SUMMARY
By diversifying the biological effector functions of antibodies, class switch DNA recombination (CSR) plays a critical role in the maturation of the immune response. It is initiated by AID-mediated dC deamination, yielding dUs, and dU glycosylation by Ung in antibody switch (S) region DNA. Here we showed that the translesion DNA synthesis polymerase Rev1 directly interacted with Ung and targeted in an AID-dependent and Ung-independent fashion the S regions undergoing CSR. Rev1–/– Ung+/+ B cells reduced Ung recruitment to S regions, DNA-dU glycosylation and CSR. This together with an S region spectrum of mutations similar to that of Rev1+/+ Ung–/– B cells suggested that Rev1 operated in the same pathway as Ung, as emphasized by further decreased CSR in Rev1–/– Msh2–/– B cells. Rescue of CSR in Rev1–/– B cells by a catalytically inactive Rev1 mutant showed that the important role of Rev1 in CSR is mediated by Rev1 scaffold, not enzymatic function.
INTRODUCTION
Class switch DNA recombination (CSR) and somatic hypermutation (SHM) are critical for the maturation of the antibody response. CSR endows antibodies with new biological effector functions by replacing the CH region with a CH region that lies downstream in the immunoglobulin heavy chain (IgH) locus (Xu et al., 2012). SHM provides the structural substrate for antigen-mediated selection of higher affinity antibody mutants by introducing mainly point-mutations with rare deletions or insertions into rearranged Ig V(D)J sequence (Casali et al., 2006). Both CSR and SHM involve activation-induced cytidine deaminase (AID)-mediated generation of DNA lesions and subsequent DNA repair (Liu and Schatz, 2009; Xu et al., 2012). AID deaminates deoxycytosine (dC) residues to yield deoxyuridine (dU):deoxyguanine (dG) mispairs. These trigger DNA repair processes that lead to introduction of mutations in V(D)J regions and generation of double-strand DNA breaks (DSBs) in switch (S) regions, in which point-mutations are also introduced (Odegard and Schatz, 2006; Xu et al., 2012). Resolution of DSBs by ligation of an upstream and a downstream S regions leads to CSR (Xu et al., 2012).
Upon induction by CD40- or TLR-signaling (Pone et al., 2012b) of specific transcription factors, including HoxC4 and NF-κB (Mai et al., 2010; Park et al., 2009; Tran et al., 2010; White et al., 2011), AID is expressed at high levels and targeted by 14-3-3 adaptors to S region DNA to generates dUs (Xu et al., 2010). These dUs can be “replicated over” to introduce dC/dG transition mutations or can be processed through the base excision repair (BER) pathway, involving glycosylation by uracil DNA glycosylase (Ung) and abasic site excision by apurinic/apyrimidinic endonucleases (APEs), thereby giving rise to DSBs (Stavnezer, 2011; Xu et al., 2012). Ung-mediated dU glycosylation initiates the major process leading to generation of DSBs and CSR (Rada et al., 2002); dU:dG mispairs can also be dealt with by the mismatch repair (MMR) machinery, which can contribute to the generation of S region DSBs (Rada et al., 2004). Accordingly, combined deficiency of Ung and Msh2, a critical MMR element, leads to ablation of CSR (Rada et al., 2004). Translesion DNA synthesis (TLS) polymerases, including polymerases η, θ, ζ and Rev1, are involved in the DNA lesion repair leading to SHM (Casali et al., 2006; Jansen et al., 2006; Weill and Reynaud, 2008; Zan et al., 2001; Zan et al., 2005); DNA polymerases β and ζ are perhaps also involved in repairing S region DSBs (Schenten et al., 2009; Wu and Stavnezer, 2007). The role of DNA polymerases in CSR, however, remains to be defined.
Among eukaryotic DNA polymerases, Rev1 is one of the few polymerases possessing both catalytic and structural functions (i.e., recruitment of other factors) for DNA repair. In its catalytic role, Rev1, a Y family TLS polymerase, inserts dCs opposite template dUs, abasic sites and other damaged bases by utilizing a unique mechanism of protein-template-directed nucleotide incorporation (Nair et al., 2005; Prakash et al., 2005; Zhang et al., 2002). The dC transferase catalytic activity as well as the N-terminal BRCA1 C terminus (BRCT) domain of Rev1 are, however, largely dispensable for efficient TLS and, therefore, tolerance of DNA damage caused by a different mutagens (Masuda et al., 2009; Otsuka et al., 2005; Ross et al., 2005). Rev1 plays a role in SHM (Jansen et al., 2006; Masuda et al., 2009). In Rev1–/– mice, the Ig Vλ gene mutation spectrum was altered: dC→dG transversions were virtually absent in the non-transcribed (coding) strand and reduced in the transcribed strand (Jansen et al., 2006). The sharing of many DNA repair factors between SHM and CSR and the multiple functions of Rev1 in DNA repair, through interactions with different DNA repair factors (Hicks et al., 2010; Sharma et al., 2011; Xie et al., 2010), prompted us to hypothesize that Rev1 is involved in CSR by interacting with other elements of the CSR machinery and enhancing their function/activity.
We have addressed the role of Rev1 in CSR in vivo and in vitro using Rev1–/– mice injected with 4-hydroxy-3-nitrophenyl acetyl (NP)-conjugated chicken gamma globulin (NP-CGG) and B cells deficient in Rev1 or other CSR factors, including AID and Ung. We adapted bimolecular fluorescence complementation (BiFC) assays to study the direct interaction of Rev1 with Ung and Ung mutants. We performed chromatin immunoprecipitation (ChIP) assays using B cells deficient in AID, Rev1 or Ung to determine the AID-dependence of Rev1 recruitment to S regions as well as the role of Rev1 in recruitment of Ung to S regions. We also enforced expression of Rev1 mutants in Rev1–/– B cells to rescue CSR in the absence of Rev1 catalytic activity. Finally, we generated double-knockout Rev1–/– Msh2–/– mice to ascertain whether Rev1 deficiency would virtually abrogate CSR when paired with Msh2 deficiency, as Ung deficiency does. Our experiments outline an important non-catalytic (scaffold) role of Rev1 in CSR; Rev1 recruits and/or stabilizes Ung to/on S regions and enhances Ung glycosylation activity.
RESULTS
If Rev1 is important in CSR, it should be expressed in germinal centers, where B cells undergo CSR at a high rate. Indeed, like AID, Rev1 protein was expressed in human tonsil germinal centers (Figure S1A). Accordingly, Rev1 expression was upregulated together with upregulation of critical CSR factors, including AID, Ung, HoxC4 and 14-3-3γ (real-time quantitative qRT-PCR) in mouse spleen and lymph nodes, which contain a large proportion of hypermutating and switching B cells but not in non-lymphoid organs (Figure S1B). Stimulation of B cells with bacterial lipopolysaccharides (LPS), LPS or CD154 plus IL-4, or LPS plus IFN-γ, which induce CSR to different Ig isotypes, upregulated Rev1 expression (Figure S1C); this was associated with upregulation of AID, Ung, HoxC4 and 14-3-3γ and CSR (not shown). Thus, Rev1 is induced by CSR-inducing stimuli and is upregulated in B cells undergoing CSR.
Rev1 deficiency impairs the class-switched antibody response to NP-CGG
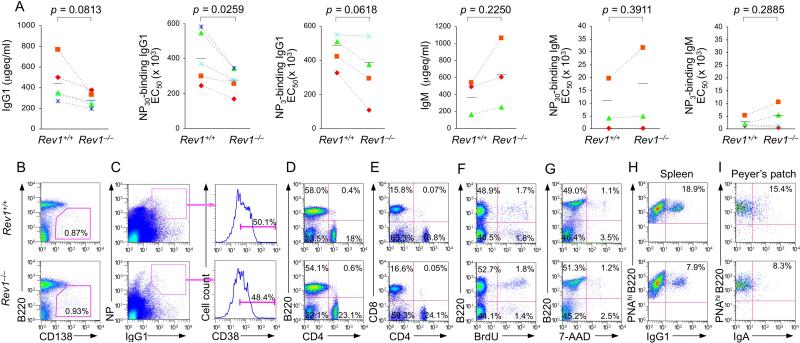
We used Rev1–/– mice (Jansen et al., 2006) to address the role of Rev1 in CSR in vivo. Rev1 plays an important role in CSR, then its deficiency should lead to a significant impairment of Ig class-switching in a specific antibody response. In non-intentionally immunized Rev1–/– mice, serum IgM titers were normal (not shown), but serum IgG1 and IgG3 concentrations were significantly lower than in their Rev1+/+ littermates and comparable to those of age- and sex-matched Ung-/- mice (Figure S2A). We injected five pairs of 8-10 week-old littermate Rev1–/– and Rev1+/+ mice with NP16-CGG and measured serum total and NP-binding IgM and IgG1 titers 7 days after boost injection. Rev1–/– mice showed reduced levels of total IgG1, NP30-binding IgG1 as well as high affinity NP3-binding IgG1, but not total IgM, NP30- or NP3-binding IgM, as compared to Rev1+/+ mice (Figure 1A).
Figure 1. Rev1 deficiency impairs the class-switched antibody response.
(A) Titers of circulating IgM and IgG1, NP30-binding IgM and IgG1, and high-affinity NP3-binding IgM and IgG1 in Rev1+/+ and Rev1–/– littermates, immunized with NP16-CGG and “boosted” 21 days later (n = 3-5 pairs of mice, each symbol represents an individual mouse), measured 7 days after the boost injection and expressed as μg equivalent/ml (μgeq/ml) or the number of dilutions needed to reach 50% of saturation binding (EC50). P values, paired t-test.
(B and C) Normal plasma cell and memory B cell differentiation in Rev1+/+ and Rev1–/– littermates 14 days after immunization with NP16-CGG (flow cytometry analysis). (B) B220loCD138+ (plasma) cells as percentage of total spleen cells. (C) CD38+ memory B cells as percentage of total NP-binding surface IgG1+ B cells. Cells were stained with PE-labeled anti-B220 mAb, FITC-PNA, PE-NP, APC-mAb to mouse IgG1 and PECy7-mAb to mouse CD38.
(D and E) Normal proportions of B cells, CD4+ T cells and CD8+ T cells in Rev1–/– mice. Spleen cells from Rev1+/+ and Rev1–/– littermates were stained for surface B220 and CD4 (D) or CD4 and CD8 (E) using PE-labeled anti-B220 mAb, FITC-labeled anti-CD4 mAb and PerCP-labeled anti-CD8 mAb.
(F) In vivo proliferation of B cells from spleens of 10-week-old Rev1+/+ and Rev1–/– mice immunized with NP16-CGG and injected 10 days later with BrdU (1 mg) twice within 16 hr. B cells were analyzed 4 hr after the final injection. Cells were stained with PE-labeled anti-B220 mAb; incorporated BrdU was detected by flow cytometry with APC-labeled mAb to BrdU.
(G) Normal viability (7-AAD–) of Rev1–/– B cells. Spleen cells from Rev1–/– and Rev1+/+ littermates were stained with 7-AAD and PE-labeled anti-B220 mAb.
(H and I) In vivo CSR in Rev1+/+ and Rev1–/– littermates. (H) Spleen cells from Rev1+/+ and Rev1–/– mice 14 days after immunization with NP16-CGG were stained with FITC-labeled PNA, PE-labeled anti-B220 mAb and APC-labeled anti-IgG1 mAb; class-switched germinal center B cells are IgG1+PNAhi B220+. (I) Cells from Peyer's patches of 12 week-old non-intentionally immunized Rev1+/+ and Rev1–/– mice were stained with Alexa Fluor® 647-labeled PNA, PE-labeled anti-B220 mAb and FITC-labeled anti-IgA mAb; class-switched B cells are sIgA+ PNAhiB220+. Data are from one representative of three independent experiments using B cells from three pairs of mice.
The defective anti-NP IgG1 antibody response in Rev1–/– mice did not result from alteration of plasma cell or NP-specific memory B cell differentiation, as the proportions of (plasma) B220loCD138+ cells and (memory) CD38hi B cells among NP-binding IgG1 B cells in Rev1–/– mice were comparable to those of their Rev1+/+ littermates (Figures 1B and 1C). In addition, it was not due to obvious defects in lymphoid differentiation, as in Rev1–/– mice, the size of the spleen and lymph nodes, and number and size of Peyer's patches were normal (not shown), so were the numbers of B and (CD4+ and CD8+) T cells (Figures 1D and 1E). In Rev1–/– mice, B cell proliferation and viability were also normal (Figures 1F and 1G). Further, after stimulation with LPS or LPS plus IL-4, Rev1–/– B lymphocytes showed cell cycle parameters and cell division rates comparable to Rev1+/+ cells (Figures S3A and S3B). Rev1–/– mice, however, showed reduced germinal center IgG1+ PNAhiB220+ B cells (by 58% in the spleen) and IgA+ PNAhiB220+ B cells (by 46% in Peyer's patches) (Figures 1H and 1I). Thus, in spite of normal B cell proliferation and normal plasma cell and memory B cell differentiation, Rev1 deficiency impairs a specific class-switched antibody response, likely because of an intrinsic impairment in the CSR machinery.
Rev1 deficiency impairs CSR
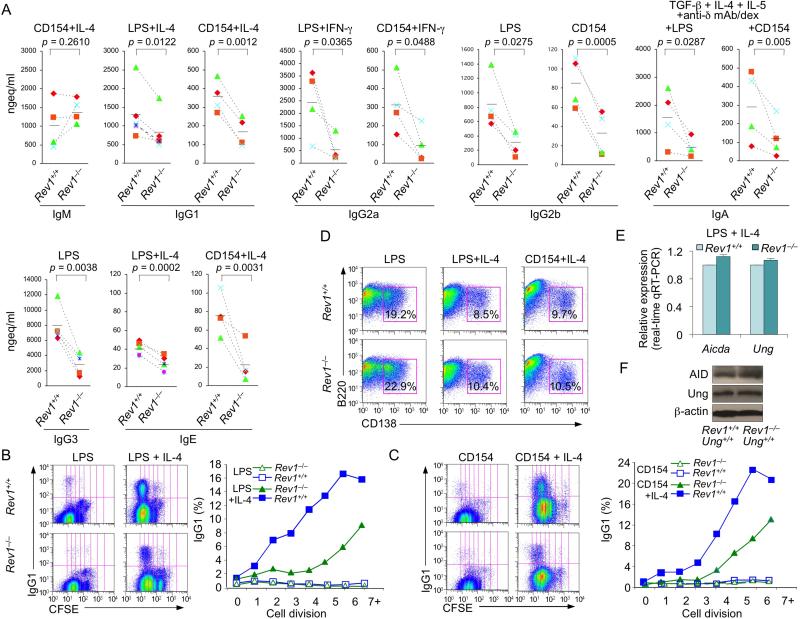
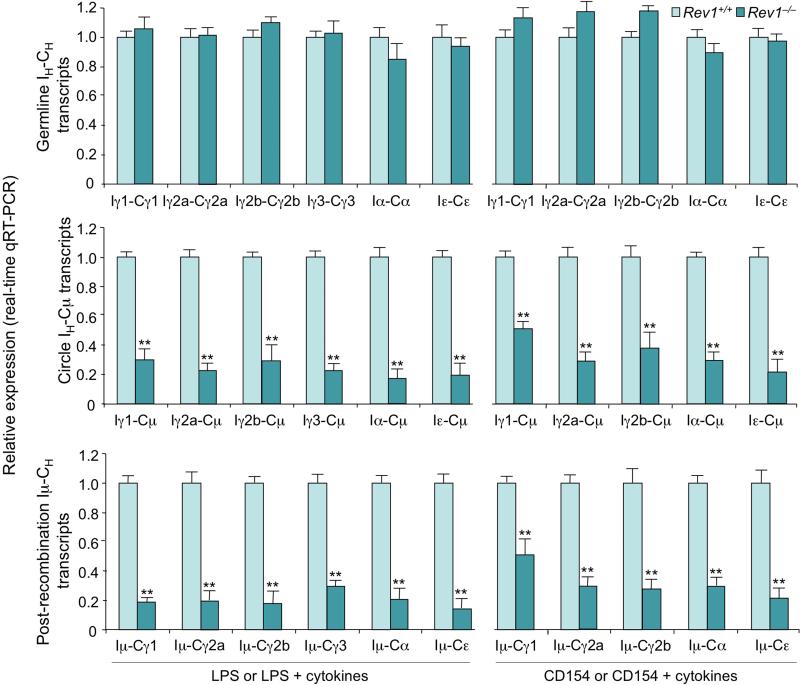
To further determine the impact of Rev1 deficiency on CSR, we stimulated Rev1–/– and Rev1+/+ (spleen) B cells with LPS (to induce switching to IgG2b and IgG3), CD154 (IgG2b), LPS or CD154 plus IL-4 (IgG1 and IgE), LPS or CD154 and IFN-γ (IgG2a) and LPS or CD154 plus TGF-β1, IL-5, IL-4 and anti–δ mAb/dex (IgA). After 4 days, switched surface Igγ1+, Igγ2a+, Igγ3+ and Igα+ B cells were fewer (by as much as 61%, 75%, 91% and 76%, respectively) among Rev1–/– than Rev1+/+ B cells (Figures S3C and S3D). Accordingly, secreted IgG1, IgG2a, IgG2b, IgG3, IgA and IgE, but not IgM were reduced in Rev1–/– B cell cultures (Figure 2A), despite Rev1–/– B cell normal cell cycle and proliferation (Figures S3A, S3B, 2B and 2C), and comparable numbers of CD138+B220lo plasma cells emerging from Rev1–/– and Rev1+/+ B cells (Figure 2D). Although profound, the CSR (to IgG1 and IgG3) impairment in Rev1–/– B cells did not match that of Ung–/– B cells, as assessed by switched surface Igγ1 and Igγ3 expression (Figure S2B). It, however, resulted in secreted IgG1 and IgG3 levels virtually as low as those of Ung–/– B cells in vivo and in vitro (IgG3) (Figure S2A and S2C). The reduced CSR in Rev1–/– B cells reflected decreased (by as much as 83%) circle IH-Cμ transcripts (byproducts of CSR) and post-recombination Iμ-CH transcripts (resulting from CSR, reduced by as much as 86%) in these B lymphocytes, in spite of the normal levels of germline IH-CH (Iγ1-Cγ1, Iγ2a-Cγ2a, Iγ2b-Cγ2b, Iγ3-Cγ3, Iα-Cα and Iε-Cε) transcripts, which precedes DNA recombination, and normal expression of Aicda and Ung transcripts as well as AID and Ung proteins (Figures 2E, 2F and 3). Thus, Rev1 deficiency impairs CSR to all class-switched Ig isotypes without affecting germline IH-CH transcription or AID and Ung expression.
Figure 2. Impaired CSR in Rev1–/– B cells.
(A) Ig titers in supernatants of cultures of Rev1+/+ and Rev1–/– B cells stimulated for 7 days with LPS (IgG2b and IgG3) or CD154 (IgG2b), or with LPS or CD154 plus IL-4 (IgM, IgG1 and IgE), IFN-γ (IgG2a) or TGF-β1, IL-4, IL-5 and anti–δ mAb/dex (IgA). P values, paired t-test. Data are from experiments with spleen B cells from three to five pairs of Rev1+/+ and Rev1–/– mice.
(B and C) Impaired CSR in Rev1–/– B cells is not due to alteration in cell proliferation. Proliferation of Rev1+/+ and Rev1–/– B cells labeled with cell division tracking fluorochrome CFSE and stimulated with (B) LPS or LPS plus IL-4, or (C) CD154 or CD154 plus IL-4. Depicted are sIgG1+ cells among B lymphocytes that had been stimulated for 4 days and completed the same number of divisions in Rev1+/+ and Rev1–/– B cells. Data are from one representative of three independent experiments.
(D) Normal plasma cell differentiation in Rev1–/– B cells. Rev1+/+ and Rev1–/– B cells were stimulated for 4 days with LPS, LPS plus IL-4 or CD154 plus IL-4, as analyzed by surface expression of B220 and CD138. Numbers in outlined areas indicate percent B220loCD138+ (plasma) cells among total cells. Data are from one representative of three independent experiments.
(E and F) Normal Aicda and Ung expression in Rev1–/– B cells. (E) Real-time qRT-PCR analysis of Aicda and Ung mRNAs in spleen Rev1+/+ and Rev1–/– B cells cultured for 60 hr with LPS plus IL-4. Expression is normalized to Cd79b expression and relative to the expression in Rev1+/+ B cells, set as 1. Data are from three independent experiments (mean and SEM). (F) Protein levels of Ung, AID and β-actin in cell lysates from spleen Rev1+/+ and Rev1–/– B cells cultured for 60 hr with LPS plus IL-4.
Figure 3. Decreased circle IH-Cμ and post-recombination Iμ-CH transcripts, but not germline IH-CH transcripts in Rev1–/– B cells.
Real-time qRT-PCR analysis of germline IH-CH transcripts (top), circle IH-Cμ transcripts (middle) and post-recombination Iμ-CH transcripts (bottom) in Rev1+/+ and Rev1–/– B cells cultured for 60 hr with LPS (Iγ2b-Cγ2b, Iγ3-Cγ3, Iγ2b-Cμ, Iγ3-Cμ, Iμ-Cγ2b and Iμ-Cγ3), CD154 (Iγ2b-Cγ2b, Iγ2b-Cμ and Iμ-Cγ2b), or LPS or CD154 plus IL-4 (Iγ1-Cγ1, Iε-Cε, Iγ1-Cμ, Iε-Cμ, Iμ-Cγ1 and Iμ-Cε), IFN-γ (Iγ2a-Cγ2a, Iγ2a-Cμ and Iμ-Cγ2a), or TGF-β1, IL-4, IL-5 and anti–δ mAb/dex (Iα-Cα, Iα-Cμ and Iμ-Cα). Expression of germline IH-CH, circle IH-Cγ and post-recombination Iμ-CH transcripts was normalized to Cd79b expression and is depicted as relative to expression in Rev1+/+ B cells, set as 1. **p < 0.01, paired t-test. Data are from three independent experiments (mean and SEM).
Rev1–/– B cells show altered mutation spectrum in S region DNA
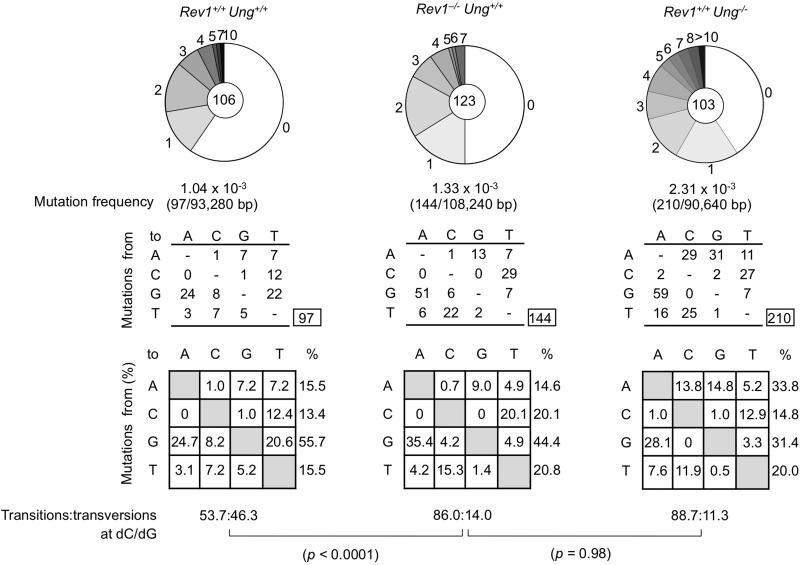
Mutations arise in S region DNA in B cells undergoing CSR (Maul and Gearhart, 2010). To ascertain whether defective CSR in Rev1–/–Ung+/+ B cells was associated with altered mutations in S regions, we sequenced the upstream region of Sμ DNA (nucleotides 4289-5168; GenBank accession no. J0040), which includes the sequence analyzed by others (Rada et al., 2002), and compared the mutations with those in B cells from Rev1+/+ Ung+/+ littermates and Rev1+/+ Ung–/– mice. In B cells from Rev1–/–Ung+/+ mice, the mutation frequency in Sμ was increased by almost 30% (p = 0.003) as compared to their Rev1+/+Ung+/+ littermates. An even greater increase in mutation frequency was detected in B cells from Rev1+/+Ung–/– mice, possibly due to a more severe defect in (error-free) DNA repair associated with Ung deficiency and/or simply reflecting inter-strain variations. Remarkably, while dC→dT or dG→dA transitions accounted for 53.7% of the mutations at dC/dG pairs in Rev1+/+Ung+/+ B cells, they accounted for 86% of the dC/dG mutations in Rev1–/–Ung+/+ B cells (p < 0.0001) (Figure 4), a phenotype resembling that of Rev1+/+Ung–/– B cells (p = 0.98), suggesting that Rev1 and Ung function in the same CSR pathway.
Figure 4. Altered spectrum of somatic mutations at dC/dG of Sμ region in Rev1–/– Ung+/+ mice.
Sμ region DNA was amplified from Peyer's patch B cells from three 12-week-old Rev1+/+Ung+/+ and three Rev1–/– Ung+/+ littermates and three age-matched Rev1+/+Ung–/– mice. Pie charts depict the proportions of sequences that carry 1, 2, 3, etc. mutations over the 880 bp Sμ DNA analyzed. The numbers of sequences analyzed are at the center of the pies. Numbers and nature of independent mutational events scored. Compilations, with the numbers indicating percentages of all mutations scored in the pool of the target sequences. Below the compilations, the ratio of transition:transversion substitutions at dC/dG is indicated. P values, Chi-squared test.
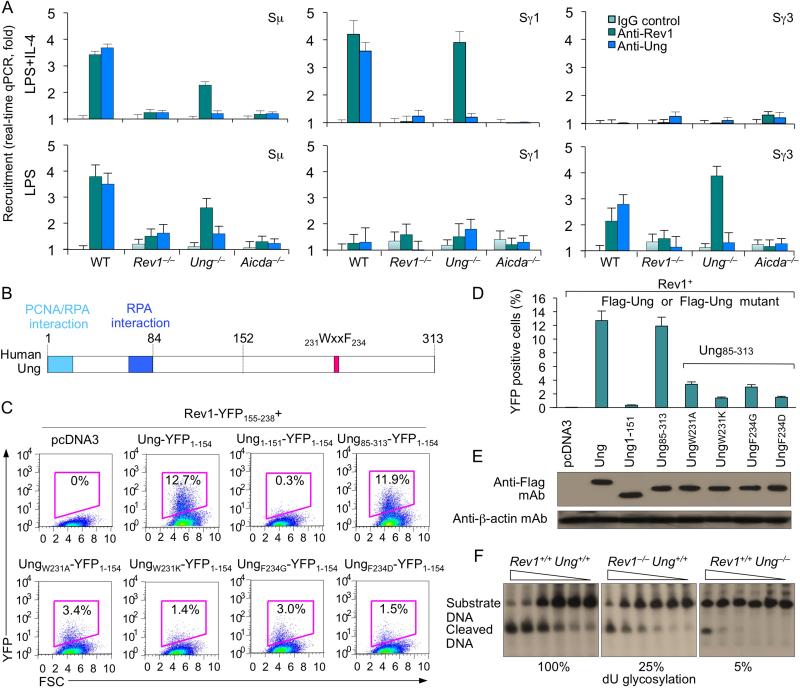
Rev1 is recruited to S regions (in an AID-dependent fashion) to recruit/stabilize Ung
We further addressed the role of Rev1 in CSR by analyzing its recruitment to S regions. In B cells stimulated with LPS plus IL-4, which induces CSR to IgG1, but not IgG3, ChIP assays showed both Rev1 and Ung bound to Sγ1, but not to Sγ3 DNA; conversely, in B cells stimulated with LPS, which induces CSR to IgG3 but not IgG1, Rev1 and Ung were recruited to Sγ3, but not to Sγ1. Both Rev1 and Ung were recruited to Sμ. Like Ung (Ranjit et al., 2011), Rev1 recruitment to S region DNA was abrogated in Aicda–/– B cells, indicating that Rev1 is recruited to S regions in response to AID-mediated lesions in DNA (Figure 5A). To determine whether Rev1 and Ung binding to S region DNA is interdependent, we performed ChIP assays utilizing anti-Rev1 and anti-Ung Abs in both Rev1–/– and Ung–/– B cells. Ung binding to Sγ1 and Sγ3 DNA was impaired in Rev1–/– B cells stimulated with LPS plus IL-4 and LPS, respectively, while Rev1 recruitment to Sγ1 and Sγ3 DNA was not affected by Ung deficiency (Ung–/– B cells), showing that the occupancy of S region DNA by Ung is dependent on Rev1, but occupancy of S region DNA by Rev1 is independent of Ung. Thus, Rev1 plays a role in recruiting and/or stabilizing Ung to/on S region DNA.
Figure 5. Rev1 is recruited to S regions in an AID-dependent fashion, directly interacts with Ung to enhance Ung catalytic activity.
(A) Rev1 deficiency impairs Ung binding to S region DNA. Wild-type, Rev1–/–, Ung–/– and Aicda–/– B cells were stimulated with LPS or LPS plus IL-4 for 48 hr. Cross-linked chromatin was precipitated using rabbit Abs specific to Rev1, Ung or preimmune control rabbit IgG. The precipitated Sγ1 and Sγ3 DNA were quantified by real-time qPCR. Data are from three experiments, each in triplicate (mean and SEM).
(B) Schematic of human Ung protein. The PCNA/RPA interaction domain, RPA interaction domain and 231WxxF234 motif are indicated.
(C) Rev1 directly interacts with Ung. BiFC analysis of human Rev1 interaction with human Ung or Ung mutants. HeLa cells were transfected with pcDNA3.1 vector or co-transfected with HA-huRev1-YFP155-238 and Flag-Ung-YFP1-154, Flag-Ung1-151-YFP1-154, Flag-Ung85-313-YFP1-154, or Ung WxxF mutant constructs Flag-UngW231A-YFP1-154, Flag-UngF234G-YFP1-154, or Flag-UngF234Q-YFP1-154, which were generated using the Rev1 interaction-proficient Ung85-313 N-deletion mutant as template. Cells were stained with 7-AAD to gate for viability. Background fluorescence, defined as the random low-level fluorescence of Flag-YFP1-154 and HA-YFP155-238, was gated out from total fluorescence to determine true positive protein-protein interactions. Data are from one representative of three experiments.
(D) Interaction of Rev1 with wild-type and different Ung mutants, as measured by specific BiFC assay.
(E) Expression levels of Flag-Ung-YFP1-154, Flag-Ung1-151-YFP1-154, Flag-Ung85-313-YFP1-154, or Ung WxxF mutant Flag-UngW231A-YFP1-154, Flag-UngF234G-YFP1-154 or Flag-UngF234Q-YFP1-154 fusion proteins in transfected cells (C), as analyzed by immunoblotting using anti-Flag mAb.
(F) Ung dU glycosylation activity in Rev1+/+Ung+/+, Rev1–/–Ung+/+ and Rev1+/+Ung–/– B cells stimulated with LPS plus IL-4 for 60 hr, as measured by incubating clarified whole-cell extract (5.0, 2.5, 1.25, 0.625, 0.31 or 0.16 μg protein) with a [32P]-labeled double-stranded oligonucleotide containing a single dU:dG mispair. The reaction products were resolved in 15% TBE-urea polyacrylamide gels. The presence of the cleaved DNA reflects glycosylation activity.
Rev1 directly interacts with Ung
If Rev1 is important for Ung recruitment/stabilization to/on S region DNA, then it must interact with Ung. To establish whether Rev1 can directly interact with Ung, rather than through a third party molecule, we adapted an ad hoc BiFC assay (Xu et al., 2010). We divided super-enhanced yellow fluorescent protein (sEYFP) coding sequence into two complementary moieties, one encoding the N-terminal 154 residues (sEYFP1–154), the other encoding the C-terminal 84 residues (sEYFP155–238); these were fused with Flag-tagged human Ung (Flag-Ungs-EYFP1–154) and influenza hemagglutinin (HA)-tagged human Rev1 (HA-Rev1-sEYFP155–238), respectively (Figures 5B). Once expressed, the two sEYFP moieties complemented each other and gave off strong yellow fluorescence, as a result of direct juxtaposition between Rev1 and Ung - had Rev1 and Ung interacted through a third “spacer” molecule, this would have prevented sEYFP1–154 and sEYFP155–238 from complementing each other and, therefore, giving off fluorescence. We then tested whether the interaction between Rev1 and Ung was through the Ung N-terminal region, which contains the PCNA/RPA interaction domain and RPA interaction domain, or the Ung C-terminal region, which contains the 231WxxF234 motif. BiFC assays involving human Rev1 and human Ung mutants containing 151 N-terminal residues (Flag–Ung1–151–sEYFP1–154) or 229 C-terminal residues (Flag–Ung85–313–sEYFP1–154) showed that Rev1 interacted with Ung through the Ung C-terminal region but not the N-terminal region (Figures 5C and 5D). Next, we determined whether the Ung 231WxxF234 motif was involved in the interaction with Rev1 - Ung mutants of the WxxF motif lacking the N-terminal region failed to restore CSR in Ung–/– B cells (Begum et al., 2009). We performed BiFC assays using human Ung molecules lacking N-terminal 1-84 amino acid residues and bearing the W231A, W231K, F234G or F234D mutation in 231WxxF234 motif. Each of these mutations reduced the ability of mouse UngΔ1-86 to mediate CSR in Ung–/– B cells, without affecting Ung protein levels (Begum et al., 2009). Accordingly, none of the four WxxF mutations (W231A, W231K, F234G or F234D) altered human Ung protein levels, but each impaired Ung interaction with Rev1 (Figures 5D and 5E). Thus, Rev1 directly interacts with Ung through the Ung 231WxxF234 motif to mediate CSR.
Rev1 deficiency impairs B cell Ung activity
To investigate whether in addition to recruiting/stabilizing Ung to/on S region DNA, Rev1 also enhances Ung function, we analyzed, using a double-strand DNA substrate containing an internal dU:dG mispair, the DNA-dU glycosylation activity of extracts from Rev1+/+Ung+/+, Rev1–/–Ung+/+ or Rev1+/+Ung–/– B cells that had been stimulated with LPS plus IL-4. While Rev1+/+Ung+/+ B cell extracts displayed abundant DNA-dU glycosylation activity, extracts from Rev1–/–Ung+/+ B cells, in which Ung mRNA and protein levels were comparable to those in Rev1+/+Ung+/+ B cells (Figures 2E and 2F), showed a 75% reduction of DNA-dU glycosylation, as compared to Rev1+/+Ung+/+ B cells (Figure 4F). Rev1+/+Ung–/– B cell extracts showed only minimal residual DNA-dU glycosylation (5%, possibly due to the expression of DNA glycosylases other than Ung, such as Smug1). Thus, Rev1 enables or enhances Ung DNA-dU glycosylation function in B cells.
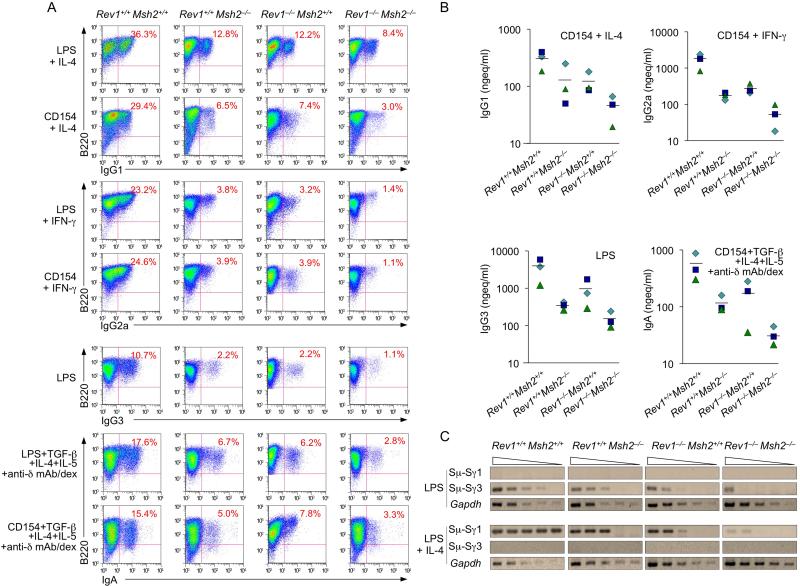
Double deficiency in Rev1 and Msh2 further impairs CSR
We reasoned that if Rev1 is important in recruitment/stabilization of Ung to/on S regions and critically enhances the activity of this enzyme, then the CSR defect brought about by Rev1 deficiency should compound that due to Msh2 deficiency, much in the same way that Ung deficiency abrogates CSR when combined with Msh2 deficiency, as in Ung–/–Msh2–/– B cells, in which both Ung-dependent and Msh2-dependent CSR pathways are aborted (Rada et al., 2004). We crossbred Rev1–/– with Msh2–/– mice to construct double knockout Rev1–/–Msh2–/– mice. In Rev1–/–Msh2–/– B cells, CSR to IgG1, IgG3, IgG2a or IgA was reduced by up to 96% as compared to Rev1+/+Msh2+/+ and 50-60% as compared to single knockout Rev1–/–Msh2+/+ or Rev1+/+Msh2–/– B cells, as shown by surface Ig expression, secreted IgG1, IgG2a, IgG3 and IgA, as well as recombinant Sμ-Sγ1 and Sμ-Sγ3 DNA detected by digestion-circularization PCR (DC-PCR) (Figure 6). Accordingly, in Rev1–/–Msh2–/– B cells, circle IH-Cμ and post-recombination Iμ-CH transcripts were decreased by as much as 98% (Iγ2a-Cμ in B cells stimulated with LPS and INF-γ) and 93% (Iμ-Cγ2a in B cells stimulated by LPS plus INF-γ), respectively, as compared to Rev1+/+ Msh2+/+ B cells (Figure S4). They were also reduced as compared to Rev1–/– Msh2+/+ B cells (by 95% and 77%, respectively) and Rev1+/+ Msh2–/– B cells (by 91% and 65%, respectively). Thus, double Rev1/Msh2 deficiency further impairs CSR due to Msh2 deficiency, suggesting that Rev1 and Ung function in the same CSR pathway.
Figure 6. Double Rev1 and Msh2 deficiency further reduces CSR.
Rev1+/+Msh2+/+, Rev1–/–Msh2+/+, Rev1+/+Msh2–/– and Rev1–/–Msh2–/– B cells were stimulated with LPS alone (for IgG3), LPS or CD154 plus IL-4 (for IgG1), IFN-γ (for IgG2a), or TGF-β1, IL-4, IL-5 and anti–δ mAb/dex (for IgA).
(A) After 4 days of culture, the cells were analyzed for surface expression of B220 and IgG1, IgG2a, IgG3 or IgA by flow cytometry. Data are form one representative of three independent experiments.
(B) After 7 days of culture, the supernatants from Rev1+/+Msh2+/+, Rev1–/–Msh2+/+, Rev1+/+Msh2–/– and Rev1–/– Msh2–/– B cells stimulated with LPS alone or CD154 plus cytokines were collected and analyzed for concentrations of different Ig isotypes. Data were derived using B cells from three sets, each of four Rev1+/+Msh2+/+, Rev1–/–Msh2+/+, Rev1+/+Msh2–/– and Rev1–/–Msh2–/– mice.
(C) Recombinant Sμ-Sγ1 or Sμ-Sγ3 DNAs analyzed by DC-PCR using serially twofold diluted HindIII digested and T4 DNA ligase-ligated genomic DNA from Rev1+/+Msh2+/+, Rev1–/–Msh2+/+, Rev1+/+Msh2-/- or Rev1–/–Msh2–/– B cells stimulated with LPS or LPS plus IL-4 for 4 days. Gapdh gene was used as a control for ligation and DNA loading. Data are from one representative of three independent experiments.
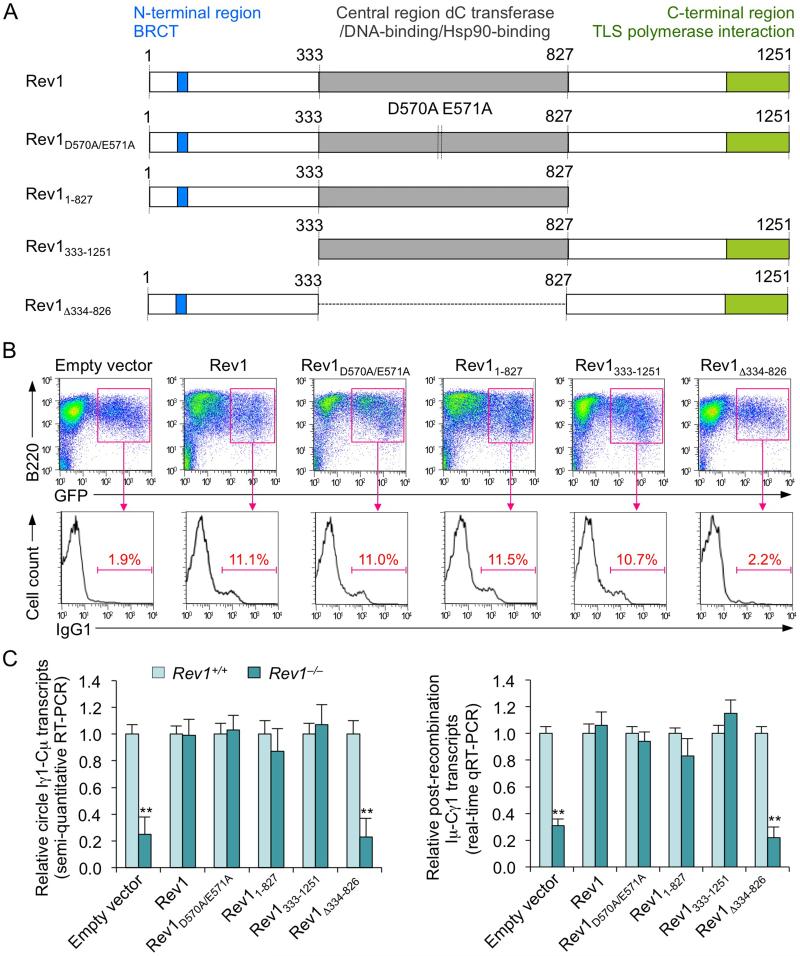
Enforced expression of catalytically inactive Rev1 in Rev1–/– B cells rescues CSR
Our experiments strongly suggested that defective CSR in Rev1–/– B cells was largely due to an impairment in recruitment/stabilization of Ung, leading us to hypothesize that Rev1 functions in CSR not as an enzyme, but rather as a structural (scaffold) element. To test our hypothesis, we enforced expression of five different human Rev1 cDNAs in Rev1–/– B cells using constructs based on the S-003 retroviral expression vector (Sayegh et al., 2003): (i) wild-type Rev1, (ii) Rev1D570A/E571A (a mutant with two point-mutations in the central region, inactivating Rev1 catalytic activity), (iii) Rev11-827 (a C-terminal region truncation mutant, retaining the central and N-terminal regions), (iv) Rev1333-1251 (a N-terminal region truncation mutant, retaining the central and C-terminal regions) or (v) Rev1Δ334-826 (a truncation mutant lacking central region and, therefore, the DNA-binding domain) (Figure 7A). Enforced expression of Rev1D570A/E571A, Rev11-827 or Rev1333-1251 in Rev1–/– B cells rescued in every case CSR to IgG1 at the level of wild-type Rev1, while enforced expression of Rev1Δ334-826 did not, as assessed by IgG1 surface fluorescence analysis and quantitation of circle Iγ1-Cμ and post-recombination Iμ-Cγ1 transcripts (Figures 7B and 7C). Thus, the Rev1 scaffold function, mediated by the central region of the molecule, but not Rev1 enzymatic activity is required for CSR.
Figure 7. Enforced expression of catalytically inactive Rev1 mutant, N-terminal region truction or C-terminal region trunction Rev1 mutant rescues CSR in Rev1–/– B cells.
CSR to IgG1 in Rev1+/+ and Rev1–/– B cells transduced with empty S-003 retrovirus or S-003 retrovirus expressing wild-type human Rev1 (includes the N-terminal region containing the BRCT domain; the central region containing the D570E571 motif critical for catalytic activity, the DNA-binding motif and the Hsp90-binding motif, and the C-terminal region containing the TLS polymerase interaction domain), catalytically inactive Rev1D570A/E571A mutant (containing two point-mutations at the D570E571 motif inactivating Rev1 catalytic activity), C-terminal region deletion Rev11-827 mutant (retaining central and N-terminal regions), N-terminal region deletion Rev1333-1251 mutant (retaining central and C-terminal regions) and central region deletion Rev1Δ334-826 mutant (retaining N-terminal and C-terminal regions). Rev1+/+ and Rev1–/– B cells were activated with LPS for 12 hr before transduction with retrovirus, then cultured for 48 hr with LPS plus IL-4.
(A) Schematic representation of human Rev1 and Rev1 mutants encoded by retroviral constructs. The N-terminal, central and C-terminal regions, BRCT domain and TLS polymerase interaction domain are indicated.
(B) CSR assessed by surface IgG1 expression in transduced (B220+GFP+) Rev1-/- B cells. Density plots show transduced B cells as a distinct (GFP+) population. Histograms show the distribution of surface IgG1+ B cells among B220+GFP+ B cells. Data are from one representative of four independent experiments.
(C) GFP+ B cells were sorted by FACS for analysis of circle Iγ1-Cμ (semi-quantitative RT-PCR) and Iμ-Cγ1 (real-time qRT-PCR) transcripts. Expression is normalized to Cd79b expression and presented as ratio of expression in Rev1–/– to Rev1+/+ B cells (set as 1). **p < 0.001, paired t-test. Data are from four independent experiments (mean and SEM).
DISCUSSION
CSR entails (i) AID deamination of dCs in the IgH locus S regions to yield dUs and dU glycosylation by Ung leading to DSBs, and (ii) DSB repair by the combined intervention of DNA replication and repair factors. As a highly regulated biological process, CSR is mediated by a multi-component complex, whose assembly would be made possible by scaffold proteins (Xu et al., 2012). As exemplified by our previous findings on 14-3-3 adaptors (Xu et al., 2010), such scaffold proteins can simultaneously interact with DNA and client proteins through different subunits or domains to greatly increase the specificity and efficiency of the CSR macromolecular machinery. Here we show that the Rev1 TLS polymerase plays an important scaffold role in CSR. Like AID and other critical CSR factors, such as Ung, HoxC4 and 14-3-3, it is induced by T-dependent (CD154:C40 engagement) and T-independent (TLR engagement) CSR-inducing stimuli and is expressed in a B cell differentiation stage-specific fashion (germinal center B cells). Rev1 is recruited to S region DNA in an AID-dependent fashion, directly interacts with Ung, recruits/stabilizes Ung to/on S regions and enhances Ung-mediated DNA dU glycosylation. Thus, the important role of Rev1 in CSR is an expression of the (non-enzymatic) scaffold function of this TLS polymerase.
As a Y family TLS polymerase, Rev1 possesses a unique and distributive dC transferase catalytic activity (Nair et al., 2005; Prakash et al., 2005; Zhang et al., 2002). Owing to its ability to interact with PCNA, ubiquitin and TLS polymerases η, ι, κ and ζ, Rev1 would recruit other TLS polymerases, thereby displaying multiple functions in different DNA repair processes (Hicks et al., 2010; Sharma et al., 2011; Xie et al., 2010). Like TLS polymerases η, θ and ζ, Rev1 is involved in SHM by bypassing DNA lesions through its catalytic activity (Jansen et al., 2006; Masuda et al., 2009). Rev1 deficiency has been associated with a strand-biased defect in dC → dG transversions in hypermutating V(D)J genes, suggesting that during SHM, Rev1 preferentially inserts dCs across from dUs and/or abasic sites introduced by AID and Ung intervention (Jansen et al., 2006; Masuda et al., 2009). As we have shown here, despite normal expression of Ung, Rev1–/– B cells greatly increased dC/dG transition mutations in S regions (point-mutations are introduced into the recombining S regions by the CSR machinery), a pattern similar to that of Ung–/– B cells, suggesting that Rev1 and Ung participate in the same CSR pathway. Thus, in the absence of Rev1 and, therefore, decreased Ung recruitment, dU residues in S regions would be replicated over, yielding dC/dG transitions, in the same way that they are in Ung-/- B cells.
The distinct scaffold and catalytic functions of Rev1 in CSR and V(D)J SHM, respectively, were also suggested by the observation that mice expressing a catalytically inactive Rev1 mutant displayed an altered mutation spectrum in IgH JH-iEμ region DNA, but B cells from these mice revealed normal CSR in vitro (Masuda et al., 2009). The two different Rev1 functions (scaffolding and catalytic) are consistent with the divergent mechanisms underpinning CSR and V(D)J SHM, which are mediated by separate AID domains (Shinkura et al., 2004), and the different requirements for the induction of these two processes (Zan et al., 1999; Zan et al., 2000; Zan et al., 2003). The Rev1 distinct functions are mediated by discrete regions (Guo et al., 2006; Mayca Pozo et al., 2011): the N-terminal region contains the BRCT domain; the central region comprises the DNA-binding, Hsp90-binding and ubiquitin-binding motif, in addition to the DE motif (D570E571 in human Rev1), which is critical for catalytic activity; and the C-terminal region includes the PCNA interaction and TLS polymerase interaction domains. As we showed here, enforced expression of our N-terminal and C-terminal Rev1 truncation mutants in Rev1–/– B cells (both truncation mutants included the full central region) restored CSR to levels comparable to that of Rev1+/+ B cells, while the construct lacking the central (DNA-binding) region did not. Rescue of CSR was not dependent on the Rev1 DE motif within this central region, strongly arguing for a non-catalytic (scaffold) function for Rev1 in CSR. Further, our BiFC assays unequivocally showed that Rev1 directly interacts with Ung and does so through the conserved Ung WxxF motif. This motif was found to be important for CSR, and was hypothesized to mediate the interaction of Ung with a putative CSR co-factor (Begum et al., 2009), which we surmise here to be Rev1.
As we have shown here, Rev1 promotes Ung recruitment/stabilization to/on S region DNA and dU glycosylation in switching B cells; Rev1 localization to S regions required AID but not Ung, while Ung recruitment to S regions required Rev1. This novel Rev1 function would be made possible by the ability of Rev1 to recognize and bind to dU-containing DNA, as suggested by Rev1-mediated insertion of dC across from dU in DNA synthesis (Zhang et al., 2002). Accordingly, our experiments proved that the central region of Rev1, which contains the DNA-binding domain, is critical for Rev1 function in CSR. In S regions, Rev1 would be recruited by DNA-bound AID, either directly, as suggested by our preliminary modeling (http://hexserver.loria.fr) (Pone and Casali, unpublished), or indirectly, through, for instance, Hsp90. Indeed, Hsp90 can bind to and stabilize AID in CSR (Orthwein et al., 2010) and can specifically bind to Rev1 to yield stable and/or functional form(s) of Rev1 (Mayca Pozo et al., 2011). In contrast to direct Ung-Rev1 interaction, Ung is unable to bind AID (Ranjit et al., 2011), further suggesting that the putative stabilization of Ung on S region DNA by AID occurs, at least in part, through Rev1. Thus, Rev1 would be recruited by AID to S region DNA and it would directly interacts with both dU and Ung, thereby forming a complex within which Ung stably associates with and glycosylates dU for CSR to unfold.
Rev1–/– B cells displayed normal cell cycle and proliferation, indicating that the CSR reduction reflected an inherent defect of the CSR machinery. The normal levels of germline IH-CH transcripts in Rev1–/– B cells suggested that the defect in CSR was not due to alterations in chromatin accessibility at S regions. As Rev1 can associate with TLS DNA polymerase ζ, which consists of the catalytic Rev3 and the regulatory Rev7 subunits, to facilitate polymerase ζ function, we could not rule out the possibility that Rev1 also contributes to CSR by promoting the activity of Rev3, which has been suggested to catalyze fill-in reactions in the course of CSR-associated DSB repair by NHEJ (Schenten et al., 2009). Nevertheless, our demonstration of: (i) further reduced CSR in double knockout Rev1–/–Msh2–/– B cells as compared to Rev1–/–Msh2+/+ B cells; (ii) predominance of dC/dG transitions at S region mutations in Rev1–/– B cells, a pattern evocative of that of Ung–/– B cells, but different from that of human and mouse B cells deficient in DNA polymerase η (loss of dA/dT mutations) (Neuberger and Rada, 2007), a TLS polymerase suggested to be recruited to Ig V(D)J region DNA by Rev1 during SHM (Hicks et al., 2010); and (iii) direct interaction of Rev1 with Ung, all support the notion that Rev1 mediates CSR mainly through its interaction with Ung. The residual CSR we detected in the Rev1–/–Msh2–/– B cells might reflect a residual recruitment/stabilization to/on S region DNA that was independent of Rev1. Indeed, as we have shown here, overall CSR in Rev1–/– B cells was significantly impaired, but not virtually abrogated, as it was in Ung-/- B cells. Accordingly, DNA dU glycosylation activity of Rev1–/–Ung+/+ B cell extracts was 25% of that in Rev1++Ung+/+ and 5% of that in Rev1+/+Ung–/– B cells.
In CSR, both generation of DSBs and DSB repair entail multiple sequential enzymatic reactions on the recombining S region DNA. They would depend on the formation of a macromolecular complex, within which (non-enzymatic) scaffold elements, such as 14-3-3 and RPA, and enzymatic elements with scaffold functions, such as Rev1 and, possibly, AID stabilize themselves and other CSR factors on S regions - after dC deamination, AID likely persists on the IgH locus (Larijani et al., 2007) and acts a scaffold for other factors, including Rev1(Xu et al., 2012). Rev1 functions as a legitimate scaffold protein in CSR: it comprises different protein-interaction domains (for Ung and possibly AID) and coordinates the assembly of at least one relevant molecular component, Ung, in a macromolecular complex to increase the efficiency of a specific biological process: dU glycosylation. Accordingly, RPA also directly interacts with Ung, although deletion of the RPA-binding domains in the N terminus of Ung does not impair CSR (Begum et al., 2009). Ung molecules with both truncation of the N-terminus and mutations of the WxxF motif, which as we show here is critical for Rev1 interaction, or of D145 and N204 in the catalytic center, failed to mediate CSR (Begum et al., 2009), suggesting that both RPA and Rev1 are required for full function of Ung in CSR. Thus, Rev1 together with Ung likely contributes to a DNA-protein macromolecular complex that also includes AID, 14-3-3 and RPA, and that is central to generation and resolution of S region DSBs.
EXPERIMENTAL PROCEDURES
Mice
Rev1–/– mice were generated by Dr. Niels de Wind (Leiden University, Netherlands) by deleting Rev1 exon 10, which encodes residues 557-609 including the conserved S(C/I)DE amino acid sequence essential for the catalytic activity (Jansen et al., 2006). Both the catalytic domain and the C-terminal domain, which interacts with other TLS polymerases, are absent in these mice (Jansen et al., 2006). Cells from Rev1–/– mice express only shortened Rev1 transcripts and no Rev1 protein. Msh2–/– mice (Nikitin et al., 2002) were obtained from Dr. Wen-Hwa Lee (University of California, Irvine). Ung–/– mice (Nilsen et al., 2000) were obtained from Dr. T. Lindahl (Cancer Research UK London Research Institute, South Mimms, UK). Rev1–/– and Ung–/– mice were on mixed C57BL/6 - 129 background, Msh2–/– mice were on C57BL/6 background. Double knockout Rev1–/–Msh2–/– mice were generated by us by crossbreeding Rev1–/– with Msh2–/– mice. The Institutional Animal Care and Use Committee (IACUC) of University of California, Irvine, approved all animal experiments.
NP16-CGG immunization and titration of total and NP-binding IgM and IgG1
Rev1–/– and Rev1+/+ mice (8-10 week-old) were immunized with 100 μg of NP16-CGG (16 molecules of 4-hydroxy-3-nitrophenyl acetyl coupled to one molecule of chicken γ-globulin (Biosearch Technologies) in Imject® alum (Pierce) for B cell and anti-NP antibody studies, as we described (Park et al., 2009; White et al., 2011).
B and T cells
B cells (B220+), CD4+ and CD8+ T cells, dead B cells, germinal center (PNAhi) B cells, plasma cells (B220lo CD138+) and NP-binding CD38hi IgG1+memory B cells were identified and analyzed in single cell suspensions using a FACSCalibur ™ flow cytometer (BD Biosciences) and specific mAbs. Germinal center (PNAhi B220+) B cells expressing IgG1 or IgA In vivo were determined by flow cytometry (Mai et al., 2010; Park et al., 2009). Cell cycle of in vitro stimulated B cells was determined by analyzing the correlated expression of total DNA (indicated by 7-AAD intercalation) and incorporated BrdU levels (Zan et al., 2003) using BD Pharmingen™ APC BrdU Flow Kit (BD Biosciences) following manufacturer's instructions. Proliferation of in vitro stimulated B cells was analyzed using the CellTrace™ CFSE Cell Proliferation Kit (Molecular Probes). Proliferation of B cells in mice immunized with NP16-CGG was measured by BrdU incorporation.
CSR
B cells were stimulated with LPS (5 μg/ml), LPS (5 μg/ml) or CD154-expressing membrane fragments of baculovirus-infected Sf21 insect cells (“CD154” here) plus appropriate cytokine(s) to induce CSR to different isotypes (Park et al., 2009; Pone et al., 2012a), and then analyzed for surface Ig. Titers of IgG1, IgG2a, IgG2b, IgG3, IgA and IgE in culture supernatants of in vitro stimulated B cells were measured using specific ELISAs, as we described (Park et al., 2009; White et al., 2011).
Quantitative real-time RT-PCR
Germline IH-CH, circle IH-Cμ, post-recombination Iμ-CH, HoxC4, Aicda, Rev1, Ung, and 14-3-3γ, Cd79b and Gapdh transcripts were analyzed by real-time quantitative RT-PCR (qRT-PCR) using specific primers as described previously (Park et al., 2009; White et al., 2011) or listed in the Supplemental Information.
DC-PCR
B cells were stimulated with LPS or LPS plus IL-4 for four days for isolation of genomic DNA and detection of recombinant Sμ-Sγ1 and Sμ-Sγ3 DNA by DC-PCR (Wang et al., 2009).
Somatic mutations in Sμ region DNA
Peyer's patch B cells switch and accumulate mutations in Sμ region at a high rate (Delbos et al., 2005). Somatic mutations in Sμ regions were analyzed using genomic DNA prepared from Peyer's patch B cells from Rev1+/+ Ung+/+ and Rev1–/– Ung+/+ littermates and age matched Rev1+/+ Ung–/– mice.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as we described (Kim et al., 2004; Park et al., 2009; Schaffer et al., 2003; Xu et al., 2010) using rabbit Abs to Rev1 (H-300; Santa Cruz Biotechnology) and Ung (FL-313; Santa Cruz Biotechnology).
Bifluorescence complementation (BiFC)
The BiFC assay to analyze the direct interaction of Rev1 with Ung was adapted from our reported method (Xu et al., 2010). Briefly, 5 × 105 HeLa cells were transiently transfected using Lipofectamine™ (Invitrogen) with plasmids (0.5 μg each) expressing HA-(human) Rev1-YFP155-238 and Flag-Ung-YFP1-154, Flag-Ung1-151-YFP1-154, or Flag-Ung85-313-YFP1-154 proteins, or Ung WxxF mutants Flag-UngW231A-YFP1-154, Flag-UngF234G-YFP1-154, or Flag-UngF234Q-YFP1-154, which were generated using the Rev1 interaction-proficient Ung85-313 truncation mutant (this human Ung molecule lacking residues 1-84 is equivalents to the mouse Ung lacking residues 1-86) as the starting construct, respectively. Cells were washed 24 hr after transfection, stained with 7-AAD and analyzed by flow cytometry.
DNA-dU glycosylation
Ung activity was measured using a 5′-[32P]-labeled double-stranded oligonucleotide with an internal dU:dG mispair as previous reported (Di Noia and Neuberger, 2002; Zan and Casali, 2008).
Rev1 and Rev1 mutant constructs and enforced expression
The human Rev1 cDNA was prepared from mRNA of human peripheral blood mononuclear cells. The catalytically inactive mutant Rev1D570A/E571A, which contained two point-mutations at the D570E571 motif (D570A and E571A) - human Rev1 D570E571 is equivalent to mouse Rev1 D568E569 (Masuda et al., 2009; Ross et al., 2005) - was generated by site-directed mutagenesis. The constructs encoding the C-terminal deletion mutant Rev11-827 (retaining the central and N-terminal regions), the N-terminal deletion mutant Rev1333-1251 (retaining the central and C-terminal regions) and the central region deletion mutant Rev1Δ334-826 (retaining the N-terminal and C-terminal regions) were generated by PCR. Wild-type and mutant Rev1 cDNAs were cloned into the S-003 retroviral expression vector (Sayegh et al., 2003) (kindly from Dr. C. Murre, University of California, San Diego). For the generation of retrovirus, the constructs were transfected along with the pCL-Eco retrovirus-packaging vector (Imgenex) into the HEK293T cell line, pre-treated with 25 mM chloroquine, using the ProFection Mammalian Transfection system (Promega). The retroviral constructs were used to transduce Rev1+/+ and Rev1–/– B cells as reported (Sayegh et al., 2003). Briefly, B cells were activated with LPS for 12 hr and then transduced with retrovirus. The transduced B cells were then stimulated with LPS plus IL-4 for 48 hr before analyzing CSR to IgG1 by semi-quantitative RT-PCR and qRT-PCR.
Immunohistochemistry
Paraffin-embedded human tonsil surgical specimens were serially sectioned with a cryostat at 6 μm onto glass slides and stained as we previously reported (Xu et al., 2010).
Statistical analyses
Differences in spectrum of mutations in Rev1+/+ and Rev1–/– mice were analyzed by Chi-squared test. Differences in Ig titers, CSR and mRNA expression were analyzed with paired or unpaired t-tests.
Supplementary Material
Highlights.
▶ Rev1 DNA polymerase plays an important role in class switch DNA recombination
▶ Such a role is mediated by Rev1 scaffold, not enzymatic function
▶ Rev1 interacts with Ung and recruits it to switch region DNA
▶ Rev1 enhances Ung-mediated dU glycosylation in DNA
ACKNOWLEDGEMENTS
This work was supported by N.I.H. grants AI 079705, AI 045011 and AI 060573 to P.C. We are grateful to Dr. E.J. Pone for his help with immunoblotting and Elliot S. Yu for his expert technical assistance. We thank Dr. Niels de Wind (Leiden University Medical Center, 2300 RC Leiden, Netherlands) and Dr. John C. Schimenti (Cornell University, Ithaca, NY 14853) for making Rev1 KO mice available to us.
H.Z. and C.A.W. contributed equally to this work; H.Z., C.A.W., J.Z., L.M.T., G.L., T.M. and Z.X. performed experiments; H.Z. designed experiments, analyzed the data and prepared the manuscript with assistance from C.A.W.; P.C. designed experiments, analyzed the data, supervised the work and prepared the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
URL link to Extended Experimental Procedures (Supplemental Information).
REFERENCES
- Begum NA, Stanlie A, Doi T, Sasaki Y, Jin HW, Kim YS, Nagaoka H, Honjo T. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc. Natl. Acad. Sci. USA. 2009;106:2752–2757. doi: 10.1073/pnas.0813252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Guéranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EC, Edmonston CR, Wu X, Schaffer A, Casali P. The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3' hs1,2 enhancer in human B cells. Relevance to class switch DNA recombination. J. Biol. Chem. 2004;279:42258–42269. doi: 10.1074/jbc.M407496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani M, Petrov AP, Kolenchenko O, Berru M, Krylov SN, Martin A. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol. Cell Biol. 2007;27:20–30. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Mai T, Zan H, Zhang J, Hawkins JS, Xu Z, Casali P. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J. Biol. Chem. 2010;285:37797–37810. doi: 10.1074/jbc.M110.169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J. Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ. Controlling somatic hypermutation in immunoglobulin variable and switch regions. Immunol. Res. 2010;47:113–122. doi: 10.1007/s12026-009-8142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayca Pozo F, Oda T, Sekimoto T, Murakumo Y, Masutani C, Hanaoka F, Yamashita T. Molecular chaperone Hsp90 regulates REV1-mediated mutagenesis. Mol. Cell Biol. 2011;31:3396–3409. doi: 10.1128/MCB.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J. Exp. Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin AY, Liu CY, Flesken-Nikitin A, Chen CF, Chen PL, Lee WH. Cell lineage-specific effects associated with multiple deficiencies of tumor susceptibility genes in Msh2-/-Rb+/- mice. Cancer Res. 2002;62:5134–5138. [PubMed] [Google Scholar]
- Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Patenaude AM, Affar el B, Lamarre A, Young JC, Di Noia JM. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J. Exp. Med. 2010;207:2751–2765. doi: 10.1084/jem.20101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka C, Kunitomi N, Iwai S, Loakes D, Negishi K. Roles of the polymerase and BRCT domains of Rev1 protein in translesion DNA synthesis in yeast in vivo. Mutat. Res. 2005;578:79–87. doi: 10.1016/j.mrfmmm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pone EJ, Xu Z, White CA, Zan H, Casali P. B cell TLRs and induction of immunoglobulin class-switch DNA recombination. Front. Biol. 2012a;17:2594–2615. doi: 10.2741/4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat. Commun. 2012b;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both immunoglobulin switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Ranjit S, Khair L, Linehan EK, Ucher AJ, Chakrabarti M, Schrader CE, Stavnezer J. AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J. Immunol. 2011;187:2464–2475. doi: 10.4049/jimmunol.1101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schaffer A, Kim EC, Wu X, Zan H, Testoni L, Salamon S, Cerutti A, Casali P. Selective inhibition of class switching to IgG and IgE by recruitment of the HoxC4 and Oct-1 homeodomain proteins and Ku70/Ku86 to newly identified ATTT cis-elements. J. Biol. Chem. 2003;278:23141–23150. doi: 10.1074/jbc.M212952200. [DOI] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, Rajewsky K. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NM, Kochenova OV, Shcherbakova PV. The non-canonical protein binding site at the monomer-monomer interface of yeast PCNA regulates the Rev1-PCNA interaction and Polzeta/REV1-dependent translesion DNA synthesis. J. Biol. Chem. 2011;286:33557–33566. doi: 10.1074/jbc.M110.206680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat. Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 2008;8:302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- White CA, Seth Hawkins J, Pone EJ, Yu ES, Al-Qahtani A, Mai T, Zan H, Casali P. AID dysregulation in lupus-prone MRL/Faslpr/lpr mice increases class switch DNA recombination and promotes interchromosomal c-Myc/IgH loci translocations: Modulation by HoxC4. Autoimmunity. 2011;44:585–598. doi: 10.3109/08916934.2011.577128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, et al. 14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Casali P. AID- and Ung-dependent generation of staggered double-strand DNA breaks in immunoglobulin class switch DNA recombination: a post-cleavage role for AID. Mol. Immunol. 2008;46:45–61. doi: 10.1016/j.molimm.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+IgD+ cell line in vitro: Definition of the requirements and the modalities of hypermutation. J. Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase ζ plays a major role in Ig and Bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. BCR engagement and T cell contact induce Bcl-6 hypermutation in human B cells: association with initiation of transcription and identity with Ig hypermutation. J. Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger IAJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.