Abstract
In honeybee societies, distinct caste phenotypes are created from the same genotype, suggesting a role for epigenetics in deriving these behaviorally different phenotypes. We found no differences in DNA methylation between irreversible worker/queen castes, but substantial differences between nurses and forager subcastes. Reverting foragers back to nurses reestablished methylation levels for a majority of genes and provided the first evidence in any organism of reversible epigenetic changes associated with behavior.
Epigenetic changes are thought to underlie lineage-specific differentiation, because the pattern of gene expression is stably changed but the DNA sequence remains the same. Recently, the epigenome of a specific differentiation pathway was mapped, defining hundreds of differentially methylated regions (DMRs) that define lineage commitment in mouse hematopoietic progenitors. DNA methylation appears critical in that system for lineage specificity, as lymphoid cells show greater global DNA methylation than myeloid cells1. However, the roles of the epigenome in global changes in organismal remodeling or in behavior have not previously been defined2.
The honeybee Apis mellifera is an ideal model organism for such studies3, as it organizes social structures from distinct individual forms that can emerge from one genome. A female embryo may become a queen by receiving a diet of royal jelly and commit her life to egg-laying (germline), or become a sterile helper ‘worker’ (soma)4. Workers follow a rich behavioral program of nursing and later undergo a transition to foraging that involves extensive gene expression changes in the brain5. In contrast to queens, worker behavior is remarkably flexible: age-matched workers can nurse or forage, and foragers may revert to nursing tasks6.
In order to investigate the potential role of DNA methylation in defining honeybee caste phenotypes, we compared the methylomes of sister queens versus workers and sister nurses versus foragers by whole genome bisulfite sequencing (WGBS) and Comprehensive High-throughput Array-based Relative Methylation (CHARM) analysis1. CHARM covers 85% and WGBS covers 92% of the CpGs in the 270 Mb genome, both revealing sparse methylation throughout the bee genome (Supplementary Fig. 1).
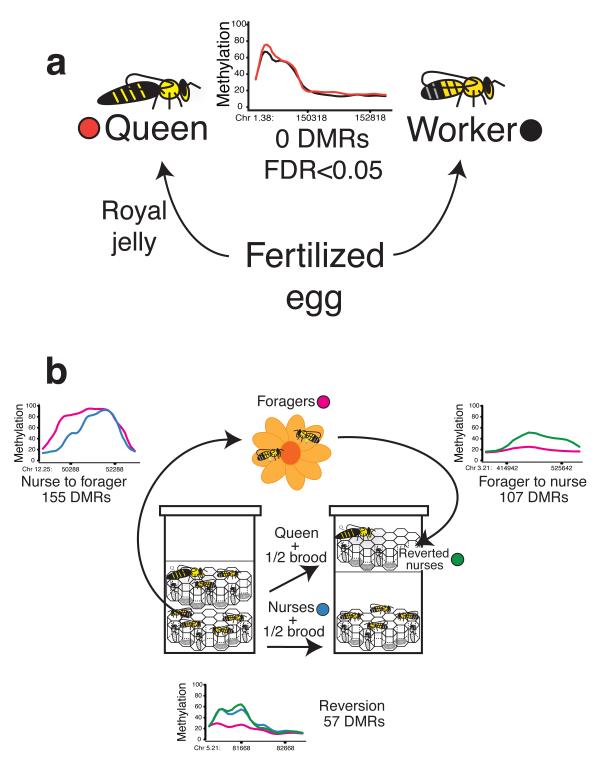
We first compared five biological replicates of queens and workers, both collected within 4 hours of adult emergence from the pupal stage (Fig. 1a). Brain was selected because of its influence on behavior and, unlike ovary, is similar in size between queens and workers. CHARM analysis found no significant DMRs by FDR test between queens and workers. WGBS of the same samples found no differences, using single CpG t-tests corrected for multiple testing. Additionally, we tested the top-ranked differences by CHARM, albeit statistically insignificant, by bisulfite pyrosequencing, an independent measure of DNA methylation at the single base level, and found no caste-specific differences (Supplementary Fig. 2a-c).
Figure 1. DNA methylation changes found between nurses and foragers, but not between queens and workers.
a, We compared newly emerged queens and workers using CHARM (n=5 per phenotype), and found no statistically significant differences. b, DNA methylation changes during nurse to forager transition and change back during forager to nurse transition (n=3 per phenotype). We found 155 DMRs associated with the nurse to forager transition, 107 DMRs associated with the forager to nurse transition, and 57 DMRs common to both lists that exhibit a nurse specific signature.
Given these negative results, we then compared subcastes of workers. Initially, most workers are nurses that care for queen and larvae inside the hive. About 2-3 weeks later, the majority switches to foraging and collect pollen, nectar and water outside5. Using CHARM, we identified 155 DMRs that distinguished nurses from foragers (Fig. 1b, Fig. 2a-b, Table 1, Supplementary Table 1, bisulfite pyrosequencing validation Supplementary Fig. 3a-e). Approximately 70% of DMRs overlap exons (Summary in Table 1, and full description of genomic location of DMRs indicated in Supplementary Table 1) similar to previous studies7,8. The genes associated with the 155 nurse-to-forager DMRs appeared to be enriched for gene regulation and development through transcriptional control and chromatin remodeling. Many histone modification writers, including LOC412350, a histone deacetylase similar to Hdac3, JIL-1 a histone phosphotransferase, and LOC411070, a histone H3 methyltransferase9 increase in methylation during the nurse to forager transition. In addition, DEAD-box helicase genes Iswi and spn-E have chromatin remodeling capacity and are involved in morphogenesis10.
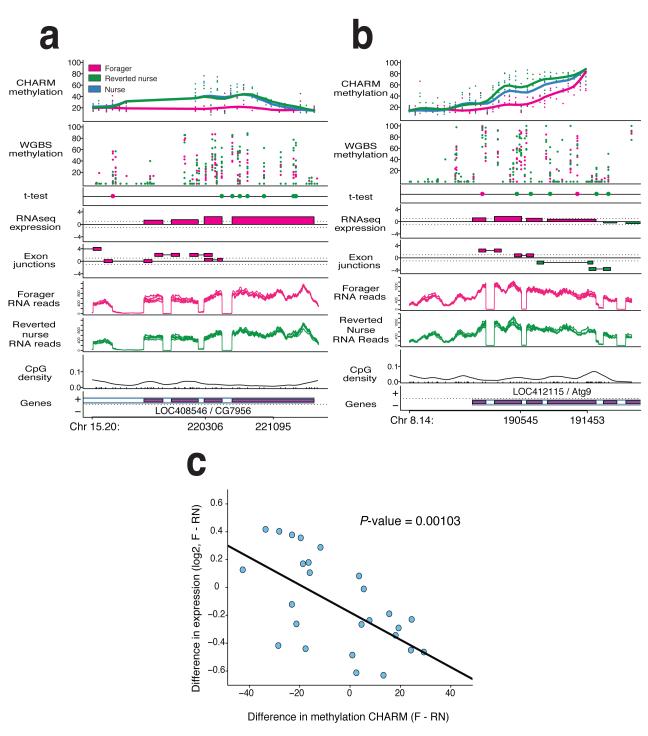
Figure 2. DNA methylation distinguishes nurses, foragers and reverted nurses.
Two examples of CHARM DMRs. a-b, Top panels show percent methylation for both CHARM and WGBS data sets, with points representing individual samples, and the smoothed lines representing the average for the phenotype. The t-test panel displays the top 1% differentially methylated CpGs by t-test. Color of the point indicates which phenotype has greater methylation at that CpG (n=6 per phenotype). The RNAseq expression panel is a t-statistic based on the number or reads detected within the annotated exons, with the color indicating the higher expressed phenotype. The Exon junctions panel is a t-statistic based on the number or reads detected spanning the exon junctions, as predicted by the TopHat program, with the color indicating the higher expressed phenotype. Switching between higher expressed nurse and forager exon junctions is indicative of alternative splicing events. The RNA reads panels indicate the number of reads per phenotype as compiled by TopHat program (n=6 per phenotype). The bottom two panels show the CpG density, and the relative position of the gene. c. Plot of relative gene expression comparing foragers to reverted nurses. 26 genes associated with DMRs were tested for gene expression differences by real-time PCR (n=12 per phenotype). The plot depicts the difference in average log2 expression versus average difference in methylation as determined by CHARM. Correlation analysis results in a P-value of 0.001.
Table 1.
Summary of Differentially Methylated Regions (DMRs)
| DMR Type | No. of DMRs |
% DMRs overlapping Exons |
GO Category examples |
Example Genes |
|---|---|---|---|---|
| Nurse to Forager Transition |
155 | 72.2% | Helicase activity, Chromatin remodeling, Neuron development |
alpha-Cat, Stat92E, Dhc64C, alpha-Spec, hts, spn-E |
| Forager to Nurse Transition |
107 | 72.9% | Nucleoside binding, Helicase activity, Cytoskeleton organization |
Hel89B, Hsc70Cb, CG7177, Upf1, CG2017, eIF-4a, kis, Dhc64C |
| Overlap | 57 | 73.7% | Nuclear import, Cell differentiation, ATP binding |
Ranbp21, Fs(2)Ket, Mtor eIF-4a, kis, poe, bur, BicD |
Iswi in particular plays a role in dendrite morphogenesis11 and may contribute to noted changes to the nurse brain prior to foraging5. In order to determine whether the DMRs that we observed during the nurse to forager transition are linked to phenotype, and not simply the result of the transition, we induced the reversion of foragers back to nurses using a strategy of hive trickery. To initiate reversion, foragers are set up to return to a hive where only queen and larvae are present (Fig. 1b). The foragers will then segregate into reverted nurses that pick up caregiver tasks, and continuing foragers that do not change behavior6. Reversion separates changes caused by nervous system development, maturation and foraging experience that are shared between reverted nurses and foragers but not nurses, from changes robustly linked to current behavior that are shared between reverted nurses and nurses but not foragers.
With CHARM, we found 107 DMRs for the forager to reverted nurse transition. The genes associated with these CHARM DMRs appeared to be enriched in transcription factors and also DEAD-box helicase genes, as seen in the nurse to forager CHARM DMRs (Fig. 2a-b, Supplementary Table 2, Supplementary Fig. 4). Of these 107 CHARM DMRs, 57 overlapped with CHARM DMRs associated with nurse to forager transition, a remarkably close concordance (P-value < 2.2 × 10−16 by Fisher’s test, P-value < 10−3 based on 1000 permutations, Supplementary Fig. 5). This subset of epigenetically reversible genes showed enrichment for development, ATP binding and nuclear pore formation (Supplementary Table 2). These genes include the ortholog to kismet, LOC726524, which regulates developmental genes such as hedgehog and affect learning and axon migration in Drosophila12,13, and might explain observed differences in learning14 between nurses and foragers. In addition, DEAD-box helicase genes LOC72530615 and LOC726524 both have roles in transcription, whereas LOC411989 is involved in translation16.
We wanted to independently validate this result, therefore we replicated the reversion experiment, and created six new pools of six brains for both foragers and reverted nurses. We performed WGBS on these 12 samples, and we found that 45/57 Reversion DMRs show the same direction of change in methylation between CHARM and WGBS. (Supplementary Fig. 6). This overlap of DMRs between replicated experiments is highly significant (P-value = 3.3 × 10−6). Further, the 45 WGBS-correlated genes show enrichment for ATP binding and nuclear pore formation (Supplementary Table 3), consistent with our analysis of the 57 CHARM reversion DMRs. These results provide evidence for a nurse-specific methylome that needs to be reestablished during the reversion.
To determine the significance of these reversible DMRs, we performed transcriptome sequencing (RNAseq) on 6 pools each of foragers and reverted nurses. We then used the TopHat program to analyze the RNA-seq data to predict the location of annotated and unannotated exons, and determine the prevalence of exon skipping. This analysis identified 22 of the 45 WGBS-correlated reversion DMRs co-localize with alternative splicing events. An example is shown in Figure 2b in the fifth panel and other examples are shown in Supplementary Figure 7a-f. These data show a high incidence of alternative splicing events within DMRs and strengthens the potential role of DNA methylation in regulating alternative splicing8. We also found a negative correlation between gene expression and levels of DNA methylation between foragers and reverted nurses for 26 genes by real-time PCR (P-value = 0.00103, Fig. 2c, Supplementary Fig. 8a-f).
Our data show a strong link between reversible DNA methylation and nurse forager transition and reversion, but no relationship to queen-worker segregation. These data stand in contrast with a study comparing 2.5 week-old mated queens and 8 day-old foraging-capable workers8, which is likely explained by the difference in timing in data, i.e. newly emerged queens and workers. While DNA methylation may play a role in distinguishing queens from workers during development3, our data clearly show that the queen and worker brain methylomes are the same at the time of emergence, despite differences in body morphology.
In summary, we found substantial DNA methylation changes that accompany phenotype switching in honeybee subcastes. Genes associated with these DMRs can potentially influence global gene expression patterns by altering chromatin structure or regulating transcriptional machinery. Profound phenotype shifts between nurses and foragers may be orchestrated by a subset of genes, they themselves regulated by DNA methylation. Key regulatory genes may either be differentially expressed, or differentially spliced, which we correlate with changes in DNA methylation. For example, the eIF-4a homolog LOC411989, which plays a critical role in translation initiation17, exhibits alternative splicing in an exon that codes for RNA binding (Supplementary Fig. 7c). Different isoforms of eIF-4a may bind to RNA with greater affinity, thereby globally affecting the rate or regulation of translation. Since this differentially spliced exon is within a DMR, methylation might be used to remember which isoform to express in nurses or foragers. Remarkably, we found that DNA methylation is able to revert, concomitant with experimental reversion of foragers back to nurses, which we demonstrated in replicated experiments. This suggests a subcaste-specific methylation signature that assists in forming subcaste phenotypes. While studies in rodents found methylation changes associated with learning, these changes disappear over several hours and do not establish a stable phenotype 18,19. Similarly, nurturing can induce long lasting methylation marks in rodents20. Our study is the first to show reversible DNA methylation corresponding to a reversible behavioral phenotype in any species.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Bee preparation
For each replicate, two colonies were initially prepared for nurse and forager rearing. They each consisted of 6000-7000 newly emerged (0-24 h old) workers born to five sister queens of our standard research stock and a wild-type queen of Californian commercial origin. All individuals were paint-marked (Testors™) on the thorax, then after a solid foraging pattern was established, foragers were paint-marked on the abdomen for the purpose of tracking their life history. The behavioral reversion was carried out essentially as described before21. This reversion resulted in two forager-derived colonies and two nurse-derived colonies. Young brood of Californian commercial origin was provided to the colonies as incentive for reversal from foraging to nursing behavior. For the replicate used in the CHARM and bisufite pyrosequencing assays, bees of three groups were collected 12-14 days after the reversion: 1. Continuous foragers: forager bees that had not reverted to nursing behavior; 2. Reverted nurse bees: forager bees that exhibited nurse-like behavior (head in brood cells, sluggish response to a flight challenge) in the brood area; 3. Continuous nurses: bees that had never been observed foraging and exhibited at least one nurse-like behavioral trait at the time of collection. For the replicate used in the WGBS and RNAseq assays, continuous foragers and reverted nurse bees were collected for the validation of the CHARM results.
Queens and workers were derived from eggs produced by a single-drone inseminated honeybee queen that belonged to a standard research stock with restricted genetic background22 and were left to develop into two-day old larvae within the hive. Subsequently, larvae at this crucial point in the divergent development of queen and worker traits, were either allowed to develop into worker pupae or, for queen pupae development, they were manually grafted into queen cells and raised as previously described23. On the day before emergence (the day of sample collection), queen and worker pupae were transferred into an incubator at 33 °C, 65-70 % relative humidity.
For all bees, individuals from both experiments were collected directly into 2ml of 80 % ice-cold ethanol and stored at 4 °C until the central brains were dissected (< 48 h). Following dissections, central brains were immediately transferred into liquid N2 and stored at −80 °C until further use.
CHARM DNA methylation analysis
gDNA was isolated from brains using the Masterpure kit from Epicentre. Brains from the nurse/forager study were pooled in groups of seven for each biological replicate, three replicates per phenotype. Brains from the queen/worker study were pooled in groups of eight for each biological replicate, five replicates per phenotype. Genome-wide methylation was assessed by CHARM24,25 and performed as previously described1. A custom designed Nimblegen 2.1 million-feature array designed for the honeybee genome which covers approximately 200 Mb of non-repeat genomic sequence and includes ~8.7 million CpG sites covering ~85% of the ~10.2 million CpG sites in the genome.
DMRs for the nurse/forager study were determined by a cutoff of at least 10% methylation difference24 and DMRs with an average methylation close to the baseline of 20% were eliminated. DMRs from pairwise comparisons were combined to determine the relative methylation of all three phenotypes. Methylation differences between Continuous Nurses and both Reverted Nurses and Continuous Foragers were determined, and these differences were plotted. Clustering analysis identified three classes of DMRs, described in Supplementary Figure 9. GO analysis26 of each class was performed by first determining the closest Apis mellifera gene to each DMR. The orthologous gene in Drosphila melanogaster was found for each Apis mellifera gene, and this new gene list was used for GO analysis using the web tools available on the DAVID bioinformatics database at david.abcc.ncifcrf.gov. DMRs for queen/worker study were determined by calculating a FDR score for each potential DMR.
Bisulfite pyrosequencing
Approximately 400ng of pooled gDNA that was also used for the CHARM analysis was bisulfite converted using the Zymo DNA-Methylation Gold kit. We used nested PCR to amplify regions of interest within DMRs and quantified the level of methylation using the Biotage PSQ HS96 pyrosequencer. The percent methylation for every CpG in our target region was calculated using the Q-CpG methylation software (Biotage). Control DNA was prepared using the Repli-G kit(Qiagen) from gDNA. Repli-G amplified DNA served as a 0% methylated control. 100% methylated control was created by treating the Repli-G amplified DNA with SSSi methyltransferase(NEB), which methylates every CpG site. 25%, 50% and 75% methylated controls were created by mixing 0% and 100% controls. All controls were bisulfite treated with same Zymo kit as test samples. Primer Sequences listed in Supplementary Table 4.
Real-time quantitative RT-PCR
RNA was extracted from single brains by first lysing cells in Chaos buffer (4.5M Guanidinium thiocyanate, 2% N-lauroylsarcosine, 50mM EDTA, 25mM Tris-HCl, 0.1M b-Mercaptoethanol) followed by phenol, chloroform, then purified with Qiagen RNeasy columns. cDNA was synthesized using Quantitect Reverse Transcription Kit(Qiagen) and 1ng of cDNA was used for each real-time PCR reaction. Fast Sybr green(Applied Biosystems) was used for real-time PCR reaction and quantified by 7900HT (Applied Biosystems ). Primer sequences are listed in Supplementary Table 5.
Whole genome bisulfite sequencing
Libraries of queen/worker and reverted nurse/forager samples were created using TruSeq DNA library preparation kits (Illumina) with some modification to the standard protocol. Genomic DNA samples were prepared by homogenizing pools of whole brains. For queen/worker samples, 5 pools of 8 brains per pool were prepared. For reverted nurse/forager samples, 6 pools of 6 brains per pool were prepared. Genomic DNA was extracted from homogenized brains using either Masterpure kit from Epicentre (queen/worker), or lysing cells in Chaos buffer (4.5M Guanidinium thiocyanate, 2% N-lauroylsarcosine, 50mM EDTA, 25mM Tris-HCl, 0.1M b-Mercaptoethanol) and purified with Qiagen DNeasy columns (reverted nurse/forager). For all samples, Genomic DNA was sheared to an average size of 350 bp using a Covaris sonicator with the following settings: Duty cycle = 10%, Intensity = 5.0, Bursts per second = 200, Duration = 60 sec. Blunt ends were created on the DNA fragments using a unique protocol to eliminate the introduction of non-genomic cytosines into the fragments, which would be falsely interpreted as unmethylated cytosines during subsequent analysis. To achieve this, we only used A, G and T nucleotides with a mixture of the enzymes T4 DNA polymerase, Klenow DNA polymerase and T4 PNK(NEB) to perform end repair of fragments. Illumina adapters were then ligated to the fragments after the addition of a single A, per TruSeq protocol. Libraries were then size selected by cutting a 400-500 bp fragment from an agraose gel (Bio-Rad-Certified Low Range Ultra Agarose, NEB-100 bp DNA Ladder, Invitrogen-SYBR® Gold nucleic acid gel stain) and purified using Qiagen MinElute Gel Extraction Kit. The purified libraries were then bisulfite converted and purified using Zymo EZ DNA Methylation Gold. Libraries were then amplified using a mixture of Uracil insensitive polymerases; Denville Choice Taq and Agilent Turbo Pfu. Queen/worker samples were amplified for 12 cycles and reverted nurse/forager for 15 cycles using the TruSeq PCR conditions.
RNA sequencing
RNA samples for the 6 pools of 6 brains of reverted nurse and foragers were derived from the same lysate that was used to create the reverted nurse and forager WGBS libraries. RNAseq libraries were created using the Illumina TruSeq RNA sample prep kit with no modifications to the standard protocol. This kit enriches for mRNA by using beads bound with a poly-T oligo to bind to the poly-A tails of mRNA.
Data analysis of queen and worker whole-genome bisulfite sequencing
We ran the Bsmooth27 bisulfite alignment pipeline (version 0.4.5-beta) on the 100-by-100 nt paired-end HiSeq 2000 sequencing reads obtained for each Queen and Worker pool. We used Bsmooth’s Bowtie 2-based alignment pipeline, which employs a version of the unbiased and efficient in-silico bisulfite conversion approach introduced by Lister et al28. We used Bowtie 229 version 2.0.0-beta5. We aligned to a reference index consisting of the Baylor Human Genome Sequencing Center A. mellifera assembly version 4.0, the A. mellifera mitochondrial sequence, and the lambda phage genome. Supplementary Table 6 summarizes alignment results.
We then used Bsmooth to extract read-level measurements. One read-level measurement corresponds to one instance where an aligned read overlapped a CpG in the reference genome. The measurement records the genomic position of the CpG, the allele observed in the read, its base quality, the alignment’s mapping quality, and other related measures.
Using Bsmooth, we filtered read-level measurements in three ways. First, we removed read-level measurements from alignments with mapping quality less than 10. Second, we remove read-level measurements where the allele in the alignment was neither C nor T. Third, we removed read-level measurements from sequencing cycles we deemed unreliable after visually inspecting the “M-bias” plot. That is, we plotted the fraction of methylated read-level measurements versus sequencing cycle. Ideally, this plot should be flat and horizontal, indicating no strong relationship between sequencing cycle and fraction of methylated read-level measurements. In practice, we found peaks and troughs at the extremes of both mates. We filtered out measurements from the affected cycles. In the case of this dataset, we filtered out read-level measurements from the 5′-most 8 nucleotides of mate 1, the 3′-most 4 nucleotides of mate 1, and the 5′-most 8 nucleotides of mate 2.
After filtering, we used Bsmooth to sort read-level measurements by genome coordinate and compile them into a table summarizing methylation measurements at each CpG in the reference genome. We used the summarized methylation measurements over the lambda genome, which we assume is entirely unmethylated, to estimate the bisulfite conversion rate. Supplementary Table 7 summarizes the read-level measurements obtained, how they were filtered, and the estimated bisulfite conversion rates. After filtering we only included evidence with a quality score greater than or equal to 20 for a particular CpG.
We then used Bsmooth to smooth the data and determine the correlation in methylation between the WGBS data and CHARM data. Since ~85% of CpGs that have greater than 25% methylation are located within genes, we compared the methylation levels between WGBS and CHARM by segmenting genes into 1000bp windows and found the average smoothed methylation value within each window. Each window was required to contain at least 4 CHARM probes and 8 CpGs. This analysis was performed for each queen and worker sample, and correlation values range from 0.691 to 0.807, mean = 0.755 (Supplementary Figure 1a-b). Methylation profiles were compared to determine the reproducibility of detecting regions of methylation (Supplementary Figure 1c-f).
In order to assess whether there was any difference between queens and workers we did the following analysis. First, we only analyzed CpGs with a coverage across the 10 samples of greater than 10. For each of these CpGs we performed a t-test for difference in methylation mean between the two groups. The t-test allows us to measure biological variability. The p-values were corrected for multiple testing using the Benjamini-Horchberg procedure and no CpGs were significant at a 0.05 false discovery rate.
Data analysis of forager and reverted nurse whole-genome bisulfite sequencing
The same methods were used to analyze the Forager and reverted Nurse sequencing data as were used to analyze the Queen and Worker sequencing data (see above). Supplementary Table 8 summarizes alignment results, while Supplementary Table 9 summarizes the read-level measurements obtained, how they were filtered, and the estimated bisulfite conversion rates.
To determine if the differences in methylation between foragers and reverted nurses that were discovered using CHARM also exist in WGBS dataset, t-tests were performed on individual CpGs within 500bp of CHARM reversion DMRs. The average difference of the top three CpGs ranked by significance within the DMR was calculated and compared to the average CHARM methylation within the DMR. These results are presented as a scatter plot in Supplementary Figure 6.
In order to asses the significance of the overlap in direction of change between CHARM and WGBS we used the following test: assuming that there is no correlation between CHARM and WGBS there should be a 50% chance that the direction of change is the same. Hence, we calculate the probability of observing a more extreme statistic by P(X >= 45) + P(X <= 11) with X being binomially distributed with 57 trials and 0.5 chance of success.
Data analysis of forager and reverted nurse RNA sequencing
We used TopHat30 v1.3.3 to align the 100-by-100 nt paired-end HiSeq 2000 sequencing reads obtained for each Nurse and reverted Forager pool. We aligned to a reference index consisting of the Baylor Human Genome Sequencing Center A. mellifera assembly version 4.0 and the A. mellifera mitochondrial sequence. Supplementary Table 10 summarizes alignment results.
TopHat alignments were overlapped with annotated exons obtained from NCBI to form a table of overlap counts per gene and per sample. An alignment was said to overlap an exon if there was any reference position covered both by the exon and by the alignment. A spliced alignment that spanned an exon without either mate overlapping the exon did not count as overlapping the exon.
Junctions and junction counts emitted by TopHat were combined to form a table of counts per junction and per sample. Two junctions that were not identical but where their boundaries differed by no more than 5 nt on one or both sides were considered identical. This was necessary because there is some variability in where exactly TopHat will place junction boundaries. Junction counts were used to determine differentially expressed exon junctions between foragers and reverted nurses. We define alternative splice events as the presence of at least two distinct exon junctions that have opposite expression within the same gene or DMR. To determine the frequency of alternative splicing events in DMRs, we expanded the DMR 500bp on each side and checked for the presence of exon junctions that were more expressed in foragers co-localizing with exon junctions that were more expressed in reverted nurses. Examples are presented in Figure 2b and Supplementary Figure 7.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Erin Fennern, Nickolas Baker, Kevin Flores and Osman Kaftanoglu for assistance with colonies, bees and brain dissections, Akiko Doi for reviewing the manuscript. We thank Georg Klein for serving as scientific schadchen to the two PI’s after hearing them lecture on different occasions at I. Ernberg’s “What is Life” series at the Karolinska Institute, without which this research would not have taken place. G.V.A. was funded by the Research Council of Norway #191699 and the PEW Charitable Trust #2009-000068-001. A.P.F. was funded by the NIH grant 1DP1OD008324.
Footnotes
Accession codes. Whole genome bisulfite and RNA sequencing data: SRA050798 CHARM data: GSE36650
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS B.R.H. performed genome-scale, gene-specific DNA methylation analysis and performed gene expression analysis; F.W. raised bees and manipulated hives for reversion experiment, collected bees and dissected brains; R.I., M.J.A., K.D.H., B.L. and B.R.H. performed statistical analysis; B.R.H. and M.J.A. generated microarray datasets; B.R.H., B.L. and K.D.H generated WGBS and RNAseq datasets. A.P.F. and G.V.A. conceived, designed, and oversaw the experiments; and A.P.F., B.R.H., and G.V.A. wrote the paper with the assistance of K.D.H., M.J.A., B.L., and F.W.
AUTHOR INFORMATION Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Ji H, et al. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf C, Linden DE. Genes Brain Behav. 2012;11:3–28. doi: 10.1111/j.1601-183X.2011.00752.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 4.Winston ML. The Biology of the Honey Bee. Harvard University Press; 1987. [Google Scholar]
- 5.Whitfield CW. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 6.Robinson GE, Page RE, Strambi C, Strambi A. Ethology. 1992;90:336–348. [Google Scholar]
- 7.Elango N, Hunt BG, Goodisman MA, Yi SV. Proc Natl Acad Sci U S A. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyko F, et al. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suganuma T, et al. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 10.Tsukiyama T, Daniel C, Tamkun J, Wu C. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 11.Barak O, et al. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terriente-Félix A, Molnar C, Gómez-Skarmeta JL, de Celis JF. Developmental Biology. 2011;350:382–392. doi: 10.1016/j.ydbio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Melicharek DJ, Ramirez LC, Singh S, Rhea T, Marenda DR. Human Molecular Genetics. 2010;19:4253–4264. doi: 10.1093/hmg/ddq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amdam GV. Aging Cell. 2011;10:18–27. doi: 10.1111/j.1474-9726.2010.00647.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldman-Levi R, Miller C, Bogoch J, Zak NB. Nucleic Acids Research. 1996;24:3121–3128. doi: 10.1093/nar/24.16.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linder P. Nucleic Acids Research. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsyan A, et al. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 18.Guo JU, et al. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller CA, Sweatt JD. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Weaver IC, et al. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 21.Amdam GV, et al. Exp Gerontol. 2005;40:939–947. doi: 10.1016/j.exger.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page RE, Fondrk MK. Behavioral Ecology and Sociobiology. 1995;36:135–144. doi: 10.1007/s00265-012-1400-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laidlaw HH, Page RE. Queen rearing and bee breeding. Wicwas Press; 1997. [Google Scholar]
- 24.Aryee MJ, et al. Biostatistics. 2011;12:197–210. doi: 10.1093/biostatistics/kxq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, et al. Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Nature Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Hansen KD, et al. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lister R, et al. Nature. 2009;462:315–322. doi: 10.1038/nature08514. doi:10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Nat Methods. 2012 doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.