Abstract
Asthma is heritable, influenced by the environment, and is modified by in utero exposures and aging; all of these features are also common to epigenetic regulation. Furthermore, the transcription factors that are involved in the development of mature T cells that are critical to the Th2 immune phenotype in asthma are regulated by epigenetic mechanisms. Epigenetic marks - DNA methylation, modifications of histone tails, and non-coding RNAs – work in concert with other components of cellular regulatory machinery to control spatial and temporal level of expressed genes. Technology to measure epigenetic marks on genomic scale and comprehensive approaches to data analysis have recently emerged and continue to improve. Alterations in epigenetic marks have been associated with exposures relevant to asthma, particularly air pollution and tobacco smoke, as well as asthma phenotypes in a few population-based studies. On the other hand, animal studies have begun to decipher the role of epigenetic regulation of gene expression associated with the development of allergic airway disease. Epigenetic mechanisms represent a promising line of inquiry that may, in part, explain the inheritance and immunobiology of asthma.
Keywords: asthma, atopy, epigenetics, gene expression, DNA methylation, histone marks, noncoding RNAs
Introduction
Asthma is a complex, heritable disease affecting more than 8% of the U.S. population, ~7 million children and ~18.7 million adults1, 2. This disease has been increasing in prevalence, incidence, and severity3 although recent evidence suggests that the prevalence of asthma and allergies may have come to a plateau in developed countries4. Asthma accounts for over $10 billion of direct health care costs in the U.S.3. In 2008, persons with asthma missed 10.5 million school days and 14.2 million work days due to their disease1. Gender and ethnic differences exist for women and African American asthmatics, both having a significantly higher rate of outpatient asthma visits, emergency room evaluations, and hospitalizations than non-Hispanic males1. Consistent with these data is an increased mortality rate in women and African American asthmatics, 45% and 200% higher respectively, than non-Hispanic white males. What is most disturbing is that ongoing increases in disease prevalence, incidence, and severity are occurring despite the intense national and international investigation into the pathobiology, genetics, and treatment of asthma. Consequently, it is essential to consider alternative explanations for the growing health problems associated with the development and persistence of asthma.
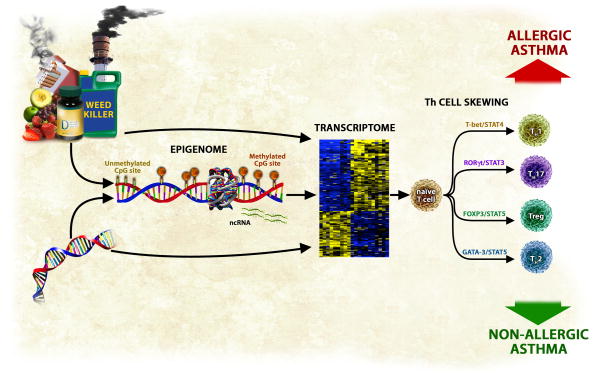
Several separate lines of evidence support a role for epigenetics in asthma (Figure 1). First, asthma, like epigenetic mechanisms, is heritable. While asthma is a strongly familial condition (36–79% heritability) with a non-Mendelian pattern of inheritance and polymorphisms in more than 100 genes5–8, these associations have infrequently been replicated and genetics has explained only a small portion of the etiology of this disease6. Second, asthma, like epigenetic mechanisms, shows a parent-of-origin transmission of inheritance with an affected mother significantly more likely to transmit the disease than an affected father9. These parent-of-origin effects may result from immune interactions between the fetus and the mother10. Alternatively, the maternal effect may be the result of epigenetically regulated genomic imprinting11. Several known genes show parent-of-origin effects on allergic disease; these genes include the FcεRI-β locus12, and the Spink5 gene13. Third, asthma, like epigenetic mechanisms14, 15, is affected by in utero exposures16, 17. Prenatal exposure to maternal and grand-maternal cigarette smoke18–20 and traffic-related air pollution21, 22 are among in utero exposures that contribute to the development of this disease. On the other hand, higher maternal fruit and vegetable intake and oily fish consumption during gestation have been associated with a lower risk of asthma 23. Fourth, asthma, like epigenetics, is influenced by the general environment24. Environmental factors are known to play important roles in the pathogenesis of asthma; both in terms of main effects, and those exerted indirectly through complex interactions with gene variants25. The dramatic increase in the prevalence, incidence, and severity of asthma over the last 20 years provides strong evidence that exposures, including diet, play an important role in the development of this disease; these changes have occurred too rapidly to be accounted for by changes in primary DNA sequence alone. While allergens are classically associated with asthma26, many other exposures, including smoking behavior25, 27, agents in the workplace28, indoor and outdoor air pollution29, viruses30, domestic31 and occupational32 exposure to endotoxin, and immunization against certain infectious diseases33 are associated with the development and progression of this disease, and several of these agents have been shown to alter epigenetic marks. Finally, asthma is an immune-mediated disease characterized mainly by skewing toward a Th2 phenotype although other T cell subtypes may be involved34. Epigenetic mechanisms regulate expression of transcription factors that are involved in T cell differentiation (Th1, Th2, and Tregs)35–42.
Figure 1.
An overview of epigenetic regulation of gene expression in asthma. Environmental exposures and dietary factors an individual is exposed to both in utero and postnatally influence epigenetic marks which in turns regulates genes expression. Underlying genetic variation can regulate gene expression by affecting epigenetic marks or by other mechanisms (alteration of transcription factor binding sites, for example). Alterations in epigenetic marks have consequences on expression of key immune genes that regulate Th subtype cell skewing, which in turn leads to development of disease. It is likely that distinct epigenomic profiles are associated with the development of allergic and non-allergic forms of asthma.
Epigenetic Mechanisms
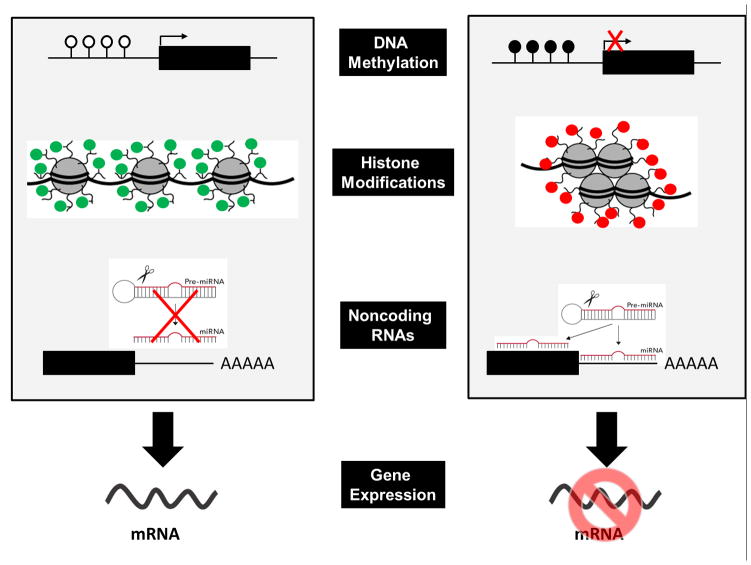
Epigenetics is traditionally defined as the study of heritable changes in gene expression caused by molecules that bind to DNA rather than changes in the underlying DNA sequence (Table 1)43. Recent evidence suggests that the epigenome is dynamic and changes in response to the environment, diet, and ageing44. In addition to a set of inherited epigenetic marks, there are likely non-heritable epigenetic marks that are more dynamic and change in response to environmental stimuli. Three main classes of epigenetic marks are DNA methylation, modifications of histone tails, and non-coding RNAs (Figure 2).
Table 1.
Key Components of the Epigenetic Regulation of Gene Expression
| Epigenetics | The word epigenetics is derived from the Greek word epi for over or above, and genetics or the science of heredity. Two key components of epigenetics (DNA methylation and histone modifications) together define chromatin state beyond the information that is encoded in the genomic DNA sequence. Together they control degree of accessibility of the genomic DNA fraction of chromatin. Tightly bound ‘closed’ chromatin state is less accessible to transcription machinery and other regulatory proteins while more accessible ‘open’ chromatin state leads to active gene transcription. Epigenetic marks work in concert with other components of cellular regulatory machinery (transcription factors, enhancer, and repressors) to control spatial and temporal level of expressed genes. |
| DNA Methylation | The extent of methylation of cytosines in ~ 30 million CpG sites in the human genome. 60–90% of CpGs across the human genome are methylated while regulatory regions containing more dense areas of CpG motifs (CpG islands; stretches of DNA >200 bp in length with >50% GC content and observed/expected CpG>0.6152) are generally unmethylated. DNA methylation also correlates with spatial organization of the chromatin (proximity of chromosomal loci)153. |
| Histone Modifications | In eukaryotes, DNA is packaged and ordered into nucleosomes, the basic structural unit of chromatin, by wrapping around the octamer consisting of 2 copies each of histone proteins (H2A, H2B, H3 and H4). Histone modifications include methylation, acetylation, phosphorylation and other modifications of specific amino acids in nucleosomal histones. Generally, histone marks are described as ‘permissive’ (active promoters), ‘repressive’ (inactive promoters) or ‘poised’ (accessible promoters). |
| Noncoding RNA | The term non-coding RNA (ncRNA) is commonly employed for RNA that does not encode a protein. ncRNAs include both small and large classes of RNA molecules that control gene expression by a variety of mechanisms. |
Figure 2.
Effect of epigenetic marks – DNA methylation, histone modifications, and miRNAs – on gene expression. White circles denote unmethylated CpGs while black circles denote methylated CpGs. Green circles refer to permissive histone modifications while red circles indicate repressive histone marks. miRNAs can affect gene expression by either RNA degradation (perfect complementarity and binding) or inhibition of protein translation (imperfect complementarity and partial binding).
Methylation of cytosine residues in CpG dinucleotides (5-methylcytosine) within the context of CpG islands is the simplest form of epigenetic regulation in eukaryotes with hypermethylation of CpG islands in gene promoters leading to gene silencing and hypomethylation leading to active transcription. CpG island methylation has long been studied in cancer with findings that hypermethylation of tumor suppressor genes and hypomethylation of oncogenes contribute to the process of carcinogenesis11, 45. More recent studies have demonstrated that methylation of less CpG dense regions near CpG islands (‘CpG island shores’) controls expression of tissue specific genes as well as genes relevant to carcinogenesis and lineage-specific cell differentiation46, 47, suggesting that DNA methylation outside of CpG islands is an important mechanism that controls gene transcription. Additionally, recent evidence suggests that DNA methylation is more prevalent within gene bodies than in promoters48. Intragenic DNA methylation functions at least in part by regulating transcription from alternative promoters49 but it is likely that other mechanisms are also involved. The DNA ‘methylome’ of the H1 human embryonic stem cell line uniquely revealed that nearly one-quarter of all methylation is in non-CpG context50, suggesting that embryonic stem cells may use different methylation mechanisms to control gene expression. 5-methylcytosine can be oxidized to 5-hydroxymethylcytosine by the recently discovered TET family of enzymes51. Although the role of 5-hydroxymethylcytosine in epigenetic regulation of gene expression is not fully elucidated, it has been suggested that 5-hyroxymethycytosine is a mark of demethylation51 and that it potentially plays a role in the regulation of specific promoters and enhancers52.
Methylation, acetylation, phosphorylation, and ubiquitylation 43 of histone tails occur at specific sites and residues, and controls gene expression by regulating DNA accessibility to RNA polymerase II and transcription factors. H3K4 trimethylation (H3K4me3), for example, is strongly associated with transcriptional activation whereas H3K27 trimethylation (H3K27me3) is frequently associated with gene silencing53. Similarly, histone tail acetylation leads to active gene transcription while deacetylation is a repressive mark and leads to gene silencing. Histone acetyltransferases (HATs) are enzymes that acetylate histone tails while histone deacetylases (HDACs) remove acetyl groups from histone tails. Analogous to DNA methylation, deregulation of these histone modifications has been linked to misregulation of gene expression in cancers54.
MicroRNAs (miRNAs) are the most studied class of noncoding RNAs that control gene expression by binding to the 3′ untranslated regions (UTRs) of messenger RNA (mRNA), which leads to either mRNA degradation or inhibition of protein translation55. Almost 2000 mature miRNAs have been identified in the human genome (http://www.mirbase.org/) but it is expected that more miRNAs will be identified in the near future. Alterations of expression of miRNAs contribute to pathogenesis of most malignancies with miRNAs acting as both oncogenes and tumor suppressor genes56 but miRNAs also have well-established roles and are therapeutic targets in cardiovascular disease57 and liver injury58. More recently, ncRNAs, such as PIWI-interacting RNAs (piRNAs), small nucleolar RNAs, (snoRNAs), transcribed ultraconserved regions (T-UCRs) and large intergenic non-coding, RNAs (lincRNAs) are emerging as a key component of deregulated transcription not only in tumorigenesis but also in many non-malignant diseases59.
An emerging paradigm for epigenetic regulation of gene expression is the relationship between DNA methylation and histone modifications. One example of these interactions is binding of DNMT3L, a regulatory factor related in sequence to mammalian de novo methyltransferases DNMT3A and DNMT3B, to the N terminus of histone H3 tail60, 61. DNMT3L recognizes unmethylated H3 tails at lysine 4 and induces de novo DNA methylation by recruitment or activation of DNMT3A2; these findings establish the N terminus of histone H3 tail with an unmethylated lysine 4 as a chromatin determinant for DNA methylation. Similarly, DNA methyltransferases preferentially target nucleosome-bound DNA62. The relationship of histones and DNA methylation is bidirectional; in addition to histones playing a role in the establishment of DNA methylation patterns, DNA methylation is important for maintaining patterns of histone modification through cell division63. Cross-talk between DNA methylation and miRNAs has also been identified64, 65.
In addition to cross-talk between different epigenetic marks, it is becoming evident that underlying genetic variation and epigenetic marks work together. The best example is allele-specific gene expression where allele-specific differences in gene expression may arise because of sequence variation that may be marked by differences in DNA methylation66–68, histone modifications or chromatin structure.
Epigenetic marks (DNA methylation and histone marks) are a key component of cell-specific gene expression and, as such, are erased during germ cell development (meiosis) and re-established following fertilization. This process is referred to as epigenetic reprogramming69 and constitutes of comprehensive erasure and re-establishment of DNA methylation and extensive remodeling of histone modifications in two steps. Epigenetic reprogramming is a key feature of inheritance of epigenetics marks. Genes that are expressed from only one parental allele, known as imprinted genes, are protected during the second reprogramming step by mechanisms that are being unraveled70.
Epigenomic Study Design
The first step in epigenomic analysis is experimental design, including the choice of tissue/cells to be profiled and study design. Challenges in selection of the material to be used include limited availability of lung tissue for asthma studies and cell heterogeneity in available samples (such as DNA from whole blood). One way to address the first challenge will be to analyze paired lung-blood samples to identify epigenetic marks that carry over from the lung to peripheral blood and test whether surrogate tissue (peripheral blood mononuclear cells, nasal epithelia, sputum, etc) adequately reflect activity in the lung. The second challenge can be addressed by collecting white blood cell count data and including the constituent cell counts in the analysis. If this information is not available, established epigenomic profiles for constituent cell cells (such as data generated by the Roadmap Epigenomic project, http://www.roadmapepigenomics.org/) can be used to estimate relative abundances of different cell types71.
Study design and power calculations based on previously collected data are important in designing studies that are able to identify significant epigenetic changes after adjustments for genome-wide comparisons. The most powerful study design for epigenomic analysis uses monozygotic twins that are essentially identical genetically so that all differences in phenotype can be contributed to environmental factors with paired-sample design allowing for better statistical power72. Another design with reasonably high power includes siblings (not necessarily twins) discordant for disease phenotypes with parental DNA also available for estimates of heritability. Case-control design with a large enough number of individuals included in the analysis is often used due to availability of samples. The final considerations in the study design are clinical and immune phenotypes of interest. Prior to sample selection from available specimens for epigenomic profiling, clinical/immune variables must be analyzed to identify those that have reliable measurements, normal distribution, and have a strong clinical or biological rationale to include in statistical models. Once epigenomic profiles are collected and data are normalized, principal components analysis (PCA) can be used to prioritize variables based on how much variance in the dataset they account for.
Epigenomic Profiling
Epigenetic marks can be studied using focused and genome-wide approaches (Table 2)73. Generally, studies begin with genome-wide approaches to identify targets followed by focused approaches to internally validate (confirmation in the same cohort) or externally validate (independent cohort) the initial findings. Microarrays have been the tool of choice for profiling epigenetic marks on a genomic scale, with several platforms and protocols available for DNA methylation (Table 2)74. Most commonly used array platforms for DNA methylation are Illumina 450k BeadChip, Comprehensive Analysis of Relative DNA Methylation (CHARM) platform75, and MeDIP arrays (Agilent Technologies and Roche Nimblegen). Array platforms have also been used to examine histone modifications by chromatin precipitation followed by hybridization on microarrays (CHIP-chip)74 and for miRNAs76.
Table 2.
Summary of Commonly Used Techniques for Epigenetic Profiling
| Type of Epigenetic Mark | Type of Sample Preparation Approach | Type of Profiling Approach |
|---|---|---|
| DNA methylation | Bisulfite conversion | Microarray (Illumina), High-throughput sequencing (BS-seq, RRBS-seq), Epityper (mass spectrometry; focused) or pyrosequencing (focused) |
| Methylated DNA immunoprecipitation (MeDIP) | Microarray or High-throughput sequencing (MeDIP-seq) | |
| Methyl-binding domain (MBD) precipitation | Microarray or high-throughput sequencing (MBD-seq) | |
| Restriction digest with methylation-sensitive restriction enzymes (McrBC, MspI, HpaII, MspJI etc) | Microarray (CHARM, HELP) or high-throughput sequencing (MRE-seq) | |
| Histone modifications | Chromatin immunoprecipitation (CHIP) | Microarray (CHIP-chip), high-throughput sequencing (CHIP-seq) or quantitative PCR (focused) |
| DNA methylation associated with chromatin modifications | CHIP followed by bisulfite conversion | High-throughput sequencing (CHIP- BS-seq, Bis-CHIP-seq) |
| Noncoding RNAs | Size selection of appropriate RNA molecules | Microarray, high-throughput sequencing (miRNA-seq and RNA-seq) or quantitative RT-PCR (focused) |
| Chromatin accessibility | DNAse I cleavage | High-throughput sequencing (DNase sensitivity-seq) |
| Chromatin accessibility Chromatin spatial organization | Formaldehyde cross-linking | High-throughput sequencing (FAIRE-seq) |
| Chromosome conformation capture | High-throughput sequencing (3C-seq, 4C-seq, 5C-seq) |
However, the most substantial advance in the area of technologies for the assessment of epigenetic marks on the genome scale in recent years has been the introduction of next-generation sequencing technologies77. Application of next-generation sequencing to epigenomic research has been recently reviewed78. These technologies have been widely used for the study of histone marks (CHIP-seq) and microRNAs (miRNA-seq) as they provide superb quality data compared to array platforms. They have also been used to identify open chromatin areas of the genome (FAIRE-seq)79 and spatial chromatin organization (3C-seq)80. The majority of methylation profiling is still done on array platforms as bisulfite-converted DNA sequencing (BS-seq) on the genomic scale is expensive. However, a number of techniques that examine only regions of the genome enriched for methylation marks have been developed and are being increasingly used81. Recent advances in the development of techniques for epigenomic profiling include attempts to define genome-wide patterns of DNA hydroxymethylation82, 83 and to study DNA methylation and histone modifications in one experiment84, 85
Pyrosequencing86 and Epityper assays on the Sequenom MassARRAY platform are commonly used techniques for interrogation of a small number of CpG sites while quantitative PCR (qPCR) methods are typically used for focused studies of histone modifications and miRNAs. In addition to site-specific methods for assessment of DNA methylation, some studies assess overall level of methylation in each sample (global methylation); this is often measured by assessing methylation in repeat regions of the genome (Alu, LINE-1, Sat2), mass spectrometry methods, or luminometric methylation assay (LUMA)87.
Epigenomic Data Analysis
The first step in analysis of collected epigenomic data is to identify statistically significant differences between disease states. Statistical methods used for microarray analysis have generally been applicable to epigenomic profiles collected on arrays or sequence data after alignments and tag counts have been performed78. Strategies for analyzing tiling arrays have also been used in epigenomic analyses (such as CHARM or CHIP-chip)88. One of the problems with this type of analysis is that it is only associative and does not demonstrate causality. Methods used in epidemiology and genetical genomics are beginning to be applied to epigenomes to identify causal relationships89.
The second and most complex step in the analysis of epigenomic data is understanding how different epigenetic mechanisms together influence gene expression. Each of the three epigenetic mechanisms is independently complex but when combined, the complexity of these interactions presents unique analytic challenges. We are just beginning to understand how one type of epigenetic mark affects gene expression90. However, the evidence for cross-talk among different types of epigenetics marks is accumulating. The complexity of epigenetic regulation of gene expression is high even when one is interested in examining only one gene or locus and there are considerable challenges associated with understanding these interactions and effect on gene regulation genome-wide. Analytical strategies for these types of integrative epigenomic analyses have not reached maturity but are starting to be applied to disease datasets91. Two types of integrative analysis will be important to apply to epigenomic data mapping strategies and network analysis. Expression quantitative trait loci (eQTL) mapping approaches92 can be applied to identify genetic variants that underlie methylation status (methyl-QTL) or methylation marks that control expression changes (methyl-expression QTL). Similarly, co-expression network analysis strategies that have been applied to expression analysis can be applied to epigenomic analysis93.
Epigenetic Marks and the Immune System
A substantial body of evidence suggests that epigenetic mechanisms affect the expression of cytokines and binding of transcription factors that control the lineage of Th1, Th2, and Treg cells. In the context of Th1/Th2 differentiation, the most extensively studied are the Th1 cytokine IFNγ, and Th2 cytokines IL-4 and IL-13. It has been shown that de novo DNA methyltransferase DNMT3A methylates CpG -53 in the Ifng promoter35 and cord blood CD4+ cells enhance the development of Th1 (but not Th2) lineage through progressive demethylation of the Ifng promoter36. Methylation of the Ifng promoter was reduced in CD8+ cells from atopic children in the age range in which hyperproduction of IFNγ occurs, suggesting that DNA methylation at this locus may be a contributing factor in the development of atopy in children. Differentiation of human CD4+ cells into the Th2 subtype is accompanied by the appearance of DNase I hypersensitive sites (DHS) and CpG demethylation around these DHS sites within IL-4 and IL-13 promoters37–39. Extensive studies of the Th2 cytokine locus control region (LCR)40 have shown that rad50 hypersensitive site 7 (RHS7) within the Th2 cytokine LCR undergoes rapid demethylation during Th2 differentiation41.
In addition to DNA methylation, histone modifications are also important in guiding T cell differentiation. T-bet and GATA-3 transcription factors control lineage-specific histone acetylation of Ifng and Il4 loci during Th1/Th2 differentiation42. Rapid methylation of H3K9 and H3K27 residues (repressive marks) at the Ifng locus are associated with differentiating Th1 cells while demethylation of H3K9 and methylation of H3K27 was associated with Th2 differentiation. In a study of human cord blood CD4+ cells, histone acetylation marks at the proximal Il13 promoter were selectively observed in Th2 cells39, suggesting that permissive histone marks together with DNA demethylation lead to expression of IL-13 in Th2 cells. In aggregate, these studies suggest that DNA methylation and histone modifications are highly dynamic and represent important determinants of Th1 and Th2 cell lineages.
Although miRNAs were discovered relatively recently, there is already a substantial body of evidence for the role of miRNAs in the development and function of the immune system94–96. A number of differentially expressed miRNAs have been identified in response to innate and adaptive immune stimuli with many commonalities in miRNA expression (miR-21, -103, -155, and -204) 94. miRNA-155 is the most often identified differentially regulated noncoding RNA in studies involving the immune system94, 95; a recent study revealed that miR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 497.
Epigenetic mechanisms controlling regulatory T cells (Tregs) development are also beginning to be explored. Tregs are a unique T-cell lineage with an important role in immunological tolerance whose development is primarily regulated by the transcription factor FOXP3. Evidence for the role of DNA methylation98 and histone modifications99 in regulation of FOXP3 expression are summarized in two recent reviews100, 101. There is also clear evidence that miRNAs are involved in Treg development and function96.
The Role of the Environment and In Utero Exposures in Modulating the Epigenome
Unlike an individual’s genetic make-up, epigenetic marks can be influenced much more easily by exposures, diet, and ageing. Randy Jirtle’s seminal experiments showed that maternal diet supplemented with methyl donors (folic acid, vitamin B12, choline and betaine) shifts coat color distribution of progeny towards the brown pseudoagouti phenotype, and that this shift in coat color resulted from an increase in DNA methylation in a transposon adjacent to the agouti gene 14, 15. These studies also revealed that mice with yellow coat color are obese and develop cancer, suggesting for the first time that changes in DNA methylation caused by diet in utero may be linked to disease development. Other studies have shown that pesticides and fungicides can alter the methylome resulting in changes in male fertility102, and that ageing is also associated with changes in DNA methylation and gene expression103. The concepts associated with environmental epigenetics were reviewed recently elsewhere44.
More recent evidence suggests that environmental exposures relevant to the development of asthma, such as air pollution and cigarette smoke, also affect the epigenome. Decreased DNA methylation in peripheral blood (as measured by LINE-1 repeats) was found to be associated with exposure to PM2.5 particles amongst 718 elderly individuals in the Boston area104, and although this correlated with time-dependent variables such as day of the week and season, there was no association with air pollution-related health effects. Another study demonstrated that hypomethylation of iNOS (Nos2) promoter in buccal cells was associated with exhaled nitric oxide (NO) levels and PM2.5 exposure among 940 participants in the Children’s Health Study105.
Several epidemiological studies have examined the relationship between exposure to cigarette smoke and epigenetic marks. Among 384 children, a global reduction in DNA methylation, as measured by the extent of methylation of Alu repeats, and differential methylation of 8 specific CpG motifs was found to be associated with in utero smoke exposure106. 15 specific genomic loci were significantly associated with current smoking, two with cumulative smoke exposure, three with time since quitting cigarettes in 1085 individuals enrolled in the International COPD Genetics Network and validated in the Boston Early-Onset COPD study (n = 369)107. Cigarette smoke exposure has also been shown to have a significant influence on expression of miRNAs108–110. Comparing current to never smokers, 28 miRNAs were differentially expressed, mostly downregulated in human bronchial airway epithelium of smokers108. miR-218 was found to be one of the strongly associated miRNA with cigarette smoke exposure and it was further shown that a change in miR-218 expression in primary bronchial epithelial cells and H1299 cell line resulted in a corresponding anti-correlated change in expression of predicted mRNA targets for miR-218.
Other studies have examined the influence of cigarette smoke exposure on epigenetic marks in vitro or in animal models. Normal human airway epithelial cells and immortalized bronchial epithelial cells exposed to cigarette smoke condensate (CSC) identified time- and dose-dependent changes in histone modifications (decrease in H4K16Ac and H4K20Me3, and increase in H3K27Me3) accompanied by decreased DNMT1 and increased DNMT3b expression; these changes are characteristic of lung cancer progression111. Two other studies also demonstrated changes in miRNA expression in lungs of mice110 and rats109 exposed to cigarette smoke with substantial overlap between mice and rats and some overlap of rodent miRNA expression changes in the lung with those observed in human airway epithelium108.
In addition to influencing epigenetic marks as a result of direct exposure, in utero exposure to components or air pollution or cigarette smoke results in changes in global and site-specific DNA methylation. Maternal exposure to benzo(a)pyrene (BaP), a representative airborne polycyclic aromatic hydrocarbon (PAH), was associated with hypermethylation of IFNγ in cord blood DNA from 53 participants in the Columbia Center for Children’s Environmental Health cohort112. In another study, global hypomethylation has been associated with maternal smoking and cotinine levels in the umbilical cord blood from 30 newborns113. In a birth cohort of 90 women born 1959-63 in New York City, prenatal tobacco exposure, measured at the time of pregnancy and not retrospectively reported, was associated with a decrease in Sat2 methylation but not LINE-1 or Alu methylation114. Examination of two differentially methylated regions (DMRs) regulating two imprinted loci (H19 and Igf2) in infants born to 418 pregnant women demonstrated that infants born to smokers had higher methylation at the Igf2 DMR than those born to never smokers or those who quit during pregnancy (no differences were seen in the H19 DMR)115. Similarly, DNA methylation in Axl, a receptor tyrosine kinase relevant in cancer and immune function, was 2.3% higher in peripheral blood of children exposed to maternal smoking in utero116. Finally, one study has demonstrated association of maternal cigarette smoking during pregnancy with downregulation of several miRNAs in placenta; expression of one of the miRNAs (miR-146a) was downregulated in dose-dependent manner in immortalized placental cell lines exposed to nicotine and BaP117.
Asthma Epigenetics – Animal Studies
Given the evidence for the strong influence of environmental exposures on epigenetic marks and the role of epigenetic regulation in T cell differentiation, it is becoming clear that epigenetic changes may be one of the factors to explain the increasing prevalence of asthma. Our group hypothesized that these dietary influences are, at least in part, mediated by the epigenome. To test this hypothesis, we conducted a study in which pregnant female mice were fed either a low or high methylation diet and progeny were sensitized and challenged with ovalbumin 17. We observed an increase in airway inflammation, serum IgE, and airway hyperresponsivness (AHR) in pups of mothers who were fed high methylation diet compared to those of mothers on low methylation diet. Furthermore, we demonstrated hypermethylation of 82 gene-associated CpG islands throughout the genome, including extensive hypermethylation of the promoter and decreased expression of Runx3, a gene known to regulate allergic airway disease in mice. Importantly, we reversed the immune phenotype by treatment with a demethylating agent (5-aza-deoxycytidine). Epidemiological evidence for association of folic acid with the development of asthma in children has been mixed118–122 but it may be that folate together with other methyl donors in the diet plays a role in this disease.
Importantly, a direct link between epigenetic control of the Th2 cytokine locus and development of allergic airway diseases was further demonstrated in mice with deficiency in the Th2 LCR123. A more recent study also identified a DNase I-hypersensitive site 2 (HS2) element in the second intron of the Il4 gene as the strongest of all known Il4 enhancers and showed that this enhancer is strictly controlled by GATA-3 binding124. Moreover, Tanaka et al. propose a new model in which independent recruitment of GATA-3 to locus-specific regulatory elements controls the status of the expression of genes encoding Th2 cytokines125.
A number of other animal studies have since examined DNA methylation in the context of allergic airway disease. Fedulov et al demonstrated DNA methylation changes in splenic CD11c+ dendritic cells (DCs) from neonate mice born to allergic mothers (mothers sensitized and challenged with ovalbumin)126. Brand and colleagues observed increased methylation of the Ifng promoter (and increased IFNγ cytokine production) in CD4+ T lymphocytes after ovalbumin sensitization challenge and demonstrated that methylation of the Ifng promoter is required for development of allergic airway disease by using 5-aza-deoxycytidine (demethylating agent) and adoptive transfer experiments of CD4+ T-cells from sensitized/challenged to naïve animals and reverse127. Although both demethylation and adoptive transfer experiments clearly demonstrate the importance of methylation marks in CD4+ cells in development of allergic airway disease, loci other than Ifng may be important in this process and should be examined. Finally, DNMT3A, but not DNMT3B, deficiency is CD4+ lymphocytes (conditional mutant mice) was shown to result in increased expression of IL-13 (and other Th2 cytokines), decreased DNA methylation and changes in H3K27 acetylation/methylation in the IL-13 promoter, increased airway inflammation and AHR in the ovalbumin model of allergic airway disease127. This study clearly demonstrates the role of DNA methylation in controlling expression of Th2 cytokines and development of allergic airway disease in mice.
Several recent studies have also begun to shed light on the role several miRNAs play in the development of allergic airway disease in animal models128. Selective blockade in miR-126 resulted in diminished Th2 response, inflammation, and AHR in the house dust mite (HDM) model; these effects were shown to be mediated by activation of the MyD88 innate immune signaling pathway. Using the same HDM model, this group also demonstrated that inhibition of miR-145 inhibited eosinophilic inflammation, mucus hypersecretion, Th2 cytokine production, and airway hyperresponsiveness, and that the anti-inflammatory effects of miR-145 antagonism were comparable to glucocorticoid treatment129. Two studies identified a controversial role for the let-7 family of miRs in the ovalbumin model of allergic airway disease130, 131. The first study showed that multiple members of the highly conserved let-7 miRNA family are the most increased lung miRNAs in response to allergen130. The authors confirmed that IL-13 is regulated by let-7a in vitro and demonstrated that inhibition of let-7 miRNAs in vivo using a locked nucleic acid profoundly inhibited allergic inflammation and AHR, suggesting a proinflammatory role for let-7d. The second independent study demonstrated that let-7 miRNAs regulate IL-13 production in A549 cells and primary cultured T cells, and that intranasal administration of mature let-7 mimic to lungs of mice with allergic inflammation resulted in decreased IL-13, AHR and mucus metaplasia, implying an anti-inflammatory role for let-7131. More studies are needed to understand the discrepancy in these findings but this illustrates the complexity of miRNA regulation of gene expression.
Finally, three recent studies have demonstrated how miRNAs play a crucial role in regulation of IFNγ and therefore T-cell polarization. Targeted ablation of miR-21 led to reduced lung eosinophilia after ovalbumin sensitization and challenge, with a broadly reprogrammed immunoactivation transcriptome and significantly increased levels of the Th1 cytokine IFNγ132. Consistent with the miR-21 binding site in IL-12p35, DCs from miR-21-deficient mice produced more IL-12 after LPS stimulation and OVA-challenged CD4+ T-cells from the same mice produced more IFNγ and less IL-4. Two studies showed that miR-29 suppresses IFNγ production133, 134. Steiner et al. performed gene expression profiling of cells that do not produce miRNAs (DGCR8-deficient cells135) transfected with a synthetic miR-29 and wild-type cells with antisense inhibitors of miR-29, respectively134. In this elegant experiment, they found reduced expression of two transcription factors that regulate IFN-γ production (Tbx21/T-bet and Eomes) under gain-of-function conditions and elevated expression of these two transcription factors under loss-of-function conditions. They further proved the role of miR-29 regulation of expression of these transcription factors in CD4+ lymphocytes in vitro and in both CD4+ and CD8+ T-cells in an in vivo virus infection model. Ma et al. demonstrated an inverse correlation between IFN-γ production and levels of miR-29 in natural killer (NK) cells and T cells from mice infected with Listeria monocytogenes or Mycobacterium bovis133. Mice lacking miR-29 infected with M. bovis showed less inflammation, lower bacterial burden and increased numbers of IFNγ-producing CD4+ T cells in their lungs compared with control mice.
Asthma Epigenetics – Human Studies
While animal studies have begun to decipher the role of epigenetic regulation of gene expression associated with the development of allergic airway disease in the lung, several recent publications in human cohorts have examined DNA methylation in cells outside of the lung - peripheral blood136, buccal cells137, 138 and nasal cells139. These early studies have only demonstrated statistical association of DNA methylation and specific exposure or asthma phenotype but have not elucidated the role of DNA methylation in the control of gene expression in human asthma. Breton et al. demonstrated that DNA methylation in promoters of two arginase genes (Arg1 and Arg2) is associated with exhaled nitric oxide in children with asthma from the Children’s Health Study and indicates a role for epigenetic regulation of nitric oxide production137. In a pilot study in the Columbia Center for Children’s Environmental Health cohort, Kuriakose and coworkers found that iNOS methylation was not significantly associated with fraction exhaled NO (FeNO) but was associated inversely with JNO (bronchial NO flux)138. This latter study emphasizes the importance of careful selection of clinical parameters used in the association study. A more recent study of DNA methylation in nasal cells from 35 asthmatic children 8–11 years old identified inverse association of FeNO and promoter methylation of both Il6 and iNOS139. Finally, data from two independent pregnancy cohorts in Spain (discovery and validation)136 showed that DNA hypomethylation in Alox12 in peripheral blood of children was associated with a higher risk of persistent wheezing at age 4. In aggregate, these studies suggest that DNA methylation in easily obtained samples (buccal, nasal or peripheral blood cells) may be a useful biomarker for airway inflammation in pediatric research.
A recent study has also examined DNA methylation in Foxp3 and Treg function in peripheral blood from children with and without asthma and with high and low exposures to air pollution140. Treg-cell suppression was impaired and Treg-cell chemotaxis was reduced as a result of exposure to air pollution. Changes in DNA methylation have also been associated with the development of asthma among older smokers in the Lovelace Smokers Cohort. Comparison of 184 smokers with asthma to 511 smoker controls with a similar smoking history (COPD cases excluded) identified an association of DNA methylation in the protocadherin-20 gene in sputum DNA with asthma as well as a significant synergistic interaction between methylation of protocadherin-20 and paired box protein transcription factor-5α on the odds for developing asthma141.
A set of earlier studies suggested that acetylation of histones may also play a role in asthma. Increased acetylation of H4 has been demonstrated in individuals with asthma and is associated with an increase in expression of several inflammatory genes in the lung142. It has also been shown that increased acetylation of histones results in decreased HDAC activity which may be responsible for enhanced expression of inflammatory genes. In addition, glucocorticoids appear to suppress inflammation by altering acetylation of histones that regulate inflammatory and anti-inflammatory genes; these studies are described in detail in a recent review143 and suggest that targeting histone acetylation (and possibly other epigenetic marks) may lead to novel anti-inflammatory therapies, especially in corticosteroid-resistant cases of asthma. A more recent study found that TGF-β2 suppresses expression of ADAM33, one of the most replicated asthma susceptibility genes, in normal or asthmatic fibroblasts and that this occurs by altering chromatin structure (deacetylation of histone H3, demethylation of lysine 4 on H3, and hypermethylation of lysine9 on H3) and not by gene silencing through DNA methylation as in epithelial cells144.
The role of miRNAs in asthma and atopy in humans is also emerging. Although no detectable differences in expression of miRNAs from airway biopsies were observed between mild asthmatics and normal subjects in an early study145, only mild asthmatics were included in this study and the number of miRNAs examined was limited. However, this study demonstrated cell-type specific expression of miRNAs in cells isolated from airways and lung tissue, suggesting a possible role for miRNAs in asthma. A more recent study has indeed identified miRNAs that play a role in specific cells in asthma. In a study of 8 controls, 4 non-severe and 12 severe asthma patients, widespread changes in mRNA and noncoding RNA expression in circulating CD8+ but not CD4+ T were associated with severe asthma146. miRNA expression profiles showed selective downregulation of miR-28-5p in CD8+ lymphocytes and reduction of miR-146a and miR-146b in both CD4+ and CD8+ T cells. It is likely that some of the other miRNAs identified in animal models play a yet uncovered role in the development of asthma in humans.
Challenges in Understanding the Asthma Epigenome
Some of the key questions in regard to future studies in asthma epigenetics revolve around understanding how the epigenome contributes to inheritance of asthma, developmental vulnerability of the epigenome, effect of the environment/diet/ageing, and the influence of asthma (and other diseases) on the epigenome. While sorting out these factors will be challenging, it is absolutely essential that the proper tissue be chosen to study the effects of the epigenome on asthma. The more pure and relevant to disease state the cell population, the more likely will the epigenetic marks regulate expression of key genes involved in pathogenesis of asthma. In the absence of airway biopsies in pediatric asthma, nasal epithelial cells or sputum may be the closest surrogate for disease relevant cells. Specific cell populations, such as CD4+ and CD8+ T lymphocytes, isolated from peripheral blood may also be informative in identifying immune genes whose expression is mis-regulated by epigenetic marks in the disease state. Despite these concerns, epigenetic marks in the peripheral blood may provide biomarkers to identify those at risk, responses to different forms of environmental stress, or likelihood of responding to specific therapeutic agents.
Analogous to asthma genetics studies, the choice of the study population will be crucial to success of future epigenetic studies. Ancestry of study subjects will likely need to be taken into account given the early evidence for the role of genetic variation and DNA methylation at asthma-associated loci, such as Ormdl3147. Moreover, a recent study suggests that DNA methylation is highly divergent between populations of European and African descent, and that this divergence may be in large part due to a combination of differences in allele frequencies and complex epistasis or gene × environment interactions148. Based on this, population stratification may be a confounder in population-based genome-wide DNA methylation studies and may have to be accounted for by using principal components from the methylation profile, whole-genome association studies (GWAS) or ancestry-specific marker panels. Given the strong influence of the environment on epigenetic marks, environmental and dietary exposures as well as medication use must be measured/recorded in the study not only for exposures of interested but also for any confounding exposures that need to be adjusted for in the analysis. Despite the differences between disease phenotypes in human cohorts and animal models of allergic airway disease, animal models with fixed genetic background and controlled exposures are likely to remain a crucial component of future studies in the field.
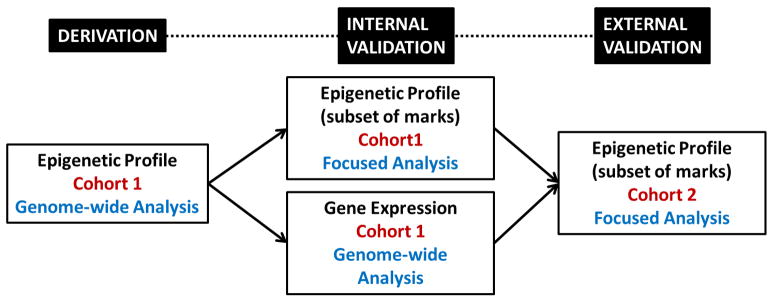
One of the major hurdles to overcome in future asthma epigenetics research will be the validation component. Necessary components of the validation process include internal validation of epigenetic marks in the same samples by a different technique, association of epigenetic marks with changes in gene expression in the study population and external validation of epigenetic marks in an independent cohort (Figure 3). Some of the difficulties encountered in the validation process are platform differences in technologies; differences in DNA methylation measurements are encountered based on the approach used to capture methylated marks (restriction digest, immunoprecipitation, bisulfite conversion) and probes to measure the extent of methylation (single CpG site vs a region covered by overlapping probes). Another major challenge in validations studies is interpretation of epigenetic marks in the context of changes in gene expression. Both cis- and trans-effects of methylation marks are likely to be important in gene regulation and this process is very complex. Depending on the site of methylation (promoter vs intron), epigenetic marks may play different roles in control of gene expression in cis. Mapping studies of methylation marks on gene expression (methyl-eQTL) will be essential in identification of cis- and trans-effects. The final major challenge will be identification of cohorts with comparable genetic background and environmental exposures to use as an independent validation step. It is likely that cohorts with similar exposures and phenotypes will be of most utility for broad validation of large number of epigenetics marks and identification of specific phenotype- and exposure-driven epigenetic changes while more divergent cohorts may still be useful in validation of a small number of epigenetic marks associated with disease regardless of other factors.
Figure 3.
An overview of the validation process for discovery of epigenetic marks associated with development of asthma. Internal validation refers to confirmation in the same cohort by a different technique and association of epigenetic marks with changes in gene expression while external validation refers to validation of epigenetic marks in an independent cohort. Genome-wide analysis of gene expression is preferable to focused approaches due to complexities in the relationship of epigenetic marks and gene expression alterations.
The Potential Impact of Epigenetics Research on Asthma
While we know that inheritance, parent-of-origin, environment, in utero exposures, and Th2 immunity play important roles in the etiology of asthma, there is no well-developed unifying mechanism accounting for these etiologic events/triggers. Although the Hygiene Hypothesis is appealing conceptually3 and ties a number of these basic etiologic events together, there are several competing hypotheses (T cell skewing, infection, diet, obesity, etc.), and none of them fully account for the complex interaction between host and environmental determinants of asthma. For example, the Hygiene Hypothesis suggests that a decrease of exposure to microbes would, through enhanced atopic immune responses, increase the incidence of allergies and allergic asthma3. However, the prevalence of atopy and asthma are not concordant, allergic mechanisms account for at most 50% of asthma cases, very high asthma rates are present in some countries where hygienic conditions are less than ideal, and although the prevalence and incidence of asthma continue to increase in inner cities in the U.S., housing conditions in these communities are becoming more hygienic.
While epigenetic mechanisms not only provide a unique cause of asthma, these basic transcriptional controls potentially serve to explain some of the prevailing hypotheses underlying the development of asthma. For example, the Hygiene Hypothesis is dependent on activation of innate immune genes, including genes activated by the Toll-like receptors; importantly, epigenetic mechanisms control the activation of these innate immune genes and, consequently, the extent of the inflammatory response149, 150. Moreover, a recent study demonstrated that microbes may also operate by means of epigenetic mechanisms151. In this animal study, prenatal administration of the farm-derived gram-negative bacterium Acinetobacter lwoffii F78A prevented the development of an asthmatic phenotype in the progeny, and this effect was IFN-γ dependent. Prenatal microbial exposure was also associated with a significant protection against loss of H4 acetylation in the promoter of Ifng, which was closely associated with IFN-γ expression in CD4+ lymphocytes as well as a decrease in H4 acetylation at the Il4 promoter. Pharmacologic inhibition of H4 acetylation in the offspring abolished the asthma-protective phenotype. So while epigenetic mechanisms have the potential of changing our basic concepts about asthma, these mechanisms may not only account for the etiologic events/triggers related to asthma but may also help to explain some of the prevailing hypotheses attributed to this disease.
Furthermore, identification of key epigenetic marks has the potential to transform asthma therapy from palliative to preventive, and may alter our recommendations for pregnancy throughout the world. Currently, other than avoidance of cigarette smoke, we are simply unable to prevent asthma. Most patients with asthma rely on chronic medications to reduce the severity of their symptoms. Understanding the importance of epigenetic mechanisms in the development of asthma and the periods of vulnerability in establishing epigenetic marks has the potential of preventing the development of this disease, not only in our offspring but in their children as well. Identification of critical epigenetic marks associated with the development of asthma and influenced by specific environmental factors at certain timepoints, in utero or postnatally, would allow us to advise on intake of dietary supplements and limiting harmful exposures during the critical windows when these dietary and environmental factors have the strongest influence on the development of disease. Understanding the complex interactions between in utero exposures and epigenetic vulnerability will provide insight into future interventions for individuals at risk for the development of allergic asthma and may lead to the prevention of this disease altogether.
However, asthma is a complex disease. And although epigenetic mechanisms may contribute to the etiology and pathogenesis of this disease, there are multiple pieces to the asthma puzzle. The challenge will be to understand how genetic variation, transcriptome, epigenetic marks, the environment, and the immune system interface with each other to result in the development of allergic and non-allergic forms of asthma.
Acknowledgments
Funding: This work was supported by the National Heart, Lung and Blood Institute (RO1-HL101251 and RC2-HL101715) and the National Institute of Allergy and Infectious Diseases (N01-AI90052).
Abbreviations
- CHIP
chromatic immunoprecipitation
- CpG
cytosine followed by guanine in DNA sequence
- DC
dendritic cell
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- GWAS
genomewide association
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- QTL
quantitative trait locus
- MBD
methyl-binding domain
- miRNA
micro RNA
- ncRNA
non-coding RNA
- PCA
principal components analysis
- qPCR
quantitative PCR
- RNA
ribonucleic acid
- SNP
single nucleotide polymorphism
- Th
T helper cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005–2009. Natl Health Stat Report. 2011:1–14. [PubMed] [Google Scholar]
- 2.Bloom B, Cohen R, Freeman G. National Center for Health Statistics D. Summary health statistics for US children: National Health Interview Survey, 2010. Hyatsville, MD: 2011. [PubMed] [Google Scholar]
- 3.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 4.Asher MI, Stewart AW, Wong G, Strachan DP, Garcia-Marcos L, Anderson HR. Changes over time in the relationship between symptoms of asthma, rhinoconjunctivitis and eczema: A global perspective from the International Study of Asthma and Allergies in Childhood (ISAAC) Allergol Immunopathol (Madr) 2012 doi: 10.1016/j.aller.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 6.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 7.March ME, Sleiman PM, Hakonarson H. The genetics of asthma and allergic disorders. Discov Med. 2011;11:35–45. [PubMed] [Google Scholar]
- 8.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–46. e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffatt MF, Cookson WO. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy. 1998;28(Suppl 1):56–61. doi: 10.1046/j.1365-2222.1998.0280s1056.x. discussion 5–6. [DOI] [PubMed] [Google Scholar]
- 10.Holt PG, Macaubas C, Stumbles PA, Sly PD. The role of allergy in the development of asthma. Nature. 1999;402:B12–7. doi: 10.1038/35037009. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 12.Cookson WO, Young RP, Sandford AJ, Moffatt MF, Shirakawa T, Sharp PA, et al. Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet. 1992;340:381–4. doi: 10.1016/0140-6736(92)91468-n. [DOI] [PubMed] [Google Scholar]
- 13.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001;29:175–8. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 14.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen RT, Raby BA, Van Steen K, Fuhlbrigge AL, Celedon JC, Rosner BA, et al. In utero smoke exposure and impaired response to inhaled corticosteroids in children with asthma. J Allergy Clin Immunol. 2010;126:491–7. doi: 10.1016/j.jaci.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008:118. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest. 2005;127:1232–41. doi: 10.1378/chest.127.4.1232. [DOI] [PubMed] [Google Scholar]
- 19.Henderson AJ, Newson RB, Rose-Zerilli M, Ring SM, Holloway JW, Shaheen SO. Maternal Nrf2 and gluthathione-S-transferase polymorphisms do not modify associations of prenatal tobacco smoke exposure with asthma and lung function in school-aged children. Thorax. 2010 doi: 10.1136/thx.2009.125856. [DOI] [PubMed] [Google Scholar]
- 20.Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:660–2. doi: 10.1513/pats.200907-065DP. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Pinkerton KE. Air pollutant effects on fetal and early postnatal development. Birth Defects Res C Embryo Today. 2007;81:144–54. doi: 10.1002/bdrc.20097. [DOI] [PubMed] [Google Scholar]
- 22.Clark NA, Demers PA, Karr CJ, Koehoorn M, Lencar C, Tamburic L, et al. Effect of early life exposure to air pollution on development of childhood asthma. Environ Health Perspect. 2010;118:284–90. doi: 10.1289/ehp.0900916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzsimon N, Fallon U, O’Mahony D, Loftus BG, Bury G, Murphy AW, et al. Mothers’ dietary patterns during pregnancy and risk of asthma symptoms in children at 3 years. Ir Med J. 2007;100(suppl):27–32. [PubMed] [Google Scholar]
- 24.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–84. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 26.Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ. Exposure to house-dust mite allergen (Der p I) and the development of asthma in childhood. A prospective study [see comments] N Engl J Med. 1990;323:502–7. doi: 10.1056/NEJM199008233230802. [DOI] [PubMed] [Google Scholar]
- 27.Oh SS, Tcheurekdjian H, Roth LA, Nguyen EA, Sen S, Galanter JM, et al. Effect of secondhand smoke on asthma control among black and Latino children. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan-Yeung M, Malo JL. Occupational asthma. N Engl J Med. 1995;333:107–12. doi: 10.1056/NEJM199507133330207. [DOI] [PubMed] [Google Scholar]
- 29.Samet JM, Lambert WE. Epidemiologic approaches for assessing health risks from complex mixtures in indoor air. Environ Health Perspect. 1991;95:71–4. doi: 10.1289/ehp.919571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folkerts G, busse W, Nijkamp F, Sorkness R, Gern J. State of the art: virus-induced airway hyperresponsiveness and asthma. Am J Respir Crit Care Med. 1998;157:1708–20. doi: 10.1164/ajrccm.157.6.9707163. [DOI] [PubMed] [Google Scholar]
- 31.Gereda J, Leung D, Thatayatikom A, Streib J, Price M, Klinnert M, et al. Relation between house-dust endotoxin exposure, type 1 T-cell development, and allergen sensitisation in infants at high risk of asthma. Lancet. 2000;355:1680–3. doi: 10.1016/s0140-6736(00)02239-x. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz DA, Donham KJ, Olenchock SA, Popendorf WJ, Van Fossen DS, Burmeister LF, et al. Determinants of longitudinal changes in spirometric function among swine confinement operators and farmers. Am J Respir Crit Care Med. 1995;151:47–53. doi: 10.1164/ajrccm.151.1.7812571. [DOI] [PubMed] [Google Scholar]
- 33.Shirakawa T, Enomoto T, Shimazu S, Hopkin JM. The inverse association between tuberculin responses and atopic disorder. Science. 1997;275:77–9. doi: 10.1126/science.275.5296.77. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol. 2010;10:838–48. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–52. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, et al. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17:557–64. doi: 10.1111/j.1399-3038.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 37.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- 38.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 39.Webster RB, Rodriguez Y, Klimecki WT, Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J Biol Chem. 2007;282:700–9. doi: 10.1074/jbc.M609501200. [DOI] [PubMed] [Google Scholar]
- 40.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–79. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A. 2007;104:17052–7. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 43.Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2009. [Google Scholar]
- 44.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 45.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 46.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010 doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer. 2012;106:248–53. doi: 10.1038/bjc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 52.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–42. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papageorgiou N, Tousoulis D, Androulakis E, Siasos G, Briasoulis A, Vogiatzi G, et al. The Role of microRNAs in Cardiovascular Disease. Curr Med Chem. 2012 doi: 10.2174/092986712800493048. [DOI] [PubMed] [Google Scholar]
- 58.Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012:9. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 60.Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci U S A. 2009;106:22187–92. doi: 10.1073/pnas.0905767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 64.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 65.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–20. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang F, Hodges E, Molaro A, Dean M, Hannon GJ, Smith AD. Genomic landscape of human allele-specific DNA methylation. Proc Natl Acad Sci U S A. 2012;109:7332–7. doi: 10.1073/pnas.1201310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 70.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 71.Milosavljevic A. Emerging patterns of epigenomic variation. Trends Genet. 2011;27:242–50. doi: 10.1016/j.tig.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–25. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med. 2011;183:1295–301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–91. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Curr Protoc Hum Genet. 2010;Chapter 20(Unit 20 1):1–19. doi: 10.1002/0471142905.hg2001s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu CG, Spizzo R, Calin GA, Croce CM. Expression profiling of microRNA using oligo DNA arrays. Methods. 2008;44:22–30. doi: 10.1016/j.ymeth.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 78.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–30. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 79.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–9. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanizawa H, Noma K. Unravelling global genome organization by 3C-seq. Semin Cell Dev Biol. 2012;23:213–21. doi: 10.1016/j.semcdb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2012;9:75–7. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robertson AB, Dahl JA, Ougland R, Klungland A. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat Protoc. 2012;7:340–50. doi: 10.1038/nprot.2011.443. [DOI] [PubMed] [Google Scholar]
- 84.Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012 doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation crosstalk. Genome Res. 2012 doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 87.Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–95. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 88.Kechris KJ, Biehs B, Kornberg TB. Generalizing moving averages for tiling arrays using combined p-value statistics. Stat Appl Genet Mol Biol. 2010;9:Article29. doi: 10.2202/1544-6115.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–76. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knight JC. Resolving the variable genome and epigenome in human disease. J Intern Med. 2012;271:379–91. doi: 10.1111/j.1365-2796.2011.02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kang HP, Yang X, Chen R, Zhang B, Corona E, Schadt EE, et al. Integration of disease-specific single nucleotide polymorphisms, expression quantitative trait loci and coexpression networks reveal novel candidate genes for type 2 diabetes. Diabetologia. 2012 doi: 10.1007/s00125-012-2568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–82. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 93.Allen JD, Xie Y, Chen M, Girard L, Xiao G. Comparing statistical methods for constructing large scale gene networks. PLoS One. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, et al. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 96.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–27. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581–9. e1–20. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 98.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–51. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9:83–9. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 101.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–35. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–9. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, et al. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129:232–9. e7. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:2319–24. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–50. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–64. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. Maternal Exposure to Polycyclic Aromatic Hydrocarbons and 5′-CpG Methylation of Interferon-gamma in Cord White Blood Cells. Environ Health Perspect. 2012 doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guerrero-Preston R, Goldman LR, Brebi-Mieville P, Ili-Gangas C, Lebron C, Witter FR, et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5:539–46. doi: 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, et al. Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomarkers Prev. 2011;20:2518–23. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012;494:36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breton CV, Salam MT, Gilliland FD. Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics. 2011;6:895–8. doi: 10.4161/epi.6.7.15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5:583–9. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Whitrow MJ, Moore VM, Rumbold AR, Davies MJ. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol. 2009;170:1486–93. doi: 10.1093/aje/kwp315. [DOI] [PubMed] [Google Scholar]
- 119.Haberg SE, London SJ, Stigum H, Nafstad P, Nystad W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch Dis Child. 2009;94:180–4. doi: 10.1136/adc.2008.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martinussen MP, Risnes KR, Jacobsen GW, Bracken MB. Folic acid supplementation in early pregnancy and asthma in children aged 6 years. Am J Obstet Gynecol. 2012;206:72, e1–7. doi: 10.1016/j.ajog.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Magdelijns FJ, Mommers M, Penders J, Smits L, Thijs C. Folic acid use in pregnancy and the development of atopy, asthma, and lung function in childhood. Pediatrics. 2011;128:e135–44. doi: 10.1542/peds.2010-1690. [DOI] [PubMed] [Google Scholar]
- 122.Bekkers MB, Elstgeest LE, Scholtens S, Haveman A, de Jongste JC, Kerkhof M, et al. Maternal use of folic acid supplements during pregnancy and childhood respiratory health and atopy: the PIAMA birth cohort study. Eur Respir J. 2011 doi: 10.1183/09031936.00094511. [DOI] [PubMed] [Google Scholar]
- 123.Koh BH, Hwang SS, Kim JY, Lee W, Kang MJ, Lee CG, et al. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc Natl Acad Sci U S A. 2010;107:10614–9. doi: 10.1073/pnas.1005383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tanaka S, Motomura Y, Suzuki Y, Yagi R, Inoue H, Miyatake S, et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol. 2011;12:77–85. doi: 10.1038/ni.1966. [DOI] [PubMed] [Google Scholar]
- 125.Van Stry M, Bix M. Explaining discordant coordination. Nat Immunol. 2011;12:16–7. doi: 10.1038/ni0111-16. [DOI] [PubMed] [Google Scholar]
- 126.Fedulov AV, Kobzik L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol. 2011;44:285–92. doi: 10.1165/rcmb.2009-0400OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brand S, Kesper DA, Teich R, Kilic-Niebergall E, Pinkenburg O, Bothur E, et al. DNA methylation of T(H)1/T(H)2 cytokine genes affects sensitization and progress of experimental asthma. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2011.12.963. [DOI] [PubMed] [Google Scholar]
- 128.Ariel D, Upadhyay D. The role and regulation of microRNAs in asthma. Curr Opin Allergy Clin Immunol. 2012;12:49–52. doi: 10.1097/ACI.0b013e32834ecb7f. [DOI] [PubMed] [Google Scholar]
- 129.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128:160–7. e4. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 130.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–49. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–85. e1–10. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 132.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–73. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 134.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–81. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, et al. DNA Hypomethylation at ALOX12 Is Associated with Persistent Wheezing in Childhood. Am J Respir Crit Care Med. 2012;185:937–43. doi: 10.1164/rccm.201105-0870OC. [DOI] [PubMed] [Google Scholar]
- 137.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med. 2011;184:191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kuriakose J, Rosa MJ, Perzanowski M, Miller R. Bronchial nitric oxide flux may be better associated with inducible nitric oxide synthase promoter methylation. Am J Respir Crit Care Med. 2012;185:460–1. doi: 10.1164/ajrccm.185.4.460. author reply 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Baccarelli A, Rusconi F, Bollati V, Catelan D, Accetta G, Hou L, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics. 2012;4:91–100. doi: 10.2217/epi.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, et al. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010;126:845–52. e10. doi: 10.1016/j.jaci.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 141.Sood A, Petersen H, Blanchette CM, Meek P, Picchi MA, Belinsky SA, et al. Methylated Genes in Sputum among Older Smokers with Asthma. Chest. 2012 doi: 10.1378/chest.11-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, et al. A molecular mechanism of action of theophylline: Induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci U S A. 2002;99:8921–6. doi: 10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:693–6. doi: 10.1513/pats.200907-071DP. [DOI] [PubMed] [Google Scholar]
- 144.Yang Y, Wicks J, Haitchi HM, Powell RM, Manuyakorn W, Howarth PH, et al. Regulation of A Disintegrin And Metalloprotease-33 Expression by Transforming Growth Factor-beta. Am J Respir Cell Mol Biol. 2012;46:633–40. doi: 10.1165/rcmb.2011-0030OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 147.Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A, et al. Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet. 2012 doi: 10.1007/s00439-012-1142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–8. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 150.Qin H, Roberts KL, Niyongere SA, Cong Y, Elson CO, Benveniste EN. Molecular mechanism of lipopolysaccharide-induced SOCS-3 gene expression in macrophages and microglia. J Immunol. 2007;179:5966–76. doi: 10.4049/jimmunol.179.9.5966. [DOI] [PubMed] [Google Scholar]
- 151.Brand S, Teich R, Dicke T, Harb H, Yildirim AO, Tost J, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. 2011;128:618–25. e1–7. doi: 10.1016/j.jaci.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 152.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 153.Espada J, Esteller M. DNA methylation and the functional organization of the nuclear compartment. Semin Cell Dev Biol. 2010;21:238–46. doi: 10.1016/j.semcdb.2009.10.006. [DOI] [PubMed] [Google Scholar]