Abstract
The LKB1 tumor suppressor is a protein kinase that controls activity of adenine monophosphate-activated protein kinase (AMPK). LKB1 activity is regulated by the pseudokinase STRADα and the scaffolding protein MO25α, through an unknown, phosphorylation-independent, mechanism. We describe the structure of the core heterotrimeric LKB1-STRADα-MO25α complex, revealing an unusual allosteric mechanism of LKB1 activation. STRADα adopts a closed conformation typical of active protein kinases and binds LKB1 as a pseudosubstrate. STRADα and MO25α promote the active conformation of LKB1, which is stabilised by MO25α interacting with the LKB1 activation loop. This previously undescribed mechanism of kinase activation may be relevant to understanding the evolution of other pseudokinases. The structure also reveals how mutations found in Peutz-Jeghers syndrome and other cancers impair LKB1 function.
Loss-of-function mutations in the LKB1 tumor suppressor gene cause the rare inherited disease Peutz-Jeghers cancer Syndrome (PJS) in humans [1] and are associated with various sporadic cancers, in particular non-small cell lung cancer (NSCLC) [2]. One prominent function of LKB1 is to ensure that growth and division are coupled to the availability of cellular energy. LKB1 phosphorylates and activates the adenosine monophosphate (AMP)-activated protein kinase (AMPK) when energy levels are low, thereby leading to inhibition of signalling pathways that promote proliferation [3]. The therapeutic effects of AMPK-activating drugs (e.g. metformin) on reducing tumor growth [4], or blood glucose levels [5] are dependent on activation of AMPK by LKB1. Another key role of LKB1 is to control cell polarity, which may be mediated by AMPK [6] as well as a group of AMPK-related protein kinases, including microtubule affinity regulating kinases (MARKs, homologous to the C. elegans kinase Par-1) [7] that are also phosphorylated and activated by LKB1 [8].
In cells, LKB1 is found in a 1:1:1 heterotrimeric complex with the pseudokinase STRAD (STe20-Related ADaptor) [9] and the scaffolding MO25 (MOuse protein 25) [10]. There are two closely related human isoforms of both STRAD (STRADα and STRADβ) and MO25 (MO25α and MO25β) that similarly interact with LKB1 [11]. Unlike the majority of protein kinases, which are regulated by phosphorylation, LKB1 is activated by binding to STRAD and MO25 [12,11] through an unknown, phosphorylation-independent, molecular mechanism. Structural analysis of MO25α reveals a helical repeat, horseshoe-shaped protein, that interacts with the C-terminal WEF (Trp-Glu-Phe) motif of STRADα through a hydrophobic pocket, located on its convex C-terminal surface [13]. The structure of STRADα complexed with MO25α reveals additional interactions between the concave surface of MO25α and the regulatory αC helix of STRADα [14]. STRADα, despite being a catalytically inactive pseudokinase, adopts a closed conformation typical of fully active protein kinases. The closed conformation of STRADα is maintained through its cooperative binding to ATP and MO25α. Mutations that inhibit binding to ATP and MO25α prevent LKB1 activation, suggesting that the active conformational state of STRADα may be required for activation of LKB1 [14].
We report the crystal structure of the LKB1-STRADα-MO25α heterotrimeric complex. We used an insect cell expression system to produce an active core LKB1-STRADα-MO25α heterotrimeric complex, comprising the kinase domain of LKB1 (residues 43-347), complexed with the pseudokinase domain of STRADα (residues 59-431) and full length MO25α (Fig. S2). The crystal structure of the heterotrimeric complex with a catalytically inactive mutant of LKB1 (Asp194Ala, preventing Mg2+ ion binding, but not assembly of the complex, Fig. S2B) in complex with the ATP analogue adenyl-5′-yl imidodiphosphate (AMP-PNP) was solved and refined to 2.65 Å (Table S1). There are two heterotrimeric complexes in the asymmetric unit displaying similar conformations (RMSD = 0.5 Å on 791 Cα atoms). Both STRADα and LKB1 are in complex with AMP-PNP, displaying binding modes typical of other protein kinases (Fig. S3) [15].
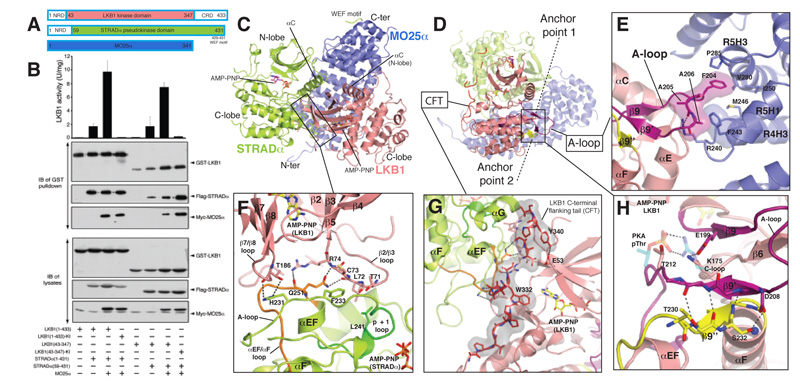
The LKB1 heterotrimer has an overall compact globular shape with considerable interactions between all of the three subunits (Figs. 1A and S4). The pseudokinase domain of STRADα binds to the kinase domain of LKB1. The horseshoe-shaped MO25α acts as a scaffold for assembly of the heterotrimer, by binding both LKB1 and STRADα through highly conserved residues on the concave face of its helical repeats (Figs. 1A and S4B). MO25α binds to STRADα through a large (2930 Å2) interface centred on the regulatory helix αC of STRADα (Fig. 1A). The structure of the STRADα-MO25α complex within the heterotrimer is similar to the binary STRADα-MO25α complex structure [14] (RMSD = 0.5 Å on 529 Cα atoms, Fig. S5), including ordered electron density for the STRADα C-terminal WEF motif interacting with a pocket on MO25α [13,14]. The remaining MO25α concave surface is engaged in contacts (1580 Å2) with the LKB1 activation loop, helix αI and the C-terminus of helix αC (Figs. 1A, S4). The interface between LKB1 and STRADα mainly involves the C-lobe of STRADα and both N- and C-lobes of LKB1 (1840 Å2, Figs. 1C, S4), and is comparable in size to the interaction between LKB1 and MO25α.
FIGURE 1. Overall structure and LKB1-STRADα-MO25α complex interactions.
A) Cartoon representation of the heterotrimeric complex and two bound AMP-PNP molecules are shown in sticks representations (LKB1, yellow carbons; STRADα, magenta carbons). The γ-P for AMP-PNP bound to LKB1 was not visible due to disorder. The WEF motif at the C-terminus of STRADα, for which connectivity could not be unambiguously identified due to disorder of the linkers, is shown in cyan.
B) Detailed view of LKB1-STRADα interaction. STRADα p+1 and αEF-αF loops are coloured green and orange respectively.
C) Interaction of the LKB1 CFTL with STRADα and LKB1 N- and C-lobes. The proline-rich CFTL is coloured red.
D) Detailed view of LKB1-MO25α interaction. LKB1 activation loop is coloured magenta.
E) Detailed view of anchor point LKB1 A-loop interactions. Backbone atoms of residues 208-210 and 230-234 are shown, whereas residues Asp208, Thr230 and Ser232 mutated in PJS are labelled and their side chains displayed. A salt bridge between Glu199 and Lys175 depicted as dashed lines, represent the interaction of LKB1 activation segment with its catalytic loop (C-loop). The corresponding interaction found in PKA (PDBID 1ATP) between the phospho-threonine 197 (pThr) and Arg165 is also shown, with PKA residues represented as transparent sticks (carbon atoms coloured cyan). The typical “activatory” threonine (Thr202) present in LKB1 A-loop is labelled. Secondary structure elements are labelled according to the structure of PKA [15].
Activation of LKB1 is thought to be mediated through a conformational change triggered by binding to STRAD and MO25 [12,11]. The structure of the core LKB1 heterotrimer is consistent with this, as LKB1 lacks phosphorylation of the activation loop, yet adopts an active conformation (Fig. S6). The LKB1 αC helix is rotated into the canonical closed conformation, forming the conserved salt bridge between Lys78 (the so-called VAIK motif in subdomain II) and Glu98 (αC-helix in subdomain III, Fig. S6). This active conformation of LKB1 appears to be achieved through contributions of both STRADα and MO25α.
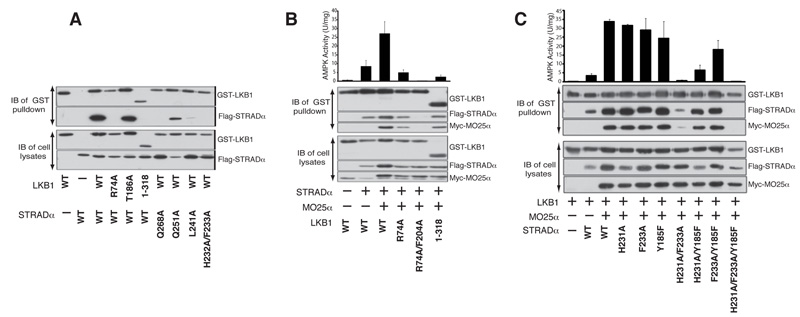
Structural elements on the STRADα C-terminal lobe that normally make up the substrate binding site in active protein kinases (i.e. the αG helix [16] and the p+1 loop [15]), interact with LKB1 (Figs. 1B and S7). Furthermore, the activation loop of STRADα interacts with both N- and C-lobes of the LKB1 kinase domain (Fig. 1B). Mutation of residues in the substrate-binding region of STRADα (Leu241 in the p+1 loop and Gln251 in the αEF-αF loop) inhibit interaction with LKB1, whereas mutation of Gln286 (αG helix) had a moderate effect (Fig. 2A). Mutation of Gln251 (αEF-αF loop) alone or in combination with a mutation on STRADα that disrupts the MO25α-STRADα interaction (Tyr185Phe) [14], suppressed LKB1 activation without affecting complex assembly (Fig. 2B). The reciprocal mutation of Arg74 on LKB1 that hydrogen bonds to Gln251 (Fig. 1B) also impairs the ability of STRADα to activate LKB1 without affecting complex assembly (Fig. 2D). These experiments suggest that binding of STRADα to the β2-β3 loop of LKB1 exerts a conformational effect that promotes LKB1 activation. Interestingly, comparison of the active and inactive structures of CDK2 and EGFR reveals that the β2-β3 loop undergoes large positional shift upon activation (Fig. S8). Furthermore, β2-β3 loop interactions of RAF are important for its dimerization-dependent [17] (Fig. S8C). Interestingly, residues on the STRADα activation loop (His231/Phe233), bind to β7-β8 (C-lobe) and the β2-β3 (N-lobe) of LKB1, respectively (Fig. 1B), perhaps aiding in the positioning of the N- and C-lobes relative to each other. In the absence of MO25α, mutation of His231, Phe233 or both prevented STRADα from binding to LKB1 (Fig. 2A). However, in the presence of MO25α, only the His231-Phe233 double mutant reduced LKB1 activation and complex assembly (Fig. 2B). Combining the His231-Phe233 double mutant with the Tyr185 mutation that disrupts interaction with MO25α [14], resulted in a mutant STRADα that did not form a complex with LKB1 and MO25α (Fig. 2B). These experiments define the regions on STRADα that interact with LKB1 and MO25α and contribute to the assembly of an active LKB1 complex.
FIGURE 2. Characterisation of the LKB1-STRADα-MO25α interactions and LKB1 activation.
A & C) The indicated constructs of GST-LKB1 and Flag-STRADα were expressed in 293 cells in the absence of MO25α. Cells at 36 h post-transfection were lysed and GST-LKB1 affinity purified on glutathione-Sepharose. The purified GST-LKB1 preparation (upper panel) as well as the cell extracts (lower panel) were immunoblotted with the indicated antibodies. Similar results were obtained in three separate experiments.
B, D & F) 293 cells were co-transfected with the indicated constructs of GST-LKB1, Flag-STRADα and Myc-MO25α. Cells at 36 h post-transfection were lysed and GST-LKB1 affinity purified and assayed for the ability to activate heterotrimeric AMPK complex expressed in E. coli as described in the Materials and Methods. Kinase activities are representative of three independent assays carried out in triplicate (error bars represent the SD for a single triplicate experiment). Affinity purified GST-LKB1 preparation (upper panel) as well as cell extracts (lower panel) were immunoblotted with the indicated antibodies.
A common feature of many protein kinase folds is a C-terminal flanking tail (CFT) that interacts with the N-terminal lobe of the kinase [18]. This tail either serves directly as an auto-activatory mechanism or provides a docking site for regulatory interacting partners [18]. LKB1 has a proline-rich CFTL (residues 311-347) that runs along the STRADα-LKB1 interface and interacts with the STRADα helix αG as well as the LKB1 N-terminal lobe (Fig. 1A and C). An LKB1 mutant, lacking part of the CFTL motif (ΔCFTL, residues 1-318) failed to interact with STRADα in the absence of MO25α (Fig. 2C). Mutation of individual residues in or interacting with the CFTL (Trp332/Tyr340/Arg74) did not affect assembly of the LKB1 complex, however LKB1(ΔCFTL) formed a complex with reduced catalytic activity when co-expressed with STRADα and MO25α (Fig. 2D). As mentioned above, mutation of Arg74 (which interacts with the CFTL, but also with STRADα, Fig. 1B and C) on LKB1, abolished interaction with STRADα in the absence of MO25α (Fig. 2C) and reduced catalytic activity of the complex (Fig. 2D). CFTL also contains two phosphorylation sites - Ser325 [19], which may be phosphorylated by ERK [20] and Thr336 is an autophosphorylation site [19]. These sites appear not to directly influence LKB1 catalytic activity [19] or complex assembly [11], but could affect association of LKB1 with substrates or regulators. These results reveal an important role for the CFTL in LKB1-STRADα interactions and LKB1 activity and are suggestive of a potential role for other, as yet unidentified, LKB1 regulators that may utilise this region.
Most protein kinases are activated by phosphorylation of their activation loop, producing a conformation competent for substrate binding [21]. Despite the lack of activating phosphorylation, the LKB1 activation loop is well-ordered (fully defined by electron density) and adopts a conformation typical of loops from active protein kinases (Fig. 1A, and S6). Key to this is the interaction of Phe204 from the LKB1 activation loop with a hydrophobic pocket on the concave surface of MO25α (Fig. 1D). Individual mutation of Phe204 did not affect LKB1 complex formation or activity (Fig. 2D). However, mutation of Phe204 together with Arg74, a residue required for LKB1-STRADα interaction (Fig. 1B and 2C), resulted in LKB1 species that were incapable of forming a heterotrimeric complex (Fig. 2D). Additional interactions occur between Arg240 and Phe243 on MO25α with the backbone of Ala205 and Ala206 of LKB1 that act as a molecular “peg”, to orient the activation loop of LKB1 and stabilise its active conformation (Fig. 1D). Although mutation of both Arg240 and Phe243 did not affect the ability of MO25α to interact with STRADα and LKB1, the resulting complex is inactive, establishing the importance of this interaction in stimulating LKB1 (Fig. 2E). Although MO25α alone is known not to form a stable complex with LKB1 [10,11], in the presence of STRADα, MO25α stabilises the activation loop of LKB1 in an optimal conformation required for phosphorylation of substrates. The position of Thr212 in the LKB1 activation loop is equivalent to that of the activation loop phospho-threonine of protein kinases that require activatory phosphorylation (Fig. 1E). However, Glu199 (β9) replaces the negative charge that would otherwise be provided by the phosphate group and is in hydrogen bonding distance with Lys175 (Fig. 1E). A PJS mutation Glu199Lys, impaired LKB1 catalytic activity, although a less severe PJS mutation (Glu199Gln) did not impair LKB1 activity (Fig. S9).
Dozens of human genes code for protein kinases that lack essential residues in their catalytic machinery and have been termed pseudokinases [22,23]. Some are in fact catalytically competent [24], but others are either incapable of binding ATP [25] or of catalysing phosphoryl transfer [14]. It is possible that STRADα evolved from a catalytically competent protein kinase that phosphorylated LKB1. This notion is supported by the observation that STRADα interacts with LKB1 through structural elements in its C-lobe that are normally used by active protein kinases to bind their substrates (e.g. the p+1 loop/αG helix). More importantly, protein kinases generally need to be in their active conformation to bind their substrates and STRADα appears to adopt an “active” conformation, stabilised through ATP and MO25 to activate LKB1 [14].
In order for LKB1 to phosphorylate AMPK, the active site cleft of LKB1 has to be accessible. Indeed, the structure of the heterotrimer shows that the C-terminal lobe of LKB1 is not engaged in interactions with STRADα or MO25α. Moreover, the region around the γ-phosphate (disordered in our structure) of ATP is solvent exposed in LKB1 (Fig. S7C).
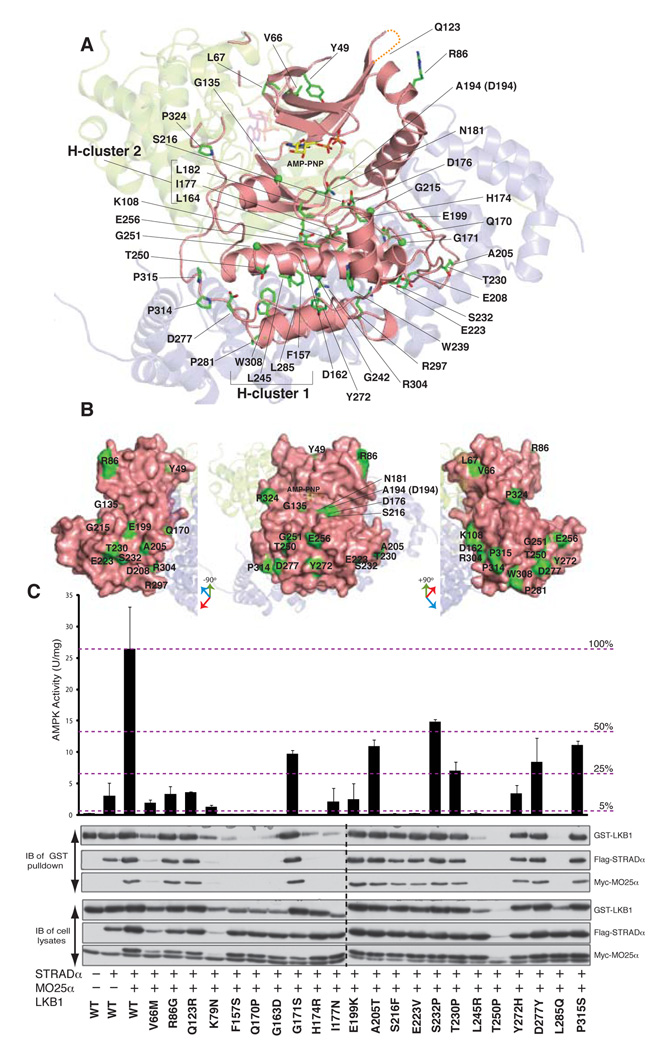
Mutations in the LKB1 gene are the main cause of Peutz-Jeghers syndrome [1] and at least 51 missense mutations have been mapped to the LKB1 kinase domain and the CFT loop (Table S2, Fig. 3 and S10). We have characterised the effects that these mutations have on the ability of LKB1 to form active heterotrimeric complexes with STRADα and MO25α [11] (Fig. S9). The majority of mutations are residues important for the structural integrity of LKB1 (Fig. 3A). There are two hydrophobic clusters, named hydrophobic cluster 1 (Phe157, Leu242, Leu285, Trp308) and hydrophobic cluster 2 (L164, I177 and L182) (Fig. 3A). Many of these mutations resulted in low LKB1 expression levels, and all of these LKB1 mutants were incapable of forming active complexes with STRADα and MO25α (Fig. 3 and Table S2). In addition, at least 10 mutations involve residues required for catalysis or substrate binding (Fig. 3A and B). Although these mutants properly assembled into complexes with STRADα and MO25α, these were devoid of catalytic activity (Fig. S9 and Table S2). Other mutations present in the activation loop (Ala205Thr and Asp208Asn), the αEF-αF loop (Thr230Pro, Ser232Pro) and CFTL region (Pro314His, Pro315Ser and Pro324Leu) did not affect the ability of LKB1 to assemble into active complexes. There are also a number of oncogenic mutations in solvent-exposed residues (Arg86Gly, Gln123Arg, Tyr272His, Asp277Tyr) that do not affect complex assembly or activity (Fig. S9 and Table S2). Thus, out of 51 mutations analysed, 18 formed complexes with STRADα and MO25α that showed LKB1 activity (Table S2). Assuming these are cancer driving rather than passenger mutations, some of these mutations may be involved in interacting with other regulators or substrates of the LKB1 pathway.
FIGURE 3. Map of oncogenic mutations on the LKB1 kinase domain and the CFTL.
A) Location of LKB1 residues that are mutated in PJS and other types of cancer. The CFT region is coloured red and dashed lines represent areas that were not well-defined by electron density.
B) Surface exposed residues that are mutated in PJS and other types of cancer.
In summary, our study reveals how LKB1 is activated. In addition to STRADα binding, MO25α plays a crucial role in stabilizing the LKB1 activation loop in a conformation required for phosphorylation of substrates. Thus, a previously unrecognized role of STRADα is to promote interaction between MO25α and LKB1. This represents a mechanism by which kinases may be regulated allosterically, independent of activation loop phosphorylation. The LKB1 complex structure also shows how cancer mutations affect LKB1 function by impairing complex assembly, catalytic activity and potential interactions with substrates or regulators. Finally, our findings provide insights into how certain pseudokinases may have evolved, by retaining active conformations that allow interactions similar to those by which active kinases bind their substrates.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility, Grenoble, for beam time at station ID14-3 and T. J. Richmond and I. Berger, ETH Zürich, Switzerland for providing the MultiBac expression vectors. This work was supported by a TENOVUS Scotland studentship (EZ), a Wellcome Trust Senior Research Fellowship (DvA), the Medical Research Council (DRA), Cancer Research UK grant C33794/A10969 and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc, Merck KgaA and Pfizer). The coordinates and structure factors have been deposited with the PDB entry 2WTK.
References
- [1].Hemminki A, et al. Nature. 1998;391:184. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- [2].Ji H, et al. Nature. 2007;448:807. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- [3].Shaw RJ. Acta Physiol (Oxf) 2009;196:65. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang X, et al. Biochem J. 2008;412:211. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- [5].Shaw RJ, et al. Science. 2005;310:1642. [Google Scholar]
- [6].Zheng B, Cantley LC. Proc Natl Acad Sci U S A. 2007;104:819. doi: 10.1073/pnas.0610157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baas AF, Smit L, Clevers H. Trends Cell Biol. 2004;14:312. doi: 10.1016/j.tcb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- [8].Lizcano JM, et al. EMBO J. 2004;23:833. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Baas AF, et al. EMBO J. 2003;22:3062. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boudeau J, et al. EMBO J. 2003;22:5102. doi: 10.1093/emboj/cdg490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boudeau J, et al. J. Cell Sci. 2004;117:6365. doi: 10.1242/jcs.01571. [DOI] [PubMed] [Google Scholar]
- [12].Hawley SA, et al. Journal of Biology. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Milburn CC, Boudeau J, Deak M, Alessi DR, van Aalten DMF. Nat. Struct. Mol. Biol. 2004;11:193. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- [14].Zeqiraj E, et al. PLoS Biol. 2009;7:e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Knighton DR, et al. Science. 1991;253:414. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- [16].Dar AC, Dever TE, Sicheri F. Cell. 2005;122:887. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- [17].Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. Nature. 2009 doi: 10.1038/nature08314. In press. [DOI] [PubMed] [Google Scholar]
- [18].Kannan N, Haste N, Taylor SS, Neuwald AF. Proc Natl Acad Sci U S A. 2007;104:1272. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sapkota GP, et al. Biochem. J. 2002;362:481. doi: 10.1042/0264-6021:3620481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zheng B, et al. Mol Cell. 2009;33:237. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nolen B, Taylor S, Ghosh G. Mol. Cell. 2004;15:661. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- [22].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- [23].Boudeau J, Mirand-Saavedra D, Barton GJ, Alessi DR. Trends Cell Biol. 2006;16:443. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- [24].Mukherjee K, et al. Cell. 2008;133:328. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure. 2009;17:128. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.