Abstract
Radiation-induced DNA double-strand breaks (DSBs) are critical cytotoxic lesions that are typically repaired by nonhomologous end joining (NHEJ) in human cells. Our previous work indicates the highly cytotoxic DSBs formed by 125I decay possess base damage clustered within 8 to 10 bases of the break, and 3′-phosphate (P) and 3′-OH ends.
This study examines the effect of such structures on NHEJ in in vitro assays employing either 125I decay-induced DSB linearized plasmid DNA, or structurally defined duplex oligonucleotides. Duplex oligonucleotides that possess either a 3′-P or 3′-phosphoglycolate (PG), or a ligateable 3′-OH end with either an AP site or an 8-oxo-dG 1 nucleotide upstream (-1n) from the 3′-terminus, have been examined for reparability. Moderate to severe end-joining inhibition was observed for modified DSB ends or 8-oxo-dG upstream from a 3′-OH end. In contrast, abolition of end joining was observed with duplexes possessing an AP site upstream from a ligateable 3′-OH end, or for a lesion combination involving 3′-P plus an upstream 8-oxo-dG. In addition, base mismatches at the -1n position are also strong inhibitors of NHEJ in this system, suggesting that destabilization of the DSB terminus as a result of base loss or improper base pairing may play a role in the inhibitory effects of these structures. Furthermore, we provide data indicating that DSB end joining is likely to occur prior to removal or repair of base lesions proximal to the DSB terminus. Our results show that base damage or base loss near a DSB end may be a severe block to NHEJ, and that complex combinations of lesions presented in the context of a DSB may be more inhibitory than the individual lesions alone. In contrast, blocked DSB 3′-ends alone, are only modestly inhibitory to NHEJ. Finally, DNA ligase activity is implicated as being responsible for these effects.
Keywords: Radiation-induced DNA damage, DNA double-strand breaks, DSB repair, non-homologous end joining, DNA repair inhibition
Introduction
Radiation-induced DNA double strand breaks (DSB) have been implicated as potentially lethal events for a cell (1, 2). Such lesions cause cell cycle arrest, and if unrepaired, they may result in cell death. However, even if rejoined, mis-repaired DSBs may result in genomic instability and ultimately lead to the development of cancer (3). The lethality of a DSB has been suggested to be dependent upon the complexity of the break, which in the case of radiation, may depend on radiation quality with high-LET radiation presumably creating DSBs of greater complexity than the majority of those produced by low-LET radiation (4–6). High-LET radiation-induced DSBs appear to be less amenable to repair than those produced by other agents, and this has been proposed to reflect greater structural complexity and to explain their high relative cytotoxicity (4–9). Even for low-LET radiation-induced DSBs, it is the subset of breaks with high structural complexity that has been suggested to be most lethal (6, 8, 10, 11). Consequently, it is the complex DSB, defined here as a double stranded DNA discontinuity of variable overhang morphology with blocked ends and flanking base damage, which has been postulated to be the most critical DSB lesion.
DNA double strand breaks (DSB) resulting from various genotoxic agents, including ionizing radiation, are most commonly repaired by nonhomologous end joining (NHEJ). The core proteins involved in this pathway have been identified by genetic studies (12–16), as well as by in vitro assays (reviewed in (17)). Numerous in vivo investigations (18–22) and cell free in vitro assays have been employed to assess the roles of individual proteins in the NHEJ pathway, but it is the in vitro assays that have been most instructive with respect to the kinetics of the repair process and the biochemical functions of specific proteins in the repair pathway (17, 23). In the case of in vitro assays designed to assess end joining of linear plasmid DNA as an end point for DSB repair, most have employed restriction enzyme cut DNA as a substrate (24–31).
Although the NHEJ machinery can easily rejoin such substrates, the substrates themselves lack structural complexity, and are a poor model for the complex DSBs typically produced by radiation and other genotoxic DNA breaking agents such as enediyne drugs (32). Consequently, in vitro assays using restriction-enzyme-cut DNA do not allow for an understanding of the relationship between authentic DSB damage and the NHEJ machinery.
In our previous work, we described the molecular characteristics of the high-LET-like DSBs that result when 125I decays in close proximity to DNA (33–36). These DSBs are highly cytotoxic and known to cause biological effects similar to those of high-LET beam radiation (37, 38). At the molecular level, we have shown that 125I decay-induced DSBs are extremely complex lesions that exhibit variable overhang end-morphology (variable length 5′ and 3′ overhangs as well as blunt ends), ends that are largely blocked by 3′-phosphate (3′-P) and other groups, but also occur with some 3′-OH. We have also shown that these DSBs possess a high frequency of base damage and abasic sites immediately upstream from the DSB end.
Although in vitro repair studies have been conducted to investigate the importance of various restriction-enzyme-induced end morphologies (i.e., overhangs and blunt ends), or end group structures on NHEJ (30, 39–41), and others have examined the effects of closely opposed base lesions in duplex DNA upon recognition and cleavage by base excision repair enzymes (42–45), it is still not clear how, and to what extent various aspects of complex DSB structure affect and/or inhibit the DSB repair process. In a recent study by Dobbs et al. (46) it was shown that 8-oxoG positioned close to a DSB end was likely to interfere with base excision repair (BER) and was a poor substrate for processing by the purified human glycosylases hOGG1 and NEIL1. It was also shown that such lesions may inhibit ligation by T4 DNA ligase and purified Ligase IV/XRCC4 complex. These results illustrate the importance of examining what effect, if any, base damage and other lesions proximal to the DSB end have upon NHEJ, and what their relative effect on the repair process is with respect to the blocked ends that are formed by radiation and other DNA damaging agents.
We have addressed this question by creating an in vitro synthetic oligonucleotide based DSB end joining assay in which each duplex oligo substrate contains defined lesions/modifications in association with the DSB end to be rejoined. The source of DSB repair enzyme activities for this assay is HeLa whole cell extracts. Using this repair system, we have compared the effects of 3′-end blocking groups, upstream base lesions, or apurinic/apyrimidinic (AP) sites on DSB repair with respect to the end joining activity observed for equivalent substrates terminated by directly ligateable DSB ends. This work confirms observations by others, that 3′-blocking groups are a hindrance to repair with respect to unblocked 3′-OH DSB ends (47) while further indicating that the presence of a base lesion 1 nucleotide upstream (-1n) from a 3′-OH terminated DSB end acts as a similar impediment to end joining. In contrast, as the structural complexity of the break is increased to contain a combination of a 3′-end blocking group (3′-P) with an 8-HO-dGua base lesion –1n from the DSB end, the end joining reaction is completely blocked. Furthermore, DSB end structural configurations that may destabilize hydrogen bonding of the 3′-end terminal nucleotide to its complement in the opposite strand, such as an AP site or a base mismatch – 1n from the 3′-end, also act as complete blocks to the end joining reaction.
Materials and Methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, non-essential amino acids (10 mM), glutamine (100 mM), penicillin/streptomycin (10,000 U/ml), Saccharomyces cerevisiae tRNA, T4 DNA ligase, and Novex™6% TBE gels were obtained from Invitrogen (Carlsbad, CA). Reagents for oligonucleotide synthesis were from Glen Research (Sterling, VA). Protease inhibitors and other chemicals were obtained from Sigma (St. Louis, MO). γ-32P-ATP (222 GBq/mmol) was obtained from Perkin Elmer Life Science (Boston, MA). T4 polynucleotide kinase (PNK) was from Fermentas (Hanover, MD). Escherichia coli endonuclease IV (endo IV), formamidopyrimidine DNA glycosylase (Fpg), and recombinant human DNA ligase IV/XRCC4 and Ku70/80 were from Trevigen (Gaithersburg, MD). Monoclonal anti-Ku70 (clone 2C3.11) antibody was from Novus Biologicals (Littleton, CO). Anti-DNA-PKcs (ab5366), anti-DNA ligase IV (ab6145), and anti-Ku80 (ab3107) antibodies were from Abcam (Cambridge, MA). The SequaGel sequencing system was obtained from National Diagnostics (Atlanta, GA). Protein assays were performed using the Bio-Rad system (Hercules, CA).
Plasmid
Plasmid pTC27 was a generous gift from Dr. Michael Seidman (NIA, Baltimore, MD). The plasmid possesses a polypurine sequence designed to permit binding of a pyrimidine-motif triple-helix-forming oligonucleotide (TFO). The features of this plasmid have been described in detail elsewhere (35). The plasmid was grown in E. coli (DH10B) and purified with a Qiagen megaprep kit (Qiagen, Valencia, CA). Purified plasmid was quantified and purity checked by spectrophotometry and 1% agarose gel electrophoresis using a High Mass DNA ladder as the calibration standard (Invitrogen).
TFO synthesis and purification, 125I mediated DSB damage induction, and purification of DSB linearized plasmid
A pyrimidine-motif TFO 3′-terminated by 125I-dC was synthesized and purified as described previously (4, 34, 35) and used to form triplexes with pTC27. Plasmid linearized by TFO targeted 125I decay-induced site-specific DSB formation was produced and characterized as described previously (4, 34, 35). As indicated in the previous work, irradiations were conducted in the presence or absence of 2M DMSO by incubation of the triplex containing plasmids in 1X TFO binding buffer (30mM CH3COONa, pH 4.5, 10mM MgCl2, and 1mM spermidine) at −80°C for one month.
Cell extracts
HeLa cells were grown as monolayers at 37°C in DMEM containing 10% (v/v) fetal bovine serum, 1% (v/v) non-essential amino acids and 1% (v/v) penicillin/streptomycin. Cells were harvested by trypsinization and a representative flask was counted. Cells were washed two times by re-suspension in complete media followed by centrifugation at 800 × g for 5 min. The resulting cell pellet was washed two times with ice cold PBS in a similar manner. The cells were re-suspended in extraction buffer (10 mM HEPES, pH 7.9, 60 mM KCl, 1 mM DTT, 1 mM EDTA, 10 mM pefabloc, 10 μg/ml aprotinin, 1.5 μg/ml leupeptin, 100 μg/ml bestatin) at a density of 8×107 cells/ml and lysed by the freeze/squeeze method using three cycles of freezing in a dry ice ethanol bath followed by immediate thawing at 37°C (48). After the third thaw cycle, cell debris was removed by centrifugation at 16000 × g for 30 minutes at 4°C. The supernatant constituted the HeLa whole cell extract (WCE) and was stored at −80°C in aliquots. WCEs of WI38 human fibroblast cells and human MO59K glioma cells were prepared in the same manner.
Plasmid based end-joining assay
End joining reactions were typically conducted in a 50 μl total volume at 17°C overnight (16 hours). Reactions contained 50 mM Tris-HCl pH 7.6, 5 mM MgCl2, 75 mM KCl, 1 mM ATP, 1 mM DTT, 5% polyethyleneglycol (PEG) 8000, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 μg/ml bestatin, 1 mM pefabloc, 100 ng DSB terminated linear plasmid DNA (damaged in the presence or absence of 2 M DMSO) and HeLa whole cell extract (15 μg total protein per reaction). In control reactions, damaged DNA was replaced with StuI cut plasmid, and in the case of positive controls, an end joined plasmid ladder was formed by ligation with T4 DNA ligase ((1U); one unit is defined as the amount of enzyme required to catalyze the exchange of 1 nmol of 32PPi into [γ/β-32P]ATP in 20 min at 37°C). The repair reactions were stopped by the addition of SDS to 0.4% followed by incubation at 65°C for 15 min. The DNA was recovered by phenol-chloroform extraction followed by ethanol precipitation and resuspension in 5 μl TE pH 8.0. The repair products were resolved in 1% agarose gels and stained with Vistra Green (Amersham Biosciences, Piscataway, NJ) at 1:20,000 dilution in TE buffer (pH 8.0) for 30 minutes at 4°C with gentle shaking. Digital images of the gels were obtained with a FluorImager 595 (Molecular Dynamics) and the data were analyzed densitometrically using GelPro 2.0 software (Media Cybernetics). The percent yield of end joined products was calculated as the sum of the end joined DNA band densities divided by the sum of all DNA band densities times 100 ([(dimer + trimer + high molecular weight multimers)/(dimer + trimer + high molecular weight multimers + linear)]×100). In some cases, repair assays were supplemented with all four dNTPs (250 μM final concentration of each) and/or with XRCC4/DNA Ligase IV ((10U);one unit is defined as the amount of enzyme required to ligate oligo (dT)24 when hybridized to poly (dA)300 at the rate of 1 pmole in 30 minutes at 37° C).
Oligonucleotides
Five 75-mer oligonucleotides, and two complementary 79-mer oligonucleotides (Table 1), were synthesized on an ABI-394 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA) and band purified from 12% preparative denaturing PAGE gels. Prior to synthesis, the oligo sequences were analyzed to avoid secondary structures by using IDT SciTools OligoAnalyzer 3.0 (available at http://scitools.idtdna.com/analyzer/Applications/OligoAnalyzer/). In the oligos containing base modifications, either an AP site (furan) or an 8-oxo-dG were positioned one nucleotide upstream from the 3′-end of the 75-mer upper-strand oligo. The individual modifications incorporated into the 75-mer oligo (b – g) are labeled as follows: the bold “p”, “pg”, “G*”, and “@”, indicate 3′-phosphate (3′-P), 3′-phosphoglycolate (3′-PG), 8-oxo-dG, and the AP site, respectively. Oligo a and d, are 3′-OH terminated 75 mer controls, and oligos h and i are the 79 mer oligos that are either complementary to the 75 mers, or can be used to form base mismatches at the – 1n position of the upper strand 3′-end when used in appropriate combinations with oligos a and e (Table 2). Oligo g will form a complex multiply damaged DSB end when annealed to oligo i. In the case of the 3′-PG terminated 75 mer, oligonucleotide synthesis and post-synthetic chemistry were performed according to the method of Urata and Akagi (49).
Table 1.
Oligonucleotides1
| Name | Sequence | Length/Modification |
|---|---|---|
| a) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTAC- 3′ | 75/5′-OH/3′-OH |
| b) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTACp - 3′ | 75/5′-OH/3′-P |
| c) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTACpg - 3′ | 75/5′-OH/3′-PG |
| d) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTGC- 3′ | 75/5′-OH/3′-OH |
| e) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTG*C - 3′ | 75/5′-OH/8-oxo-G |
| f) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTT@C - 3′ | 75/5′-OH/AP site |
| g) | 5′-TAGAGACGGGATGAGTGGAATTAGGACTGAGACTATGGTTGCTGACTAATCGAGACCCATCATTAGCATAGTTG*Cp - 3′ | 75/5′-OH/8-oxo-G+3′-P |
| h) | 5′-GTACGTAACTATGCTAATGATGGGTCTCGATTAGTCAGCAACCATAGTCTCAGTCCTAATTCCACTCATCCCGTCTCTA -3′ | 79/5′-32P/3′-OH |
| i) | 5′-GTACGCAACTATGCTAATGATGGGTCTCGATTAGTCAGCAACCATAGTCTCAGTCCTAATTCCACTCATCCCGTCTCTA -3′ | 79/5′-32P/3′-OH |
The 3′-32P labeled modification of oligo h, designated h*, is not shown. Oligo h* has the same sequence as oligo h, but is labeled at the -2n position from the 3′-end (see Materials and Methods for labeling details) and possesses a non-radioactive PO4 at its 5′-end.
Table 2.
DSB End Structures of Duplex Oligo NHEJ Substrates
| Duplex | Sequence | Modification |
|---|---|---|
| a/h | …TAC-OH-3′ …ATGCATG-32P-5′ |
3′-OH/5′-32P (Unmodified ligateable control) |
| b/h | …TAC-P-3′ …ATGCATG-32P-5′ |
3′-P/5′-32P |
| c/h | …TAC-PG-3′ …ATGCATG-32P-5′ |
3′-PG/5′-32P |
| d/i | .…TGC-OH-3′… ACGCATG-32P-5′ |
3′-OH/5′-32P (Unmodified ligateable -1n G:C control) |
| e/h | …TG*C-OH-3′ …ATGCATG-32P-5′ |
3′-OH/5′-32P + -1n 8-oxo-G:T mismatch |
| f/h | …T@C-OH-3′ …ATGCATG-32P-5′ |
3′-OH/5′-32P + - 1n AP site (furan) |
| a/i | …TAC-OH-3′ …ACGCATG-32P-5′ |
3′-OH/5′-32P + -1n A:C mismatch |
| e/i | …TG*C-OH-3′ …ACGCATG-32P-5′ |
3′-OH/5′-32P + -1n8-oxo-G |
| g/i | …TG*C-P-3′ …ACGCATG-32P-5′ |
3′-P/5′-32P + -1n8-oxo-G |
The presence of the modifications in the 75 mer oligos were confirmed by lesion-specific enzymatic treatment of 75 mer/79 mer duplex oligos in which the individual 75 mer oligos were typically 5′-32P-end labeled and subsequently tested for their ability to serve as substrates for appropriate enzymes. The presence and location of the AP site in oligo f was confirmed by cleavage with E. coli endo IV (1U), while the 8-oxo-dG of oligo e was confirmed by cleavage with E. coli Fpg (1U) following extension of the 3′-end with exo− Klenow DNA polymerase (5U). The presence of the 3′-P in oligo b was confirmed by the ability of exo− Klenow polymerase to extend this oligo only after 3′-phosphatase treatment with T4 PNK (10U). The presence of 3′-PG in oligo c was confirmed in two ways. Pretreatment of the duplex oligo with endo IV was required to permit extension of the 3′-end by exo− Klenow polymerase (no extension was observed following 3′-phosphatase treatment with T4 PNK), and production of a 25 mer oligo with a migration pattern characteristic of 3′-PG was observed by 20% denaturing PAGE following Taq1 restriction enzyme digestion of the duplex oligo and 5′-32P-end labeling.
Oligonucleotide based end-joining assay
As described previously (50), we have developed an in vitro NHEJ assay based on a 75 base pair duplex oligonucleotide with a 4 base 5′ overhang (a 75 mer upper strand and a 79 mer lower strand) as an end-joining substrate. As employed here, the 79 mer strand is either 5′ end labeled by γ-32P-ATP using T4 PNK (10U) in the forward reaction (10 pmole oligo in 50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT, 0.1 mM spermidine, 0.1 mM EDTA, and 2.22 MBqγ32P-ATP in 10 μl reaction volume at 37°C for 45 min), or 3′-32P end labeled by extension of a 4 base 3′-shortened version of the 79 mer strand by using exo− Klenow DNA polymerase (5U) mediated incorporation of α-32P-dCTP in the presence of TTP and dATP. Klenow mediated 3′-end labeling results in incorporation of a single α-32P-dCTP 2 nucleotides upstream from the fully extended 3′-end of the 79 mer strand and we have designated this modification as oligo h*. To form duplexes, equimolar amounts of the appropriate 32P-labeled 79 mer and cold 75 mer are mixed in 10 mM Tris-HCl, pH 8.0 in a reaction volume of 20 μl, incubated at 90°C for 5 minutes and allowed to gradually cool to room temperature (RT). The resulting duplex oligonucleotide possesses an end with a 4 base complementary 5′-overhang, and a non-ligateable blunt end in which both the 3′- and the 5′-ends are OH. Duplex formation was confirmed by 6% non-denaturing PAGE (data not shown). The end joining reaction using these substrates was essentially similar to the plasmid based assay, except the reaction volume for the oligonucleotide assay was 10 μl, and incubation was 30 min at 17°C followed by enzyme inactivation at 65°C for 15 min, also, positive control reactions used 0.2U T4 DNA ligase. The reaction products were then immediately loaded onto 6% non-denaturing, or 8% denaturing PAGE gels as indicated in the figure legends.
EMSA assays
Electrophoretic mobility shift assays (EMSA) were performed as described previously for the undamaged duplex substrate (50) to determine the ability of the damage-bearing duplex oligonucleotides to support NHEJ protein-complex formation. To generate a supershift upon electrophoresis in 6% non-denaturing PAGE gels, anti-Ku 70 (2 μl), anti-DNA PKcs (2 μl), or anti-DNA ligase IV antibodies (2 μl) were added to individual reaction mixtures (20 μl) after addition of the cell extract (15 μg). Gels were visualized with a Fuji BAS-2500 phosphoimager and analyzed using FujiFilm Image Gauge v3.45 software.
Results
Complex DSB end joining by human cell extracts
To determine what aspect of the DSB structure, as exemplified by the structures associated with the high-LET-like DSBs produced by 125I decay (3′-end structure or other lesions upstream of the DSB end) affects the ability of NHEJ to process the break and end-join the complex-DSB-terminated linear plasmids, we initially examined the ability of HeLa cell extracts to end join 125I-TFO linearized plasmid DNA that had undergone either T4 PNK 3′-phosphatase treatment, or was left untreated prior to end joining.
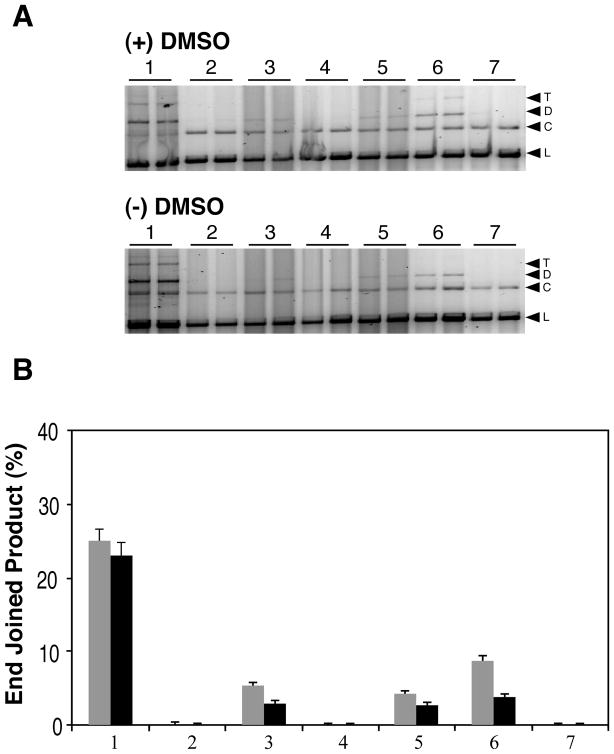
DNA from both (+) and (−) DMSO samples were subjected to HeLa cell extract mediated end joining assays (Fig. 1). Both irradiation conditions produce highly complex DSBs with near equal amounts of base loss (AP sites) proximal to the break ends, and although substantial under both conditions, up to ~2-fold greater yields of oxidative base lesions occur in this region following irradiation in the absence of DMSO (33, 34). Furthermore, while both irradiation conditions result in DSBs with variable overhang structures, T4 PNK 3′-phosphatase treatment of (+) DMSO samples converts up to 92% of the DSB 3′-ends to 3′-OH, whereas similar treatment of (−) DMSO samples only results in conversion of 33% of DSB ends to 3′-OH (36). Therefore, even though the HeLa extract is expected to possess hPNK 3′-phosphatase activity and should be capable of dephosphorylating DSB 3′-ends, to ensure 3′-dephosphorylation of the damaged plasmids prior to end joining, the substrates were pretreated with T4 PNK. In surprising contrast to our previous T4 DNA-Ligase-mediated direct ligation results (36), 3′-phosphatase treatment prior to the end joining reactions did not alter the yield of end-joined products formed by the HeLa extract with either substrate. Furthermore, the product yields were small for both DNA samples with only 6% of the (+) DMSO sample (Fig. 1A upper panel lanes 3 & 5, and 1B grey bars) and 3% of the (−) DMSO sample (Fig. 1A lower panel lanes 3 & 5, and 1B black bars) converted to end joined products regardless of the 3′-phospatase pretreatment conditions. Therefore, although these DSBs are theoretically capable of being directly ligated to a large extent, and the HeLa extract was capable of efficiently end-joining StuI restriction enzyme induced blunt ends (Fig. 1A and B, lanes 1), the extract could not efficiently end join the complex DSBs. One possible explanation for these results may be a deficiency in the repair reactions, such as a need for additional dNTPs to support DNA polymerase mediated fill-in reactions, or, as has been reported by others (51), the HeLa WCE might have low DNA ligase IV levels even though it is capable of efficiently end joining the restriction enzyme induced DSBs. Alternatively, these results may indicate that the 3′-end blocking groups of the DSB are not the primary DSB structural feature that is impeding end joining, but that other structural features, such as upstream base damage/loss, may be responsible for the blockage of end joining by the extract.
Figure 1.
HeLa extract mediated end joining of 125I decay-induced DSB terminated linear plasmid DNA. A) Typical 1% agarose gel results for HeLa cell extract mediated end joining of linear plasmid DNA. All reactions were run in duplicate. The upper panel shows end-joining results for damaged linear plasmid DNA irradiated (+) 2 M DMSO, while the lower panel presents results for plasmid linearized by irradiation (−) DMSO. The contents of the lanes in both gels are as follows: lanes 1, cell extract end-joining activity control using StuI cut plasmid DNA (blunt end); lanes 2–7 present results for the end-joining reactions using the 125I-linearized damaged plasmids as substrates; lanes 2, damaged DNA + heat inactivated WCE; lanes 3, damaged DNA + WCE; lanes 4, T4 PNK 3′-phosphatase pretreated damaged DNA alone; lanes 5, T4 PNK 3′ phosphatase pretreated damaged DNA + WCE; lanes 6, damaged DNA + T4 DNA ligase; lanes 7, damaged DNA alone. The bands on the gels are identified as follows: L, linear plasmid; C, Open circular; D, dimer; T, trimer. B) Densitometric analysis of the data presented in panel A. The column numbering of the graph corresponds to the lane numbering described for panel A. Gray bars indicate DNA irradiated (+) DMSO, black bars indicate DNA irradiated (−) DMSO. The mean of the replicates is plotted and the error bars indicate the standard deviation.
The effect of dNTPs and DNA ligase IV/XRCC4 addition on complex DSB end joining
To address the possibility that the poor end joining efficiency exhibited by the HeLa extracts for the complex-DSB-terminated plasmid may be a function of co-factor or DNA ligase availability, the end joining reactions were modified to include additional dNTPs to further support DNA polymerase mediated fill-in reactions (Fig. 2A), or recombinant human DNA ligase IV/XRCC4 complex, or both (Fig. 2B). Since the damaged plasmid from the (+) DMSO samples was somewhat more amenable to end joining by the HeLa extract than was the (−) DMSO DNA sample (Fig. 1), this DNA was chosen to test these modifications of the reaction conditions.
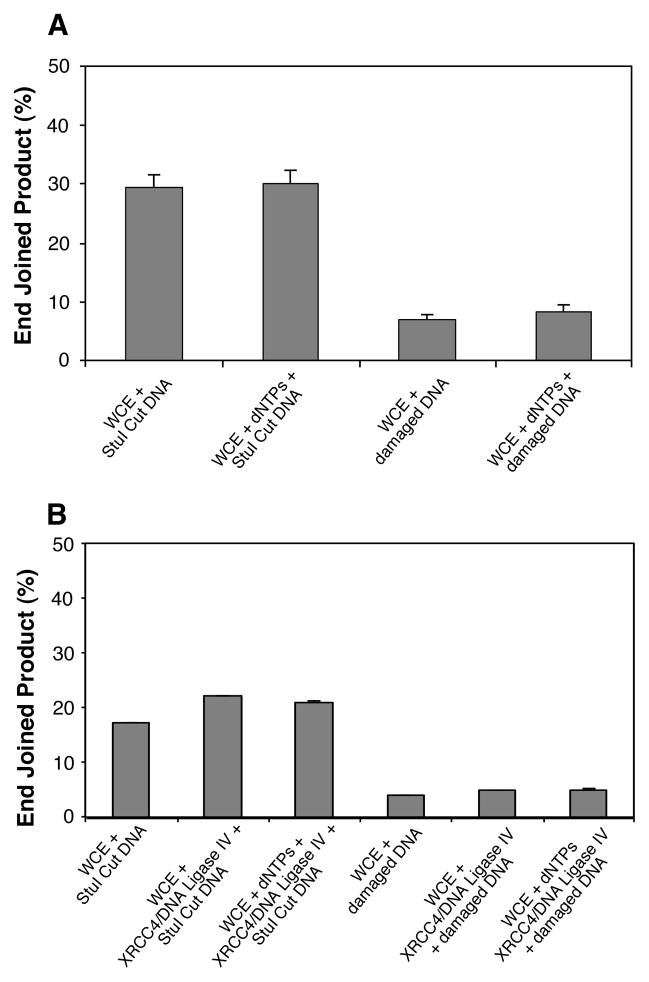
Figure 2.
Effect of reaction-component supplementation on end joining. A) The effect of 250 μM dNTP supplementation on HeLa WCE mediated end-joining reactions conducted with either the undamaged StuI-cut positive-control linear plasmid (blunt end), or damaged linear-plasmid DNA from the (+) DMSO irradiated sample. B) HeLa WCE mediated end-joining reactions supplemented with 10 units of recombinant human DNA ligase IV/XRCC4 complex alone, or the DNA ligase IV/XRCC4 complex and 250 μM dNTPs. As in panel A, reactions were conducted with the linear StuI-cut positive-control plasmid, or the (+) DMSO irradiated damaged linear-plasmid substrates. Reactions were run in duplicate and the mean of the replicates is plotted. The error bars indicate standard deviation.
As shown in Fig. 2A, addition of 250 μM of each dNTP (dGTP, dATP, dCTP, and TTP) to the repair reactions resulted in only a marginal increase in end-joined product over reactions performed without dNTP addition. This result indicates that the paucity of end joining by the HeLa extract with this substrate (in comparison to the level observed with the StuI restriction enzyme cut substrate) is not due to the need for a DNA polymerase mediated fill-in step during, or prior to, the end joining reaction.
To determine if the low end-joining efficiency was due to an inadequate level of DNA ligase IV activity, and/or a concomitant lack of dNTPs to support DNA polymerase activity, reactions were run with the addition of these components (Fig. 2B). Addition of 10 units of recombinant human DNA ligase IV/XRCC4 complex to the HeLa extract mediated end-joining reactions produced up to a 29% increase in end joined product with the directly ligateable blunt ends of the StuI cut plasmid substrate. However, addition of the DNA ligase IV complex to end joining reactions employing the damaged DNA substrate failed to enhance end joining more than a nominal amount, and further addition of dNTPs had no effect on the yield of product. Consequently, the inability of the HeLa extract to efficiently end join the complex DSBs of the damaged DNA substrate is not due to any of the known potential deficiencies of either the extract or the repair reaction conditions.
Taken together, these results suggest that rather than being an artifact or due to some deficiency in the WCE, the poor end-joining efficiency of the damaged DNA is a function of DSB structure that is independent of the DSB end group structure. Consideration of these results and the structural characteristics we have determined for the 125I-induced DSB, leads us to the hypothesis that base damage (including base loss) upstream, but in close proximity to a DSB end, may have a greater impact upon the ability of the NHEJ pathway to rejoin the break than does DSB end group chemistry.
The effect of upstream base damage vs. blocked 3′-ends on NHEJ
In order to compare the effects of base damage or base loss proximal to the DSB end with respect to 3′ blocking groups at the DSB end on NHEJ, we modified our previously described in vitro duplex-oligonucleotide-based NHEJ assay (50) by incorporating the structurally defined oligonucleotides depicted in Table 1. Individually annealing oligos a-g with oligo h, or in three cases i, forms a panel of nine 75-mer duplex oligonucleotides with 5′-OH/3′-OH blunt ends and an opposite end lower-strand with a 5′-32P-labeled self-complementary 4 nucleotide overhang. The upper strand 3′-end of these duplexes are modified with a terminal blocking group, or with a nucleobase alteration and/or a mismatch such that it occurs –1n upstream from the upper-strand 3′-end. A combination of these structures may also be formed (Table 2).
The impact on the ability of HeLa WCE mediated NHEJ to rejoin 3′-blocked DSB ends in contrast to DSBs possessing ligateable 3′-OH ends but with a damaged base, an AP site, or, a damaged or undamaged base mismatch –1n from the DSB 3′-end was assessed in repair reactions using the appropriately modified duplex oligo substrates. To determine the effect of 3′-blocked ends on HeLa cell extract mediated NHEJ, the extract’s ability to end-join the undamaged a/h oligo duplex was compared to the yield of end joined products in parallel repair reactions with the 3′-P blocked b/h oligo and the 3′-PG blocked c/h oligo (Fig. 3A and Table 2). Although less than the T4 DNA ligase positive control (Fig. 3A, lane 2), a substantial yield (~28%) of end-joined oligonucleotide dimers was typically obtained in reactions with the WCE and the undamaged a/h oligo substrate (Fig. 3A, lane 3). In contrast, the yield (~15%) of end joined product from the 3′-P blocked b/h oligo was typically about half that formed with the undamaged a/h oligo (Fig. 3A, lane 5). Furthermore, although the extract was capable of forming product with the 3′-PG blocked c/h oligo, end joining activity with this substrate was even less than with the 3′-P blocked oligo (compare Fig. 3A, lane 4 and 5). These results are consistent with previous observations of NHEJ mediated repair of 3′-end blocked DSBs (47, 52), in which, as observed here, the blocked DSB ends are capable of supporting end joining, but at a lower efficiency than directly ligateable DSB ends.
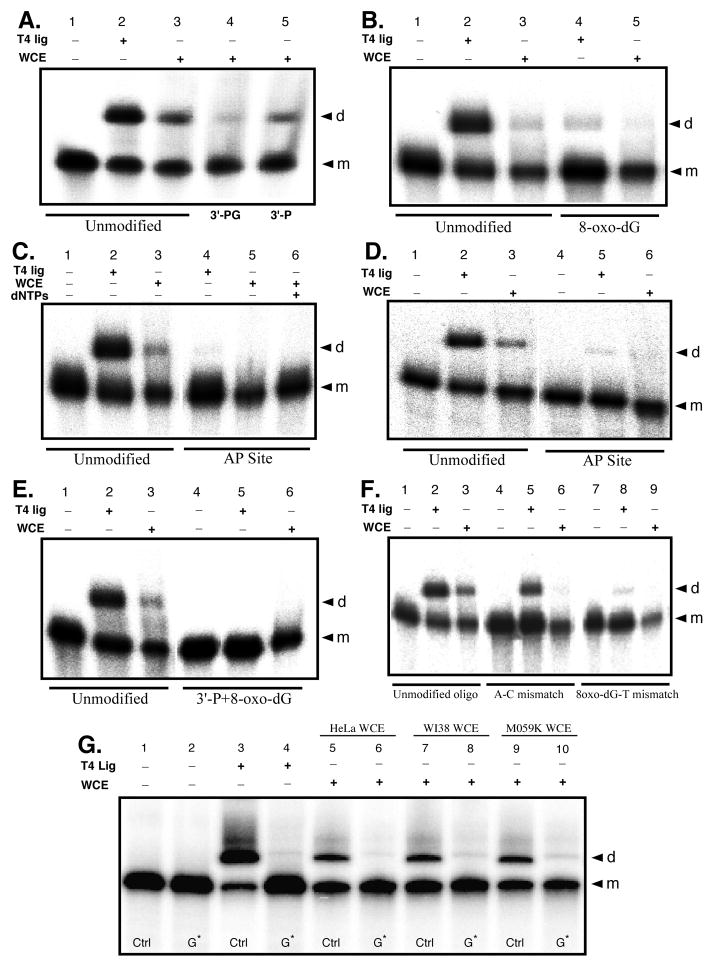
Figure 3.
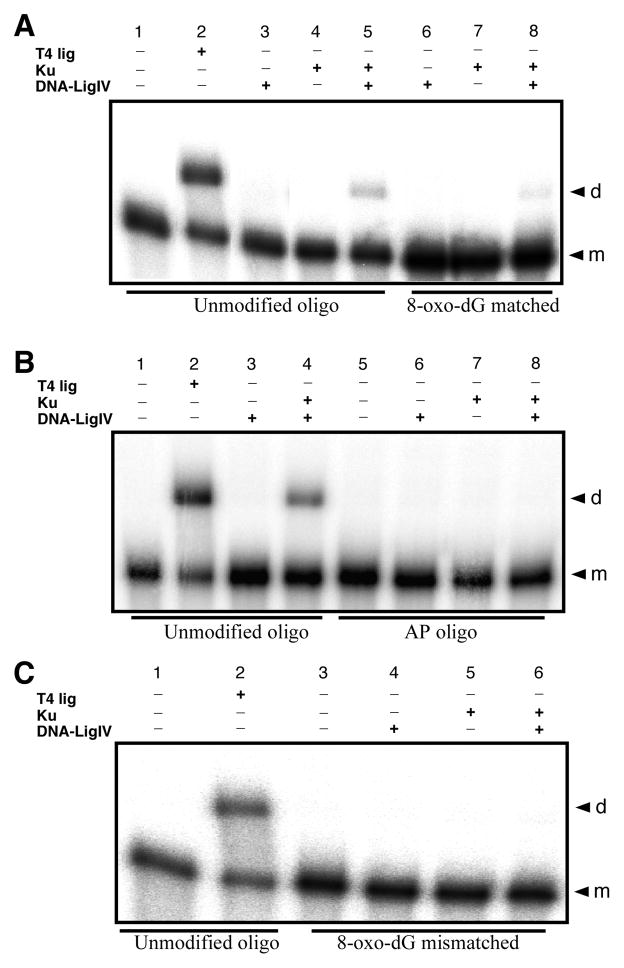
Effects of 3′-end group structure on HeLa WCE mediated end joining. Duplex oligonucleotide substrates with defined end group chemistry were formed by individually annealing oligos a – g to complementary 5′-32P labeled oligo h or i (Table 1), or in one case to the 3′-32P labeled (see Materials and Methods) oligo h*. All end joining reactions were run with 15 μg WCE or 0.2 unit T4 DNA ligase as indicated. Reaction products were resolved and identified by 8% denaturing PAGE and phosphorimaging, and gels representative of 3 experiments are shown. A) Typical end joining results with the unmodified a/h control oligo duplex and the 3′-P b/h and 3′-PG c/h terminated oligo duplexes. Lane 1, negative control; lane 2, T4 DNA ligase positive control; lanes 3–5 WCE mediated reactions. B) Typical end joining results for the –1n 8-oxo-dG e/i oligo duplex. C) Typical end joining results for the –1n AP site f/h oligo duplex. D) End joining reactions complementary to those depicted in panel C, only employing the 3′-32P end labeled lower-strand in the a/h* control, and f/h* –1n AP site modified, oligo duplex substrates to assess for potential 5′-32P label loss in the previous experiments. E) Typical end joining results for the multiply damaged g/i duplex oligo structure containing a –1n 8-oxo-dG in combination with a 3′-P blocked upper-strand 3′-end. F) Typical end joining reaction results for assays using the mismatch oligo substrates exhibiting either a –1n A:C mismatch (duplex a/i), or a –1n mismatch involving the 8-oxo-dG base lesion opposite thymine (duplex e/h). G) Typical end joining results for the –1n 8-oxo-dG e/i oligo duplex (G*) matched with the d/i -1n G:C base sequence matched control (Ctrl) and WCEs as indicated. In all panels, the “d” to the right of the gel indicates dimer end joining products and the “m” indicates monomer substrate oligonucleotide.
Surprisingly, although not a complete block to end joining, HeLa extract mediated NHEJ is inhibited to a similar or even greater extent when the reaction is run using a duplex oligo substrate with ligateable 3′-OH ends, but with an 8-oxo-dG modification at the –1n position of the upper strand 3′-end (duplex e/i; Fig. 3B, lane 5; also see Fig. 3G, lanes 5 and 6). Furthermore, although the e/i oligo duplex supports some direct ligation by T4 DNA ligase, the –1n 8-oxo-dG modification does appear to substantially inhibit its ligation (Fig. 3B, compare lane 2 and lane 4), but not to the same degree as the inhibition observed for HeLa extract NHEJ. In contrast, when presented with a duplex oligo substrate possessing an AP site at the –1n position of the upper-strand’s 3′-OH end (duplex f/h), HeLa extract mediated NHEJ is completely blocked (Fig. 3C, lane 5). During quality control testing of the f/h oligo substrate, we demonstrated cleavage of the AP site by E. coli endo IV. Therefore, it was possible that the AP site in the f/h duplex might be cut by a HeLa extract AP endonuclease such as APE1 prior to initiation of the NHEJ ligation reaction, if so, this might account for the complete lack of end joining with this substrate if insufficient dNTPs were present in the extract to support DNA polymerase mediated fill-in reactions subsequent to incision at the AP site. To test this, we supplemented the end-joining reaction with 250 μM of all four dNTPs. However, dNTP supplementation did not rescue WCE mediated end joining activity for the –1n AP site oligo duplex substrate (Fig. 3C, Lane 6). Once again, although the –1n AP site duplex supported direct ligation by T4 DNA ligase, this modification was an even stronger inhibitor of direct ligation than was the –1n 8-oxo-dG lesion (Fig. 3C, lane 4).
Unlike reactions involving the unmodified substrate, the AP site modified substrate (and to a lesser extent the –1n 8-oxo-dG) appears to be susceptible to 5′-dephosphorylation or degradation as evidenced by the reduced signal of the substrate in Fig. 3C, lane 5, and this may mask dimer product formation since the yield of end-joined dimers would be expected to be proportionately less than the remaining unreacted oligo substrate. To differentiate between the inability of the HeLa extract to form end joined products with the AP site modified oligo duplex and simple loss of the 32P label from the 5′-end of the duplex’s lower strand, end joining reactions were run in which the 79-mer lower strand of the duplex was 32P 3′-end labeled by Klenow DNA polymerase extension as described in the Materials and Methods (oligo h*, Table 1). End joining reactions employing the duplex formed between the 3′-end labeled lower strand and the AP site modified upper strand (duplex f/h*), produced essentially identical results to those obtained with the 5′-32P end labeled duplex f/h substrate (compare Fig. 3D and C). Furthermore, the end joining reactions employing the 3′-end labeled lower strand, indicated that the AP site modified oligo duplex is not detectably exonucleolytically degraded by the HeLa extract (Fig. 3D, lane 6), therefore the signal loss observed in the end joining reactions using the 5′-32P end labeled duplex lower strand is most likely due to loss of the 5′-32P label, either through the action of a phosphatase, or possibly as a result of 5′-end trimming which would be consistent with the known structure/function characteristics of Artemis activity at 5′-overhangs (53, 54). Consequently, while 3′-blocking groups have a negative impact on the ability of the human NHEJ pathway to rejoin a DSB, it appears that damage proximal to the DSB end may be an even greater obstacle for the NHEJ pathway to overcome.
Neither 3′-P, 3′-PG, nor –1n 8-oxo-dG alone, cause complete inhibition of HeLa extract mediated NHEJ. However, based upon numerous reports (1, 8, 33, 36, 55, 56), lesions such as these might be expected to exist concurrently in combination at complex radiation-induced DSBs, and thus constitute a form of multiply damaged site. Furthermore, as we have shown here and elsewhere (4, 57), highly complex DSBs like those formed by 125I decay, at which such multiply damaged sites do exist, are largely unrejoinable by the human NHEJ pathway. Therefore, we sought to determine if a multiply damaged DSB end consisting of a combination of lesions that are not complete blocks to NHEJ when presented individually at a DSB end, might act as a stronger block to NHEJ when presented together. To test this, we employed the multiply modified g/i duplex oligo substrate (Table 2; Fig. 3E). The g/i oligo duplex possesses an unligatable upper strand 3′-P end in combination with a –1n 8-oxo-dG. This multiply damaged DSB structure acts as a complete block to end joining, and is thus a more effective inhibitor of NHEJ than either modification alone when presented for end joining individually at a DSB end (Fig. 3E, lane 6). Also, as expected the g/i duplex does not support direct ligation by T4 DNA ligase due to its terminal 3′-P (Fig. 3E, lane 5).
Placement of 8-oxo-dG or AP site modifications at the –1n position of the DSB 3′-end may act as an end destabilizer. Therefore, the inhibition observed during end joining of these DSBs may not reflect specific NHEJ responses to specific base lesion structures, but may reflect a more generalized inability of the NHEJ pathway to efficiently join DSBs that possess unstable or poorly hydrogen bonded and base stacked configurations near the DSB end. To investigate this possibility, we generated two oligo duplex end-joining substrates (oligos e/h and a/i, Table 2) that possess mismatches at the –1n position from the DSB 3′-end. Duplex a/i contains a mismatch between unmodified adenosine and cytosine bases at the –1n position of the upper strand 3′-end, whereas oligo e/h contains a mismatch between a modified 8-oxo-dG and a thymine. T4 DNA ligase mediated direct ligation is considerably less efficient for the –1n 8-oxodG:T mismatch than it is for the A:C mismatch, suggesting that the mismatch containing the base lesion is more difficult to align and ligate (compare Fig. 3F lane 5 and lane 8). Regardless of their ability to be directly ligated, neither mismatch substrate supports end joining by the WCE (Fig. 3F, lanes 6 and 9). The inability of both duplex e/h and a/i to support end joining by the HeLa extract suggests that destabilization of the DSB ends in these substrates may be a factor influencing inhibition of NHEJ, as opposed to the specific base lesion structure that is present near the DSB end.
To confirm that inhibition of NHEJ by base damage proximal to a DSB end is a broad-based property of NHEJ and not just an effect limited to HeLa cell extracts, freshly prepared WCE of HeLa, WI38, and MO59K cells were assayed for their ability to end join the e/i duplex containing 8-oxo-dG at the -1n position. Furthermore, to establish that the NHEJ inhibition observed to this point was not simply an effect of the terminal sequence of the substrate oligonucleotides used previously, reactions assessing the ability of the extracts to end join the e/i duplex were run in conjunction with the undamaged control duplex d/i, which contains the sequence matched canonical G:C Watson-Crick base pair at the -1n position of the upper strand 3′-end (Fig. 3G). End joining of the control duplex was efficient for all three WCEs, ranging between ~30 – 40% (Fig. 3G, lanes 5, 7, and 9). In contrast, end joining by all three WCEs was strongly inhibited (≥80%) by 8-oxo-dG at the -1n position of the duplex (Fig. 3G, lanes 6, 8 and 10). Furthermore, direct ligation of the -1n G:C base paired d/i undamaged control duplex by T4 DNA ligase occurred at an efficiency similar to, if not better than that of the -1n A:T base-pair-containing a/h undamaged control duplex (compare Fig. 3A–F lanes 2, to Fig. 3G lane 3). Again, as seen previously (Fig. 3B, lane 4), the 8-oxo-dG containing e/i duplex only supported minimal amounts (~4%) of direct ligation by T4 DNA ligase (Fig. 3G, lane 4).
Effects of lesion structures on binding and activity for HeLa cell extract nonhomologous end joining proteins on damaged duplex substrates
We have previously shown that HeLa cell extract mediated dimer formation in the in vitro end joining reaction with the unmodified a/h oligo duplex is a function of NHEJ, Ku dependent, and not simply a result of direct ligation (50). To establish that the lesion modifications of our oligo duplex substrates do not interfere with the ability of the oligos to support binding and protein-complex formation by the NHEJ pathway core proteins, we conducted electrophoretic mobility shift/supershift assays (EMSA) under the same conditions used previously with the a/h oligo.
The unmodified (duplex a/h), or damage containing duplex oligos (duplexes e/i or f/h; 10 pmole/reaction) were permitted to form complexes with HeLa cell extract proteins (15 μg) either in the presence or absence of antibodies to NHEJ core-pathway protein components as indicated in Fig. 4A through 4C. All of the duplexes displayed mobility shifts in the presence of HeLa extract protein, as well as antibody-specific supershifts following incubation with anti-Ku70, anti-DNA-PKcs, or anti-DNA ligase IV antibodies. Similar results were also obtained with EMSA assays in which the oligo duplexes contained 3′-P or 3′-PG blocked ends (data not shown). The demonstration of NHEJ core protein dependent EMSA supershifts regardless of the duplex oligo substrate used in the EMSA reaction indicates that not only are the 75-mer duplex oligo substrates long enough to permit binding of these proteins, but that under our reaction conditions NHEJ protein complexes form on the oligos.
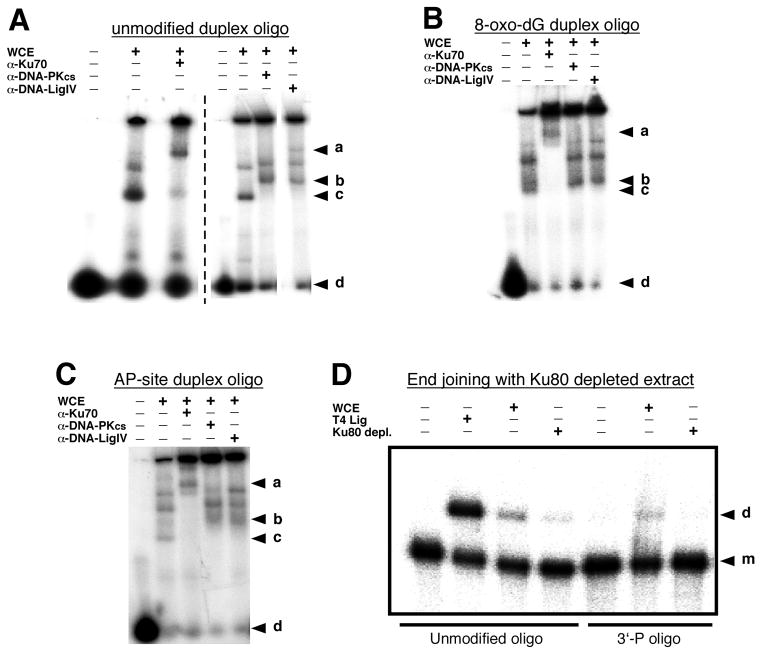
Figure 4.
Electrophoretic mobility shift assays (EMSA) and Ku80 immunodepletion effects. EMSAs were conducted as described in the Materials and Methods with the HeLa cell extract and the 5′-32P end labeled undamaged a/h oligo duplex (A), the 8-oxo-dG modified e/i oligo duplex (B), or the AP site modified f/h oligo duplex (C) as the protein-binding targets. The protein-bound duplex oligonucleotides were resolved in a 6% non-denaturing PAGE gel and data was obtained by phosphorimaging. Panel A is a compilation of data from two separate experiments as indicated by the vertical dashed line. In panels A – C, the letters to the right of the gel images represent the following: “a” identifies the position of the anti-Ku70 antibody mediated supershift; bands migrating between a and b in the 6th & 7th lanes of panel A and 4th & 5th lanes of panels B and C are the anti-DNA-Pkcs and anti-DNA ligase IV supershift bands as indicated. The “c” indicates the position of the HeLa extract mediated band shift, and “d” indicates the position of the unshifted duplex oligo targets. The functional effects of Ku80 immunodepletion on HeLa WCE end joining was assessed as indicated in panel D using either the unmodified a/h oligo duplex or the 3′-P terminated b/h duplex as the end joining substrates. In this case, the “d” to the right of the gel indicates the end joined dimer product and the “m” indicates the monomer substrate duplex.
To further establish the role of the NHEJ pathway in this assay, end-joining reactions were performed with Ku80 immunodepleted HeLa extract. Following immunodepletion, the yield of end-joined dimer products with the unmodified a/h and 3′-P terminated b/h oligo duplexes was decreased (Fig. 4D), and the reduction in product yield was proportional to the extent of Ku80 protein reduction in the depleted extract (data not shown). Taken together, demonstration of NHEJ pathway core-protein complex formation on the 75-mer duplex oligo substrates, along with inhibition of end joining after HeLa extract Ku80 immunodepletion, indicates that the oligonucleotide dimer products formed in the assay are produced via a Ku dependent NHEJ reaction. This conclusion is further supported by results obtained by replacing HeLa WCE in the oligo end-joining assay with purified recombinant Ku70/80 heterodimer, and/or DNA ligase IV/XRCC4 tetramer (Fig. 5). End joining of the unmodified a/h oligo duplex is only supported in the presence of both Ku70/80 and DNA ligase IV/XRCC4 (Fig. 5A, lane 5), and DNA ligase IV/XRCC4 alone is not sufficient to support direct ligation of this duplex (Fig. 5A&B, lane 2). Furthermore, when presented with the same –1n 8-oxo-dG (duplex e/i), -1n AP site (duplex f/h), and –1n 8-oxo-dG mismatch (duplex e/h) modified oligo duplexes used earlier, levels of dimer formation were observed that were consistent with, and similar to, those obtained for these substrates when the HeLa WCE was used as the source of NHEJ proteins and enzyme activities (Fig. 5A through C).
Figure 5.
End joining of modified and unmodified duplex oligonucleotide substrates with purified recombinant human Ku70/80 heterodimer and/or DNA ligase IV/XRCC4 tetrameric protein complexes. A) Comparison of end joining for the unmodified a/h control oligo duplex with either T4 DNA ligase (as an end joining positive control (0.2 U)) or Ku 70/80 (1.5 U) + DNA ligase IV/XRCC4 (1.5 U), and the equivalent reactions using the e/i –1n 8-oxo-dG oligo duplex substrate. B) Comparison of end joining for the unmodified a/h control oligo duplex with either T4 DNA ligase (0.2 U) or Ku 70/80 (1.5 U) + DNA ligase IV/XRCC4 (1.5 U), and the equivalent reactions using the f/h –1n AP site modified oligo duplex. C) Attempted end joining of the e/h –1n 8-oxo-dG:T mismatch containing oligo duplex with Ku 70/80 (1.5 U) + DNA ligase IV/XRCC4 (1.5 U). In all cases, the “d” to the right of the gel indicates the end joined dimer product and the “m” indicates the monomer substrate duplex.
Temporal sequencing of DNA repair events at the DSB end during NHEJ of complex DSBs
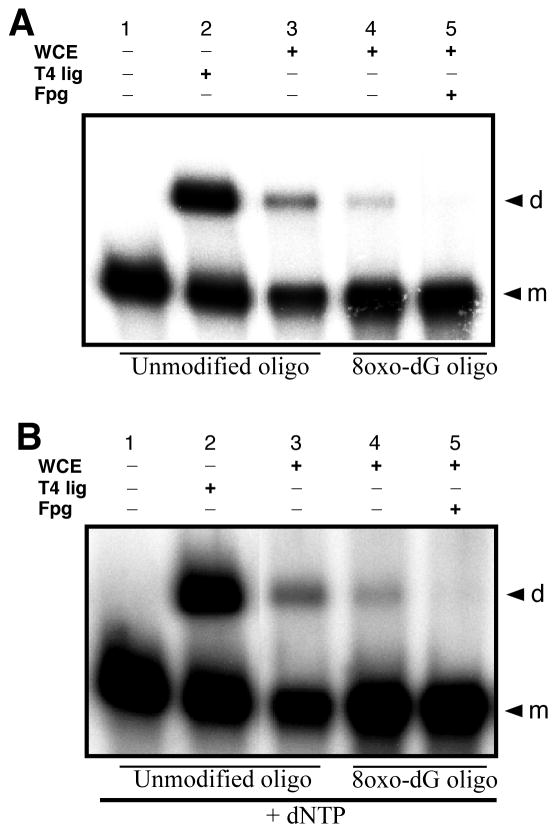
As discussed above for HeLa WCE mediated NHEJ of the f/h oligo duplex containing an AP site in the upper-strand 3′-end at the –1n position, it appears that the AP site is not being incised and repaired by the BER proteins in the WCE (Fig. 3C and D). This finding suggests that base lesions near the DSB end may still be present in the dimer products of the end joining reaction. The continued presence of base lesions in the products of the NHEJ reaction would suggest base damage processing may not be linked to NHEJ and does not occur concurrent with NHEJ, but rather is an independent process, which if it occurs, acts subsequent to the end-joining event. Thus, the continued presence of base lesions in the end joined dimer products of the NHEJ reactions employing oligo duplex e/i, in which 8-oxodG is positioned at the –1n position from the upper strand’s 3′-end, would indicate that the NHEJ reaction may take priority over BER in such structures. To test this, we subjected the e/i oligo duplex to the standard in vitro end joining reaction using the HeLa WCE as the source of NHEJ activities. The products of the end joining reaction were then treated with E. coli Fpg to probe for the continued presence of the 8-oxo-dG modification (Fig. 6). The HeLa WCE was demonstrated to be active for NHEJ mediated dimer formation (Fig. 6A and B, lanes 3 and 4) for both the unmodified control a/h oligo duplex and the –1n 8-oxo-dG modified e/i oligo duplex, and the dimer products of these reactions were susceptible to cleavage by Fpg, indicating the continued presence of the base lesion in the dimer products (Fig. 6A, lane 5). Furthermore, addition of 250 μM of all four dNTPs did not rescue the BER reaction, nor did dNTP addition appear to rescue 8-oxo-dG removal prior to the NHEJ reaction and thus support subsequent dimer formation (Fig. 6B, lane 5). All dimer products of the NHEJ reaction were equally susceptible to Fpg cleavage, suggesting that if BER occurs at this position, it occurs subsequent to the end joining reaction, and with the diminished kinetics that would be expected for such closely opposed base lesions (42, 45, 58–61).
Figure 6.
Temporal sequence of DNA repair reactions at complex DNA DSB ends. HeLa WCE mediated NHEJ reactions were performed using either the unmodified a/h oligo duplex as a positive control for end joining, or the –1n 8-oxo-dG modified e/i oligo duplex. After stopping the standard reaction, the end-joined dimer products of the assay were treated with Fpg (1U, 37°C for 30 min) to test for the continued presence of 8-oxo-dG. A) Fpg treatment of the e/i oligo duplex dimer end-joining products. B) End joining and Fpg treatment of the e/i oligo duplex dimer end-joining products in the presence of 250 μM dNTPs.
Discussion
The work presented in the current study was designed to determine what aspect(s) of the complex DSB structure induced by 125I have the greatest impact on DSB repair and thus may be important contributing factors to radiation cytotoxicity. The results presented here, as well as those of previous studies (4, 57, 62), indicate that the high-LET-like site-specific 125I DSBs produced in our irradiation system are refractory to repair. The inability of HeLa WCE to efficiently end join the 125I-induced DSBs does not appear to be due to a cofactor or enzyme deficiency of the extract, since supplementation of end joining reactions with either all four dNTPs or recombinant human DNA ligase IV/XRCC4, or both, did not improve the end joining ability of the extract (Fig. 2). These results suggested that the poor end joining activity of the HeLa extract for the 125I-induced DSBs was not due to dNTP starvation of a DNA polymerase mediated DSB repair step (63–65), or DNA polymerase dependent secondary repair process such as base excision repair (BER). Furthermore, although our restriction-enzyme-cleaved control DNA was efficiently end joined by the WCE, it has been reported by others that extracts of HeLa cells may be somewhat deficient in DNA ligase IV/XRCC4 complex and that this complex may also be involved in the polymerase mediated gap filling step of NHEJ (64, 66). Therefore, we also conducted DNA ligase IV/XRCC4 supplementation reactions. However, the end joining activity of the supplemented reactions only improved marginally, and did not approach the high efficiency end joining observed for the restriction-enzyme-cleaved control DNA. Nor did concurrent supplementation of dNTPs and DNA ligase IV/XRCC4 result in better end joining activity than observed in their absence or with each component individually. These results suggest that the inability of the HeLa cell extract to support efficient end joining of the 125I-induced DSBs is not due to one of the known potential deficiencies of the extract, but is a function of the DSB structure.
There are three basic categories of structural complexity that we have determined for the 125I-induced DSB; these are DSB end morphology (i.e., DSB-end overhang configurations and polarities), DSB end group structure (i.e., DSB-end terminal structures such as 3′-P and 3′-PG), and nucleotide damage in close proximity to the DSB end. Our examination of end group structure indicated that many of the DSB ends consist of either 3′-OH or 3′-P, particularly in DNA irradiated in the presence of 2 M DMSO. Furthermore, plasmid DNA linearized by 125I decay supported direct ligation by T4 DNA ligase and the yield of this reaction (8%) was significantly improved (to 45%) by prior T4 PNK 3′-phosphatase treatment (36). These results not only indicated that the end group structure of the 125I-induced DSBs was relatively simple and might be expected to be capable of supporting ligation by the HeLa extract, but that the overhang morphology of the breaks were also not a major impediment to ligation. However, as we have shown, this is not the case. The 125I-induced DSBs are refractory to NHEJ regardless of the reaction conditions or DNA pretreatments used. These results suggested that the base damage that occurs proximal to the 125I-induced DSBs might be a greater inhibitor of end joining than the DSB end structure or overhang morphology.
The question of which lesion structural configuration (damage upstream from the DSB end or blocked 3′-ends) is most detrimental to DSB processing by the NHEJ machinery was addressed using the panels of defined duplex oligo nucleotide end-joining substrates depicted in Table 2. Our results indicate that a base lesion or base loss immediately upstream from the DSB end may be a more potent inhibitor of NHEJ than is an unligatable 3′-end blocked by 3′-P or 3′-PG. Although a specific effect on NHEJ imparted by the lesion chemistry cannot be ruled out, our results may reflect an incapacity of the NHEJ system to efficiently process DSB ends that are unstable with respect to base pairing or base stacking. Such a conclusion is supported by the observation that an A:C base mismatch was equally inhibitory to the NHEJ reaction as were a mismatch involving and 8-oxodG base lesion, or an AP site at the same –1n position in the substrate. Thus, lesions that would appear to prevent normal hydrogen bonding and/or base stacking at or near the DSB end may be strong inhibitors of human NHEJ. Furthermore based upon the results obtained for direct ligation of such substrates with T4 DNA ligase (Fig. 3), as well as the similarity of the WCE mediated NHEJ results to those obtained with purified Ku and DNA ligase IV/XRCC4 (Fig. 3 and 5), it appears likely that inhibition of the NHEJ reaction by the damaged DSB structures most probably occurs at the ligation step of the reaction, and may reflect the ability/inability of DNA Ligase IV/XRCC4 to act on such structures.
Furthermore, the results of the temporal sequencing assay (Fig. 6) suggest that ligation of the DSB ends by DNA Ligase IV/XRCC4 may be a prerequisite for repair of a base lesion in close proximity to the DSB end. Thus, base damage processing mechanisms that might be expected to reduce the DSB end complexity and result in increased DSB end-joining efficiency, do not appear to be capable of effective activity prior to the ligation step of the NHEJ reaction. This is consistent with a recent study indicating that base lesions in close proximity to a DSB end are poor substrates for purified human DNA glycosylases (46). Similar results have been obtained in a plasmid based system in which an AP site has been situated in a 2 nucleotide 5′-overhang adjacent to the duplex terminus, and juxtaposed to a blunt end at the opposite end of the plasmid (67). Following repair processing in human cells, although poorly repaired, nearly half of the products of this reaction involved a translesion synthesis mechanism and therefore, retained the AP site in place during the end joining process.
If there is in fact, interaction between the NHEJ and the BER pathways during the processing of complex DSB damage, our results seem to suggest that the presence of NHEJ pathway proteins bound to the DSB ends may further restrict access and the opportunity for BER proteins and other potential end processing enzymes to act at DSB ends.
In contrast, in the context of clustered base lesions, where attempted repair of two closely opposed lesions could give rise to a DSB, the role of NHEJ in preventing DSB formation has already been examined. In one study (68), it was found that NHEJ is only partially able to prevent conversion of clustered lesions to potentially lethal DSBs due to an incomplete or inaccurate repair carried out by Ku-dependent as well as Ku-independent pathways. Several other studies have suggested a role for the key components of the NHEJ pathway, especially the DNA-PK holoenzyme in the processing of non-DSB clustered DNA damage. This is in addition to the indirect role that DNA-PKcs exhibits in the regulation of expression for two key DSB processing proteins, ATM and Artemis (69). Previous work had already suggested a potential role of the Ku heterodimer in the repair of closely opposed base lesions in DNA (70). Recent work from the Georgakilas laboratory also describe a potentially important role for DNA-PKcs during the processing of oxidative base-lesion clusters in DNA. These studies (71, 72), demonstrate a significant decrease in processing of base-lesion clusters and enhanced cell death in the absence of DNA-PKcs or in the presence of chemically inactivated or partially deficient DNA-PKcs indicating that it may have an important role in clustered base damage processing.
Such findings suggest that DNA-PKcs, and possibly other component proteins of the NHEJ pathway, may have complex multidimensional roles in the processing of structurally complex DNA damage. These proteins may be required for the processing of some classes of complex clustered base lesions, while in the case of the DSB lesions described here, they may be incapable of acting or possibly even interfere and/or compete with proteins that might be required to process these DSBs into a reparable form. In support of this, our results suggest that the presence of NHEJ pathway proteins bound to the DSB ends may further restrict access and the opportunity for BER proteins and other potential end processing enzymes to act at DSB ends.
Consequently, the exact interaction of DNA ligase IV/XRCC4 with the damaged DSB structures, as well as its precise role in the NHEJ inhibition observed with these DSB structures, is of considerable interest for future studies, as is the exact nature of the damaged DSB structure required to induce strong inhibition of NHEJ (i.e. with respect to the sequence position and type of base lesion in relation to the DSB end).
Knowledge of the roles played by component structures of a complex DSB during the repair process may help us to better understand this process as well as some of the mechanisms that may underlie radiation cytotoxicity. This information may also provide insights supporting new approaches to enhance the effectiveness of radiation therapy by developing agents and/or methods that selectively increase the yield of those DSB-associated lesions that make the DSB difficult to repair and hence toxic to the cell. Alternatively, insights into these aspects of DSB structure-dependent inhibition of NHEJ may provide direction for the development of highly effective and specific small molecule inhibitors of human NHEJ that also might be used for therapeutic advantage in a wide range of cancer treatment protocols. Furthermore, understanding the biological consequences of critical complex DSB-lesion components may also be applicable to questions as diverse as establishing better risk estimates for long duration space travel in which astronauts traveling beyond the earth’s magnetic field will be exposed to even greater doses of high-LET radiation than in low earth orbit (73).
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, through the Warren Grant Magnuson Clinical Center.
References
- 1.Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]
- 2.Iliakis G. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. Bioessays. 1991;13:641–648. doi: 10.1002/bies.950131204. [DOI] [PubMed] [Google Scholar]
- 3.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 4.Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Radford IR. DNA lesion complexity and induction of apoptosis by ionizing radiation. Int J Radiat Biol. 2002;78:457–466. doi: 10.1080/09553000110120337. [DOI] [PubMed] [Google Scholar]
- 6.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int J Radiat Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 7.Ward JF, Blakely WF, Joner EI. Mammalian cells are not killed by DNA single-strand breaks caused by hydroxyl radicals from hydrogen peroxide. Radiat Res. 1985;103:383–392. [PubMed] [Google Scholar]
- 8.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 9.Leatherbarrow EL, Harper JV, Cucinotta FA, O’Neill P. Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int J Radiat Biol. 2006;82:111–118. doi: 10.1080/09553000600599783. [DOI] [PubMed] [Google Scholar]
- 10.Burkart W, Jung T, Frasch G. Damage pattern as a function of radiation quality and other factors. C R Acad Sci III. 1999;322:89–101. doi: 10.1016/s0764-4469(99)80029-8. [DOI] [PubMed] [Google Scholar]
- 11.Sachs RK, Hahnfeld P, Brenner DJ. The link between low-LET dose-response relations and the underlying kinetics of damage production/repair/misrepair. Int J Radiat Biol. 1997;72:351–374. doi: 10.1080/095530097143149. [DOI] [PubMed] [Google Scholar]
- 12.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeggo PA. DNA breakage and repair. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 14.Lieber MR. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 15.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche M-C, Sanal O, Plebani A, Stephan J-L, Hufnagel M, le Deist F. Cernunnos, a Novel Nonhomologous End-Joining Factor, Is Mutated in Human Immunodeficiency with Microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 16.Ahnesorg P, Smith P, Jackson SP. XLF Interacts with the XRCC4-DNA Ligase IV Complex to Promote DNA Nonhomologous End-Joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Pastwa E, Somiari RI, Malinowski M, Somiari SB, Winters TA. In vitro non-homologous DNA end joining assays-The 20th anniversary. Int J Bioch & Cell Biol. 2009;41:1254–1260. doi: 10.1016/j.biocel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munz PL, Young CS. End-joining of DNA fragments in adenovirus transfection of human cells. Virol. 1991;183:160–169. doi: 10.1016/0042-6822(91)90129-y. [DOI] [PubMed] [Google Scholar]
- 20.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehman CW, Jeong-Yu S, Trautman JK, Carroll D. Repair of heteroduplex DNA in Xenopus laevis oocytes. Genetics. 1994;138:459–470. doi: 10.1093/genetics/138.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labhart P. Nonhomologous DNA end joining in cell-free systems. Eur J Biochem. 1999;265:849–861. doi: 10.1046/j.1432-1327.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- 24.Pfeiffer P, Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988;16:907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.North P, Ganesh A, Thacker J. The rejoining of double-strand breaks in DNA by human cell extracts. Nucleic Acids Res. 1990;18:6205–6210. doi: 10.1093/nar/18.21.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairman MP, Johnson AP, Thacker J. Multiple components are involved in the efficient joining of double stranded DNA breaks in human cell extracts. Nucleic Acids Res. 1992;20:4145–4152. doi: 10.1093/nar/20.16.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thacker J, Chalk J, Ganesh A, North P. A mechanism for deletion formation in DNA by human cell extracts: the involvement of short sequence repeats. Nucleic Acids Res. 1992;20:6183–6188. doi: 10.1093/nar/20.23.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derbyshire MK, Epstein LH, Young CS, Munz PL, Fishel R. Nonhomologous recombination in human cells. Mol Cell Biol. 1994;14:156–169. doi: 10.1128/mcb.14.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffer P, Thode S, Hancke J, Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol Cell Biol. 1994;14:888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci U S A. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HC, Perrault AR, Takeda Y, Qin W, Wang HY, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 33.Datta K, Jaruga P, Dizdaroglu M, Neumann RD, Winters TA. Molecular Analysis of Base Damage Clustering Associated with a Site-Specific Radiation-Induced DNA Double-Strand Break. Radiat Res. 2006;166:767–781. doi: 10.1667/RR0628.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datta K, Neumann RD, Winters TA. Characterization of complex apurinic/apyrimidinic-site clustering associated with an authentic site-specific radiation-induced DNA double-strand break. Proc Natl Acad Sci USA. 2005;102:10569–10574. doi: 10.1073/pnas.0503975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datta K, Neumann RD, Winters TA. Characterization of a complex 125I-induced DNA double-strand break: implications for repair. Int J Radiat Biol. 2005;81:13–21. doi: 10.1080/09553000400017713. [DOI] [PubMed] [Google Scholar]
- 36.Datta K, Weinfeld M, Neumann RD, Winters TA. Determination and Analysis of Site-Specific 125I Decay-Induced DNA Double-Strand Break End-Group Structures. Radiat Res. 2007;167:152–166. doi: 10.1667/rr0629.1. [DOI] [PubMed] [Google Scholar]
- 37.Kassis AI, Sastry KS, Adelstein SJ. Kinetics of uptake, retention, and radiotoxicity of 125IUdR in mammalian cells: implications of localized energy deposition by Auger processes. Radiat Res. 1987;109:78–89. [PubMed] [Google Scholar]
- 38.Sedelnikova OA, Panyutin IG, Thierry AR, Neuman RD. Radiotoxicity of iodine-125-labeled oligodeoxyribonucleotides in mammalian cells. J Nucl Med. 1998;39:1412–1418. [PubMed] [Google Scholar]
- 39.Beyert N, Reichenberger S, Peters M, Hartung M, Gottlich B, Goedecke W, Vielmetter W, Pfeiffer P. Nonhomologous DNA end joining of synthetic hairpin substrates in Xenopus laevis egg extracts. Nucleic Acids Res. 1994;22:1643–1650. doi: 10.1093/nar/22.9.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3 ′-overhangs and 3 ′-phosphoglycolate termini - Effect of Ku on repair fidelity. J Biol Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 41.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3 ′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 42.David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Bioch. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 43.David-Cordonnier MH, Boiteux S, O’Neill P. Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res. 2001;29:1107–1113. doi: 10.1093/nar/29.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David-Cordonnier MH, Laval J, O’Neill P. Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Bioch. 2001;40:5738–5746. doi: 10.1021/bi002605d. [DOI] [PubMed] [Google Scholar]
- 45.Lomax ME, Salje H, Cunniffe S, O’Neill P. 8-OxoA inhibits the incision of an AP site by the DNA glycosylases Fpg, Nth and the AP endonuclease HAP1. Radiat Res. 2005;163:79–84. doi: 10.1667/rr3284. [DOI] [PubMed] [Google Scholar]
- 46.Dobbs TA, Palmer P, Maniou Z, Lomax ME, O’Neill P. Interplay of two major repair pathways in the processing of complex double-strand DNA breaks. DNA Repair. 2008;7:1372–1383. doi: 10.1016/j.dnarep.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Gu XY, Weinfeld MA, Povirk LF. Implication of DNA-dependent protein kinase in an early, essential, ocal phosphorylation event during end-joining of DNA double-strand breaks in vitro. Bioch. 1998;37:9827–9835. doi: 10.1021/bi980198o. [DOI] [PubMed] [Google Scholar]
- 48.Hendrickson CL, Purkayastha S, Pastwa E, Neumann RD, Winters TA. Coincident In Vitro Analysis of DNA-PK-Dependent and -Independent Nonhomologous End Joining. J Nucleic Acids. 2010:Article ID 823917. 11. doi: 10.4061/2010/823917. 823910.824061/822011/823917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urata H, Akagi M. A Convenient Synthesis of Oligonucleotides with a 3′-Phosphoglycolate and 3′-Phosphoglycaldehyde Terminus. Tetrahedron Lett. 1993;34:4015–4018. [Google Scholar]
- 50.Datta K, Neumann RD, Winters TA. An in vitro nonhomologous end-joining assay using linear duplex oligonucleotides. Anal Bioch. 2006;358:155–157. doi: 10.1016/j.ab.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 51.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pastwa E, Neumann RD, Winters TA. In vitro repair of complex unligatable oxidatively induced DNA double-strand breaks by human cell extracts. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 54.Ma YM, Schwarz K, Lieber MR. The Artemis: DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 55.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 56.Goodhead DT, Thacker J, Cox R. Weiss Lecture. Effects of radiations of different qualities on cells: molecular mechanisms of damage and repair. Int J Radiat Biol. 1993;63:543–556. doi: 10.1080/09553009314450721. [DOI] [PubMed] [Google Scholar]
- 57.Mezhevaya K, Winters TA, Neumann RD. Gene targeted DNA double-strand break induction by 125I-labeled triplex-forming oligonucleotides is highly mutagenic following repair in human cells. Nucleic Acids Res. 1999;27:4282–4290. doi: 10.1093/nar/27.21.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J Mol Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 59.Lomax ME, Cunniffe S, O’Neill P. 8-oxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 60.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 61.Malyarchuk S, Castore R, Harrison L. Apex1 can cleave complex clustered DNA lesions in cells. DNA Repair. 2009;8:1343–1354. doi: 10.1016/j.dnarep.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Odersky A, Panyutin IV, Panyutin IG, Schunck C, Feldmann E, Goedecke W, Neumann RD, Obe G, Pfeiffer P. Repair of Sequence-specific 125I-induced Double-strand Breaks by Nonhomologous DNA End Joining in Mammalian Cell-free Extracts. J Biol Chem. 2002;277:11756–11764. doi: 10.1074/jbc.M111304200. [DOI] [PubMed] [Google Scholar]
- 63.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda - Implications for function. J Biol Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 64.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 65.Mahajan KN, McElhinny SAN, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: Role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee JW, Yannone SM, Chen DJ, Povirk LF. Requirement for XRCC4 and DNA ligase IV in alignment-based gap filling for nonhomologous DNA end joining in vitro. Cancer Res. 2003;63:22–24. [PubMed] [Google Scholar]
- 67.Covo S, de Villartay JP, Jeggo PA, Livneh Z. Translesion DNA synthesis-assisted non-homologous end-joining of complex double-strand breaks prevents loss of DNA sequences in mammalian cells. Nucleic Acids Res. 2009;37:6737–6745. doi: 10.1093/nar/gkp703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng YL, Woods RG, Beamish H, Ye RQ, Lees-Miller SP, Lavin MF, Bedford JS. Deficiency in the catalytic subunit of DNA-Dependent protein kinase causes down-regulation of ATM. Cancer Res. 2005;65:1670–1677. doi: 10.1158/0008-5472.CAN-04-3451. [DOI] [PubMed] [Google Scholar]
- 70.Hashimoto M, Donald CD, Yannone SM, Chen DJ, Roy R, Kow YW. A possible role of Ku in mediating sequential repair of closely opposed lesions. J Biol Chem. 2001;276:12827–12831. doi: 10.1074/jbc.M010906200. [DOI] [PubMed] [Google Scholar]
- 71.Peddi P, Francisco DC, Cecil AM, Hair JM, Panayiotidis MI, Georgakilas AG. Processing of clustered DNA damage in human breast cancer cells MCF-7 with partial DNA-PKcs deficiency. Cancer Lett. 2008;269:174–183. doi: 10.1016/j.canlet.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 72.Peddi P, Loftin CW, Dickey JS, Hair JM, Burns KJ, Aziz K, Francisco DC, Panayiotidis MI, Sedelnikova OA, et al. DNA-PKcs deficiency leads to persistence of oxidatively induced clustered DNA lesions in human tumor cells. Free Rad Biol Med. 2010;48:1435–1443. doi: 10.1016/j.freeradbiomed.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Setlow RB. The hazards of space travel. EMBO Rep. 2003;4:1013–1016. doi: 10.1038/sj.embor.7400016. [DOI] [PMC free article] [PubMed] [Google Scholar]