Abstract
Carbohydrates and carbohydrate-containing biomolecules engage in binding events that underlie many essential biological processes. Yet these carbohydrate-mediated interactions are often poorly characterized, due to their low affinities and heterogenous natures. The use of photocrosslinking functional groups offers a way to photochemically capture carbohydrate-containing complexes, which can be isolated for further analysis. Here we survey progress in the synthesis and use of carbohydrate-based photoprobes, reagents that incorporate carbohydrates or their analogs, photocrosslinking moieties, and affinity purification handles. Carbohydrate photoprobes, used in combination with modern mass spectrometry methods, can provide important new insights into the cellular roles of carbohydrates and glycosylated molecules.
I. Introduction
Carbohydrates are key mediators of biological interactions. The glycan components of glycoproteins and glycolipids are critical to the function of these molecules and regulate their ability to interact with binding partners. The interactions of glycosylated molecules play essential roles in diverse areas of biology, notably in immunology, developmental processes, and infectious disease. Despite their essential roles, glycan-mediated interactions are typically low-affinity, often displaying equilibrium dissociation constants in the micromolar or even millimolar range. Thus, isolating and defining the binding partners that engage in glycan-mediated binding interactions is challenging because the labile complexes often fail to withstand the necessary purification steps. One possible solution to this challenge is to use photocrosslinking reagents to introduce a covalent bond between the binding partners. Once covalently constrained, complexes can be isolated and the binding partners characterized using routine analytical methods.
Photocrosslinking functional groups must possess certain key features in order to function in the capture of biologically relevant interactions. These functional groups must be bioorthogonal to cellular functional groups, so that they do not engage in spurious reactions. They should be stable until activated by application of light of an appropriate wavelength. Ideally, photocrosslinking groups will be excited by a long wavelength of light, to minimize cellular damage, and should have a high crosslinking efficiency, with minimal propensity for side reactions. Finally, the size of the photocrosslinking group is important, as larger functional groups have the potential to sterically interfere with the interactions under study. To date, four classes of photocrosslinking groups have been widely used in biological settings: benzophenones, aryl azides, alkyl diazirines, and trifluoromethyl phenyl diazirines. Further discussion of the relative merits and weaknesses of these photocrosslinking groups can be found elsewhere.[1]
This review article describes a variety of photoactivatable probes that have been prepared and used to study carbohydrate biology. Each of the probes contains some type of targeting feature, typically a glycan or glycan analog, that drives association with the carbohydrate-binding pocket of an enzyme or glycan-binding protein. In the figures shown in this article, these interaction motifs are depicted in blue. Each photoactivable probe also contains a light-activated photocrosslinking group, typically a benzophenone, aryl azide, or diazirine, that enables the probe to form covalent bonds with the targeted enzyme or protein. In the figures shown in this article, photocrosslinking groups are diagrammed in red. Many, although not all, of the probes discussed here also include handles that may be used for detection (radiolabels, fluorophores) or affinity purification (biotin, alkynes, azides); these functionalities are indicated in green.
II. Reagents for studying carbohydrate-active enzymes
A. Glycoside hydrolases
Glycoside hydrolases, or glycosidases, are hydrolytic enzymes that cleave glycosidic bonds. Constituting a large enzyme family found in all domains of life, glycosidases vary in glycoconjugate specificity and function.[2–4] Glycosidases play essential roles in biomass degradation, nutrient acquisition, viral and bacterial pathogenesis, and host defense. They also catalyze key steps required in glycoconjugate biosynthesis (for example, N-linked glycan assembly). As described below, photoaffinity probes have potential applications in defining the active sites of glycosidases and in isolating glycosidases with particular glycan specificities from complex mixtures.
Achieving a specific reaction between a small molecule probe and a glycosidase active site is challenging because of the transient nature of glycosidase-substrate binding interactions. In some cases, enzymatically-activated probes, which generate reactive species such as quinone methide, have successfully been used to covalently label the active sites of glycosidases.[5] One intrinsic disadvantage with the use of these latently reaction probes is that, upon activation, the targeting reagent and the reactive species separate, resulting in the potential for non-specific labeling. Mechanism-based inactivating probes based on fluorosugars[6,7] are another powerful approach, although their utility can be limited by slow reaction kinetics or incomplete knowledge of reaction mechanism. Photocrosslinking probes offer another alternative to the existing approaches. Since the recognition (carbohydrate) element remains attached to the photoreactive moiety throughout the crosslinking process, photocrosslinking probes are expected to result in less non-specific labeling than enzymatically-activated probes. In addition, once photoactivated, photocrosslinking probes have very rapid reaction kinetics, so high efficiency labeling can be achieved. A limitation of most photocrosslinking probes is that they are not enzyme-activated, so they may also label glycan-binding proteins, rather than being selective for glycosidases.
Below, we describe examples of photocrosslinking reagents designed to selectively label glycosidases, and the utility of these reagents in defining enzyme active sites and in isolating glycosidases with specificity for particular glycans. Key features of these reagents include the enzyme-binding motifs, typically glycans, the selection of photoreactive functional group, and the choice of labeling or affinity probes. In many cases, glycosidases have tightly-packed active sites that do not allow space for the affinity label. Thus, ideal photoaffinity group and reporter group placement through a well-designed linker are important factors in the successful use of these reagents.
1. Bacterial β-N-acetylglucosaminidase, NagZ
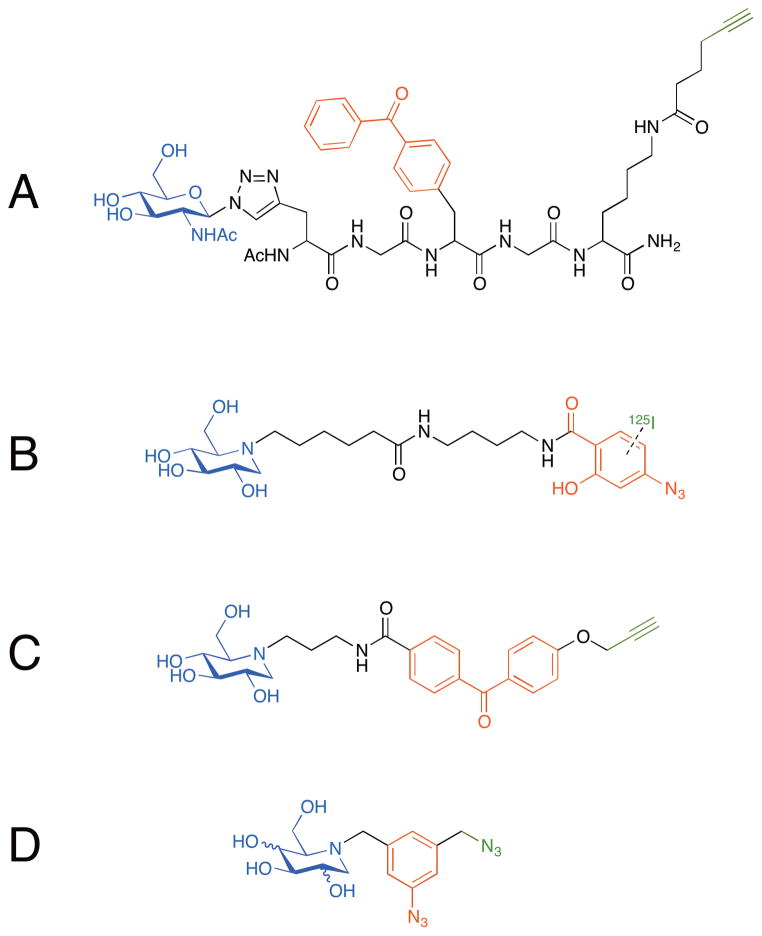
Orth et al. reported a modular synthesis of photoprobes designed for labeling and affinity purification of bacterial glycosidases.[8] Using the 1,3-dipolar Huisgen cycloaddition reaction, they coupled β-azidosugars (glucose, galactose, or N-acetylglucosamine) through a non-hydrolyzable linkage (triazole) to a peptide linker modified with a photocrosslinker (benzophenone) and an alkyne handle to be used for subsequent isolation (Figure 1A). Using these three carbohydrate probes, the researchers confirmed the glycan specificity of recombinant NagZ, a β-N-acetylglucosaminidase from E. coli: only the N-acetylglucosamine probe was efficiently crosslinked to the enzyme. While this initial report did not go on to investigate whether the photoprobes can be used in a more complex mixture for glycosidase isolation and identification, the modular route by which these reagents were assembled could be generally useful since complex carbohydrates with an azido functional group at the reducing end are readily available and could be assembled to the similar type of photoprobes
Figure 1.
Photoaffinity probes for labeling glycosidases. (a) An N-acetylglucosamine probe with a benzophenone photocrosslinker and an alkyne purification handle on a peptide linker via a non-hydrolyzable (triazole) linkage. This probe labels recombinant NagZ while similar glucose and galactose-containing probes do not. (b) A radiolabeled (125I) 1-deoxynojirimycin probe with an aryl azide photocrosslinker. This probe preferentially labels glucosidase I over glucosidase II. (c) A 1-deoxynojirimycin probe with a benzophenone photocrosslinker and an alkyne handle. This probe fluorescently labels the lysosomal glucocerebrosidase in the presence of mammalian cell lysates. (d) Imino-sugar based probes with a bifunctional azidomethyl aryl azide for photocrosslinking and conjugation. These probes are designed for isolation of glycosidases from complex mixtures.
2. Glucosidases
Glucosidases, enzymes that hydrolytically release glucose residues, are found in a variety of organisms and play diverse functional roles. For example, in mammals, glucosidase I is a regulator of N-linked glycan maturation, while lysosomal β-glucosidase catalyzes an essential step in glucosylceramide degradation. Termites and some microorganisms rely on glucosidases to degrade the cellulose components of plant cell walls. 1-Deoxynojirimycin, an iminosugar initially discovered in the root bark of mulberry tree,[9] and its N-alkyl derivatives are potent glucosidase inhibitors. As a non-hydrolyzable analog of glucosidase substrates, 1-deoxynojirimycin functions as an excellent affinity probe for glucosidases. In 2004, Romaniouk et al. synthesized a probe that contained deoxynojirimycin conjugated to an aryl azide photocrosslinker with an 125I label (Figure 1B). In an experiment conducted in bovine mammary gland extract, this reagent enabled selective labeling of a 24 kDa peptide containing the potential active site of glucosidase I.[10] Also, consistent with the inhibitory activity of 1-deoxynojirimycin, this probe showed selective labeling of glucosidase I over glucosidase II. While the crosslinked complex could be visualized by autoradiography, the radiolabeled probe did not have an affinity handle to enable isolation and identification of the crosslinked species. More recently, Pieters’s group reported another deoxynojirimycin-based probe containing both a photocrosslinking functionality and handle for fluorescent labeling.[11] These probes were prepared by conjugating propargylated photocrosslinkers to 1-deoxynojirimycin or glucose; the propargyl group was employed for fluorescent labeling through a 1,3-dipolar cycloaddition with a fluorescein-azide adduct. The benzophenone-conjugated deoxynojirimycin (Figure 1C) yielded efficient labeling of a lysosomal glucocerebrosidase in the presence of HeLa cell lysate, while a glucose analog suffered from poorly labeling efficiency, likely because the probe was hydrolyzed by the enzyme. Taken together, these results demonstrate that sugar analogs can provide useful starting points for photoaffinity probes.
3. Proteomic profiling of glycosidases
One important application for glycosidase probes is to isolate and identify glycosidases present in complex mixtures. To achieve this, probes must achieve efficient and selective enzyme labeling, as described above, but also contain an affinity tag that enables effective purification of crosslinked species. With the goal of identifying exoglycosidases, glycosidases that remove terminal monsaccharides, Gandy et al. prepared a series of iminosugar-based photocrosslinkable probes.[12] They synthesized these probes by coupling 1-deoxynojirimycin, 1-deoxymannozirimycin, or 1-deoxygalactonojirimycin to a bifunctional azidomethyl aryl azide (Figure 1D). These probes could be first photocrosslinked to a target glycosidase through the aryl azide and then conjugated, through Staudinger ligation, to a phosphine-FLAG reagent, to be used for subsequent isolation of the crosslinked complex using an anti-FLAG antibody. Model reactions with probes based on each of the three iminosugars demonstrated successful crosslinking to purified enzymes. Furthermore, the 1-deoxynojirimycinbased reagent was successfully used to isolate two proteins from a bacterial lysate. The molecular weights of the isolated species were consistent with known exo-α-glycosidases of the source bacterium, although further verification of identity of the protein bands was not described in this initial report.
B. Enzymes involved in carbohydrate metabolism
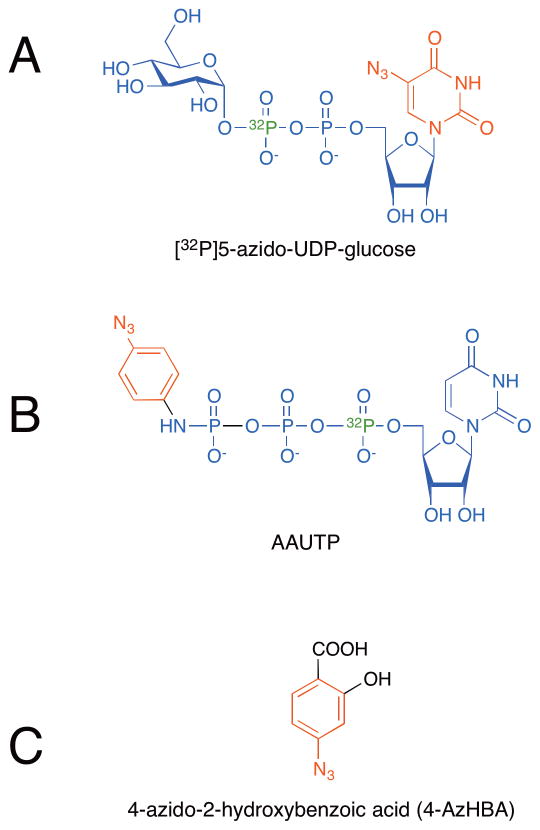
There are relatively few reports of the use of photocrosslinking reagents to study enzymes involved in carbohydrate metabolism. In one recent example, Huh et al. reported use of [32P]5-azido-UDP-glucose (Figure 2A) to identify the UDP-glucose binding site of human UDP-glucose dehydrogenase (UGDH).[13] This approach employed a compact photocrosslinking probe, in which the modified base, 5-azidouridine, functioned as the photocrosslinking moiety, while the phosphates provided an affinity purification handle. The 32P radiolabel provided an additional reporter to track the formation and isolation of the crosslinked adduct. After photoaffinity labeling of recombinant human UGDH with [32P]5-azido-UDP-glucose and subsequent tryptic digest of the enzyme, a labeled peptide was isolated through phosphate-based affinity capture. The site of photoaffinity labeling was determined by sequencing the peptide and confirmed by additional experiments conducted with point mutants of UGDH. This elegant use of a modified nucleotide sugar provided new information about the ligand-binding site of UGDH; however, the approach is likely limited to cases where purified enzyme is available since complex mixtures will contain other many other phosphopeptides that would complicate analysis.
Figure 2.
Photoaffinity probes for labeling carbohydrate-binding enzyme active sites. (a) [32P]5-azido-UDP-glucose was used to identify the nucleotide sugar binding sites in human UDP-glucose dehydrogenase (UGDH) and in glucosyltransferases. (b) A radiolabeled (32P) photoaffinity probe based on UTP with an azido aniline photocrosslinker (AAUTP). This probe was used to confirm the membrane orientation of ER-resident glycosyltransferases, as well as to label the GlcNAc-phosphatidylinositol synthetic complex. (c) A surrogate acceptor substrate, 4-azido-2-hydroxylbenzoic acid (4-AzHBA). This probe was used to label the active site of human UDP-glucuronosyltransferase 1A10.
C. Glycosyltransferases
Glycosyltransferases are enzymes that form new glycosidic bonds by transferring monosaccharides from activated nucleotide sugar donors, such as UDP-glucose, to acceptor molecules. Glycosyltransferases are found in all kingdoms of life and produce a diverse area of glycosylated species, including bacterial natural products, peptidoglycan, glycolipids, glycoproteins, glycosaminoglycans, glycogen, and cellulose. Photoaffinity probes based on nucleotide sugars have been used to study various glycosyltransferases.
In particular, the [32P]5-azido-UDP-glucose probe (Figure 2A and described above) has been widely used to identify the nucleotide sugar binding sites of glucosyltransferases. In 1990, Frost et al. used [32P]5-azido-UDP-glucose to photolabel red beet (Beta vulgaris L.) storage tissue. They identified a 57-kDa polypeptide proposed to be the UDP-glucose-binding subunit of callose ((1,3)-β-glucan) synthase.[14] In the same year, Lin et al. used [32P]5-azido-UDP-glucose to identify the UDP-glucose-binding subunit of cellulose synthase in Acetobacter xylinum.[15] More recently, [32P]5-azido-UDP-glucose was used to identify the UDP-glucose binding region of glycogenin, the autoglucosyltransferase responsible for initiating glycogen biosynthesis in animals. Glycogenin isolated from purified rabbit skeletal muscle proteoglycogen was photolabeled with [32P]5-azido-UDP-glucose, then proteolyzed. Sequence analysis identified two labeled fragments corresponding to glycogenin’s UDP-glucose-binding domain.[16]
The same [32P]5-azido-UDP-glucose probe was used to determine the membrane orientation of active sites of ER- and Golgi-resident glycosyltransferases.[17] Drake et al. performed photoaffinity labeling of microsomal preparations using the [32P]5-azido-UDPglucose reagent to determine whether glycosyltransferase active sites were in the interior or exterior of microsomal vesicles. UDP-glucose:dolichol phosphate glucosyltransferase (Glc-P-Dol synthase), an enzyme involved in the early steps of N-linked glycan biosynthesis, was labeled equally well in intact versus disrupted vesicles, indicating that the catalytic domain was located on the outside of the microsomal vesicles, corresponding to a cytoplasmic orientation in the originating ER membrane. In contrast, glucuronosyltransferases, which transfer glucuronic acid to a variety of substrates, were more efficiently labeled in disrupted vesicles, indicative of a lumenal orientation for the catalytic domains of these enzymes. Similarly, Rancour and Menon reported enzyme orientation studies using a novel photoaffinity probe based on UTP. This probe contains the 32P radiolabel at the α-phosphate and an azidoaniline photocrossinker attached to the γ-phosphate (AAUTP, Figure 2B). This reagent was used to confirm the membrane orientation of ER-resident enzymes and has the advantage that it does not include modifications to the uridine portion of the molecule, which is a key recognition element for many glycosyltransferases.[18] The AAUTP probe has also been used to selectively label and identify the UDP-GlcNAc binding subunit of the GlcNAc-phosphatidylinositol synthetic complex.[19]
Another approach to mapping glycosyltransferase active sites involved the use of a photocrosslinking reagent as a surrogate acceptor substrate, which binds the active site of the enzyme with high affinity. Xiong et al. used the novel photocrosslinker 4-azido-2-hydroxybenzoic acid (4-AzHBA, Figure 2C) to label the active site of human UDP-glucuronosyltransferase 1A10.[20] Subsequent mass spectrometry analysis of the trypsinized protein identified the crosslinked peptide (EFMVFHAQWK) and was followed by analysis of site-directed mutants to determine that two phenylalanine residues (Phe90 and Phe93) are associated with acceptor binding in this enzyme.
D. Sugar transporters
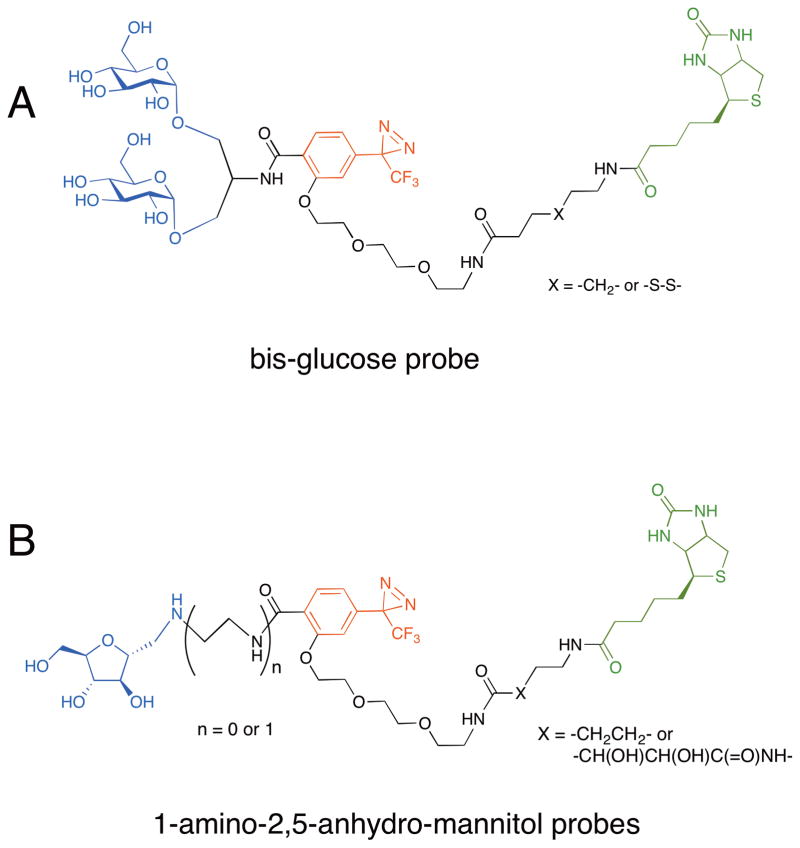
Energy-independent transport of hexoses, including glucose, galactose and fructose, across membranes is mediated by a family of glucose transporters (GLUTs). The GLUT family comprises five primary members, as well as additional isoforms. Over the past 25 years, the Holman group has reported a variety of photocrosslinking carbohydrate reagents, many of which have been employed in the study of GLUTs. The most advanced probes can be added to living cells, where they crosslink to extracellularly oriented GLUTs and can be used for isolation and characterization of these transporters. In early studies, this group reported bis-mannose based photoaffinity probes for labeling erythrocyte and adipocyte hexose transporters, and have shown the usefulness of radiolabeled bis-glycoside photoaffinity probes to crosslink glucose transporters in erythrocyte or adipocytes.[21,22] In 2001, the group constructed a bis-glucose probe that includes a 3-trifluoromethyl-3-phenyl-diazirine crosslinker and a biotin tag for purification (Figure 3A).[23] They incubated the bisglucose probe with rat adipocytes and irradiated the cells with UV light. After precipitating the biotinylated proteins from the lysed cells, they found the GLUT4 was isolated, although the possible existence of copurifying proteins was not investigated. The Holman group has also prepared probes selective for GLUT5, which permits transport of D-fructose. These probes incorporate 1-amino-2,5-anhydro-mannitol, a potent inhibitor of GLUT5, an aryl diazirine, and biotinylated linker (Figure 3B).[24,25] Interestingly, the affinities of the probes were significantly enhanced when there was an ethylenediamine spacer between the furanose and the biotinylated photoaffinity group. The best probe was capable of capturing GLUT5 from CHO cells expressing this protein. While all of these probes seem to be effective at capturing the intended sugar transporter targets, their selectivities have not yet been assessed.
Figure 3.
Photoaffinity probes for labeling glucose transporters. (a) A bis-glucose probe with an aryl diazirine crosslinker and a biotin purification tag. This probe successfully captured GLUT4 transporter expressed in rat adipocytes. (b) 1-Amino-2,5-anhydro-mannitol probe with an aryl diazirine crosslinker and a biotin purification tag. This probe was used to captured the GLUT5 transporter expressed in CHO cells.
III. Reagents for capturing glycan-binding interactions
Glycans, which may be attached to proteins or lipids, are critical mediators of macromolecular recognition events, playing key roles in biological processes such as cell adhesion or endocytosis. However, the transient nature of these interactions (low binding affinities and rapid dissociation rates) often precludes their characterization, since biologically relevant complexes may not be able to withstand traditional purification. As will be described in the following section, the utilization of photocrosslinkers that can create a covalent bond between glycoconjugates and their binding partners offers a powerful strategy to stabilize glycoconjugate complexes and thereby enable their characterization.
A. Important early examples
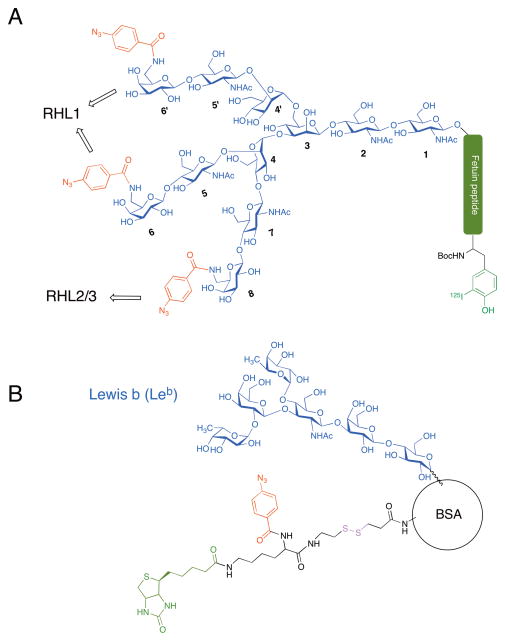
In the mid-1980s, Lee and coworkers reported the preparation of photoaffinity glycopeptides that were used to interrogate the binding properties of the asialoglycoprotein receptor. Asialoglycoprotein receptor is abundant in mammalian livers and named for its function as a lectin that interacts with galactose residues on desialylated glycoproteins.[26–28] The photoaffinity glycopeptides used in these studies were prepared from naturally occurring sources. In one such study, the authors isolated an asialoglycopeptide from bovine fetuin. This peptide contained a single N-linked glycan with a triantennary structure, upon which they introduced a single 6-amino-Gal residue at one of three positions, for subsequent incorporation of an aryl azide photocrosslinking functionality as well as a 125I label for detection (Figure 4A).[29] Using these asialoglycopeptide ligands, the authors were able to demonstrate that the regiochemical placement of the photocrosslinker resulted in differential labeling of the major (RHL1) and minor (RHL2/3) asialoglycoprotein receptor subunits, thus defining the geometry of the galactose residues in the binding site. In subsequent studies, the same group developed “glycoprobes” for a more facile synthesis of reagents for lectin labeling in complex biological samples.[30,31] These glycoprobes incorporated a digoxin tag between the photocrosslinking group and the variable glycan chain so that a general commercially available antibody could be used for visualization of the crosslinked species formed upon photoirradiation. Using this system, the authors were able to successfully label all 3 subunits of the asialoglycoprotein receptor from preparations of rat liver membranes.
Figure 4.
Early examples of photoaffinity probes designed to label glycan-binding proteins. (a) A desialylated fetuin glycopeptide differentially labeled with an aryl azide photocrosslinking agent on the terminal galactose residue of each antenna and an 125I-label on the fetuin peptide. (b) A semisynthetically-prepared glycoprotein modified with the fucosylated Lewis b (Leb) blood group antigen. This reagent was used to biotinylate its binding partner, BabA, via an aryl azide photocrosslinker conjugated through a cleavable disulfide linker to allow for subsequent enrichment.
Another important early example comes from the work of Borén and coworkers. They identified the adhesion factor BabA present on the surface of the pathogenic bacteria Helicobacter pylori through the use of a semisynthetically prepared glycoprotein with both an affinity-tag and photocrosslinker handle.[32] H. pylori is the causative agent for peptic ulcer disease. Adherence of this bacterium to the human gastric epithelium for subsequent colonization was known to be mediated by a fucosylated Lewis b (Leb) blood group antigen. Therefore, the authors conjugated the Leb glycan to human serum albumin, which was also modified with a disulfide-linked, multifunctional reagent containing a biotin purification handle and an aryl azide photocrosslinker. Thus, interactions between Leb glycans and their binding partners on the surface of bacteria brought the bacteria in proximity for efficient photocrosslinking to the biotinylated tag. Subsequent reduction of the disulfide bond liberated the albumin conjugate, freeing the now biotinylated bacterial receptor (Figure 4B). Biotinylation enabled detection of the receptor by immunoblot with peroxidase-streptavidin or purification of the protein using streptavidin-coated magnetic beads. Using N-terminal sequencing, the purified receptor was identified as BabA. This process was termed receptor activity-directed affinity tagging or ReTagging, and is an important early example of the power of photocrosslinking approaches applied in a cellular setting. Moreover, this example illustrates important challenges such as the utility of an affinity tag for effective purification and the requirement for ample sample to facilitate identification. It is also noteworthy that BabA’s identity was determined by N-terminal sequencing. In the intervening years since this study, advances in mass spectrometry have enabled identification of proteins with much smaller quantities of sample, suggesting that approaches like ReTagging are worth revisiting since they may now be even more effective and easier to implement.
B. Probe optimization
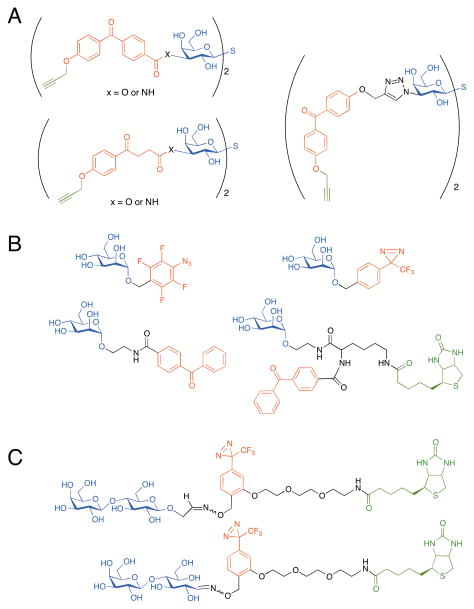
The design of effective photocrosslinking reagents is greatly aided by a priori knowledge of the structure of the glycan-binder/glycan pair to be studied. However, even in cases where this information is available, optimization is often essential. The process of probe optimization is demonstrated in work by the Pieters group aimed at developing glycan-based photoprobes to selectively label galectin-3 in different sample types (i. e., different cancerous states). The intended application for these probes is in the determination of galectin-3 levels in clinical tissues for diagnosis and prognosis assessment.[33] Recent work has focused on a new series of photoprobes based on a thiodigalactoside inhibitor of galectin-3.[34] In this work, the linker connecting the thiodigalactoside and the photoaffinity label was varied and the effects on binding affinity and specificity of labeling the target protein were assessed. Acetophenone or benzophenone photocrosslinking groups were connected through either an ester, amide, or triazole linkage to the C3 positions of the thiodigalactoside (Figure 5A). Probes also incorporated a terminal alkyne for fluorescent labeling with click chemistry. The compound with the best affinity and labeling efficiency for galectin-3 was identified experimentally and included a flexible acetophenone moiety and an amide linkage. However, rigidity seemed to be beneficial for maximizing labeling selectivity in the complex mixture of mammalian cell lysates, with the compound possessing the benzophenone moiety and triazole linkage exhibiting the highest specificity for galectin-3 labeling. This example illustrates the difficulties associated with predicting which attributes will lead to the desired properties, and how testing the compounds in a cellular system is the ultimate determinant.
Figure 5.
Optimization of glycan-based photoprobes. (a) A series of thiodigalactosides in which a photocrosslinker (acetophenone or benzophenone) was attached to the C3 position through a variable linkage (ester, amide, or triazole) were tested for their abilities to label galectin-3 with an alkyne. (b) A series of α-mannosides modified with a variable photocrosslinker (diazirine, aryl azide, or benzophenone) at the C1 position were tested for their label efficiencies of amino acids and model peptides. (c) Photoaffinity probes possessing either a closed-ring or open-ring structure at the 43 reducing terminus of the glycan were compared for their abilities to label lectins with a biotin tag.
Similarly, the best photocrosslinking functionality for a particular application may also need to be determined experimentally. The commonly used photocrosslinking groups vary in size, solubility, and reactivity.[1] The feature most important for a particular application may be difficult to predict. For example, Lindhorst and coworkers developed a series of photoreactive α-mannosides possessing either a diazirine, an aryl azide, or a benzophenone group attached to the C1 position (Figure 5B). These photoprobes generate carbenes, nitrenes, and radicals, respectively, upon UV irradiation.[35] The authors performed a comparison of their photocrosslinking efficiency by carrying out in vitro labeling experiments with a series of six amino acids (Thr, Ser, Tyr, Ile, Lys, and Arg) and three model peptides, and analyzing the products by mass spectrometry. While all photoprobes were able to form insertion products with at least one of the amino acids and peptides investigated, the diazirine was deemed the most useful for crosslinking to biomolecules in future studies. In addition to its favorable properties in terms of water solubility and time of irradiation required for activation, the preference of the diazirine for insertion into polar groups (such as amines and hydroxyl groups) is predicted to be advantageous for studies involving protein targets under physiological conditions.
Finally, the conformational properties of the glycan itself can influence crosslinking efficiencies. Sakushima and co-workers compared the behavior of photoprobes in which the attached sugar (at the reducing terminus of the glycan) was in the open chain or cyclic form (Figure 5C). A probe containing the closed ring sugar displayed improved photocrosslinking to a cognate lectin.[36] However, as with the previous example, these photocrosslinking experiments were performed in defined reaction mixtures. Examination of photocrosslinking efficiency and specificity in more complex biological samples will be required to fully assess the relative utility of the different photoprobe structures.
C. Toward identification of binding sites
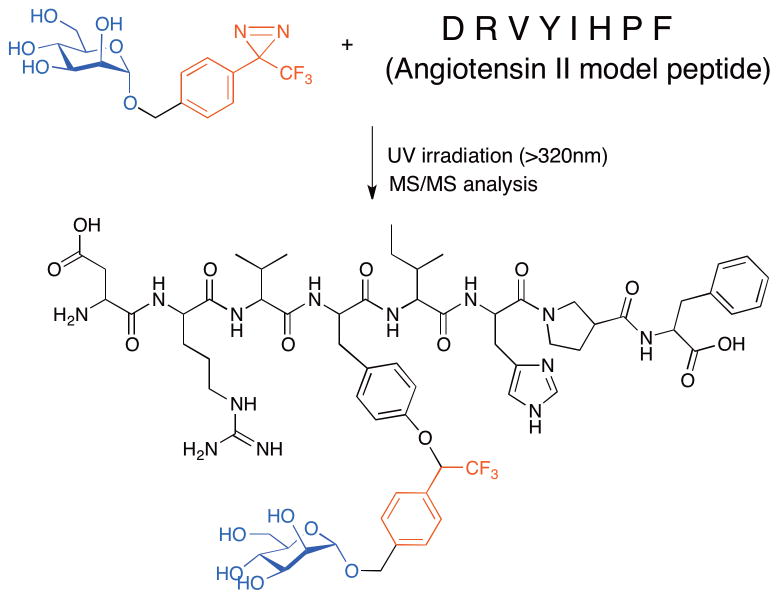
One intended application for carbohydrate-based photocrosslinking reagents is their use in defining functionally relevant carbohydrate binding sites on protein targets. Advances toward this goal have been reported by Lindhorst and coworkers. As described above, this group determined that photoreactive α-mannosides possessing a diazirine ring attached to C1 seemed to be the most suitable for labeling peptides (Figure 5B).[35] This photoprobe was incubated with an octapeptide from angiotensin II (H-Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-OH) and then UV irradiated to induce crosslinking. The researchers observed a 1:1 crosslinked product by MALDI-TOF-MS and ESI-MS. Further MS/MS studies revealed a preference for carbene insertion into the tyrosine residue, demonstrating that these reagents can be used to define preferential crosslinking sites (Figure 6).[37] In the future, such α-mannoside probes may be used for the analysis of more functionally relevant binding partners, such as the bacterial lectin FimH. For instance, it is still unknown if there is an auxiliary binding site utilized by this lectin that is important for host binding, and this photocapture/MS approach is well-suited to address that question. To this end, Lindhorst and coworkers have used MS to show that their diazirine-modified α-mannoside probe (Figure 5B) can crosslink and form a 1:1 adduct with the isolated N-terminal CRD of FimH.[37] They propose purifying the proteolytically digested fragments in analogous studies using a biotin-modified probe for analysis by MS/MS to identify critical amino acids mediating this interaction. As will be described in the next section, similar work using photocrosslinking to identify binding site residues in an isolated bacterial toxin has been performed with glycolipid probes as well.
Figure 6.
Toward identification of carbohydrate binding sites on protein targets. The α-mannoside with a diazirine photocrosslinker at the C1 position preferentially inserts into the angiotensin octapeptide at the tyrosine residue as determined by MS/MS.
D. Glycolipid binding
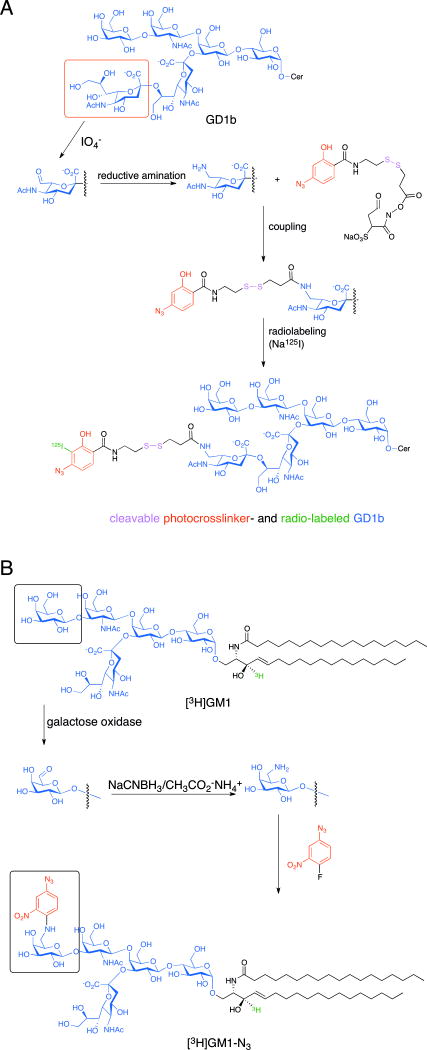
In addition to simpler sugars, glycans, and glycopeptides described thus far, scientists have also introduced photocrosslinkers into glycolipids to search for their interaction partners. For instance, Schnaar and coworkers prepared glycolipid probes with the goal of more precisely defining the regions within Clostridial neurotoxins, botulinum and tetanus, that can target glycolipids as receptors for endocytosis. One of the novel aspects of this approach was the way that Schnaar and coworkers synthesized a ganglioside-based photocrosslinking reagent by introducing an aryl azide onto the C7 position of the terminal sialic acid of GD1b via a disulfide-containing linker (Figure 7A).[38] To do so, the authors first treated the GD1b with periodic acid under mild conditions to selectively oxidize the glycerol side chain of terminal sialic acids resulting in aldehyde formation. Secondly, this aldehyde was converted to an amine by a reductive amination reaction. Thirdly, they coupled the arylazido group to this amine by using a commercial photoaffinity intermediate sulfosuccinimidyl 2-(p-azidosalicylamido) ethyl-1,3′-dithiopropionate. Finally, this photoaffinity labeled GD1b probe was radioiodinated by sodium iodide (125I). The tetanus toxin C fragment (residues ~864–1315) was incubated with 125I-azido-GD1b and irradiated to induce crosslinking. Following reduction for cleavage of disulfide bond and proteolytic digestion, they observed that a single peptide fragment was 125I-labeled, and this labeling event was inhibited by preincubation of the toxin with the b-series ganglioside GT1b. The unlabeled polypeptide was sequenced and shown to encompass the 34-amino acid C-terminal domain of tetanus toxin (residues 1282–1315); further MALDI-MS analysis revealed a single modification site at His1293. With this information, the authors could then perform a sequence alignment of the C-termini of the 7 homologous Clostridial toxins (tetanus and botulinum types A-F) which possess different abilities to bind to GT1b and speculate on which amino acids had diverged to yield different glycolipid specificities over time.
Figure 7.
Preparation of glycolipid photoprobes. (a) Synthetic route to modify ganglioside GD1b on the terminal sialic acid with an aryl azide photocrosslinker via a cleavable disulfide-containing linker, followed by its radioiodination (125I). (b) Synthetic route to modify a radiolabeled [3H]GM1 on the terminal galactose with a nitrophenylazide photocrosslinker.
Another example of a glycolipid photoprobe comes from the work of the Sonnino group. These researchers synthesized photoreactive derivatives of gangliosides (GM3, GM1, GD1b) to capture their protein binding partners found in cultured cells.[39] In particular, one such derivative of GM1 was designed to study molecules interacting with the hydrophilic end of the ganglioside. This photoprobe consisted of a photoreactive nitrophenylazide group attached via the terminal galactose on the sialyloligosaccharide (Figure 7B).[40] To prepare this probe, they oxidized a radiolabeled [3H]GM1 by galactose oxidase to produce an aldehyde, followed by a reductive amination that resulted in an amine derivative of [3H]GM1. Finally, this amine derivative was treated with 4-fluoro-3-nitrophenylazide to obtain a photoactivatable aryl azide conjugate, i. e. [3H]GM1-N3. Upon exogenous addition of [3H]GM1-N3 to cultured neuronal cells, a minor portion was recovered from sphingolipid-enriched membrane fractions (SEMFs) as determined by sucrose gradient ultracentrifugation, thus suggesting their insertion into the lipid bilayer in a physiologically relevant manner mimicking that of endogenous GM1. By biotin labeling of cell surface proteins followed by UV irradiation of treated cells, the SEMF obtained could be further affinity purified with streptavidin beads to enrich for those GM1-crosslinked proteins that possess exoplasmic domains. While the majority of biotin-labeled proteins recovered by this method were in the 50 to 70 kDa range, two radioactive bands at 135 and 35 kDa were also detected. The 135 kDa species was immunologically identified as the GPI-anchored adhesion protein TAG-1, a typical component of the SEMF from cerebellar neurons previously identified as a GM1 interaction partner. This exciting result suggests that targets can be labeled in complex systems with this approach.
IV. Metabolic incorporation of photocrosslinkers into cellular glycoconjugates
Another approach for creating photoreactive glycoconjugates is to exploit the naturally occurring metabolic pathways of a cell. Simple sugars modified with a photoreactive crosslinker can be added to the media surrounding cultured cells. These precursors can be transformed by the cells, producing a pool of photocrosslinking nucleotide sugars that can be utilized by the cell’s glycosyltransferases in the assembly of complex glycoconjugates. Due to the numerous possible glycan structures that are capable of being modified in the cell, advantages include being able to use the same simple sugar precursor to address various biological questions (i. e., to study different target biomolecules). As described below, this approach has been successfully applied to the examination of multiple sialic acid-modified glycoconjugates.
A. Metabolic incorporation of aryl azide into sialosides
In 2005, Paulson and coworkers demonstrated the first use of metabolically incorporated photocrosslinking sugars to define glycan-mediated interactions. To determine the glycan-dependent cis ligands of CD22 in B cells, they prepared a photoreactive metabolite comprised of N-acetylneuraminic acid modified with an aryl azide at the C9 position (9-AAz-NeuAc).[41] BJAB K20, a B-cell line that is deficient in sialic acid biosynthesis, were used to monitor the efficiency of 9-AAz-NeuAc incorporation into cell-surface glycoconjugates. These cells were cultured in serum-free media containing 9-AAz-NeuAc. Flow cytometry analysis with a fluorescently labeled sialic acid-binding lectin (SNA, Sambucus nigra agglutinin) or a CD22-Fc chimera revealed that the cell-surface sialic acid levels of the K20 cells were similar to those of sialic acid producing B cells. Furthermore, deployment of the Staudinger ligation with a triphenylphosphine-biotin probe revealed cell surface display of the aryl azide (AAz) functional group. Upon UV irradiation and immunoblotting for CD22, cells supplemented with NeuAc gave the expected CD22 band at 120 kDa, while those cells cultured with 9-AAz-NeuAc yielded an additional higher molecular weight band of >250 kDa. Subsequent immunoprecipitation of CD22 and immunoblotting for probable interaction partners such as CD45, PMCA, IgM, and CD19 yielded no positive results. However, the authors were able to decipher that, in fact, a CD22 homomultimeric complex was being observed by performing a co-immunoprecipitation of lysates from cells simultaneously expressing both Flag- and myc-tagged versions of CD22. This method allowed a clear demonstration of the homomeric, glycan-mediated interaction of CD22 that masks this protein from other potential cis binding partners.
In more recent work, these authors combined the same photoreactive reagent with a quantitative proteomics mass spectrometry approach, enabling identification of the trans ligands of CD22 in homotypic B-cell interactions.[42] As often encountered in mass spectrometry analysis of crosslinked samples,[43] a primary challenge is to obtain a suitable quantity of crosslinked material for mass spectrometry analysis. To circumvent this challenge, the authors first generated a candidate list of interaction partners, by crosslinking a CD22-Fc chimera to 9-AAz-NeuAc-labeled B cells (K20) and immunoaffinity-capturing the complexes. The higher molecular weight crosslinked complexes were then subjected to in-gel trypsin digestion and nano-LC-MS/MS, whereby (based on the spectral count values compared with that of non-irradiated lysates) 24 candidate proteins were identified, 18 of which were known glycoproteins and carried on for further analysis. In an alternative approach, the authors performed similar experiments under SILAC conditions with stable isotopes of lysine and arginine. Thus, differentially labeled heavy or light 9-AAz-NeuAc-labeled B cells (K20) were both incubated with the CD22-Fc chimera, but only one set was subjected to UV irradiation. The cell lysates were subsequently combined and purified by immunoaffinity capture. The samples were then subjected to on-bead trypsin digestion and MuDPITMS/MS, whereby 19 candidate proteins were identified, all of which were membrane glycoproteins. Taken together, these two proteomics approaches yielded a total 27 candidate glycoproteins (10 candidates were identified by both approaches). In order to determine which of these candidates are bone fide trans ligands for CD22 in the context of contacting cells, 9-AAz-NeuAc-labeled B cells (K20) were overlaid on a CHO cell monolayer expressing CD22 with a C-terminal V5 tag and UV irradiated to induce photocrosslinking (Figure 8). Anti-V5 immunoprecipitates were subjected to immunoblot analysis with antibodies for 11 of the 27 candidate proteins, and of these only IgM, CD45, and Basigin appeared in crosslinked complexes. Further support for these findings came from the observation that redistribution of the most robust ligand, IgM, to sites of contacting B cells was decreased in mice lacking CD22 or the sialyltransferase responsible for its sialylation. This work illustrates challenges associated with characterizing photocrosslinked complexes and the importance of developing methods to verify that interaction partners observed in vitro are biologically relevant.
Figure 8.
Strategy to capture the in situ trans ligands of CD22. N-acetylneuraminic acid modified with an aryl azide at the C9 position (9-AAz-NeuAc) was metabolically incorporated into cellular glycoproteins. B cells deficient in sialic acid biosynthesis (BJAB K20) convert exogenously added 9-AAz-NeuAc into its CMP-activated form inside the nucleus. Upon transport into the Golgi, CMP-9-AAz-NeuAc can be utilized as a substrate by glycosyltransferases for incorporation into cell-surface glycoproteins. These 9-AAz-NeuAc-modified B cells are then overlaid onto a CHO cell monolayer expressing CD22 with a V5 tag and UV irradiated to induce photocrosslinking. Anti-V5 immunoprecipitated complexes can be immunoblotted for candidate proteins identified previously by mass spectrometry-based proteomics.
B. Metabolic incorporation of diazirine into sialosides
The Kohler group also captured CD22 homooligomerization in B cells, this time utilizing a diazirine photocrosslinker located on the N-acyl side chain of sialic acid (SiaDAz). The diazirine can be incorporated into glycoconjugates by culturing cells with a protected form of SiaDAz, or its mannosamine precursor (ManNDAz) (Figure 9).[44] SiaDAz has been demonstrated to effectively compete with endogenous sialic acid for incorporation into glycoconjugates in several cell types, including B cells (BJAB K88) and T cells (Jurkat).[44,45] The Kohler group has gone on to use SiaDAz to crosslink ganglioside-protein interactions.[46] First, they used MALDI-TOF-MS to confirm that Jurkat T cells cultured with Ac4ManNDAz produce SiaDAz-modified glycolipids, including GM1 and GM2. In addition, GM1-SiaDAz extracted from these cells could still be recognized by a known binding partner, cholera toxin subunit B (CTxB). Furthermore, when Jurkat T cells were cultured with Ac4ManNDAz, incubated with CTxB, and UV-irradiated, a new toxin-containing species was formed, which was identified by SAMDI-TOF-MS to be covalently crosslinked CTxB-GM1-SiaDAz.[45] This group is currently pursing use of the SiaDAz probe, in concert with mass spectrometry approaches, to identify novel sialoside-mediated binding interactions. In addition, efforts are underway to introduce photocrosslinkers into classes of glycans other than sialosides.[47]
Figure 9.
Strategy to capture ganglioside binding partners using diazirine-modified sialic acid. Cells are cultured with Ac5-5-SiaDAz or a mannosamine precursor (Ac4ManNDAz). Diazirine-modified sialic acid competes with endogenous sialic acid for incorporation into cell surface glycoconjugates. UV irradiation of cells produces covalent complexes containing SiaDAz-containing glycoconjugates and interaction partners. This method has been implemented to capture glycoprotein interactions (CD22 homooligomerization) and glycolipid interactions (CTxB-GM1 binding).
C. Effects of photoaffinity labels on carbohydrate recognition
The incorporation of a photocrosslinker on a carbohydrate unavoidably results in a structural change that can potentially alter recognition events. Therefore, the identity of the photocrosslinking group and its position of attachment onto the sugar are key aspects of experimental design. In the case of sialic acid, modifications are known to have a substantial impact on glycan recognition. An important concern is that modifications might sterically interfere with recognition events, so the use of smaller photocrosslinking moieties (diazirine rather than a benzophenone) may be advised. In particular, modifications to the O9 or Na-acyl positions of sialosides have been shown to result in significant alterations in the affinity and selectivity of recognition by sialoside binding proteins, such as sialoadhesin, myelin associated glycoprotein, and CD22.[48] Surprisingly, in some cases, aromatic modification of sialosides at the C9 position of sialic acid can significantly increase binding affinity.[49] Indeed, the 9-AAz-NeuAc reagent described above [41] likely enhances CD22 recognition events.
Examination of structural data for sialoside recognition complexes reveals that the C9-modified 9-AAz-NeuAc and the C5-modified SiaDAz are likely to have complementary applications. For example, the crystal structures of murine sialoadhesin show that tryptophan 2 is in van der Waals contact with the acetamido methyl group, likely hindering recognition of sialosides modified on sialic acid’s N-acyl position.[50,51] Indeed, sialoadhesin preferentially binds NeuAc, over both natural and unnatural N-acyl variants.[52,53] On the other hand, a small hydrophobic pocket enables sialoadhesin to recognize sialosides with aromatic modifications at the C9 position of sialic acid. Thus, the 9-AAz-NeuAc is likely the preferred reagent to examine sialoadhesin binding partners. The crystal structure of cholera toxin complexes with the GM1 glycan shows a somewhat contrasting picture.[54] In this complex, the N-acyl position of sialic acid is highly accessible, while O9 engages in a hydrogen bound with glutamine 56. In this case, SiaDAz may be the preferred reagent to capture the binding interaction.
In addition, in nature, sialic acids are subject to a wide variety of post-glycosylational modifications that can dramatically alter their binding properties and functions.[55] Introduction of photocrosslinking groups may prevent these modifications and interfere with study of interactions that require variant forms of sialic acid. Again, the 9-AAz-NeuAc and SiaDAz reagents offer complementary uses. C9-modified compounds can be used to examine interactions in which N-glycolyl or deaminated sialic acids are required. N-acyl-modifed compounds can be used in situations where O9-acetylation, - lactylation, or –phosphorylation is physiologically important.
For the discovery of unknown binding events in physiological samples, which is likely the most important application for these approaches, data describing sialic acid recognition and modifications will often be unavailable or incomplete. In these cases, rational selection of the preferred photocrosslinking reagent is impossible. Therefore, the best strategy will likely be to empirically determine the best photocrosslinking reagent for a particular application, using the panel of reagents that have been shown to have at least reasonably efficient metabolic incorporation.
V. Conclusions
As described above, the current arsenal of carbohydrate photoprobes comprises a diverse class of reagents. The carbohydrate portions of these reagents have been based on naturally occurring glycoconjugates, synthetic glycans, and carbohydrate analogs. These targeting motifs enable binding to all classes of carbohydrate-binding proteins and metabolic enzymes. Biocompatible photocrosslinking groups enable selective crosslinking, even in complex lysates or cellular settings. Finally, the use of affinity purification tags allows isolation of crosslinked complexes for further characterization. While early work in this area demonstrated effective crosslinking of carbohydrate photoprobes, characterization of the crosslinked complexes remained a challenge. However, new developments in analytical methods are providing solutions to this difficulty. Recently, the use of Huisgen 1,3-dipolar cycloaddition chemistry has enabled the deployment of small functional groups (alkynes and azides) as handles for subsequent attachment of affinity purification tags. This strategy allows for effective purification of crosslinked species from complex mixtures. In addition, new mass spectrometry methods facilitate identification of crosslinked molecules and are beginning to allow delineation of crosslinking sites. Taken together, the use of carbohydrate photoprobes, in combination with modern analytical methods, is poised to provide new essential new insights into the biological roles of carbohydrates.
Acknowledgments
We acknowledge support from the NIH (R01GM090271 and R21AI094427) and from the Welch Foundation (I-1686). J. J. K. is an Alfred P. Sloan Research Fellow.
References
- 1.Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol Biosyst. 2008;4:473–480. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 2.Bourne Y, Henrissat B. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Op Struct Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 3.Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Op Chem Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Zechel DL, Withers SG. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc Chem Res. 2000;33:11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- 5.Tsai CS, Li YK, Lo LC. Design and synthesis of activity probes for glycosidases. Org Lett. 2002;4:3607–3610. doi: 10.1021/ol0265315. [DOI] [PubMed] [Google Scholar]
- 6.Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of glycosidase activity from cell lysates. Angewandte Chemie. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- 7.Stubbs KA, Scaffidi A, Debowski AW, Mark BL, Stick RV, Vocadlo DJ. Synthesis and use of mechanism-based protein-profiling probes for retaining beta-D-glucosaminidases facilitate identification of Pseudomonas aeruginosa NagZ. J Amer Chem Soc. 2008;130:327–335. doi: 10.1021/ja0763605. [DOI] [PubMed] [Google Scholar]
- 8.Orth R, Pitscheider M, Sieber SA. Chemical probes for labeling of the bacterial glucosaminidase NagZ via the Huisgen cycloaddition. Synthesis. 2010:2201–2206. [Google Scholar]
- 9.Yagi M, Kouno T, Aoyagi Y, Murai H. Structure of moranoline, a piperidine alkaloid from Morus species. J Agric Chem Soc Japan. 1976;50:571–572. [Google Scholar]
- 10.Romaniouk AV, Silva A, Feng J, Vijay IK. Synthesis of a novel photoaffinity derivative of 1-deoxynojirimycin for active site-directed labeling of glucosidase I. Glycobiology. 2004;14:301–310. doi: 10.1093/glycob/cwh044. [DOI] [PubMed] [Google Scholar]
- 11.van Scherpenzeel M, van den Berg RJ, Donker-Koopman WE, Liskamp RM, Aerts JM, Overkleeft HS, Pieters RJ. Nanomolar affinity, iminosugar-based chemical probes for specific labeling of lysosomal glucocerebrosidase. Bioorg Med Chem. 2010;18:267–273. doi: 10.1016/j.bmc.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 12.Gandy MN, Debowski AW, Stubbs KA. A general method for affinity-based proteomic profiling of exo-alpha-glycosidases. Chem Comm. 2011;47:5037–5039. doi: 10.1039/c1cc10308c. [DOI] [PubMed] [Google Scholar]
- 13.Huh JW, Lee HJ, Choi MM, Yang SJ, Yoon SY, Kim DW, Kim SY, Choi SY, Cho SW. Identification of a UDP-glucose-binding site of human UDPglucose dehydrogenase by photoaffinity labeling and cassette mutagenesis. Bioconjug Chem. 2005;16:710–716. doi: 10.1021/bc0500387. [DOI] [PubMed] [Google Scholar]
- 14.Frost DJ, Read SM, Drake RR, Haley BE, Wasserman BP. Identification of the UDP-glucose-binding polypeptide of callose synthase from Beta vulgaris L. by photoaffinity labeling with 5-azido-UDP-glucose. J Biol Chem. 1990;265:2162–2167. [PubMed] [Google Scholar]
- 15.Lin FC, Brown RM, Jr, Drake RR, Jr, Haley BE. Identification of the uridine 5′-diphosphoglucose (UDP-Glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-Glc. J Biol Chem. 1990;265:4782–4784. [PubMed] [Google Scholar]
- 16.Carrizo ME, Curtino JA. Identification of two uridine binding domain peptides of the UDP-glucose-binding site of rabbit muscle glycogenin. Biochem Biophys Res Comm. 1998;253:786–789. doi: 10.1006/bbrc.1998.9856. [DOI] [PubMed] [Google Scholar]
- 17.Drake RR, Igari Y, Lester R, Elbein AD, Radominska A. Application of 5-azido-UDP-glucose and 5-azido-UDP-glucuronic acid photoaffinity probes for the determination of the active site orientation of microsomal UDP-glucosyltransferases and UDP-glucuronosyltransferases. J Biol Chem. 1992;267:11360–11365. [PubMed] [Google Scholar]
- 18.Rancour DM, Menon AK. Identification of endoplasmic reticulum proteins involved in glycan assembly: synthesis and characterization of P3-(4-azidoanilido)uridine 5′-triphosphate, a membrane-topological photoaffinity probe for uridine diphosphate-sugar binding proteins. Biochem J. 1998;333:661–669. doi: 10.1042/bj3330661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostova Z, Rancour DM, Menon AK, Orlean P. Photoaffinity labelling with P3-(4-azidoanilido)uridine 5′-triphosphate identifies Gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem J. 2000;350:815–822. [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y, Bernardi D, Bratton S, Ward MD, Battaglia E, Finel M, Drake RR, Radominska-Pandya A. Phenylalanine 90 and 93 are localized within the phenol binding site of human UDP-glucuronosyltransferase 1A10 as determined by photoaffinity labeling, mass spectrometry, and site-directed mutagenesis. Biochemistry. 2006;45:2322–2332. doi: 10.1021/bi0519001. [DOI] [PubMed] [Google Scholar]
- 21.Holman GD, Parkar BA, Midgley PJ. Exofacial photoaffinity labelling of the human erythrocyte sugar transporter. Biochim Biophys Acta. 1986;855:115–126. doi: 10.1016/0005-2736(86)90195-1. [DOI] [PubMed] [Google Scholar]
- 22.Holman GD, Karim AR, Karim B. Photolabeling of erythrocyte and adipocyte hexose transporters using a benzophenone derivative of bis(D-mannose) Biochim Biophys Acta. 1988;946:75–84. doi: 10.1016/0005-2736(88)90459-2. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto M, Hatanaka Y, Yang J, Dhesi J, Holman GD. Synthesis of biotinylated bis(D-glucose) derivatives for glucose transporter photoaffinity labelling. Carb Res. 2001;331:119–127. doi: 10.1016/s0008-6215(01)00025-8. [DOI] [PubMed] [Google Scholar]
- 24.Tatibouet A, Yang J, Morin C, Holman GD. Synthesis and evaluation of fructose analogues as inhibitors of the D-fructose transporter GLUT5. Bioorg Med Chem. 2000;8:1825–1833. doi: 10.1016/s0968-0896(00)00108-5. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Dowden J, Tatibouet A, Hatanaka Y, Holman GD. Development of high-affinity ligands and photoaffinity labels for the D-fructose transporter GLUT5. Biochem J. 2002;367:533–539. doi: 10.1042/BJ20020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee RT, Lee YC. Affinity labeling of the galactose/N-acetylgalactosamine-specific receptor of rat hepatocytes: preferential labeling of one of the subunits. Biochemistry. 1987;26:6320–6329. doi: 10.1021/bi00394a005. [DOI] [PubMed] [Google Scholar]
- 27.Rice KG, Lee YC. Modification of triantennary glycopeptide into probes for the asialoglycoprotein receptor of hepatocytes. J Biol Chem. 1990;265:18423–18428. [PubMed] [Google Scholar]
- 28.Lee RT, Lee YC. Preparation of a high-affinity photolabeling reagent for the Gal/GalNAc lectin of mammalian liver: demonstration of galactose-combining sites on each subunit of rabbit hepatic lectin. Biochemistry. 1986;25:6835–6841. doi: 10.1021/bi00370a016. [DOI] [PubMed] [Google Scholar]
- 29.Rice KG, Weisz OA, Barthel T, Lee RT, Lee YC. Defined geometry of binding between triantennary glycopeptide and the asialoglycoprotein receptor of rat heptocytes. J Biol Chem. 1990;265:18429–18434. [PubMed] [Google Scholar]
- 30.Lauc G, Dumic J, Flogel M, Lee YC. A new synthetic route for the preparation of glycoprobes. Anal Biochem. 2003;320:306–309. doi: 10.1016/s0003-2697(03)00387-7. [DOI] [PubMed] [Google Scholar]
- 31.Lauc G, Lee RT, Dumiae J, Lee YC. Photoaffinity glycoprobes-a new tool for the identification of lectins. Glycobiology. 2000;10:357–364. doi: 10.1093/glycob/10.4.357. [DOI] [PubMed] [Google Scholar]
- 32.Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 33.Ballell L, van Scherpenzeel M, Buchalova K, Liskamp RM, Pieters RJ. A new chemical probe for the detection of the cancer-linked galectin-3. Org Biomol Chem. 2006;4:4387–4394. doi: 10.1039/b611050a. [DOI] [PubMed] [Google Scholar]
- 34.van Scherpenzeel M, Moret EE, Ballell L, Liskamp RM, Nilsson UJ, Leffler H, Pieters RJ. Synthesis and evaluation of new thiodigalactoside-based chemical probes to label galectin-3. Chembiochem. 2009;10:1724–1733. doi: 10.1002/cbic.200900198. [DOI] [PubMed] [Google Scholar]
- 35.Wiegand M, Lindhorst TK. Synthesis of photoactive alpha-mannosides and mannosyl peptides and their evaluation for lectin labeling. Europ J Org Chem. 2006:4841–4851. [Google Scholar]
- 36.Ohtsuka I, Sadakane Y, Higuchi M, Hada N, Hada J, Kakiuchi N, Sakushima A. The effect of structural differences in the reducing terminus of sugars on the binding affinity of carbohydrates and proteins analyzed using photoaffinity labeling. Bioorg Med Chem. 2011;19:894–899. doi: 10.1016/j.bmc.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 37.Lindhorst TK, Marten M, Fuchs A, Knight SD. En route to photoaffinity labeling of the bacterial lectin FimH. Beilstein J Org Chem. 2010;6:810–822. doi: 10.3762/bjoc.6.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro RE, Specht CD, Collins BE, Woods AS, Cotter RJ, Schnaar RL. Identification of a ganglioside recognition domain of tetanus toxin using a novel ganglioside photoaffinity ligand. J Biol Chem. 1997;272:30380–30386. doi: 10.1074/jbc.272.48.30380. [DOI] [PubMed] [Google Scholar]
- 39.Loberto N, Prioni S, Prinetti A, Ottico E, Chigorno V, Karagogeos D, Sonnino S. The adhesion protein TAG-1 has a ganglioside environment in the sphingolipid-enriched membrane domains of neuronal cells in culture. J Neurochem. 2003;85:224–233. doi: 10.1046/j.1471-4159.2003.01655.x. [DOI] [PubMed] [Google Scholar]
- 40.Prioni S, Mauri L, Loberto N, Casellato R, Chigorno V, Karagogeos D, Prinetti A, Sonnino S. Interactions between gangliosides and proteins in the exoplasmic leaflet of neuronal plasma membranes: a study performed with a tritiumlabeled GM1 derivative containing a photoactivable group linked to the oligosaccharide chain. Glycoconj J. 2004;21:461–470. doi: 10.1007/s10719-004-5536-4. [DOI] [PubMed] [Google Scholar]
- 41.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 42.Ramya TN, Weerapana E, Liao L, Zeng Y, Tateno H, Yates JR, 3rd, Cravatt BF, Paulson JC. In situ trans ligands of CD22 identified by glycan-protein photocross-linking-enabled proteomics. Mol Cell Proteomics. 2010;9:1339–1351. doi: 10.1074/mcp.M900461-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamurthy M, Dugan A, Nwokoye A, Fung YH, Lancia JK, Majmudar CY, Mapp AK. Caught in the act: Covalent cross-linking captures activator-coactivator interactions in vivo. ACS Chem Biol. 2011;6:1321–1326. doi: 10.1021/cb200308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Amer Chem Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 45.Bond MR, Zhang H, Kim J, Yu SH, Yang F, Patrie SM, Kohler JJ. Metabolism of diazirine-modified N-acetylmannosamine analogues to photo-crosslinking sialosides. Bioconj Chem. 2011;22:1811–1823. doi: 10.1021/bc2002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bond MR, Whitman CM, Kohler JJ. Metabolically incorporated photocrosslinking sialic acid covalently captures a ganglioside-protein complex. Mol Biosys. 2010;6:1796–1799. doi: 10.1039/c0mb00069h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu S-H, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1114356109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelm S, Brossmer R, Isecke R, Gross HJ, Strenge K, Schauer R. Functional groups of sialic acids involved in binding to siglecs (sialoadhesins) deduced from interactions with synthetic analogues. Eur J Biochem. 1998;255:663–672. doi: 10.1046/j.1432-1327.1998.2550663.x. [DOI] [PubMed] [Google Scholar]
- 49.Kelm S, Gerlach J, Brossmer R, Danzer CP, Nitschke L. The ligandbinding domain of CD22 is needed for inhibition of the B cell receptor signal, as demonstrated by a novel human CD22-specific inhibitor compound. J Exp Med. 2002;195:1207–1213. doi: 10.1084/jem.20011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.May AP, Robinson RC, Vinson M, Crocker PR, Jones EY. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 51.Zaccai NR, Maenaka K, Maenaka T, Crocker PR, Brossmer R, Kelm SR, Jones EY. Structure-gGuided design of sialic acid-based Siglec inhibitors and crystallographic analysis in complex with sialoadhesin. Structure. 2003;11:557–567. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 52.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. Sialoside specificity of the Siglec family assessed using novel multivalent probes. J Biol Chem. 2003;278:31007–31019. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- 53.Luchansky SJ, Bertozzi CR. Azido sialic acids can modulate cell-surface interactions. Chembiochem. 2004;5:1706–1709. doi: 10.1002/cbic.200400148. [DOI] [PubMed] [Google Scholar]
- 54.Merritt EA, Sarfaty S, Akker FVD, L’Hoir C, Martial JA, Hol WGJ. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Prot Sci. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Chen X. Carbohydrate post-glycosylational modifications. Org Biomol Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]