Variant repeats interspersed throughout ALT telomeres recruit nuclear receptors, leading to the destabilized telomere architecture and enhanced telomeric recombination.
Abstract
Telomeres in cells that use the recombination-mediated alternative lengthening of telomeres (ALT) pathway elicit a DNA damage response that is partly independent of telomere length. We therefore investigated whether ALT telomeres contain structural abnormalities that contribute to ALT activity. Here we used next generation sequencing to analyze the DNA content of ALT telomeres. We discovered that variant repeats were interspersed throughout the telomeres of ALT cells. We found that the C-type (TCAGGG) variant repeat predominated and created a high-affinity binding site for the nuclear receptors COUP-TF2 and TR4. Nuclear receptors were directly recruited to telomeres and ALT-associated characteristics were induced after incorporation of the C-type variant repeat by a mutant telomerase. We propose that the presence of variant repeats throughout ALT telomeres results from recombination-mediated telomere replication and spreading of variant repeats from the proximal regions of the telomeres and that the consequent binding of nuclear receptors alters the architecture of telomeres to facilitate further recombination.
Introduction
Telomeres are specialized nucleoprotein complexes at the ends of linear chromosomes that function to maintain chromosomal integrity (Palm and de Lange, 2008). Human telomeres consist of tandem arrays of the sequence 5′-TTAGGG-3′ (Moyzis et al., 1988) and gradually shorten as a consequence of cellular proliferation (Harley et al., 1990). To bypass the proliferative barrier this imposes, and thereby become immortal, cells require a means to replenish their telomeres. Although the majority of human cancers do so by activating the enzyme telomerase (Shay and Bacchetti, 1997), many of the remainder use a homologous recombination (HR)–dependent alternative lengthening of telomeres (ALT) mechanism (Henson and Reddel, 2010). Telomeric DNA is associated with a protein complex called shelterin, which prevents the telomere from being recognized as a DNA double-strand break (d’Adda di Fagagna et al., 2003) and suppresses chromosomal end-to-end fusions (van Steensel et al., 1998; Hockemeyer et al., 2005) as well as HR (Wang et al., 2004; Celli et al., 2006; Sfeir et al., 2010). In addition, a lariat structure termed a t-loop formed by sequestration of the telomere end within more proximal sequences of the same telomere is thought to safeguard the chromosome terminus from detection by the DNA damage repair machinery (Griffith et al., 1999).
The most proximal 2-kb region of human telomeres contains a nonrandom distribution of variant (for example TCAGGG, TGAGGG, and TTGGGG) and canonical repeats, which are in linkage disequilibrium and have evolved along haploid lineages, whereas the distal ends comprise homogeneous arrays of TTAGGG sequence (Allshire et al., 1989; Baird et al., 1995, 2000; Coleman et al., 1999). To date little is known regarding the identity, abundance, and distribution of these variants within the telomere. Presumably, however, this would vary considerably depending on the cell type or chromosome analyzed. Cells that use ALT exhibit both intra- and inter-telomeric HR-mediated replication (Dunham et al., 2000; Muntoni et al., 2009), and telomeric exchange events have been detected within variant repeats in the proximal ends of telomeres (Varley et al., 2002). ALT cells display phenotypic characteristics suggestive of recombination-mediated telomere replication, including telomere length heterogeneity (Bryan et al., 1995, 1997; Perrem et al., 2001), an abundance of extrachromosomal circular and linear telomeric DNA (Tokutake et al., 1998; Cesare and Griffith, 2004; Wang et al., 2004; Nabetani and Ishikawa, 2009; Henson et al., 2009), a specific class of promyelocytic leukemia (PML) nuclear bodies containing telomeric DNA and telomere binding proteins, as well as proteins involved in recombination, known as ALT-associated PML bodies (APBs; Yeager et al., 1999), and elevated levels of telomere sister chromatid exchange (T-SCE) events (Bechter et al., 2004; Londoño-Vallejo et al., 2004). Moreover, the ALT mechanism is repressed when the function of proteins involved in HR is inhibited (Jiang et al., 2005; Zhong et al., 2007).
Many telomeres in ALT cells elicit a DNA damage response that is partly independent of telomere length and occurs in the absence of chromosomal end-to-end fusions (Cesare et al., 2009), suggesting the existence of underlying telomeric structural defects. Here we investigated telomeric structural abnormalities associated with the ALT mechanism. We show for the first time that variant repeats are interspersed throughout ALT telomeres. The presence of variant repeats leads to the ALT-specific telomeric recruitment of a group of nuclear receptors and may also contribute to shelterin undersaturation. Although nuclear receptors are capable of binding directly to the canonical telomere repeat in vitro, we show that the nuclear receptor TR4 holds a greater affinity than the shelterin component telomeric repeat-binding factor 2 (TRF2) for the most common variant telomeric repeat. In ALT cells, nuclear receptors bind along the entirety of the telomere array and are not limited to the proximal regions. Finally, we demonstrate that incorporation of variant repeats into the telomeres of telomerase-positive cells results in the recruitment of nuclear receptors to telomeres as well as the induction of numerous ALT-associated phenotypic characteristics. Therefore, we propose that the presence of variant sequences in ALT telomeres and the consequent binding of nuclear receptors may destabilize the telomere architecture through competitive inhibition of shelterin binding and facilitation of telomeric recombination.
Results
ALT telomeres contain abundant variant repeat sequence
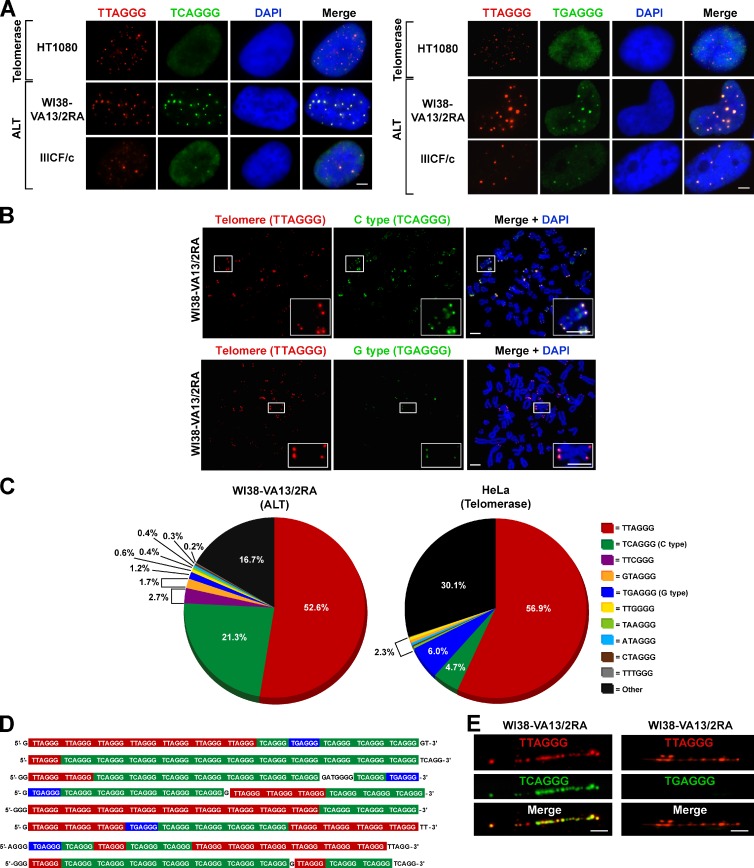
Telomeric recombination as a consequence of ALT activity can occur at variant repeats within the proximal region of the telomere array (Varley et al., 2002). We hypothesized that this may result in interspersion of variant sequences throughout the telomeric repeat array, the dissolution of linkage disequilibrium, and consequent telomere structural ramifications. To test this hypothesis, we used FISH to determine whether the variant repeats TCAGGG (C type) and TGAGGG (G type) are present throughout ALT telomeres. Using fluorophore-conjugated peptide nucleic acid (PNA) probes, we identified C- and G-type variant repeats colocalizing with telomeric DNA in ALT cells (Figs. 1 A and S1 A), but variant repeats were not detectable in telomerase-positive or mortal cells (Table 1 and Fig. S1, A and B). The low abundance and restriction of these variant repeats to the proximal regions of the telomeres of telomerase-positive and mortal cell types presumably limits their detection by FISH analysis. To verify that the variant repeats were present at individual telomeres and not restricted to large telomeric foci (predominantly APBs), we performed FISH to detect variant repeats on metaphase spreads (Fig. 1 B). Although FISH detected these variants only in ALT telomeres, their abundance varied considerably across different ALT cell lines (Fig. S1 A). For instance, the C-type variant was abundant at the telomeres of WI38-VA13/2RA chromosomes (Fig. 1 B), but lower levels were detected at the telomeres of the IIICF/c cell line. The C-type variant was detected at individual telomeres of all ALT cells analyzed, whereas the G type was less abundant, only detectable within APBs and at the telomeres of a subset of WI38-VA13/2RA chromosomes (Table 1). Although these variants were detected within the telomeres of the ALT cell lines WI38-VA13/2RA, JFCF-6/T.1M, JFCF-6/T.1R, and GM847, they were not found in their respective mortal cell precursors (WI38, JFCF-6, and GM02063, respectively; Table 1), further demonstrating that these variant repeats are a specific feature of ALT telomeres.
Figure 1.
Variant repeats are interspersed throughout the telomeres of ALT cells. (A) FISH performed on interphase nuclei using both a Texas red–conjugated (TTAGGG)3 telomeric probe (red) and Alexa 488–OO-(TCAGGG)3 or (TGAGGG)3 variant probe (green) counterstained with DAPI (blue). (B) Representative metaphase spreads of the WI38-VA13/2RA ALT cell line processed for FISH as described in A showing localization of the C- and G-type variant repeat at individual telomeres. Magnified examples are shown in the bottom right. (C) Percentage of each telomeric repeat type generated from deep sequencing analysis of WI38-VA13/2RA and HeLa cells. Variant repeats were counted within reads containing more than six nonconsecutive TBAGGG telomeric repeats. (D) Examples of various telomeric 75-nucleotide reads from sequencing data of WI38-VA13/2RA cells. (E) Distribution of the C- and G-type variant repeats along the length of the telomere in the WI38-VA13/2RA cell line, visualized on chromatin fibers. Bars, 5 µm.
Table 1.
Variant repeats and nuclear receptors present at ALT telomeres
| Cell line | Cell type | Immortalization | C-type variant | G-type variant | TR2 | TR4 | COUP-TF1 | COUP-TF2 | EAR-2 | Telomere maintenance |
| WI38 | Fibroblast; lung | NA | − | − | − | − | − | − | − | Mortal |
| JFCF-6 | Fibroblast; jejunal | NA | − | − | − | − | − | − | − | Mortal |
| GM02063 | Fibroblast; skin | NA | − | − | − | − | − | − | − | Mortal |
| HT1080 | Fibrosarcoma | Tumor | − | − | − | − | − | − | − | Telomerase |
| HeLa | Cervical carcinoma | Tumor | − | − | − | − | − | − | − | Telomerase |
| JFCF-6/T.1F | Fibroblast; jejunal | SV40 | − | − | − | − | − | − | − | Telomerase |
| GM639 | Fibroblast; skin | SV40 | − | − | − | − | − | − | − | Telomerase |
| WI38-VA13/2RA | Fibroblast; lung | SV40 | + | + | + | + | + | + | + | ALT |
| U-2 OS | Osteosarcoma | Tumor | + | APBs onlya | + | + | − | + | − | ALT |
| IIICF/c | Fibroblast; breast, LFSb | Spontaneous | + | APBs only | + | + | + | + | + | ALT |
| JFCF-6/T.1M | Fibroblast; jejunal | SV40 | + | APBs only | + | + | + | + | + | ALT |
| JFCF-6/T.1R | Fibroblast; jejunal | SV40 | + | APBs only | + | + | + | + | + | ALT |
| GM847 | Fibroblast, skin | SV40 | + | APBs only | + | + | + | + | + | ALT |
| SUSM-1 | Fibroblast; liver | Chemical | + | APBs only | + | + | + | + | − | ALT |
| Saos-2 | Osteosarcoma | Tumor | + | APBs only | + | + | − | + | − | ALT |
Variant repeat only detected within APBs, not at individual telomeres, by FISH analysis performed on metaphase spreads.
LFS, Li-Fraumeni syndrome.
A potential caveat to using variant repeat PNA probes for the detection of isolated interspersed variant repeats is the lack of a linear signal output. For this reason, we used next generation sequencing to quantitatively determine the identity and extent of variant sequence within telomere arrays of WI38-VA13/2RA as well as the telomerase-positive HeLa cell line. Samples were paired-end sequenced, and reads containing greater than six nonconsecutive telomeric repeats in the format TBAGGG were considered to be reads derived from telomeres. This criterion was used to enrich for telomeric sequence rather than interstitial repeats, to assess the frequency of variant repeats in the telomeres, and to qualitatively establish variant repeat interspersion patterns. It is important to note that the criterion is somewhat biased toward canonical sequence and variants of the format TBAGGG, although interspersed variants such as CTAGGG or TTGGGG and nonhexameric sequences are detected. Numerous variant repeat types were identified at varying proportions (Figs. 1 C and S1 C). Interestingly, the C-type variant constituted ∼21.3% of the total extracted telomeric repeats in WI38-VA13/2RA compared with 4.7% in HeLa. In contrast, the G type accounted for only 1.2% of the telomeres in WI38-VA13/2RA and 6.0% of HeLa telomeres (Fig. 1 C). The representation of variant repeats is expressed as a proportion of the total telomeric repeats and is therefore dependent on telomere length. Variant repeats present in the proximal regions of the telomeres will therefore be overrepresented in mortal and telomerase-positive cell lines with short telomeres. Consequently, the abundance of G-type repeats in HeLa cells is high in comparison to WI38-VA13/2RA cells, which in general display substantially longer telomeres (see Fig. S1 D for quantitation of total telomeric DNA content in the complete panel of cell lines). Similarly, the abundance of C-type repeats in HeLa cells is likely to be overrepresented. Examples of the 75-nucleotide telomeric reads in the WI38-VA13/2RA cell line clearly show that variant repeats are interspersed among the canonical telomeric repeats (Fig. 1 D). Other sequences include incomplete repeats at either end of the sequence read and nonhexameric repeats (Fig. 1 D), as well as other variants and additional nontelomeric sequences from interstitial or subtelomeric sites. Interestingly, the percentage and abundance of numerous variant sequences within the telomeres of both cell types was high regardless of ALT status, suggesting that the proximal region of human telomeres may contain more variant repeats in terms of both diversity and abundance than previously considered. Variant repeat integration along the entire length of ALT telomeres was further confirmed by telomere and variant repeat-specific FISH on chromatin fibers, demonstrating that the C-type variant repeats are not restricted to the proximal region of the telomere (Fig. 1 E). G-type variant repeats were not detected in elongated chromatin fibers, presumably because of their low abundance in WI38-VA13/2RA telomeres.
Nuclear receptors bind preferentially to C-type variant repeats
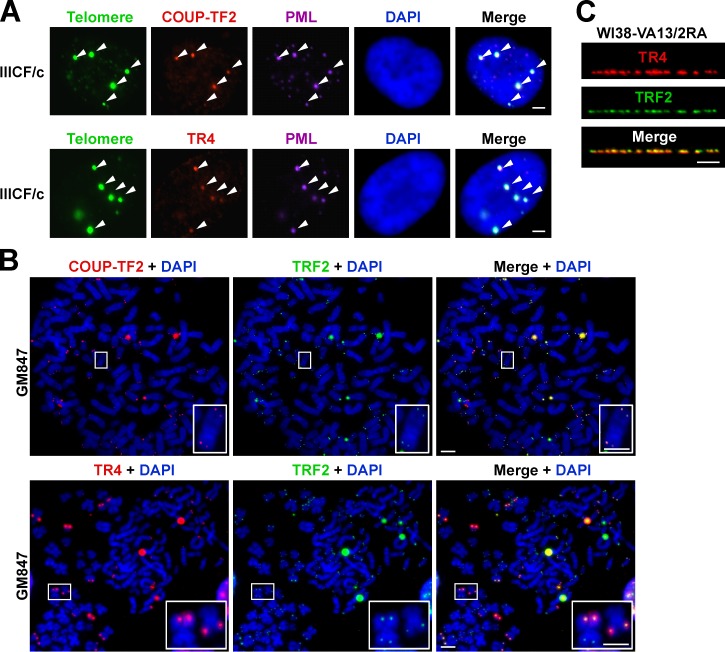
Numerous nuclear receptors have been identified at the telomeres of ALT cells (Déjardin and Kingston, 2009), but the mechanism by which these proteins are recruited to telomeres remains to be determined. Using indirect immunofluorescence against five nuclear receptors (TR2, TR4, COUP-TF1, COUP-TF2, and EAR-2) coupled with telomere FISH on interphase nuclei of a variety of cell types, we confirmed that nuclear receptor binding to telomeres is ALT specific (Table 1). We chose to focus our further investigations on COUP-TF2 and TR4, as they were the most abundant of the five nuclear receptors at ALT telomeres and were detectable at the telomeres of all ALT cell lines examined but not mortal and telomerase-positive cell lines (Fig. S2 and Table 1). As for the variant repeats, we identified COUP-TF2 and TR4 nuclear receptors both in APBs (Fig. 2 A) and at chromosome termini (Fig. 2 B). Immunostaining of chromatin fibers further demonstrated the nuclear receptors to be interspersed along the length of the telomere (Fig. 2 C).
Figure 2.
Nuclear receptors are detected at ALT telomeres. (A) Indirect immunofluorescence staining of IIICF/c nuclei at interphase. COUP-TF2 or TR4 (red) and PML (purple) coupled with telomere FISH (green) and DAPI (blue) counterstaining shows that nuclear receptors are present within APBs (arrowheads). (B) Representative metaphase spreads of the GM847 ALT cell line stained with COUP-TF2 or TR4 (red) and TRF2 (green) immunofluorescence and DAPI (blue) show localization of nuclear receptors to telomeres. Magnified examples are shown in the bottom right. (C) Distribution of TR4 (red) among TRF2 (green) along the length of the telomere in the WI38-VA13/2RA cell line visualized at chromatin fibers. Bars, 5 µm.
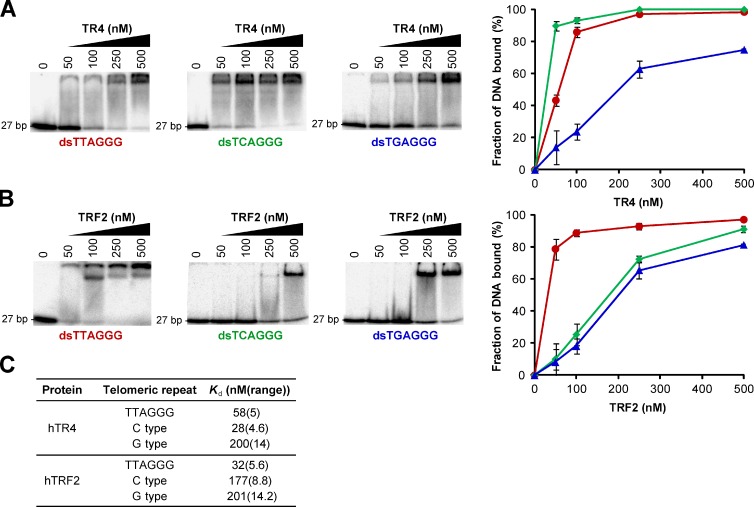
Nuclear receptors are predicted to bind to the direct repeat 5′-RGGTCA-3′, with spacings of zero to eight nucleotides (Kato et al., 1995; Sandelin and Wasserman, 2005). In light of this and the abundance of the C-type variant within ALT telomeres, we performed electrophoretic mobility shift assays (EMSAs) to determine whether nuclear receptors were capable of binding directly to variant repeats in vitro. TR4 was capable of binding to both the C- and G-type variants as well as the canonical telomeric repeat and, as predicted, had a significantly higher affinity for the C-type variant (Fig. 3, A and C). We also conducted EMSAs to determine the binding affinity of the shelterin component TRF2 for the variant repeats. As expected, TRF2 displayed a considerably higher affinity for the canonical telomeric repeat than for each of the two variant repeats (Fig. 3, B and C). We found no significant differences in global nuclear receptor protein levels in ALT versus telomerase-positive cell lines by Western blotting, which showed that nuclear receptor expression was not elevated in ALT cells nor was their lack of recruitment to the telomeres of telomerase-positive cells the result of insufficient expression (Fig. S3). We then used the telomeric subset of reads described in the previous section to calculate the percentage of C-type repeats that are present in pairs, forming the canonical nuclear receptor binding site (RGGTCA(n0–8)RGGTCA, where n is the number of nucleotide spacers). Nuclear receptor binding sites were found to occur at 89.5% of C-type repeats in WI38-VA13/2RA telomeres and 76.9% of C-type repeats in HeLa telomeres, demonstrating that nuclear receptor binding sites are enriched after C-type repeat interspersion in ALT telomeres.
Figure 3.
Binding of TR4 and TRF2 to variant telomeric repeat sequences. (A) EMSA titration experiments conducted using double-stranded γ32P-radiolabeled C- and G-type variant and canonical telomeric repeat probes incubated with purified human TR4. A binding curve generated from two independent titration experiments is also shown (average ± range). (B) EMSA titration experiments and binding curve for human TRF2. (C) Estimation of dissociation constants (Kd) obtained from titration displayed in A and B. The Kd was calculated by quantifying the disappearance of the band corresponding to free DNA and determining the concentration at which 50% of the probe is bound by protein.
Telomeric localization of nuclear receptors contributes to the ALT phenotype
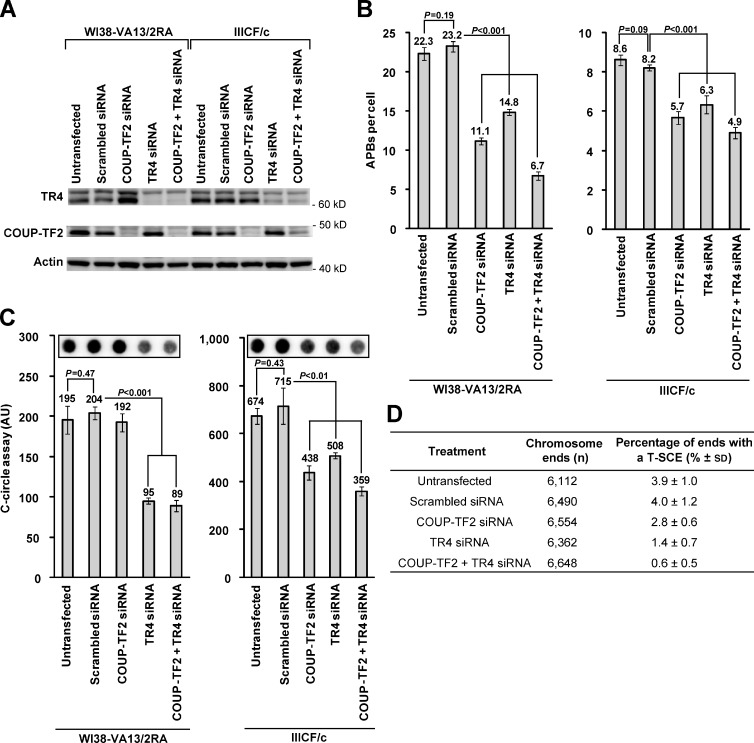
Nuclear receptors have high sequence homology to each other, bind to similar sequences, and are frequently found to have similar physiological and biochemical functions. In agreement with previous observations (Déjardin and Kingston, 2009), knockdown of COUP-TF2 resulted in an increase in TR4 expression in WI38-VA13/2RA cells (Fig. 4 A). In case these receptors possess redundant functions, we therefore performed knockdown experiments of COUP-TF2 and TR4 individually and simultaneously in the WI38-VA13/2RA and IIICF/c ALT cell lines to investigate the extent of their connection to the ALT mechanism (Fig. 4 A). These cell lines were selected because they represent the two extremes of both nuclear receptor recruitment to the telomere and variant repeat content, with WI38-VA13/2RA having the most and IIICF/c having the least. We then investigated the effects of knockdown on a variety of ALT-associated characteristics. Knockdown of COUP-TF2 and TR4 with siRNA caused a significant reduction in APBs after 72 h, both individually and in combination (Fig. 4 B). In addition, depletion of nuclear receptors led to a decrease in partially single-stranded C-rich telomere circles known as C-circles (Fig. 4 C), which are a marker of cells that use the ALT mechanism (Henson et al., 2009). Although knockdown of both nuclear receptors led to a reduction in C-circles, we did not observe a decrease in WI38-VA13/2RA cells upon depletion of COUP-TF2 alone (Fig. 4 C), which may be caused by functional redundancy of these receptors. We then sought to establish whether removal of nuclear receptors from the telomere would influence telomere capping. However, nuclear receptor depletion did not affect the DNA damage response at ALT telomeres (Fig. S4 A) nor the rate of chromosome end-to-end fusions (Fig. S4 B). Finally, we determined whether depletion of nuclear receptor expression in ALT cells altered the frequency of intra-allelic telomere recombination events. A significant reduction in the levels of T-SCEs was observed by chromosome orientation FISH (CO-FISH) analysis (Londoño-Vallejo et al., 2004) upon transient knockdown of COUP-TF2 and TR4 individually and to a greater extent after simultaneous knockdown in WI38-VA13/2RA cells (Fig. 4 D). These data demonstrate that nuclear receptor recruitment to the telomere contributes to the ALT phenotype.
Figure 4.
Nuclear receptor depletion results in suppression of ALT phenotypic characteristics. (A) Western blot analysis of whole-cell extracts (105 cell equivalents) from WI38-VA13/2RA and IIICF/c cells 72 h after transfection with siRNA against COUP-TF2 and TR4 shows knockdown of proteins. (B) Quantitation of APBs 72 h after transfection of WI38-VA13/2RA and IIICF/c cells with siRNA against COUP-TF2 and TR4 via indirect immunofluorescence against the PML protein and telomere FISH (mean ± SD; n = 3 independent experiments, quantifying 50 nuclei per replicate). (C) Quantitation of C-circle levels 72 h after transfection of WI38-VA13/2RA and IIICF/c cells (mean ± SD; n = 3). (D) Quantitation of T-SCEs observed via CO-FISH after knockdown of nuclear receptor expression in WI38-VA13/2RA cells (mean ± SD; n = 3, quantifying >2,000 chromosome ends or ∼20 metaphases per replicate). Quantitation of T-SCEs was restricted to chromosome ends with clearly distinguishable sister telomeres.
Telomeric incorporation of C-type variant repeats results in nuclear receptor recruitment
Exogenous expression of a human telomerase RNA (hTR) containing mutations in the template sequence reconstitutes active telomerase enzyme in vivo that is capable of adding mutant telomeric repeats onto the ends of chromosomes (Marusíc et al., 1997). Because variant telomeric repeats are common in ALT cells and C-type variant repeats specifically provide preferential binding sites for nuclear receptors, we adapted this approach to determine whether incorporation of the C- and G-type variant repeats in telomerase-positive cells directly causes nuclear receptor recruitment to the telomere. We found overexpression of both C- and G-type hTR to be detrimental to the viability of numerous telomerase-positive cell types. Indeed, mutant hTR expression has been previously shown to lead to the formation of chromosomal end-to-end fusions, inhibit cell proliferation, and induce apoptosis (Guiducci et al., 2001; Kim et al., 2001; Li et al., 2004; Stohr and Blackburn, 2008). To both circumvent this limitation and to emulate the variant interspersion pattern we have identified in ALT telomeres, we exogenously expressed wild-type hTR in addition to wild-type, C-type, or G-type variant hTR in the telomerase-positive HT1080 cell line (referred to as HT1080 hTR wild type, HT1080 hTR C type, and HT1080 hTR G type, respectively). Cells were passaged for more than 150 population doublings (pds) to achieve sufficient variant repeat incorporation and potential cell line contamination was excluded by 16-locus short tandem repeat profiling.
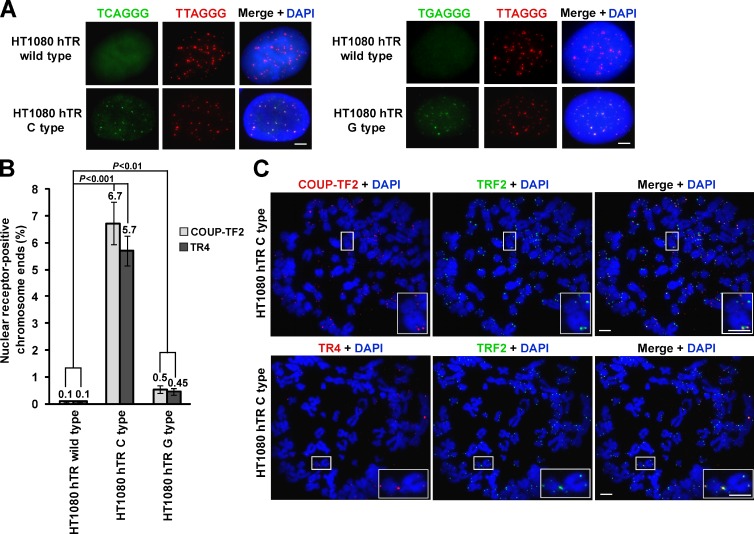
We first confirmed that the C- and G-type variant repeats were incorporated into the telomere by terminal restriction fragment analysis using γ32P-radiolabeled probes specific to each of the three repeat types (Fig. S5 A). In addition, FISH against both the canonical telomeric repeat and either the C- or G-type variant was conducted to confirm their incorporation specifically into telomeres (Fig. 5 A). Indirect immunofluorescence performed on metaphase spreads showed that both COUP-TF2 and TR4 were recruited to the telomeres as a result of the addition of the C-type repeat (Fig. 5, B and C) and to a considerably lesser extent after incorporation of either the wild-type or G-type repeat (Figs. 5 B and S5 B).
Figure 5.
Nuclear receptor recruitment upon variant telomeric repeat incorporation. (A) FISH performed on interphase nuclei using both a Texas red–conjugated (TTAGGG)3 telomeric probe (red) and Alexa 488–OO-(TCAGGG)3 or (TGAGGG)3 variant probe (green) counterstained with DAPI (blue). (B) Quantitation of the percentage of nuclear receptor-positive chromosome ends stained with either COUP-TF2 or TR4 on one or both chromatids (mean ± SD; n = 3, quantifying >2,000 chromosome ends or ∼20 metaphases per replicate). (C) Representative metaphase spreads of the HT1080 hTR C-type cell line stained with COUP-TF2 or TR4 (red) and TRF2 (green) immunofluorescence and DAPI (blue). Magnified examples are shown on the bottom right. Bars, 5 µm.
Variant repeat incorporation induces ALT-associated characteristics but not inter-telomeric recombination
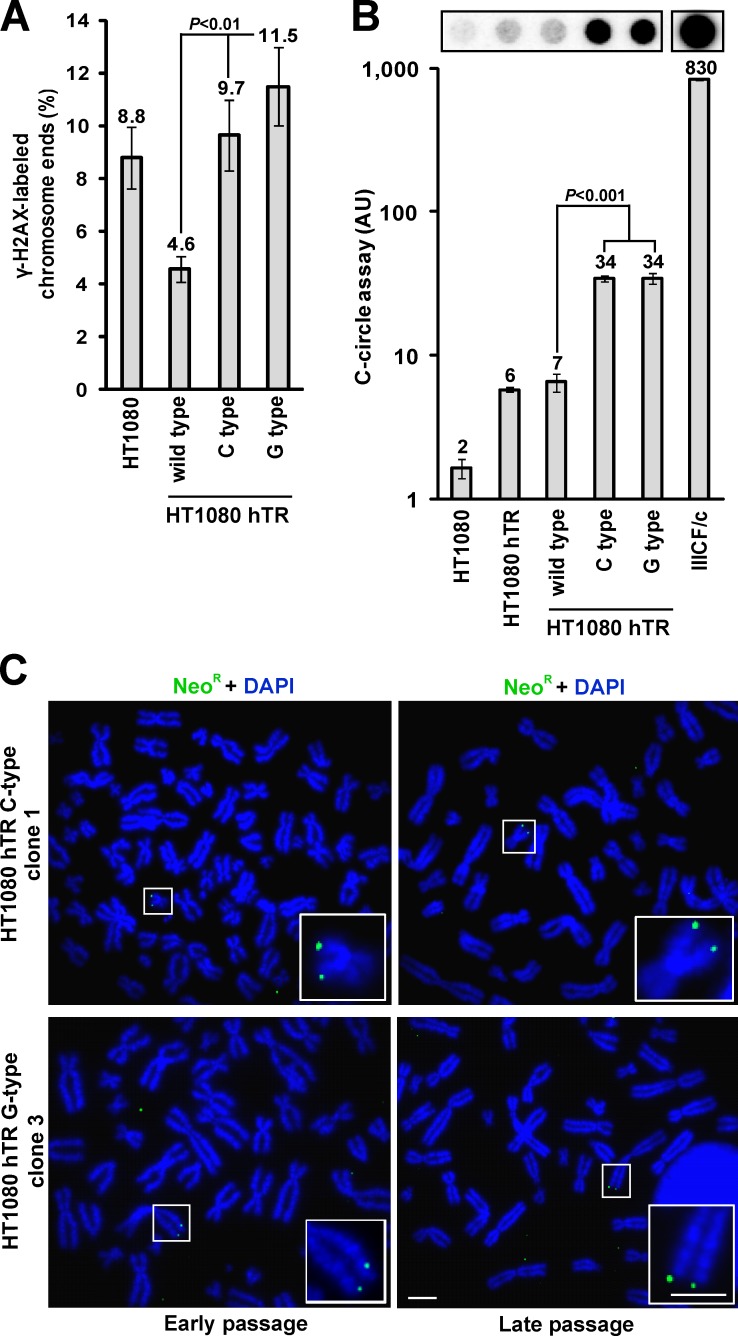
As insertion of mutant repeats into the telomere would be expected to compromise shelterin binding, we examined the DNA damage response at the telomeres. Telomere dysfunction-induced foci (TIFs) were detected at the telomere by combining telomere FISH with immunofluorescence against the DNA damage response protein H2AX phosphorylated at serine 139 (γ-H2AX) on metaphase spreads (meta-TIFs; Cesare et al., 2009). Introduction of either C- or G-type variant repeats into the telomeres led to an elevation in the number of meta-TIFs (Fig. 6 A). This is in contrast to the HT1080 cells expressing only wild-type hTR, which have previously been reported to show decreased meta-TIFs, presumably caused by a reduction in the number of very short telomeres (Pickett et al., 2009). Moreover, HT1080 cells expressing either the C- or G-type variant hTR exhibited a marked increase in C-circles (Fig. 6 B). These cells also exhibited heterogeneous telomeres as well as a marked increase in telomere length compared with those expressing wild-type hTR alone as demonstrated by terminal restriction fragment analysis (Fig. S5 A). Interestingly, we also noticed that the amount of the C-type variant repeat decreased at the latest time point (pd +150), suggesting that there may be some selection against telomeric incorporation of this variant. Most ALT cells lack functional p53 and can consequently sustain a greater level of DNA damage (Cesare and Reddel, 2008). For this reason, we similarly incorporated C- and G-type variant repeats into the HT1080-6TG cell line because of its nonfunctional p53 status (Anderson et al., 1994). However, despite similar telomere lengths (regardless of whether the cells were expressing wild-type, C-type, or G-type hTR), similar results were obtained in the absence of p53 (unpublished data).
Figure 6.
Incorporation of variant repeats into the telomeres induces ALT-associated characteristics. (A) Quantitation of meta-TIF analysis showing the percentage of chromosome ends stained with γ-H2AX on one or both chromatids in HT1080 cells expressing variant hTR compared with wild-type (mean ± SD; n = 3, quantifying >2,000 chromosome ends or ∼20 metaphases per replicate). (B) Quantitation of C-circle levels in HT1080 cells expressing wild-type and variant hTR in comparison to the IIICF/c ALT cell line (mean ± SD; n = 3) plotted on a logarithmic scale. (C) Representative metaphase spreads of the HT1080 hTR C- and G-type cells processed for neoR-FISH showing no copying of the neoR tag at early and late pds. Magnified examples are shown in the bottom right. Bars, 5 µm.
Finally, we determined whether the incorporation of variant repeats could instigate inter-telomeric recombination-mediated replication, which is characteristic of ALT cells and is proposed to be a component of the ALT mechanism. We inserted a telomere-targeting tag (Dunham et al., 2000), composed of a neomycin resistance gene (neoR) cassette flanked on either side by 800 bp of telomeric DNA, into HT1080 cells containing C- and G-type variant repeat telomeres. Single locus integration of the telomeric neoR tag was confirmed by FISH in two clonal populations of both C- and G-type variant HT1080 cells; however, no copying of the tag was detected after culture for a further 60 pds (Fig. 6 C), which has previously been demonstrated to be sufficient to identify copying of the tag by ALT (Dunham et al., 2000). Thus, integration of variant repeats in a telomerase-positive cell line can elicit several ALT-associated characteristics but is not sufficient to activate detectable levels of the inter-telomeric recombination-mediated copying that is a hallmark of ALT.
Discussion
In this study, we have described telomeric structural abnormalities associated with ALT cells and provided evidence for their role in the ALT pathway. The data presented here demonstrate for the first time that ALT telomeres are enriched with variant telomeric repeats that differ from the canonical form, the most common of which is the C-type variant. In mortal and telomerase-positive cells, variant repeats are restricted to the proximal 2 kb of the telomere where they evolve along haploid lineages by intra-allelic mutational mechanisms, such as replication errors and slippage, and persist as a result of low selective pressure and inaccessibility to telomerase (Baird et al., 1995, 2000; Coleman et al., 1999). However, in ALT cells, telomeres engage in complex HR-mediated replication events (Dunham et al., 2000; Varley et al., 2002), which presumably result in the dispersion of proximal repeats throughout the telomeres that we have observed here. The specific location and percentage (relative to telomere length) of these variant repeats within the telomere may determine whether they contribute to the ALT phenotype. We have demonstrated that the C-type variant repeat is highly abundant within ALT telomeres, existing in small interspersed blocks of variant sequences throughout the entire canonical repeat arrays. It is unclear why the C-type repeat specifically dominates. Presumably any variant repeat present in the proximal telomere region can be copied and spread throughout the telomeres by ALT activity, but the functional properties of some variants may provide a selective advantage. We therefore investigated potential binding partners of telomeric variant repeats and found that the interspersion of C-type repeats through telomere repeat arrays provides frequent high affinity binding sites for nuclear receptors.
Although the variant repeat content and corresponding extent of nuclear receptor recruitment to the telomere were found to vary considerably between cell lines, we have demonstrated that this phenomenon is specific to ALT cells, as both variant repeats and nuclear receptors were not detectable at the telomeres of mortal or telomerase-positive cell lines. Subsequently, we demonstrated that nuclear receptors have a higher affinity for the C-type variant repeat than the canonical and G-type repeats, whereas TRF2 binds more strongly than TR4 to the canonical telomeric repeat. Also, the DNA binding domain of its shelterin partner TRF1 is known to hold an even greater affinity for double-stranded telomeric DNA than that of TRF2 (Hanaoka et al., 2005). This suggests that recruitment of nuclear receptors to canonical telomeric repeats is hindered by robust shelterin binding; however, upon the introduction of variant repeats, nuclear receptors preferentially bind to the C-type repeat, altering the protein stoichiometry at the telomere.
Nuclear receptor depletion was shown to substantially diminish numerous ALT phenotypic characteristics, including the formation of APBs, generation of extrachromosomal C circles and T-SCE events, indicating that the recruitment of these proteins to the telomeres significantly contributes to the ALT mechanism. In view of their many known biological functions, the effect of nuclear receptor depletion upon ALT may possibly be indirect. However, in further support of the hypothesis that it results from the lack of their recruitment to the telomere, nuclear receptor knockdown had a greater effect on the ALT phenotype within the WI38-VA13/2RA cell line compared with IIICF/c, which is in accordance with our observations that the C-type variants (and nuclear receptors) are more abundant at WI38-VA13/2RA telomeres. Knockdown of nuclear receptors did not, however, affect the number of meta-TIFs or rate of chromosome fusions, implicating the inability of shelterin to bind to variant repeats, rather than the presence of nuclear receptors themselves, as the primary source of these telomeric abnormalities. Alternatively, nuclear receptor redundancy may require the removal of more than two nuclear receptors to affect the DNA damage response at the telomere or the level of chromosomal end-to-end fusions.
Notably, we have shown that insertion of the C-type variant into the telomeres of telomerase-positive cells resulted in the recruitment of nuclear receptors to the telomere. This directly demonstrates sequence-specific telomeric binding of nuclear receptors. In addition, both C- and G-type variant repeat incorporation led to the induction of ALT-associated characteristics such as telomere lengthening, telomere length heterogeneity, meta-TIFs, and C circles. These observations are comparable to previous studies in which mutant telomeric repeat incorporation was found to produce an ALT-like phenotype in yeast (Bechard et al., 2009) and, recently, insertion of TTTGGG mutant repeats into the telomeres of a telomerase-positive human breast cancer cell line with nonfunctional p53 induced several ALT-associated characteristics (Brault and Autexier, 2011). However, we were unable to detect inter-telomeric replication-mediated copying of an individual telomere-inserted tag to multiple chromosome ends, indicating that the insertion of variant repeats and the recruitment of nuclear receptors is insufficient to activate an ALT mechanism in an immortalized telomerase-positive cell line. Nonetheless, it remains possible that nuclear receptor recruitment contributes to ALT activation within mortal cell lines. Telomerase-positive cells already have a telomere maintenance mechanism and may not possess the genetic changes that are fully permissive for the activation of ALT. For instance, further genetic alterations such as loss of ATRX, which has recently been shown to be widely mutated in ALT tumors and immortalized cell lines (Heaphy et al., 2011; Lovejoy et al., 2012; Schwartzentruber et al., 2012), may be required for complete ALT activation. Furthermore, the variant repeat content and distal arrangement of artificially generated repeats may be insufficient to initiate HR-mediated telomeric replication.
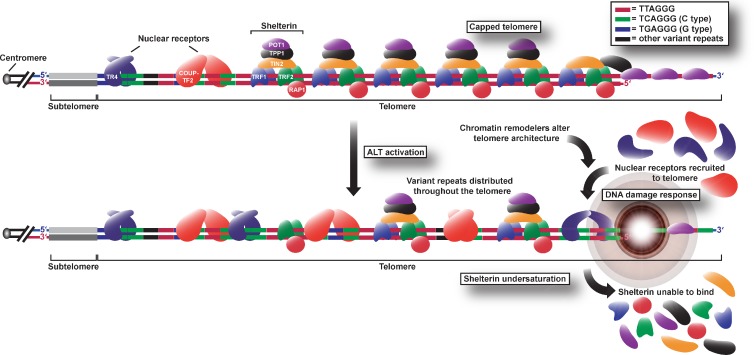
In summary, we have demonstrated that ALT telomeres display high levels of interspersed variant repeats, which have profound implications for telomere architecture and function. We propose that the presence of variant sequences throughout the telomeres of ALT cells, and the consequent binding of nuclear receptors, destabilize telomeres by attenuating shelterin binding (Fig. 7). In addition, incorporation of variant repeats and nuclear receptor recruitment may cause the telomeric chromatin structure to become more permissive to recombination, creating a positive feedback loop that leads to the incorporation of more variant repeats and further destabilization of the telomere. Although the prevalence of the C-type variant at ALT telomeres may be more functionally relevant in certain cell lines than in others, it is possible that the telomeres of these cells are enriched with other variant repeat sequences that could similarly prevent shelterin binding and cause recruitment of nuclear receptors as well as other proteins not generally associated with the telomere. The presence of variant repeats may also result in replication-dependent defects such as stalled replication forks and may disrupt strand invasion and the formation of the t loop by means of both sequence disruption and protein binding affinity. Furthermore, the interspersion of variant repeats may influence telomeric heterochromatin and nucleosome positioning. Nuclear receptors have been shown to be capable of altering expression of target genes via recruitment of chromatin remodeling complexes (Cui et al., 2011), so we speculate that recruitment of such factors may also change the heterochromatic state of ALT telomeres, thus contributing to derepression of telomeric recombination. Alternatively, nuclear receptors may cause telomeres to interact with one another and/or APBs via their oligomerization or recruitment of common binding partners (Déjardin, 2012), thus facilitating HR-dependent telomere elongation by the ALT mechanism.
Figure 7.
Model of alterations in telomere architecture during ALT activation. Dispersal of variant sequences among distal telomeric repeats leads to changes in telomere structure such as removal of shelterin, recruitment of nuclear receptors, and elicitation of a DNA damage response that facilitates telomere elongation via HR. Nuclear receptors may also recruit chromatin remodeling complexes capable of altering the heterochromatic state of ALT telomeres. Although this schematic shows only COUP-TF2 and TR4, additional nuclear receptors are likely recruited to the telomeres as either homodimers or heterodimers or even monomers.
Materials and methods
Cell culture
All cell lines were cultured in DMEM (Gibco) supplemented with 10% (vol/vol) FBS without antibiotics in a humidified incubator at 37°C with 5% CO2. Cell lines were authenticated by 16-locus short tandem repeat profiling and confirmed to be free of Mycoplasma species by CellBank Australia (Children’s Medical Research Institute).
Antibodies
The following primary antibodies were used in this study: TR2, TR4, COUP-TF1, EAR-2 (all obtained from Perseus Proteomics), COUP-TF2 (Perseus Proteomics or Abcam), PML (Santa Cruz Biotechnology, Inc.), γ-H2AX (BioLegend), TRF2 (Novus Biologicals), and actin (Sigma-Aldrich). Fluorophore-conjugated secondary antibodies (Life Technologies) were used for indirect immunofluorescence analyses. For Western blotting, secondary antibodies used were polyclonal goat anti–mouse and anti–rabbit immunoglobulin conjugated to horseradish peroxidase (Dako).
Indirect immunofluorescence and FISH
Cell cultures were treated with 20 ng/ml colcemid for 1–4 h when required, harvested by trypsinization, resuspended in 0.2% (wt/vol) KCl and 0.2% (wt/vol) trisodium citrate hypotonic buffer at room temperature for 10 min, and cytocentrifuged onto SuperFrost Plus glass slides (Menzel-Glaser) at 1200 rpm for 10 min in a Shandon Cytospin 4 at high acceleration. Indirect immunofluorescence and FISH were then conducted as described previously, with minor alterations (Cesare et al., 2009). Slides were subjected to preextraction by incubation in permeabilization solution (20 mM Hepes-KOH, pH 7.9, 20 mM NaCl, 5 mM MgCl2, 300 mM sucrose, and 0.5% [vol/vol] NP-40) for 10 min. For microscopic analysis of interphase, nuclei cells were cultured on sterile glass coverslips for 24–48 h and washed twice in PBS before preextraction. Slides were then washed in PBS, fixed at room temperature with 4% (vol/vol) formaldehyde in PBS for 10 min, and blocked with 100 µg/ml DNase-free RNase A (Sigma-Aldrich) in antibody dilution buffer (ABDIL; 20 mM Tris-HCl, pH 7.5, 0.2% [vol/vol] fish gelatin, 2% [wt/vol] BSA, 0.1% [vol/vol] Triton X-100, 150 mM NaCl, and 0.1% [wt/vol] sodium azide) for 30 min at 37°C. Slides were incubated with primary antibody in ABDIL for 1 h at 37°C, washed in PBST (PBS with 0.1% [vol/vol] Tween 20), incubated with secondary antibody in ABDIL for 30 min at 37°C, washed in PBST, and fixed with 4% (vol/vol) formaldehyde in PBS for 10 min at room temperature. Slides were subjected to a graded ethanol series (70% [vol/vol] for 3 min, 90% [vol/vol] for 2 min, and 100% for 2 min) to dehydrate and then allowed to air dry. Dehydrated slides were then overlaid with either 0.3 µg/ml Alexa 488–OO-(CCCTAA)3 or Texas red–OO-(TTAGGG)3 telomeric or Alexa 488–OO-(TCAGGG)3 or (TGAGGG)3 PNA probes (Panagene) in PNA hybridization solution (70% [vol/vol] deionized formamide, 0.25% [vol/vol] NEN blocking reagent [PerkinElmer], 10 mM Tris-HCl, pH 7.5, 4 mM Na2HPO4, 0.5 mM citric acid, and 1.25 mM MgCl2), denatured for 3 min at 80°C, and hybridized for 2 h at room temperature. Slides were washed in PNA wash A (70% [vol/vol] formamide and 10 mM Tris-HCl, pH 7.5), and then in PNA wash B (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.08% [vol/vol] Tween 20) and DAPI at 50 ng/ml added to the final wash. Finally, slides were rinsed briefly with deionized water and mounted in DABCO (90% [vol/vol] glycerol, 2.3% [vol/vol] 1,4-diazabicyclo(2.2.2)octane (Sigma-Aldrich), and 50 mM Tris-HCl, pH 8.0).
Preparation of chromatin fibers
Chromatin fibers were prepared as described previously with minor alterations (Sullivan, 2010). In brief, cells were harvested by trypsinization and resuspended in hypotonic solution (25 mM KCl and 0.27% [wt/vol] trisodium citrate) at a concentration of 6.1 × 104/ml for 5 min at room temperature. Cells were cytocentrifuged onto SuperFrost Plus glass slides (Menzel-Glaser) at 1,350 rpm for 4 min in a Shandon Cytospin 4 at high acceleration. Slides were then immediately immersed in fiber lysis buffer (2.5 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 1% [vol/vol] Triton X-100, and 0.5 M urea) for 16 min, then slowly removed at a steady rate of ∼20 s/slide. Fibers were then fixed at room temperature in PBS with 4% (vol/vol) formaldehyde for 10 min and extracted in with 0.1% (vol/vol) Triton X-100 in PBS at room temperature for 10 min. RNase A treatment, indirect immunofluorescence, or FISH was then performed as described in the previous paragraph.
Chromosome-orientation telomere-FISH
Cells were cultured in fresh medium supplemented with 7.5 µM BrdU and 2.5 µM 5-bromo-2′-deoxycytidine (BrdC; 3:1 ratio; Sigma-Aldrich) for 16–24 h depending on the mitotic index of the cell line. Cell cultures were treated with 20 ng/ml colcemid (Life Technologies) for the last 4 h of incubation to accumulate mitotic cells. Chromosome preparations were then obtained according to standard cytogenetic methods and CO-FISH was conducted as described previously with minor modifications (Pickett et al., 2009). In brief, cells were harvested by trypsinization and incubated in hypotonic buffer for no longer than 15 min at 37°C. Cells were then fixed in 3:1 methanol/acetic acid, spun at 1,200 rpm for 8 min, and washed three times in fixative. Concentrated cells in fixative were then dropped onto clean, dry microscope slides, left for ∼20 s, held over a water bath at 75°C for 3 s, and left to dry overnight. Slides were then RNase A treated as described in the Indirect immunofluorescence and FISH section, rinsed in PBS, and subjected to a postfix in 4% (vol/vol) formaldehyde in PBS at room temperature for 10 min. Slides were then dehydrated again, and then stained in 0.5 µg/ml Hoechst 33258 (Sigma-Aldrich) in 2× SSC for 15 min at room temperature. Slides were then flooded with 50 µl 2× SSC, coverslip applied, and exposed to long wave (∼365 nm) UV light (Stratalinker 1800 UV irradiator; Agilent Technologies) for 40 min. The BrdU/BrdC-substituted DNA strands were then digested in 10 U/µl Exonuclease III solution (New England Biolabs, Inc.) in buffer supplied by the manufacturer for 15 min at 37°C. Slides were then rinsed in PBS and subjected to another ethanol series. Dehydrated slides were then overlaid with 0.3 µg/ml Texas red–OO-(TTAGGG)3 telomeric probe in PNA hybridization solution and hybridized at room temperature for 2 h. Slides were then washed in PNA wash A for 15 min then overlaid with 0.3 µg/ml Alexa 488–OO-(CCCTAA)3 and hybridized again at room temperature for 2 h. Finally, slides were washed in PNA wash A and B, stained with DAPI, and mounted in DABCO as described in the Indirect immunofluorescence and FISH section. Only T-SCE events observed with both leading and lagging strand probes simultaneously were scored as positive.
NeoR-FISH
HT1080 hTR C- and G-type cells at ∼150 pds after expression of mutant hTR were transfected with linearized telomere-targeting plasmid, Tel (Dunham et al., 2000), using siPORT NeoFX Transfection Agent (Ambion). Approximately 48 h after transfection, cells were seeded at low density and selected in 500 µg/ml Geneticin. Isolated clones were then assayed at early (pd +175) and late (pd +210) pds for integration of the telomeric plasmid sequence by neoR-FISH. The plasmid backbone pSXneo was labeled with biotin-16-dUTP using the Biotin-Nick Translation Mix (Roche) according to the manufacturer’s instructions. NeoR-FISH was then performed as described previously (Dunham et al., 2000; Fasching et al., 2005; Pickett et al., 2009). In brief, chromosomes were prepared according to standard cytogenetic methods, RNase A treated, postfixed, and dehydrated with an ethanol series as explained in the previous section. Chromosomes were then denatured in 70% (vol/vol) formamide/2× SSC for 2 min at 72°C and subjected to a cold ethanol dehydration series. Approximately 10 ng/ml of pSXneo probe was denatured at 90°C for 5 min, added to slides, and hybridized overnight in a humidified chamber at 37°C. Slides were then washed three times in 50% (vol/vol) formamide/2× SSC, pH 7.0, for 5 min at 42°C followed by another three washes in 2× SSC for 5 min at room temperature. The probe was then detected by incubating slides with FITC-conjugated avidin DCS (1:400; Vector Laboratories) with 3% (wt/vol) skim milk in 4× SSC/0.1% (vol/vol) Tween 20 for 30 min in the dark at 37°C. Slides were then washed three times in 4× SSC/0.1% (vol/vol) Tween 20 for 5 min at 42°C. Finally, slides were counterstained with DAPI and mounted in DABCO as described in the section Indirect immunofluorescence and FISH. We analyzed ∼100 metaphases for each clone. Individual chromosome ends were scored as positive for the neoR signal if a signal could be detected on both sister chromatids of the respective chromosome in multiple metaphases.
Imaging
All cells were visualized using a microscope (Axio-Imager M1; Carl Zeiss) at room temperature, with a Plan-Apochromat 63× or 100× objective (NA 1.4) and a digital camera (AxioCam MRm; Carl Zeiss). Metaphase cells were imaged in 12 Z planes with 0.3-µm increments for the green and red filters and as a single focal plane in the blue filter, and then merged using the extended-focus setting (Metafer4 software; MetaSystems). Images of metaphase spreads were also analyzed with Isis software.
Deep sequencing analysis
DNA was extracted from WI38-VA13/2RA and HeLa cell lines and underwent indexed library construction. In brief, the genome was fragmented by sonication and the fragment ends were phosphorylated and A tailed. Indexed forked adapters were then ligated, and the resultant libraries were amplified and applied to a lane of an Illumina HiSeq. Results were aligned to the genome using Burrows-Wheeler Alignment tool (Li and Durbin, 2009). The telomeric subset of reads were extracted using a custom script (motif_counter) incorporating functions from SAMtools (Li et al., 2009) and standard Linux pattern-matching programs (http://sourceforge.net/projects/motifcounter/).
RNAi
The following Stealth RNAi siRNA were designed and synthesized by Life Technologies: TR4, COUP-TF2, and the Stealth RNAi siRNA Negative Control Med GC Duplex #2. Cells were seeded at 3.5 × 105/T-75 flask and reverse transfected using Lipofectamine RNAiMAX (Life Technologies) with an siRNA concentration of 20 µM as per the manufacturer’s instructions and harvested for Western blotting, C-circle assay, and microscopic analysis 72 h after transfection.
EMSA
The G strand consensus telomeric (5′-ACATGTTAGGGTTAGGGTTAGGGTTAG-3′), C- (5′-ACATGTCAGGGTCAGGGTCAGGGTCAG-3′), and G-type (5′-ACATGTGAGGGTGAGGGTGAGGGTGAG-3′) sequences and their respective complementary C strands were used to form a double-stranded oligonucleotide. G strands were labeled at the 5′ end with γ-[32P]-ATP using T4 polynucleotide kinase (Promega). Double-stranded probes were assembled by the addition of labeled G strand and unlabeled C strand (200 nM) in TEN buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 250 mM NaCl), followed by heating at 90°C for 3 min and slowly cooling for 1 h at room temperature. EMSA reactions were performed in 13% (vol/vol) glycerol, 10 mM Hepes-KOH, pH 8.0, 100 mM KCl, 0.5 mM EDTA, 10 mM MgCl2, 100 µg/ml BSA, 1 mM dithiothreitol, and 0.004% (wt/vol) xylene cyanol incubated with either purified human TR4 or TRF2 (OriGene). Increasing concentrations of TR4 (0, 50, 100, 200, and 300 nM) and TRF2 (0, 100, 200, 400, and 600 nM) were added to the double-stranded substrate and incubated at room temperature for 30 min. The 5% (vol/vol) native polyacrylamide gel (using 1.5-mm spacers) was run in 0.5× Tris-Borate-EDTA (TBE) at 200 V for 1 h at 4°C. The gel was dried on Whatman paper under vacuum at 70°C for 1 h, visualized by a phosphorimager (Typhoon), and analyzed using ImageQuant TL software (Molecular Dynamics). DNA probes containing less than four telomeric repeats were used for the EMSA experiments because of their inability to form G-quadruplexes.
Western blotting
Cells were harvested via trypsinization, washed in PBS, and resuspended in ice-cold RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [vol/vol] NP-40, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, 1 mM EDTA, complete protease inhibitors, and 1 mM PMSF) at 100 µl per 106 cells, rocking on ice for 30 min. Cell lysates were then centrifuged at 14,000 g for 15 min at 4°C and the supernatant was removed immediately. Samples were prepared and run on NuPAGE Novex Bis-Tris Mini Gels (Life Technologies) according to the manufacturer’s instructions. The membrane was stained with Ponceau S for 5 min and blocked for 1 h at room temperature in PBST containing 5% (wt/vol) skim milk. The membrane was then incubated with primary antibody containing 0.5% (wt/vol) skim milk in PBST overnight at 4°C. The membrane was washed in PBST followed by incubation for 1 h at room temperature with secondary antibody (mouse and rabbit anti–goat immunoglobulin horseradish peroxidase; Dako) diluted 1:1,000 in PBST. The membrane was rinsed with PBS before incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) for 5 min at room temperature. The blot was then exposed to a Luminescent Image Analyzer (LAS-4000; Fujifilm) and quantitated using Multigauge software.
Generation and expression of wild-type and mutant hTR constructs
The pBABEpuroU3-hTR (Wong and Collins, 2006) and pApex-U3-hTR (Stern et al., 2012) plasmids were used to overexpress hTR encoding wild-type and variant telomeric repeats. Variant repeat-encoding hTR plasmids were produced by site-directed mutagenesis (Agilent Technologies). Specifically, positions three and nine of the template region of hTR were modified using the following forward (F) and reverse (R) primers and mutagenesis was confirmed by DNA sequencing (Australian Genome Research Facility): C-type F, 5′-GGCCATTTTTTGTCTGACCCTGACTGAGAAGGGCGTAG-3′; C-type R, 5′-CTACGCCCTTCTCAGTCAGGGTCAGACAAAAAATGGCC-3′; G-type F, 5′-GGCCATTTTTTGTCTCACCCTCACTGAGAAGGGCGTAG-3′; G-type R, 5′-CTACGCCCTTCTCAGTGAGGGTGAGACAAAAAATGGCC-3′.
The telomerase-positive human fibrosarcoma cell line HT1080 stably overexpressing wild-type hTR (designated HT1080 hTR [Pickett et al., 2009]) was transfected at early passage with either wild-type, C-type, or G-type pApex-U3-hTR plasmids using siPORT NeoFX transfection agent (Ambion). HT1080-6TG cells were stably transfected in the same manner with wild-type pApex-U3-hTR. These cells, designated HT1080-6TG hTR, were then retrovirally transduced with wild-type, C-type, or G-type pBABEpuroU3-hTR. Stable mass populations were selected and maintained with 0.4 µg/ml puromycin (Sigma-Aldrich) and 150 µg/ml hygromycin B (Roche).
Genomic DNA extraction
Cells were harvested via trypsinization, washed in PBS, and resuspended in lysis solution (100 mM Tris-HCl,pH 7.5, 10 mM EDTA, 100 mM NaCl, and 1% [wt/vol] N-lauroylsarcosine). Lysates were then subjected to RNase A (50 µg/ml) treatment for 20 min at room temperature followed by Proteinase K (100 µg/ml) treatment for 6 h at 55°C. DNA was extracted using equal volumes of phenol/chloroform/isoamyl alcohol (25:24:1) and the resulting phases were separated by centrifugation at 2,000 rpm for 10 min at room temperature using Phase Lock Gel Light tubes (5 PRIME). The aqueous layer was discarded and the DNA was ethanol precipitated (0.1 vol of sodium acetate and 2.5 vol of 100% ethanol) and resuspended in TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA).
Terminal restriction fragment analysis
Telomeric restriction fragments were prepared by HinfI and RsaI digestion of genomic DNA and separated by pulsed-field gel electrophoresis as described previously with some alterations (Perrem et al., 1999). In brief, genomic DNA was prepared via phenol/chloroform extraction described in the previous paragraph and digested with 4 U/µg HinfI and RsaI overnight at 37°C. The DNA was then ethanol precipitated and 1 µg was loaded on a 1% (wt/vol) agarose gel in 0.5× TBE. Pulsed-field gels were run at 6 V for 14 h at 14°C with an initial switch time of 1 and a final switch time of 6. The gels were dried for 2 h at 60°C, denatured in 0.5 M NaOH/1.5 M NaCl for 1 h, and neutralized in 0.5 M Tris-HCl (pH 8.0)/1.5 M NaCl for 1 h. Gels were then rinsed in 2× SSC and prehybridized in Church buffer (250 mM sodium phosphate buffer, pH 7.2, 7% [wt/vol] SDS, 1% [wt/vol] BSA fraction V grade, and 1 mM EDTA) for 2 h at 37°C. Finally, gels were hybridized overnight with a γ-[32P]-ATP–labeled (CCCTAA)3, (CCCTGA)3, or (CCCTCA)3 oligonucleotide probe specific to one of the three telomeric repeat types, washed three times in 0.1× SSC for 15 min at 37°C, and exposed to a PhosphorImager screen overnight.
C-circle assay
The C-circle assay was conducted using 20 ng of genomic DNA as described previously with some modifications (Henson et al., 2009). Genomic DNA was prepared via phenol/chloroform extraction as described in the Genomic DNA extraction section and resuspended in TE. For the C-circle reaction, 10 µl of DNA was added to 10 µl 200 µg/ml BSA, 0.1% (vol/vol) Tween 20, 1 mM each dNTP, 1× Φ29 buffer (New England Biolabs, Inc.), and 7.5 U Φ29 DNA polymerase (New England Biolabs, Inc.) and incubated for 8 h at 30°C, and then for 20 min at 65°C. The reaction products were then dot blotted onto a 2× SSC-soaked Biodyne B 0.45-µm nylon membrane (Pall). The DNA was then UV cross-linked onto the membrane (∼254 nm) and prehybridized in PerfectHyb Plus hybridization buffer (Sigma-Aldrich) for 1 h at 37°C. The signal was then detected by hybridization with end-labeled γ32P-(CCCTAA)3 overnight at 37°C. The membrane was then washed three times in 2× SSC for 20 min shaking at room temperature. The membrane was then exposed to a PhosphorImager screen for 8 h and scanned on a Typhoon imager with ImageQuant software, using edge subtraction for background correction.
Telomere dot blot
The analysis of total telomeric DNA content was performed by adding 400 ng of genomic DNA digested with HinfI and RsaI to a dot blot as described in the previous paragraph with the C-circle assay.
Statistical analyses
Microsoft Excel was used to generate graphs and perform statistical analysis by calculating P values using unpaired Student’s two-tailed t tests. To statistically compare multiple groups, one-way analysis of variance tests were conducted using Prism 4 (GraphPad Software).
Online supplemental material
Fig. S1 shows the abundance of C- and G-type variant telomeric repeats in a variety of cell types. The figure also tabulates the mean numbers of each telomeric repeat generated from deep sequencing analysis of WI38-VA13/2RA and HeLa cells and also displays the relative abundance of telomeric DNA in the entire panel of mortal, telomerase-positive, and ALT cell lines. Fig. S2 shows the localization of COUP-TF2 and TR4 to telomeric DNA in a variety of cell types. Fig. S3 shows the global expression of COUP-TF2 and TR4 in a panel of ALT and telomerase-positive cell lines. Fig. S4 shows that both the number of meta-TIFs and rate of chromosomal end-to-end fusions do not change after COUP-TF2 and TR4 knockdown. Fig. S5 shows the terminal restriction fragment analysis of the HT1080 hTR cell lines for each of the three telomeric repeat types. The figure also shows examples of the immunofluorescence experiments conducted on metaphase spreads of the HT1080 hTR wild-type and HT1080 hTR G-type cell lines. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207189/DC1.
Acknowledgments
This work was supported by an Australian Postgraduate Award (to D. Conomos), a Cancer Institute NSW Research Scholar Award (to D. Conomos), a Cancer Council NSW program grant (to R.R. Reddel), and a National Health and Medical Research Council project grant (#1009231; to R.R. Reddel and H.A. Pickett).
Footnotes
Abbreviations used in this paper:
- ALT
- alternative lengthening of telomeres
- APB
- ALT-associated PML body
- CO-FISH
- chromosome orientation FISH
- EMSA
- electrophoretic mobility shift assay
- HR
- homologous recombination
- hTR
- human telomerase RNA
- pd
- population doubling
- PML
- promyelocytic leukemia
- PNA
- peptide nucleic acid
- TIF
- telomere dysfunction-induced foci
- TRF2
- telomeric repeat-binding factor 2
- T-SCE
- telomere sister chromatid exchange
References
- Allshire R.C., Dempster M., Hastie N.D. 1989. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 17:4611–4627 10.1093/nar/17.12.4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.J., Casey G., Fasching C.L., Stanbridge E.J. 1994. Evidence that wild-type TP53, and not genes on either chromosome 1 or 11, controls the tumorigenic phenotype of the human fibrosarcoma HT1080. Genes Chromosomes Cancer. 9:266–281 10.1002/gcc.2870090407 [DOI] [PubMed] [Google Scholar]
- Baird D.M., Jeffreys A.J., Royle N.J. 1995. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 14:5433–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D.M., Coleman J., Rosser Z.H., Royle N.J. 2000. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 66:235–250 10.1086/302721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechard L.H., Butuner B.D., Peterson G.J., McRae W., Topcu Z., McEachern M.J. 2009. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol. Cell. Biol. 29:626–639 10.1128/MCB.00423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W. 2004. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 64:3444–3451 10.1158/0008-5472.CAN-04-0323 [DOI] [PubMed] [Google Scholar]
- Brault M.E., Autexier C. 2011. Telomeric recombination induced by dysfunctional telomeres. Mol. Biol. Cell. 22:179–188 10.1091/mbc.E10-02-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271–1274 10.1038/nm1197-1271 [DOI] [PubMed] [Google Scholar]
- Celli G.B., Denchi E.L., de Lange T. 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8:885–890 10.1038/ncb1444 [DOI] [PubMed] [Google Scholar]
- Cesare A.J., Griffith J.D. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24:9948–9957 10.1128/MCB.24.22.9948-9957.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A.J., Reddel R.R. 2008. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129:99–108 10.1016/j.mad.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Cesare A.J., Kaul Z., Cohen S.B., Napier C.E., Pickett H.A., Neumann A.A., Reddel R.R. 2009. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 16:1244–1251 10.1038/nsmb.1725 [DOI] [PubMed] [Google Scholar]
- Coleman J., Baird D.M., Royle N.J. 1999. The plasticity of human telomeres demonstrated by a hypervariable telomere repeat array that is located on some copies of 16p and 16q. Hum. Mol. Genet. 8:1637–1646 10.1093/hmg/8.9.1637 [DOI] [PubMed] [Google Scholar]
- Cui S., Kolodziej K.E., Obara N., Amaral-Psarris A., Demmers J., Shi L., Engel J.D., Grosveld F., Strouboulis J., Tanabe O. 2011. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic β-type globin promoters in differentiated adult erythroid cells. Mol. Cell. Biol. 31:3298–3311 10.1128/MCB.05310-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Déjardin J. 2012. How chromatin prevents genomic rearrangements: locus colocalization induced by transcription factor binding. Bioessays. 34:90–93 10.1002/bies.201100122 [DOI] [PubMed] [Google Scholar]
- Déjardin J., Kingston R.E. 2009. Purification of proteins associated with specific genomic Loci. Cell. 136:175–186 10.1016/j.cell.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447–450 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- Fasching C.L., Bower K., Reddel R.R. 2005. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 65:2722–2729 10.1158/0008-5472.CAN-04-2881 [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. 1999. Mammalian telomeres end in a large duplex loop. Cell. 97:503–514 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Guiducci C., Cerone M.A., Bacchetti S. 2001. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 20:714–725 10.1038/sj.onc.1204145 [DOI] [PubMed] [Google Scholar]
- Hanaoka S., Nagadoi A., Nishimura Y. 2005. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 14:119–130 10.1110/ps.04983705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C.B., Futcher A.B., Greider C.W. 1990. Telomeres shorten during ageing of human fibroblasts. Nature. 345:458–460 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S., et al. 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 333:425 10.1126/science.1207313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.D., Reddel R.R. 2010. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 584:3800–3811 10.1016/j.febslet.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Henson J.D., Cao Y., Huschtscha L.I., Chang A.C., Au A.Y., Pickett H.A., Reddel R.R. 2009. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27:1181–1185 10.1038/nbt.1587 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Sfeir A.J., Shay J.W., Wright W.E., de Lange T. 2005. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 24:2667–2678 10.1038/sj.emboj.7600733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.Q., Zhong Z.H., Henson J.D., Neumann A.A., Chang A.C., Reddel R.R. 2005. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 25:2708–2721 10.1128/MCB.25.7.2708-2721.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Sasaki H., Suzawa M., Masushige S., Tora L., Chambon P., Gronemeyer H. 1995. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 15:5858–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.M., Rivera M.A., Botchkina I.L., Shalaby R., Thor A.D., Blackburn E.H. 2001. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. USA. 98:7982–7987 10.1073/pnas.131211098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rosenberg J.E., Donjacour A.A., Botchkina I.L., Hom Y.K., Cunha G.R., Blackburn E.H. 2004. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 64:4833–4840 10.1158/0008-5472.CAN-04-0953 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo J.A., Der-Sarkissian H., Cazes L., Bacchetti S., Reddel R.R. 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 64:2324–2327 10.1158/0008-5472.CAN-03-4035 [DOI] [PubMed] [Google Scholar]
- Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H., Sung P.A., Jasin M., et al. ; ALT Starr Cancer Consortium 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8:e1002772 10.1371/journal.pgen.1002772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusíc L., Anton M., Tidy A., Wang P., Villeponteau B., Bacchetti S. 1997. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol. Cell. Biol. 17:6394–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. 1988. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA. 85:6622–6626 10.1073/pnas.85.18.6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni A., Neumann A.A., Hills M., Reddel R.R. 2009. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum. Mol. Genet. 18:1017–1027 10.1093/hmg/ddn436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Ishikawa F. 2009. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 29:703–713 10.1128/MCB.00603-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Perrem K., Bryan T.M., Englezou A., Hackl T., Moy E.L., Reddel R.R. 1999. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene. 18:3383–3390 10.1038/sj.onc.1202752 [DOI] [PubMed] [Google Scholar]
- Perrem K., Colgin L.M., Neumann A.A., Yeager T.R., Reddel R.R. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21:3862–3875 10.1128/MCB.21.12.3862-3875.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett H.A., Cesare A.J., Johnston R.L., Neumann A.A., Reddel R.R. 2009. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28:799–809 10.1038/emboj.2009.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Wasserman W.W. 2005. Prediction of nuclear hormone receptor response elements. Mol. Endocrinol. 19:595–606 10.1210/me.2004-0101 [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.A., Tönjes M., et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 482:226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Sfeir A., Kabir S., van Overbeek M., Celli G.B., de Lange T. 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 327:1657–1661 10.1126/science.1185100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W., Bacchetti S. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer. 33:787–791 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
- Stern J.L., Zyner K.G., Pickett H.A., Cohen S.B., Bryan T.M. 2012. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell. Biol. 32:2384–2395 10.1128/MCB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr B.A., Blackburn E.H. 2008. ATM mediates cytotoxicity of a mutant telomerase RNA in human cancer cells. Cancer Res. 68:5309–5317 10.1158/0008-5472.CAN-08-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A. 2010. Optical mapping of protein-DNA complexes on chromatin fibers. Methods Mol. Biol. 659:99–115 10.1007/978-1-60761-789-1_7 [DOI] [PubMed] [Google Scholar]
- Tokutake Y., Matsumoto T., Watanabe T., Maeda S., Tahara H., Sakamoto S., Niida H., Sugimoto M., Ide T., Furuichi Y. 1998. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun. 247:765–772 10.1006/bbrc.1998.8876 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell. 92:401–413 10.1016/S0092-8674(00)80932-0 [DOI] [PubMed] [Google Scholar]
- Varley H., Pickett H.A., Foxon J.L., Reddel R.R., Royle N.J. 2002. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 30:301–305 10.1038/ng834 [DOI] [PubMed] [Google Scholar]
- Wang R.C., Smogorzewska A., de Lange T. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 119:355–368 10.1016/j.cell.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Wong J.M., Collins K. 2006. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 20:2848–2858 10.1101/gad.1476206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager T.R., Neumann A.A., Englezou A., Huschtscha L.I., Noble J.R., Reddel R.R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- Zhong Z.H., Jiang W.Q., Cesare A.J., Neumann A.A., Wadhwa R., Reddel R.R. 2007. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 282:29314–29322 10.1074/jbc.M701413200 [DOI] [PubMed] [Google Scholar]