Abstract
Scorpion venoms have been studied for over fifty years; however, the majority of research has focussed primarily on medically important Buthidae species. Additionally, venoms of the estimated 200 species of scorpion native to Australia have received very little attention. The first venom mass profiles of six non-buthid and one buthid scorpion species are presented herein, four of which are endemic to Australia. While masses under 5 kDa dominated the venoms of all species, the buthid venom contained considerably more masses between 7 and 8 kDa than those of the non-buthids, corroborating the emergent trend that buthids are richer in long-chain neurotoxins than non-buthids. The Australian scorpion venom fractions were also analysed with the relatively new MALDI-ToF matrix 1,5-DAN. Over forty partial sequences were obtained, the majority of which are homologous to scorpion antimicrobials such as opistoporin and IsCT2. Overall, this study is the single most comprehensive mass spectrometric analysis of scorpion venom landscapes to date and provides an insight into untapped Australian species.
Keywords: scorpion venom, mass landscapes, mass spectrometry
INTRODUCTION
Since the early 1960s, scientists have been aware that scorpion venoms are comprised of a complex heterogeneity of substances and have sought to deconvolute them to their constituents (Rodriguez de la Vega et al, 2010). Initially, scorpion venoms were studied to ascertain the components responsible for human morbidity. It was soon discovered that the venoms contained neurotoxic peptides and that many of these toxins act via one of two modes of action-blockage of potassium channels or modulation of sodium channel activity (Rodriguez de la Vega and Possani, 2004). It was also observed that these peptides contained three or four disulfide bonds, with the potassium channel blockers being around 35 amino acids in length and the sodium channel toxins consisting of about 65 residues. Based on their size, the potassium channel toxins were termed ‘short-chain’ and the sodium channel toxins ‘long-chain’ (Rodriguez de la Vega and Possani, 2004; Rodriguez de la Vega and Possani, 2005). The selectivity of these disulfide-rich peptides made them ideal tools for the study of ion channels and their use as pharmacological probes became widespread. Hence, most scorpion venom research has focused on the discovery and characterisation of such functionally and structurally related neurotoxins, accounting for the dominance of peptides that target sodium or potassium channels in current scorpion toxin literature. To date, over 750 scorpion venom peptides have been sequenced, of which more than 500 are classified as sodium or potassium channels ligands (Jungo et al, 2012). Additionally, over 85% of known scorpion venom peptides are derived from less than 50 species of Buthidae, which reflects the historical bias of studying the venom of medically important species (Jungo et al, 2012). Worldwide, only about 30 of the 1900 scorpion species are considered hazardous to people and nearly all belong to the Buthidae family (Isbister et al, 2003). Thus, current literature is ‘buthid-centric’ and focuses on peptides that affect only two main receptor targets (Rodriguez de la Vega et al, 2010). A more accurate depiction of the diversity of peptides in animal venoms can be ascertained through proteomic profiling (Escoubas et al, 2008; Gutierrez et al, 2009). Comprehensive mass profiles have been reported for only about fifteen scorpion species, seven of which are buthids (for a review, see (Rodriguez de la Vega et al, 2010).
This study presents the mass profiles of seven additional species, including six non-buthids. Four of the species are endemic to Australia, whose estimated 200 species have received very little research attention (Isbister et al, 2004). Venoms from the Australian species were analysed using 1,5-diaminonaphthalene (1,5-DAN), a compound whose use as a MALDI matrix has only recently been described (Fukuyama et al, 2006). It was previously reported that 1,5-DAN causes partial reduction of cystines in the MALDI laser plume, thereby providing important information on the number of disulfide bonds in a peptide (Fukuyama et al, 2006). 1,5-DAN also enhances the in-source decay (ISD) fragmentation of peptides, resulting in the observation of c- and z- ions that are useful for de novo sequencing (Juhasz, 1993; Fukuyama et al, 2006). In addition, peptide fragmention via ISD has the advantage of theoretically retaining post-translational modifications such as phosphorylation, which are degraded with the traditional collision-induced dissociation (CID) method (Asakawa and Takayama, 2012). This study is the first to use 1,5-DAN to examine scorpion venom peptides. It is also only the second reported use of this matrix for the examination of crude venom fractions, the first being that of the cone snail Conus textile, in which 1,5-DAN enabled the detection of 140 peptides containing between zero and three disulfides (De Pauw et al, 2007). Also, the mass profiles in this report were obtained by analysis of venoms using both nanospray and MALDI-ToF methods. It is known that masses yielded by the two methods are complementary and result in increased coverage of the peptidome (Bodnar et al, 2003). The combination of 1,5-DAN analysis and traditional tandem mass spectrometry (MS/MS), as well as mass profiling, has provided a useful glimpse into the components of several scorpion venoms, with a focus on Australian species.
MATERIALS AND METHODS
Venom sources
Crude scorpions and venom from the Australian species Urodacus elongatus, Urodacus yaschenkoi, Urodacus armatus and Lychas marmoreus obscurus were obtained by inducing live scorpions (purchased from The Green Scorpion, Port Macquarie, NSW, Australia) to repeatedly sting an Eppendorf tube with the top covered in parafilm. The clear prevenom of the initial defensive stings (Inceoglu et al, 2003) and the proceeding opaque venom deposits were pooled together. A minimum of two weeks elapsed between milkings and venom was pooled, lyophilised and stored at -20oC. Lyophilised venoms electrically milked from scorpion species Vaejovis spinigerus, Opistophthalmus glabrifrons and Pandinus imperator were purchased from SpiderPharm (Yarnell, AZ, United States). Venoms were reconstituted in a minimal volume of 50:50 solvent A (0.05%, v/v, TFA): solvent B (90%, v/v, ACN/0.043%, v/v, TFA), sonicated for 10min and diluted to their final concentrations with solvent A.
Fractionation of venoms
Four milligram reconstituted U. elongatus and U. yaschenkoi venoms were rpHPLC fractionated using a semipreparative reversed-phase C18 column (Thermo Aquasil C18, 250x10mm, 5μM pore size). Venom components were eluted at a flow rate of 6ml/min using a linear gradient of 5-70% solvent B over 110min preceded by 5% solvent B for 5min. Approximately 250μg of reconstituted venom from all other species were rpHPLC fractionated using a narrow bore reversed-phase C8 column (Zorbax C8, 2.1x150mm, 300Å), flow rate 0.35ml/min, elution gradient varied as required. A Shimadzu VP system coupled to a SPD-10AVP UV detector monitoring at 280nm and either 217nm for semipreparative or 214nm for narrow bore separations was used for all fractionations. Fractions were manually collected and lyophilised.
Mass spectrometric analysis using MALDI-ToF
Lyophilised rpHPLC fractions were dissolved in 10μl of a 50:50 mixture of solvent A and B and 0.5μl each fraction was spotted twice with 0.5μl different matrices on a MALDI-ToF plate. The matrices used were α-cyano-4-hydroxycinnamic acid (αCHCA) and sinapinic acid (SA). U. yaschenkoi, U. armatus, U. elongatus and L. m. obscurus fractions were also spotted with 0.5μl 1,5-diaminonaphthalene (1,5-DAN). All matrices were dissolved in 60%, v/v, acetonitrile/0.05%, v/v, TFA at 10mg/ml with 10min sonication. Spots were analysed with an ABI 4700 Voyager mass spectrometry system in positive reflector mode. The instrument was calibrated using the following standards (monoisotopic masses listed): des-Arg bradykinin (903.45 Da), angiotensin 1 (1295.68 Da), adrenocorticotropic hormone (ACTH) clip 1-17 (2092.08 Da), ACTH clip 18-39 (2464.19 Da), ACTH clip 7-38 (3656.92 Da) and bovine ubiquitin (8564.80 Da). Masses corresponding to the addition of common adducts K+, Na+ and oxidation were manually excluded from mass lists.
Nanospray analysis
Crude venoms were dissolved in a minimal amount of a 50:50 mixture of solvent A and B and then diluted to to 0.5mg/ml in solvent A. 0.75μl of dissolved venom was fractionated by reversed-phased HPLC (rpHPLC) using a Vydac C18 MS column (300Å, 5μm, 150x0.3mm). Venom components were eluted at a flow rate of 4μl/min using a gradient of 2% buffer B (90%, v/v, acetonitrile, 0.1%, v/v, formic acid) for 10min, followed by 10-40% buffer B over 110min, then 40-80% buffer B over 10min. Online nanospray analysis was performed using a QSTAR Elite mass spectrometer.
RESULTS AND DISCUSSION
Venom mass landscapes
The results of this study significantly increases the number of scorpion venom mass profiles reported in current literature (Newton et al, 2007; Ma et al, 2010; Rodriguez de la Vega et al, 2010; Caliskan et al, 2012; Diego-Garcia et al, 2012; Gurrola et al, 2012). Over 100 unique masses were present in the venom of each species (Supp. Tables 1-7). Of the seven species analysed, peptides have previously only been described for P. imperator. Masses corresponding to the following P. imperator venom peptides were present: Imperatoxin-A, α-KTx 6.1, α-KTx 6.4, α-KTx 6.5, α-KTx 7.1, scorpine and pandinin-1. Pandinin-2, tetrapandin 1, 2 and α-KTx 7.2 were not seen. The absence of these peptides may be due to several factors, including intraspecific and individual variation in the venom (Abdel-Rahman et al, 2009; Ruiming et al, 2010) and differences in the venom extraction and handling procedures compared to other studies (Martin et al, 1987; Zerrouk et al, 1991). Additionally, 22 masses (within 1 a.m.u) were common between the mass lists of P. imperator and a previous study of venom from Pandinus cavimanus (Diego-Garcia et al, 2012), a scorpion of the same genus. A comparison of V. spinigerus and Vaejovis mexicanus (Gurrola et al, 2012) venoms also revealed over 40 similar masses.
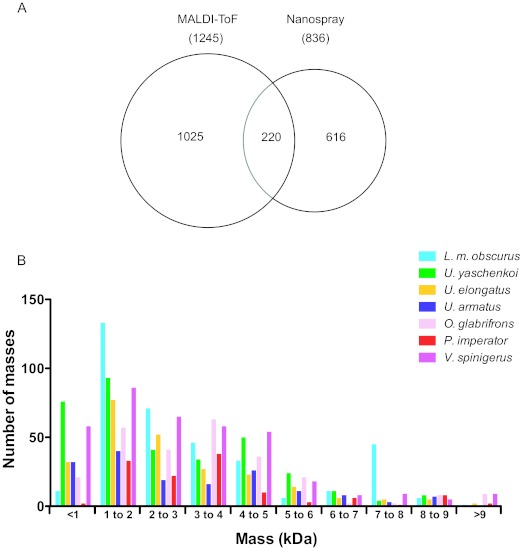
The importance of using a combination of nanoelectrospray and MALDI-ToF instruments was emphasised, since only about 12% of the total observed masses were present in both methods (Figure 1A). Several previous studies of complex mixtures derived from a range of venomous and non-venomous sources have also highlighted the complementarity of both methods (Pimenta et al, 2001; Batista et al, 2006; Molle et al, 2009; Smith et al, 2011). Therefore, future venom profiling investigations should avoid utilising only one mass acquisition method, as this will result in an underestimation of components.
Figure 1.
Analysis of the venom peptidome mass landscapes of scorpion venoms. A. Venn diagram showing overlap of peptide masses determined using electrospray and MALDI-ToF mass spectrometry. B. Histogram showing the abundance of peptide toxins in the venom of seven scorpion species, sorted into 1kDa molecular weight bins.
The mass spectrometric analyses reported herein highlights emerging trends in scorpion venom mass landscapes. Firstly, a large proportion of peptides present in the venoms are of low molecular weight (Figure 1B). In this study, masses under 2kDa accounted for 39% of the total number of venom peptides seen, ranging from 27% in P. imperator to 48% in U. yaschenkoi. The high number of small peptides observed is consistent with other scorpion venom peptidomic studies (Newton et al, 2007; Ma et al, 2010; Caliskan et al, 2012; Diego-Garcia et al, 2012; Gurrola et al, 2012) and reviewed in (Rodriguez de la Vega et al, 2010). Even though these lower molecular weight peptides are dominant components in the venoms of both non-buthid and buthid scorpions from around the world, they have not been studied as extensively as the longer neurotoxins. Of the ∼750 scorpion venom peptides sequenced to date, only about 100 are such small peptides (Jungo et al, 2012). These peptides are typically linear, adopt an alpha helical structure and possess antimicrobial, cytolytic, immune modulatory or antifungal properties (Zeng et al, 2005; Remijsen et al, 2010). It is hypothesised that the activities of these peptides are due to their ability to form pores in cell walls, however their precise mechanisms of action have not been delineated (Zeng et al, 2005). Compared to many spider species, the other major group of arachnids, as well as other venomous animals such as cone snails, scorpion venoms contain far greater numbers of these small linear peptides and are thus an excellent source of potentially novel antimicrobials (Rates et al, 2011). It is also a possibility that these small peptides are ligands to molecular targets that are yet to be ascertained.
Secondly, previous reports have shown that venoms from non-buthids contain few peptides with masses between 6kDa to 8kDa, in contrast to buthid venoms where peptides in this mass range appear to be ubiquitous with activities targeting sodium channel toxins (Rodriguez de la Vega and Possani, 2005). This trend was also observed in the present report, with masses in the 6kDa to 8kDa range accounting for 15% of total masses seen in the buthid L. m. obscurus venom. The highest percentage of masses in the venom of a non-buthid in this range was only 6.3% from U. armatus. It is hypothesised that buthids evolved a more potent, neurotoxin rich venom as they are generally smaller and have delicate chelae compared to non-buthids, who rely more on crushing prey with their powerful chelae rather than subduing them with venom (van der Meijden et al, 2010). The lack of research into non-buthids, combined with the intrinsically low number of peptides between 6kDa to 8kDa in their venoms has resulted in only three sodium channel toxins sequenced from one non-buthid species to date, compared to over 300 sodium channel toxins sourced from buthid venoms (Jungo et al, 2012). Furthermore, recent transcriptomic analyses have revealed that many non-buthid species are actually missing transcripts that encode for sodium channel toxins (Schwartz et al, 2007; Ma et al, 2009; Cao et al, 2010). Much of the current literature on the evolution of scorpions is contradictory and there are conflicting views on classification, even at the family level (Fet and Soleglad, 2005; Prendini and Wheeler, 2005). The presence or absence of long-chain sodium channel toxins in non-buthid species may be an important factor that could aid in the delineation of the scorpion evolutionary tree.
MALDI-ToF analysis of Australian scorpion venoms
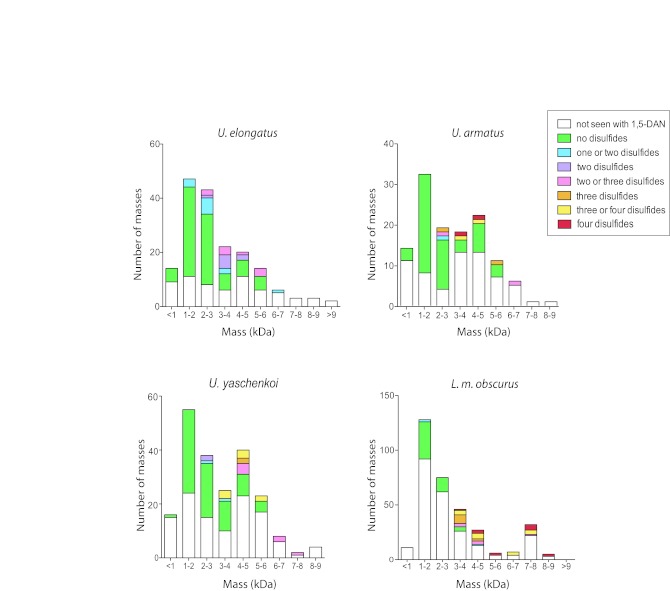
A useful attribute of 1,5-DAN as a MALDI-ToF matrix is its ability to cause partial reduction of disulfide bonds in the laser plume. When used to analyse disulfide-bond containing peptides, this property results in the appearance of a shifted isotopic mass distribution pattern from which the number of cystines can be delineated (De Pauw et al, 2007). Since biologically interesting peptides in animal venoms are often cystine rich, 1,5-DAN can be used as a tool to easily screen for such peptides. As mentioned previously, scorpion venom peptides of low molecular weight are usually linear and the larger short- and long-chain peptides generally contain numerous disulfides. Analysis with 1,5-DAN matrix revealed this was also the case for the Australian scorpion venom fractions (Figure 2). Notably, the number of peptides seen with two or more disulfides in the buthid L. m. obscurus venom was double than of the non-buthid Urodacus species. This could indicate the buthid venom contains a greater number of archetypical sodium and potassium channel scorpion toxins than the non-buthid venoms.
Figure 2.
Histogram of the abundance of peptide masses in four Australian scorpion species. Bars represent the total masses observed using αCHCA and SA matrices on MALDI-ToF. Number of disulfide bonds determined using 1,5-DAN matrix. Masses sorted into 1kDa molecular weight bins.
De novo sequencing of peptides in the Australian scorpion venom fractions produced 41 partial sequences homologous to 11 known scorpion venom peptides and one unknown, with 24 sequences obtained by MS/MS and 18 using 1,5-DAN matrix (Table 1). Despite the isolation of the Australia-New Guinea landmass from its neighbours for the past 80 million years, a high degree of conservation appears to exist between peptides found in the venom of the Australian scorpion species studied and other scorpion venom peptides from around the world. The vast majority of peptide sequences obtained was homologous to linear antimicrobial scorpion peptides, with peptides related to opistoporin 4 and IsCT2 found in all Urodacus sp. venoms. It was also evident that many of the lower molecular weight sequences were fragments of the larger scorpine and opistoporin 4 peptide homologues. Peptide fragments had been observed and sequenced previously in Tityus serrulatus scorpion venom and accounted for the majority of the total sequences found (Pimenta et al, 2008). Whether these peptide fragments serve any function or if they are merely degradation products remains to be determined. In some cases it appears these peptide fragments are biologically active, as evidenced by the antimalarial activity of an N-terminal fragment of MeuTXKβ3, a long-chain scorpion peptide related to scorpines (Gao et al, 2010).
Table 1.
List of peptide sequences from Australian scorpion venoms obtained by MS/MS and 1,5-DAN. Numbers in brackets refer to residue masses (monoisotopic) at the N or C terminus for which sequence was not obtained. A hyphen indicates the last residue listed is not terminal. ‘J’ represents the isobaric amino acids isoleucine and leucine. ‘X’ represents instances where lysine and glutamine could not be differentiated. An asterisk represents a C-terminal amide. Lowercase ‘a’ indicates average mass. Square brackets contain the accession number.
| Species | Sequence | Method | Precursor mass (M+H)+ | Homologous peptide |
| U. elongatus | (355.9)DWJXXTAXXVWNSDTANXJQSXAJNAAXNF | MS/MS | 3774.8 | Opistoporin-4 [Q5VJS9] |
| U. elongatus | (947.6)XTAXXVWNSDTANXJXSXAJNAAXNF- | 1,5-DAN | N/A | |
| U. elongatus | XXVWNSDTANX | MS/MS | 1290.5 | |
| U. elongatus | VWNSDTANXJXSAX | MS/MS | 1561.7 | |
| U. elongatus | VWNSDTANX | MS/MS | 1034.4 | |
| U. elongatus | FVAEXJGATPA | MS/MS | 1103.5 | |
| U. elongatus | (3436.0)NFVAEX- | 1,5-DAN | N/A | |
| U. armatus | (1529.8)WNSDVAXXJXGKAINAAKD- | 1,5-DAN | N/A | |
| U. armatus | (1714.8)NSDVAXXJXGKPSNAAKDFVA- | 1,5-DAN | N/A | |
| U. armatus | VAEXJGATPAEAGX | MS/MS | 1341.7 | |
| U. yaschenkoi | (1715.9)JXGXAJNAAXDF- | 1,5-DAN | N/A | |
| U. yaschenkoi | (1158.6)XJXGXAJNAAXDA- | 1,5-DAN | N/A | |
| U. yaschenkoi | (2130.0)AKKJKGKAJNAAKD- | 1,5-DAN | N/A | |
| U. elongatus | SSJFSTJWKGJKSJF* | MS/MS | 1713.0 | IsCT2 [Q8MTX2] |
| U. elongatus | JFSTJWKGJKSJF* | MS/MS | 1538.9 | |
| U. elongatus | JFSAJWSGJKSJF* | MS/MS | 1467.9 | |
| U. elongatus | FFSTJWSGJXSJF* | MS/MS | 1531.9 | |
| U. elongatus | (558.1)JWGGJKSVJGKR(440.3) | MS/MS | 2312.2 | |
| U. yaschenkoi | JJSAJWSGJKSJF* | MS/MS | 1433.9 | |
| U. armatus | JJSAJWSGJXS | MS/MS | 1174.7 | |
| U. elongatus | (935.5)GJJKAAAKAJ(485.2) | MS/MS | 2376.4 | Scorpine [P56972] |
| U. elongatus | (1108.6)GJJKAAAKAJ(485.2) | MS/MS | 2549.5 | |
| U. elongatus | (1159.7)AJDEKJPNGFJXGAAKAJVHXJAXSEYGCMM- | 1,5-DAN | 8594a | |
| U. armatus | (1952.0)NGFJXGGGKAVVHKN- | 1,5-DAN | 8548a | |
| U. yaschenkoi | (1500.8)XSAAXJVHXJSXJXXJ- | 1,5-DAN | N/A | |
| U. yaschenkoi | (1166.5)JJXAAAXAJ(520.3) | MS/MS | 2584.4 | |
| U. yaschenkoi | GFWGXJWEGV | MS/MS | 1178.6 | OcyC2 [C5J887] and Con13 [C7C1L2] |
| U. yaschenkoi | GFWGXJWE | MS/MS | 1022.6 | |
| U. yaschekoi | GFWGXJWEG | MS/MS | 1079.6 | |
| U. armatus | GFWGXJWEGV | MS/MS | 1178.5 | |
| U. armatus | GFWGKJWEGVKNAJ* | MS/MS | 1603.8 | |
| U. armatus | JWEGVKNAJ* | MS/MS | 1028.5 | |
| L. m. obscurus | (1487.9)KKAWQSKJAK | 1,5-DAN | N/A | Parabutoporin [P83312] |
| U. armatus | (934.4)JAKKAWKSKJAKDJRXTAGXAJRNYA- | 1,5-DAN | 5199.3 | |
| U. elongatus | (881.6)JAKKAWKSKJAKKJRXTAGXAJRNPH | 1,5-DAN | 5088.9 | |
| L. m. obscurus | (950.6)GFVSKVVPDGJVKX- | 1,5-DAN | N/A | β-KTx31.1 [P0CI49] |
| U. elongtaus | (1089.6)JVPVGXXXJDSSTCTJYXCSJN- | 1,5-DAN | 6343.7 | La1 [P0C5F3] |
| L. m. obscurus | (1384.6)JTCGJNNE- | 1,5-DAN | N/A | BotIT2 [P59863] |
| L. m. obscurus | FFSJJPSJJGGIASJKK(302.2) | MS/MS | 2093.3 | Imcroporin [C7B247] |
| L. m. obscurus | KHGYPJVR | MS/MS | 969.6 | LmNaTx64.1 [P0CI58] |
| L. m. obscurus | (1005.5)DSDVRGCXFSCMFN- | 1,5-DAN | N/A | Putative NaTx [C9X4J9] |
| L. m. obscurus | (1066.5)TGNNCQRKCSSCD- | 1,5-DAN | N/A | Unknown |
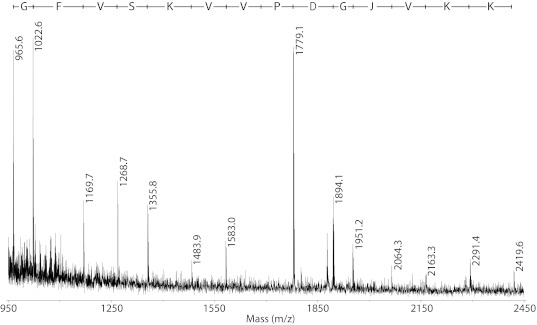
Peptides with intact disulfide bonds are generally not amenable to de novo sequencing by MS/MS, however the presence of cystines does not affect the ability of 1,5-DAN to promote formation of ions useful for sequencing. The use of the 1,5-DAN matrix therefore permitted sequence information to be obtained from native disulfide bonded peptides without the need for prior treatment with a reducing agent. Partial sequences of peptides homologous to cystine-containing potassium and sodium channel scorpion toxins were found in the buthid L. m. obscurus venom, with two peptides previously only detected at the transcript level. One of these sequences shows similarity to the long chain potassium channel toxin 31.1 (βKTx31.1), reported by (Ruiming et al, 2010) as an mRNA transcript containing a predicted propeptide region. The partial sequence obtained can be aligned to the first 14 residues of the mature βKTx31.1 peptide, however the fragment ion series from which the sequence is read begins at 965.6 kDa (see Figure 3). This mass corresponds exactly to the proposed propeptide region of β-KTx31.1 and indicates the putative propeptide region has been retained. Another βKTx-like peptide with the identical proposed propeptide sequence attached had been previously isolated from venom of the buthid Tityus costatus, along with the N-terminally processed form (Diego-Garcia et al, 2005). It has not been determined if the N-terminally extended form is a true propeptide, i.e., removal of the pro-region is required for production of the mature functional peptide, or whether both forms are biologically active. The retainment of this particular pro-region may prove to be of some functional or structural significance since it has now been observed in peptides from two scorpion genera. Further analyses should be performed to explore the function, if any, of this region.
Figure 3.
Partial sequence of βKTx31.1 homologue obtained with 1,5-DAN matrix using MALDI-ToF. Deduced sequence written above spectrum in single letter amino acid code, J represents the isobaric residues isoleucine/leucine.
De novo sequencing of peptides from the Urodacus venoms did not yield sequences corresponding to any known sodium or potassium channel toxins, reflecting the observation that toxins targeting these receptors may be of low abundance or absent in non-buthid venoms. Instead, several of the larger peptides in the mass range typically associated with sodium or potassium channel toxins were found to be homologous to long chain disulfide-containing scorpion antimicrobials such as scorpine and parabutoporin. Unlike MS/MS, sequencing with 1,5-DAN is not limited to small peptides and its use was advantageous in attaining sequences from these larger masses without the need for prior enzymatic digestion. It should be mentioned that several attributes of 1,5-DAN matrix noted in this study were also observed in a previous report on cone snail venom fractions analysed with 1,5-DAN (De Pauw et al, 2007). While 1,5-DAN was useful for ascertaining the presence of disulfide bonds in peptides, fewer masses were detected with 1,5-DAN compared to the total number seen using other matrices, with 42% observed in this study and 23% seen in the cone snail venom report (De Pauw et al, 2007). All masses distinguishable by 1,5-DAN were observed with αCHCA or SA. As with the cone snail study, there were also numerous peptides where the number of disulfide bonds could not be resolved by 1,5-DAN. Despite these limitations, the use of 1,5-DAN for the analysis of scorpion venom fractions has proved to be a valuable tool in addition to the methods and matrices commonly used for current peptidomic studies.
CONCLUSIONS
Venom mass profiles of the six non-buthid and one buthid species reported herein highlights the abundance of small linear peptides under 2kDa in scorpion venoms and the lower number of masses corresponding to long chain sodium channel toxins in non-buthids compared to buthids.
1,5-DAN matrix is a useful matrix for rapid analyses of complex peptide mixtures such as scorpion venoms.
The majority of sequences obtained from the Australian scorpion venoms studied were homologous to known scorpion antimicrobial peptides from both buthid and non-buthid species.
ACKNOWLEDGEMENTS
We acknowledge financial support from an Institute for Molecular Bioscience (IMB) Postgraduate Award and support from an NHMRC Program grant 569927.
ABBREVIATIONS
- 1,5-DAN
1,5-diaminonaphthalene
- ACN
acetonitrile
- αCHCA
α-cyano-4-hydroxycinnamic acid
- MALDI-ToF
matrix assisted laser desorption ionisation-time of flight
- MS/MS
tandem mass spectrometry
- rpHPLC
reversed-phase high performance liquid chromatography
- SA
sinapinic acid
- TFA
trifluoroacetic acid
COMPETING INTERESTS
None declared.
REFRENCES
- Abdel-Rahman MA, Omran MA, Abdel-Nabi IM, Ueda H, McVean A. Intraspecific variation in the Egyptian scorpion Scorpio maurus palmatus venom collected from different biotopes. Toxicon. 2009;53:349–359. doi: 10.1016/j.toxicon.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Asakawa D, Takayama M. Mass spectrometric characterization of phosphorylated peptides using MALDI in-source decay via redox reactions. J Mass Spectrometry. 2012;47:180–187. doi: 10.1002/jms.2052. [DOI] [PubMed] [Google Scholar]
- Batista CV, D’Suze G, Gomez-Lagunas F, et al. Proteomic analysis of Tityus discrepans scorpion venom and amino acid sequence of novel toxins. Proteomics. 2006;6:3718–3727. doi: 10.1002/pmic.200500525. [DOI] [PubMed] [Google Scholar]
- Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J Am Soc Mass Spectrom. 2003;14:971–979. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- Caliskan F, Quintero-Hernandez V, Restano-Cassulini R, et al. Turkish scorpion Buthacus macrocentrus: general characterization of the venom and description of Bu1, a potent mammalian Na-channel alpha-toxin. Toxicon. 2012;59:408–415. doi: 10.1016/j.toxicon.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Cao ZJ, Ma YB, Zhao Y, et al. Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics. 2010;10:2471–2485. doi: 10.1002/pmic.200900763. [DOI] [PubMed] [Google Scholar]
- De Pauw E, Quinton L, Demeure K, et al. New method for characterizing highly disulfide-bridged peptides in complex mixtures: Application to toxin identification from crude venoms. J Proteome Res. 2007;6:3216–3223. doi: 10.1021/pr070142t. [DOI] [PubMed] [Google Scholar]
- Diego-Garcia E, Batista CV, Garcia-Gomez BI, et al. The Brazilian scorpion Tityus costatus Karsch: genes, peptides and function. Toxicon. 2005;45:273–283. doi: 10.1016/j.toxicon.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Diego-Garcia E, Peigneur S, Clynen E, et al. Molecular diversity of the telson and venom components from Pandinus cavimanus (Scorpionidae Latreille 1802): transcriptome, venomics and function. Proteomics. 2012;12:313–328. doi: 10.1002/pmic.201100409. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Quinton L, Nicholson GM. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J Mass Spectrometry. 2008;43:279–295. doi: 10.1002/jms.1389. [DOI] [PubMed] [Google Scholar]
- Fet V, Soleglad ME. Contributions to scorpion systematics. I. On recent changes in high-level taxonomy. Euscorpius. 2005;31:1–13. [Google Scholar]
- Fukuyama Y, Iwamoto S, Tanaka K. Rapid sequencing and disulfide mapping of peptides containing disulfide bonds by using 1,5-diaminonaphthalene as a reductive matrix. J Mass Spectrometry. 2006;41:191–201. doi: 10.1002/jms.977. [DOI] [PubMed] [Google Scholar]
- Gao B, Peigneur S, Tytgat J, Zhu S. A potent potassium channel blocker from Mesobuthus eupeus scorpion venom. Biochimie. 2010;92:1847–1853. doi: 10.1016/j.biochi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Gurrola GB, Hernandez-Lopez RA, Rodriguez de la Vega RC, et al. Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: a novel potent blocker of Kv1.3 potassium channels of human T lymphocytes. Biochemistry. 2012;51:4049–4061. doi: 10.1021/bi300060n. [DOI] [PubMed] [Google Scholar]
- Gutierrez JM, Lomonte B, Leon G, et al. Snake venomics and antivenomics: Proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J Proteomics. 2009;72:165–182. doi: 10.1016/j.jprot.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Inceoglu B, Lango J, Jing J, et al. One scorpion, two venoms: prevenom of Parabuthus transvaalicus acts as an alternative type of venom with distinct mechanism of action. Proc Natl Acad Sci USA. 2003;100:922–927. doi: 10.1073/pnas.242735499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbister GK, Graudins A, White J, Warrell D. Antivenom treatment in arachnidism. J Toxicol-Clin Toxicol. 2003;41:291–300. doi: 10.1081/clt-120021114. [DOI] [PubMed] [Google Scholar]
- Isbister GK, Volschenk ES, Seymour JE. Scorpion stings in Australia: five definite stings and a review. Intern Med J. 2004;34:427–430. doi: 10.1111/j.1445-5994.2004.00625.x. [DOI] [PubMed] [Google Scholar]
- Juhasz P. Selection of matrix for MALDI. Proceeding of Workshop of 41st American Society of Mass Spectrometry (ASMS) Conference; Boston, MA. 1993. [Google Scholar]
- Jungo F, Bougueleret L, Xenarios I, Poux S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon. 2012;60:551–557. doi: 10.1016/j.toxicon.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhao Y, Zhao R, et al. Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics. 2010;10:2471–2485. doi: 10.1002/pmic.200900763. [DOI] [PubMed] [Google Scholar]
- Ma YB, Zhao RM, He Y, et al. Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: implication for the evolution of the scorpion venom arsenal. BMC Genomics. 2009;10:290. doi: 10.1186/1471-2164-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MF, Rochat H, Marchot P, Bougis PE. Use of high performance liquid chromatography to demonstrate quantitative variation in components of venom from the scorpion Androctonus australis Hector. Toxicon. 1987;25:569–573. doi: 10.1016/0041-0101(87)90293-5. [DOI] [PubMed] [Google Scholar]
- Molle D, Jardin J, Piot M, et al. Comparison of electrospray and matrix-assisted laser desorption ionization on the same hybrid quadrupole time-of-flight tandem mass spectrometer: application to bidimensional liquid chromatography of proteins from bovine milk fraction. J Chromatogr A. 2009;1216:2424–2432. doi: 10.1016/j.chroma.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Newton KA, Clench MR, Deshmukh R, Jeyaseelan K, Strong PN. Mass fingerprinting of toxic fractions from the venom of the Indian red scorpion, Mesobuthus tamulus: biotope-specific variation in the expression of venom peptides. Rapid Commun Mass Spectrom. 2007;21:3467–3476. doi: 10.1002/rcm.3240. [DOI] [PubMed] [Google Scholar]
- Pimenta AM, Stocklin R, Favreau P, Bougis PE, Martin-Eauclaire MF. Moving pieces in a proteomic puzzle: mass fingerprinting of toxic fractions from the venom of Tityus serrulatus (Scorpiones, Buthidae) Rapid Commun Mass Spectrom. 2001;15:1562–1572. doi: 10.1002/rcm.415. [DOI] [PubMed] [Google Scholar]
- Pimenta AMC, Rates B, Ferraz KKF, et al. Tityus serrulatus venom peptidomics: Assessing venom peptide diversity. Toxicon. 2008;52:611–618. doi: 10.1016/j.toxicon.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Prendini L, Wheeler WC. Scorpion higher phylogeny and classification, taxonomic anarchy, and standards for peer review in online publishing. Cladistics. 2005;21:446–494. doi: 10.1111/j.1096-0031.2005.00073.x. [DOI] [PubMed] [Google Scholar]
- Rates B, Verano-Braga T, Santos DM, et al. From the stretcher to the pharmacy's shelf: drug leads from medically important brazilian venomous arachnid species. Inflamm Allergy Drug Targets. 2011;10:411–419. doi: 10.2174/187152811797200614. [DOI] [PubMed] [Google Scholar]
- Remijsen Q, Verdonck F, Willems J. Parabutoporin, a cationic amphipathic peptide from scorpion venom: much more than an antibiotic. Toxicon. 2010;55:180–185. doi: 10.1016/j.toxicon.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega RC, Possani LD. Current views on scorpion toxins specific for K+-channels. Toxicon. 2004;43:865–875. doi: 10.1016/j.toxicon.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega RC, Possani LD. Overview of scorpion toxins specific for Na+ channels and related peptides: biodiversity, structure-function relationships and evolution. Toxicon. 2005;46:831–844. doi: 10.1016/j.toxicon.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega RC, Schwartz EF, Possani LD. Mining on scorpion venom biodiversity. Toxicon. 2010;56:1155–1161. doi: 10.1016/j.toxicon.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Ruiming Z, Yibao M, Yawen H, et al. Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genomics. 2010;11:452. doi: 10.1186/1471-2164-11-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EF, Diego-Garcia E, Rodriguez de la Vega RC, Possani LD. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones) BMC Genomics. 2007;8:119. doi: 10.1186/1471-2164-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Hill JM, Little MJ, et al. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc Natl Acad Sci USA. 2011;108:10478–10483. doi: 10.1073/pnas.1103501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden A, Herrel A, Summers A. Comparison of chela size and pincer force in scorpions; getting a first grip. J Zool. 2010;280:319–325. [Google Scholar]
- Zeng XC, Corzo G, Hahin R. Scorpion venom peptides without disulfide bridges. Iubmb Life. 2005;57:13–21. doi: 10.1080/15216540500058899. [DOI] [PubMed] [Google Scholar]
- Zerrouk H, Bougis PE, Ceard B, Benslimane A, Martin-Eauclaire MF. Analysis by high-performance liquid chromatography of Androctonus mauretanicus mauretanicus (black scorpion) venom. Toxicon. 1991;29:951–960. doi: 10.1016/0041-0101(91)90078-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.