An apolar patch and a polar cluster in the protein translocation channel cooperate to discriminate between signal sequences and less hydrophobic segments of cytosolic proteins.
Abstract
A critical event in protein translocation across the endoplasmic reticulum is the structural transition between the closed and open conformations of Sec61, the eukaryotic translocation channel. Channel opening allows signal sequence insertion into a gap between the N- and C-terminal halves of Sec61. We have identified a gating motif that regulates the transition between the closed and open channel conformations. Polar amino acid substitutions in the gating motif cause a gain-of-function phenotype that permits translocation of precursors with marginally hydrophobic signal sequences. In contrast, hydrophobic substitutions at certain residues in the gating motif cause a protein translocation defect. We conclude that the gating motif establishes the hydrophobicity threshold for functional insertion of a signal sequence into the Sec61 complex, thereby allowing the wild-type translocation channel to discriminate between authentic signal sequences and the less hydrophobic amino acid segments in cytosolic proteins. Bioinformatic analysis indicates that the gating motif is conserved between eubacterial and archaebacterial SecY and eukaryotic Sec61.
Introduction
An initial step in the biogenesis of most integral membrane and secreted proteins is the transport of the protein through the Sec61 protein translocation channel in the rough ER. The Sec61 complex is an evolutionarily conserved heterotrimer. Archaebacterial and eubacterial organisms transport proteins through the homologous SecYEβ and SecYEG complexes.
High-resolution structures of archaebacterial and eubacterial translocation channels have provided detailed views of the closed and partially open channel conformations (Van den Berg et al., 2004; Tsukazaki et al., 2008; Zimmer et al., 2008; Egea and Stroud, 2010). In the closed conformation, the ten transmembrane (TM) spans of SecY are arranged in two five-helix bundles (TM spans 1–5 and 6–10) to form an hourglass-shaped transport pore with a central constriction termed the pore ring (Van den Berg et al., 2004). A reentrant loop preceding TM2 forms the exoplasmic plug domain. The pore ring and plug domain prevent ion flow through the channel in the closed conformation (Park and Rapoport, 2011), and, respectively, must expand and be displaced to permit protein transport through the pore (Zimmer et al., 2008). Opening of the lateral gate of the channel, which is formed by TM2, TM3, TM7, and TM8 (Van den Berg et al., 2004), allows signal sequence insertion into a signal sequence binding site formed by TM2 and TM7 (Plath et al., 1998). SecYEG translocons that are locked in the closed conformation by formation of a disulfide between TM2 and TM7 are inactive in protein translocation (du Plessis et al., 2009). One interpretation of the partially open SecA–SecYE complex is that binding of a cytosolic effector (e.g., SecA in eubacteria) initiates lateral gate opening (Zimmer et al., 2008). The partially open or preopen conformation of the translocation channel is primed for signal sequence insertion into the signal sequence-binding site (Zimmer et al., 2008). In the fully open conformation, the gap in the lateral gate is sufficient to allow an α-helical TM span of a nascent integral membrane protein to move laterally from the central pore of SecY/Sec61 through the lateral gate to integrate into the membrane bilayer. Channel opening is thought to occur by a rigid body separation of the N- and C-terminal halves of the channel, with loop 5 serving as a flexible hinge (Van den Berg et al., 2004; Gumbart and Schulten, 2007; Egea and Stroud, 2010; Trueman et al., 2011).
There are distinct co- and posttranslational pathways in Saccharomyces cerevisiae. The Sec61 complex and the nonessential Ssh1 complex (Finke et al., 1996) serve as cotranslational translocation channels in S. cerevisiae. Substrates with more hydrophobic signal sequences, including integral membrane proteins, are primarily translocated by a cotranslational, signal recognition particle (SRP)–dependent pathway (Ng et al., 1996) that mediates binding of the ribosome-nascent chain complex to Sec61 or Ssh1 (Cheng et al., 2005; Becker et al., 2009). Posttranslational translocation of proteins across the yeast rough ER is mediated by the heptameric Sec complex, which is composed of a Sec61 heterotrimer plus the Sec62–Sec63 complex (Deshaies et al., 1991; Panzner et al., 1995).
The prl class of Escherichia coli SecY and S. cerevisiae sec61 mutations cause a gain-of-function phenotype, resulting in more efficient translocation of precursors bearing signal sequence mutations (Emr et al., 1981; Junne et al., 2007). Most point mutations that cause the prl phenotype map to the plug domain or pore ring of SecY or Sec61 (Smith et al., 2005; Junne et al., 2007). Mutations that cause the prl phenotype are thought to reduce the fidelity of signal sequence recognition by altering the equilibrium between the closed and open conformations of the translocation channel (Smith et al., 2005; Bondar et al., 2010). Several loss-of-function sec61 alleles, including sec61-2, which maps to an invariant glycine residue in the hinge loop (Nishikawa et al., 2001), may reduce translocation activity by stabilizing the closed conformation of the channel (Trueman et al., 2011).
Despite the insights provided by the SecYEG and SecYEβ structures, it is not clear how lateral and lumenal gate opening are coordinated. A structural link between the lateral and lumenal gates would allow a rapid and efficient response of the Sec61 complex to the cytosolic effectors. Here, we have identified a cluster of residues in the lumenal and lateral gate domains that function in a coordinated manner to regulate the transition between the closed and open conformations of the translocation channel. A cluster of three polar amino acid residues that link the N- and C-terminal sides of the lateral gate is sandwiched between an apolar patch in the plug domain and the hydrophobic core of the membrane bilayer. Point mutations in the polar cluster cause either a translocation defect or the prl phenotype depending on the hydrophobicity of the substituted amino acid. Point mutations in the apolar patch or polar cluster alter the hydrophobicity threshold for signal sequence gating of the protein translocation channel. The hydrophobicity threshold for wild-type Sec61 is poised to allow exquisite discrimination between authentic signal sequences and the marginally hydrophobic segments of cytosolic proteins.
Results
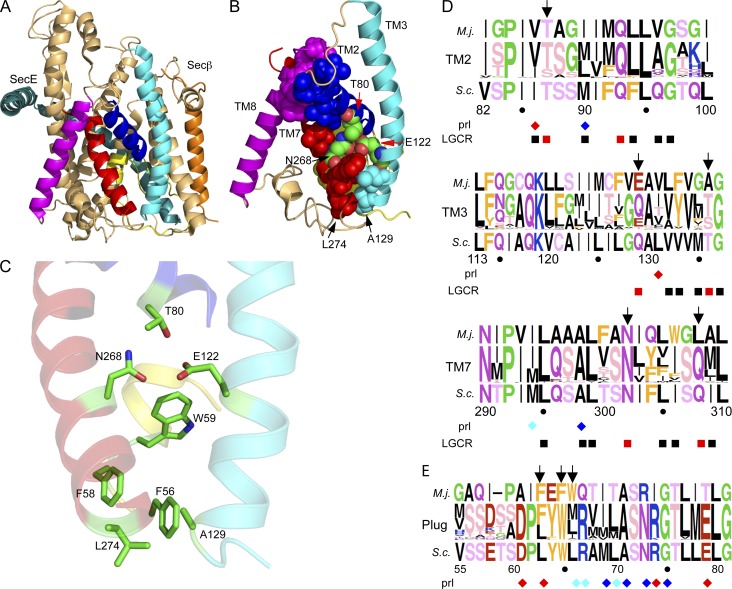
The structure of Methanococcus jannaschii SecYEβ (Van den Berg et al., 2004) was examined to identify conserved residues in the lumenal and lateral gates that might regulate the transition between the open and closed channel conformations (Fig. 1 A). As a first step, we identified residues in the N- (TM2 and TM3) and C-terminal (TM7 and TM8) sides of the lateral gate that make contacts across the gate in the closed conformation of the channel. These lateral gate contact residues (LGCRs) are shown as spheres in Fig. 1 B. Although the majority of LGCRs have hydrophobic side chains, three polar amino acids (T80, E122, and N268) reside in an unusual location in the center of the membrane bilayer (Fig. 1 B). It has long been recognized that unsatisfied hydrogen bond donors and acceptors are energetically unfavorable on the lipid-exposed surfaces of integral membrane proteins (Engelman et al., 1986; Rees et al., 1989). Although the 3.2-Å resolution of the M. jannaschii SecYEβ crystal structure is insufficient to unambiguously determine hydrogen bond geometries, the side chains of T80, E122, and N268 are within hydrogen bonding distance (Fig. 1 C) as reported previously (Bondar et al., 2010). Because of the location of T80, E122, and N268 in the center of the membrane bilayer, protein-bound water molecules are unlikely to form hydrogen bonds to T80, E122, and N268 in the closed conformation of SecYEβ. Molecular dynamics simulations of M. jannaschii SecYEβ provide support for the formation of H bonds between these three residues (Gumbart and Schulten, 2007; Bondar et al., 2010). M. jannaschii SecY residues T80, E122, and N268 align with T87, Q129, and N302 in S. cerevisiae Sec61. We will refer to these three Sec61 residues (T87, Q129, and N302) as the lateral gate polar cluster. Alignment of 264 nonredundant eukaryotic Sec61 sequences revealed that the lateral gate polar cluster is a strikingly conserved motif (Fig. 1 D). Chloroplast SecY sequences, because of their eubacterial origin, and fungal Ssh1 sequences, because of their extensive divergence from Sec61, were excluded from the Sec61 alignment. All eukaryotic Sec61 sequences have polar amino acid residues at all three positions (T or S at T80, Q or E >> T at E122, and N >> Q at N268). As expected, most of the other LGCRs have hydrophobic side chains (Fig. 1 D, black squares). Two additional LGCRs have polar side chains in all eukaryotic Sec61 proteins (T136 in TM3 and Q308 in TM7). These LGCRs align with A129 and L274 of M. jannaschii SecY, both of which are near F56 from the plug domain (Fig. 1 C).
Figure 1.
Lateral gate polar cluster and plug domain apolar patch. The M. jannaschii SecYEβ channel (A) and lateral gate views (B and C) are color coded as follows: TM2, TM3, TM7, and TM8 are blue, cyan, red, and magenta, respectively; the plug domain is yellow; the remainder of SecY is tan. Residue numbers correspond to M. jannaschii SecY. (B) The lateral gate and plug domains of SecY LGCRs are shown as spheres. (C) Polar cluster and apolar patch residues are shown as color coded sticks. TM2, TM3, and TM7 are rendered semitransparent. (D and E) Sequence logos of TM2, TM3, TM7, and the plug domain derived from 264 Sec61 sequences. Residue numbers correspond to S. cerevisiae Sec61. Residues are color coded by side chain property; letter height is proportional to frequency. The M. jannaschii and S. cerevisiae sequences flank the logo. Color-coded diamonds (red, S. cerevisiae; blue, E. coli; cyan, both) designate sites of previously described prl alleles. Polar (red) and nonpolar (black) LGCRs are shown below the logos. A–C were made using PYMOL v1.3 software and PDB file 1RHZ. Sequence logos were made using WebLogo.
Three aromatic residues (F56, F58, and W59) in the plug domain form an apolar patch that contacts the polar cluster (T80, E122, and N268) or residues in TM3 and TM7 that are one helical turn away from E122 and N268. The sequence alignment indicated that the three apolar patch positions are occupied by aromatic (F) or bulky aliphatic (M, I, L, or V) residues in more than 98% of Sec61 sequences including S. cerevisiae Sec61 (L63, W65, and L66; Fig. 1 E). We will refer to these three Sec61 residues (L63, W65, and L66) as the apolar patch. In the closed conformation of Sec61, the lateral gate polar cluster is clamped between the apolar patch in the plug domain and the hydrophobic core of the membrane bilayer.
We used the S. cerevisiae experimental system to test whether the polar cluster and apolar patch cooperate as a gating motif to coordinate the opening of the protein translocation channel. A plasmid shuffle procedure was used to replace wild-type Sec61 with a sec61 mutant in a haploid yeast strain that lacks the nonessential Ssh1 translocation channel (Cheng et al., 2005). Ssh1 expression can suppress certain classes of Sec61 mutants (Cheng et al., 2005).
Point mutations in the apolar patch cause the prl phenotype
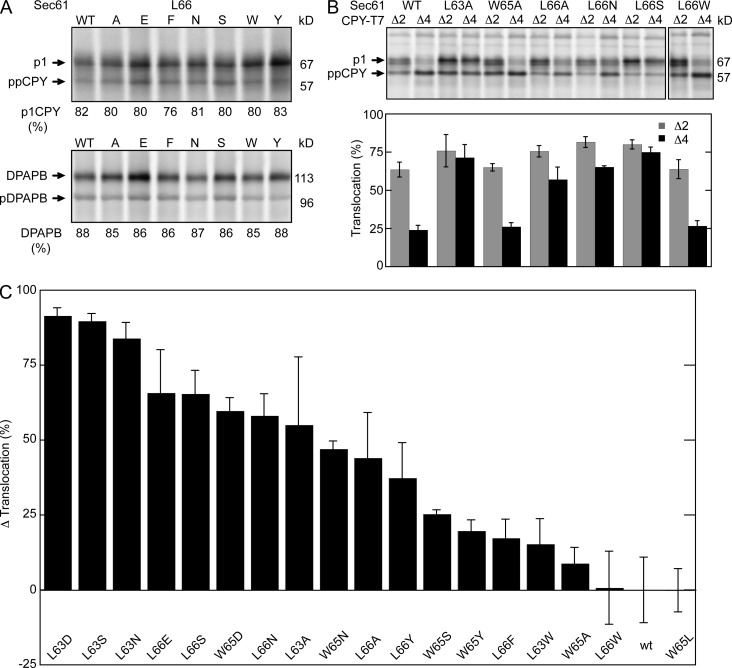
The three apolar patch residues (L63, W65, and L66) were replaced with either aliphatic or aromatic residues to alter side chain volume or with polar residues to introduce hydrogen bond donors or acceptors. All of the resulting strains were viable and did not display growth rate defects relative to the parental ssh1Δ strain (unpublished data). The complete collection of sec61 apolar patch mutants were assayed for translocation of carboxypeptidase Y (CPY) or integration of dipeptidylaminopeptidase B (DPAPB) by pulse labeling. Posttranslational translocation of CPY is detected by the N-glycosylation–induced gel mobility difference between the cytosolic precursor (ppCPY) and the ER form of proCPY (p1CPY). Representative translocation assays for seven of the sec61 L66 mutants are displayed in Fig. 2 A. None of the sec61 apolar patch mutants showed a significant reduction in translocation of CPY relative to the SEC61 ssh1Δ control strain. Cotranslational integration of DPAPB, as detected by the glycosylation-dependent decrease in gel mobility, was not altered by substitutions at L63, W65, or L66 (Fig. 2 A and Table S1).
Figure 2.
Mutations in the apolar patch cause the prl phenotype. (A) Translocation assays of sec61 L66 mutants. Translocation of CPY and integration of DPAPB was assayed by 7-min pulse labeling of wild-type and mutant yeast cells. CPY and DPAPB were immunoprecipitated from pulse-labeled cell extracts using CPY- and DPAPB-specific antisera. The glycosylated ER forms of CPY (p1) and DPAPB were resolved from nontranslocated precursors (ppCPY and pDPAPB) by SDS-PAGE. The p1CPY doublet is caused by p1CPY glycoforms that have three or four N-linked glycans. The percentage of translocation (CPY) or integration (DPAPB) is the mean of four or more determinations, one of which is shown here. (B) The prl phenotype of sec61 mutants was assayed by pulse labeling using the ppCPYΔ2-T7 (Δ2) and ppCPYΔ4-T7 (Δ4) reporters. Precursors (ppCPYΔ2-T7 or ppCPYΔ4-T7) and the translocated product (p1CPY-T7) that were immunoprecipitated using anti-T7 antisera were resolved by SDS-PAGE. The percentage of translocation is the mean of four determinations; error bars designate standard deviations. (C) The Δ translocation value for a sec61 allele corresponds to the percentage of increase or decrease in p1CPY relative to wild-type Sec61 for the ppCPY, ppCPYΔ2, and ppCPYΔ4 reporters. Wild-type and mutant strains were all assayed two or more times. Table S1 includes the assay values (CPY translocation, DPAPB integration, and prl reporter translocation) and Δ translocation value for all mutants that are displayed in this panel. The error bars correspond to the sum of the individual errors for the three assays that were used to calculate the Δ translocation value. The indicated molecular masses shown in this and subsequent figures are the apparent molecular masses of the observed protein species relative to prestained molecular mass markers that were electrophoresed on all gels.
Many previously characterized point mutations that cause the prl phenotype map to the plug domain of SecY or Sec61 (Fig. 1 E, diamonds; Smith et al., 2005; Junne et al., 2007). The prl phenotype can be assayed by pulse-labeling yeast cells that express CPY derivatives that lack two or four residues of the signal sequence (Trueman et al., 2011). Representative assays using the prl reporters (CPYΔ2 and CPYΔ4) are shown in Fig. 2 B. Translocation of CPYΔ4 by the wild-type Sec complex is very inefficient. Reducing the side chain volume of the apolar patch residues (L63A and L66A but not W65A) or increasing side chain polarity (L66S and L66N) causes an increase in translocation of CPYΔ2 and CPYΔ4 (Fig. 2 B), whereas aromatic or bulky aliphatic substitutions do not (Fig. 2 B). The prl reporter assay results for the complete collection of sec61 apolar patch mutants are shown in Table S1.
To allow a graphical comparison of the sec61 apolar patch mutants, we combined the percentage of the translocated product (p1CPY) for the ppCPY, ppCPYΔ2, and ppCPYΔ4 reporters. For wild-type Sec61 this value was 170. We then calculated a Δ translocation value, which corresponds to the percentage of increase or decrease in the sum of the translocated products (p1CPY for the ppCPY, ppCPYΔ2, and ppCPYΔ4 reporters) in a given sec61 mutant relative to wild-type Sec61 (Table S1). A positive Δ translocation value is indicative of the gain-of-function prl phenotype, whereas a negative value corresponds to a translocation defect. Although the Δ translocation value could in principle range between +130 (100% p1CPY for each reporter) and −170 (0% p1CPY for each reporter), the observed range of Δ translocation values for viable yeast strains is between +90 and −150. Polar substitutions at L63 or L66, including those that are isosteric (L63D), caused a strong prl phenotype (Fig. 2 C). Aromatic substitutions (e.g., L66F or L66W) did not cause the prl phenotype unless the substitution introduced a new hydrogen bond donor (i.e., L66Y) near the polar cluster. Mutations at L63 and L66, which contact both the N- and C-terminal sides of the lateral gate, cause stronger prl phenotypes than point mutations at W65 (Fig. 2 C).
Point mutations in the polar cluster cause gain- or loss-of-function phenotypes
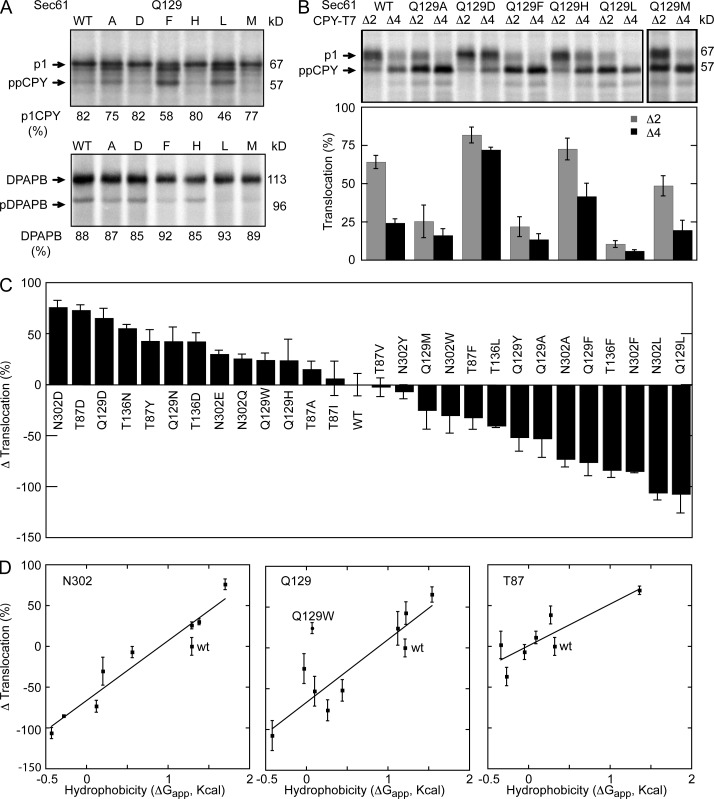
The role of the lateral gate polar cluster was investigated by introducing point mutations at residues T87, Q129, and N302. Point mutations at T136 and Q308 were also tested to determine whether other polar LGCRs are also important for channel gating. None of the lateral gate polar cluster mutations caused an obvious growth defect at 30°C; minor reductions in colony diameter were observed for a subset of the mutants at 37°C (Fig. S1). The complete collection of sec61 polar cluster mutants was assayed for translocation of ppCPY, ppCPYΔ2, and ppCPYΔ4 and integration of DPAPB (Table S2). Assays of selected Q129 mutants are shown in Fig. 3 (A and B) to illustrate several points. Replacement of Q129 with an aliphatic residue or an aromatic residue caused a reduction in translocation of CPY (Fig. 3 A). When assayed for translocation of the prl reporters (Fig. 3 B), sec61 mutants that displayed a minor defect in translocation of wild-type CPY (e.g., Q129A or Q129M) showed more dramatic reductions in translocation of ppCPYΔ2 and ppCPYΔ4 (Fig. 3, A and B; and Table S2). Remarkably, point mutations at N302 or Q129 that caused a reduction in translocation of CPY did not cause a reduction in the integration of DPAPB (Fig. 3 A, Q129L and Q129F; and Table S2). Polar substitutions at Q129 caused substantial increases in translocation of the prl reporters (Fig. 3 B, Q129D and Q129H).
Figure 3.
Loss- and gain-of-function polar cluster mutations. Integration of DPAPB (A) and translocation of CPY (A) and the prl reporters (B) were assayed as described in Fig. 2. Quantified translocation activity is the mean of two or more determinations. (C) The Δ translocation value was calculated as described in Fig. 2. Table S2 contains the assay values (CPY translocation, DPAPB integration, and prl reporter translocation) and Δ translocation value for all mutants displayed in this panel. (D) The Δ translocation values from C for the N302, Q129, and T87 series of mutants are plotted versus the hydrophobicity (ΔGapp) of the amino acid residue. The lines are linear regression fits with the Q129W mutant excluded from the Q129 series. Error bars in B–D designate individual data points or standard deviations.
To allow a comparison of the polar cluster mutants, we calculated the Δ translocation value as described for the sec61 apolar patch mutants (Fig. 3 C and Table S2). Polar substitutions at four of the selected residues (T87, Q129, T136, and N302) caused a prl phenotype even when the altered residue was an isosteric (N302D) or conservative (Q129N) substitution. Aromatic or bulky aliphatic substitutions at three residues (Q129, T136, and N302) caused translocation defects. The effects of LGCR mutagenesis are residue specific, as point mutations at Q308 (e.g., Q308D and Q308L) caused minor reductions in translocation of the prl reporters regardless of side chain polarity (Table S2).
Point mutations at the polar cluster residues caused gain- or loss-of-function phenotypes that correlated with the hydrophobicity of the substituted amino acid (Fig. 3 C). To obtain a better understanding of this relationship we used a biological hydrophobicity scale (Hessa et al., 2007) to obtain a membrane location–specific hydrophobicity value (ΔGapp) for each tested amino acid substitution at T87, Q129, and N302. When the Δ translocation values for the polar cluster mutants were plotted versus the hydrophobicity of the substituted amino acid, we observed a remarkable correlation for the N302 and Q129 series of mutants (Fig. 3 D). Linear regression analysis indicated that the two lines are nearly coincident. This analysis also revealed that the Q129W mutation does not cause the anticipated reduction in translocation activity, perhaps because the larger tryptophan side chain can perturb lateral gate–lumenal gate contacts for steric reasons. Importantly, the data points for the wild-type residues (N302 and Q129) fall below the plotted lines because polar amino acid substitutions of similar hydrophilicity (i.e., N302Q and Q129N) alter polar cluster residue interactions by changing side chain length by one methylene unit. The slope of the line for the T87 series of mutants is lower because aliphatic substitutions (e.g., T87A, T87I, and T87V) did not cause a significant translocation defect (Fig. 3 C). The correlation between residue hydrophobicity and Δ translocation activity indicates that polar amino acid substitutions destabilize the lateral gate, whereas nonpolar substitutions stabilize the closed conformation of the channel by enhancing lateral gate–lumenal gate interactions.
Lateral gate polar cluster mutations affect substrates of both translocation pathways
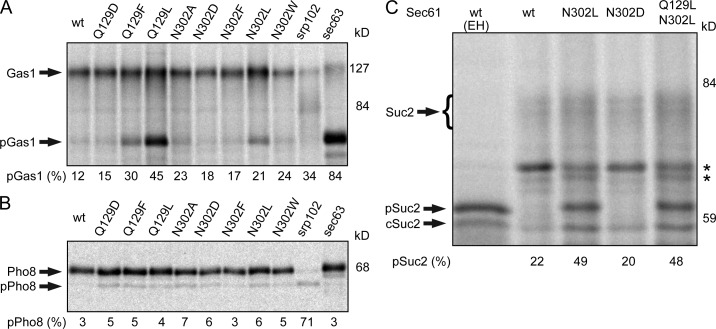
Replacing N302, Q129, or T136 with nonpolar amino acids reduced posttranslational translocation of CPY, CPYΔ2, and CPYΔ4 but did not reduce cotranslational integration of DPAPB. Three additional substrates were tested to determine whether the polar cluster mutants cause a pathway-specific defect in translocation channel function. Translocation of the glycosylphosphatidylinositol-anchored protein Gas1p was assayed by the N-glycosylation–induced mobility decrease (Fig. 4 A). As reported previously (Ng et al., 1996), posttranslational translocation of Gas1p is blocked by a mutation in the Sec63 subunit of the Sec complex (sec63-201). Point mutations that caused the most severe defects in CPY translocation (Q129L, Q129F, and N302L) caused readily detectable increases in the Gas1p precursor (pGas1). Cotranslational integration of Pho8p (Fig. 4 B) was blocked by a mutation in the SRP receptor (srp102 K51I), but was not reduced by sec61 translocation-defective alleles (e.g., Q129L) or enhanced by sec61 prl alleles (e.g., sec61 N302D). Invertase (Suc2p) was assayed as an example of a secreted protein that is translocated by a cotranslational pathway (Johnsson and Varshavsky, 1994; Cheng and Gilmore, 2006; Trueman et al., 2011). The cytoplasmic form of invertase (cSuc2) and core-glycosylated secretory invertase (Suc2) are the major products detected by pulse labeling in cells expressing wild-type Sec61 (Fig. 4 C). The nontranslocated precursor of secretory invertase (pSuc2), which comigrates with Endo H–digested Suc2p, accumulated in the sec61 N302L and the sec61 Q129L N302L double mutant but not in a sec61 prl allele (sec61 N302D).
Figure 4.
Loss-of-function polar cluster mutations reduce translocation of several substrates. Translocation of Gas1p (A), Pho8p-T7 (B), and Suc2p (C) was assayed by pulse labeling. The srp102 K51I mutant was labeled 3 h after the cells were shifted to 37°C. The glycosylated forms of Gas1p, Pho8p, and Suc2p were resolved from nontranslocated precursors (pGas1, pPho8, and pSuc2) and from cytoplasmic invertase (cSuc2) by SDS-PAGE. Samples digested with Endo H are designated (EH). Percentage of precursor is the mean of two determinations, one of which is shown here. The asterisks designate nonspecific bands.
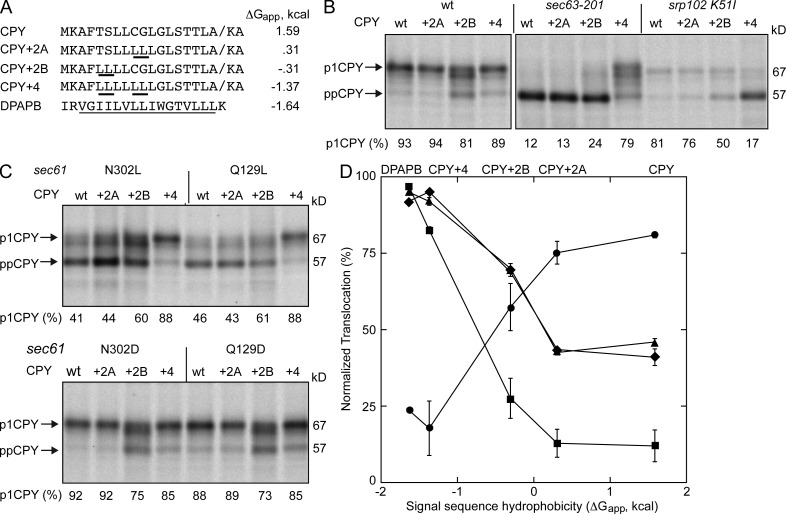
Nonpolar substitutions at Q129 and N302 reduced the translocation of secretory proteins (CPY and Suc2) and the glycosylphosphatidylinositol-anchored protein (Gas1p) but did not inhibit the integration of type 2 integral membrane proteins (Pho8p and DPAPB). TM spans are more hydrophobic than cleavable signal sequences, so our results suggest that the difference in the hydrophobicity of the targeting sequence rather than the targeting pathway might explain why certain proteins are not efficiently translocated by the sec61 N302L and sec61 Q129L mutants. To address this hypothesis, we increased the hydrophobicity of the CPY signal sequence by replacing weakly hydrophobic (C9 and G10) or hydrophilic (T5 and S6) residues with leucine residues in the CPY+2A and CPY+2B mutants (Fig. 5 A). The leucine substitutions, particularly at T5 and S6, increase the hydrophobicity of the signal sequence (Fig. 5 A and Fig. S2). When all four residues are replaced with leucine, the calculated hydrophobicity (ΔGapp; Hessa et al., 2007) of the CPY+4 signal sequence and the TM span of DPAPB are similar (Fig. 5 A). First, we tested whether the signal-enhanced CPY derivatives were translocated by wild-type cells, and observed that CPY+2B is translocated somewhat less efficiently than CPY, CPY+2A, or CPY+4 (Fig. 5 B). The leucine substitutions in the signal sequence of the CPY mutants do not alter the predicted signal sequence cleavage site (Fig. S2) as determined using SignalP 4.0 (Petersen et al., 2011). We did not observe a reduction in gel mobility of p1CPY that would suggest that the signal sequence was not cleaved. Based on a previous signal sequence-exchange experiment (Ng et al., 1996), we anticipated that CPY+4 would be redirected into the cotranslational translocation pathway and this was indeed the case (Fig. 5 B) as detected by the complete block of CPY+4 translocation in the SRP receptor mutant (srp102 K51I; Ogg et al., 1998). Although translocation of CPY+2B was reduced in the sec63-201 and srp102 K51I mutants relative to the wild-type strain, the more severe impact of the sec63-201 mutation indicates that CPY+2B is primarily translocated by the posttranslational targeting pathway.
Figure 5.
Signal sequence hydrophobicity alters translocation efficiency. (A) The sequence and calculated hydrophobicity (ΔGapp) of the signal sequence of CPY, CPY+2A, CPY+2B, CPY+4, and the TM span of DPAPB. The underlined residues in the CPY mutants indicate the position of leucine substitutions. The underlined residues in the DPAPB sequence designate the TM span. (B and C) Translocation of ppCPY-T7 and derivatives thereof (+2A, +2B, and +4) was assayed by pulse labeling. All strains are ssh1Δ except for the sec63-201 mutant. The srp102 K51I mutant was labeled 3 h after cells were shifted to 37°C. (D) Translocation activity of the sec61 N302L (triangles), sec61 Q129L (diamonds), srp102 K51I (circles), and sec63-201(squares) mutants are plotted versus the calculated hydrophobicity of the signal sequence. Translocation activity for the CPY derivatives was normalized using the assay results for wild-type cells to correct for the lower efficiency of the ppCPY+2B signal sequence. Percentage of translocation is the mean of two determinations, one of which is shown in B and C. Error bars designate individual data points.
The signal-enhanced CPY derivates were then expressed in the sec61 polar cluster mutants that have loss- (Q129L and N302L) or gain-of-function (Q129D and N302D) translocation phenotypes. The CPY+4 substrate was efficiently translocated by both the sec61 N302L and sec61 Q129L mutants (Fig. 5 C). Remarkably, the CPY+2B mutant also showed reduced accumulation of the precursor relative to wild-type CPY or CPY+2A. Assay results for the four translocation-defective yeast strains (Fig. 5, B and C) were normalized using assay results for the wild-type strain (Fig. 5 B) and plotted versus signal sequence hydrophobicity (Fig. 5 D). The improved translocation efficiency of the CPY+2B signal sequence in a polar cluster mutant (e.g., sec61 Q129L) exceeded the extent of precursor redirection into the cotranslational targeting pathway. Collectively with Fig. 4, these results indicate that signal sequence hydrophobicity, not targeting pathway, is primarily responsible for the substrate-specific differences in translocation efficiency of the sec61 N302L and sec61 Q129L mutants.
The apolar patch–polar cluster is a gating motif that regulates channel function
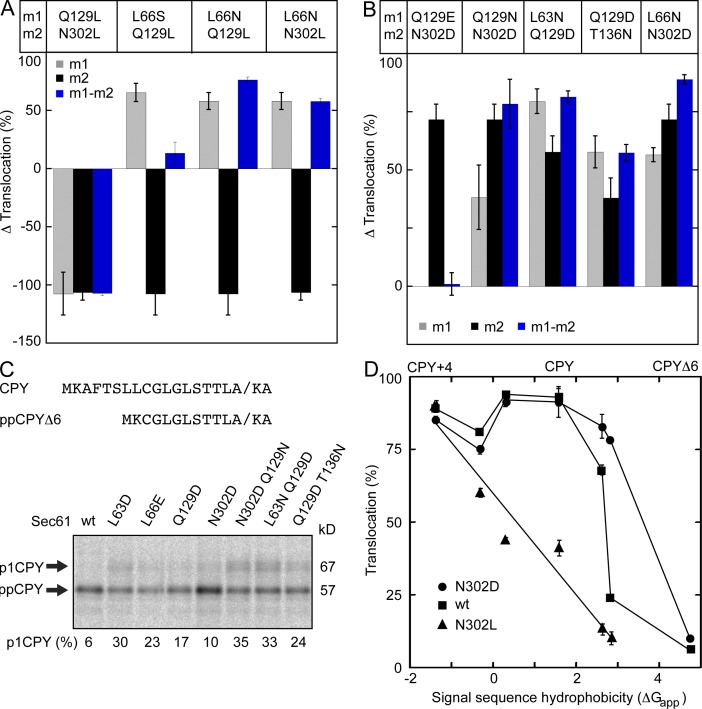
Substitution of residues in the lateral gate polar cluster and plug domain apolar patch alter interactions between the lateral and lumenal gates and cause loss- or gain-of-function translocation phenotypes. We constructed several sec61 double mutants to determine whether the observed phenotypes were either additive or mutually suppressive when combined. Translocation assay results for the double mutants are shown in Table S3. The combination of two mutations that eliminate polar residues on both sides of the lateral gate (Q129L and N302L) yielded a double mutant that was remarkably similar to the two single mutants in terms of Δ translocation activity (Fig. 6 A). Thus, elimination of both polar residues does not further stabilize the closed conformation of Sec61. Substitution of L66 with a polar residue (L66N) or a small residue (L66S or L66A) caused a moderately strong prl phenotype. We propose that polar substitutions at L66 provide additional hydrogen bond donors or acceptors adjacent to the polar cluster, thereby altering the critical interaction between N302 and Q129. The observation that the L66S or L66N mutations suppress the translocation defect of a sec61 Q129L or N302L mutation supports the hypothesis that the N302L or Q129L mutations stabilize the closed conformation of Sec61 because of enhanced hydrophobic contact between the lateral gate and plug domain (Fig. 6 A). Many protist and plant Sec61 sequences have a glutamic acid rather than glutamine at the residue that aligns with Q129 (Fig. 1 E). Surprisingly, the sec61 Q129E N302D double mutant resembled wild-type Sec61 (Fig. 6 B), indicating that normal channel gating could be restored with acidic residues at both positions. Because of their local environment in the membrane, acidic residues in the polar cluster are expected to be protonated at neutral pH. This assumption is strongly supported by the finding that the sec61 Q129E N302D double mutant does not cause a strong prl phenotype, as would be expected if the acidic side chains were charged.
Figure 6.
The polar cluster and apolar patch cooperate to regulate channel gating. (A and B) The Δ translocation value was calculated as in Fig. 2 (see Table S3 for CPY, DPAPB, and prl reporter assay values). Gray, black, and blue bars designate the Δ translocation value for the m1, the m2, and the m1m2 double mutant, respectively. (C) The signal sequence of the ppCPYΔ6-T7 reporter. Translocation of ppCPYΔ6-T7 was assayed by pulse labeling. (D) The signal sequence hydrophobicity for the tested substrates was plotted against translocation (percentage) for the following strains: N302D (circles), wild type (squares), and N302L (triangles). Percentage of translocation (C and D) and Δ translocation (A and B) are the means of two or more determinations; error bars designate individual data points or standard deviations.
We next asked whether the combination of two sec61 prl alleles would yield a double mutant with an enhanced prl phenotype. In most cases, the Δ translocation value for the double mutant resembled the stronger of the two initial single mutants (Fig. 6 B). Because the strongest sec61 prl alleles translocate ppCPYΔ4 and ppCPY with nearly equal efficiency (Tables S1–S3), we constructed a reporter with an even weaker signal sequence (ppCPYΔ6; Fig. S2) to characterize stronger sec61 prl alleles (Fig. 6 C). We assayed several of the stronger sec61 prl alleles using the ppCPYΔ6 reporter and observed that all tested sec61 prl mutants translocate ppCPYΔ6 more efficiently than the wild-type strain. Although two moderate sec61 prl alleles can be combined to obtain a strong double mutant (e.g., sec61 N302D Q129N), translocation of ppCPYΔ6 did not exceed 35% for any of the single or double sec61 mutants.
The hydrophobicity of the signal sequence of all tested CPY derivates was calculated using the in vivo hydrophobicity scale (Fig. 6 D). Wild-type Sec61 efficiently translocates precursors with marginally hydrophobic signal sequences like ppCPY, yet effectively discriminates against precursors with less hydrophobic signals like ppCPYΔ4 or ppCPYΔ6. The sec61 prl alelles cause an expansion of the allowed hydrophobicity window for a functional signal sequence. In contrast, replacement of Q129 or N302 with a leucine residue (sec61 N302L) caused a reduction in translocation efficiency that correlates strongly with the decreasing hydrophobicity of the signal sequence.
The gating motif is evolutionarily conserved
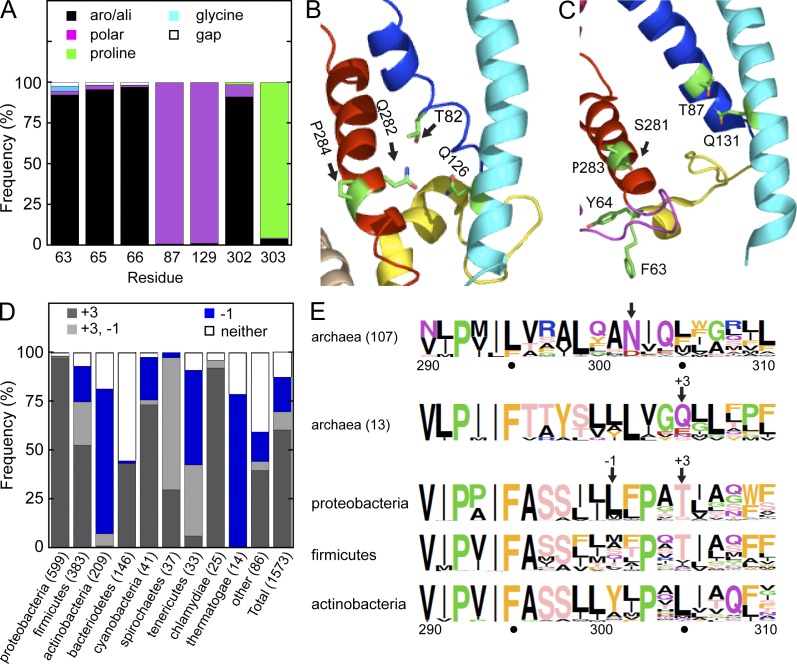
To determine if the gating motif is an evolutionarily conserved structural feature of protein translocation channels, we aligned currently available eubacterial and archaebacterial SecY sequences. An alignment of 1,646 unique eubacterial SecY sequences showed that more than 95% of eubacterial SecY sequences have an aliphatic or aromatic residue aligned with all three apolar patch residues (L63, W65, and L66; Fig. 7 A). Although two of the three lateral gate polar cluster residues (T87 and Q129) are strongly conserved in eubacterial SecY sequences (Fig. S3, eubacterial TM2 and TM3 sequence logos), the residue that aligns with N302 is almost always aliphatic or aromatic, as reported previously (Bondar et al., 2010). Intriguingly, the next residue in 95% of eubacterial SecY sequences is a proline (Fig. 7 A). Examination of the lateral gate in the closed conformation of Thermus thermophilus SecYEG (Tsukazaki et al., 2008) indicated that the conserved proline residue (P284) in the +1 position alters the helical register of TM7 so that I283 faces phospholipid, whereas Q282 projects toward T82 and Q126 (Fig. 7 B). In T. thermophilus SecY, the polar residue (Q282) located at the −1 position relative to N302 could provide the missing hydrogen bond donor or acceptor for the polar cluster. Of 120 archaebacterial SecY sequences, 113 have a polar residue (N or D) that aligns with N302 in S. cerevisiae Sec61 (Fig. 7 E). The remaining 13 archaebacterial SecY sequences have a polar residue (Q, E, N, or S) located one helical turn forward on TM7 (+3 position). We propose that polar residues at the −1 and +3 positions relative to N302 serve as alternative hydrogen bond donors or acceptors in TM7 that could contact the conserved glutamine residue in TM3.
Figure 7.
Evolutionary conservation of the apolar patch–polar cluster motif. (A) A collection of 1,646 unique eubacterial SecY sequences were aligned. Quantification of amino acid side chain properties for the polar cluster and apolar patch residues. Residues are numbered with respect to S. cerevisiae Sec61. Aromatic and aliphatic residues are abbreviated aro/ali. (B and C) Isolated lateral gate views of the closed conformation of T. thermophilus SecYEG and the SecA-gated structure of T. maritima SecYE. TM spans and the plug domain are color coded as in Fig. 1. (D) Quantification of the frequency of −1 and/or +3 polar residues in TM7 of eubacterial organisms relative to the location of N302 in S. cerevisiae Sec61. Numbers in parentheses indicate the number of sequences. (E) Sequence logos of TM7 for select groups of archaebacterial and eubacterial organisms. Residues are color coded by side chain property, and residue height is proportional to frequency. Residue numbers correspond to S. cerevisiae Sec61.
The 1573 eubacterial SecY sequences that have the proline residue at the +1 position were aligned and examined for the presence of a polar residue at the −1 or +3 positions. We found that 87% of eubacterial SecY sequences have a polar residue in one or both of these positions (Fig. 7 D). When the eubacterial SecY sequences were sorted with respect to taxonomy it became clear that polar cluster patterns correlate with eubacterial phyla. For example, Proteobacteria almost always have a polar residue at the +3 position, whereas Actinobacteria and Thermatogae have the polar residue at the −1 position, as in T. thermophilus SecY. Sequence logos for Proteobacteria, Firmicutes, and Actinobacteria are shown in Fig. 7 E.
Although the gating motif is not universally conserved in eubacterial SecY sequences, many of the most striking outliers include closely related organisms. For example, the 75 eubacterial SecY sequences that lack the conserved proline residue in TM7 include 50 species from the phylum Phytoplasma. Phytoplasma SecY proteins also have a truncated plug domain and account for all eubacterial SecY sequences that had an alignment gap at the apolar patch residues (L63, W65, and L66; Fig. 7 A).
We examined the partially open conformation of the Thermatoga maritima SecA–SecYE complex (Zimmer et al., 2008) to determine whether the interaction between the apolar patch and the lateral gate persists upon channel opening. Interestingly, T87 in TM2 remains adjacent to Q131 in TM3 when the channel is open (Fig. 7 C). A partial counterclockwise rotation of the TM7 helix that occurs during channel gating by SecA causes the −1 residue (S281) to face the channel interior rather than toward the lipid bilayer. The two apolar patch residues (F63 and Y64) that were resolved in the T. maritima SecA–SecYE structure no longer contact the lateral gate, supporting the conclusion that lateral gate separation and plug domain movements are coordinated and occur before signal sequence insertion.
Discussion
The opening of the lumenal and lateral gates of the translocation channel is a critical early event in the translocation of secretory proteins and integration of membrane proteins. Here we have identified a conserved gating motif in Sec61 that links the N- and C-terminal halves of the lateral gate to the lumenal gate. Mutagenesis of residues in the gating motif revealed that the polar cluster–apolar patch interaction regulates the hydrophobicity threshold for signal sequence function.
Previous random mutagenesis studies in yeast or in E. coli did not lead to the identification of prl alleles that mapped to T87, Q129, or N302 (Smith et al., 2005; Junne et al., 2007). Unexpectedly, relatively conservative substitutions that change the position of hydrogen bond donors and acceptors (e.g., sec61 Q129N) caused the prl phenotype, whereas mutations that replace Q129 or N302 with a bulky hydrophobic amino acid (e.g., sec61 Q129L) caused a translocation defect phenotype because of an enhanced nonpolar interaction between the lateral and lumenal gates. Support for this conclusion was provided by double mutant analysis. The translocation defects of the sec61 Q129L and sec61 N302L mutants were completely suppressed by replacing L66 with a polar residue. Contact between the apolar patch and lateral gate polar cluster is necessary for normal translocation channel function because a reduction in side chain volume of two apolar patch residues causes the gain-of-function prl phenotype (sec61 L66A or sec61 L63A mutants).
The role of the polar cluster in channel gating
The precise structure of the lateral gate polar cluster appears to be critical for normal channel gating as polar amino acid substitutions in either the apolar patch or polar cluster cause the prl phenotype. The closed conformation of SecYEβ is thought to be stabilized by hydrogen bond networks between well-conserved polar residues (Gumbart and Schulten, 2007; Bondar et al., 2010). Molecular dynamics simulations suggest that elimination of critical hydrogen bonds in SecY promotes movement of the plug domain and increased hydration of the transport pore in the vicinity of the pore ring (Bondar et al., 2010). Importantly, point mutations in the lateral gate polar cluster were not tested in these simulations. Although hydrogen bonds between amino acid side chains in integral membrane proteins might be expected to be unusually strong, the experimental evidence obtained in several model systems indicates that they are similar in strength to hydrogen bonds in water-soluble proteins (Bowie, 2011). Although the Q129–N302 hydrogen bond likely stabilizes the closed conformation of the channel, an unusually strong H bond network in the lateral gate would have a deleterious impact on the ability of the channel to open in response to cytosolic effectors. Unsatisfied hydrogen bond donors or acceptors are energetically unfavorable in the hydrophobic interior of the membrane bilayer. Consequently, the polar cluster residues (T87, Q129, and N302) might promote channel closing if Q129 and N302 were in contact with phospholipid acyl chains. Examination of the structure of a partially open channel (T. maritima SecYE; Fig. 7 C) suggests that these hydrogen bond donors and acceptors either remain satisfied in the open conformation (T87 and Q129) or project toward the interior of the translocation channel (N302).
Hydrophobicity threshold for signal sequence function
The SecYE structure of T. maritima provides evidence that the translocation channel adopts a partially open conformation in response to the cytosolic effector SecA (Zimmer et al., 2008). This partial gating results in lateral gate separation and plug domain movement away from the pore ring. We propose that the apolar patch and polar cluster provide a structural link between the lateral and lumenal gates that responds to the binding of cytosolic effectors to Sec61 or SecY. Once SecYEG or Sec61 are in a partially open conformation, the signal sequence can be inserted into the signal sequence-binding site between TM2 and TM7.
Point mutations in the polar cluster that caused loss-of-function phenotypes (e.g., N302L) did not reduce translocation of all substrates in an equivalent manner, but instead had a more profound impact upon substrates with less hydrophobic signal sequences. Our initial hypothesis was that the cotranslational gating mechanism was relatively insensitive to perturbation of the polar cluster. After testing several additional substrates, it became clear that the translocation efficiency of the sec61 N302L mutant was almost linearly dependent on the hydrophobicity of the signal sequence and not directly dependent on the targeting pathway. What could explain the reduced translocation efficiency of the sec61 N302L translocation channel? The excessive stability of the lateral gate–lumenal gate contact may diminish lateral gate separation in response to a cytosolic effector. Less hydrophobic signal sequences, unlike the TM span of DPAPB, may have a reduced probability of inserting into the signal sequence-binding site to promote the transition between the partially open and fully open conformations of the protein translocation channel.
Wild-type protein translocation channels respond to signal sequences that vary considerably in terms of hydrophobicity, ranging between the very hydrophobic TM spans of integral membrane proteins like DPAPB or Pho8p to the weakly hydrophobic cleaved signal sequences of proteins like CPY or Gas1p. By testing a series of CPY derivatives that cover a broad range of signal sequence hydrophobicity, we observed that sec61 prl alleles do not simply allow signal sequence-independent translocation but instead reduce the hydrophobicity threshold for a functional signal sequence. Proteins with less hydrophobic signal sequences (e.g., ppCPYΔ4 or ppCPYΔ6) are rejected by wild-type Sec61 in a reaction step that must occur after precursor targeting to the Sec complex because ppCPYΔ4 can be efficiently translocated by the strongest sec61 prl alleles in our collection (e.g., sec61 L63D). Evidence that the mammalian Sec61 complex can discriminate between an authentic signal sequence and a signal sequence mutant has been obtained by analyzing SRP-independent binding of ribosome nascent chain complexes to ribosome-stripped microsomal membranes (Jungnickel and Rapoport, 1995). An authentic signal sequence, but not a mutant signal sequence, inserts into the signal sequence-binding site, whereas the mutant signal does not. Additional in vitro evidence that signal-deficient precursors are rejected by the Sec61 complex was obtained by initiating translation on ribosomes that were prebound to the translocation channel (Potter and Nicchitta, 2000). The ability to discriminate between authentic signal sequences and marginally hydrophobic segments of cytosolic proteins is an important property of Sec61 or SecY that minimizes aberrant secretion of cytosolic proteins. In this regard it is worth noting that the hydrophobic core (underlined residues) of the ppCPYΔ6 signal sequence (MKCGLGLSTTA/KA) is shorter and less hydrophobic than many segments in cytosolic proteins.
Conservation of the gating motif
The lateral gate polar cluster (T87, Q129, and N302) was a previously recognized feature of archaebacterial and eukaryotic Sec61 sequences. However, the polar cluster was paradoxically absent from essentially all eubacterial SecY sequences despite the pronounced conservation of polar residues at the T87 and Q129 positions (Bondar et al., 2010). Here, we present evidence that the majority of eubacterial SecY sequences in the protein sequence database have alternative H bond donors or acceptors in TM7 that are provided by residues located at the −1 or +3 positions relative to N302. Structural confirmation for the proposed role of a +3 hydrogen bond donor, which is most frequently a threonine or serine residue, must await crystallization of a SecYEG complex that belongs to this category.
The apolar patch and lateral gate polar cluster form a conserved gating motif that regulates the transition between the closed and open conformations of the Sec61 complex. Although the crystal structure of the SecA–SecYEG complex provides insight into one mechanism of channel gating, it seems likely that other cytosolic effectors (ribosome nascent chain complexes or the Sec62–Sec63 complex) will promote channel opening via different contacts than those used by SecA. An important objective for future research will be to understand how the ribosome–nascent chain complex or the Sec62–Sec63 complex promotes separation of the lateral gate and movement of the plug domain.
Materials and methods
Plasmid and strain construction
Standard yeast media (YPAD, YPAEG, and SD), supplemented as noted, were used for growth and strain selection (Sherman, 1991). To evaluate growth rates, yeast strains were grown in YPAEG media (YP media supplemented with adenine, 2% ethanol, and 3% glycerol) at 30°C to mid-log phase. After dilution of cells to 0.1 OD at 600 nm, 5-µl aliquots of fivefold serial dilutions were spotted onto YPAD plates (YP media with adenine and dextrose) that were incubated at 30 or 37°C for 2 d.
Oligonucleotides encoding amino acid substitutions were used as primers together with the template plasmid pBW11 (pRS315 LEU2 SEC61; Wilkinson et al., 1996) in recombinant PCR reactions to produce the sec61 mutants alleles that were subcloned into the LEU2 marked low-copy plasmid pRS315 (Sikorski and Hieter, 1989). The sec61 mutants were characterized in an ssh1Δ yeast strain (RGY400; Cheng et al., 2005). A plasmid shuffle procedure (Sikorski and Boeke, 1991) was used to replace the plasmid pEM324 (pRS316 URA3 SEC61-V5) with the LEU2 marked plasmids encoding the sec61 mutants. In brief, RGY400 was transformed with the pRS315 derivatives encoding mutant sec61 alleles and Leu+ prototrophs were selected on SD (synthetic defined media with dextrose) plates supplemented with adenine, tryptophan, and uracil. Several transformants for each sec61 mutant were streaked onto plates containing 5-fluoro-orotic acid and grown for 2 d at 30°C to select for colonies that had lost the pEM324 plasmid.
The plasmid pEM497 (pRS316 PRC1 URA3; Trueman et al., 2011), which contains the PRC1 gene encoding ppCPY appended with a T7 epitope tag, was further modified by deleting six codons from the signal sequence to obtain pEM820 (ppCPYΔ6-T7). The other prl reporters (ppCPYΔ2-T7 and ppCPYΔ4-T7) were described previously (Trueman et al., 2011). Alternatively, the coding sequence for ppCPY-T7 was modified to replace residues in the signal sequence (T5 and S6; C9 and G10; or T5, S6, C9, and G10) with leucine residues to obtain pEM517, pEM518, and pEM519. The PHO8 gene was amplified by PCR and cloned into pDN317 (pRS316 URA3 DAP2; Ng et al., 1996) using BamHI–XbaI sites to replace the Dap2p coding sequence with the Pho8p coding sequence. DNA encoding the T7 epitope tag was inserted before the Pho8 termination codon to obtain plasmid pEM807. Pho8p expression from pEM807 is under control of the yeast glyceraldehyde 3-phosphate dehydrogenase promoter.
Immunoprecipitation of radiolabeled proteins
All cell labeling, lysis, and subsequent immunoprecipitation of yeast proteins was performed as described previously (Jiang et al., 2008). In brief, yeast were grown to mid-log phase (0.4–0.6 A600) at 30°C in SD media, collected by centrifugation, resuspended in fresh SD media at a concentration of 4 A600 ml−1, and allowed to recover at 30°C for 10 min. For the strain containing the SRP receptor mutation (srp102 K51I; Ogg et al., 1998), yeast cells were initially grown to mid-log phase (0.4–0.6 A600) at 25°C in SD media, and then shifted to 37°C for 3 h before being collected, resuspended in fresh SD media, and allowed to recover at 37°C before pulse labeling. To induce invertase (Suc2p) expression, 4 A600 units of cells were collected by centrifugation and resuspended in 5 ml of SD media containing 0.1% dextrose and incubated for 30 min at 30°C. Cells were pulse labeled for 7 min with 100 µCi/OD Tran-35S-label. Radiolabeling experiments were terminated by dilution of the culture with an equal volume of ice-cold 20 mM NaN3, followed by freezing in liquid nitrogen. Rapid lysis of cells with glass beads and immunoprecipitation of yeast proteins was performed as described previously (Rothblatt and Schekman, 1989). The prl reporters (ppCPYΔ2-T7, ppCPYΔ4-T7, and ppCPYΔ6-T7) and Pho8-T7 were immunoprecipitated using antisera specific for the T7 epitope tag (Covance). CPY, DPAPB, Suc2p, and Gas1p were immunoprecipitated using protein-specific antisera validated in previous studies (Silberstein et al., 1995; Cheng et al., 2005; Trueman et al., 2011). Antisera specific for Suc2p and Gas1p were provided by R. Schekman (University of California, Berkeley, Berkeley, CA) and D. Ng (University of Singapore, Singapore), respectively. Immunoprecipitated proteins were resolved by PAGE in SDS and quantified using ImageQuantL software from scans obtained with the FLA-9000 Fluorescent Image Analyzer (GE Healthcare). Molecular masses of protein products on SDS-PAGE gels were estimated relative to prestained molecular mass markers (Prestained Protein Marker, Broad Range; New England Biolabs, Inc.).
Bioinformatics analysis and generation of sequence logos
Eukaryotic Sec61 sequences were retrieved from the National Center for Biotechnology Information (NCBI) database by searching for Sec61 and SecY. After sequence alignment, duplicate sequences or very closely related sequences from a single species were eliminated to obtain a single Sec61 sequence for each eukaryote. Chloroplast SecY sequences and fungal Ssh1 sequences were excluded from the eukaryotic Sec61 alignment. Eubacterial and archaebacterial SecY sequences were retrieved from the NCBI database by searching for SecY and sequence lengths of 350 to 550 residues. From the obtained sequences, we removed duplicate sequences, severely truncated sequences, and a small number of sequences derived from unclassified eubacteria. SecY2 proteins, which have few in vivo substrates, were also removed from the eubacterial alignment. Alignment of sequences was achieved using MUSCLE, and the eubacterial sequences were sorted based on taxonomy. Sequence alignment and analysis were performed using the following software packages: Jalview, BioEdit sequence alignment editor (version 7.1.3), and BioPython (Cock et al., 2009). Sequence logos were made using WebLogo.
The hydrophobicity of the signal sequence of CPY and derivatives thereof was estimated using an in vivo hydrophobicity scale (Hessa et al., 2007) as implemented using the ΔG server (http://dgpred.cbr.su.se/). A window corresponding to residues 1–20 of CPY or its derivatives was used to apply a similar calculation method for CPY deletion mutants and point mutants. The position-specific hydrophobicity of amino acid substitutions at T87, Q129, and N302 was also calculated using the ΔG server and the amino sequences of TM2, TM3, and TM7 of S. cerevisiae Sec61.
Online supplemental material
Fig. S1 shows a comparison of growth rates of selected sec61 lateral gate polar cluster mutants as determined by serial dilution analysis. Fig. S2 shows hydropathy plots for CPY, several CPY signal sequence mutants, and DPAPB. Fig. S3 shows sequence logos for TM2 and TM3 of eubacterial SecY proteins. Tables S1–S3 show the translocation assay values for the sec61 apolar patch mutants (Table S1), the sec61 lateral gate polar cluster mutants (Table S2), and the sec61 double mutants (Table S3). Tabulated values in these supplemental tables was used to calculate the Δ translocation values that are displayed in Figs. 2 C, 3 (C and D), and 6 (A and B). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207163/DC1.
Acknowledgments
The authors thank Randy Schekman for providing antisera to Sec61p and Suc2p and Davis Ng for providing antisera to Gas1p.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number GM35687.
Footnotes
Abbreviations used in this paper:
- CPY
- carboxypeptidase Y
- DPAPB
- dipeptidylaminopeptidase B
- LGCR
- lateral gate contact residue
- SRP
- signal recognition particle
- TM
- transmembrane
References
- Becker T., Bhushan S., Jarasch A., Armache J.P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., et al. 2009. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 326:1369–1373 10.1126/science.1178535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar A.N., del Val C., Freites J.A., Tobias D.J., White S.H. 2010. Dynamics of SecY translocons with translocation-defective mutations. Structure. 18:847–857 10.1016/j.str.2010.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J.U. 2011. Membrane protein folding: how important are hydrogen bonds? Curr. Opin. Struct. Biol. 21:42–49 10.1016/j.sbi.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Gilmore R. 2006. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat. Struct. Mol. Biol. 13:930–936 10.1038/nsmb1146 [DOI] [PubMed] [Google Scholar]
- Cheng Z., Jiang Y., Mandon E.C., Gilmore R. 2005. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 168:67–77 10.1083/jcb.200408188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25:1422–1423 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Sanders S.L., Feldheim D.A., Schekman R. 1991. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 349:806–808 10.1038/349806a0 [DOI] [PubMed] [Google Scholar]
- du Plessis D.J., Berrelkamp G., Nouwen N., Driessen A.J. 2009. The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284:15805–15814 10.1074/jbc.M901855200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea P.F., Stroud R.M. 2010. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA. 107:17182–17187 10.1073/pnas.1012556107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S.D., Hanley-Way S., Silhavy T.J. 1981. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 23:79–88 10.1016/0092-8674(81)90272-5 [DOI] [PubMed] [Google Scholar]
- Engelman D.M., Steitz T.A., Goldman A. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15:321–353 10.1146/annurev.bb.15.060186.001541 [DOI] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T.A., Hartmann E., Sommer T. 1996. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 15:1482–1494 [PMC free article] [PubMed] [Google Scholar]
- Gumbart J., Schulten K. 2007. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 46:11147–11157 10.1021/bi700835d [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N.M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S.H., von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 450:1026–1030 10.1038/nature06387 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Cheng Z., Mandon E.C., Gilmore R. 2008. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J. Cell Biol. 180:1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A. 1994. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 13:2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B., Rapoport T.A. 1995. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 82:261–270 10.1016/0092-8674(95)90313-5 [DOI] [PubMed] [Google Scholar]
- Junne T., Schwede T., Goder V., Spiess M. 2007. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J. Biol. Chem. 282:33201–33209 10.1074/jbc.M707219200 [DOI] [PubMed] [Google Scholar]
- Ng D.T.W., Brown J.D., Walter P. 1996. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134:269–278 10.1083/jcb.134.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S.-I., Fewell S.W., Kato Y., Brodsky J.L., Endo T. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153:1061–1070 10.1083/jcb.153.5.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S.C., Barz W.P., Walter P. 1998. A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor β-subunit. J. Cell Biol. 142:341–354 10.1083/jcb.142.2.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S., Dreier L., Hartmann E., Kostka S., Rapoport T.A. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 81:561–570 10.1016/0092-8674(95)90077-2 [DOI] [PubMed] [Google Scholar]
- Park E., Rapoport T.A. 2011. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature. 473:239–242 10.1038/nature10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8:785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B.M., Stirling C.J., Rapoport T.A. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 94:795–807 10.1016/S0092-8674(00)81738-9 [DOI] [PubMed] [Google Scholar]
- Potter M.D., Nicchitta C.V. 2000. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 275:33828–33835 10.1074/jbc.M005294200 [DOI] [PubMed] [Google Scholar]
- Rees D.C., DeAntonio L., Eisenberg D. 1989. Hydrophobic organization of membrane proteins. Science. 245:510–513 10.1126/science.2667138 [DOI] [PubMed] [Google Scholar]
- Rothblatt J., Schekman R. 1989. A hitchhiker’s guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 32:3–36 10.1016/S0091-679X(08)61165-6 [DOI] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Boeke J.D. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302–318 10.1016/0076-6879(91)94023-6 [DOI] [PubMed] [Google Scholar]
- Silberstein S., Collins P.G., Kelleher D.J., Gilmore R. 1995. The essential OST2 gene encodes the 16-kD subunit of the yeast oligosaccharyltransferase, a highly conserved protein expressed in diverse eukaryotic organisms. J. Cell Biol. 131:371–383 10.1083/jcb.131.2.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Clemons W.M., Jr, DeMars C.J., Flower A.M. 2005. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J. Bacteriol. 187:6454–6465 10.1128/JB.187.18.6454-6465.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman S.F., Mandon E.C., Gilmore R. 2011. Translocation channel gating kinetics balances protein translocation efficiency with signal sequence recognition fidelity. Mol. Biol. Cell. 22:2983–2993 10.1091/mbc.E11-01-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T., Mori H., Fukai S., Ishitani R., Mori T., Dohmae N., Perederina A., Sugita Y., Vassylyev D.G., Ito K., Nureki O. 2008. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 455:988–991 10.1038/nature07421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W.M., Jr, Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44 10.1038/nature02218 [DOI] [PubMed] [Google Scholar]
- Wilkinson B.M., Critchley A.J., Stirling C.J. 1996. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J. Biol. Chem. 271:25590–25597 10.1074/jbc.271.41.25590 [DOI] [PubMed] [Google Scholar]
- Zimmer J., Nam Y., Rapoport T.A. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 455:936–943 10.1038/nature07335 [DOI] [PMC free article] [PubMed] [Google Scholar]