Abstract
To better understand the response to mitochondrial dysfunction, we examined the mechanism by which Activating Transcription Factor associated with Stress-1 (ATFS-1) senses mitochondrial stress and communicates with the nucleus during the mitochondrial unfolded protein response (UPRmt). We found that the key point of regulation was the mitochondrial import efficiency of ATFS-1. In addition to a nuclear localization sequence, ATFS-1 has an amino-terminal mitochondrial targeting sequence, which was essential for UPRmt repression. Normally, ATFS-1 is imported into mitochondria and degraded. However, during mitochondrial stress, import efficiency was reduced allowing a percentage of ATFS-1 to accumulate in the cytosol and traffic to the nucleus. Our results show that cells monitor mitochondrial import efficiency via ATFS-1 to coordinate the level of mitochondrial dysfunction with the protective transcriptional response.
Mitochondria import ~99% of their proteome through the TOM (Translocase of the Outer Membrane) and TIM (Translocase of the Inner Membrane) complexes (1, 2). The mitochondrial protein-folding environment is maintained by mitochondrial molecular chaperones whose expression levels are coupled to the state of mitochondrial protein homeostasis by a mitochondria-to-nuclear signaling pathway termed the UPRmt (3, 4). Evidence in Caenorhabditis elegans implicates the mitochondrial inner membrane peptide transporter, HAF-1, and the bZip transcription factor ATFS-1 in UPRmt signaling (5).
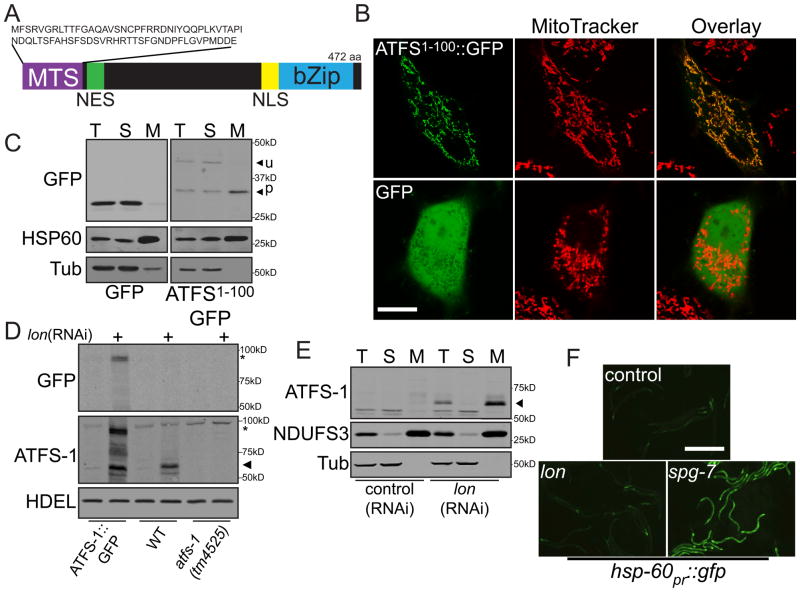
During mitochondrial stress, ATFS-1 accumulates in the nucleus, because of a nuclear localization signal (NLS). A protein sequence prediction algorithm, Mitoprot II, predicted the presence of an amino-terminal mitochondrial targeting sequence (MTS) as well (6) (Fig. 1A). Indeed, amino acids 1-100 of ATFS-1 were sufficient to target green fluorescent protein (GFP) to HeLa cell mitochondria (Figs. 1B & S1). Consistent with cleavage of the MTS, the mitochondrial enriched form of ATFS-11-100::GFP was smaller than the unprocessed form found in the postmitochondrial supernatant (Fig. 1C).
Figure 1. In the absence of stress, ATFS-1 is imported into mitochondria and degraded.
A. ATFS-1 schematic.
B. Photomicrographs of HeLa cells expressing ATFS-11-100::GFP or GFP stained with MitoTracker. Scale bar, 0.25 mm.
C. Immunoblots of HeLa cells expressing GFP or ATFS-11-100::GFP following fractionation into total lysate (T), postmitochondrial supernatant (S) and mitochondrial pellet (M). Longer exposure of the ATFS-11-100::GFP panel was required due to toxicity and weak expression.
D. Immunoblots of atfs-1pr::atfs-1::gfp, wild-type or atfs-1(tm4525) worms raised on control or lon(RNAi). ATFS-1 (➤) and ATFS-1::GFP (*) are marked.
E. Immunoblots of wild-type worms fed control or lon(RNAi) following cellular fractionation. Endogenous NDUFS3 serves as a mitochondrial marker and α-tubulin as a cytosolic marker.
F. Photomicrographs of hsp-60pr::gfp transgenic worms raised on control, lon or spg-7(RNAi). Scale bar, 0.5 mm.
To understand UPRmt regulation in C. elegans, we sought to determine the localization of ATFS-1 in the absence of UPRmt-activation. We were unable to detect endogenous ATFS-1 or the ATFS-1::GFP fusion protein using ATFS-1 or GFP-specific antibodies (Figs. 1D & S2). Additionally, atfs-1pr::gfp worms expressed GFP strongly in all cells indicative of an active promoter, while the ATFS-1::GFP fusion protein was nearly undetectable (fig. S3A). These data suggest that ATFS-1 is rapidly degraded.
We hypothesized that ATFS-1 was degraded by a mitochondrial matrix protease such as the caseinolytic peptidase ClpP or Lon (7). Animals fed lon(RNAi), but not clpp(RNAi) accumulated endogenous ATFS-1 as well as ATFS-1::GFP (Figs. 1D, S3B & S3C). ATFS-1 was absent in lysates from atfs-1(tm4525) worms (Figs. 1D & S3D), which were unable to activate the UPRmt (fig. S4). lon(RNAi) did not affect atfs-1 transcription (fig. S5A). Furthermore, in lon(RNAi)-treated worms, ATFS-1 co-fractionated with a known mitochondrial protein (Figs. 1E & S3E). Unlike spg-7(RNAi), which impairs a mitochondrial protease required for electron transport chain (ETC) quality control and mitochondrial ribosome biogenesis (8), lon(RNAi) did not activate the UPRmt transcriptional reporter hsp-60pr::gfp or impair worm development (5) (Fig. 1F). Therefore, in the absence of UPRmt-activation, ATFS-1 is imported into mitochondria and degraded.
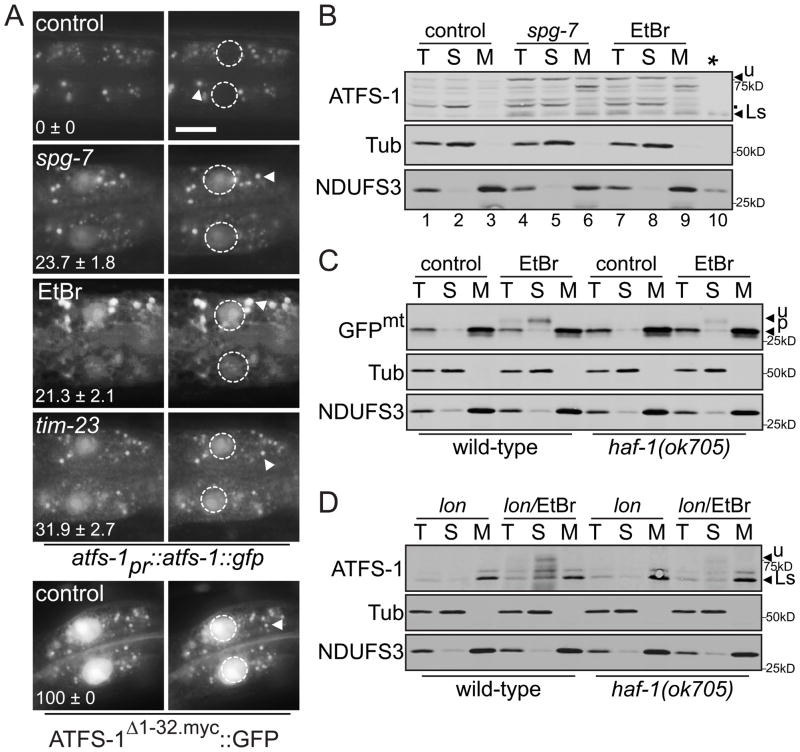
During UPRmt activation, ATFS-1::GFP accumulated in nuclei (Fig. 2A) (5). The predominant form of ATFS-1 that accumulated during spg-7(RNAi) or ethidium bromide (EtBr) treatment was of a higher molecular weight than the form detected in mitochondria of worms raised on lon(RNAi) and was enriched in the postmitochondrial supernatant (Figs. 2B & S4C) suggesting that during UPRmt activation, a percentage of ATFS-1 remains in the cytosol, thus maintaining its MTS.
Figure 2. In the presence of mitochondrial stress, unprocessed ATFS-1 accumulates in nuclei.
A. Photomicrographs of two intestinal cells in atfs-1pr::atfs-1::gfp or hsp-16pr::atfs-1Δ1-32.myc::gfp transgenic animals raised on control, spg-7 or tim-23(RNAi) or 100 μg/ml EtBr with the nuclei outlined (right panels). The punctae (arrowhead) are endogenous autofluorescence from intestinal cell lysosomes. The mean percentage ± SEM of worms with nuclear accumulation of ATFS-1::GFP is indicated (N = 3). Scale bar, 15 μm.
B. Immunoblots of fractionated lysates from wild-type worms raised on control, spg-7(RNAi) or EtBr (100 μg/ml). Lanes 1–9 are 100 μg from the described fractions and lane 10 (*) is 3 μg from the mitochondrial pellet of worms raised on lon(RNAi) for size comparison. Unprocessed and lon(RNAi) stabilized (Ls) ATFS-1 are indicated as are non-specific bands (.).
C. Immunoblots of fractionated extracts from wild-type or haf-1(ok705) worms raised on control(RNAi) in the absence or presence of 30 μg/ml EtBr expressing hsp-16pr::gfpmt.
D. Immunoblots of fractionated extracts from wild-type or haf-1(ok705) worms raised on lon(RNAi) in the absence or presence of 30 μg/ml EtBr expressing hsp-16pr::atfs-1FL.
Mitochondrial toxins such as paraquat, which activated the UPRmt (Fig. 3A), are known to impair mitochondrial import causing the accumulation of MTS containing proteins in the cytosol (9). To determine if a general impairment of import occurs during UPRmt activation, we generated a transgenic strain expressing GFP with a MTS (GFPmt) (10, 11). Because steady-state detection of unprocessed MTS containing proteins is very difficult (12), we expressed GFPmt via the inducible hsp-16 promoter (fig. S6). Only in the presence of UPRmt-activating stress was unprocessed GFPmt detected in the post-mitochondrial supernatant consistent with impaired import (Fig. 2C). Similarly, when ATFS-1 was expressed via the hsp-16 promoter, unprocessed ATFS-1 was only detectable in the postmitochondrial supernatant during UPRmt-activating stress (Figs. 2D & S6). Import was not completely blocked during UPRmt activation because the processed forms of ATFS-1 (revealed by lon(RNAi)) and GFPmt were detected in mitochondria (Figs. 2C & 2D).
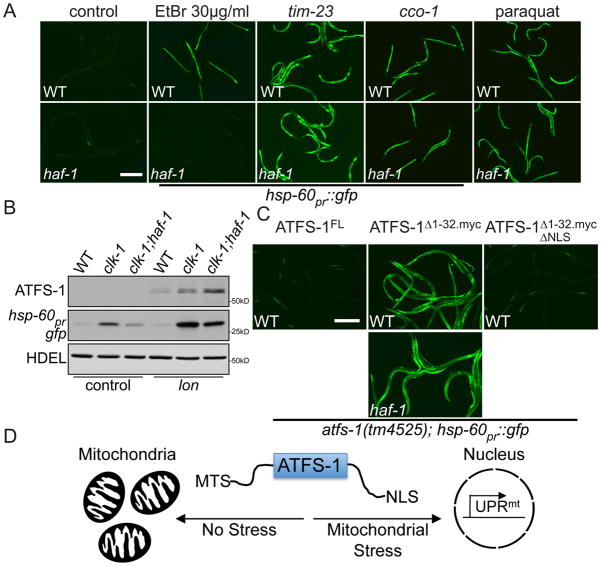
Figure 3. HAF-1 modulates UPRmt signaling by slowing mitochondrial import of ATFS-1.
A. Photomicrographs of wild-type and haf-1(ok705); hsp-60pr::gfp worms raised on control, tim-23, cco-1(RNAi), EtBr or 0.5 mM paraquat. Scale bar, 0.5 mm. The images for cco-1(RNAi) and paraquat were exposed longer because of smaller worm size.
B. Immunoblots of wild-type, clk-1(qm30) or clk-1(qm30); haf-1(ok705) worms raised on control or lon(RNAi).
C. Photomicrographs of atfs-1(tm4525); hsp-60pr::gfp worms expressing wild-type (FL) ATFS-1, ATFS-1Δ1-32.myc or ATFS-1Δ1-32.myc.ΔNLS raised on control(RNAi). The lower panel harbors the haf-1(ok705) allele. Scale bar, 0.5 mm.
D. Schematic illustrating ATFS-1 regulation.
Import into the matrix requires the TOM complex, the TIM23 complex, the ETC and the matrix-localized molecular chaperone mtHsp70 (1). tim-23(RNAi) caused ATFS-1::GFP to accumulate within nuclei (Fig. 2A) and strongly induced hsp-60pr::gfp expression (Figs. 3A & S7). Furthermore, impairment of mtHsp70 (3) or the ETC by the isp-1(qm150) mutation (13) or cco-1(RNAi) also activated the UPRmt suggesting that ATFS-1 responds to mitochondrial protein import perturbations (Fig. 3A).
We next considered how HAF-1, the previously identified UPRmt regulator (5), affected ATFS-1. As expected, haf-1(ok705) worms were unable to induce hsp-60pr::gfp expression caused by the clk-1(qm30) mutation (13) (Fig. 3B) or when raised on 30 μg/ml EtBr (Fig. 3A). However, hsp-60pr::gfp induction caused by mitochondrial stresses that arrest worm development such as 100 μg/ml EtBr or spg-7(RNAi) treatment did not require haf-1 (fig. S8). Additionally, UPRmt activation caused by conditions that directly inhibited mitochondrial import such as tomm-40(RNAi) (fig. S8A), tim-23(RNAi), cco-1(RNAi) or paraquat (9), also did not require haf-1 (Fig. 3A), suggesting that HAF-1 affects UPRmt signaling by modulating the mitochondrial import of ATFS-1. During mitochondrial stress, steady state measurements indicated that more ATFS-1 accumulated within mitochondria of haf-1(ok705) worms as revealed by lon(RNAi) (Figs. 3B & S6B). Thus, in the absence of haf-1, ATFS-1 partitions to mitochondria during stress reducing UPRmt activation.
To more directly examine the effect of haf-1(ok705) on ATFS-1 mitochondrial import efficiency, ATFS-1 was expressed from the inducible hsp-16 promoter (fig. S6). As discussed above, unprocessed ATFS-1 accumulated in the postmitochondrial supernatant during UPRmt-activating stress. However, in haf-1(ok705) worms, much less ATFS-1 was detected in the cytosol (Fig. 2D). Similarly, the slowed import of GFPmt during mitochondrial stress was not observed in haf-1(ok705) animals (Fig. 2C) suggesting that HAF-1 is a general attenuator of mitochondrial protein import during stress and is probably how HAF-1 modulates UPRmt signaling.
To determine if prevention of ATFS-1 import into mitochondria is sufficient to cause ATFS-1 nuclear accumulation and UPRmt activation, we generated a series of transgenic lines that expressed wild-type ATFS-1(full-length (FL)), ATFS-1Δ1-32.myc, an engineered variant of ATFS-1 unable to be imported into mitochondria, and ATFS-1Δ1-32.mycΔNLS which lacked the NLS (fig. S9). Removal of the MTS was sufficient to cause nuclear accumulation of ATFS-1Δ1-32.myc::GFP (Fig. 2A) and expression of ATFS-1Δ1-32.myc caused constitutive expression of hsp-60pr::gfp indicating that impaired mitochondrial import of ATFS-1 is sufficient for UPRmt activation. Activation of hsp-60pr::gfp by ATFS-1Δ1-32.myc did not require haf-1 (Fig. 3C) further supporting a role for HAF-1 in mitochondrial import regulation. Mutating the NLS in ATFS-1 lacking the MTS prevented hsp-60pr::gfp expression indicating that when ATFS-1 cannot be imported into mitochondria, ATFS-1 requires the NLS for UPRmt activation (Figs. 3C & 3D).
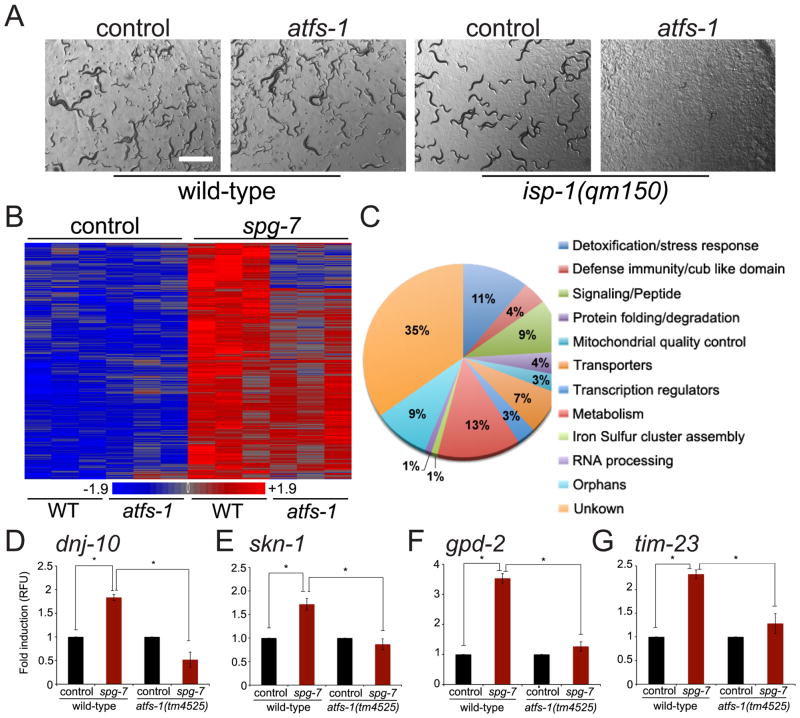
To examine the physiological role of ATFS-1 during mitochondrial dysfunction, wild-type, clk-1(qm30) or isp-1(qm150) (13) worms were raised on control or atfs-1(RNAi). While unstressed worms were unaffected by atfs-1(RNAi), the mitochondrial stressed worms were unable to develop (Figs. 4A & S13A). Because ATFS-1 is regulated by mitochondrial import efficiency, a process linked to mitochondrial function, we sought to identify the entire ATFS-1 mediated response. Transcripts from wild-type and atfs-1(tm4525) worms raised in the presence and absence of stress were compared. A broad transcriptional response totaling 685 genes was induced during mitochondrial stress (Fig. 4B & Table S2), of which 391 required atfs-1 (Table S3).
Figure 4. ATFS-1 mediates a broad and protective transcriptional program.
A. Representative photomicrographs of wild-type or isp-1(qm150) worms raised on control or atfs-1(RNAi). Scale bar, 1mm.
B. Heat map comparing gene expression patterns of wild-type or atfs-1(tm4525) worms raised on control or spg-7(RNAi).
C. Functional categories of the 391 ATFS-1-dependent genes identified by hierarchical clustering.
D–G. Expression levels of dnj-10, skn-1, gpd-2, and tim-23 mRNA in wild-type or atfs-1(tm4525) worms raised on control or spg-7(RNAi) determined by qRT-PCR (N = 3, ± SD, p* (student t-test) < 0.05).
Included in the ATFS-1 program were many genes with roles in protecting against mitochondrial dysfunction (Fig. 4C) including the mitochondrial chaperones dnj-10 (14) (Fig. 4D) and hsp-60 (Fig. S13B). Numerous components involved in reactive oxygen species detoxification required ATFS-1 for induction during stress including the transcription factor skn-1 (15) (Fig. 4E). Several glycolysis genes including gpd-2 (glyceraldehyde-3-phosphate dehydrogenase) were also induced (Fig. 4F) suggesting the UPRmt may contribute to a shift in ATP production from respiration to glycolysis. ATFS-1 was also required for the induction of tim-23 and tim-17 (Figs. 4G & S13C); core components of the TIM23 complex (1).
The presence of a NLS and a MTS in a single transcriptional activator allows the cell to monitor global mitochondrial import efficiency and determine the level of mitochondrial dysfunction. If mitochondria are functioning properly the constitutively synthesized ATFS-1 partitions into mitochondria where it is degraded. As mitochondrial dysfunction increases, mitochondrial import efficiency is reduced favoring translocation of ATFS-1 to the nucleus. Thus, mitochondrial homeostasis is maintained by the stress-dependent partitioning of a transcriptional activator between an inactive state in mitochondria and an active state in the nucleus.
Supplementary Material
Acknowledgments
This work was supported by the Louis V. Gerstner, Jr. Young Investigators Fund, the Alfred W. Bressler Scholar Fund, the Ellison Medical Foundation and the NIH (R01AG040061). We thank Kelly Rainbolt for lon(RNAi) advice, the National BioResource Project, the CGC and the Genomics Facility at MSKCC. The microarray data have been submitted in MIAME-compliant format to the Gene Expression Omnibus database (accession number GSE38196).
Footnotes
Materials and Methods
Figs. S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13
References and Notes
- 1.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donzeau M, Kaldi K, Adam A, Paschen S, Wanner G, Guiard B, et al. Tim23 links the inner and outer mitochondrial membranes. Cell. 2000;101(4):401. doi: 10.1016/s0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- 3.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. Embo J. 2002;21(17):4411. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37(4):529. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241(3):779. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 7.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. Embo J. 2008;27(2):306. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123(2):277. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263(1):107. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 10.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13(4):467. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 12.Ades IZ, Butow RA. The transport of proteins into yeast mitochondria. Kinetics and pools. J Biol Chem. 1980;255(20):9925. [PubMed] [Google Scholar]
- 13.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1(5):633. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 14.Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, et al. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77(2):249. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira RP, Abate J Porter, Dilks K, Landis J, Ashraf J, Murphy CT, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8(5):524. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24(1):34. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 17.Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Assaying metabolic activity in ageing Caenorhabditis elegans. Mech Ageing Dev. 2002;123(2–3):105. doi: 10.1016/s0047-6374(01)00331-1. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427(6975):645. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki CK, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264(5161):891. doi: 10.1126/science.8178144. [DOI] [PubMed] [Google Scholar]
- 20.Hill AA, Hunter CP, Tsung BT, Tucker-Kellogg G, Brown EL. Genomic analysis of gene expression in C. elegans. Science. 2000;290(5492):809. doi: 10.1126/science.290.5492.809. [DOI] [PubMed] [Google Scholar]
- 21.Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6(2):e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174(1):229. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.