Abstract
Changes in gene expression induced by toxic levels of Al were characterized to investigate the nature of Al stress. A cDNA library was constructed from Arabidopsis thaliana seedlings treated with Al for 2 h. We identified five cDNA clones that showed a transient induction of their mRNA levels, four cDNA clones that showed a longer induction period, and two down-regulated genes. Expression of the four long-term-induced genes remained at elevated levels for at least 48 h. The genes encoded peroxidase, glutathione-S-transferase, blue copper-binding protein, and a protein homologous to the reticuline:oxygen oxidoreductase enzyme. Three of these genes are known to be induced by oxidative stresses and the fourth is induced by pathogen treatment. Another oxidative stress gene, superoxide dismutase, and a gene for Bowman-Birk protease inhibitor were also induced by Al in A. thaliana. These results suggested that Al treatment of Arabidopsis induces oxidative stress. In confirmation of this hypothesis, three of four genes induced by Al stress in A. thaliana were also shown to be induced by ozone. Our results demonstrate that oxidative stress is an important component of the plant's reaction to toxic levels of Al.
The mechanism by which Al inhibits plant root growth is not known, despite extensive physiological investigation of Al-treated roots (for review, see Delhaize and Ryan, 1995; Kochian, 1995). A large number of hypotheses for Al toxicity have been suggested, including alteration of the cation-exchange capacity of cell walls (Horst, 1996), changing the membrane potential of the cell, directly affecting uptake of the cations Ca2+ and/or Mg2+, induction of oxidative stress via lipid peroxidation, replacement of Mg2+ or Fe3+ in cellular reactions, interference with signal transduction (Jones and Kochian, 1995), and binding directly to DNA and/or RNA. There are suggestive arguments and indirect evidence supporting each of these possibilities (Delhaize and Ryan, 1995; Kochian, 1995), but to date there is little direct evidence favoring one over the others.
To help elucidate the mechanism of Al toxicity, several groups have examined the molecular response of Al-treated cells. Seven genes that are induced by Al in wheat roots have been cloned (Snowden and Gardner, 1993; Richards et al., 1994). The most highly induced genes included a metallothionein-like protein and two Bowman-Birk protease inhibitors. These genes were also induced by toxic levels of all other metals tested and by physical wounding of roots (Snowden et al., 1995). An acidic PR protein, PR-2, was found to be induced in wheat by Al, as well as by a wide range of other stresses (Cruz-Ortega and Ownby, 1993). More recently, a second PR protein, β-glucanase, was observed to be induced by Al in wheat (Cruz-Ortega et al., 1997). Three additional genes induced by Al in tobacco cell cultures were identified by Ezaki et al. (1995, 1996); they are anionic peroxidase and the auxin-induced genes parA and parB (encoding GST). A common feature of these Al-induced genes is that all are known to be induced by a wide range of other stresses in addition to Al. However, to date they have little in common with what specific cellular stress they are responding to.
Here we identify additional genes induced by Al treatment of Arabidopsis thaliana. The identity of some of these genes, including peroxidase and GST, suggested that there is a strong connection between Al stress and oxidative stress. This connection was confirmed by showing that SOD was induced by Al treatment and by showing that three of the new Al-induced genes were also induced by ozone stress.
MATERIALS AND METHODS
Arabidopsis thaliana cv Columbia seeds were surface sterilized by a 20-min incubation in 1.5% (w/v) sodium hypochlorite containing 2% (v/v) Tween 20 per milliliter as a wetting agent. After three washes with water, seeds (5000 per bottle) were added to 1-L Schott bottles containing 400 mL of low-ionic-strength Ruakura medium (pH 4.3; Snowden et al., 1995). The bottles were aerated in a growth chamber under conditions of 16 h of light (190 μmol m−2 s−1) at 22°C and 8 h of dark at 18°C. Medium was replaced every 1 to 2 d. After 7 d of submerged growth, the seedlings were treated by adding Al2(SO4)3 to a final concentration of 25 μm (50 μm Al3+). Seedlings were harvested at various times with a combination taken from at least two bottles for each sample.
For experiments that required that only the roots be exposed to Al3+, seeds were germinated on black muslin (Putterill et al., 1991) supported by stainless steel mesh (2 mm). The mesh was supported above (and in contact with) 1.5 L of aerated Arabidopsis medium (5 mm KNO3, 2.5 mm KH2PO4, 2 mm MgSO4, 2 mm Ca[NO3]2, 12.5 μm FeEDTA, 7 μm H3BO3, 14 μm MnCl2, 0.5 μm CuSO4, 1 μm ZnSO4, 10 μm NaCl, and 0.1 μm CoCl2, pH 5.8; Haughn and Somerville, 1986) and the seeds were allowed to germinate in a growth chamber under conditions of 16 h of light (90–150 μmol m−2 s−1), 8 h of dark at 20°C. After 5 d the seeds had germinated and the medium was changed to low-ionic-strength Ruakura medium (pH 4.3; Snowden et al., 1995). Nine days later the medium was treated with Al2(SO4)3, exposing the seedling roots to 50 μm Al3+ for a range of times (0, 0.5, 2, and 8 h), and the roots were harvested.
RNA Isolation
For the Al treatments, tissue was ground to a fine powder in liquid nitrogen using a mortar and pestle and the RNA was extracted using the following method. Each 0.5 to 2 g of powdered tissue was added to 5 mL of extraction buffer (300 mm NaCl, 50 mm Tris [pH 8.0], 5 mm EDTA, 5% SDS, and 10 mm β-mercaptoethanol [added just before use]). The solution was vortexed, 0.7 mL of 3 m KCl was added, and the mixture was incubated on ice for 20 min. After the sample was centrifuged at 6000g for 15 min (4°C), 10 m LiCl was added to the supernatant (final concentration of 2 m). The RNA was left to precipitate overnight at −20°C, pelleted by centrifugation at 8000g for 20 min (4°C), and resuspended in 1 mL of water. The RNA was extracted with phenol:ChCl3 (1:1, v/v) and ChCl3 and precipitated with ethanol.
cDNA Library Construction and Screening
The RNA from six bottles of seedlings treated with Al3+ (50 μm) for 2 h was extracted and the mRNA was isolated using an mRNA purification kit (Pharmacia). A cDNA library was constructed using the Superscript Plasmid System (BRL) with clones inserted into NotI and SalI sites in the pSPORT plasmid. Eight filters containing approximately 2000 clones each were differentially screened (Sambrook et al., 1989) with cDNA probes produced using RNA isolated from untreated seedlings and from seedlings treated with Al3+ for 2 h. cDNA was labeled with 32P using a random primers DNA-labeling system (BRL).
Northern Hybridization and Densitometry
Induction of cDNA clones by Al was confirmed by northern hybridization using RNA isolated from submerged seedlings harvested over time. RNA was denatured using glyoxal and DMSO, electrophoresed, and transferred to a Hybond N+ membrane (Amersham) using the alkali procedure (Sambrook et al., 1989). Labeled cDNA inserts were hybridized to membranes (Snowden and Gardner, 1993). The relative amount of RNA in each lane was verified by probing with an asparagus rRNA probe (King and Davies, 1992). Blots were washed, exposed, and then stripped as described by Snowden et al. (1995), with each blot used up to seven times. Analysis of northern hybridizations was performed by scanning autoradiographs with a ScanJet 3C (Hewlett-Packard) and using NIH Image 1.57 software to normalize expression levels using the rRNA level in each lane. Induction of genes was calculated relative to the basal level seen in the control samples.
Sequencing
Nucleotide sequencing was performed using a Catalyst robotic work station and 373 DNA sequencer (Applied Biosystems). Results were edited by visual inspection of the output. Sequence comparisons against the databases used the BLAST algorithm (Altschul et al., 1990), and sequence similarities were calculated using the subroutine GAP in the GCG program (Devereux et al., 1984).
Ozone Treatment
The ozone induction experiments shown in Figure 6 were as described by Sharma and Davis (1994). Briefly, plants were exposed to 300 nL L−1 ozone for 6 h, leaves were harvested at intervals thereafter, RNA was prepared, and northern hybridizations were quantified, using a PhosphorImager and ImageQuant software (Molecular Dynamics), relative to the rRNA signal.
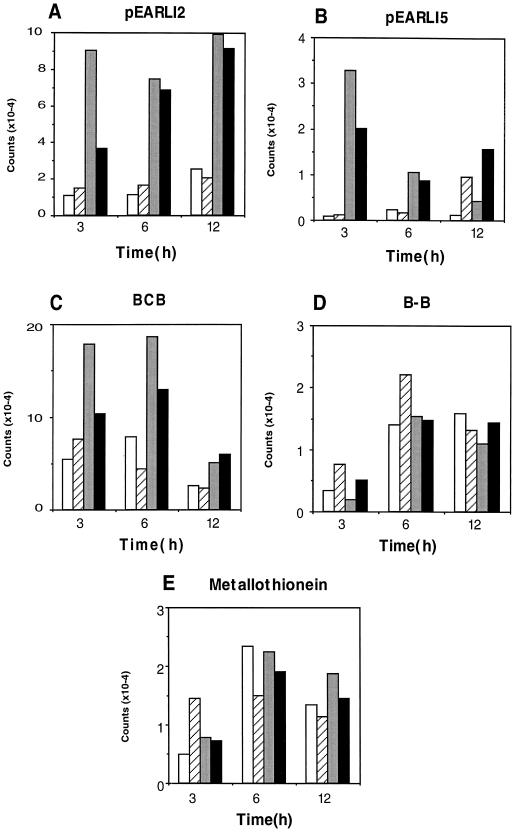
Figure 6.
Gene expression in A. thaliana leaves after ozone stress. The graphs show duplicate results for counts hybridizing to the five probes indicated after 3, 6, and 12 h of exposure to ambient air (white and hatched bars) or to 300 nL L−1 ozone for the first 6 h (gray and black bars).
RESULTS
Isolation of Al-Induced Genes in Arabidopsis
A. thaliana is sensitive to low levels of Al (Putterill et al., 1991; Wheeler et al., 1992; Larsen et al., 1996). Intact seedlings of A. thaliana were grown submerged in low-ionic-strength medium and aerated by bubbling. Under these growth conditions, with seedling densities of approximately 10,000 L−1, 10 μm Al3+ is slightly inhibitory to growth and 50 μm Al3+ is sufficient to block A. thaliana root growth completely (T. Richardson and L. Boyd, unpublished results).
A cDNA library was constructed from mRNA isolated from 7-d-old seedlings that had been treated with 50 μm Al3+ for 2 h. Approximately 16,000 clones were screened differentially for genes induced by Al using labeled cDNA extracted 0 and 2 h after the start of Al treatment. Clones that showed increased expression after 2 h of Al treatment were purified, their Al induction was confirmed by preliminary northern hybridization, and a partial cDNA sequence was obtained. We also identified several clones with expression that appeared to decrease after 2 h of Al treatment.
Nucleotide Sequencing of the Clones
A summary of the clones from which a sequence was obtained is provided in Table I. Where clones could be identified on the basis of sequence identity with known A. thaliana genes, they are referred to by name. The nomenclature pEARLI (for early Arabidopsis aluminium-induced genes) is used for the remaining cDNAs.
Table I.
Characteristics of clones with expression affected by Al
| Clone | Insert Size | Transcript Size | Time of Induction (Repression) | Homology |
|---|---|---|---|---|
| kb | ||||
| pEARLI8 | 2.2 | 2.2 | 15 min–1 h | EST h36573 |
| pEARLI1 | 0.85 | 1.0 | 15 min–2 h | Richards and Gardner (1995) |
| pEARLI2 | 0.9 | 1.4 | 15 min–4 h | None detected |
| Aldolase | 0.6 | 1.6 | 30 min–4 h | EST t43001 |
| pEARLI4 | 2.5 | 2.5 | 2–4 h | Richards et al. (1995) |
| pEARLI5 | 1.9 | 2.0 | 2 h + | Reticuline oxidoreductase (S65550) |
| BCB | 0.85 | 1.3 | 2 h + | z15058 |
| GST | 1.0 | 1.4 | 2 h + | d17672 |
| Peroxidase | 1.3 | 1.3 | 1 h + | x71794 |
| Ala aminotransferase | 1.75 | 2.0 | (8 h) | EST t41718, |
| CAB | 1.2 | 1.0 | (4 h) | x64459 |
Clone nomenclature, time of induction, or repression of expression and homology are as described in the text (clones showing repression are indicated by italics inside parentheses). The approximate size of each cloned insert was measured by mobility of DNA restriction fragments on agarose gels, and the approximate size of the gene transcript was measured by the mobility of the hybridizing band following northern hybridizations.
The sequences of two genes, pEARLI1 and pEARLI4, have been determined in their entirety and reported elsewhere. pEARLI1 belongs to a group of highly conserved, Pro-rich hydrophobic proteins of unknown function (Richards and Gardner 1995), whereas pEARLI4 is a hydrophilic, Pro-rich, repetitive protein, also of unknown function (Richards et al., 1995).
The remaining nine clones were sequenced in one direction from their 5′ end. Four of the sequences show >98% identity over this region to the published A. thaliana sequences for the BCB (van Gysel et al., 1993), GST (Kiyosue et al., 1993), peroxidase (Intapruk et al., 1994), and the CAB (McGrath et al., 1992). The clone referred to as aldolase shows >98% identity to published A. thaliana EST sequence t43001 and 60% amino acid similarity to the pea (Pisum sativum) gene for chloroplastic aldolase (Razdan et al., 1992), whereas Ala aminotransferase shows >98% identity to EST clone t41718 and 46% identity to the first 103 amino acids of the Ala aminotransferase gene of proso millet (Panicum miliaceum; Son and Sugiyama, 1992).
pEARLI5 shows 27% amino acid identity (56% similarity) to amino acids 4 to 131 in the sequence of the reticuline:oxygen oxidoreductase enzyme (also called the berberine bridge enzyme) from California poppy (Escholtzia californica; Dittrich and Kutchan, 1991). This enzyme is involved in the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Of the remaining genes, pEARLI8 is >98% identical to EST clone h36573 of unknown function, whereas pEARLI2 showed no significant similarity to any genes in the database.
Timing of Gene Induction
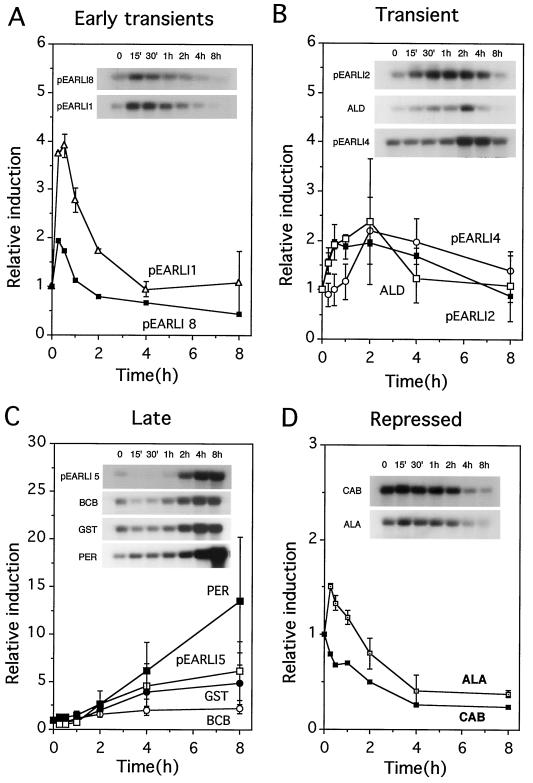
Figure 1 shows the results of a northern hybridization with these clones over an 8-h time course of Al treatment and quantitation of northern analyses using rRNA bands as an internal control. In these and subsequent quantifications, the time zero point for each probe was given an arbitrary value of 1 against which to measure subsequent changes in expression; however, the absolute transcript level for each probe at time zero varied considerably.
Figure 1.
Time course of gene induction by 50 μm Al3+ treatment of submerged seedlings of A. thaliana. A, Transcripts showing a transient peak of induction in the 1st h of Al stress; B, transcripts showing a later transient induction by Al stress; C, transcripts showing late induction with no decrease by 8 h; and D, transcripts with abundance decreasing during Al stress. Northern autoradiographs were used to quantify the levels of hybridizing RNA present, using rRNA as an internal standard (see Methods). For each individual probe, the value for time zero was set at 1, and all subsequent times for that probe were calculated relative to the time zero value. The points shown are the average of two data sets, except for GST (average of four northern hybridizations), BCB (three), and pEARLI8 and CAB (each from the single hybridization shown). Error bars denote sds. The results of one northern hybridization for each probe (all from the same Al stress experiment) are shown as an inset. ALD, Aldolase; PER, peroxidase; ALA, Ala aminotransferase.
The mRNAs detected by five clones were transiently induced early in the Al treatment. Transcripts pEARLI8 and pEARLI1 (Fig. 1A) were induced very rapidly by the treatment, with increases detected by the first sampling time (15 min). Three other transiently induced genes, aldolase, pEARLI4, and pEARLI2, were induced slightly later and for a longer period (Fig. 1B).
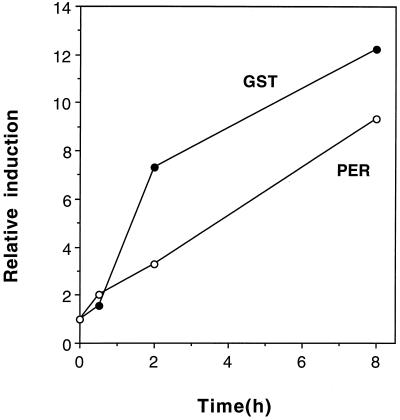
A second category, termed late-induced genes, included four clones that had transcripts that increased after 2 h and stayed induced throughout the 8-h treatment (Fig. 1C). Clones in this category included GST, BCB, peroxidase, and pEARLI5.
Finally, the transcript levels of two genes, CAB and Ala aminotransferase, decreased during Al treatment (Fig. 1D). Ala aminotransferase appeared to undergo a slight transient induction (approximately 1.5 times above background expression) before declining.
Induction of the Late Genes
We carried out a series of additional induction experiments using the four late-induced genes. RNA was isolated from the plants grown in a range of treatment conditions and northern blots were probed sequentially with the four genes.
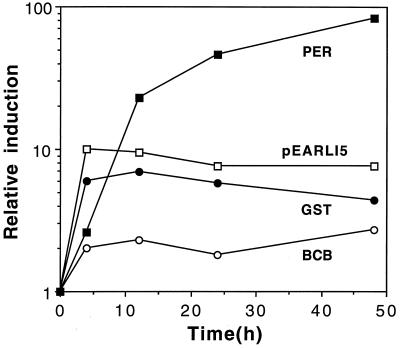
Intact seedlings were grown in submerged culture and induced with Al for a longer time (48 h) to examine the duration of induction. The results are shown in Figure 2. Peroxidase mRNA levels continued to increase over the 48 h of Al treatment, whereas the other three genes appeared to maintain the level of transcript seen at 4 h. The degree of induction was lowest for BCB (2- to 3-fold), intermediate for pEARLI5 and GST (5- to 10-fold), and highest for peroxidase (up to 80-fold induction).
Figure 2.
Induction of BCB, GST, peroxidase (PER), and pEARLI5 during long-term (48 h) exposure to Al. Experimental treatment, symbols, and abbreviations are as for Figure 1.
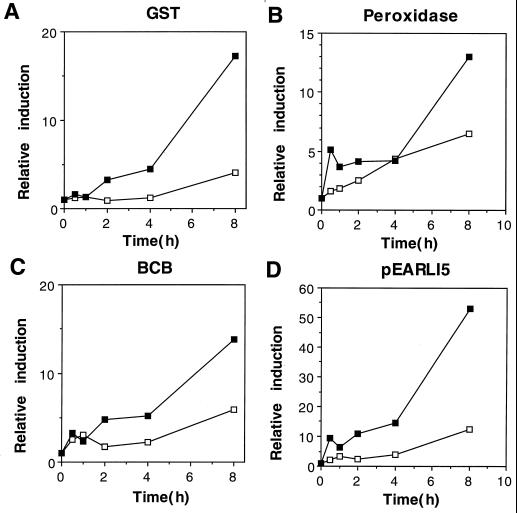
Plants were also grown in the light without Al and in the dark both with and without Al. The aim of these treatments was to confirm that induction was due to the effect of Al and was not controlled by diurnal rhythms or by light. An 8-h light treatment without Al showed no significant difference in transcript levels for any of the genes (data not shown), suggesting that diurnal rhythms did not affect their steady-state mRNA levels. However, all four genes showed induction after 4- to 8-h growth in the dark without Al (Fig. 3, A-D). When the dark-treated seedlings were also exposed to Al, additional induction was seen for all four genes.
Figure 3.
Induction of BCB, GST, peroxidase, and pEARLI5 during extended dark treatment of A. thaliana seedlings, grown with (▪) or without (□) Al3+ (50 μm). Symbols and abbreviations are as for Figure 1.
Plants were grown with their roots submerged in aerated, low-ionic strength, hydroponic medium, but with their tops in air. The roots were then exposed to Al. This experiment was performed to see whether the submerged growth of the seedlings was affecting the induction of the four late genes. However, low yields of root RNA were obtained and interpretable signals were seen only from peroxidase and GST probes (Fig. 4). Both were clearly induced in the roots, with kinetics similar to the induction in submerged seedlings.
Figure 4.
Effect of Al treatment of hydroponically grown A. thaliana roots. The roots of hydroponically grown A. thaliana plants were exposed to 50 μm Al3+. RNA was isolated from plant roots and used in northern hybridizations. Symbols and abbreviations are as for Figure 1.
Induction of Known Stress Genes by Al
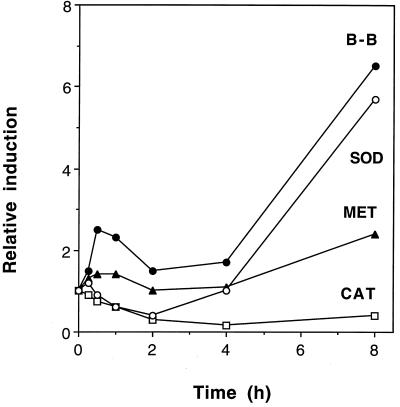
The four genes identified above as being induced at later times by Al all potentially fall into the category of oxidative stress genes (see Discussion). We therefore set out to test whether other genes known to be induced by oxidative stresses were induced by Al. Clones encoding a cytosolic Cu/Zn SOD (EST corresponding to GenBank no. t04184) and catalase (EST corresponding to x64271, which is homologous to maize catalase; Chevalier et al., 1992) were obtained from the Arabidopsis Stock Center at The Ohio State University (Columbus). The clones were labeled and used to probe a northern blot representing a 0- to 8-h time course of Al treatment (Fig. 5). The results showed that SOD mRNA was induced by Al treatment but catalase mRNA levels decreased.
Figure 5.
Time course of Al induction for mRNAs of SOD, catalase (CAT), metallothionein-like protein (MET), and Bowman-Birk proteinase inhibitor (B-B). RNA from the experiment shown in Figure 1 was hybridized to the four probes in northern blots and the bands were quantified relative to rRNA bands, as described for Figure 1.
Genes for metallothionein-like proteins and Bowman-Birk proteinase inhibitors were previously identified as being induced in wheat by Al (Snowden and Gardner, 1993; Richards et al., 1994). We obtained A. thaliana EST clones homologous to these two genes (z26416 and z17665, respectively) and used them to probe northern blots. Transcripts of the A. thaliana Bowman-Birk proteinase inhibitor gene were clearly induced by Al after 8 h (Fig. 5) and also showed signs of an early transient induction. Induction of the metallothionein transcript was minimal, with only a 2-fold induction seen at the 8-h point (Fig. 5).
Induction of Al-Induced Genes by Ozone
Since many of the Al-induced genes correspond to previously identified oxidative stress genes, we next tested whether some of the other genes induced by Al are also induced by ozone stress. The five genes tested included pEARLI2, BCB, and pEARLI5, which we showed above are induced by Al in A. thaliana. We also tested the two A. thaliana EST clones that have homologs that are induced by Al treatment in wheat: metallothionein-like protein and the Bowman-Birk protease inhibitor.
The results (Fig. 6) showed clear evidence for induction of transcripts hybridizing to pEARLI2, BCB, and pEARLI5 in A. thaliana leaves exposed to 300 nL L−1 ozone for 6 h. However, there was no significant change in transcripts hybridizing to the Bowman-Birk protease and metallothionein-like protein clones. The timing of induction by ozone differed considerably compared with the Al results for two of the induced genes. pEARLI2 was only transiently induced by Al (Fig. 1) but remained at elevated levels for at least 6 h after the 6-h ozone treatment (Fig. 6). Conversely, pEARLI5 was stably induced during 4 to 48 h of Al treatment but gave its highest induction during the initial 3 h of ozone treatment, with some reduction thereafter. BCB showed 2- to 3-fold elevated levels compared with control leaves throughout the ozone treatment, similar to its kinetics during Al treatment.
DISCUSSION
We have isolated and characterized a number of genes that appear to be induced or repressed by Al treatment of submerged A. thaliana seedlings. Five genes showed a rapid, transient increase in the levels of their mRNA transcripts, four showed a longer-term increase in their transcript levels, and two genes were repressed by exposure of the seedlings to Al. The proposed identity of some of these genes (based on their sequence) led us to examine the relationship of Al and oxidative stress.
Many Al-Induced Genes Are Also Induced by Oxidative Stress
The four late Al-induced genes isolated here are all induced by oxidative stress. GST (Sharma and Davis, 1994; Conklin and Last, 1995) and peroxidase (Sharma and Davis, 1994) were already known to be induced in plants by ozone treatment, whereas pEARLI5 and BCB were shown to be ozone induced (Fig. 6). The biological role of these latter two genes is unclear. pEARLI5 is homologous to a gene that is induced by infection with a plant pathogen (Dittrich and Kutchan, 1991) and by treatment with methyl jasmonate and fungal elicitors (Facchini et al., 1996). The BCB gene was previously isolated because it is induced by extended dark treatment (van Gysel et al., 1993).
Al stress also induced transcripts hybridizing to cytoplasmic Cu/Zn SOD, another known oxidative stress gene (Sharma and Davis, 1994; Willekens et al., 1994). In our hands, the mRNA for catalase declined during Al stress. Willekens et al. (1994) previously found that two types of catalase gene in Nicotiana plumbaginifolia were induced by ozone, whereas a third declined. It is not known which of the Nicotiana sp. catalase genes (if any) corresponds to the catalase gene studied here. Sharma and Davis (1994) found no effect of ozone on levels of a different catalase mRNA in A. thaliana, using a probe quite distant (79.5% amino acid identity) to the one used here.
These results suggest a link between Al and oxidative stress. The genes that have been shown to be induced by Al are listed in Table II and what is known about their response to ozone, a known inducer of oxidative stress in plants, is summarized. In addition to the five genes above, Phe ammonia lyase, β-glucanase, and pEARLI2 are induced by both stresses. No ozone induction data are available for PR2 and parA. However, PR2 is a defense-related gene, which would be consistent with induction by oxidative stress (Sharma and Davis, 1997). The parA gene is also induced by Al, phosphate starvation (Ezaki et al., 1995), and high auxin (Takahashi et al., 1989). The final two genes in Table II that are induced by Al but not ozone are considered below.
Table II.
Induction of Al stress genes by ozone
| Gene | Induction by Al
|
Induction by ozone | ||
|---|---|---|---|---|
| Wheat | Tobacco | Arabidopsis | ||
| Phe ammonia lyase | Snowden and Gardner (1993) | Sharma and Davis (1994) | ||
| Metallothionein-like protein | Snowden and Gardner (1993) | This paper | This paper | |
| Bowman-Birk proteinase inhibitors | Snowden and Gardner (1993), Richards et al. (1995) | This paper | This paper | |
| PR2 | Cruz-Ortega and Ownby (1993) | |||
| β-glucanase | Cruz-Ortega et al. (1997) | Schraudner et al. (1992) | ||
| Auxin-induced gene (parA) | Ezaki et al. (1995) | |||
| GST | Ezaki et al. (1995) | This paper | Sharma and Davis (1994), Conklin and Last (1995) | |
| Peroxidase | Ezaki et al. (1996) | This paper | Sharma and Davis (1994) | |
| SOD | This paper | Sharma and Davis (1994), Willekens et al. (1994) | ||
| pEARLI2 | This paper | This paper | ||
| BCB | This paper | This paper | ||
| Reticuline:oxygen oxidoreductase (pEARLI5) | This paper | This paper | ||
Genes are listed that have been shown to be induced by Al stress in wheat, tobacco, or Arabidopsis. References for their induction by ozone stress are listed.
In summary, most plant genes so far known to be induced by Al are either known oxidative stress genes or are induced by a range of conditions that are likely to involve oxidative stress. In addition to these commonalities in gene induction, we showed here that CAB mRNA decreased during Al stress. CAB mRNA has been shown to be repressed during both UV-B (Strid, 1993) and ozone stress (Conklin and Last, 1995; Glick et al., 1995).
The Relationship between Al Stress and Oxidative Stress
A simple conclusion that can be drawn from these similarities might be that Al stress corresponds to a form of oxidative stress. One of the suggested mechanisms of Al toxicity is that it causes lipid peroxidation (Gutteridge et al., 1985; Kochian, 1995). In vivo studies have shown that Al treatment increases the activation of several oxidative stress enzyme activities (Cakmak and Horst, 1991). Moreover, Al3+ exacerbates oxidative stress induced by Fe2+ (Ono et al., 1995) and the antioxidant N,N′diphenyl-p-phenylenediamine protected against Al toxicity (Yamamoto et al., 1996). However, the interaction between Fe2+ and Al3+ does not occur in all medium conditions (for discussion, see Yamamoto et al., 1996), and no lipid peroxidation was found in Al-treated soybean cultures (Stass and Horst, 1995). An alternative mechanism for Al toxicity was suggested recently that may also involve oxidative stress. Lukazewski and Blevins (1996) showed that Al treatment impairs ascorbate metabolism, perhaps by inducing B deficiency.
Our results suggest at least two differences between Al stress and ozone stress. First, the Bowman-Birk proteinase inhibitor mRNA was induced by Al stress but not by ozone. The metallothionein-like mRNA may fall into a similar category. Second, the relative levels of induction of the various genes varied between stresses. For example, subjecting A. thaliana to ozone stress led to very high levels of GST mRNA (induced 30- to 60-fold over background) compared with SOD (3- to 4-fold; Sharma and Davis 1994; Conklin and Last, 1995). In contrast, both genes were induced 4- to 8-fold by Al stress.
Two explanations could account for the differences. One is that the oxidative stresses imposed by Al and ozone are different and that genes such as the Bowman-Birk proteinase inhibitors and metallothionein-like proteins form part of an Al-induced oxidative stress response but not of an ozone-induced response. It is known that different oxidative stress genes show variability in their level and time of induction following treatment with different oxidative stresses (Willekens et al., 1994; Sharma and Davis, 1997).
Alternatively, Al may induce a complex of stress genes in A. thaliana, only some of which combat oxidative stress. For example, it is possible that oxidative stress is induced by Al as a secondary response to the block in root elongation, an idea put forward previously by Horst (1996). One aspect of our results that supports this hypothesis is that, with the exception of peroxidase and pEARLI2, there was a 2-h lag before any significant change was seen in mRNA levels of the oxidative-stress-related genes (Fig. 1C). Current evidence suggests that Al blocks root elongation within seconds or minutes and that cell division in the root tip stops immediately thereafter (Delhaize and Ryan, 1995; Kochian, 1995).
Dark Induction of Oxidative Stress Genes
We observed induction of four oxidative-stress-related genes during a prolonged dark treatment. Induction of the BCB gene during extended dark periods has been observed previously (van Gysel et al., 1993). In effect, our “dark” treatment amounted to an unexpectedly long night for these seedlings; the “normal” 8-h dark period was followed by an additional 8.5 h of darkness (harvesting started half an hour into what would have been the light period). We suggest that extended dark treatment at a time when the plant's internal clock is “expecting” light may subject the plant to oxidative stress.
Genes Induced Early during Al Stress
The five genes that are transiently induced very early after Al stress clearly need to be characterized in more detail. Northern blots using RNA from dissected tops and roots from submerged seedlings showed that transcripts of the four late-induced genes all increased in roots, but signals from three of the five early-induced genes were detected only in tops (data not shown). We assume that the bias toward genes expressed in tops occurred because we treated whole seedlings with Al; RNA yields may have been higher from the tops than the roots. Since roots are the biologically relevant organ for Al stress studies, the five early-induced genes were not initially pursued further. The subsequent demonstration that one of the early genes, pEARLI2, is induced in leaves during ozone stress suggests that they may be worthy of further investigation. In situ hybridization using the early transiently induced genes as probes would clearly establish their site of expression and help to show their biological relevance for Al stress. Monitoring the response of the remaining early-induced genes during ozone treatment would determine whether they are also responsive to oxidative stress.
Additional expression studies are also needed to confirm that induction of the early transiently induced genes is due to the Al treatment rather than being part of a diurnal rhythm. A diurnal rhythm may explain the early increase seen with the Ala aminotransferase probe, the mRNA of which is known to be light induced (Son and Sugiyama, 1992). The subsequent shut-down of this transcript, like that of CAB, may reflect the response of a central metabolic pathway to Al stress.
CONCLUSIONS
We have isolated five new genes that are transiently expressed in response to Al treatment of A. thaliana seedlings. An additional six genes were shown to be induced for an extended period during Al stress. The identity of these genes led us to suggest that oxidative stress is a central feature of the plant's response to inhibitory levels of Al. We predict that other oxidative stress genes (e.g. ascorbate oxidase, glutathione peroxidase) will also prove to be induced by Al. It is possible that transgenic plants that overexpress oxidative stress enzymes and that are tolerant to a range of oxidative stresses (Bowler et al., 1991; McKersie et al., 1993; van Camp et al., 1994) may also show increased tolerance to Al.
ACKNOWLEDGMENT
We are grateful to the Arabidopsis Stock Center for supplying DNA probes.
Abbreviations:
- BCB
blue copper-binding proteinCAB chlorophyll a/b binding protein
- EST
expressed sequence tag
- GST
glutathione S-transferase
- PR
pathogenesis-related
- SOD
superoxide dismutase
Footnotes
Funding for this research came from the New Zealand Foundation for Research Science and Technology (grant no. C10310) and from the U.S. Department of Agriculture, Cooperative State Research Service, under agreement no. 96-35100-3214.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bowler C, Slooten L, Vandenbranden S, De Rycke R, Botterman J, Sybesma C, van Montagu M, Inze D. Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J. 1991;10:1723–1732. doi: 10.1002/j.1460-2075.1991.tb07696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I, Horst WJ. Effect of aluminum on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max L.) Physiol Plant. 1991;834:463–468. [Google Scholar]
- Chevalier C, Yamaguchi J, McCourt P. Nucleotide sequence of a cDNA for catalase from Arabidopsis thaliana. Plant Physiol. 1992;99:1726–1728. doi: 10.1104/pp.99.4.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Last RL. Differential accumulation of antioxidant mRNAs in Arabidopsis thaliana exposed to ozone. Plant Physiol. 1995;109:203–212. doi: 10.1104/pp.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ortega R, Cushman JC, Ownby JD. cDNA clones encoding 1,3-β-glucanase and a fimbrin-like cytoskeletal protein are induced by Al toxicity in wheat roots. Plant Physiol. 1997;114:1453–1460. doi: 10.1104/pp.114.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ortega R, Ownby JD. A protein similar to PR (pathogenesis-related) proteins is elicited by metal toxicity in wheat roots. Physiol Plant. 1993;89:211–219. [Google Scholar]
- Delhaize E, Ryan PR. Aluminum toxicity and tolerance in plants. Plant Physiol. 1995;107:315–321. doi: 10.1104/pp.107.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich H, Kutchan TM. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA. 1991;88:9969–9973. doi: 10.1073/pnas.88.22.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki B, Tsugita S, Matsumoto H. Expression of a moderately anionic peroxidase is induced by aluminum treatment in tobacco cells: possible involvement of peroxidase isozymes in aluminum ion stress. Physiol Plant. 1996;96:21–28. [Google Scholar]
- Ezaki B, Yamamoto Y, Matsumoto H. Cloning and sequencing of the cDNAs induced by aluminium treatment and Pi starvation in cultured tobacco cells. Physiol Plant. 1995;93:11–18. [Google Scholar]
- Facchini PJ, Penzes C, Johnson AG, Bull D. Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 1996;112:1669–1677. doi: 10.1104/pp.112.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick RE, Schlagnhaufer CD, Arteca RN, Pell EJ. Ozone-induced ethylene emission accelerates the loss of ribulose-1,5-bisphosphate carboxylase/oxygenase and nuclear-encoded mRNAs in senescing potato leaves. Plant Physiol. 1995;109:891–898. doi: 10.1104/pp.109.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge JMC, Quinlan GJ, Clark I, Halliwell B. Aluminium salts accelerate peroxidation of membrane lipids stimulated by iron salts. Biochim Biophys Acta. 1985;835:441–447. doi: 10.1016/0005-2760(85)90113-4. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;202:430–434. [Google Scholar]
- Horst WJ. The role of the apoplast in aluminium toxicity and resistance of higher plants: a review. Z Pflanzenernahr Bodenk. 1996;158:419–428. [Google Scholar]
- Intapruk C, Takano M, Shinmyo A. Plant Physiol. 1994;104:285–286. doi: 10.1104/pp.104.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Kochian LV. Aluminum inhibition of the inositol 1,4,5-triphosphate signal transduction pathway in wheat roots: a role in aluminum toxicity. Plant Cell. 1995;7:1913–1922. doi: 10.1105/tpc.7.11.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA, Davies KM. Identification, cDNA cloning, and analysis of mRNAs having altered expression in tips of harvested asparagus spears. Plant Physiol. 1992;100:1661–1669. doi: 10.1104/pp.100.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD11 and ERD13) for dehydration-inducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993;335:189–192. doi: 10.1016/0014-5793(93)80727-c. [DOI] [PubMed] [Google Scholar]
- Kochian LV. Cellular mechanisms of aluminium toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:237–260. [Google Scholar]
- Larsen PB, Tai C-Y, Kochian LV, Howell SH. Arabidopsis mutants with increased sensitivity to aluminum. Plant Physiol. 1996;110:743–751. doi: 10.1104/pp.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukazewski KM, Blevins DG. Root growth inhibition in boron-deficient or aluminum-stressed squash may be a result of impaired ascorbate metabolism. Plant Physiol. 1996;112:1135–1140. doi: 10.1104/pp.112.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JM, Terzaghi WB, Sridhar P, Cashmore AR, Pichersky E. Sequence of the fourth and fifth photosystem II type I chlorophyll a/b-binding protein genes of Arabidopsis thaliana and evidence for the presence of a full complement of the extended CAB gene family. Plant Mol Biol. 1992;19:725–733. doi: 10.1007/BF00027069. [DOI] [PubMed] [Google Scholar]
- McKersie BD, Chen Y, de Beus M, Bowley SR, Bowler C, Inze D, D'Halluin K, Botterman J. Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.) Plant Physiol. 1993;103:1155–1163. doi: 10.1104/pp.103.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yamamoto Y, Hachiya A, Matsumoto H. Synergistic inhibition of growth by aluminum and iron of tobacco (Nicotiana tabacum L.) cells in suspension culture. Plant Cell Physiol. 1995;36:115–125. [Google Scholar]
- Putterill JJ, Richards KD, Boyd L, Konigstorfer A, Richardson TE, Gardner RC. Molecular approaches to aluminium tolerance in plants. Curr Top Plant Biochem Physiol. 1991;10:142–147. [Google Scholar]
- Razdan KKR, Heinrikson RL, Zurcher-Neely HA, Morris P, Anderson LE. Biophys. 1992;298:192–197. doi: 10.1016/0003-9861(92)90112-a. [DOI] [PubMed] [Google Scholar]
- Richards KD, Donaldson S, Gardner RC. Nucleotide sequence of pEARLI4 ( L40381) from Arabidopsis. Plant Physiol. 1995;109:1497. [Google Scholar]
- Richards KD, Gardner RC. pEARLI1 ( L40380) an Arabidopsis member of a conserved gene family. Plant Physiol. 1995;109:1497. [Google Scholar]
- Richards KD, Snowden KC, Gardner RC. wali6 and wali7: genes induced by aluminum in wheat (Triticum aestium L.) roots. Plant Physiol. 1994;105:1455–1456. doi: 10.1104/pp.105.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schraudner M, Ernst D, Langebartels C, Sandermann H., Jr Biochemical plant responses to ozone. III. Activation of the defense-related proteins β-1,3-glucanase and chitinasein tobacco leaves. Plant Physiol. 1992;99:1321–1328. doi: 10.1104/pp.99.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma YK, Davis KR. Ozone-induced expression of stress-related genes in Arabidopsis thaliana. Plant Physiol. 1994;105:1089–1096. doi: 10.1104/pp.105.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma YK, Davis KR. The effects of ozone on antioxidant responses in plants. Free Radical Biol Med. 1997;23:480–488. doi: 10.1016/s0891-5849(97)00108-1. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Gardner RC. Five genes induced by aluminum in wheat (Triticum aestivum) roots. Plant Physiol. 1993;103:855–861. doi: 10.1104/pp.103.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Richards KD, Gardner RC. Aluminum induced genes. Induction by toxic metals, low Ca and wounding, and pattern of expression in root tips. Plant Physiol. 1995;107:341–347. doi: 10.1104/pp.107.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son D, Sugiyama T. Molecular cloning of an alanine aminotransferase from NAD-malic enzyme type C4 plant Panicum miliaceum. Plant Mol Biol. 1992;20:705–713. doi: 10.1007/BF00046455. [DOI] [PubMed] [Google Scholar]
- Stass A, Horst WJ. Effect of aluminium on membrane properties of soybean (Glycine max) cells in suspension culture. Plant Soil. 1995;171:113–118. [Google Scholar]
- Strid A. Alteration in expression of defence genes in Pisum sativum after exposure to supplementary ultraviolet-B radiation. Plant Cell Physiol. 1993;34:949–953. [Google Scholar]
- Takahashi Y, Kuroda H, Tanaka T, Michida K, Takebe I, Nagata T. Isolation of an auxin-regulated gene cDNA expressed during the transition from Go to S phase in tobacco mesophyll protoplasts. Proc Natl Acad Sci USA. 1989;86:9279–9283. doi: 10.1073/pnas.86.23.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Camp W, Willekens W, Bowler C, van Montagu M, Inze D, Peupold-Popp P, Sandermann H, Jr, Langebartels C. Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Biotechnology. 1994;12:165–168. [Google Scholar]
- van Gysel A, van Montagu M, Inze D. A negatively light-regulated gene from Arabidopsis thaliana encodes a protein showing high similarity to blue copper-binding proteins. Gene. 1993;136:79–85. doi: 10.1016/0378-1119(93)90450-h. [DOI] [PubMed] [Google Scholar]
- Wheeler DM, Edmeades DC, Christie RA, Putterill J, Gardner RC. Effect of aluminium and pH on relative yield and plant chemical concentrations of 7 dicotyledonous species grown in solution culture at low ionic strength. J Plant Nutr. 1992;15:419–433. [Google Scholar]
- Willekens H, van Camp W, van Montagu M, Inze D, Langebartels C, Sandermann H., Jr Ozone, sulfur dioxide, and ultraviolet B have similar effects on mRNA accumulation of antioxidant genes in Nicotiana plumbaginafolia L. Plant Physiol. 1994;106:1007–1014. doi: 10.1104/pp.106.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Masamoto K, Rikiishi S, Hachiya A, Yamaguchi Y, Matsumoto H. Aluminum tolerance acquired during phosphate starvation in cultured tobacco cells. Plant Physiol. 1996;112:217–227. doi: 10.1104/pp.112.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]