PI4KIIIα is targeted to the plasma membrane via an evolutionarily conserved complex comprised of EFR3 and TTC7 to control PtdIns4P synthesis and the selective enrichment of PtdIns(4,5)P2 in this membrane.
Abstract
Plasma membrane phosphatidylinositol (PI) 4-phosphate (PtdIns4P) has critical functions via both direct interactions and metabolic conversion to PI 4,5-bisphosphate (PtdIns(4,5)P2) and other downstream metabolites. However, mechanisms that control this PtdIns4P pool in cells of higher eukaryotes remain elusive. PI4KIIIα, the enzyme thought to synthesize this PtdIns4P pool, is reported to localize in the ER, contrary to the plasma membrane localization of its yeast homologue, Stt4. In this paper, we show that PI4KIIIα was targeted to the plasma membrane as part of an evolutionarily conserved complex containing Efr3/rolling blackout, which we found was a palmitoylated peripheral membrane protein. PI4KIIIα knockout cells exhibited a profound reduction of plasma membrane PtdIns4P but surprisingly only a modest reduction of PtdIns(4,5)P2 because of robust up-regulation of PtdIns4P 5-kinases. In these cells, however, much of the PtdIns(4,5)P2 was localized intracellularly, rather than at the plasma membrane as in control cells, along with proteins typically restricted to this membrane, revealing a major contribution of PI4KIIIα to the definition of plasma membrane identity.
Introduction
Phosphoinositides are minor components of cellular membranes but play key regulatory roles in cell physiology (Di Paolo and De Camilli, 2006). Their phosphorylated head groups, which protrude from the cytosolic leaflet of membranes, bind with variable affinity and specificity to proteins located in or exposed to the cytosolic space (Lemmon, 2008). Through these interactions, the heterogeneous distribution of the seven phosphoinositides in different membranes helps to generate a code of membrane identity that plays a fundamental role in orchestrating the processes occurring on such membranes. Some phosphoinositides (in particular phosphatidylinositol [PI] 4,5-bisphosphate [PtdIns(4,5)P2]) also act as precursors of other intracellular messenger molecules.
Among phosphoinositides, PI 4-phosphate (PtdIns4P) plays numerous fundamental roles. It has critical functions in the Golgi complex, the endosomal system, and the plasma membrane, which are mediated by its direct interaction with PtdIns4P-binding proteins (D’Angelo et al., 2008). Additionally, phosphorylation of PtdIns4P at the 5 position by type I PI monophosphate kinases (PIPKIs) represents the major pathway for the synthesis of PtdIns(4,5)P2 (Doughman et al., 2003), which, in turn, is the precursor of other important signaling metabolites. PtdIns4P also has major regulatory actions on the localization and metabolism of other membrane lipids. For example, several oxysterol-binding protein–related proteins contain PtdIns4P binding sites that target them to PtdIns4P-containing membranes. One yeast oxysterol-binding protein–related protein (Osh4) was recently shown to bind either PtdIns4P or cholesterol via the same lipid-binding pocket and to exchange these two lipids on artificial membranes (Lehto et al., 2001; Raychaudhuri and Prinz, 2010; de Saint-Jean et al., 2011). Finally, genetic and biochemical interactions between enzymes that control PtdIns4P levels (both PI 4-kinases and PI 4-phosphatases), and enzymes of the sphingolipid metabolic pathways have been reported (Tabuchi et al., 2006; Breslow et al., 2010).
In view of these considerations, and of numerous findings that link PtdIns4P, its downstream metabolites, and the lipids indirectly controlled by PtdIns4P to human disease (Vicinanza et al., 2008; McCrea and De Camilli, 2009), it is critically important to understand the mechanisms controlling PtdIns4P levels and metabolism on different membranes, in particular the plasma membrane, whose PtdIns4P pool serves as the precursor of the bulk of cellular PtdIns(4,5)P2. However, information on these mechanisms in metazoan cells is not as advanced as in yeast. The mammalian genome encodes four PI 4-kinase isoforms. PI4KIIIα and PI4KIIIβ (encoded by PI4KA and PI4KB, respectively) are the homologues of the two major yeast PI 4-kinases, Stt4 and Pik1, which mediate PtdIns4P generation in the plasma membrane and Golgi complex, respectively, in this organism (Audhya et al., 2000; Balla and Balla, 2006). PI4KIIα and PI4KIIβ, two very similar proteins encoded by PI4K2A and PI4K2B, respectively, represent a duplication of yeast Lsb6 and function in the Golgi complex, the endosomal system, and also to some extent at the plasma membrane (Balla and Balla, 2006).
In yeast, Stt4 has been shown to be localized at plasma membrane hotspots termed PI kinase (PIK) patches in a complex with the accessory proteins Efr3 and Ypp1 (Baird et al., 2008). Interestingly, considering that the bulk of the cortical ER is tightly apposed to the plasma membrane in this organism, Stt4 contains a so-called FFAT (two phenylalanines [FF] in an acidic tract) motif that enables binding to the integral ER membrane proteins Scs2/22. In fact, contact sites between the ER and the plasma membrane serve as hubs controlling PtdIns4P metabolism in this organism (Stefan et al., 2011).
Although a pharmacological and knockdown study has suggested that, as in the case of yeast Stt4, PI4KIIIα functions in the generation of PtdIns4P at plasma membrane (Balla and Balla, 2006), surprisingly little is known about this enzyme. Based on immunofluorescence, a localization of PI4KIIIα in the ER has been reported (Wong et al., 1997), but such a localization remains highly debated, in spite of the partial conservation of its FFAT motif that would be expected to bind VAP-A/B, the mammalian Scs2/22 homologues. The exogenous expression of tagged PI4KIIIα also failed to provide consistent results, and no evidence existed for its targeting to the plasma membrane (Balla and Balla, 2006). In mammalian cells, ER–plasma membrane contacts are much less extensive than in yeast, and the bulk of the ER is not localized in close proximity to the plasma membrane. How the mechanisms of PtdIns4P generation have adjusted to the different architecture of cells of higher eukaryotes is an important open question.
An issue of special interest is the link between the functions of Stt4/PI4KIIIα and of Efr3 (Baird et al., 2008). Efr3 is the yeast homologue of Drosophila melanogaster rolling blackout (RBO), a protein expressed at high levels in the nervous system of this organism whose mutation is responsible for a temperature-sensitive paralysis (Huang et al., 2004). RBO was proposed to be a transmembrane lipase, and its potential role in lipid metabolism was supported by changes in phosphoinositide and diacylglycerol levels in temperature-sensitive rbo mutants at the restrictive temperature (Huang et al., 2004).
Additional interest in the localization and properties of PI4KIIIα came from the recent identification by several groups of this enzyme as a critical host factor for the hepatitis C virus life cycle (Berger et al., 2009; Borawski et al., 2009; Tai et al., 2009; Trotard et al., 2009; Alvisi et al., 2011), pointing to PI4KIIIα as to a potential target for anti–hepatitis C virus therapies (Altan-Bonnet and Balla, 2012).
Collectively, these findings highlight the importance of addressing the still unclear and controversial subcellular localization of PI4KIIIα and the function of the PtdIns4P pools regulated by this enzyme. Here, we show recruitment of PI4KIIIα to the plasma membrane and show that its targeting requires presence in this membrane of EFR3/RBO, which we demonstrate to be a palmitoylated peripheral scaffold protein, not a transmembrane lipase. We also show that loss of PI4KIIIα leads to a striking compensatory increase of PIPKIs that minimizes a global decrease in cellular levels of PtdIns(4,5)P2, in spite of the dramatic decrease of PtdIns4P. However, the bulk of PtdIns(4,5)P2 produced by this compensatory response is localized not in the plasma membrane but, surprisingly, on intracellular vesicles. Proteins and lipids normally segregated in the plasma membrane are also found on these vesicles, emphasizing how targeting of PI4KIIIα to the plasma membrane is a key upstream event in defining the proteomic and lipidomic identity of this membrane.
Results
Full-length PI4KIIIα visits the plasma membrane dynamically
In contrast to yeast Stt4, which is localized at the plasma membrane (Audhya and Emr, 2002), a GFP-tagged PI4KIIIα construct based on the published ORF (Nakagawa et al., 1996) displayed diffuse cytosolic localization with no discernable enrichment on any cellular membrane (Fig. 1 A, left), as reported previously (Balla and Balla, 2006). We noted, however, that the mobility by SDS-PAGE of a construct corresponding to untagged PI4KIIIα was slightly faster than the mobility of the endogenous protein, as assessed by Western blotting (Fig. 1 B, compare first and third lanes). We thus considered the possibility of an incorrect annotation and reexamined the reported translational start site of PI4KA (Nakagawa et al., 1996). An alignment of vertebrate PI4KA homologues (Fig. 1 C) revealed substantial conservation upstream of the reported translation start site (hence defined as M1*; Nakagawa et al., 1996), including an additional conserved Met residue (defined as M1). Western blot analysis demonstrated that exogenously expressed M1-PI4KIIIα co-migrated with endogenous PI4KIIIα, in contrast to the smaller M1*-PI4KIIIα (Fig. 1 B). Furthermore, determination of PI4KA translation initiation sites using ribosome footprinting after harringtonine treatment (Ingolia et al., 2011) suggested that M1, but not M1*, was a strong translation start site (Fig. 1 D). This analysis also revealed a potential non-AUG start site (ACG, encoding Thr and indicated as T* in Fig. 1 D) between M1 and M1*.
Figure 1.
Full-length PI4KIIIα visits the plasma membrane dynamically. (A) Confocal imaging of HeLa cells transfected with GFP-M1*-PI4KIIIα (left) or GFP-M1-PI4KIIIα (right). (B) Anti-PI4KIIIα Western blot comparing electrophoretic mobility of endogenous (Endog.) PI4KIIIα and either exogenously expressed M1-PI4KIIIα or M1*-PI4KIIIα in lysates of HEK cells. Molecular masses are given in kilodaltons. (C) Alignment of the N-terminal region of vertebrate PI4KIIIα orthologues. The black shading denotes conservation of amino acid identity, and the gray shading denotes conservation of amino acid similarity. (D) Ribosome footprinting reveals M1 and T*, but not M1*, as likely PI4KIIIα translation start sites. The data shown were pooled from two biological replicate experiments. (E) TIRF microscopic analysis shows highly dynamic spots of GFP-M1-PI4KIIIα, but not of GFP-M1*-PI4KIIIα, at the plasma membrane, revealing very transient visits of the enzyme to this membrane. COS-7 cells were transfected with GFP-M1-PI4KIIIα (top row) or GFP-M1*-PI4KIIIα (bottom row) and imaged by TIRF microscopy, with a 1-s interval between each image acquisition. Bars: (A) 20 µm; (E) 2 µm.
We considered the possibility that an incorrectly assigned start site may have affected previous studies of PI4KIIIα and next investigated whether M1-PI4KIIIα had a different subcellular localization than M1*-PI4KIIIα. The majority of GFP-M1-PI4KIIIα fluorescence was still localized in the cytosol (Fig. 1 A, right). However, total internal reflection fluorescence (TIRF) microscopy revealed that GFP-M1-PI4KIIIα, but not GFP-M1*-PI4KIIIα, transiently visited the plasma membrane (Fig. 1 E and Video 1; also see Fig. 3). Although this dynamic behavior of PI4KIIIα appeared to differ from that of its yeast homologue, Stt4, which forms stable PIK patches at the plasma membrane (Baird et al., 2008), our finding is consistent with a role for PI4KIIIα at the plasma membrane. It was therefore of interest to determine how PI4KIIIα is recruited to the plasma membrane.
Figure 3.
Endogenous EFR3 and TTC7 are required for recruitment of GFP-M1-PI4KIIIα to the plasma membrane. HeLa cells were treated with a negative control siRNA or siRNA against EFR3A and EFR3B (siEFR3) and TTC7A and TTC7B (siTTC7) either alone or in combination. After 24 h, the cells were transfected with GFP-M1*-PI4KIIIα or GFP-M1-PI4KIIIα, and after an additional 24 h, the cells were imaged by TIRF microscopy, with a 1-s interval in between each image acquisition. (A) Representative images for each of the samples quantified in B. (B) Quantification of the number of GFP puncta observed at the plasma membrane, normalized per unit area. Error bars indicate standard deviations. *, P < 0.005 and **, P < 0.02, relative to GFP-M1-PI4KIIIα with control siRNA (n = 4–6). Bar, 5 µm.
PI4KIIIα recruitment to the plasma membrane requires its interaction with EFR3 and TTC7
Immunoprecipitation (IP) of endogenous PI4KIIIα from mouse brain extracts, followed by mass spectrometry analysis, identified two interactors, EFR3B and TTC7B, that, interestingly, are the yeast homologues of Efr3 and Ypp1, the other constituents of yeast PIK patches (Fig. S1; Baird et al., 2008). The interaction of EFR3B and of TTC7B with PI4KIIIα was confirmed using IP followed by Western blotting with endogenous and exogenously expressed proteins (Fig. 2, A and B). To assess whether EFR3B and TTC7B have a role in PI4KIIIα recruitment, we expressed tagged versions of these proteins and performed knockdown experiments.
Figure 2.
PI4KIIIα, EFR3B, and TTC7B form a complex at the plasma membrane. (A) Western blot analysis of anti-EFR3B and control immunoprecipitates from mouse brain homogenates. (B) Western blot analysis of anti-GFP, anti-HA, and anti-FLAG immunoprecipitates from lysates of HeLa cells transfected with GFP-M1-PI4KIIIα, EFR3B-HA, and TTC7B-FLAG. (C) Confocal imaging of live HeLa cells transfected with TTC7B-GFP (top) or EFR3B-tdTomato (bottom). (D) Imaging analysis of HeLa cells triple transfected with GFP-M1-PI4KIIIα, TTC7B-mCherry, and EFR3B-HA. EFR3B-HA immunofluorescence was performed with an anti-HA antibody followed by an Alexa Fluor 647–conjugated anti–rat secondary antibody. (E and F) Imaging analysis of HeLa cells double transfected with EFR3B-HA and either TTC7B-mCherry (E) or GFP-M1-PI4KIIIα (F) after anti-HA immunofluorescence labeling. Insets show the region outlined with dotted lines at higher magnification. Bars: (full size images) 20 µm; (insets) 2 µm.
Individually, TTC7B was localized in the cytosol, whereas EFR3B was found constitutively at the plasma membrane (Fig. 2 C). Coexpression of GFP-M1-PI4KIIIα with EFR3B and TTC7B resulted in significant colocalization of all three proteins at the plasma membrane (Fig. 2 D). Control experiments demonstrated that although EFR3B could recruit TTC7B to the plasma membrane (Fig. 2 E), EFR3B alone was not sufficient to recruit GFP-M1-PI4KIIIα to this membrane (Fig. 2 F). We note that the stable association of a large fraction of the cellular GFP-M1-PI4KIIIα pool with the plasma membrane when it is cooverexpressed with TTC7B and EFR3B stands in contrast the puncta observed by TIRF microscopy in cells expressing only GFP-M1-PI4KIIIα (Fig. 1 E). Thus, most likely, when M1-PI4KIIIα is overexpressed alone, its predominant localization in the cytosol as well as its dynamic behavior at the plasma membrane (Fig. 1 E) are explained by limiting amounts of endogenous EFR3B and TTC7B at the plasma membrane. Consistent with this possibility, siRNA-mediated knockdown of EFR3B (and its close homologue EFR3A) and/or TTC7B (and its close homologue TTC7A) resulted in a striking reduction of the dynamic GFP-M1-PI4KIIIα puncta at the plasma membrane (Fig. 3). Collectively, these experiments demonstrate that PI4KIIIα, like its yeast homologue, is at least partially targeted to the plasma membrane and requires both EFR3 and TTC7 for efficient recruitment to this membrane.
EFR3 is a peripheral membrane protein whose N-terminal palmitoylation is essential for plasma membrane localization
To gain further understanding of the mechanisms that localize PI4KIIIα to the plasma membrane, we investigated the membrane targeting of EFR3, as this protein is a necessary factor for recruiting both TTC7 and PI4KIIIα to this membrane. At the molecular level, EFR3/RBO was proposed to function as an integral membrane lipase (Huang et al., 2004). Our bioinformatic analysis, however, predicted that none of the EFR3/RBO homologues from yeast to humans contains transmembrane regions and that the protein, instead, comprises multiple HEAT (Huntington, elongation factor 3, regulatory subunit A of protein phosphatase 2A, and target of rapamycin) repeats. Further sequence analysis identified a Cys-rich motif at the N terminus of EFR3 that is conserved in all metazoans and could potentially act as a palmitoylation site (Fig. 4 A). Accordingly, a bioorthogonal metabolic labeling approach (Hang et al., 2011) revealed that FLAG-tagged EFR3A and EFR3B were palmitoylated at the N-terminal Cys-rich motif (Fig. 4 B). Importantly, palmitoylation-deficient mutants (C6S/C7S/C8S/C9S for EFR3A and C5S/C7S/C8S for EFR3B) were localized in the cytosol (Fig. 4 C), and the EFR3B palmitoylation-deficient mutant was unable to recruit tagged TTC7B and M1-PI4KIIIα to the plasma membrane (Fig. S2).
Figure 4.
EFR3 is palmitoylated at a conserved N-terminal Cys-rich motif. (A) Alignment of N terminus of EFR3 orthologues in several species reveals a conserved Cys-rich motif. Red, Cys residues; blue, basic residues; green, hydrophobic residues. (B) EFR3A-FLAG and EFR3B-FLAG are palmitoylated at an N-terminal Cys-rich motif. Recombinant EFR3A-FLAG (wild type [WT] or C6S/C7S/C8S/C9S quadruple mutant, denoted C6–9S) or EFR3B-FLAG (wild type or C5S/C7S/C8S triple mutant, denoted C5,7,8S) were immunoprecipitated from lysates of HeLa cells metabolically labeled with alkynyl palmitate (alk-16) or vehicle and transfected with the appropriate EFR3-FLAG construct. The samples were subsequently labeled with biotin-azide using click chemistry, treated with or without hydroxylamine (NH2OH) to cleave thioester linkages, and analyzed by Western blotting, using streptavidin or anti-FLAG antibody. (C) Confocal imaging of live HeLa cells transfected with wild-type EFR3A-tdTomato, C6–9S EFR3A-tdTomato, or C5,7,8S EFR3B-tdTomato. Bars, 20 µm.
These data suggest that EFR3 is a peripheral membrane protein whose posttranslational lipidation enables its localization to membranes and its function in PI4KIIIα-mediated PtdIns4P synthesis at the plasma membrane. Interestingly, the N-terminal Cys-rich motif is absent from yeast Efr3, indicating that, in spite of the evolutionary conservation of the PI4KIIIα–EFR3–TTC7 complex, the mechanism that targets it to the plasma membrane is not conserved between yeast and higher eukaryotes.
Impact of the lack of PI4KIIIα on PtdIns4P and on PtdIns4P- and PtdIns(4,5)P2-dependent processes at the plasma membrane
To assess the precise contribution of PI4KIIIα to the plasma membrane pool of PtdIns4P and more generally the role of this enzyme in cell metabolism and physiology, a gene knockout (KO) approach was used. A conventional KO of mouse PI4KA, which encodes PI4KIIIα, resulted in early embryonic lethality (Fig. S3). Thus, we established a conditional PI4KA KO mouse that also harbored a Cre recombinase-estrogen receptor fusion transgene (Badea et al., 2003). Addition of (Z)-4-hydroxytamoxifen (4-OHT) to primary cultures of mouse embryonic fibroblasts (MEFs) derived from these mice induced gene recombination, leading to complete disappearance of PI4KIIIα within 10 d (Fig. 5 A) and a dramatic depletion of cellular PtdIns4P (to 25% of control cell levels; Figs. 5 B and S4).
Figure 5.
PI4KIIIα and PtdIns4P production. (A) Western blot analysis of the four PI 4-kinase isoforms in lysates from control (Ctrl) and PI4KIIIα KO MEFs. (B and C) Quantification of PtdIns4P and PtdIns(4,5)P2 (shown as a percentage of total inositol phospholipids, n = 3) in control and PI4KIIIα KO MEFs, respectively. *, P < 0.0001. (D) Imaging of the plasma membrane PtdIns4P pool in control and PI4KIIIα KO MEFs using GFP-PHOsh2. (E) Quantification of the plasma membrane/cytosol (PM/cyt) GFP-PHOsh2 fluorescence ratio in D based on line scans (control, n = 29; KO, n = 30; *, P < 0.0001). (F) Imaging of the Golgi complex PtdIns4P pool in control and PI4KIIIα KO MEFs using GFP-GOLPH3. (G) Imaging of Golgi complex morphology in control and PI4KIIIα KO MEFs using anti-GM130 immunofluorescence. Error bars indicate standard deviations. Bars, 20 µm.
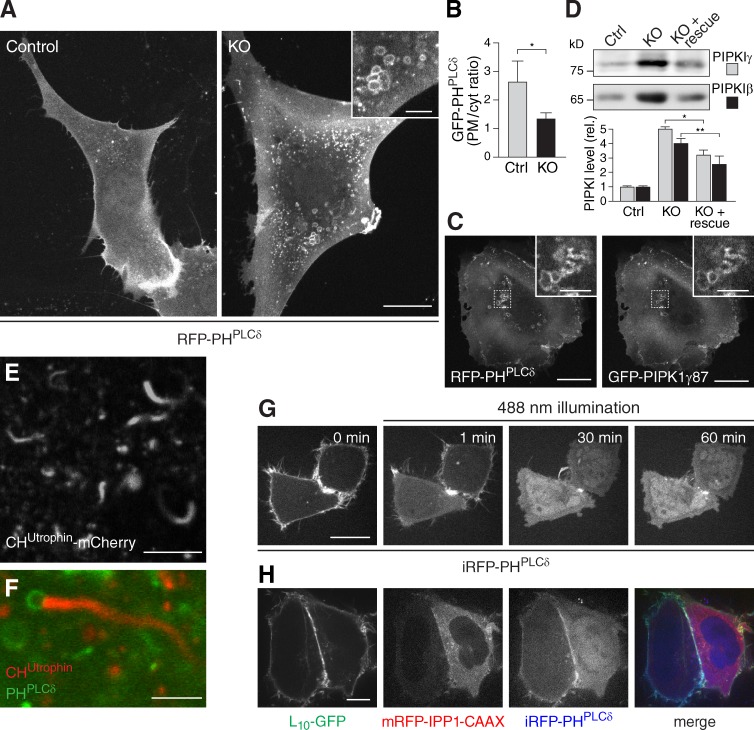
The subcellular distribution of PtdIns4P in KO MEFs was probed via PtdIns4P biosensors that selectively bind to either the plasma membrane or the Golgi pool of this phosphoinositide (GFP-PHOsh2 [Balla et al., 2008] and GFP-GOLPH3 [Wood et al., 2009], respectively). These experiments revealed a selective loss of plasma membrane PtdIns4P in KO MEFs (Fig. 5, D–F). The morphology of the Golgi complex was not affected (Fig. 5, F and G), in contrast to the dramatic changes in Golgi complex structure and function observed in cells lacking PI4KIIIβ or PI4KIIα, two PI 4-kinases known to synthesize PtdIns4P in this region (Wang et al., 2003; Balla et al., 2005; Balla and Balla, 2006). Additionally, no changes were observed in the levels of these other PI 4-kinases as well as of PI4KIIβ, a further confirmation of the nonredundant functions of the PI 4-kinase isoforms (Fig. 5 A).
Consistent with a decrease of plasma membrane PtdIns4P and thus of a predicted decrease also of plasma membrane PtdIns(4,5)P2, a physiological phenomenon known to require the presence of these lipids in this membrane, namely the formation of STIM1-dependent ER–plasma membrane contacts upon thapsigargin-induced depletion of intracellular Ca2+ stores (Korzeniowski et al., 2009; Walsh et al., 2010; Calloway et al., 2011), was impaired (Fig. 6, A–D; and Videos 2 and 3). Rescue experiments to confirm the specificity of this phenotype were successful using GFP-M1-PI4KIIIα but not GFP-M1*-PI4KIIIα (Fig. 6, E–H; and Video 4), demonstrating that only GFP-M1-PI4KIIIα is a fully functional protein.
Figure 6.
Loss of PI4KIIIα causes perturbations of PtdIns4P- and PtdIns(4,5)P2-dependent processes at the plasma membrane. (A–H) Control (A and B) and PI4KIIIα KO MEFs (C–H) were transfected with STIM1-mRFP. For rescue experiments, KO MEFs were transfected with GFP-M1*-PI4KIIIα (E and F) or GFP-M1-PI4KIIIα (G and H). Plasma membrane–associated STIM1-mRFP fluorescence was imaged by TIRF microscopy before (−Tg) or 8 min after (+Tg) the addition of 1 µm thapsigargin (Tg). (I) F-actin in control and PI4KIIIα KO MEFs as revealed by staining with fluorescently labeled phalloidin. (J and K) Altered assembly of the endocytic clathrin machinery in control and PI4KIIIα KO MEFs as assessed by immunofluorescence labeling of AP-2 (J) and epsin 1 (K). Insets show a representative region at higher magnification. Bars: (A–H) 10 µm; (I–K, full size images) 20 µm; (J and K, insets) 5 µm.
Other PtdIns(4,5)P2-mediated processes occurring at the plasma membrane were clearly altered in PI4KIIIα KO MEFs. A drastic disruption of cortical actin, with a major loss of stress fibers, was observed, as assessed by phalloidin staining (Fig. 6 I). The organization of endocytic clathrin coat components was also affected. The normal fine puncta of immunoreactivity for the endocytic clathrin adaptors (and PtdIns(4,5)P2-binding proteins) AP-2 and epsin 1, which represent clathrin-coated pits selectively localized at the plasma membrane, were replaced in PI4KIIIα KO MEFs by more coarse puncta (Fig. 6, J and K), many of which were localized on intracellular structures (also see Fig. 8 A). Further direct assessment of plasma membrane clathrin organization confirmed the abnormal size and distribution of clathrin puncta in PI4KIIIα KO MEFs (Fig. S5 A). Additionally, most of these puncta were stable and did not turn over as in control MEFs, as visualized by TIRF microscopy (Fig. S5 B).
Figure 8.
Loss of plasma membrane identity in PI4KIIIα KO MEFs. Proteins typically restricted to the plasma membrane are found on intracellular PtdIns(4,5)P2-positive vesicles, which are negative for a variety of endosomal and Golgi markers. (A) PtdIns(4,5)P2-positive intracellular vesicles, visualized with GFP- or RFP-PHPLCδ, colocalize with endocytic clathrin machinery components (endogenous AP-2, epsin 1, and dynamin 2–GFP) but not with markers of endosomes (endogenous EEA1, GFP-Rab5, and GFP-Rab9), the endosomal phosphoinositide PtdIns3P (GFP-2×FYVEHrs), and Golgi-localized PtdIns4P (GFP-PHOSBP). For clarity, PHPLCδ is false colored green, and the second marker is false colored red in all images. (B) Mislocalization of plasma membrane proteins in cells lacking PI4KIIIα. PI4KIIIα KO MEFs were transfected with M1R-YFP, L10-GFP, or EFR3B-GFP and imaged by confocal microscopy. Note the accumulation of all markers in intracellular vesicles highlighted in the inset images. (C) Measurement of total and plasma membrane fraction of free cholesterol in control and PI4KIIIα KO MEFs. Total free cholesterol levels were measured by HPLC, after lipid extraction, and normalized to total cellular phospholipid levels. *, P < 0.0001 (n = 12 for control and KO). Plasma membrane free cholesterol was measured by treatment of cells with 10 mM MβCD for 1 min followed by quantification of extracted cholesterol using an enzyme-coupled assay and normalization to total protein content. Error bars indicate standard deviations. *, P < 0.02 (n = 6 for control and KO). Bars: (A and B, insets) 5 µm; (B, full size images) 20 µm.
Ectopic PtdIns(4,5)P2 synthesis and localization in PI4KIIIα KO cells
Expression of RFP-PHPLCδ, a biosensor for PtdIns(4,5)P2, revealed a robust reduction of PtdIns(4,5)P2 levels at the plasma membrane as expected (Fig. 7, A and B). Most surprisingly, however, global levels of cellular PtdIns(4,5)P2, as assessed biochemically, were only moderately reduced in KO MEFs (to 80% of control cell levels; Fig. 5 C). A potential explanation for this modest change, which contrasts with the much stronger reduction in global PtdIns4P levels (to 25% of control; Figs. 5 B and S4), in spite of the loss of PtdIns(4,5)P2 at the plasma membrane, came from a further analysis of the localization of PtdIns(4,5)P2 and of the expression levels and localization of PIPKIs (the enzymes that generate PtdIns(4,5)P2 from PtdIns4P; Doughman et al., 2003) in PI4KIIIα KO cells. Although both PtdIns(4,5)P2, as detected by RFP-PHPLCδ, and exogenously expressed PIPKIs are typically segregated at the plasma membrane (Fairn et al., 2009), numerous vesicles that were highly heterogeneous in size and positive for RFP-PHPLCδ and for GFP-PIPKIγ were detected intracellularly in KO cells (Fig. 7, A [inset] and C). Furthermore, PIPKIs (PIPKIβ and PIPKIγ) were found to be massively up-regulated (Fig. 7 D), and numerous actin comets were observed in the cytoplasm (Fig. 7 E and Video 5), a known effect of the ectopic accumulation of PtdIns(4,5)P2 on intracellular organelles (Rozelle et al., 2000). Accordingly, PtdIns(4,5)P2-positive vesicles were detected at the tip of comets (Fig. 7 F). Thus, loss of PI4KIIIα in MEFs leads to initiation of homeostatic mechanisms by which cells attempt to restore PtdIns(4,5)P2 levels through a more efficient conversion of PtdIns4P to PtdIns(4,5)P2 in KO MEFs, but much of the PtdIns(4,5)P2 generated under such conditions is ectopically localized.
Figure 7.
Ectopic PtdIns(4,5)P2 localization and up-regulation of PIPKIs in PI4KIIIα KO MEFs. (A) Imaging of PtdIns(4,5)P2 distribution in control and PI4KIIIα KO MEFs using RFP-PHPLCδ. (inset) Note the presence of PtdIns(4,5)P2-positive vesicles only in KO MEFs. (B) Quantification of the plasma membrane/cytosol (PM/cyt) GFP-PHPLCδ fluorescence ratio in control and PI4KIIIα KO MEFs based on line scans (control, n = 22; KO, n = 23; *, P < 0.0001). (C) PIPKIγ and PtdIns(4,5)P2 colocalize on intracellular vesicles in cells lacking PI4KIIIα. PI4KIIIα KO MEFs were transfected with RFP-PHPLCδ and GFP-PIPKIγ87 and imaged by confocal microscopy. Insets show the region outlined with dotted lines at higher magnification. (D) Western blot analysis of PIPKIβ and PIPKIγ levels in control MEFs, PI4KIIIα KO MEFs, and PI4KIIIα KO MEFs transfected with GFP-M1-PI4KIIIα (KO + rescue). *, P < 0.0002 (n = 3); **, P < 0.003 (n = 3). rel., relative. (E) Actin comet tails in PI4KIIIα KO MEFs visualized by CHUtrophin-mCherry (a marker of F-actin). (F) Actin comet tail in PI4KIIIα KO MEFs double transfected with CHUtrophin-mCherry and GFP-PHPLCδ. Note the presence of a PtdIns(4,5)P2-positive vesicle at the head of the comet. (G) Acute, 488-nm light-mediated depletion of plasma membrane PtdIns(4,5)P2 from HeLa cells (see Materials and methods for details) results in diffuse intracellular fluorescence of the PtdIns(4,5)P2 probe iRFP-PHPLCδ but no detectable PtdIns(4,5)P2-positive intracellular vesicles. (H) Chronic depletion of plasma membrane PtdIns(4,5)P2 from HeLa cells by transfection with a membrane-anchored inositol 5-phosphatase (mRFP-IPP1-CAAX; see cell on the right) results in diffuse intracellular fluorescence of the PtdIns(4,5)P2 probe iRFP-PHPLCδ, but no PtdIns(4,5)P2-positive intracellular vesicles are apparent 16 h after transfection. Note that the myristoylated/palmitoylated plasma membrane marker L10-GFP is retained at the plasma membrane. Ctrl, control. Error bars indicate standard deviations. Bars: (A [inset], C [inset], E, and F) 5 µm; (H) 10 µm; (A [full size], C [full size], and G) 20 µm.
To test whether the mere loss of plasma membrane PtdIns(4,5)P2 in wild-type cells could cause a similar ectopic intracellular PtdIns(4,5)P2 phenotype, we acutely depleted this lipid from the plasma membrane of HeLa cells using a recently developed optogenetic approach (Idevall-Hagren et al., 2012). Although light-mediated recruitment of an inositol 5-phosphatase to the plasma membrane resulted in a rapid and massive loss of PtdIns(4,5)P2 in this membrane, as revealed by shedding of infrared RFP (iRFP)-PHPLCδ, no formation of intracellular PHPLCδ-positive vesicles was observed, even when the illumination was continually applied over a 1-h period (Fig. 7 G). To chronically prevent PtdIns(4,5)P2 accumulation at the plasma membrane, we expressed an inositol 5-phosphatase construct with a C-terminal CAAX motif that ensures constitutive plasma membrane targeting (Milosevic et al., 2005). Even 16 h after transfection, no PtdIns(4,5)P2-positive intracellular vesicles similar to those found in PI4KIIIα KO MEFs were observed (Fig. 7 H), suggesting that more complex homeostatic mechanisms are involved in their formation.
Loss of plasma membrane identity in PI4KIIIα KO cells
Further characterization of the PtdIns(4,5)P2-positive intracellular vesicles in PI4KIIIα KO MEFs revealed that they were positive for many proteins typically restricted to the plasma membrane. For example, they were decorated by AP-2–, epsin-, and dynamin-positive puncta (Fig. 8 A). Dynamin was also localized on actin comet tails (Video 6), as previously shown in cells overexpressing PIPKI (Lee and De Camilli, 2002). In contrast, the PtdIns(4,5)P2-positive vesicles did not colocalize with early or late endosomal markers (EEA1, Rab5, Rab9, and the PtdIns3P biosensor FYVEHrs) or a Golgi/PtdIns4P marker (PHOSBP; Fig. 8 A). Additionally, integral and peripheral membrane proteins normally selectively segregated at the plasma membrane, including fluorescently tagged M1 muscarinic receptor (M1R-YFP; Wess et al., 2007) and myristoylated/palmitoylated Lck membrane anchor (L10-GFP; Fig. 7 H; Rodgers, 2002), were found at these intracellular sites (Fig. 8 B). Likewise, wild-type EFR3B partially relocalized to the intracellular structures positive for PtdIns(4,5)P2 (Fig. 8 B), suggesting a reciprocal requirement of both EFR3 and PI4KIIIα for correct targeting of the PI4KIIIα–EFR3–TTC7 complex to the plasma membrane. Additionally, plasma membrane levels of cholesterol, a lipid normally concentrated in this membrane, were ∼20% lower in PI4KIIIα KO MEFs relative to controls, whereas intracellular levels were increased (Fig. 8 C). Thus, PI4KIIIα appears to be a critical determinant of the lipidomic and proteomic identity of the plasma membrane.
Discussion
This study provides answers to long-standing questions concerning the generation of PtdIns4P at the plasma membrane. It has long been known that an important pool of PtdIns4P exists in this membrane, and studies in both yeast and higher eukaryotes had implicated Stt4/PI4KIIIα in its generation (Audhya et al., 2000; Balla et al., 2005). However, the localization in cells of higher eukaryotes of this enzyme, which is expected to be of fundamental importance in cell physiology because its product is an essential precursor of key regulatory messenger molecules, had remained highly controversial (Balla and Balla, 2006). The prevailing view was that it was predominantly localized in the ER (e.g., Wong et al., 1997; Balla and Balla, 2006; Altan-Bonnet and Balla, 2012; Bianco et al., 2012), and no evidence has been reported for its targeting to the plasma membrane.
Because of lack of suitable antibodies for the detection of the endogenous protein by immunocytochemistry (despite repeated attempts), the subcellular targeting of PI4KIIIα can only be explored by the expression of tagged PI4KIIIα. Here, we show that the failure of a previous study to reveal an association of PI4KIIIα with the plasma membrane is explained by the incorrect identification of the N-terminal sequence of this enzyme (Balla and Balla, 2006). We demonstrate that EFR3/RBO and TTC7, the mammalian homologues of yeast Efr3 and Ypp1, play a key role in the recruitment of PI4KIIIα to the plasma membrane. As in yeast, EFR3 is constitutively localized at the plasma membrane, but it is not capable of recruiting PI4KIIIα in the absence of Ypp1/TTC7. TTC7 is a soluble protein that requires interaction with EFR3 for membrane recruitment and likely acts as a bridge between PI4KIIIα and EFR3.
In yeast, in which fluorescently tagged proteins were expressed under the regulation of the endogenous promoter (and thus at physiological levels), Stt4 was shown to be stably associated with the plasma membrane at “PIK patches.” In our experiments, the recruitment of transfected GFP-tagged PI4KIIIα to the plasma membrane was highly dynamic unless its binding partners EFR3B and TTC7B were also overexpressed, possibly reflecting its stoichiometric excess relative to EFR3 and TTC7 when expressed alone. Whether endogenous PI4KIIIα, EFR3, and TTC7 form stable patches in mammalian cells remains an open question.
A large fraction of the ER is constitutively closely apposed to the plasma membrane in yeast, and these contact sites play an important role in PtdIns4P metabolism, as the ER-localized PtdIns4P phosphatase Sac1 is thought to act in trans to dephosphorylate PtdIns4P in the plasma membrane (Stefan et al., 2011). Although the relationship between yeast PIK patches and ER–plasma membrane contact sites remains unclear (Baird et al., 2008), it is of interest that Stt4 contains an amino acid sequence matching the so-called FFAT motif, which enables binding of cytosolic proteins to the ER-resident proteins Scs2/Scs22. This motif is partially conserved in PI4KIIIα and thus could mediate the association of a small PI4KIIIα pool with the ER via the mammalian Scs2/Scs22 homologue VAP-B (Kaiser et al., 2005), although contacts between the cortical ER and the plasma membrane are less abundant and smaller in mammalian cells compared with in yeast. No evidence for a major association of PI4KIIIα with the ER was obtained in our study.
Importantly, our results shed new light on EFR3/RBO. They demonstrate that EFR3 is not a transmembrane lipase as originally proposed by a study in Drosophila (Huang et al., 2004). First, improved structural predictions do not support the presence of transmembrane regions or of a lipase-type fold. Instead, our bioinformatic analysis predicts that EFR3 is a cytosolic protein consisting entirely of α-helical HEAT repeats. Second, the protein is palmitoylated at a conserved Cys-rich motif at its N terminus, and mutation of the Cys residues in this motif abolishes the plasma membrane targeting of EFR3. Interestingly, the palmitoylation motif is conserved in all metazoan EFR3 homologues but is absent from yeast Efr3, demonstrating a conservation of function in spite of potentially divergent membrane-targeting mechanisms. In view of this revised function of EFR3/RBO, it will be of interest to reassess and further investigate the mechanisms of the temperature-sensitive paralytic phenotype in Drosophila rbo mutants.
The association of PI4KIIIα with the plasma membrane that we show here and the results of our gene KO experiments provide conclusive evidence for a role of PI4KIIIα as a key contributor to plasma membrane PtdIns4P, as previously suggested by pharmacological and knockdown studies (Balla et al., 2005, 2008). Interestingly, in the absence of PI4KIIIα, cells very strongly up-regulate the expression of PIPKIs, which convert PtdIns4P to PtdIns(4,5)P2, so that PtdIns(4,5)P2 levels are much less drastically decreased than PtdIns4P levels. The intracellular signaling pathways underlying this compensatory change remain to be determined and represent an important question to be addressed by future work. Additionally, PtdIns(4,5)P2, which is normally primarily concentrated at the plasma membrane, was surprisingly mislocalized on intracellular structures in PI4KIIIα KO MEFs, leading to a partial relocation onto these vesicles of actin regulatory proteins and endocytic proteins normally restricted to the plasma membrane. Because binding to PtdIns4P contributes to the membrane targeting of PIPKIs (Kunz et al., 2002), PtdIns4P generated on endosomes by type II PI 4-kinases (Balla and Balla, 2006) may play a role in the ectopic localization of the PIPKIs and may function as a substrate for PtdIns(4,5)P2 generation. Thus, it remains possible that the very robust reduction in total PtdIns4P levels observed in PI4KIIIα KO MEFs may be caused not only by lack of PtdIns4P production at the plasma membrane but also by increased consumption of the PtdIns4P pool generated by type II PI 4-kinases.
Importantly and strikingly, the selective localization of specific intrinsic membrane proteins of the plasma membrane is disrupted by lack of PI4KIIIα. In PI4KIIIα KO MEFs, proteins normally segregated primarily in this membrane, including the M1 muscarinic receptor, a prototypical transmembrane protein (Wess et al., 2007), and the myristoylated/palmitoylated N-terminal anchor of Lck (Rodgers, 2002), were also found and enriched on PtdIns(4,5)P2-positive intracellular structures, which adopted a plasma membrane–like identity. Even EFR3, whose plasma membrane targeting is mediated by palmitoylation and thus at least partially via interactions with plasma membrane lipids (potentially cholesterol-enriched membrane rafts; Foster et al., 2003), localizes to such intracellular sites in PI4KIIIα KO MEFs, indicating that the unique protein composition of the plasma membrane relative to other membranes is perturbed. Effects produced by the deficiency of plasma membrane PtdIns4P on other lipids in this membrane, notably cholesterol, may contribute to these changes. Thus, impairing the production of PtdIns4P at the plasma membrane drastically affects the proteomic and lipidomic composition of the plasma membrane, pointing to PI4KIIIα as a key upstream factor in determining the “identity” of this membrane. We note that an independent study published just after our manuscript was submitted also reported that generation of PtdIns4P at the plasma membrane is a critical determinant of the identity of this membrane and that loss of plasma membrane PtdIns4P is not matched by a corresponding reduction in PtdIns(4,5)P2 levels (some discrepancies between the two studies may depend upon the different experimental models; Hammond et al., 2012).
In conclusion, our study demonstrates that, in contrast to previous studies, at least a pool of PI4KIIIα is targeted to the plasma membrane. Furthermore, we provide mechanistic insight concerning the recruitment of PI4KIIIα to this membrane and show that the PtdIns4P pool produced by this enzyme plays essential roles not only as a precursor to a variety of key regulatory metabolites but also, either directly or via its downstream metabolites, as a critical determinant of plasma membrane homeostasis and identity.
Materials and methods
Plasmids
The sources of cDNAs were as follows: STIM1-mRFP obtained from B. Baird (Cornell University, Ithaca, NY), GFP-PHOsh2 obtained from T. Balla (National Institutes of Health, Bethesda, MD), GFP-GOLPH3 obtained from C. Burd (Yale University, New Haven, CT), CHUtrophin-mCherry obtained from W. Bement (University of Wisconsin-Madison, Madison, WI), GFP-PHPLCδ obtained from A. De Matteis (Telethon Institute of Genetics and Medicine, Naples, Italy), RFP-PHPLCδ obtained from S. Grinstein (University of Toronto, Toronto, Ontario, Canada), M1R-YFP obtained from B. Hille (University of Washington, Seattle, WA), clathrin light chain-GFP obtained from J. Keen (Thomas Jefferson University, Philadelphia, PA), dynamin 2–GFP obtained from M. McNiven (Mayo Clinic, Rochester, MN), GFP-Rab9 obtained from S. Pfeffer (Stanford University, Stanford, CA), GFP-2×FYVEHrs obtained from H. Stenmark (Oslo University, Oslo, Norway), and L10-GFP, GFP-PIPK1γ87, iRFP-PHPLCδ, CIBN-CAAX, mCherry-CRY2-5PtaseOCRL, mRFP-IPP1-CAAX, GFP-Rab5, and GFP-PHOSBP obtained from the De Camilli Laboratory.
M1*-PI4KIIIα and M1-PI4KIIIα were amplified by PCR from the following cDNA clones, respectively: NM_058004.1 (GenBank accession number; OriGene) and AB210002 (GenBank accession number; Kazusa DNA Research Institute) using the following primer pairs: M1*-PI4KIIIα, 5′-GCACAGAATTCGCCATGTGTCCAGTGGATTTCCATG-3′ and 5′-GCACACTCGAGTCAGTAGGGGATGTCATTCTGA-3′; and M1-PI4KIIIα, 5′-GCACAGAATTCATGGCGGCGGCCCCGGC-3′ and 5′-GCACACTCGAGTCAGTAGGGGATGTCATTCTGA-3′. The PCR amplicons were cloned into pcDNA3.0 (Invitrogen) using EcoRI and XhoI cut sites. GFP-tagged constructs were generated by amplification from the appropriate original PI4KIIIα-containing vector and cloning into the pEGFP-C2 vector (Takara Bio Inc.) for GFP-M1-PI4KIIIα and the pEGFP-C1 vector (Takara Bio Inc.) for GFP-M1*-PI4KIIIα. We note that, during the course of our study, an NCBI revision of human PI4KA (available from GenBank under accession no. NM_058004.3) expanded the predicted ORF to include the sequence in between M1* and M1. EFR3B was amplified from mouse brain cDNA using the following primers: 5′-GCTCGAGGCCACCATGTACGGTGTGTGTGGCTGC-3′ and 5′-CGAATTCGGTATACACACAGATCAGGAAACTTCATC-3′. The PCR amplicon was digested with XhoI and EcoRI and cloned into the pEGFP-N1 vector (Takara Bio Inc.). EFR3B was subsequently subcloned into the ptdTomato-N1 vector (Takara Bio Inc.) using XhoI and EcoRI restriction sites to generate the EFR3B-tdTomato construct and into the pcDNA3.0 vector (Invitrogen) with C-terminal FLAG (DYKDDDDKI) or HA (YPYDVPDYA) tags incorporated into the antisense primers, using EcoRI and NotI restriction sites, to generate the EFR3B-FLAG and EFR3B-HA constructs. Mouse EFR3A (GenBank accession no. BC007482) was obtained from Thermo Fisher Scientific and cloned into the ptdTomato-N1 vector (Takara Bio Inc.) using XhoI and EcoRI restriction sites to obtain the EFR3A-tdTomato construct and into the pCDNA3.0 vector (Invitrogen) with a C-terminal FLAG (DYKDDDDKI) tag incorporated into the antisense primers, using EcoRI and NotI restriction sites, to generate the EFR3A-FLAG construct. The C6–9S mutants of EFR3A-FLAG and EFR3A-tdTomato and the C5,7,8S mutants of EFR3B-FLAG and EFR3B-tdTomato were generated using a mutagenesis kit (QuikChange II XL; Agilent Technologies). Human TTC7B (GenBank accession no. BC148529) was obtained from Thermo Fisher Scientific and cloned into the pEGFP-N1 vector using XhoI and SalI restriction sites. TTC7B was subsequently subcloned into the pmCherry-N1 vector (Takara Bio Inc.) using a single digestion with XhoI (proper orientation was assessed using a test digestion with BamHI) and into the pcDNA3.0 vector (Invitrogen) with a C-terminal FLAG (DYKDDDDKI) tag incorporated into the antisense primers, using HindIII and XbaI restriction sites, to generate the TTC7B-FLAG construct.
Transfection and other reagents
MEFs were electroporated using the Amaxa Nucleofector method (Lonza), and HeLa and COS-7 cells were transfected with plasmids using transfection reagent (FuGENE HD; Promega) and with siRNA duplexes (obtained from Integrated DNA Technologies) using Lipofectamine RNAiMAX (Invitrogen).
Antibodies
Antibodies against mouse PI4KIIIα and EFR3B were generated by immunization of rabbits with the following constructs: PI4KIIIα, GST fusion to residues 1,021–1,207 (based on translation initiation at M1), and EFR3B, a keyhole limpet hemocyanin conjugate of the peptide Ac-TDEDRLSKRKSIGETISLQVC-NH2 (Cocalico Biologicals, Inc.). Sera were affinity purified using the antigenic peptide immobilized on SulfoLink resin following the manufacturer’s instructions (Thermo Fisher Scientific). A full list of antibodies used in this study appears in Table S1. All Western blots were developed by chemiluminescence using the SuperSignal West Pico, Femto, or Dura reagents (Thermo Fisher Scientific) unless otherwise noted. Sources of all antibodies used in this study are provided in Table S1.
Ribosome profiling to determine PI4KIIIα translation start sites
HEK 293 cells were treated for 90 s with 2 µg/ml harringtonine (LKT Laboratories) to specifically immobilize initiating ribosomes while permitting runoff elongation. Cells were washed once in PBS and harvested in lysis buffer consisting of ribosome buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 100 µg/ml cycloheximide) with 0.5% Triton X-100. Cells were incubated for 10 min on ice and triturated, and the lysate was clarified by centrifugation. The clarified lysate was treated with 2.5 U/µL RNase I (Ambion) for 45 min at room temperature, and ribosomes were harvested by ultracentrifugation (265,000 g for 4 h) through a 1-M sucrose cushion. RNA was extracted directly from ribosomal pellets and used to generate a deep sequencing library as previously described (Ingolia et al., 2011). Footprint sequencing libraries were analyzed on a sequencing system (HiSeq 2000; Illumina), and sequences were mapped to the human genome (hg19) using the TopHat short read aligner. Ribosome peptidyl site positions were identified as the location 12 nt (for 28- and 29-nt footprints) or 13 nt (for 30- and 31-nt footprints) downstream of the 5′ end of the footprint (Ingolia et al., 2011).
TIRF microscopy
TIRF microscopy was performed at 37°C using an objective-type inverted microscope (IX-70; Olympus) fitted with a 60×, NA 1.45 TIRF microscopy lens (Olympus) and controlled by iQ software (Andor Technology). Laser lines (488 and 568 nm) from argon and argon/krypton lasers (CVI Melles Griot) were coupled to the TIRF microscopy condenser through a single optical fiber. The calculated evanescent field depth was ∼100 nm. Cells were typically imaged without binning and with 0.2–0.5-s exposures and detected with a back-illuminated electron-multiplying charge-coupled device camera (512 × 512 pixels; 16 bit; iXon 887; Andor Technology).
Spinning-disc confocal microscopy
All imaging experiments with the exception of TIRF microscopy were performed on a spinning-disc confocal microscope, using the UltraVIEW VoX system (PerkinElmer) including an inverted microscope (Ti-E Eclipse; Nikon) equipped with Perfect Focus, temperature-controlled stage, 14-bit electron-multiplying charge-coupled device camera (C9100-50; Hamamatsu Photonics), and spinning disc-confocal scan head (CSU-X1; Yokogawa Corporation of America) controlled by Volocity software (PerkinElmer). All images were acquired through a 60 or 100× oil objective (1.4 NA, CFI Plan Apochromat VC). Green fluorescence was excited with a 488-nm/50-mW diode laser (Coherent) and collected by a band pass (BP) 527/55-nm filter. Red fluorescence was excited with a 561-nm/50-mW diode laser (Cobolt) and collected by a BP 615/70-nm filter. Near-infrared fluorescence was excited with a 640-nm/50-mW diode laser (CVI Melles Griot) and collected by a BP 705/90-nm filter. Multicolor images were acquired sequentially.
IP experiments
For mouse brain IP experiments, mouse brain homogenates were generated by ∼50 pulses in a Dounce homogenizer of ∼20 frozen mouse brains thawed into IP lysis buffer (150 mM NaCl, 20 mM Tris, 1 mM EDTA, and 1% NP-40, pH 7.4, supplemented with an EDTA-free inhibitor cocktail tablet [cOmplete; Roche]). The suspension was centrifuged at 16,000 g for 30 min. The supernatant was subjected to IP as described in this paragraph. For IP experiments from transfected HeLa cells, HeLa cells were harvested 24 h after transfection with the appropriate combinations of GFP-M1-PI4KIIIα, TTC7B-FLAG, and EFR3B-HA, resuspended in IP lysis buffer, sonicated briefly, and centrifuged for 10 min at 16,000 g. The supernatant was immunoprecipitated in IP lysis buffer by addition of the appropriate primary antibody (or no primary antibody as a negative control) and rocking for 1 h at 4°C followed by the addition of 20 µl protein G–conjugated Sepharose (GE Healthcare) and an additional 1-h incubation at 4°C, rocking. The resin was then isolated by centrifugation at 1,000 g, rinsed three times with IP lysis buffer, and analyzed either by SDS-PAGE or Western blotting. In the case of the PI4KIIIα IP, candidate bands were excised from the Coomassie-stained gel, and protein identification by liquid chromatography–mass spectrometry was performed at the Keck Research Facility at Yale University.
Biochemical analysis of EFR3 palmitoylation
HeLa cells were transfected with the appropriate EFR3-FLAG construct using FuGENE HD, and palmitoylated proteins were labeled using the protocol of Yount et al. (2011). 5 h after transfection, the media were exchanged for media made with dialyzed serum that was supplemented with 10 µM alk-16 (17-octadecynoic acid; Cayman Chemicals), and the cells were incubated for an additional 18 h. The cells were then rinsed with PBS. Lysis, IP with M2 anti-FLAG–conjugated agarose (Sigma-Aldrich), and rinses were performed using alk-16 IP buffer (150 mM NaCl, 50 mM triethanolamine, and 1% Triton X-100, pH 7.4, supplemented with an EDTA-free protease inhibitor cocktail tablet [cOmplete]). After the rinses, the supernatant was aspirated, and biotin-azide (a gift from M. Boyce and C. Bertozzi, University of California, Berkeley, Berkeley, CA) was conjugated to the resin-bound EFR3-FLAG using copper-catalyzed click chemistry. Where indicated, an aliquot of this solution was incubated with 2.5% neutral hydroxylamine (NH2OH). The samples were then boiled and analyzed by Western blotting, detecting with an infrared imager (Odyssey; LI-COR Biosciences).
Generation of PI4KIIIα conditional KO mice
The PI4KA conditional targeting strategy flanked exons 48–52 with loxP sites and inserted a neomycin resistance gene flanked by flippase recognition target sites into intron 47 (Fig. S3). The targeting vector (Gene Dynamics) was electroporated into hybrid C57BL/6J-129S1/Sv mouse embryonic stem cells (Yale Cancer Center Animal Genomics Shared Resource), and positive clones were identified by Southern blotting (Gene Dynamics; Fig. S3). Targeted embryonic stem cells were injected into blastocysts of C57BL/6J mice and transferred to the uteri of pseudopregnant recipient females. Chimeric male offspring were subsequently crossed to a enhanced flippase recombinase-expressing deleter strain (B6.Cg-Tg(ACTFLPe)9205Dym/J; The Jackson Laboratory) to remove the neomycin resistance gene. Obtained conditional mutant mice were then crossed with either a β-actin-Cre mouse (FVB/N-Tg(ACTB-cre)2Mrt/J; The Jackson Laboratory) to delete the PI4KA gene ubiquitously or a strain expressing an Esr1-Cre fusion protein (B6;129-Gt(ROSA)26Sortm1(cre/ERT)Nat/J; The Jackson Laboratory) to generate mutants in which PI4KA gene ablation could be induced pharmacologically by the addition of 4-OHT. Animal care and use was performed in accordance with our institutional guidelines. Genotyping of the mutant mice was performed by PCR using the following primers: wild-type allele, 5′-CCGCCAGACTGACTTTGGCATGTATG-3′ and 5′-CCAGCATGATGTTGCCATTGTGCCTG-3′; KO allele, 5′-GGATGCTGTGCAGCACCACTCCCTC-3′ and 5′-GGCCTGTGTCCAACATGAGTGTAACCAG-3′; and floxed allele, 5′-GGCAGAGGCAGGTGAATCTCTGAGTTCAAG-3′ and 5′-GCAAGGGAGCATCAGGCTCTGGC-3′.
Generation of PI4KIIIα conditional KO MEFs
Primary MEFs were generated from embryonic day 15 to postnatal day 1 from the appropriate strains. Some populations of MEFs were immortalized using the standard NIH 3T3 protocol (Ferguson et al., 2009). To generate cells lacking PI4KIIIα, recombination was induced by treatment of cells with 0.3–1.5 µM 4-OHT for 24–36 h followed by replacement of the media and subsequent growth for a total period of 7–10 d. Cells were grown in DMEM containing glutamine, 10% fetal bovine serum, and 1% penicillin/streptomycin supplement. In this study, three different sets of control and KO cells were used interchangeably, as indicated in Table S2, and no differences were observed between sets of control/KO pairs in all assays in which multiple sets were tested.
Quantification of phosphoinositide levels
For the quantification of PtdIns4P and PtdIns(4,5)P2 shown in Fig. 5, control and PI4KIIIα KO MEFs were metabolically labeled with [3H]myo-inositol for days 7–10 after treatment with 4-OHT (in the case of PI4KIIIα KO MEFs). Cells were lysed in 4.5% perchloric acid, and the pellet was rinsed three times with 0.1 M EDTA. The pellet was deacylated with a mixture of methylamine/water/n-butanol/methanol (36:8:9:47) for 1 h at 50°C and dried in a SpeedVac (SVC 100; Savant), and the residue was extracted using 500 µl of a mixture of n-butanol/petroleum ether/ethyl formate (20:40:1) and 500 µl of water. The aqueous phase was resolved using anion-exchange HPLC with an ammonium phosphate gradient (LC-20AT ultra-fast liquid chromatography [Shimadzu] equipped with a Partisil 5 SAX 4.6 × 125–mm column [Whatman]) and identified using a radiometric detector (β-RAM 4B; LabLogic). The identity of each peak was determined by comparison with known compounds, and the values reported in Fig. 5 are shown as the amount of the desired lipid as a percentage of total detectable inositol phospholipids (PtdIns, PtdIns4P, and PtdIns(4,5)P2). For the time course of phosphoinositide measurements shown in Fig. S4, total anionic lipids were isolated by lysis in 1:1 methanol/aqueous 1 M HCl and 2 mM AlCl3 followed by addition of chloroform, extraction of the organic layer with 1:1 methanol/2 mM oxalic acid, and evaporation of the solvent. Deacylation was performed as described previously in this paragraph, and the resultant glycerol-inositol phosphate peaks were analyzed by an HPLC system (Dionex; Thermo Fisher Scientific) equipped with a conductivity detector (Nakatsu et al., 2010).
Acute depletion of PtdIns(4,5)P2 using an optogenetic approach
PtdIns(4,5)P2 was depleted from HeLa cells essentially as recently described (Idevall-Hagren et al., 2012). In brief, HeLa cells were transfected with CIBN-CAAX, mCherry-CRY2-5PtaseOCRL, and iRFP-PHPLCδ and kept in the dark before the experiment. PtdIns(4,5)P2 depletion was initiated by illumination with 488-nm light after a baseline image of the mCherry and iRFP channels was acquired. Constant PtdIns(4,5)P2 depletion was ensured by repeated 200-ms illuminations with 488-nm light at 1-min intervals for 1 h, whereas mCherry and iRFP images were acquired at 15-min intervals to minimize photobleaching.
Chronic depletion of PtdIns(4,5)P2
Chronic depletion of PtdIns(4,5)P2 was achieved by transfection of HeLa cells with a constitutively membrane-anchored 5-phosphatase (CAAX-tagged catalytic domain of synaptojanin 1; Milosevic et al., 2005), iRFP-PHPLCδ to monitor PtdIns(4,5)P2, and the myristoylated/palmitoylated anchor L10-GFP to monitor the plasma membrane. Images were acquired 16 h after transfection by confocal microscopy.
Cholesterol measurements
Total free cholesterol was analyzed as described previously (Shui et al., 2011). Only HPLC-grade solvents were used, and the deuterated cholesterol standard (cholesterol-26,26,26,27,27,27-d6) was obtained from C/D/N Isotopes Inc. In brief, total cellular lipid extracts were dissolved in a 1:1 mixture of chloroform/methanol, spiked with 2.5 µg/ml cholesterol-d6, and analyzed using an HPLC system (1100 Series; Agilent Technologies) coupled to a mass spectrometer (3200 QTRAP; Applied Biosystems). The multiple reaction monitoring transitions for endogenous cholesterol (369.4/161.0) and cholesterol-d6 (375.4/161.0) were monitored in positive atmospheric pressure chemical ionization mode. Measured cholesterol levels were first normalized to the cholesterol-d6 standard and finally presented relative to the measured phospholipid concentration to account for sample-to-sample variation. The values shown in Fig. 8 C represent the mean free cholesterol shown as a percentage of control cell levels. Error bars represent the standard deviation (n = 12 each for control and KO). Plasma membrane cholesterol levels were quantified by brief exposure of cells to methyl-β-cyclodextrin (MβCD; Sigma-Aldrich) followed by measurement of the amount of extracted cholesterol using an enzyme-coupled assay, normalizing to total protein content determined after cell lysis (Zidovetzki and Levitan, 2007). In brief, control and PI4KIIIα KO MEFs were rinsed three times with PBS and incubated with 10 mM MβCD (in PBS) at room temperature for 1 min. The solution was then collected, and the concentration of free cholesterol was determined using the enzyme-coupled assay kit (Amplex Red; Invitrogen) according to the manufacturer’s instructions, with fluorescence detection on a fluorescence microplate reader (Infinite M1000; Tecan). The cells were lysed in lysis buffer (150 mM NaCl, 20 mM Tris, 1% Triton X-100, and 1 mM EDTA, pH 7.4, supplemented with an EDTA-free protease inhibitor cocktail tablet [cOmplete]), and protein concentration was determined using the Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific). The relative cholesterol concentration for each sample was normalized to protein concentration (n = 6 each for control and KO).
Image analysis
Image analysis was performed using Volocity and ImageJ (National Institutes of Health). For the quantification of GFP-PHOsh2 and GFP-PHPLCδ, a ratio of plasma membrane to cytosolic signal (derived from a line scan) was computed for each cell and averaged (n = 29 cells for control; n = 30 cells for KO for GFP-PHOsh2 and n = 22 cells for control; n = 23 cells for KO for GFP-PHPLCδ). For quantification of GFP-M1-PI4KIIIα plasma membrane puncta (Fig. 3), samples were blinded, a random 100 × 100–pixel area was cropped, and puncta were counted manually (n = 4–6 cells per condition).
Statistical analysis
In all cases throughout the paper, statistical significance was calculated using an unpaired two-tailed Student’s t test with unequal variance, and error bars represent standard deviation.
Online supplemental material
Fig. S1 shows mass spectrometry data supporting the interaction of EFR3B and TTC7B with PI4KIIIα. Fig. S2 shows that palmitoylation of EFR3B is required for recruitment of TTC7B and PI4KIIIα to the plasma membrane. Fig. S3 depicts the strategy for generation of a PI4KIIIα KO mouse. Fig. S4 shows a time course of PtdInsP and PtdInsP2 levels in PI4KIIIα KO MEFs after gene recombination. Fig. S5 shows altered clathrin dynamics in PI4KIIIα KO MEFs. Table S1 provides a list of antibodies used in this study. Table S2 shows the sets of control/KO pairs of MEF cells used in this study. Video 1 shows dynamic behavior of GFP-M1-PI4KIIIα at the plasma membrane. Video 2 shows induction of STIM1-mRFP–containing ER–plasma membrane contact sites upon thapsigargin stimulation in control MEFs. Video 3 shows impaired induction of STIM1-mRFP–containing ER–plasma membrane contact sites upon thapsigargin stimulation in PI4KIIIα KO MEFs. Video 4 shows that transfection of PI4KIIIα KO MEFs with GFP-M1-PI4KIIIα restores induction of STIM1-mRFP–containing ER–plasma membrane contact sites upon thapsigargin stimulation. Video 5 shows ectopic actin nucleation and dynamic actin comet tails in PI4KIIIα KO MEFs. Video 6 shows localization of dynamin 2 on dynamic actin comet tails in PI4KIIIα KO MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206095/DC1.
Acknowledgments
We thank Lijuan Liu, Louise Lucast, Dayna Morel, and Frank Wilson for technical assistance, Timothy Nottoli (Yale Animal Genomics Services) for help with mouse gene targeting, Stacy Wilson for help with image analysis, and Bruno Antonny, Jonathan Bogan, Shawn Ferguson, Howard Hang, Olof Idevall-Hagren, Stanimir Ivanov, Ira Milosevic, and Tobias Walther for helpful discussions.
This work was supported in part by National Institutes of Health grants R37NS036251 (to P. De Camilli), DK082700 (to P. De Camilli), DK45735 (Yale Diabetes Endocrinology Research Center; to P. De Camilli and M. Hao), T32GM007223 (to J. Chung), DA018343 (Yale/ National Institute on Drug Abuse Neuroproteomics Center), DK078625 (to M. Hao), and DK075772 (principal investigator: Jonathan Bogan); the Simons Foundation (to P. De Camilli); the Singapore National Research Foundation (to M.R. Wenk); the Biomedical Research Council of Singapore (to M.R. Wenk); the Searle Scholars Program (to N.T. Ingolia); and fellowships from the Japanese Society for the Promotion of Science (to F. Nakatsu), the American Diabetes Association (to F. Nakatsu), and the Jane Coffin Childs Fund (to J.M. Baskin).
Footnotes
Abbreviations used in this paper:
- 4-OHT
- (Z)-4-hydroxytamoxifen
- BP
- band pass
- IP
- immunoprecipitation
- iRFP
- infrared RFP
- KO
- knockout
- MβCD
- methyl-β-cyclodextrin
- MEF
- mouse embryonic fibroblast
- PI
- phosphatidylinositol
- PIK
- PI kinase
- PIPKI
- type I PI monophosphate kinase
- PtdIns(4, 5)P2
- PI 4,5-bisphosphate
- PtdIns4P
- PI 4-phosphate
- RBO
- rolling blackout
- TIRF
- total internal reflection fluorescence
References
- Altan-Bonnet N., Balla T. 2012. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 37:293–302 10.1016/j.tibs.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvisi G., Madan V., Bartenschlager R. 2011. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol. 8:258–269 10.4161/rna.8.2.15011 [DOI] [PubMed] [Google Scholar]
- Audhya A., Emr S.D. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2:593–605 10.1016/S1534-5807(02)00168-5 [DOI] [PubMed] [Google Scholar]
- Audhya A., Foti M., Emr S.D. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 11:2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea T.C., Wang Y., Nathans J. 2003. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23:2314–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D., Stefan C., Audhya A., Weys S., Emr S.D. 2008. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 183:1061–1074 10.1083/jcb.200804003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A., Balla T. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351–361 10.1016/j.tcb.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Tsiomenko A., Várnai P., Balla T. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 16:1282–1295 10.1091/mbc.E04-07-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A., Kim Y.J., Varnai P., Szentpetery Z., Knight Z., Shokat K.M., Balla T. 2008. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIalpha. Mol. Biol. Cell. 19:711–721 10.1091/mbc.E07-07-0713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.L., Cooper J.D., Heaton N.S., Yoon R., Oakland T.E., Jordan T.X., Mateu G., Grakoui A., Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. USA. 106:7577–7582 10.1073/pnas.0902693106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Reghellin V., Donnici L., Fenu S., Alvarez R., Baruffa C., Peri F., Pagani M., Abrignani S., Neddermann P., De Francesco R. 2012. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 8:e1002576 10.1371/journal.ppat.1002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C.J., Myer V., Cornellataracido I., et al. 2009. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 83:10058–10074 10.1128/JVI.02418-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., Weissman J.S. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463:1048–1053 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloway N., Owens T., Corwith K., Rodgers W., Holowka D., Baird B. 2011. Stimulated association of STIM1 and Orai1 is regulated by the balance of PtdIns(4,5)P2 between distinct membrane pools. J. Cell Sci. 124:2602–2610 10.1242/jcs.084178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G., Vicinanza M., Di Campli A., De Matteis M.A. 2008. The multiple roles of PtdIns(4)P — not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 121:1955–1963 10.1242/jcs.023630 [DOI] [PubMed] [Google Scholar]
- de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., Drin G. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195:965–978 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Doughman R.L., Firestone A.J., Anderson R.A. 2003. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 194:77–89 10.1007/s00232-003-2027-7 [DOI] [PubMed] [Google Scholar]
- Fairn G.D., Ogata K., Botelho R.J., Stahl P.D., Anderson R.A., De Camilli P., Meyer T., Wodak S., Grinstein S. 2009. An electrostatic switch displaces phosphatidylinositol phosphate kinases from the membrane during phagocytosis. J. Cell Biol. 187:701–714 10.1083/jcb.200909025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.M., Raimondi A., Paradise S., Shen H., Mesaki K., Ferguson A., Destaing O., Ko G., Takasaki J., Cremona O., et al. 2009. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev. Cell. 17:811–822 10.1016/j.devcel.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster L.J., De Hoog C.L., Mann M. 2003. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. USA. 100:5813–5818 10.1073/pnas.0631608100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond G.R.V., Fischer M.J., Anderson K.E., Holdich J., Koteci A., Balla T., Irvine R.F. 2012. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 337:727–730 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang H.C., Wilson J.P., Charron G. 2011. Bioorthogonal chemical reporters for analyzing protein lipidation and lipid trafficking. Acc. Chem. Res. 44:699–708 10.1021/ar200063v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.-D., Matthies H.J.G., Speese S.D., Smith M.A., Broadie K. 2004. Rolling blackout, a newly identified PIP2-DAG pathway lipase required for Drosophila phototransduction. Nat. Neurosci. 7:1070–1078 10.1038/nn1313 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., De Camilli P. 2012. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 109:E2316–E2323 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., Weissman J.S. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S.E., Brickner J.H., Reilein A.R., Fenn T.D., Walter P., Brunger A.T. 2005. Structural basis of FFAT motif-mediated ER targeting. Structure. 13:1035–1045 10.1016/j.str.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Korzeniowski M.K., Popovic M.A., Szentpetery Z., Varnai P., Stojilkovic S.S., Balla T. 2009. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J. Biol. Chem. 284:21027–21035 10.1074/jbc.M109.012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Fuelling A., Kolbe L., Anderson R.A. 2002. Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J. Biol. Chem. 277:5611–5619 10.1074/jbc.M110775200 [DOI] [PubMed] [Google Scholar]
- Lee E., De Camilli P. 2002. Dynamin at actin tails. Proc. Natl. Acad. Sci. USA. 99:161–166 10.1073/pnas.012607799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M., Laitinen S., Chinetti G., Johansson M., Ehnholm C., Staels B., Ikonen E., Olkkonen V.M. 2001. The OSBP-related protein family in humans. J. Lipid Res. 42:1203–1213 [PubMed] [Google Scholar]
- Lemmon M.A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9:99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- McCrea H.J., De Camilli P. 2009. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda). 24:8–16 10.1152/physiol.00035.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic I., Sørensen J.B., Lang T., Krauss M., Nagy G., Haucke V., Jahn R., Neher E. 2005. Plasmalemmal phosphatidylinositol-4,5-bisphosphate level regulates the releasable vesicle pool size in chromaffin cells. J. Neurosci. 25:2557–2565 10.1523/JNEUROSCI.3761-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Goto K., Kondo H. 1996. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 271:12088–12094 10.1074/jbc.271.20.12088 [DOI] [PubMed] [Google Scholar]
- Nakatsu F., Perera R.M., Lucast L., Zoncu R., Domin J., Gertler F.B., Toomre D., De Camilli P. 2010. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 190:307–315 10.1083/jcb.201005018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Prinz W.A. 2010. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 26:157–177 10.1146/annurev.cellbio.042308.113334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers W. 2002. Making membranes green: construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. Biotechniques. 32:1044–1051 [DOI] [PubMed] [Google Scholar]
- Rozelle A.L., Machesky L.M., Yamamoto M., Driessens M.H., Insall R.H., Roth M.G., Luby-Phelps K., Marriott G., Hall A., Yin H.L. 2000. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10:311–320 10.1016/S0960-9822(00)00384-5 [DOI] [PubMed] [Google Scholar]
- Shui G., Cheong W.F., Jappar I.A., Hoi A., Xue Y., Fernandis A.Z., Tan B.K.-H., Wenk M.R. 2011. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: applications to a rabbit model for atherosclerosis. J. Chromatogr. A. 1218:4357–4365 10.1016/j.chroma.2011.05.011 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., Emr S.D. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A.B., Boone C., Emr S.D. 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26:5861–5875 10.1128/MCB.02403-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai A.W., Benita Y., Peng L.F., Kim S.-S., Sakamoto N., Xavier R.J., Chung R.T. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 5:298–307 10.1016/j.chom.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotard M., Lepère-Douard C., Régeard M., Piquet-Pellorce C., Lavillette D., Cosset F.L., Gripon P., Le Seyec J. 2009. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 23:3780–3789 10.1096/fj.09-131920 [DOI] [PubMed] [Google Scholar]
- Vicinanza M., D’Angelo G., Di Campli A., De Matteis M.A. 2008. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell. Mol. Life Sci. 65:2833–2841 10.1007/s00018-008-8353-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.M., Chvanov M., Haynes L.P., Petersen O.H., Tepikin A.V., Burgoyne R.D. 2010. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem. J. 425:159–168 10.1042/BJ20090884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.J., Wang J., Sun H.Q., Martinez M., Sun Y.X., Macia E., Kirchhausen T., Albanesi J.P., Roth M.G., Yin H.L. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 114:299–310 10.1016/S0092-8674(03)00603-2 [DOI] [PubMed] [Google Scholar]
- Wess J., Eglen R.M., Gautam D. 2007. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat. Rev. Drug Discov. 6:721–733 10.1038/nrd2379 [DOI] [PubMed] [Google Scholar]
- Wong K., Meyers R., Cantley L.C. 1997. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272:13236–13241 10.1074/jbc.272.20.13236 [DOI] [PubMed] [Google Scholar]
- Wood C.S., Schmitz K.R., Bessman N.J., Setty T.G., Ferguson K.M., Burd C.G. 2009. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187:967–975 10.1083/jcb.200909063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount J.S., Zhang M.M., Hang H.C. 2011. Visualization and identification of fatty acylated proteins using chemical reporters. Curr. Protoc. Chem. Biol. 3:65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R., Levitan I. 2007. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta. 1768:1311–1324 10.1016/j.bbamem.2007.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]