diseases where components of DNA repair or checkpoint signaling are lost or mutated; examples include hereditary nonpolyposis colorectal cancer syndrome (HNPCC; mutations in MLH1, MSH2, MSH6, or PMS2; Spry et al., 2007; Hoeijmakers, 2009), hereditary breast and ovarian cancer syndrome (mutations in BRCA1 and BRCA2; Fackenthal and Olopade, 2007), Fanconi anemia (caused by mutations in any of a number of Fanconi genes important for DNA repair; Moldovan and D’Andrea, 2009), and Li-Fraumeni syndrome (mutations in TP53; Varley et al., 1997).
Independent of DNA damage and mutation, whole chromosome and segmental aneuploidies can also dramatically alter gene copy number of relevant oncogenes and tumor suppressors. Recent mouse models demonstrate that merely elevating the rates of chromosome missegregation is sufficient to promote tumor development in vivo, at least in part by facilitating loss of heterozygosity of known tumor suppressor genes (Weaver et al., 2007; Baker et al., 2009; Baker and van Deursen, 2010). A number of cellular defects are known to generate whole chromosome aneuploidy, including atypical mitotic spindle assembly, inefficient chromosome congression, abnormal microtubule dynamics, cohesion and condensation defects, supernumerary centrosomes, and a defective spindle assembly checkpoint (Schvartzman et al., 2010; Compton, 2011; Gordon et al., 2012; Holland and Cleveland, 2012). The common factor among these defects is that they all manifest during mitosis, when chromosomes physically separate. Thus, it is widely accepted that abnormal mitosis can contribute to tumorigenesis via the generation of aneuploidy.
One unresolved question concerns the extent to which abnormal mitosis and DNA damage, the two key promoters of genomic instability, are linked. Although it has been known for some time that DNA damage adversely affects the efficacy of mitosis, the reciprocal possibility—that abnormal mitosis promotes DNA damage—has been largely overlooked in studies of cancer cell biology. However, several recent reports demonstrate that abnormal mitosis alone is sufficient to generate DNA damage. Thus, impaired mitosis may negatively effect genome stability in two ways: not only by causing genome destabilizing whole chromosome aneuploidy, but also by promoting the acquisition of potentially growth-promoting mutations.
The damaging effects of prolonged mitosis
In proliferating cells, the phases of the cell cycle exist to accomplish one specific task: to accurately replicate all chromosomes so that they can be efficiently and equally partitioned into two daughter cells during mitosis. Numerous checkpoints have evolved to ensure that mitosis only proceeds when growth conditions are ideal and chromosomes are efficiently replicated and free of damage. This level of quality control takes time, and, generally speaking, proliferating mammalian somatic cells require 12–30 h to properly prepare for division. By contrast, mitosis itself is relatively rapid, typically lasting only 20–60 min, depending on chromosome number and the efficiency of spindle assembly (Yang et al., 2008). It may seem surprising that cells are programed to move so swiftly through mitosis given its importance and the amount of time and energy invested in preparing for the event. This begs the question: Why the rush?
Ironically, the simplest explanation is that mitosis is both destructive and stressful for the dividing cell, and is therefore a process best finished quickly. During mitosis, among other things, the nuclear envelope is torn apart (Gerace et al., 1978), the Golgi and ER membrane systems undergo dramatic reorganization (Hetzer, 2010; Robbins and Gonatas, 1964), vesicle trafficking ceases (Sager et al., 1984), chromosomes condense and transcription is disabled (Taylor, 1960; Prescott and Bender, 1962), translation is slowed (Prescott and Bender, 1962; Bonneau and Sonenberg, 1987), and both the actin and microtubule cytoskeletons are reshaped to facilitate cell rounding and assembly of the bipolar mitotic spindle (Saxton et al., 1984; Kunda and Baum, 2009). Such dramatic perturbations to the normal cellular architecture during mitosis cannot be tolerated indefinitely, and we are just beginning to understand the consequences of extending such an abnormal state: the infrastructure of mitotic chromosomes, slowly but surely, begins to break down during prolonged mitosis, ultimately giving rise to DNA breaks.
The first hints that prolonged mitotic arrest might promote DNA damage came from early studies that used microtubule poisons (e.g., nocodazole, colchicine) to arrest cells in mitosis. Such drug-treated cells were unable to satisfy the spindle assembly checkpoint (SAC) and were maintained in mitotic arrest until they either died in mitosis or “slipped” back into interphase without anaphase or cytokinesis, becoming tetraploid (Rieder and Palazzo, 1992; Lanni and Jacks, 1998; Rieder and Maiato, 2004; Quignon et al., 2007). These tetraploid cells arrested in the subsequent G1 phase in a p53-dependent manner that was shown to be a result of DNA damage, though whether the damage occurred during the prolonged mitosis or was a consequence of slippage was unclear (Lanni and Jacks, 1998; Rieder and Maiato, 2004; Quignon et al., 2007). More robust evidence that DNA damage arises during mitosis quickly followed, after both drug as well as genetic treatments were used to prolong mitosis in a variety of cell lines. DNA damage, as identified by detection of the phosphorylated histone variant H2AX (γ-H2AX; Rogakou et al., 1998), was observed to subtly emerge beginning ∼6 h after mitotic arrest and gradually accumulate with sustained mitosis (Dalton et al., 2007). Many groups, using alternative methods to prolong mitosis in a wide variety of cell types, have confirmed the generality of this finding (Uetake and Sluder, 2010; Crasta et al., 2012; Hayashi et al., 2012; Orth et al., 2012).
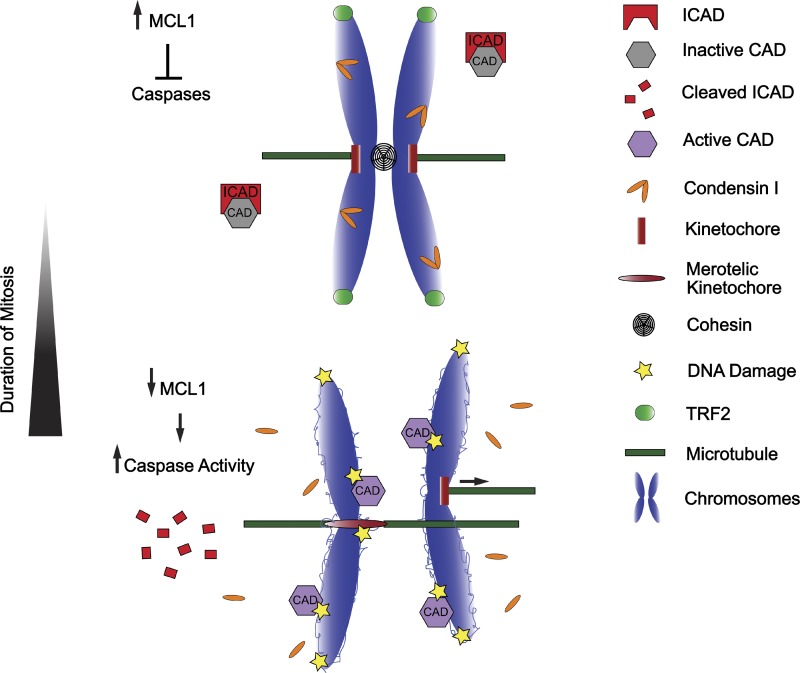
Why prolonged mitotic arrest causes DNA damage remains an open area of investigation, but some mechanisms have recently become apparent. Orth et al. (2012) observed that cells arrested in mitosis for an extended period of time (∼16 h) showed outer mitochondrial membrane permeabilization and subtle leakage of cytochrome c concomitant with the emergence of DNA damage (Orth et al., 2012). Release of cytochrome c into the cytosol is a well-known initiator of apoptosis, and activates a family of cysteine proteases termed caspases (the “executioners” of the cell). Among the numerous protein targets that caspases cleave is ICAD, an inhibitor of the DNase enzyme CAD (Enari et al., 1998; Sakahira et al., 1998). Cleaved ICAD thus frees CAD, which in turn proceeds to cleave chromosomal DNA, once presumed to be a “point of no return” for cells (Enari et al., 1998; Sakahira et al., 1998). However, the authors propose that the low levels of cytochrome c release induced by prolonged mitosis may trigger only a partial apoptotic response with limited CAD DNase activation and, instead of shearing chromosomes entirely, simply induce limited DNA breaks (Fig. 1; Orth et al., 2012). This view is supported by the finding that addition of caspase inhibitors significantly reduces the occurrence of γ-H2AX and mitotic cell death during prolonged mitosis, as does suppression of CAD DNase activity by expression of a noncleavable version of ICAD (Orth et al., 2012). Destruction of another caspase target protein during mitosis, Cap-H, also facilitates chromosomal cleavage by CAD. Cap-H is a member of the condensin I complex, which maintains chromosome structure during mitosis. Cleavage of Cap-H by partially activated caspases abolishes the condensin I complex, disrupts the integrity of compacted mitotic chromosomes, and exposes highly accessible decondensed DNA loops to CAD nuclease activity (Lai et al., 2011). This cleavage of Cap-H by caspases is critical for the induction of DNA breaks: expression of a caspase-resistant form of Cap-H protects mitotic chromosome structure during prolonged mitosis, and prevents chromosomal fragmentation (Lai et al., 2011).
Figure 1.
Prolonged mitosis gives rise to DNA damage through multiple mechanisms. Prolonged mitosis leads to depletion of many proteins, including anti-apoptotic proteins such as MCL1. This induces a partial caspase activation and destruction of ICAD and CAP-H, which frees the DNase CAD to cleave decondensed loops on chromosomal DNA. In addition, the telomere-protecting protein TRF2 loses its telomeric localization, exposing the linear ends of chromosomes that are recognized as DSBs. Finally, sister chromatid cohesion is gradually lost during prolonged mitosis, and this “cohesion fatigue” promotes premature sister chromatid separation and merotelic attachment. The forces generated by merotelic attachment at kinetochores may combine with preexisting DNA damage at the underlying centromeres to generate arm-level chromosome breaks, an anomaly commonly observed in human cancers.
Why cells that undergo protracted mitosis exhibit mitochondrial outer membrane permeabilization and cytochrome c release remains unknown. At least part of the explanation comes from the fact that anti-apoptotic proteins of the BCL-2 family, such as MCL1 and BCLxl, which antagonize mitochondrial outer membrane permeabilization and cytochrome c release, are gradually lost during prolonged mitosis. Several studies have now identified mitotically active E3 ligases, such as APCCdc20 and SCFFBW7, which target MCL1 for proteasomal destruction (Harley et al., 2010; Sánchez-Pérez et al., 2010; Inuzuka et al., 2011; Millman and Pagano, 2011). Consequently, prolonged mitosis may eventually reduce MCL1 protein to levels that are insufficient to completely suppress mitochondrial permeability. Collectively, these data reinforce the view that cytochrome c release and caspase activation do not necessarily ignite an amplifiable “all or nothing” cellular termination program (Goldstein et al., 2000; Vaughan et al., 2002; Abraham and Shaham, 2004; Khodjakov et al., 2004; Larsen et al., 2010); rather, under certain conditions such as prolonged mitosis, subtle activation of components of the apoptotic machinery can lead to DNA damage without a requisite death sentence.
These data also demonstrate that a number of complex factors regulate the susceptibility of cells to acquiring DNA damage during prolonged mitosis, and suggest that some cells may be more prone to such damage than others. As an example, several studies have demonstrated that efficient loading of components of the condensin II complex to chromosomes requires functional pRb, and that pRb loss, or mutations in pRb that abolish its ability to efficiently load condensin II, lead to chromosome condensation and mitotic defects (Longworth et al., 2008; Manning et al., 2010). It would be interesting to examine whether the less compact chromosomes that lack condensin II are more susceptible to caspase-induced nuclease activity and DNA breaks during the abnormally prolonged mitosis. Perhaps an increased susceptibility to DNA breaks during mitosis may help explain the finding that pRb mutations, which abolish condensin II loading without disrupting the normal G1–S transition, promote tumor formation and aggressiveness in mouse models (Coschi et al., 2010).
Additional pathways, which are independent of partial caspase activation, also promote DNA damage during prolonged mitosis. This is best illustrated by the observation that caspase inhibitors are not always sufficient to prevent the onset of DNA damage during abnormal cell division (Dalton et al., 2007). One recent study observed that a large portion of DNA damage that stems from prolonged mitosis initially appears at telomeres, suggesting that telomere-capping proteins, which act to suppress the DNA damage response at the truncated ends of linear chromosomes, might become functionally inactivated (Hayashi et al., 2012). Indeed, TRF2, one such capping component, has been shown to leave the ends of telomeres during prolonged mitosis, even though its overall levels remain unaltered (Fig. 1). How this comes about is unclear, but it has been postulated that Aurora B kinase directly or indirectly plays a functional role in regulating the delocalization of TRF2 from telomere ends. One possibility is that TRF2 loss from telomeres is due to enhanced Aurora B activity during prolonged mitosis, perhaps because of a gradual loss of the phosphatase activity that opposes Aurora B (Hayashi et al., 2012). Indeed, declining steady-state levels of unstable proteins may be a major contributing factor to the accumulation of DNA damage during prolonged mitosis, as mitotic cells are transcriptionally silenced and do not demonstrate cap-dependent translation (only IRES-mediated translation of a limited number of proteins persists; Bonneau and Sonenberg, 1987). It would be interesting to know if reducing enzymes (e.g., catalases, peroxidases) are one such family of limiting factors. Reducing enzymes convert DNA-damaging reactive oxygen species (ROS), which are produced by mitochondria during normal aerobic respiration, into byproducts that are harmless to the cell. It is tempting to speculate that reducing enzymes may diminish during prolonged mitosis to levels that can no longer adequately neutralize ROS, thus enabling an attack on DNA.
Although a comprehensive understanding of the mechanisms underlying DNA damage remains to be elucidated, prolonged mitosis clearly poses a substantial threat to the genomic stability and viability of daughter cells. Supporting this notion is the remarkable finding that cells may actually have evolved a “clock” to time the duration of mitosis, thereby furnishing a mechanism to identify potentially dangerous cells that took too long to complete division. If mitosis takes even a little longer to complete than normal (for instance, lasting longer than ∼1.5 h) then the resulting daughter cells activate a durable p53-dependent G1 arrest that culls them from the proliferating population (Uetake and Sluder, 2010).
This observation raises several questions regarding the trigger for p53-dependent G1 arrest in daughter cells that are born from just slightly prolonged mitosis. The most likely culprit, DNA damage, does not appear to be responsible for this phenomenon, as no obvious DNA damage can be detected in cells that are arrested in mitosis for such relatively short periods of time, consistent with previous studies which show that γ-H2AX staining is detectable starting only after ∼5–16 h of mitosis depending on cell type (Dalton et al., 2007; Orth et al., 2012). Alternatively, it has been postulated that p53 gradually accumulates during prolonged mitosis, enforcing subsequent arrest in G1 (Blagosklonny, 2006); however, this has not yet been experimentally observed (Minn et al., 1996; Orth et al., 2012). Nevertheless, whatever the stress from prolonged mitosis may be, it appears to persist, or perhaps even permanently mark cells as being defective. This was demonstrated in an elegant experiment in which daughter cells generated from a slightly prolonged mitosis, which would normally arrest, were treated transiently with a p38 inhibitor to allow them to bypass the p53-induced arrest, proceed through a second cell cycle, and reenter mitosis. Remarkably, despite the fact that the second mitosis completed with normal timing, the daughter cells once again rearrested in G1 (Uetake and Sluder, 2010). This confirms that when it comes to prolonged mitosis, nontransformed cells don’t take any chances: strong anti-proliferative mechanisms prevent the progeny of these abnormal cells, and whatever genetic anomalies they may harbor, from further division.
Effects of merotely on genome stability
In addition to prolonged mitosis, other aspects of abnormal cell division may play significant roles in generating DNA damage. Recent sequencing efforts have revealed that tumor cells are highly enriched in chromosome whole-arm amplifications, indicative of chromosome breaks at centromeres, yet the mechanisms through which these breaks occur remain completely unresolved (Beroukhim et al., 2010). One distinct possibility is that DNA replication and/or repair mechanisms are inefficient through the highly repetitive α-satellite regions of centromeres, which predisposes to breakage. However, it is also plausible that such shearing events occur during abnormal mitosis as a consequence of merotelic attachments.
Merotelic attachments are a specific type of kinetochore–microtubule attachment error that occurs when a single kinetochore from one chromosome is attached to microtubules from more than one spindle pole (Salmon et al., 2005; Cimini, 2008; Gregan et al., 2011). This type of attachment error is particularly dangerous because it satisfies the spindle assembly checkpoint and permits anaphase, even if left uncorrected (Cimini et al., 2001). As a consequence, during anaphase the merotelically attached chromosome is simultaneously pulled toward opposite poles via its lone kinetochore. In most cases, one spindle pole has more microtubules attached to the kinetochore than the other, causing segregation of the offending chromosome to one daughter cell, albeit to the same one as its sister chromosome at some low frequency (Cimini et al., 2001, 2002, 2003, 2004; Thompson and Compton, 2011). Occasionally, however, both poles pull the single kinetochore with equal strength, thus stalling the progression of the chromosome to one daughter or another. This gives rise to what is referred to as an anaphase “lagging chromosome” (Fig. 2 A), not to be confused with bridging chromosomes, which are formed by distinct mechanisms (discussed in the next section). Lagging chromosomes can end up in either daughter cell, depending on where the cytokinetic furrow ingresses, though they most frequently segregate to the correct cell, which is the cell opposite to where the sister chromosome segregated (Cimini et al., 2004; Thompson and Compton, 2011). Nevertheless, anaphase lagging chromosomes often lag so severely behind the other chromosomes that upon telophase they form their own nuclear envelope, creating a micronucleus, which has its own repercussions (discussed later).
Figure 2.
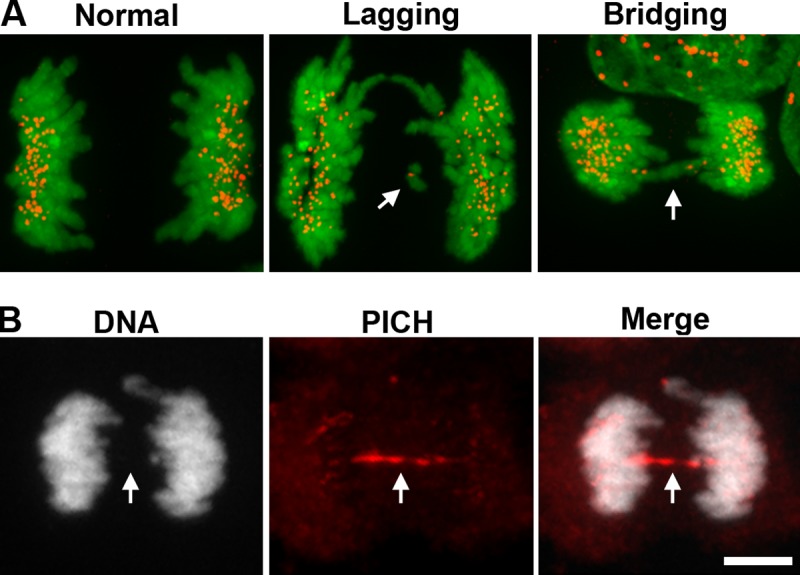
Anaphase lagging chromosomes versus chromosome bridges. (A) Anaphase lagging chromosomes (“Lagging”, white arrow) are identified as single, kinetochore-positive chromosomes that lag between the two masses of segregating chromosomes during anaphase. Lagging chromosomes are commonly caused by merotelic attachments. By contrast, pathological chromosome bridges (“Bridging”, white arrow) completely span the segregating masses of chromosomes during anaphase. Pathological bridges are caused by condensation and cohesion defects, or by dicentric chromosomes being pulled to opposite poles, and are readily visualized with DNA-intercalating dyes (kinetochores, red; chromosomes stained with the DNA intercalating dye Hoechst, green). Images reproduced from Ganem et al. (2009). (B) Ultra-fine chromosome bridges (white arrow) result from inefficient decatenation of sister chromosomes and cannot be detected using DNA dyes, requiring instead detection with specific protein markers (PICH, red; chromosomes stained with the DNA intercalating dye Hoechst, white). Bar, 10 µm. Images courtesy of Taruho Kuroda (Harvard Medical School, Boston, MA).
Anaphase lagging chromosomes experience a microtubule-generated pulling force that is strong enough to lead to the dramatic physical stretching and deformation of their kinetochores and underlying centromeric DNA (Cimini et al., 2001, 2004), though whether this force is capable of physically breaking the DNA at the centromere remains unresolved. Consideration of the force required to break the phosphodiester backbone of naked DNA makes the idea somewhat plausible: the estimated force required to rupture a single covalent bond (∼2 nN; Grandbois et al., 1999) is in the ballpark of a generous estimate of force produced by a mature kinetochore fiber containing ∼20 microtubules (∼0.2–1.5 nN; Alexander and Rieder, 1991; Nicklas, 1988). However, arguing against this idea is the fact that breaking highly condensed chromosomal DNA requires a force (∼100 nN) that is 2–3 orders of magnitude stronger than what is produced by a kinetochore fiber (Houchmandzadeh et al., 1997). This is due in part to the high elasticity of chromosomes, which can return to their normal shape after being stretched more than 10 times, and the force-diffusing pliability of kinetochores (Houchmandzadeh et al., 1997; Dong et al., 2007; Loncarek et al., 2007; Bloom, 2008). Practically, it also seems illogical that microtubule-generated forces that evolved to push and pull chromosomes in order to facilitate congression and segregation during mitosis would exhibit forces anywhere near strong enough to actually break the chromosomes. Supporting this view, several groups have shown that experimentally induced merotelically attached lagging chromosomes at early anaphase do not display any obvious signs of DNA damage (Thompson and Compton, 2010; Uetake and Sluder, 2010; Crasta et al., 2012).
Nevertheless, the possibility that certain perturbations may predispose cells to centromere breakage in conjunction with merotely during mitosis cannot be discounted. Evidence for one such situation comes from observations on a mouse cell line that lacks the Dido gene product. Dido is a centrosome-localized protein whose loss gives rise to multiple mitotic defects including centrosome amplification and lagging chromosomes (Trachana et al., 2007). Dido-null cells reportedly exhibit γ-H2AX foci adjacent to merotelically attached kinetochores, which suggests that forces from merotelic attachments may generate sufficient force to break centromeric DNA, at least in certain genetic contexts (Guerrero et al., 2010).
That cells with preexisting DNA damage are more susceptible to the pulling forces generated by merotelic attachment should also be considered. For example, merotelic attachment may unravel DNA from single-strand nicks, thus promoting conversion to a double-stranded break (DSB). If so, then conditions that promote both DNA damage and merotely might combine to generate breaks specifically at centromeres. One such condition arises during prolonged mitosis: as detailed extensively already, DNA damage accumulates during abnormally protracted mitosis, but occurring concurrently is the gradual loss of cohesion between sister centromeres, a phenomenon termed cohesion fatigue (Daum et al., 2011). A consequence of cohesion fatigue is the disassociation of sister chromatids, which promotes merotelic attachment (Fig. 1). Thus, it is tempting to speculate that subtle, perhaps imperceptible, DNA damage caused by prolonged mitosis (e.g., single nicks not identified by γ-H2AX localization) may synergize with excess merotely generated after cohesion fatigue or other mitotic defects to promote chromosomes breaks at centromeres.
Rather than breaking chromosomes directly, merotelic attachments could also facilitate the physical separation of chromosomes that already possess breaks at or near centromeres. For example, the MRN complex (Mre11–Rad50–Nbs1), which is a primary responder to DNA DSBs, detects and binds DSBs during mitosis. Mre11 is a component of the MRN complex that is believed to physically tether broken ends of chromosomes, keeping them in close proximity to facilitate repair (Chen et al., 2001; Hopfner et al., 2002). This raises the possibility that the forces generated by merotelic attachment are sufficient to overcome the tethering forces applied by the MRN complex on broken chromosomes, thus making preexisting DSBs located at or near centromeres significantly more susceptible to being completely torn apart by strong microtubule-generated forces.
The acquisition of DNA damage during cytokinesis
Cells experiencing abnormal mitosis that progress from prometaphase to anaphase without acquiring DNA lesions are not “out of the woods,” especially if they encounter problems in efficiently segregating their chromosomes. Awaiting cells after anaphase is cytokinesis, where a contracting actin-myosin ring, which generates sufficient force to cleave one cell into two, looms. A number of studies in budding and fission yeast, as well as in plants, have established that failure to clear chromosomes from the central spindle and out of the oncoming path of the cytokinetic ring and enclosing cell wall results in chromosomal cleavage, which has dire consequences for cells (McClintock, 1941; Hirano et al., 1986; Baxter and Diffley, 2008). Consequently, in yeast a quality control mechanism termed the “NoCut” pathway has been proposed that delays cytokinesis when chromatin fails to segregate out of the spindle midzone—this mechanism buys additional time for resolving the defect (Norden et al., 2006; Mendoza et al., 2009).
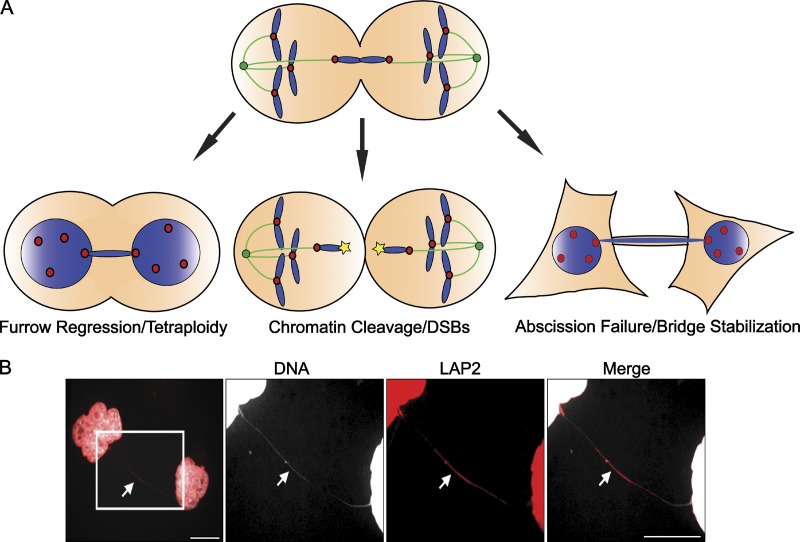
In contrast to the findings in yeast and plants, the consequences of having chromatin trapped within the cleavage plane during the ingression of the cytokinetic furrow is much more variable in mammals, and can give rise to several distinct fates including cleavage furrow regression, abscission delay, and/or chromatin cleavage (Fig. 3 A). Cleavage furrow regression is a well-documented consequence of having chromatin trapped under the furrow during mammalian cytokinesis (Mullins and Biesele, 1977), and in contrast to the obligatory chromosome breakage experienced by yeast, frequently occurs without any visible signs of DNA damage (Steigemann et al., 2009). This may be due to the fact that mammalian cytokinesis, unlike yeast and plants, does not require potentially damaging cell wall deposition after actin-myosin ring contractility. However, inducing tetraploidy to prevent chromosomal breaks may not be a positive long-term strategy for cells: tetraploid cells possess inherent stresses that typically limit their long-term proliferation (Andreassen et al., 2001; Ganem and Pellman, 2007; Krzywicka-Racka and Sluder, 2011), but they also have an increased capacity to promote transformed growth (Duelli et al., 2005; Fujiwara et al., 2005; Ganem et al., 2007; Davoli and de Lange, 2011, 2012).
Figure 3.
Consequences of chromosome bridges. (A) Chromosome bridges can promote cytokinetic furrow regression, chromosomal cleavage, or abscission failure. However, the factors governing which of these outcomes will occur remain entirely unknown. One possibility is that previously nicked or otherwise damaged chromosomes may be more susceptible to cleavage, whereas undamaged whole chromosomes may be more likely to promote furrow regression or abscission delays. (B) Chromosome bridges (white arrow) that are not cleaved or resolved during cytokinesis persist well into the following cell cycle, even stretching extensively as cells move apart (DNA, white; LAP2, red). Bars, 10 µm. Images courtesy of Taruho Kuroda (Harvard Medical School, Boston, MA).
Alternatively, it has been reported that an Aurora B–dependent mechanism similar to the yeast NoCut pathway exists in mammalian cells to stabilize the cytokinetic bridge after furrow ingression and prevent abscission in the presence of trapped chromatin (Steigemann et al., 2009). This pathway potentially provides time for cells to resolve segregation errors during the subsequent interphase while preventing the deleterious effects of tetraploidy. Remarkably, despite the fact that the daughters in this condition remain physically linked by the bridging chromatin, they behave as two discrete entities with independent cell cycle dynamics (Steigemann et al., 2009). However, virtually nothing is known about how these cells mechanistically resolve the stabilized chromatin that links them, or whether this can be accomplished without chromosomal cutting. Furthermore, chromosome bridges can persist throughout the ensuing cell cycle, and even stretch extensively as cells move far apart (Fig. 3 B), raising the possibility that no specific mechanisms exist to accurately resolve such bridges once they have been stabilized.
Finally, imaging of mammalian cells confirms that chromatin trapped in the spindle midzone does occasionally break during cytokinesis in mammalian cells (Hoffelder et al., 2004; Samoshkin et al., 2009; Janssen et al., 2011), producing recombinagenic fragments that can generate chromosome translocations in the next cell cycle (Janssen et al., 2011). Inhibition of cytokinetic ring furrowing can rescue such chromatin from acquiring DNA damage, supporting the idea that cytokinesis plays a direct role in breaking chromosomes (Janssen et al., 2011).
What remains entirely unclear are the molecular underpinnings dictating whether or not the trapped chromatin will break or cause furrow regression. A potential source of variability certainly derives from the nature of the trapped chromatin, which can be in the form of a pathological chromosome bridge, an ultra-fine chromosome bridge, or an anaphase lagging chromosome. Pathological chromosome bridges frequently result from dicentric chromosomes, which originate from inappropriately repaired DSBs or from fusions of critically shortened telomeres regions, which are then pulled to opposing poles during anaphase (Figs. 2 A and 3 A). Condensation and cohesion defects also promote these bridges, which are readily identifiable with common DNA intercalating dyes (Hauf et al., 2001; Hetzer, 2010). Ultra-fine chromosome bridges (UFBs), by contrast, are so subtle as to be virtually invisible using standard chromosome staining protocols; they arise from incomplete decatenation of entangled DNA, frequently at centromeres (Chan and Hickson, 2011), and can only be detected by visualizing protein components such as the nuclear membrane protein LAP2 or the helicases PICH and BLM that specifically localize to these bridges (Fig. 2 B; Baumann et al., 2007; Chan et al., 2007; Ke et al., 2011). Unlike pathological bridges, UFBs may play an important structural role in normal mitosis by physically linking sister centromeres during early anaphase in order to maintain tension and prevent SAC reactivation, then resolved in a regulated manner later in anaphase to prevent bridging in the spindle midzone (Baumann et al., 2007; Chan and Hickson, 2011). However, such resolution is often deregulated in cancer cells and can give rise to numerous stable UFBs that stem from both centromeres as well as from chromosome arms, after mitosis (Chan et al., 2009). Both pathological and ultra-fine chromosome bridges physically span the entire midzone, and cannot avoid the cytokinetic ring: this makes them fundamentally different from anaphase lagging chromosomes, which are capable of “sidestepping” the ingressing furrow. Bridged chromosomes are therefore significantly more likely to undergo cytokinesis-induced damage, or cause furrow regression, though anaphase lagging chromosomes are not immune (Janssen et al., 2011).
Whether or not the cytokinetic furrow cleaves the bridged chromosomes may also reflect whether or not the trapped DNA possesses any additional damage. Bridged chromosomes, weakened by DNA lesions, may be prone to breakage by the combined forces of the anaphase spindle and the cytokinetic ring, whereas undamaged chromosomes may be more resistant, and promote furrow regression. Chromosome bridges may also avoid breakage by finding strength in their numbers: because of their physical properties, chromosome bridges align parallel to the anaphase spindle during cytokinesis, and if more than one chromosome bridge exists in a given cell then the pinching cleavage furrow stacks the separate bridges upon one another. This raises the possibility that a single bridging chromosome is more susceptible to breakage than multiple bridged chromosomes, which may together form a chromatin bundle that is better able to fend off the ingressing furrow.
Imaging studies document that when breakage of bridging chromosomes occurs during cytokinesis, it frequently does so adjacent to centromeres (Hoffelder et al., 2004). Although this may be a direct consequence of centromeres experiencing more pulling forces at anaphase, it is also tempting to speculate that cleavage may be the consequence of a more biochemical reaction, such as nuclease-mediated chromosomal cutting. In this light, it is interesting to note that topoisomerase-IIα, an enzyme important for decatenating sister chromatids, localizes to centromeres during mitosis (Spence et al., 2007; Wang et al., 2008, 2010). This raises the possibility that centromere-localized topoisomerase-IIα, or perhaps another cellular nuclease, may play a key role in resolving chromosome bridges by inducing breakage at centromeres.
The cellular response to DNA damage acquired during mitosis
Cells rapidly respond to DNA damage acquired during interphase by engaging signaling pathways that disable cell cycle progression and promote DNA repair (Ciccia and Elledge, 2010). In contrast, mitotic cells do not activate any checkpoints in response to DNA damage, per se, as studies using laser irradiation or drug treatments to induce minor DNA damage during mitosis show no delays in mitotic progression despite the presence of obvious DNA fragments (Rieder and Cole, 1998). Robust DNA damage can delay mitotic progression, but this only happens when massive damage occurs right at centromeric DNA and alters kinetochore function, thus preventing normal spindle assembly checkpoint silencing (Mikhailov et al., 2002; Nitta et al., 2004; Dotiwala et al., 2010).
The discovery that no DNA damage checkpoint exists to delay mitotic progression led to speculation that cells might be “blind” to mitotic chromosomal damage and might fail to mount any sort of response, let alone try and repair the lesions. However, the mere fact that γ-H2AX appears at sites of DNA damage in mitotic cells demonstrates the existence of at least a partial response, as γ-H2AX requires phosphorylation by the kinase ATM, which is recruited to sites of DNA damage by the DNA break-sensing MRN complex (Ciccia and Elledge, 2010; Giunta and Jackson, 2011). Although it is clear that mitotic cells are capable of mounting such a primary DNA damage response, Giunta et al. (2010) demonstrate that the second phase of the DNA damage response, the recruitment of downstream proteins important for chromosome unwinding and subsequent repair (such as RNF8, RNF168, 53BP1, and BRCA1), is absent until cells exit from mitosis into the subsequent G1 phase. Several groups have observed that many of these proteins (e.g., 53BP1), which localize to spontaneously arising DNA breaks during interphase, are removed from chromosomes once cells enter mitosis (Jullien et al., 2002; Nelson et al., 2009; van Vugt et al., 2010).
Other aspects of the DNA damage response are similarly impaired during mitosis. For example, the checkpoint kinase Chk2, which is a downstream target of ATM, fails to become activated during mitosis despite the presence of active ATM. This is due at least in part to inhibitory phosphorylation by the mitotically active kinase Plk1 (van Vugt et al., 2010). Thus, mitotic cells have developed numerous mechanisms to prevent a complete DNA damage response during cell division. It has been speculated that attempting DNA repair in mitosis would be catastrophic, as repair would require disruption of the integrity of the highly compacted chromosome structure and perhaps lead to a multitude of segregation defects, not to mention potentially prolonging stressful mitosis and increasing the risk of acquiring even more damage (Giunta and Jackson, 2011). Consequently, it has been posited that mitotic cells simply mark sites of DNA damage during mitosis so that these sites can be more quickly identified and dealt with during the subsequent G1 phase, when repair is less threatening (Giunta et al., 2010). This still represents a risky proposition, as unrepaired chromosome fragments may ultimately get missegregated during the next mitosis.
Indirect consequences of abnormal mitosis
In addition to the direct damage to chromosomal integrity, abnormal mitosis also exerts indirect effects on the future stability of the genome. For example, chromosome missegregation and the generation of aneuploidy are common byproducts of abnormal mitosis and may occur without any immediate acquisition of DNA damage. Nevertheless, aneuploidy is not without consequences: missegregation of even a single chromosome in mammalian cells can lead to the deregulated expression of hundreds, or even thousands, of individual genes, including many that are involved in critical processes such as DNA replication and repair (Williams et al., 2008; Stingele et al., 2012). Consequently, it has been observed that spontaneously arising nontransformed aneuploid cells are prone to p53-mediated cell cycle arrest (Thompson and Compton, 2010). However, cells capable of escaping this arrest are subject to increased mutation rates, recombination frequencies, and further chromosome missegregation (Thompson and Compton, 2010; Sheltzer et al., 2011), supporting the long-standing belief that aneuploidy imparts a “mutator phenotype” (Duesberg et al., 1998; Holliday, 1989). Thus, missegregation of a single chromosome caused by abnormal mitosis has the capacity to set in motion a self-propagating storm of genomic instability.
Another outcome of abnormal mitosis is the generation of micronuclei, which form when anaphase lagging chromosomes reassemble nuclear envelopes independent from the spatially separated primary nucleus during telophase. Micronuclei are conspicuous structures that have long been observed in genomically unstable tumor cells, yet their biological consequences were controversial (Terradas et al., 2010). It has recently been demonstrated that micronuclei possess a lower density of nuclear pores relative to primary nuclei, and exhibit inefficient nuclear import; as a consequence, recruitment of DNA licensing, replication, and repair factors is impaired, and duplication of chromosomes residing in micronuclei during S phase is markedly perturbed, thereby inducing replication stress and DNA breaks (Crasta et al., 2012). Furthermore, DNA replication in micronuclei is also asynchronous with primary nuclei, with replication in micronuclei persisting in otherwise G2 cells. Upon mitotic entry, chromosomes in micronuclei condense even though they are still in S phase, a phenomenon termed premature chromosome compaction (PCC), which drastically disrupts the normal structural integrity of chromosomes and gives rise to a “pulverized” appearance (Johnson and Rao, 1970; Crasta et al., 2012). The extent to which pulverized chromosomes are broken remains unclear, as studies suggest that prematurely compacted chromosomes from normal S phase cells often have thin, unreplicated double-stranded DNA connecting regions of condensed, replicated chromosome masses (Hanks et al., 1983; Gollin et al., 1984; Gotoh, 2007). However, PCC may have a significantly more detrimental effect on the massively damaged chromosomes in micronuclei, which lack a normal complement of DNA replication and repair machinery. In addition, micronuclei often persist for several cell cycles before reincorporating into the primary nucleus, providing multiple opportunities for chromosomal damage to occur and accumulate.
The isolation of a single chromosome within a micronuclei offers an appealing mechanistic explanation for “chromothripsis,” the recently discovered phenomenon in which a massively reorganized whole chromosome, or arm-level chromosome, is generated in a single catastrophic event (Stephens et al., 2011). It appears that one mechanism, or a combination of two distinct mechanisms, generate chromothripsis: chromosome pulverization followed by restitching via nonhomologous end-joining, and abortive replication with fork stalling and template switching or microhomology-mediated break induced repair (Liu et al., 2011, 2012; Stephens et al., 2011; Forment et al., 2012). Both types of defect can occur in micronuclei because chromosomes trapped in micronuclei are prone to both replication stress and fragmentation. Thus, chromothripsis may be another mutagenic outcome of abnormal mitosis.
Conclusions
At any given moment, an estimated 100 million cells in our bodies are undergoing mitosis (Alberts et al., 2002). Given the overall complexity and large number of proteins required to successfully accomplish cell division, it is not surprising that the process is not foolproof: roughly 1 out of every 100 cell divisions gives rise to a chromosome missegregation event, indicating an underlying mitotic abnormality (Cimini et al., 1999; Thompson and Compton, 2008). Mitotic defects can lead to aneuploidy as well as DNA damage, either of which can severely alter the genetic landscape of a cell and impart strong growth-promoting properties. Thus, both genetic and environmental factors that disrupt mitosis may play a more significant role in generating tumorigenesis than previously appreciated.
Nevertheless, it is interesting to consider whether exploiting the consequences of abnormal mitosis may have therapeutic value, especially given that such events preferentially occur in tumor, and not normal, cells. As an example, two common byproducts of abnormal mitosis—extra centrosomes and aneuploidy—have been identified as potentially cancer-specific drug targets (Rebacz et al., 2007; Kwon et al., 2008; Leber et al., 2010; Tang et al., 2011; Raab et al., 2012). Other defects arising from chaotic mitosis, such as tetraploidy, micronuclei, and bridging chromosomes, also represent potential therapeutic targets.
Moreover, a detailed understanding of how cells respond to DNA damage during mitosis may also help to improve existing anti-mitotic therapies that target rapidly dividing mitotic cells (e.g., Taxol). Although this class of drugs has proven beneficial, and often even curative, many cancers remain refractory. This is due in large part to the fact that cells are capable of escaping mitosis through slippage before a full apoptotic response can be achieved (Gascoigne and Taylor, 2008). Consequently, it has recently been proposed that targeting mitotic exit, and thus forcing cells to persist in a state of stressful and damaging mitosis, may have a stronger therapeutic effect, especially in conjunction with treatments that potentiate the apoptotic response (Huang et al., 2009; Rieder and Medema, 2009; Zeng et al., 2010; Shi et al., 2011; Tan et al., 2011).
In sum, abnormal mitosis disrupts genome stability through a variety of mechanisms and has the capacity to empower cells with growth advantages that promote the development of cancer. However, abnormal mitosis also imparts specific vulnerabilities. Identifying these weaknesses, as well as novel ways to exploit them, remains a primary objective of future cell biological research.
Acknowledgments
We apologize to the many authors whose work we were unable to cite due to space constraints. We would like to thank Duane Compton, Sonal Jhaveri-Schneider, and members of the Pellman laboratory for critical reading of the manuscript; and Taruho Kuroda for images in Figs. 2 B and 3 B.
N. Ganem is supported by a K99 award (K99CA154531-01) from the National Cancer Institute. D. Pellman is supported by the Howard Hughes Medical Institute and awards from the National Institutes of Health.
References
- Abraham M.C., Shaham S. 2004. Death without caspases, caspases without death. Trends Cell Biol. 14:184–193 10.1016/j.tcb.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. 2002. Molecular Biology of the Cell. 4th ed Garland Science, NY: 1548 pp [Google Scholar]
- Alexander S.P., Rieder C.L. 1991. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 113:805–815 10.1083/jcb.113.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen P.R., Lohez O.D., Lacroix F.B., Margolis R.L. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell. 12:1315–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., van Deursen J.M. 2010. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 9:1711–1716 10.4161/cc.9.9.11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jin F., Jeganathan K.B., van Deursen J.M. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 16:475–486 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C., Körner R., Hofmann K., Nigg E.A. 2007. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 128:101–114 10.1016/j.cell.2006.11.041 [DOI] [PubMed] [Google Scholar]
- Baxter J., Diffley J.F.X. 2008. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 30:790–802 10.1016/j.molcel.2008.04.019 [DOI] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M.V. 2006. Prolonged mitosis versus tetraploid checkpoint: how p53 measures the duration of mitosis. Cell Cycle. 5:971–975 10.4161/cc.5.9.2711 [DOI] [PubMed] [Google Scholar]
- Bloom K.S. 2008. Beyond the code: the mechanical properties of DNA as they relate to mitosis. Chromosoma. 117:103–110 10.1007/s00412-007-0138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau A.M., Sonenberg N. 1987. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 262:11134–11139 [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Chan K.L., Hickson I.D. 2011. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 22:906–912 10.1016/j.semcdb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Chan K.-L., North P.S., Hickson I.D. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397–3409 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11:753–760 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A.E. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 8:1105–1115 10.1016/S1097-2765(01)00388-4 [DOI] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta. 1786:32–40 [DOI] [PubMed] [Google Scholar]
- Cimini D., Tanzarella C., Degrassi F. 1999. Differences in malsegregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis. 14:563–568 10.1093/mutage/14.6.563 [DOI] [PubMed] [Google Scholar]
- Cimini D., Howell B., Maddox P., Khodjakov A., Degrassi F., Salmon E.D. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527 10.1083/jcb.153.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Fioravanti D., Salmon E.D., Degrassi F. 2002. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 115:507–515 [DOI] [PubMed] [Google Scholar]
- Cimini D., Moree B., Canman J.C., Salmon E.D. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213–4225 10.1242/jcs.00716 [DOI] [PubMed] [Google Scholar]
- Cimini D., Cameron L.A., Salmon E.D. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155 10.1016/j.cub.2004.11.029 [DOI] [PubMed] [Google Scholar]
- Compton D.A. 2011. Mechanisms of aneuploidy. Curr. Opin. Cell Biol. 23:109–113 10.1016/j.ceb.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi C.H., Martens A.L., Ritchie K., Francis S.M., Chakrabarti S., Berube N.G., Dick F.A. 2010. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 24:1351–1363 10.1101/gad.1917610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton W.B., Nandan M.O., Moore R.T., Yang V.W. 2007. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 67:11487–11492 10.1158/0008-5472.CAN-07-5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J.R., Potapova T.A., Sivakumar S., Daniel J.J., Flynn J.N., Rankin S., Gorbsky G.J. 2011. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21:1018–1024 10.1016/j.cub.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., de Lange T. 2011. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27:585–610 10.1146/annurev-cellbio-092910-154234 [DOI] [PubMed] [Google Scholar]
- Davoli T., de Lange T. 2012. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 21:765–776 10.1016/j.ccr.2012.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K.J., Meng X., Khodjakov A., McEwen B.F. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F., Harrison J.C., Jain S., Sugawara N., Haber J.E. 2010. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr. Biol. 20:328–332 10.1016/j.cub.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli D.M., Hearn S., Myers M.P., Lazebnik Y. 2005. A primate virus generates transformed human cells by fusion. J. Cell Biol. 171:493–503 10.1083/jcb.200507069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P., Rausch C., Rasnick D., Hehlmann R. 1998. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA. 95:13692–13697 10.1073/pnas.95.23.13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 391:43–50 10.1038/34112 [DOI] [PubMed] [Google Scholar]
- Fackenthal J.D., Olopade O.I. 2007. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer. 7:937–948 10.1038/nrc2054 [DOI] [PubMed] [Google Scholar]
- Forment J.V., Kaidi A., Jackson S.P. 2012. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat. Rev. Cancer. 12:663–670 10.1038/nrc3352 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 437:1043–1047 10.1038/nature04217 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Pellman D. 2007. Limiting the proliferation of polyploid cells. Cell. 131:437–440 10.1016/j.cell.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17:157–162 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Taylor S.S. 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 14:111–122 10.1016/j.ccr.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Gerace L., Blum A., Blobel G. 1978. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J. Cell Biol. 79:546–566 10.1083/jcb.79.2.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Jackson S.P. 2011. Give me a break, but not in mitosis: the mitotic DNA damage response marks DNA double-strand breaks with early signaling events. Cell Cycle. 10:1215–1221 10.4161/cc.10.8.15334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Belotserkovskaya R., Jackson S.P. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190:197–207 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J.C., Waterhouse N.J., Juin P., Evan G.I., Green D.R. 2000. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2:156–162 10.1038/35004029 [DOI] [PubMed] [Google Scholar]
- Gollin S.M., Wray W., Hanks S.K., Hittelman W.N., Rao P.N. 1984. The ultrastructural organization of prematurely condensed chromosomes. J. Cell Sci. Suppl. 1:203–221 [DOI] [PubMed] [Google Scholar]
- Gordon D.J., Resio B., Pellman D. 2012. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13:189–203 [DOI] [PubMed] [Google Scholar]
- Gotoh E. 2007. Visualizing the dynamics of chromosome structure formation coupled with DNA replication. Chromosoma. 116:453–462 10.1007/s00412-007-0109-5 [DOI] [PubMed] [Google Scholar]
- Grandbois M., Beyer M., Rief M., Clausen-Schaumann H., Gaub H.E. 1999. How strong is a covalent bond? Science. 283:1727–1730 10.1126/science.283.5408.1727 [DOI] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. 2011. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21:374–381 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A.A., Gamero M.C., Trachana V., Fütterer A., Pacios-Bras C., Díaz-Concha N.P., Cigudosa J.C., Martínez-A C., van Wely K.H.M. 2010. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc. Natl. Acad. Sci. USA. 107:4159–4164 10.1073/pnas.0912143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hanks S.K., Gollin S.M., Rao P.N., Wray W., Hittelman W.N. 1983. Cell cycle-specific changes in the ultrastructural organization of prematurely condensed chromosomes. Chromosoma. 88:333–342 10.1007/BF00285856 [DOI] [PubMed] [Google Scholar]
- Harley M.E., Allan L.A., Sanderson H.S., Clarke P.R. 2010. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 29:2407–2420 10.1038/emboj.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Waizenegger I.C., Peters J.M. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323 10.1126/science.1061376 [DOI] [PubMed] [Google Scholar]
- Hayashi M.T., Cesare A.J., Fitzpatrick J.A.J., Lazzerini-Denchi E., Karlseder J. 2012. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat. Struct. Mol. Biol. 19:387–394 10.1038/nsmb.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W. 2010. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000539 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. 1986. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 5:2973–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J.H.J. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Hoffelder D.R., Luo L., Burke N.A., Watkins S.C., Gollin S.M., Saunders W.S. 2004. Resolution of anaphase bridges in cancer cells. Chromosoma. 112:389–397 10.1007/s00412-004-0284-6 [DOI] [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2012. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 13:501–514 10.1038/embor.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. 1989. Chromosome error propagation and cancer. Trends Genet. 5:42–45 10.1016/0168-9525(89)90020-6 [DOI] [PubMed] [Google Scholar]
- Hopfner K.-P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A.L., Karcher A., Henderson B., Bodmer J.-L., McMurray C.T., et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 418:562–566 10.1038/nature00922 [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B., Marko J.F., Chatenay D., Libchaber A. 1997. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 139:1–12 10.1083/jcb.139.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-C., Shi J., Orth J.D., Mitchison T.J. 2009. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 16:347–358 10.1016/j.ccr.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R.S., Zhai B., Wan L., Gutierrez A., Lau A.W., et al. 2011. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 471:104–109 10.1038/nature09732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., van der Burg M., Szuhai K., Kops G.J.P.L., Medema R.H. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 333:1895–1898 10.1126/science.1210214 [DOI] [PubMed] [Google Scholar]
- Johnson R.T., Rao P.N. 1970. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 226:717–722 10.1038/226717a0 [DOI] [PubMed] [Google Scholar]
- Jullien D., Vagnarelli P., Earnshaw W.C., Adachi Y. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115:71–79 [DOI] [PubMed] [Google Scholar]
- Ke Y., Huh J.-W., Warrington R., Li B., Wu N., Leng M., Zhang J., Ball H.L., Li B., Yu H. 2011. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 30:3309–3321 10.1038/emboj.2011.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C., Mannella C.A., Kinnally K.W. 2004. Laser micro-irradiation of mitochondria: is there an amplified mitochondrial death signal in neural cells? Mitochondrion. 3:217–227 10.1016/j.mito.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Krzywicka-Racka A., Sluder G. 2011. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J. Cell Biol. 194:199–207 10.1083/jcb.201101073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Baum B. 2009. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 19:174–179 10.1016/j.tcb.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., Pellman D. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.-K., Wong C.-H., Lee Y.-P., Li H.-Y. 2011. Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death. Cell Death Differ. 18:996–1004 10.1038/cdd.2010.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni J.S., Jacks T. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18:1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B.D., Rampalli S., Burns L.E., Brunette S., Dilworth F.J., Megeney L.A. 2010. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc. Natl. Acad. Sci. USA. 107:4230–4235 10.1073/pnas.0913089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B., Maier B., Fuchs F., Chi J., Riffel P., Anderhub S., Wagner L., Ho A.D., Salisbury J.L., Boutros M., Krämer A. 2010. Proteins required for centrosome clustering in cancer cells. Sci. Transl. Med. 2:ra38 10.1126/scitranslmed.3000915 [DOI] [PubMed] [Google Scholar]
- Liu P., Erez A., Nagamani S.C.S., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A., et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 146:889–903 10.1016/j.cell.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Carvalho C.M.B., Hastings P.J., Lupski J.R. 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22:211–220 10.1016/j.gde.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Kisurina-Evgenieva O., Vinogradova T., Hergert P., La Terra S., Kapoor T.M., Khodjakov A. 2007. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 450:745–749 10.1038/nature06344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth M.S., Herr A., Ji J.-Y., Dyson N.J. 2008. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 22:1011–1024 10.1101/gad.1631508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Longworth M.S., Dyson N.J. 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24:1364–1376 10.1101/gad.1917310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1941. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 26:234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M., Norden C., Durrer K., Rauter H., Uhlmann F., Barral Y. 2009. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 11:477–483 10.1038/ncb1855 [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., Rieder C.L. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Millman S.E., Pagano M. 2011. MCL1 meets its end during mitotic arrest. EMBO Rep. 12:384–385 10.1038/embor.2011.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn A.J., Boise L.H., Thompson C.B. 1996. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10:2621–2631 10.1101/gad.10.20.2621 [DOI] [PubMed] [Google Scholar]
- Moldovan G.-L., D’Andrea A.D. 2009. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43:223–249 10.1146/annurev-genet-102108-134222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins J.M., Biesele J.J. 1977. Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73:672–684 10.1083/jcb.73.3.672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V.G., Halazonetis T.D. 2010. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11:220–228 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Nelson G., Buhmann M., von Zglinicki T. 2009. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle. 8:3379–3383 10.4161/cc.8.20.9857 [DOI] [PubMed] [Google Scholar]
- Nicklas R.B. 1988. The forces that move chromosomes in mitosis. Annu. Rev. Biophys. Biophys. Chem. 17:431–449 10.1146/annurev.bb.17.060188.002243 [DOI] [PubMed] [Google Scholar]
- Nitta M., Kobayashi O., Honda S., Hirota T., Kuninaka S., Marumoto T., Ushio Y., Saya H. 2004. Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents. Oncogene. 23:6548–6558 10.1038/sj.onc.1207873 [DOI] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., Barral Y. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 125:85–98 10.1016/j.cell.2006.01.045 [DOI] [PubMed] [Google Scholar]
- Orth J.D., Loewer A., Lahav G., Mitchison T.J. 2012. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell. 23:567–576 10.1091/mbc.E11-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D.M., Bender M.A. 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26:260–268 10.1016/0014-4827(62)90176-3 [DOI] [PubMed] [Google Scholar]
- Quignon F., Rozier L., Lachages A.-M., Bieth A., Simili M., Debatisse M. 2007. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 26:165–172 10.1038/sj.onc.1209787 [DOI] [PubMed] [Google Scholar]
- Raab M.S., Breitkreutz I., Anderhub S., Rønnest M.H., Leber B., Larsen T.O., Weiz L., Konotop G., Hayden P.J., Podar K., et al. 2012. GF-15, a novel inhibitor of centrosomal clustering, suppresses tumor cell growth in vitro and in vivo. Cancer Res. 72:5374–5385 10.1158/0008-5472.CAN-12-2026 [DOI] [PubMed] [Google Scholar]
- Rebacz B., Larsen T.O., Clausen M.H., Rønnest M.H., Löffler H., Ho A.D., Krämer A. 2007. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res. 67:6342–6350 10.1158/0008-5472.CAN-07-0663 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W. 1998. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J. Cell Biol. 142:1013–1022 10.1083/jcb.142.4.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Maiato H. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 7:637–651 10.1016/j.devcel.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Medema R.H. 2009. No way out for tumor cells. Cancer Cell. 16:274–275 10.1016/j.ccr.2009.09.021 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Palazzo R.E. 1992. Colcemid and the mitotic cycle. J. Cell Sci. 102:387–392 [DOI] [PubMed] [Google Scholar]
- Robbins E., Gonatas N.K. 1964. The ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol. 21:429–463 10.1083/jcb.21.3.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Sager P.R., Brown P.A., Berlin R.D. 1984. Analysis of transferrin recycling in mitotic and interphase HeLa cells by quantitative fluorescence microscopy. Cell. 39:275–282 10.1016/0092-8674(84)90005-9 [DOI] [PubMed] [Google Scholar]
- Sakahira H., Enari M., Nagata S. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 391:96–99 10.1038/34214 [DOI] [PubMed] [Google Scholar]
- Salmon E.D., Cimini D., Cameron L.A., DeLuca J.G. 2005. Merotelic kinetochores in mammalian tissue cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:553–568 10.1098/rstb.2004.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoshkin A., Arnaoutov A., Jansen L.E.T., Ouspenski I., Dye L., Karpova T., McNally J., Dasso M., Cleveland D.W., Strunnikov A. 2009. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS ONE. 4:e6831 10.1371/journal.pone.0006831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez T., Ortiz-Ferrón G., López-Rivas A. 2010. Mitotic arrest and JNK-induced proteasomal degradation of FLIP and Mcl-1 are key events in the sensitization of breast tumor cells to TRAIL by antimicrotubule agents. Cell Death Differ. 17:883–894 10.1038/cdd.2009.176 [DOI] [PubMed] [Google Scholar]
- Saxton W.M., Stemple D.L., Leslie R.J., Salmon E.D., Zavortink M., McIntosh J.R. 1984. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 99:2175–2186 10.1083/jcb.99.6.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.-M., Sotillo R., Benezra R. 2010. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 10:102–115 10.1038/nrc2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J.M., Blank H.M., Pfau S.J., Tange Y., George B.M., Humpton T.J., Brito I.L., Hiraoka Y., Niwa O., Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science. 333:1026–1030 10.1126/science.1206412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhou Y., Huang H.-C., Mitchison T.J. 2011. Navitoclax (ABT-263) accelerates apoptosis during drug-induced mitotic arrest by antagonizing Bcl-xL. Cancer Res. 71:4518–4526 10.1158/0008-5472.CAN-10-4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.M., Phua H.H., Mills W., Carpenter A.J., Porter A.C.G., Farr C.J. 2007. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 120:3952–3964 10.1242/jcs.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spry M., Scott T., Pierce H., D’Orazio J.A. 2007. DNA repair pathways and hereditary cancer susceptibility syndromes. Front. Biosci. 12:4191–4207 10.2741/2380 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H.A., Held M., Guizetti J., Maar S., Gerlich D.W. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 144:27–40 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S., Stoehr G., Peplowska K., Cox J., Mann M., Storchova Z. 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8:608 10.1038/msb.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N., Malek M., Zha J., Yue P., Kassees R., Berry L., Fairbrother W.J., Sampath D., Belmont L.D. 2011. Navitoclax enhances the efficacy of taxanes in non-small cell lung cancer models. Clin. Cancer Res. 17:1394–1404 10.1158/1078-0432.CCR-10-2353 [DOI] [PubMed] [Google Scholar]
- Tang Y.-C., Williams B.R., Siegel J.J., Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell. 144:499–512 10.1016/j.cell.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.H. 1960. Nucleic acid synthesis in relation to the cell division cycle. Ann. N. Y. Acad. Sci. 90:409–421 10.1111/j.1749-6632.1960.tb23259.x [DOI] [PubMed] [Google Scholar]
- Terradas M., Martín M., Tusell L., Genescà A. 2010. Genetic activities in micronuclei: is the DNA entrapped in micronuclei lost for the cell? Mutat. Res. 705:60–67 10.1016/j.mrrev.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188:369–381 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2011. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. USA. 108:17974–17978 10.1073/pnas.1109720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana V., van Wely K.H.M., Guerrero A.A., Fütterer A., Martínez-A C. 2007. Dido disruption leads to centrosome amplification and mitotic checkpoint defects compromising chromosome stability. Proc. Natl. Acad. Sci. USA. 104:2691–2696 10.1073/pnas.0611132104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt M.A.T.M., Gardino A.K., Linding R., Ostheimer G.J., Reinhardt H.C., Ong S.-E., Tan C.S., Miao H., Keezer S.M., Li J., et al. 2010. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 8:e1000287 10.1371/journal.pbio.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley J.M., Evans D.G., Birch J.M. 1997. Li-Fraumeni syndrome—a molecular and clinical review. Br. J. Cancer. 76:1–14 10.1038/bjc.1997.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A.T.M., Betti C.J., Villalobos M.J. 2002. Surviving apoptosis. Apoptosis. 7:173–177 10.1023/A:1014374717773 [DOI] [PubMed] [Google Scholar]
- Wang L.H.-C., Schwarzbraun T., Speicher M.R., Nigg E.A. 2008. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 117:123–135 10.1007/s00412-007-0131-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H.-C., Mayer B., Stemmann O., Nigg E.A. 2010. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 123:806–813 10.1242/jcs.058255 [DOI] [PubMed] [Google Scholar]
- Weaver B.A.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Williams B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 322:703–709 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Loncarek J., Khodjakov A., Rieder C.L. 2008. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 10:748–751 10.1038/ncb1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Sigoillot F., Gaur S., Choi S., Pfaff K.L., Oh D.-C., Hathaway N., Dimova N., Cuny G.D., King R.W. 2010. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 18:382–395 10.1016/j.ccr.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]