Abstract
Interneurons comprise approximately one third of the total cortical neurons in the mammalian cerebral cortex. Studies have revealed many details in the generation of this cell type. However, the mechanism that defines interneuron-lineage specific gene expression is not well understood. Gene regulatory elements, e.g., promoters, enhancers, and trans-acting factors, are essential for the proper control of gene expression. Here, we report that a novel evolutionarily conserved cis-element in the second intron of the Notch1 locus play an important role in regulating gene expression in interneuron progenitors. The spatiotemporal activity of the cis-element in the developing central nervous system (CNS) was determined by both transient reporter expression in the developing chick and a transgenic mouse model. Its activity is well correlated with neurogenesis in both the chick and mouse and restricted to neural progenitor cells in the ganglionic eminence that are fated to differentiate into GABAergic interneurons of the neocortex. We further demonstrate that the cis-element activity requires the binding motif for trans-acting factors Gsh1/Barx2/Brn3. Deletion of this binding motif abolishes reporter gene expression. Together, these data provide new insights into the regulatory mechanisms of interneuron development in the vertebrate CNS.
INTRODUCTION
The differentiation of neural stem cells and the acquisition of their cell-fate are fundamental issues facing the field of developmental neurobiology and stem cell biology. Radial glia are multipotential stem cells that give rise to neurons and then glia (Ever and Gaiano, 2005; Kriegstein and Alvarez-Buylla, 2009). Neurogenesis occurs from embryonic day 9.5 (E9.5) to birth in the mouse (Noctor et al., 2001) and from E3 to E9 in the chick (Cheung et al., 2007; Ferretti and Whalley, 2008; Ono et al., 1995). The elongated processes of radial glia enable their own migration (Angevine and Sidman, 1961; Brittis et al., 1995; Norris and Kalil, 1991) and guide the migration of nascent neurons from the ventricular zone (VZ) to their site of differentiation and maturation (Rakic, 1971a, b, 1972). Notch1 is a focal gene in the genetic network related to the specification of neural stem cells to neural progenitors and then mature cells (Gaiano et al., 2000; Louvi and Artavanis-Tsakon as, 2006; Lowell et al., 2006; Pierfelice et al., 2011), and it plays an integral role in maintaining the radial glial state as well as regulating neurogenesis and gliogenesis (Bao and Cepko, 1997; Gaiano et al., 2000; Noctor et al., 2001). In addition to Notch1, brain lipid binding protein (BLBP) and transcription factors Sox2 are expressed in neural stem cells (Feng et al., 1994; Zappone et al., 2000). Pax6 is a transcription factor present in ventral progenitor cell populations (Ericson et al., 1997). In the developing neuroepithelium during neurogenesis, neural stem cells undergo asymmetric division and give rise to a progenitor cell and a differentiated daughter cell (Nowakowski and Hayes, 1999; Zhong and Chia, 2008), and proteins such as nuclear mitotic apparatus protein (NuMa) are involved during the asymmetric division process (Bowman et al., 2006; Van Ness and Pettijohn, 1983).
It is now established that different types of neurons in the brain originate from separate progenitor pools in distinct regions (Marin and Rubenstein, 2001). Excitatory glutamatergic neurons are born in the proliferative VZ of the dorsal pallium and migrate radially to form the cortical plate (Sidman and Rakic, 1973), while interneurons are born from the VZ of the ganglionic eminence (GE) and tangentially migrate into the neocortex (Batista-Brito and Fishell, 2009; Miyoshi et al., 2010; Parnavelas et al., 2000). Interneurons contain the neurotransmitter γ-aminobutyric acid (GABA), which is synthesized by glutamate decarboxylase (GAD) (Soghomonian and Martin, 1998). GAD65/67 marks GABAergic interneurons and their progenitor population (Lopez-Bendito et al., 2004; Riccio et al., 2012). The medial ganglionic eminence (MGE) is a major source of interneurons originating from the striatum (Anderson et al., 2001; Lavdas et al., 1999; Wichterle et al., 1999; Wichterle et al., 2001), while the lateral ganglionic eminence (LGE) gives rise to interneurons of the olfactory bulb (Wichterle et al., 1999). Several known transcription factor-coding genes are essential for the existence of interneurons including Gsh1/2 (Toresson and Campbell, 2001; Yun et al., 2003) and Dlx1/2 (Anderson et al., 1997). Gsh1/2 are expressed in the GE and have demonstrated their roles in interneuron progenitors (Toresson and Campbell, 2001; Yun et al., 2003).
Neurogenesis, amongst other basic developmental processes, is highly conserved across a wide range of species (Finlay and Darlington, 1995; Gomez-Skarmeta et al., 2006). The conservation is also believed to be present in non-protein coding regions that regulate gene expression during development (Gomez- Skarmeta et al., 2006; Long and Miano, 2007). Cis-regulatory elements (segments of DNA involved in gene regulation through the binding of trans-acting factors) enhance or repress transcription in a spatiotemporal specific manner independent of orientation or position relative to the transcription site (Blackwood and Kadonaga, 1998; Jeziorska et al., 2009; Khoury and Gruss, 1983). However, there are only a few known gene regulatory elements for neural progenitor cells (Visel et al., 2007; Wahlbuhl et al., 2012), and no known cis-regulatory elements within Notch1 or for Notch1 have been identified. In this study, we report for the first time that an evolutionarily conserved, 399-base pair, noncoding cis-element in the second intron of Notch1 gene locus (Notch1CR2 or CR2) is active throughout the duration of neurogenesis, primarily in interneuron progenitors of the developing CNS. We also present evidence that the binding motif for trans-acting factors Gsh1/Brn3/Barx2 is required for the activity this cis-element.
EXPERIMENTAL METHODS
Genomic sequence analysis and reporter plasmid CR2
The genomic sequences containing Notch1 from various vertebrate species were obtained using a non-coding sequence retrieval system (Doh et al., 2007). Comparative genomic analysis using sequence alignment method (LAGAN/VISTA) (Brudno et al., 2003) was performed to reveal evolutionarily conserved regions. A 399-base pair region in the second intron of the Notch1 locus (Notch1CR2, or CR2) was found highly conserved among various species including human, mouse and chicken. Further analysis by MatInspector shows that (Cartharius et al., 2005; Quandt et al., 1995; Werner, 2000), CR2 contains 164 predicted transcription factor binding sites (TFBS).
The plasmid reporter constructs were designed to identify gene regulatory activity of CR2. CR2 was subcloned into an expression vector containing a minimal β-globin promoter (βGP) and a GFP reporter gene. When this 399-base pair region is activated by endogenous transcription factors, detectable levels of GFP are expressed. CR2.1 (ACA GCA TTA ATC GCC TCC C) was cloned into the βGP-GFP backbone. Clones were confirmed with colony PCR and sequencing (forward primer: GCA ACG TGC TGG TTA TTG TGC TGT; reverse primer: GTG GTA TTT GTG AGC CAG GGC ATT).
Negative controls for in ovo transfections, which consist of GFP driven by the β-globin promoter alone or with a random sequence, were also created. The transfection control is reporter DsRed driven by the constitutively active CAG promoter, which drives DsRed expression in all cell types.
In ovo electroporation and detection of enhancer activity
Fertilized eggs were purchased from Sunrise Farms, Inc. and incubated at 38 °C with 60% humidity. The developmental stages of the chicks were determined according to stages established by Hamilton and Hamburger (Hamburger and Hamilton, 1992). In ovo electroporation was performed as described previously (Blank et al., 2007; Doh et al., 2010; Islam et al., 2012) with minor modifications. Briefly, chick embryos at Hamburger-Hamilton stage 11 (about 40–44 hours in incubation, ~ embryonic day 2, E2). DNA was injected into the neural tube of the embryo at the telencephalic/diencephalic boundary. The DNA mix consisted of CR2-GFP (6 μg/μl), CAG-DsRed (6 μg/μl), and 0.025% Fast Green for visualization. Gold-plated, L-shaped electrodes (Model 512, Genetrodes) were placed along the right and left sides of the embryo. An electric pulse at 15 V for 50 ms was applied 4 times at 950 ms intervals using an ECM 830 Electro Square Porator (BTX Harvard Apparatus). After the electroporation, the aperture was sealed with tape, and the eggs were returned to the incubator. Embryos were harvested at the desired time points (E4, E6, and E8) and examined under a fluorescent dissecting microscope (Leica, MZ16FA). Transfection patterns of enhancer activity were highly reproducible (20 embryos at E4, 15 embryos at E6, and 10 at E8). The prosencephalon was inaccessible during transfection, and therefore, the optic lobes of mesencephalic regions were the main region of focus for characterization.
Generation of transgenic mice
Digested DNA (Notch1CR2-GFP) was gel purified using Seakem GTG agarose gel. 3–5 pg of purified DNA was introduced by microinjection in 0.5 dpc (days post coitum) fertilized F1 (C57Bl/6J x CBA) mouse embryos and transferred to pseudopregnant recipient females. At P10, tissue samples were collected from tail tip and DNA was prepared and digested by EcoRI restriction enzyme, analyzed on 0.8% agarose gel for 18 hours at 35 volts in TAE buffer, and transferred to a HybondN (Amersham) membrane. The 32P labeled probe was hybridized at 65 °C for 20 hours and was washed with 2X SSC/0.1% SDS for 20 minutes, 1X SSC/0.1%SDS for 20 minutes, and then 0.5X SSC/0.1%SDS for 20 minutes. The washed blot was exposed to KODAK X-ray film. The transmission of the transgene in following generations was verified by PCR genotyping (forward primer: GCA ACG TGC TGG TTA TTG TGC TGT; reverse primer GTG GTA TTT GTG AGC CAG GGC ATT).
Tissue processing
For immunohistochemical analysis, three embryos at each time point were processed. Whole embryos were fixed in 4% paraformaldehyde for 4–6 hours and subsequently washed in cold, sterile PBS (three times, 15 minutes each), and soaked in sterile 30% sucrose in PBS overnight until embryos sunk to the bottom. The tissue was mounted in Tissue Tek® OCT Compound, frozen at −80 °C, and sectioned at 12–15 μm thickness using the CryotomeE (Thermo Electron Corporation). Slide-mounted sections immersed in blocking buffer containing 10% donkey serum, 0.1% TritonX, and 0.1% Tween for 30 minutes at room temperature.
Immunohistochemistry
Slide-mounted sections were incubated overnight in primary antibodies: Brn3a (mlgG1, 1:100, Millipore), Doublecortin (RblgG, 1:250, Cell Signaling), NeuN (mlgG1, 1:500, Millipore), Notch1 (RblgG, 1:400, Cell Signaling), Pax6 (mlgG1, DSHB, 1:10), Tbr1 (RblgG, 1:250, Santa Cruz Biotechnologies), Tbr2 (RblgG, 1:300, Abcam), and Sox2 (mlgG2b, 1:200, Millipore). A co-staining with anti-GFP antibody (RblgG, 1:1000, Millipore GtlgG, 1:300 AbCam) was used to help visualize CR2-GFP. Primary antibodies were diluted in blocking buffer and then washed three times (10mn each) with PBS containing 0.1% Tween. For immunofluorescent staining, tissue sections were incubated with appropriate secondary antibodies conjugated to different fluorophores (donkey anti-RblgG Alexa 488 or donkey anti-GtlgG Alexa 388, 1:300, Jackson ImmunoResearch Labs) (donkey anti-mlgG Alexa 647 or donkey anti-RblgG Alexa647, 1:150, Jackson ImmunoResearch Labs). Secondary antibodies were prepared in blocking buffer and applied at room temperature for one hour, followed by three 10mn washes with PBS and a 5mn rinse in distilled water to remove salt crystals. After air-drying for 5mn, slides were mounted with 40 μl of mounting media with Dapi (Vector Laboratories).
The immunostained samples were visualized and imaged on a Zeiss Axio Imager M1 (AxioVision 4.8 software), Zeiss 200-M fluorescence microscope (Zeiss 510 LSM software), which has four available filters for capturing green, red, blue, and far red fluorescence, and a FluoView FV10i confocal microscope (Olympus, Inc.). For transfected chick embryos samples, cell counts were performed on cells marked by Dapi-stained nucleus. For each embryo in a sample size of 3, 100 CR2-GFP+ cells and 400 CAG-DsRed+ cells were counted from each individual embryo. Percent colocalizations of CR2 active cells were calculated by number of (GFP+ plus far red cells)/total GFP+ cells. Percent colocalizations in transfected cells were calculated by the number of (DsRed+ plus far red cells)/total DsRed+ cells. In transgenic mouse tissue, at least 200 CR2-GFP+ cells were counted for each antibody stain. The regions of focus were the lateral/medial ganglionic eminence at E15.5 and the lateral ventricle at P0. Percent colocalization was calculated by (CR2-GFP+ cells plus antibody+ cells) / CR2-GFP+ cells.
Data quantification
In all experiments, percentages of cells represent the averages calculated from at least three independently samples. All values are shown as a mean ± standard error of the mean. Error bars in figures represent the standard error of the mean. In cases where results were tested for statistical significance, a student’s t-test was applied with a significance determined as p-value < 0.05.
Site-directed mutagenesis
Mutations were generated by deleting one or four base pairs or core-binding sequences. According to Table S3, primers were designed based on the 399 bp sequence of CR2 with deletions that targeted the core-binding site of Brn3/Barx2/Gsh1 (position 91–94) and Dlx2 (position 76–79) as well as the peripheral binding sequence of Brn3/Barx2/Gsh1 (position 95–96). The deletion of position 235–239 served as a negative control since this region contains no known core-binding sequence. PCR was performed using a high fidelity enzyme ExTaq with the following program: initial heat inactivation = 95 °C, 5 minutes; 16 cycles of heat inactivation [95 °C, 30 seconds]; annealing [Tm, 1 minute], extension [72 °C, 10 minutes]. The PCR product was transformed into NEB5α competent cells (NEB), colonies were analyzed by cPCR, and their sequences were confirmed at the DNA Sequencing Core Facility. Confirmed sequences were amplified and purified using MidiPrep and expression vectors were tested in chick embryos using in ovo electroporation method.
RESULTS
Regulatory activity of a novel cis-element in the Notchl locus is exclusive to the CNS of developing chick embryos
Based on sequence conservation among vertebrate species (e.g., human, mouse, and chick), we identified a 399-base pair region in the second intron of the Notch1 locus (CR2) as a cis-regulatory element (Fig. S1A). The ability of CR2 to direct tissue-specific gene expression in the vertebrate CNS was explored using a GFP reporter assay (Fig. S1B). A plasmid construct, CR2-GFP (containing CR2, a minimum beta-globin promoter, and a reporter GFP, Fig. S1B), along with a transfection control construct CAG-DsRed were injected at equal concentrations and electroporated into the chick neural tube to transfect neural progenitor cells at Hamburg-Hamilton stage 12 (HH12), which is approximately embryonic day 2 (E2). CR2-dependent GFP expression was found exclusively in the developing CNS (Fig. 1A). In contrast, reporter gene expression in the transfection control (CAG-GFP) was also observed in the visceral arch in addition to the CNS, suggesting that the activity of CR2 is specific to the CNS (Fig. 1B).
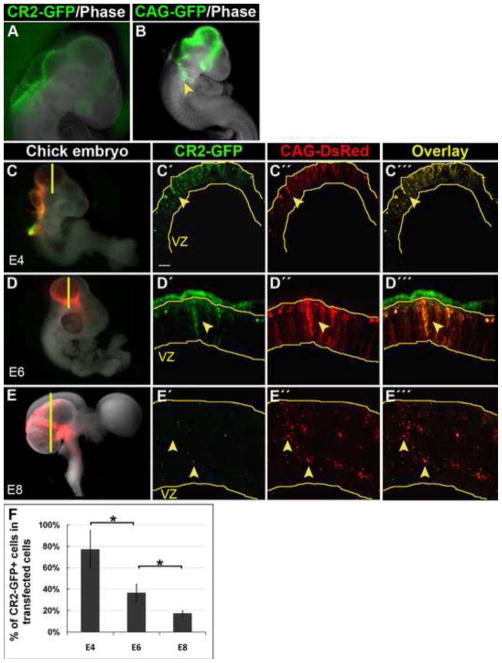
Figure 1. CR2 directs reporter GFP expression in embryonic chick CNS.
(A–B) Whole mount images of transfected chick embryos at E4 two days after electroporation at E2 (HH12). CR2-GFP expression was observed only in the CNS. CAG-GFP expression (the transfection controls) was observed in both the CNS and non-CNS tissues, e.g., the visceral arch (arrowhead). (C–E) Fluorescent microphotographs of chick brain sections showing CR2-GFP+ and CAG-DsRed+ cells in developing chick midbrain. Chick embryos were co-transfected with CAG-DsRed control and CR2-GFP at E2 and harvested at E4, E6, and E8. CR2-GFP expression decreased dramatically from E4 to E8, while control DsRed expression remained relatively constant. (F) Histograph showing that the percentage of CR2-GFP+ cells in the total number of transfection control CAG-DsRed+ cells decreased from 78% at E4, to 38% at E6, to 15% at E8. For all three developmental stages, n = 3; p<0.05. Scale bar = 10 μm.
To determine temporal CR2 activity, CR2-GFP expression was compared with the expression of the transfection control CAG-DsRed at E4 (Fig. 1C), E6 (Fig. 1D), and E8 (Fig. 1E) after electroporation at E2. These three stages were analyzed because they cover most of the neurogenic period in chick (Cheung et al., 2007; Ferretti and Whalley, 2008; Ono et al., 1995). The expression of the transfection control CAG-DsRed remained strong from E4 to E8. However, CR2-GFP expression diminished modestly from E4 to E6 and dramatically by E8. This decrease in CR2-GFP expression was reflected in the percentage of CR2- GFP+ cells in the transfected cell populations (CAG-DsRed+ cells), which decreased progressively from E4 (78±17%, n = 3) to E6 (38±8%, n = 3) and to E8 (15±2%, n = 3) (Fig. 1F). This finding indicates that CR2 activity is well correlated with the decreasing trend of neural progenitor cells during chick neurogenesis (Myat et al., 1996).
To ensure that the expression of GFP reporter is solely due to activity of CR2, we performed negative control experiments with a beta-globin promoter. DNA constructs containing a minimal beta-globin promoter alone without a conserved element or with a random DNA sequence were injected and electroporated to transfect the neural progenitors in the developing chick neural tube. No GFP expression was observed in transfected tissues at any of the three stages (i.e., E4, E6, and E8), indicating that the minimal promoter alone or a random DNA sequence does not possess the ability to independently direct GFP expression (n = 6, data not shown).
CR2 is active in early neural progenitor cells but not in differentiated neurons in the developing chick CNS
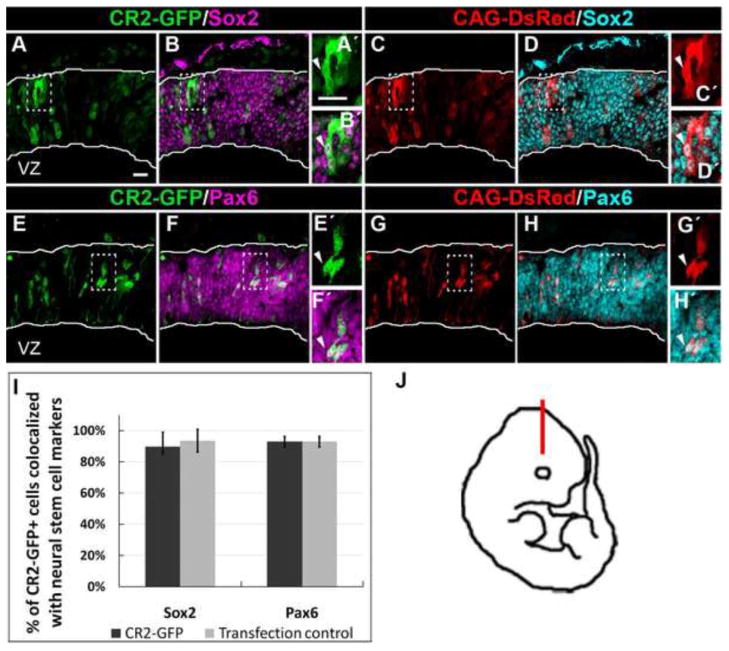
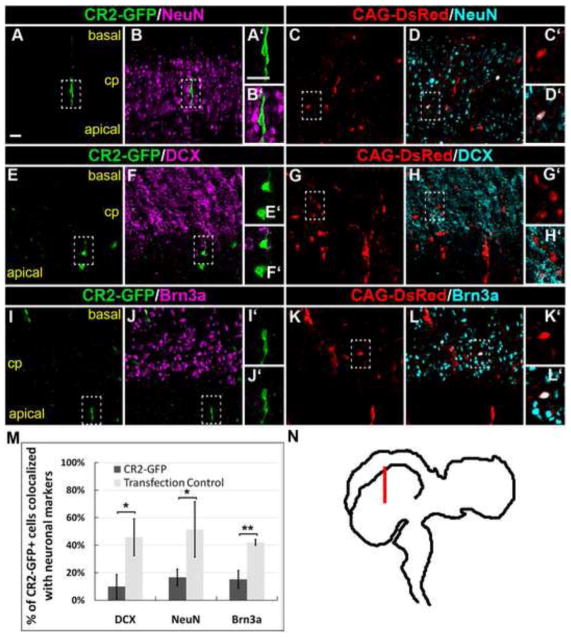
Next, we determined the identity of CR2-GFP+ cells by immunohistochemistry. Sections of transfected chick brains were stained with neural progenitor markers. Cell counts of E4 samples found that 90±9% (n = 3) and 93±3% (n = 3) of CR2-GFP+ cells co-localized with Sox2 (Fig. 2A–D) and Pax6 (Fig. 2E–G), respectively, indicating that the vast majority of the CR2-GFP+ cells at E4 were neural progenitors. Similarly, 94±7% and 93±3% of CAG-DsRed+ cells co-localized with Sox2 and Pax6, respectively. This result indicates that there is no significant difference between the expression of CR2-GFP and CAG- DsRed, since chick neural tube at this stage consists entirely of undifferentiated neural progenitor cells. However, differences were observed at E8, a later stage of neurogenesis (Fig. 3). Immunostaining of neuronal markers, NeuN (Fig. 3A–D), doublecortin (DCX) (Fig. 3E–H), and Brn3a (Fig. 3I–L), showed that approximately one half of the control CAG-DsRed+ cells differentiated into neurons (NeuN = 52±20%; DCX = 46±13%; Brn3a = 42±2%), while in CR2-GFP+ cells, these percentages significantly decreased (NeuN = 17±6%; DCX = 10±9%; Brn3a = 15±6%; for all cell counts, n = 3) (Fig. 3M). This result indicates that CR2 activity is significantly decreased in differentiated neurons.
Figure 2. CR2 activity is in neural progenitor cells at early chick neurogenesis.
Confocal images of CR2-GFP+ cells in early chick neurogenesis at E4 with immunofluorescence staining of neural progenitor cell markers: Sox2 (A–D) and Pax6 (E–H). (A–B) At E4, 90±9% (n = 3) and 93±3% (n = 3) of CR2-GFP+ cells co-localized with Sox2 and Pax6, respectively. (C-D) 94±7% and 93±3% of CAG-DsRed+ cells were co-stained with Sox2 or Pax6. (I) Histograph showing the percentage of CR2-GFP+ cells co-localized with neural progenitor markers. There was no significant difference between the experimental CR2-GFP and control CAG-DsRed groups (n = 3). (J) Immunostained coronal sections were located in the dorso-lateral optic tectum. Scale bar = 20 μm.
Figure 3. CR2 activity is dramatically decreased in differentiated neurons at late chick neurogenesis.
Confocal images of CR2-GFP+ cells in late chick neurogenesis at E8 with immunofluorescence staining of neuronal markers, NeuN (A–D), DCX (E–H), and Brn3a (I–L). The vast majority of CR2-GFP+ cells were not labeled with a neuronal marker (cells in dash-lined box). (M) Histograph showing the percentage of CR2-GFP+ cells co-localized with neuronal markers. The percentage of CR2-GFP+/neuronal marker+ cells was significantly lower than that in the control CAG-DsRed+ cells, i.e., 10±9% vs. 46±13% for DCX, 17±6% vs. 52±20% for NeuN, and 15±6% vs. 42±2% for Brn3a, respectively (n > 3; *p<0.05, **p<0.005). (N) Immunostained coronal sections were located in the dorso-lateral optic tectum. Scale bar = 20 μm.
CR2 activity is prominent in the embryonic ganglionic eminence and the neonatal rostral migratory stream of the transgenic mice
Since electroporation experiments in the developing chick only provide transient gene expression, transgenic mouse lines (n = 2) containing the CR2-GFP construct were generated. The two founder lines were analyzed for a complete spatiotemporal expression profile of CR2 activity. No obvious differences were detected in phenotypes with CR2-GFP expression between the two lines.
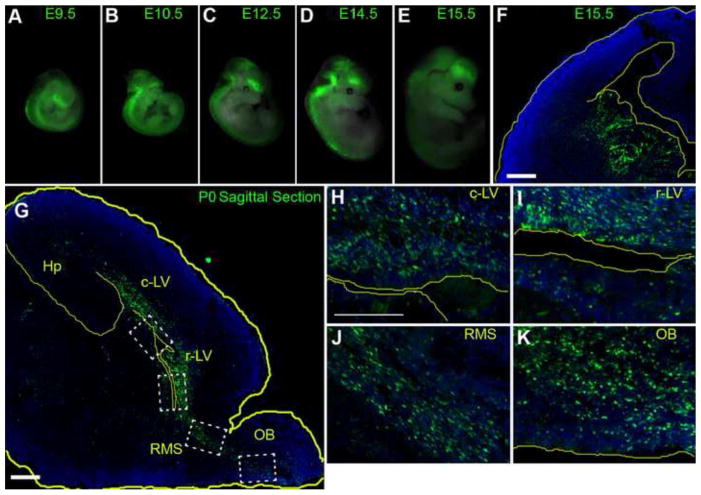
Similar to the GFP expression generated by in ovo electroporation experiments, CR2-GFP expression in transgenic mice was also found exclusive to the developing CNS (Fig. 4A–E). CR2-GFP expression in whole-mount transgenic mice was detected starting from E9.5 (n = 3) until postnatal day 0 (P0) pups (n = 3) (Fig. 4,S2), and prominently in the ventricular zone (VZ) and subventricular zones (SVZ) of the ganglionic eminences (GE) at E15.5 (Fig. 4F). Although GFP+ cells were not directly observed in brain sections at P7 and later stages under fluorescence scope, low level or residual GFP expression can be retrieved after staining with anti-GFP antibody in regions known to contain neural progenitor cells in the postnatal mouse brain, e.g., the VZ and SVZ of the lateral ventricles of the neocortex (Fig. S2), the hippocampus at P7 (Fig. S3), the rostral migratory stream (RMS) and the olfactory bulb (Fig. 4,S4). Since interneurons are born from the VZ/SVZ of the GE (Batista-Brito and Fishell, 2009; Miyoshi et al., 2010; Parnavelas et al., 2000), this well correlated spatiotemporal GFP expression pattern suggests that CR2 activity is in the interneuron progenitors.
Figure 4. CR2-GFP+ cells are predominant in the embryonic ganglionic eminence and in VZ/SVZ of the mouse neonatal cortex.
(A–E) Whole mount images of transgenic mouse embryos at various stages: E9.5 (n=3), E10.5 (n = 3), E11.5 (n = 4), E12.5 (n = 4), E14.5 (n = 5), and E15.5 (n=10). GFP expression was exclusive to the CNS ranging from the telencephalon to the sacral spinal cord. (F) Fluorescent microphotographs of coronal brain sections of the transgenic mouse at E15.5 showing CR2-GFP+ cells were predominantly located in the VZ/SVZ of the ganglionic eminences (GE). (G–K) CR2-GFP+ cells in saggittal brain sections of transgenic mouse at P0 were primarily located in regions surrounding the lateral ventricle, the olfactory bulb, the rostral migratory stream, and hippocampus. Blue represent nuclear marker Dapi staining. c-LV = caudal lateral ventricle, Hp = hippocampus, r-LV rostral-lateral ventricle, SVZ = subventricular zone, VZ = ventricular zone, OB = olfactory bulb, rms = rostral migratory stream. Scale bars = 20 μm.
CR2-GFP+ cells in transgenic mice are interneuron progenitors
To determine the cellular identity of CR2-GFP+ cells in the transgenic mice, immunostaining was performed on E15.5 and P0 tissue sections because E15.5 is near the peak of neurogenesis and P0 is the end of neurogenesis (Angevine and Sidman, 1961; Cai et al., 2002; Eccles, 1970; Sidman et al., 1959).
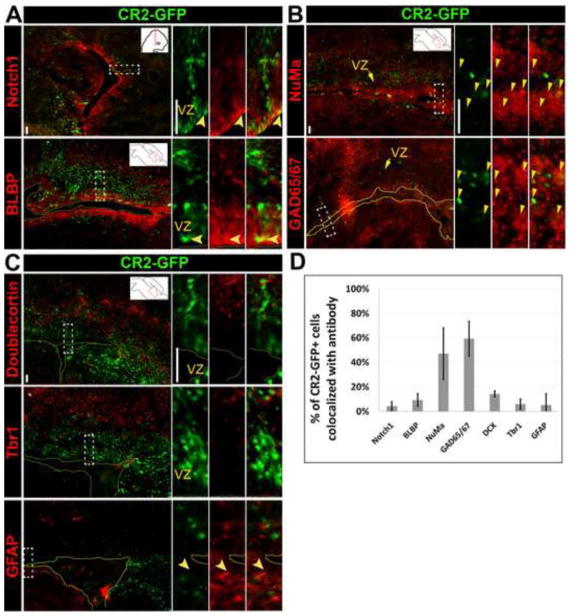
At E15.5, less than 10% of CR2-GFP+ cells were co-stained with radial glial markers, Notch1 (Fig. 5A) and BLBP (Fig. 5B), suggesting there were few neural stem cells at this time. However, 56±9% of CR2-GFP+ cells co-stained with a nuclear mitotic antigen (NuMa) (Fig. 5C), which is unevenly distributed between two asymmetrically dividing cells (Knoblich, 2008; Siller and Doe, 2009). In addition, only 5±4% of CR2-GFP+ cells were co-stained with DCX newborn neurons (Fig. 5D). These results indicate that the majority of GFP+ cells are not post-mitotic neurons but rather asymmetrically dividing neural progenitor cells.
Figure 5. CR2-GFP+ cells are in neural progenitors undergoing asymmetric division in E15.5 transgenic mouse.
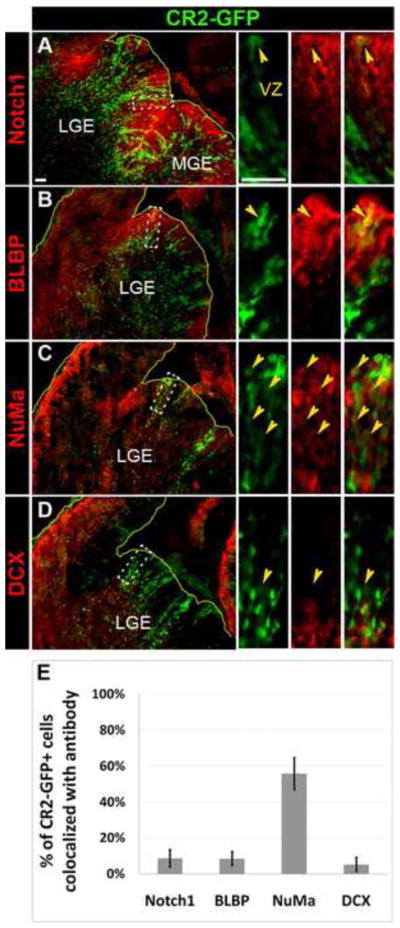
(A–D) Fluorescent microphotographs of coronal brain sections of the transgenic mouse at E15.5 showing CR2-GFP+ cells co-stained with radial glial markers, Notch1 (A) and BLBP (B), a marker for asymmetric division, NuMa (C), and an early neuronal marker, DCX (D). (E) Histograph showing the percentage of CR2-GFP+ cells co-localized with cell-specific markers. Less 10% of CR2-GFP+ cells were co-stained with Notch1 (9±5%), BLBP (8±4%), and DCX (5±4%). 56±9% of CR2-GFP+ cells were co-stained with NuMa. Scale bar = 20 μm.
At P0, CR2-GFP+ cells were largely found in the VZ/SVZ of the neocortex (Fig. 4G, 6, S2). Similar to E15.5, few of these GFP+ cells were co-stained with Notch1 (4±3%) and radial glial marker BLBP (9±5%) (Fig. 6A,S2B). Approximately one half of CR2-GFP+ cells (47±21%) around the VZ co-stained with NuMa and 59±14% co-localized with GAD65/67 (Fig. 6B), suggesting that they are asymmetrically dividing progenitor cells with GABAergic potential. Thus, GAD65/67+/GFP+ cells possessed a phenotype of interneurons or interneuron progenitors, since some GAD65/67+ neurons have been shown to be dividing progenitor cells (Riccio et al., 2012).
Figure 6. CR2-GFP+ cells are asymmetrically dividing interneuron precursors in P0 transgenic mouse.
Fluorescent microphotographs of coronal (A) and sagittal (B,C) brain sections of the transgenic mouse at P0. CR2-GFP+ cells co-stained with cell specific markers. (D) Histograph represents the percentage of CR2-GFP+ cells co-stained with radial glial markers Notch1 (4±3%) and BLBP (9±5%) (A); an asymmetric division marker NuMa (47±21%) and a marker for GABAergic interneuron progenitor GAD65/67 (59±14%) (B); and an early neuronal marker DCX (14±2%), a layer VI neuronal marker Tbr1 (6±4%), and an astrocyte marker GFAP (5±9%) (C). Scale bar = 20 μm.
To determine whether CR2 activity is present in the differentiated cells of the transgenic CNS, immunostaining recovered GFP+ cells were co-stained with DCX for early differentiated neurons, Tbr1 for layer VI projection (excitatory) neurons in the neocortex (Hevner et al., 2001), and GFAP for astrocytes. Only about 14±2%, 6±4%, and 5±9% of CR2-GFP+ cells were co-stained with DCX, Tbr1, and GFAP, respectively (Fig. 6C–D,S2). In addition, CR2-GFP+ cells did not co-stain well with Tbr2 (Fig. S2C), an intermediate progenitor marker []. The boundary between CR2-GFP+ cells and Tbr1+ cells was distinct, suggesting that CR2 activity was not prevalent in differentiated cortical layer VI projection neurons or astrocytes.
CR2-GFP+ cells are GABAergic interneuron progenitors as revealed by GFP immunofluorescence staining
Protein expression (e.g., GFP) is a dynamic process dictated by rates of protein expression and degradation. Since the half-life of GFP is 26 hours (Corish and Tyler-Smith, 1999), it takes approximately 7 days for 99% of the GFP protein to degrade after initial expression. Immunofluorescence staining on transgenic brain sections thus allows the retrieval of GFP signals that may not be observed directly due to decreased/degraded GFP fluorescence. Thus, a CR2-GFP+ cell may be “tracked” for approximately 7 days after GFP is no longer expressed and thus provides an extended period of time for tracking the fate of GFP+ cells.
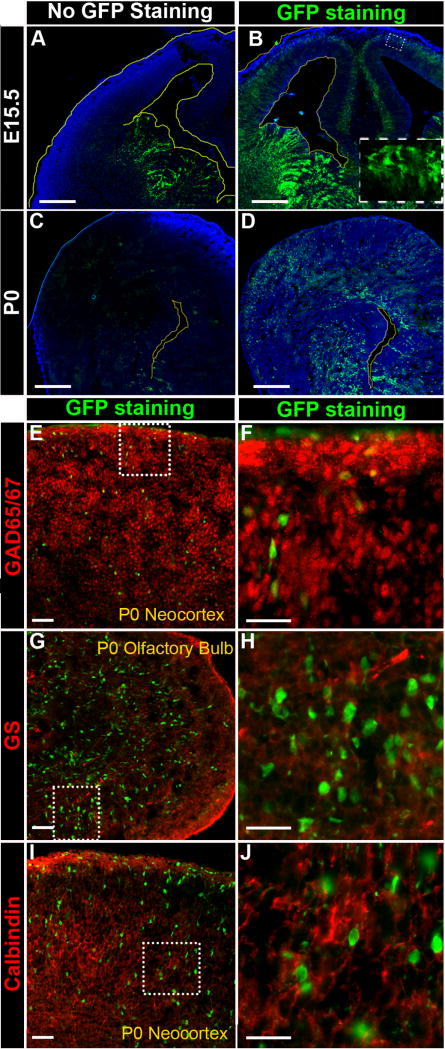
When stained with anti-GFP antibody, additional GFP+ cells were unveiled in new regions of the CNS. At E15.5, bright GFP+ cells were detected in the ganglionic eminence below the VZ/SVZ and the basal (top) layer of the neocortex (Fig. 7B), as well as faint GFP+ cells stretching across the neocortex at P0 (Fig. 7D). This new pattern contrasts with GFP fluorescence without anti-GFP antibody staining, which is only observed in the VZ/SVZ layers of the ganglionic eminence at E15.5 (Fig. 7A) and the cortical lateral ventricles at P0 (Fig. 7C).
Figure 7. Immunostaining recovered CR2-GFP+ cells are GABAergic interneuron progenitors in the transgenic neocortex.
Fluorescent microphotographs of coronal brain sections of the transgenic mouse at E15.5 (A–B) and P0 (C–D). (A) At E15.5, without anti-GFP staining, the CR2-GFP+ cells were prominently seen in the VZ/SVZ of the GE. (B) With anti-GFP staining, three additional populations of CR2-GFP+ cells were revealed: the ventral ganglionic eminence (GE) below the VZ/SVZ, the basal (upper) layers of the neocortex, and VZ/SVZ of the neocortex. (C) At P0, without anti-GFP staining, CR2-GFP+ cells were only seen mostly VZ/SVZ surrounds the lateral ventricle of the ventral forebrain, and a few GFP+ cells in the cortical plate. (D) With anti-GFP staining, more than twice as many CR2-GFP+ cells were revealed, and they were located in the VZ/SVZ of the entire lateral ventricle and widely spread over the cortical plate. (E-J) Sagittal brain sections of P0 transgenic mouse were co-stained with anti-GFP antibody and cell specific markers, GAD65/67 (E–F), glutamine synthetase (GS) (G–H), and Calbindin (I–J). The majority of immunostained GFP+ cells were co-stained with GAD65/67 but not with GS or calbindin. Scale bars = 20 μm.
At P0, immunostaining recovered GFP+ cells continued to co-stain with GAD65/67 (Fig. 7E–F) but were not co-labeled with either glutamine synthetase (GS) for glutamatergic neurons (Fig. 7G-H) or calbindin interneurons (Fig. 7I–J). At P7, immunostaining recovered GFP+ cells were detected in the ependyma, cortical plate, hippocampus, and midbrain (Fig. S3A), and they were co-stained with NuMa and GAD65/67 in some cells but not with BLBP, Tbr2, or Tbr1 (Fig. S3B). These results further confirmed that CR2-GFP+ cells are dividing interneuron progenitor cells with GABAergic potential.
Binding motif of Gsh1/Brn3/Barx2 is required for CR2 gene regulatory activity
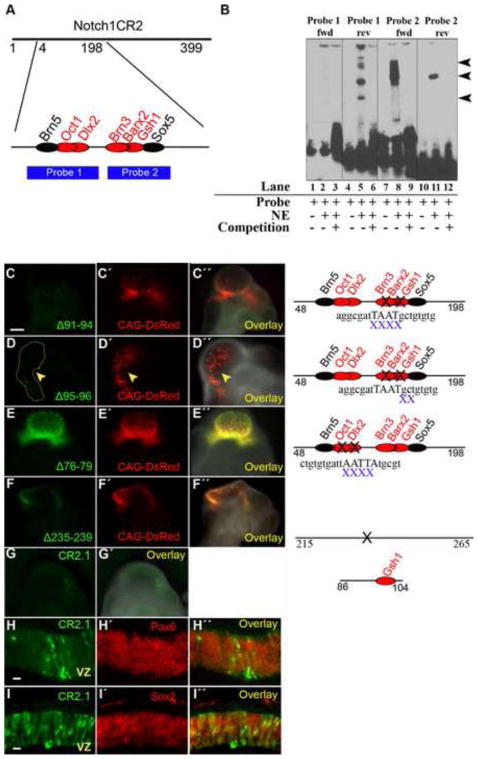
To determine the mechanism by which CR2 regulates specific gene expression in interneuron progenitors, we searched for trans-acting factor binding sites (TFBSs) of CR2 that were conserved between chick and mouse using MatInspector (Genomatix, Germany). The search result contained 164 predicted TFBSs (supplemental Table S1). Further sequence analysis showed that a 150 bp region of the mouse CR2 sequence (position 48–198) shared 83% similarity with a chick Notch1 locus. In this 150 bp DNA fragment, 10 known TFBSs were predicted, which are common to both M. musculus and G. gallus (supplemental Table S2). We then determined sequence-specific protein factor binding to narrow down active subregions of CR2. Electrophoretic mobility shift assays (EMSAs) were performed using DNA probes designed to cover the 150 bp conserved region (Fig. 8A,B). Probe 1 spanned the region of CR2 with TFBSs for Brn5, Oct1, and Dlx2; while Probe 2 contained TFBSs for Brn3, Barx2, Gsh1, and Sox5 (Fig. 8A). After testing the forward and reverse strands separately, results showed binding of Probe 1 (reverse only) and Probe 2 (forward and reverse) with nuclear extract from the E8 chick brain (Fig. 8B). The potential TFBSs located on these probes include Oct1 and Dlx2 (Probe 1-reverse), Sox5 (Probe 2-forward), and Brn3, Barx2, and Gsh1 (Probe 2-reverse).
Figure 8. Gsh1 binding site in CR2 is important in directing GFP expression.
(A) Two probes (1 and 2) were designed between position 48 and 198 of CR2 which possessed TFBSs that are conserved between mouse and chick. TFBSs on forward and reverse strands are represented by red and black font, respectively. (B) Electrophoretic mobility shift assays (EMSA) show that DNA probes containing the Brn5/Oct1/Dlx2 (Probe 1) or Brn3/Barx2/Gsh1 (Probe 2) have CR2 sequence-specific binding activity with nuclear extract (NE) from E8 chick brain. The probes were run as forward strands (lanes 1–3, 7–9) and as reverse strands (lanes 4–6, 10–12). Lanes 1, 4, 7, and 10 are controls consisting of the probe alone without nuclear extract. Probe 1-rev, Probe 2-fwd, and Probe 3-rev showed band shift (lanes 5, 8, 11) that was competed away by the unlabeled competition probes (lanes 6, 9, 12). (C–F) Reporter assay using in ovo electroporation/transfection method with mutant CR2-GFP constructs generated by site-directed mutagenesis. (C) Deletion of position 91–94 (Δ91–94) removed the core binding sequence for the trans-acting factors of Brn3, Barx2, and Gsh1. The transfection of CR2Δ91–94 –GFP construct was not able to produce GFP expression in the E4 chick embryo. (D) Deletion of position 95–96 (Δ95–96) disturbed the peripheral binding sequence for Brn3, Barx2, and Gsh. The transfection of CR2-GFPΔ95–96 –GFP construct resulted in diminished GFP expression. (E) Deletion of position 76–79 removed the TFBS for Dlx2, which is a transcription factor of inhibitory neurons but is not conserved. The transfection of CR2-GFPΔ76–79 –GFP construct was able to drive GFP expression. (F) As a negative control, the deletion of position 235–239 (containing no known TFBS) on CR2 did not affect GFP expression. (G) Plasmid constructs containing subregions of CR2 were electroporated into the developing chick neural tube at E2, and GFP expression was detected at E4. CR2.1 (position 86–104) is an 18 bp fragment containing the conserved binding motif for Gsh1. (H–I) CR2.1 co-localizes with Pax6 and Sox2.
To determine whether regions with strong binding as determined by EMSA were important for gene regulatory activity of CR2, mutant reporter constructs were generated by site-directed mutagenesis (primers listed in the supplemental Table S3). A deletion of the 4 bp core-binding motif of Gsh1/Barx2/Brn3 (position 91–94) abolished the ability of CR2 to drive GFP expression (n = 2) in the E4 chick CNS (Fig. 8C). A two bp deletion in the peripheral binding sequences of Gsh1/Barx2/Brn3 (position 95–96) greatly diminished GFP expression (n = 2) (Fig. 8D), suggesting that this region is crucial for CR2 activity. However, deletion of the Dlx2 binding motif (position 76–79) did not affect GFP expression (n = 2) (Fig. 8E). The Dlx2 binding motif was also examined due to its role in inhibitory interneurons (Mizuguchi et al., 2006; Toresson and Campbell, 2001; Yun et al., 2003), although it is not conserved between mouse and chicken. As a negative control, deletion of position 235–239, a region without known TFBS, did not affect CR2-GFP expression (Fig. 8F). Therefore, these results suggest that TFBSs for Brn3/Barx2/Gsh1 are important for reporter GFP expression.
To further determine the minimum sequence requirement for gene regulatory activity, CR2.1, a subregion of CR2, was designed to contain the 18 bp DNA fragment at position 86–104 with the whole conserved binding motif of Gsh1 and the core 4 bp binding motif of Gsh1/Brn3/Barx2 (Fig. S1B). Gsh1 is known to be an important transcription factor for GABAergic interneurons (Mizuguchi et al., 2006; Stenman et al., 2003; Toresson and Campbell, 2001), Then, CR2.1 was tested for its ability to direct reporter GFP expression in chick embryos using in ovo electroporation technique. The CR2.1 subregion was shown to direct GFP expression in E4 chick CNS (n = 2) (Fig. 8G). CR2.1-GFP+ cells were shown to stain with neural progenitor markers Pax6 (Fig. 8H) and Sox2 (Fig. 8I). Thus, the data indicates that CR2.1 possesses gene regulatory activity and supports the role of Gsh1/Barx2/Brn3 binding motif in regulating CR2 activity in neural progenitor cells.
DISCUSSION
CR2 activity is well correlated with neurogenesis in both the chick and mouse
This study identified that CR2, an evolutionary conserved non-coding DNA sequence in the second intron of Notch1 locus, is involved in regulating gene expression in interneuron progenitors. Its exclusivity in the CNS during neurogenesis is indicative of cis-regulatory characteristics. CR2-GFP expression patterns in chick (E4-E8) and in mouse (E9.5 to P0) correspond to their respective periods of neurogenesis and Notch1 expression during CNS development. These results also support the known role of Notch1 in inhibiting neuronal differentiation during development (Ishibashi et al., 1994; Nye et al., 1994). Notch1 maintains the stem cell state through lateral inhibition, a process by which binding of the Notch1 ligand in one cell prevents its own differentiation through interactions with an inhibitory ligand (i.e., Delta) of an adjacent cell (Chitnis, 1995).
However, it appears that CR2 activity is not entirely related to the radial glial phenotype or Notch1 expression despite its location in the Notch1 locus. There were three striking phenomena of CR2 activity in the transgenic mouse cortex that may not be consistent with a radial glial phenotype. (1) Expression of CR2-GFP was limited to the ventral CNS as seen in the whole mounts. Long, radial GFP+ cells illuminate the VZ/SVZ of primarily the ganglionic eminences at E15.5. (2) GFP is not limited to the VZ/SVZ at P0 but rather includes the rostral migratory stream and olfactory bulb. (3) Few CR2-GFP+ cells co-localize with radial glial markers BLBP and Notch1. Since the majority of CR2-GFP+ cells were not radial glia, neurons, or glia, then the phenotype was expected to be in an intermediary state such as of a neuronal progenitor.
CR2 is a regulatory element for GABAergic interneuron progenitors
Our interpretation that CR2 is preferentially active in GABAergic progenitors is based on the following two observations: 1) during embryonic development at E15.5, CR2-GFP+ cells (without anti-GFP antibody staining) were predominantly found in the VZ/SVZ of the ganglionic eminence where interneuron progenitors reside (Anderson et al., 1997; Ang et al., 2003; Lavdas et al., 1999; Lopez-Bendito et al., 2004; Morozov et al., 2009; Nery et al., 2002; Wichterle et al., 1999); but not the dorsal pallium where progenitors of excitatory glutamatergic projection neurons reside (Fig. 4F); and 2) the majority of CR2-GFP+ cells at P0 (with anti-GFP antibody staining) were co-labeled with an interneuron marker GAD65/67 (Fig. 6B), and GFP+ cells were not co-labeled with Tbr1, a marker for glutamatergic neurons (Fig. 6C).
Embryonic and postnatal CR2-GFP+ cells were not newborn neurons or glia (negative for NeuN, Tbr1, and GFAP staining), but rather mitotic cells that continue to undergo asymmetric division (positive for NuMa staining indicative of mitotic progenitors (Knoblich, 2008; Siller and Doe, 2009)). Recently, a radial glia-like progenitor cell has been reported to arise from asymmetric divisions of radial glia, to reside in the outer SVZ, and to generate neurons (Wang and Kriegstein, 2009). Studies have shown that GAD65/67 marks not only GABAergic interneurons but also interneuron precursors (Lopez-Bendito et al., 2004; Riccio et al., 2012). Thus, the fact that the majority of CR2-GFP+ cells co-stained with GAD65/67 indicates that CR2 is active in interneuron progenitors.
After CR2 activity subsides, cells may differentiate into interneurons that tangentially migrate into the neocortex. Immunostaining recovered GFP signals allowed tracking of the fate of cells after losing GFP fluorescence. Although there was no CR2-GFP expression in adolescent and adult mice, immunostaining recovered GFP+ cells were present in the hippocampus, the rostral migratory stream and the olfactory bulb. These regions contain GABAergic inhibitory cells, therefore, these results indicate that CR2 activity is in cells with GABAergic potential. Since a very small population of GFP+ cells were found in the VZ of the developing cerebral cortex where progenitors of glutamatergic projection neurons reside, it is possible that CR2 may be active in glutamatergic lineage. If this were the case, immunostaining with anti-GFP antibody would have revealed GFP+ glutamatergic neurons. However, no immunostaining recovered GFP+ cells were found to be glutamatergic neurons or calbindin-binding interneurons. Thus, the possibility that CR2 might be active in progenitors of these two neuronal types was ruled out. Overall, it is not surprising that gene expression in GABAergic interneuron progenitors is regulated by a conserved regulatory element because GABA signaling is conserved in different species during neurogenesis (Abdel-Mannan et al., 2008; Cheung et al., 2007).
Gsh1/Barx2/Brn3 binding motif is required for CR2 gene regulatory activity
Among the conserved TFBSs on CR2, several are directly relevant to the CNS development. For example, Brn3 is essential for retinal and auditory sensory progenitors (Bryant et al., 2002), Barx2 is involved in the brain and the floor plate of the spinal cord (Jones et al., 1997; Meech et al., 1999), and Gsh1 plays an important role in interneuron progenitors of the brain and spinal cord (Mastick et al., 1997; Mizuguchi et al., 2006). In particular, several studies have demonstrated that Gsh1 is expressed in the VZ of the ganglionic eminence and plays an important role in the development of the striatum and olfactory bulb (Toresson and Campbell, 2001; Yun et al., 2003). The observed CR2-GFP expression pattern in the ganglionic eminence, the rostral migratory stream, and the olfactory bulb was tightly correlated to the Gsh1 expression. The absence of Gsh1 binding motif on CR2 abolishes its ability to independently drive GFP expression in the reporter assays, and a subregion representing the whole binding sequence of Gsh1 demonstrates activity. Gsh1 is expressed in areas of CR2 activity including the MGE and LGE (Toresson and Campbell, 2001; Valerius et al., 1995; Yun et al., 2003). In addition, Gsh1 has been shown to promote the maturation of progenitors towards neurogenesis (Pei et al., 2011). These observations strongly support the role of Gsh1 in CR2 regulation of interneuron progenitors. Thus, we propose that a mechanism of CR2 activation and regulation of interneuron gene expression is through the binding of transcription factor Gsh1.
In summary, we identified a 399-bp cis-element in the second intron of the Notch1 locus that functions in GABAergic interneuron progenitors. CR2 and its regulatory mechanism contribute to the comprehensive knowledge of the genetic network in neural progenitor cells for interneuron development in the CNS. For instance, CR2-GFP transgenic mouse can be utilized as a model for tracking/studying interneuron progenitors and their cell fate in normal development as well as other neurological disease/injury models. Comprehensive knowledge of CR2 can also serve bioengineering purposes. CR2, Gsh1 and possibly other factors including Brn3, and Barx2 are factors to consider when engineering interneuron progenitors for transplant in patients with GABAergic interneuron deficiencies. Our findings provide a new approach for the future investigation of interneuron progenitors in the CNS.
Supplementary Material
Highlights.
a cis-element in Notch1 locus regulates gene expression in interneuron progenitors
activity of this cis-element is correlated with vertebrate neurogenesis
spatiotemporal activity of this cis-element is in GABAergic progenitors
cis-element activity requires the binding motif for Gsh1/Barx2/Brn3
CR2-GFP transgenic mice are useful for study of interneuron development and defects
Acknowledgments
The authors would like to thank Dr. Connie Cepko for plasmid DNA reporter constructs (pCAG-GFP and pCAG-DeRed); Drs. Bonnie Firestein, RenpingZhou, Mladen-Roko Rasin, Gabriella D’Arcangelo, and the members of the Cai laboratory for insightful discussion and proof-reading. We would also like to acknowledge the assistance of the Transgenic and Knockout Mouse Shared Resource of the Cancer Institute of New Jersey, which is supported by the NCI CCSG grant (P30CA072720), and by the Robert Wood Johnson Foundation support of the Child Health Institute of New Jersey. This work was supported in part by the grants from the National Institute of Health (EY018738), the New Jersey Commission on Spinal Cord Research grants (10A-3091-SCR-E-0 and 08-3074-SCR-E0), and the Busch Biomedical Research Awards to LC. ET was a graduate fellow of the National Science Foundation Integrative Graduate Education Research Traineeship Grant (IGERT - DGE 0801620) and the New Jersey Commission on Spinal Cord Research (08-2936-SCR-E0).
Footnotes
AUTHOR CONTRIBUTIONS
LC conceived and designed the experiments. ET performed the experiments. SS contributed to EMSA and site-directed mutagenesis. SD performed computational comparative analysis and generated the CR2-GFP reporter construct. HH contributed to troubleshooting experiments. YL and AW collected some transgenic mouse samples. ET, MG and LC analyzed the data. ET and LC wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Mannan O, Cheung AF, Molnar Z. Evolution of cortical neurogenesis. Brain research bulletin. 2008;75:398–404. doi: 10.1016/j.brainresbull.2007.10.047. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Ang ES, Jr, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Fishell G. The developmental integration of cortical interneurons into a functional network. Current topics in developmental biology. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Blank MC, Chizhikov V, Millen KJ. In ovo electroporations of HH stage 10 chicken embryos. Journal of visualized experiments. JoVE. 2007:408. doi: 10.3791/408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Developmental cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Brittis PA, Lemmon V, Rutishauser U, Silver J. Unique changes of ganglion cell growth cone behavior following cell adhesion molecule perturbations: a time-lapse study of the living retina. Mol Cell Neurosci. 1995;6:433–449. doi: 10.1006/mcne.1995.1032. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J, Goodyear RJ, Richardson GP. Sensory organ development in the inner ear: molecular and cellular mechanisms. Br Med Bull. 2002;63:39–57. doi: 10.1093/bmb/63.1.39. [DOI] [PubMed] [Google Scholar]
- Cai L, Hayes NL, Takahashi T, Caviness VS, Jr, Nowakowski RS. Size distribution of retrovirally marked lineages matches prediction from population measurements of cell cycle behavior. J Neurosci Res. 2002;69:731–744. doi: 10.1002/jnr.10398. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Cheung AF, Pollen AA, Tavare A, DeProto J, Molnar Z. Comparative aspects of cortical neurogenesis in vertebrates. Journal of anatomy. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis AB. The role of Notch in lateral inhibition and cell fate specification. Mol Cell Neurosci. 1995;6:311–321. [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Doh ST, Hao H, Loh SC, Patel T, Tawil HY, Chen DK, Pashkova A, Shen A, Wang H, Cai L. Analysis of retinal cell development in chick embryo by immunohistochemistry and in ovo electroporation techniques. BMC developmental biology. 2010;10:8. doi: 10.1186/1471-213X-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh ST, Zhang Y, Temple MH, Cai L. Non-coding sequence retrieval system for comparative genomic analysis of gene regulatory elements. BMC Bioinformatics. 2007;8:94. doi: 10.1186/1471-2105-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles JC. Neurogenesis and morphogenesis in the cerebellar cortex. Proceedings of the National Academy of Sciences of the United States of America. 1970;66:294–301. doi: 10.1073/pnas.66.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Ever L, Gaiano N. Radial ‘glial’ progenitors: neurogenesis and signaling. Current opinion in neurobiology. 2005;15:29–33. doi: 10.1016/j.conb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Ferretti P, Whalley K. Successful neural regeneration in amniotes: the developing chick spinal cord. Cell Mol Life Sci. 2008;65:45–53. doi: 10.1007/s00018-007-7430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Lenhard B, Becker TS. New technologies, new findings, and new concepts in the study of vertebrate cis-regulatory sequences. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:870–885. doi: 10.1002/dvdy.20659. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Developmental dynamics : an official publication of the American Association of Anatomists. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Doh ST, Cai L. In ovo electroporation in embryonic chick retina. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeziorska DM, Jordan KW, Vance KW. A systems biology approach to understanding cis-regulatory module function. Semin Cell Dev Biol. 2009;20:856–862. doi: 10.1016/j.semcdb.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Jones FS, Kioussi C, Copertino DW, Kallunki P, Holst BD, Edelman GM. Barx2, a new homeobox gene of the Bar class, is expressed in neural and craniofacial structures during development. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2632–2637. doi: 10.1073/pnas.94.6.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annual review of neuroscience. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Miano JM. Remote control of gene expression. J Biol Chem. 2007;282:15941–15945. doi: 10.1074/jbc.R700010200. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Davis NM, Andrew GL, Easter SS., Jr Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- Meech R, Kallunki P, Edelman GM, Jones FS. A binding site for homeodomain and Pax proteins is necessary for L1 cell adhesion molecule gene expression by Pax-6 and bone morphogenetic proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2420–2425. doi: 10.1073/pnas.96.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Morozov YM, Torii M, Rakic P. Origin, early commitment, migratory routes, and destination of cannabinoid type 1 receptor-containing interneurons. Cereb Cortex. 2009;19(Suppl 1):i78–89. doi: 10.1093/cercor/bhp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat A, Henrique D, Ish-Horowicz D, Lewis J. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol. 1996;174:233–247. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Norris CR, Kalil K. Guidance of callosal axons by radial glia in the developing cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1991;11:3481–3492. doi: 10.1523/JNEUROSCI.11-11-03481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski RS, Hayes NL. CNS development: an overview. Development and psychopathology. 1999;11:395–417. doi: 10.1017/s0954579499002126. [DOI] [PubMed] [Google Scholar]
- Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- Ono K, Bansal R, Payne J, Rutishauser U, Miller RH. Early development and dispersal of oligodendrocyte precursors in the embryonic chick spinal cord. Development. 1995;121:1743–1754. doi: 10.1242/dev.121.6.1743. [DOI] [PubMed] [Google Scholar]
- Parnavelas JG, Anderson SA, Lavdas AA, Grigoriou M, Pachnis V, Rubenstein JL. The contribution of the ganglionic eminence to the neuronal cell types of the cerebral cortex. Novartis Foundation symposium. 2000;228:129–139. doi: 10.1002/0470846631.ch10. discussion 139–147. [DOI] [PubMed] [Google Scholar]
- Pei Z, Wang B, Chen G, Nagao M, Nakafuku M, Campbell K. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1675–1680. doi: 10.1073/pnas.1008824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Guidance of neurons migrating to the fetal monkey neocortex. Brain research. 1971a;33:471–476. doi: 10.1016/0006-8993(71)90119-3. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. The Journal of comparative neurology. 1971b;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. The Journal of comparative neurology. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Riccio O, Murthy S, Szabo G, Vutskits L, Kiss JZ, Vitalis T, Lebrand C, Dayer AG. New Pool of Cortical Interneuron Precursors in the Early Postnatal Dorsal White Matter. Cereb Cortex. 2012 doi: 10.1093/cercor/bhr086. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Miale IL, Feder N. Cell proliferation and migration in the primitive ependymal zone: an autoradiographic study of histogenesis in the nervous system. Exp Neurol. 1959;1:322–333. doi: 10.1016/0014-4886(59)90024-x. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain research. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Campbell K. A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development. 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- Valerius MT, Li H, Stock JL, Weinstein M, Kaur S, Singh G, Potter SS. Gsh-1: a novel murine homeobox gene expressed in the central nervous system. Dev Dyn. 1995;203:337–351. doi: 10.1002/aja.1002030306. [DOI] [PubMed] [Google Scholar]
- Van Ness J, Pettijohn DE. Specific attachment of nuclear-mitotic apparatus protein to metaphase chromosomes and mitotic spindle poles: possible function in nuclear reassembly. Journal of molecular biology. 1983;171:175–205. doi: 10.1016/s0022-2836(83)80352-0. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Research. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlbuhl M, Reiprich S, Vogl MR, Bosl MR, Wegner M. Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 2012;40:88–101. doi: 10.1093/nar/gkr734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DD, Kriegstein AR. Defining the role of GABA in cortical development. J Physiol. 2009;587:1873–1879. doi: 10.1113/jphysiol.2008.167635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol Biol. 2000;132:337–349. doi: 10.1385/1-59259-192-2:337. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;2:461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Yun K, Garel S, Fischman S, Rubenstein JL. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. The Journal of comparative neurology. 2003;461:151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- Zhong W, Chia W. Neurogenesis and asymmetric cell division. Current opinion in neurobiology. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.