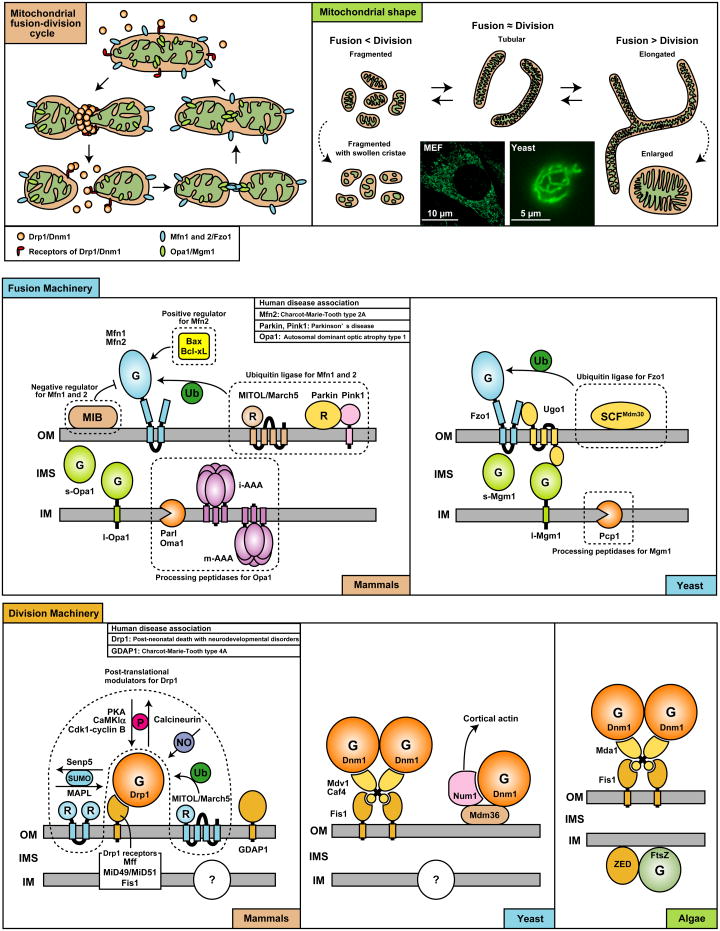
Mitochondrial fusion and division
Mitochondria are tubular, highly dynamic organelles that continuously fuse and divide in a regulated manner. A balance of fusion and division controls mitochondrial morphology; imbalanced dynamics leads to altered morphology, which is associated with a variety of pathological conditions. When fusion is decreased, mitochondria fragment into small, spherical mitochondria that are often characterized by swollen cristae and impaired respiratory functions. When division is inhibited, tubular mitochondria fuse, generating elongated mitochondrial tubules with increased connectivity. In some neurons, however, decreased division leads to enlarged, spherical mitochondria.
Highlighting the importance of mitochondrial fusion and division in human health and disease, mutations in mitochondrial dynamics components have recently been linked to several neurodevelopmental and neurodegenerative diseases including a birth defect with multiple neurological disorders (Drp1), Parkinson's disease (Parkin and Pink1), autosomal dominant optic atrophy type 1 (Opa1) and Charcot-Marie-Tooth neuropathies (Mfn2 and GDAP1). In addition, Alzheimer's and Huntington's diseases are not associated with such mutations, but show altered activity and abundance of mitochondrial dynamics components.
Core machineries for mitochondrial fusion and division
Dynamin-related GTPases feature prominently in mitochondrial fusion and division. Complexes of mitofusin 1 and 2 (Fzo1) control outer membrane fusion, while Opa1 (Mgm1) mediates inner membrane fusion. In addition to its role in fusion, Opa1 (Mgm1) has been implicated in direct control of cristae junctions. For mitochondrial division, Drp1 (Dnm1) is recruited to the organelle surface where it assembles into spiral filaments that are thought to generate mechanical force, constricting and pinching off the mitochondria.
Regulation of fusion
Although outer and inner membrane fusion events are coordinated, these processes require separate machineries. In the outer membrane, levels of mitofusins (Fzo1), which are regulated by ubiquitin proteasome pathway, influence the amount of organelle fusion. In mammals, mitofusins are ubiquitinated by two E3-ligases, MITOL/March5 and Parkin. MITOL/March5 is located in the outer membrane and ubiquitinates mitofusin 1. Parkin is translocated to dysfunctional, depolarized mitochondria by the Pink1 kinase. Ubiquitination and proteasomal degradation of mitofusins inhibit re-fusion of damaged mitochondria and promote autophagic degradation of mitochondria. The ubiquitin proteasome pathway plays a similar role in yeast with the ubiquitin ligase, SCFMdm30, regulating the levels of Fzo1.
Mitochondrial fusion can be modulated independently of the amount of fusion proteins. For instance, the proapoptotic Bcl-2 family members, Bax and Bcl-xL stimulate mitofusin 2 activity, while a mitofusin binding protein (MIB) inhibits mitofusin 1 and 2.
In the inner membrane, Opa1 (Mgm1) exists in two forms: l-Opa1 (l-Mgm1) is integral to the inner membrane while s-Opa1 (s-Mgm1) is soluble as a result of proteolytic cleavage of the integrated membrane form. Both are required for mitochondrial fusion. In mammals, several inner membrane-localized proteases cleave Opa1, including PARL, i-AAA, m-AAA, and Oma1. In yeast, a homolog of PARL, Pcp1, cleaves Mgm1. Changes in matrix ATP levels and the membrane potential across the inner membrane affect Opa1 (Mgm1) processing.
Because mitochondrial have two membranes, efficient fusion of the organelle requires coordination of outer and inner membrane fusion. Ugo1, an outer membrane protein binds Fzo1 and Mgm1, linking these two fusion events in yeast. The mammalian homolog of Ugo1 remains unidentified.
Regulation of division machinery
Multiple integral outer membrane proteins recruit Drp1 (Dnm1) to mitochondria. In mammals, Mff and MiD49/51, interact directly with Drp1, anchoring it to the mitochondrial surface. Fis1 was the first outer membrane protein identified as a tether for Drp1, but its function has been challenged recently. One other integral outer membrane protein, GDAP1, has also been implicated in Drp1-dependent mitochondrial division in mammals; however, the exact function of this protein awaits elucidation.
Several types of posttranslational modifications regulate Drp1 in mammals. Similar to mitofusins, the activity of Drp1 appears to be regulated by MITOL/March5-dependent ubiquitination. In addition, SUMOylation, phosphorylation, and N-nitrosylation of Drp1 also control its functions.
Although Drp1 appears to bind directly to outer membrane receptors to promote division in mammalian cells, Dnm1 is tethered to the outer membrane by two functionally redundant protein complexes in yeast. Fis1 recruits Dnm1 via two WD40 domain-containing adaptor proteins, Mdv1 and Caf4. Parallel to this mechanism, Mdm36 and the cortical protein Num1 retain Dnm1 at the mitochondrial surface. The Num1-Mdm36 mechanism connects mitochondria to the cell cortex and promotes appropriate segregation and inheritance of mitochondria during cytokinesis.
In algae, Dnm1 may also associate with Fis1 and the WD40 domain-containing protein Mda1 on the outer membrane. Additionally, two proteins related to bacterial division components, FtsZ and ZED, are located on the matrix side of the inner membrane and form a ring structure, which potentially mediates inner membrane division. The machinery necessary for inner membrane division has yet to be identified in mammals and yeast.
Acknowledgments
We thank M. Iijima for helpful discussions and Y. Kageyama for providing the image of mitochondria in mouse embryonic fibroblasts. Y.T. is supported by a Japan Society for the Promotion of Science fellowship. H.S. is supported by an National Institutes of Health grant (GM89853).
Abbreviations
- Mfn
mitofusin
- G
GTPase
- R
ring-domain
- Ub
ubiquitin
- P
phosphate
- SUMO
small ubiquitin-like modifier
- OM
outer membrane
- IMS
intermembrane space
- IM
inner membrane
- SCF
Skp1-cullin-F-box
- MEF
mouse embryonic fibroblast. When the names for mammalian and their yeast orthologues differ, the yeast name is in parentheses
References
- Campello S, Scorrano L. Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 2010;11:678–84. doi: 10.1038/embor.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–716. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Sesaki H. Mitochondrial division: molecular machinery and physiological functions. Curr Opin Cell Biol. 2011 doi: 10.1016/j.ceb.2011.04.009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. Mechanisms of organelle division and inheritance and their implications regarding the origin of eukaryotic cells. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:455–471. doi: 10.2183/pjab.86.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H, Soubannier V. Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Curr Biol. 2010;20:R274–276. doi: 10.1016/j.cub.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011 doi: 10.1038/embor.2011.54. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol. 2010;21:542–549. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]