Abstract
We introduce two large-scale resources for functional analysis of microRNA—a decoy/sponge library for inhibiting microRNA function and a sensor library for monitoring microRNA activity. To take advantage of the sensor library, we developed a high-throughput assay called Sensor-seq, which permits the activity of hundreds of microRNAs to be quantified simultaneously. Using this approach, we show that only the most abundant microRNAs within a cell mediate significant target suppression. Over 60% of detected microRNAs had no discernible activity, indicating that the functional ‘miRNome’ of a cell is considerably smaller than currently inferred from profiling studies. Moreover, some highly expressed microRNAs exhibit relatively weak activity, which in some cases correlated with a high target-to-microRNA ratio or increased nuclear localization of the microRNA. Finally, we show that the microRNA decoy library can be used for pooled loss-of-function studies. These tools provide valuable resources for studying microRNA biology and for microRNA-based therapeutics.
Introduction
microRNAs (miRNAs) are important regulators of gene expression. More than 400 different miRNAs are encoded in the human genome, and each cell type and cell state has been reported to express a unique battery of miRNAs1. miRNAs regulate gene expression by guiding Argonaute (Ago) proteins to specific sequences in transcripts2. If the target site is perfectly complementary to the miRNA, Ago2 can cleave or ‘slice’ the target transcript. In mammals, the vast majority of natural target sites are not perfectly complementary, and regulation occurs through a non-slicing mechanism in which the miRNA/Ago complex inhibits translation and/or promotes destabilization of the transcript. Because miRNAs only need as few as 7 nucleotides of complementarity to bind to their target, thousands of different genes can be subject to regulation by a single miRNA or miRNA family3. Although much has been learned about miRNA biology, fundamental questions remain, and the function of many miRNAs is still unknown. A critical limitation has been the lack of high-throughput approaches to study miRNA function.
In previous work, we and others incorporated synthetic target sites for a specific miRNA into a gene expression vector to make the transcript a substrate of the endogenous miRNA4. When these targets are expressed at physiological levels they can ‘sense’ miRNA activity within a cell5,6, or even provide a means to eliminate vector or virus expression from unwanted cells types for emerging therapies7–10. Conversely, when the target sites are expressed at supraphysiological levels (>10,000 transcripts/cell) they can act as a ‘sponge’ or ‘decoy’ that sequesters the miRNA, thereby preventing regulation of its natural targets and providing a platform for loss-of-function studies11–15.
Until now, miRNA sensor and decoy studies have been performed using individual vectors. Here, we developed a library of miRNA sensor and decoy vectors, and established a rapid and global means to study miRNA behavior. We used these libraries to determine the relationship between miRNA activity and concentration, and report that a high miRNA concentration is required for target suppression, but that even highly abundant miRNAs may have relatively weak activity.
Results
Sensor-seq provides a means to profile cellular miRNA activity
In order to profile miRNA activity, we first generated a library of miRNA sensor vectors. Target sites for 291 miRNAs conserved between mice and humans were synthesized as five tandem copies with either perfect complementarity (PT), or mismatches at nucleotides 10 and 11 of the miRNA (BT). The BT configuration creates a bulge that prevents Ago2-mediated slicing and results in the transcript being regulated in the more common non-slicing pathway3. All 582 target sites were cloned downstream of eGFP in a bidirectional lentiviral vector (BdLV, Supplementary Fig. 1a) that includes a truncated form of the nerve growth factor receptor (NGFR) reporter gene, which is co-expressed as a distinct transcript. The expression level of target-bearing eGFP depends on the activity of the cognate miRNA, while NGFR serves as an internal control15. The vector utilizes a ubiquitously active mammalian promoter that mediates expression of 27±5 transgene transcripts/cell at single vector copy (Supplementary Fig. 1b), which is within the range of expression of most endogenously expressed genes16. Importantly, at this level of target expression we do not see any evidence of target-mediated miRNA saturation or decay13,15,17 (Supplementary Fig. 1c). The sensor library was produced as a pool, and deep sequencing showed a relatively even distribution of vectors (Supplementary Fig. 1d).
To obtain a global profile of miRNA activity in a single experiment, we devised Sensor-seq, which pairs high throughput sequencing with fluorescence-based sorting of sensor-bearing cells (Fig. 1a). Monocyte, macrophage, and kidney cell lines were transduced at low concentration with the sensor library to achieve 1 vector/cell. The cells were analyzed by FACS to enable quantitative detection of eGFP and NGFR at the single-cell level. The range of NGFR expression was tight, as expected at 1 vector/cell. In contrast, there was a 3 log range in eGFP expression (Fig. 1b). Because a highly active miRNA will result in lower eGFP expression whereas low miRNA activity will result in higher eGFP expression, this pattern reflects the distribution of miRNA activity within the cells.
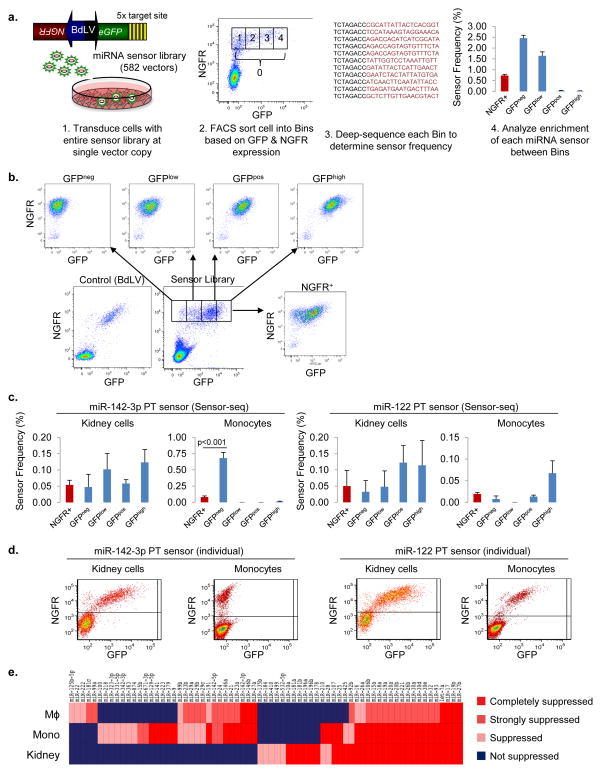
Figure 1. Sensor-seq provides a rapid, high-throughput means to assess miRNA activity.
(a) Schematic of the Sensor-seq assay. BdLV, bidirectional lentiviral vector. GFP, green fluorescent protein, NGFR, truncated nerve growth factor receptor. (b) Representative FACS plots from sorted THP1 cells. (c) Expression pattern of specific sensors as determined by Sensor-seq. Values are mean ± s.d.; n = 3. The P value was generated from a t-test comparison of GFPneg and NGFR+ bins. (d) Representative FACS plots for individually transduced sensors; n = 3. (e) Comparative analysis of target suppression between monocyte, macrophage (MΦ), and kidney cell lines based on Sensor-seq. miRNA sensors were classified based on significant enrichment (≥2-fold, P < 0.05, t-test) in GFPneg:completely suppressed, GFPneg and GFPlow: strongly suppressed, GFPlow: suppressed, and GFPpos and GFPhigh: Not suppressed.
To determine which sensors were being suppressed, and thus which miRNAs were most active, we FACS-sorted the cells into GFPnegative or GFPlow bins, which contain cells in which the sensor was suppressed, and GFPpositive and GFPhigh bins, which contain cells in which the sensor was not suppressed (Fig. 1a). We also sorted the total population of transduced (NGFR+) cells. The gating of each bin was based on a standard approach that utilizes negative and positive controls for each sort (see Online Methods). After sorting, DNA was extracted, and the portion of the vector encoding the target sites were amplified with barcoded primers for multiplexing and subjected to deep sequencing.
Sensor levels were normalized by calculating the frequency of each sensor within a sample (sensor reads/total reads). There was a strong correlation in sensor frequency between replicates from the same bin (from 0.83–0.98) indicating good reproducibility (Supplementary Fig. 1e). There was also a high correlation within the suppressed bins, but not between suppressed and unsupressed bins, consistent with there being a different complement of sensors in cells falling into the different categories.
To validate Sensor-seq, we examined the profiles of several well known miRNAs, such as miR-142-3p, a pan-hematopoietic miRNA, and miR-122, a hepatocyte-specific miRNA1. In monocytes, the miR-142-3p PT sensor was found in 0.08% of the total population. However, in the GFPneg bin, 0.68% of the cells carried the miR-142-3p PT sensor (an 8.5-fold enrichment), compared with less than 0.01% in the other three bins (Fig. 1c). This distribution indicates that 98% of the cells carrying the miR-142-3p PT sensor were in the GFP-negative fraction. Strikingly, when we transduced monocytes with an individual miR-142-3p BdLV, 100% of NGFR+ (i.e. vector transduced) cells were GFP-negative by FACS (Fig. 1d). In kidney cells, neither Sensor-seq nor the individual miR-142-3p PT sensor exhibited enrichment in the GFPneg or GFPlow fraction, consistent with the lack of miR-142-3p expression in these cells1. The Sensor-seq profile of the miR-122 sensor also corresponded to the expression pattern of the individual miR-122 sensor (Fig. 1c,d), and we further confirmed the correspondence between the Sensor-seq profile and individual sensor profile for 10 other miRNAs (see below and data not shown).
These results demonstrate that Sensor-seq can be used as a rapid, high-throughput means for measuring the cellular expression of a library of miRNA sensor vectors, and by inference, miRNA activity, within a population of cells.
Sensor-seq can identify sequences for miRNA-based targeting
A major utility of the library is as an off-the-shelf source of target sites for miRNA-based targeting, which is currently being used to improve the safety and efficacy of a number of novel therapies, including cancer oncolytics and viral vaccines4,7–10. Until now, identifying the most suppressive miRNAs within a cell type or cell state has required a trial and error approach using individual sensors. Sensor-seq provides an unbiased, high-throughput approach for target site selection that allows the identification of targets that are uniquely suppressed in a given cell type (Fig. 1e). For example, among the three cell types we studied, the miR-9 sensor was only suppressed in the monocytes, whereas the miR-99a sensor was only suppressed in the macrophages, as indicated by Sensor-seq, and confirmed using individual sensors for these miRNAs (Supplementary Fig. 2). Thus, the target sites from these sensors can be incorporated into a vector or virus to selectively de-target transgene expression or viral replication from the specific cell type.
Only the most abundant miRNAs mediate target suppression
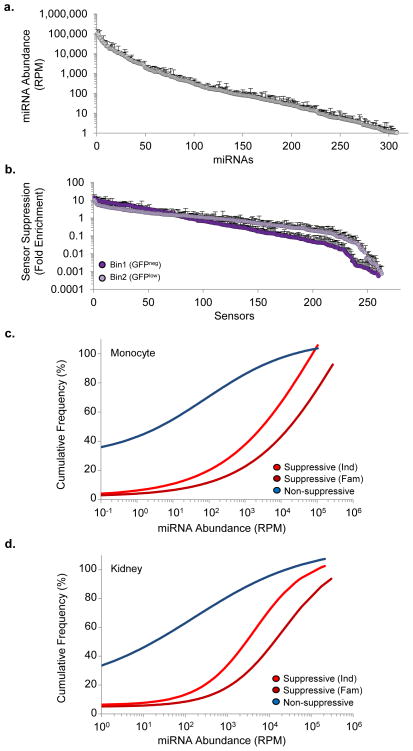
An outstanding question in miRNA biology is how miRNA concentration relates to target suppression3. Deep sequencing indicates that hundreds of miRNAs are expressed within a cell, but how many of these are functional is not known. We and others have suggested that only the most highly expressed miRNAs are able to significantly regulate a target15,18, but formally demonstrating this has not been possible due to the limited sensitivity or scope of current approaches.
To address this issue, we examined the expression of the monocyte miRNome by deep-sequencing and by quantitative PCR17,19. We detected expression of more than 310 miRNAs (Fig. 2a). Our library included sensors for 165 of these miRNAs (188 when considering families), but we detected suppression of only 67 sensors (Fig. 2b). Thus, 59% of the expressed miRNAs that we sampled did not have suppressive activity. For the majority of sensors (>80%) that were significantly suppressed the corresponding miRNA was expressed above 100 reads per million (RPM, Fig. 2c). The cognate miRNAs of some suppressed sensors were not highly expressed, but many of these were part of a miRNA family in which one of the family members was highly expressed, such as the miR-17 or Let-7 family (Supplementary Fig. 3). Since the targets of one miRNA are subject to regulation by all family members3, we refined our analysis by considering the cumulative concentration of an entire miRNA family. We found that the majority of suppressed sensors corresponded to miRNA or miRNA families expressed above 1000 RPM. Of 582 sensors, only 10 with evidence of suppression corresponded to miRNAs that were expressed below 100 RPM. Since none of these sensors contained a cryptic seed-matching site for a highly expressed miRNA, these sensors may be regulated by a non-miRNA-mediated process, or represent false positives. A comparison of the miRNA expression profile of the kidney cells19 with their Sensor-seq profile also found that miRNAs expressed below 100 – 1000 RPM did not have suppressive activity (Fig. 2d).
Figure 2. Correlating miRNA abundance and target suppression.
(a) miRNA expression levels in monocytes determined by deep sequencing. Values are the mean reads per million (RPM) ± s.d.; n = 3 shown for all miRNAs over 1 RPM. (b) Fold enrichment in the indicated bin over the total NGFR+ population for each miRNA sensor, as determined by Sensor-seq. Values are mean ± s.d.; n = 3 (c,d) The concentration of each miRNA as a function of whether the its sensor was suppressed or not suppressed in (c) THP1 monocytes and (d) 293T embryonic kidney cells. A sensor was deemed suppressed if the frequency of the sensor was significantly enriched (P<0.05 t-test) by 2-fold or more in GFPneg or GFPlow bins compared to the total NGFR+ population. Ind, individual miRNA. Fam, miRNA family.
To appreciate how regulation of our sensors compares to a natural target, we inserted into the BdLV a 130 nucleotide sequence from the GSTM4 3′UTR, which contains a single sequence that pairs with nucleotides 2–9 of miR-142-3p (BdLV.GSTM4.NT). In parallel, we generated a version of this 3′UTR sequence that would also allow base-pairing with nucleotides 10–19 of miR-142-3p (BdLV.GSTM4.PT). In this way the natural target context is maintained, but GSTM4.PT becomes subject to slicing. Transduction of monocytes, in which miR-142-3p is highly active (see Fig. 1c,d), resulted in a 2-fold suppression of GSTM4.NT compared to the control, whereas GSTM4.PT was suppressed by more than 10-fold (Supplementary Fig. 4a). The sensitivity of the perfect target to miRNA regulation is further supported by the fact that in more than 70% of target pairs in Sensor-seq, the PT sensor was better regulated than the BT sensor (Supplementary Fig. 4b).
While the observed activity threshold may reflect the sensitivity of Sensor-seq, by incorporating multiple, sometimes perfectly complementary, target sites, and by expressing the reporter at physiological levels, our sensors are more sensitive to regulation than a natural target. Thus, our results strongly suggest that miRNAs expressed below 100 – 1000 RPM would not themselves be capable of mediating significant regulation of a natural target.
Widespread post-biogenesis control of miRNA activity
Above 1000 RPM, the majority of miRNAs were active (>80%), but the degree of suppression did not correlate with miRNA concentration (Fig. 3a), implying some degree of post-biogenesis and pre-decay control of individual miRNA activity. While several such mechanisms have been reported20–22, our large-scale comparison of miRNA activity and concentration demonstrates that this level of control is widespread.
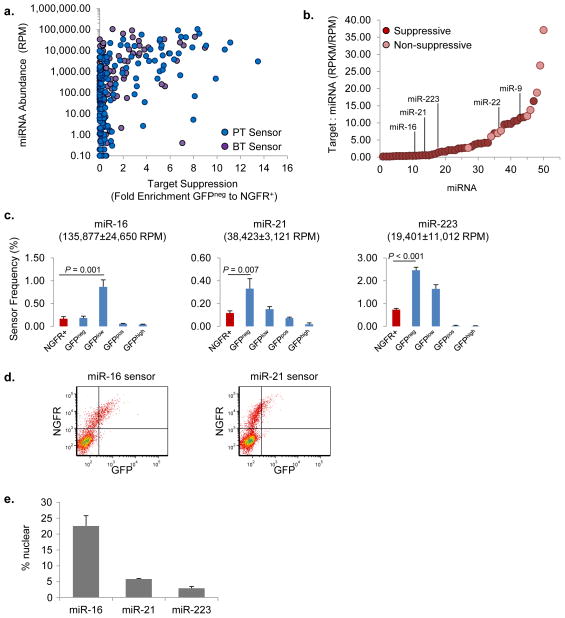
Figure 3. miRNAs have different effective concentrations.
(a) Mean miRNA abundance as a function of the mean fold enrichment of corresponding PT or BT sensors in the GFPneg bin over the NGFR+ population; n = 3. Note that a sensor not enriched in the GFPneg fraction may still be enriched in the GFPlow fraction, and thus not all the points in this graph that are <2-fold correspond to a non-suppressive miRNA. (b) Ratio of the sum of predicted target transcript abundance to miRNA abundance for sensor library miRNAs in THP1 cells. n = 3. RPKM, average reads per kilobase of exon per million mapped reads. Only miRNAs expressed at >1,000 RPM are shown; miRNAs that were non-suppressive are highlighted. (c) Sensor-seq profiles of miR-16, miR-21 and miR-223 BT sensors in THP1 cells. Sensor frequencies are mean ± s.d.; n = 3. The frequency of each sensor in the total population of transduced cells is highlighted in red. The mean ± s.d. concentration of the corresponding miRNA is in parentheses; n = 3. (d) Representative FACS plots of THP1 monocytes 1 week after transduction with miR-16 or miR-21 BT sensors; n = 3. (e) Percentage of miRNAs in the nucleus relative to the entire cell for THP1 cells determined by quantitative PCR. Values are mean ± s.d.; n = 3.
One extrinsic factor that can influence miRNA activity post-transcriptionally is the cumulative concentration of a miRNA’s targets within the cell20,23,24. To investigate this, we quantified the polyA+ transcriptome of monocytes by deep sequencing (Supplementary Fig. 5a,b), and summed the quantity of predicted target transcripts for each miRNA and miRNA family expressed above 1000 RPM. We then compared the concentration of each miRNA to the cumulative concentration of its targets by calculating the target-to-miRNA ratio (mRNA RPKM:miRNA RPM), and found a trend between weaker miRNA activity and higher target abundance. Five out of six of the miRNAs with the highest target-to-miRNA ratios had little to no detectable activity (Fig. 3b), whereas all but one with a target-to-miRNA ratio below 5 had strong activity. There were exceptions to this correlation; for example, miR-9 had a higher target-to-miRNA ratio than miR-22, but miR-9 mediated target suppression whereas miR-22 did not (Fig. 3b). However, because many predicted targets are not bona fide targets of a miRNA25, this difference may be due to inaccurate estimates of the target-to-miRNA ratio, or an alternate mechanism. Overall, our analysis supports the notion that target abundance negatively affects miRNA activity, and demonstrates how Sensor-seq can be used to measure this phenomenon.
Another miRNA that exhibited weaker activity relative to its concentration was miR-16, which was the most abundant miRNA in the monocytes (Fig. 2a). However, the miR-16 sensor was less suppressed than those of miR-21 and miR-223, which were expressed at 3 and 7-fold lower levels, respectively (Fig. 3c–d). This was not dependent on weaker seed-pairing stability (SPS) since miR-21 has the weakest SPS of the three miRNAs24, nor target abundance since miR-223 had the highest target-to-miRNA ratio. It was miRNA dependent, since the miR-223 sensor was not suppressed in kidney cells where the cognate miRNA was not expressed (see Fig. 1e).
Sequence analysis indicated that 10% of miR-16 molecules had a non-templated 3′ adenine, whereas <3% of miR-21 or miR-223 had non-templated modifications (Supplementary Fig. 5c and ref. 16); and such modifications may affect miRNA regulatory capacity21. Interestingly, when we analyzed miRNA abundance in the nucleus compared to the total cell, we found a high portion of miR-16 molecules, but not miR-21 or miR-223, in the nucleus (Fig. 3e). This suggests that subcellular localization of miR-16 is a contributing factor to its weaker activity in THP1 monocytes.
A miRNA decoy library that functions at single vector copy
A major goal of miRNA biology is to determine the function of each miRNA. As noted, overexpression of miRNA target sites can be used to sponge or decoy a miRNA to study miRNA function11–13,26, or potentially for therapeutics27. To enable large-scale loss-of-function studies, we generated a pool of miRNA decoy vectors by incorporating a library of miRNA target sites, designed as ‘tough decoys’14, downstream of the strong, constitutive U6 promoter in a lentiviral vector (Supplementary Fig. 6a). This configuration mediates more efficient miRNA inhibition than tandem targets expressed from a RNA polymerase II promoter14; possibly because the decoys better promote miRNA degradation27.
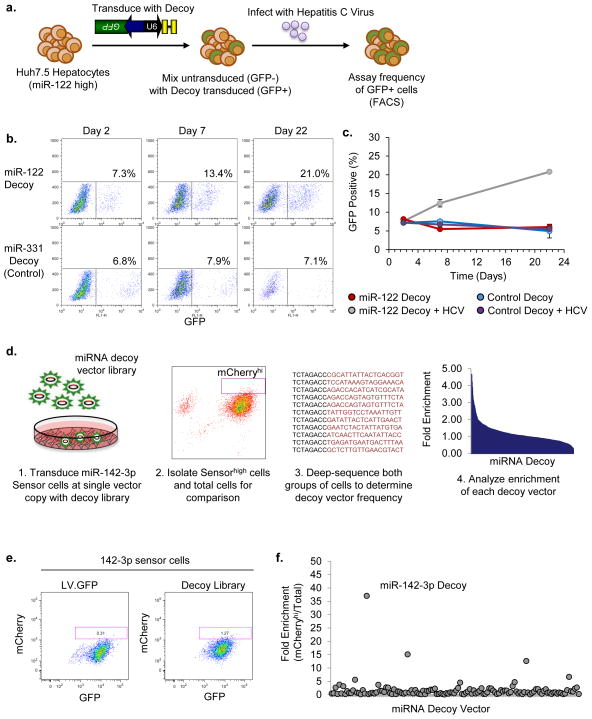
To test the efficacy of the decoy design, we investigated whether we could inhibit miR-122, the most abundant miRNA in hepatocytes1. Interestingly, hepatitis C virus (HCV) exploits miR-122 in order to replicate in hepatocytes, and knockdown of miR-122 has been shown to block HCV infection28. To determine whether the miR-122 decoy vector in our library could inhibit miR-122 function, we transduced human hepatoma 7.5 (Huh-7.5) cells with the miR-122 decoy vector or a control vector that expresses a decoy to miR-331, which is not expressed in Huh-7.5 cells. The transduced cells were marked by eGFP expressed from the vectors. We mixed the transduced (GFP+) with untransduced (GFP−) Huh-7.5 cells, and infected them with HCV. HCV is not a lytic virus, but viral replication does reduce the fitness of infected cells, which allows us to use cell survival as an indicator of resistance to infection (Fig. 4a). The frequency of cells expressing the miR-122 decoy steadily increased from 8.0% at 2 days to 14.7% at 7 days, finally reaching 26.0% by 22 days post-infection (Fig. 4b,c), compared with no change in the frequency of cells expressing the control vector. These results demonstrate that the miR-122 decoy vector can effectively inhibit miR-122 to prevent HCV replication, and provide cells with a selective advantage.
Figure 4. miRNA decoy library enables pooled loss-of-function screens.
(a) Schematic of the approach used to assess miR-122 loss of function. (b) Representative FACS plots showing frequency of GFP-positive cells transduced with miR-122 decoy or control miR-311 decoy at three time points after hepatitis C virus (HCV) infection of Huh-7.5 cells. Note that due to differences in titer the miR-331 transduced cells had a higher vector copy per cell than the miR-122 decoy transduced cells. (c) The frequency of Huh-7.5 cells encoding the miR-122 decoy (GFP+), as determined by FACS, in the presence or absence of HCV infection plotted over time. Values are mean ± s.d.; n = 3 biological replicates. (d) Schematic of the approach used to assess miR-142-3p loss-of-function in a pooled decoy screen. (e) FACS analysis of 142-3p sensor cells transduced at low multiplicity of infection with the decoy vector library. (f) Ratio of mCherryhighGFP+ to mCherry+GFP+ decoy vector frequencies in miR-142-3p sensor cells based on normalized read frequencies from deep sequencing. Representative of two experiments is shown.
In order to permit pooled screens, the decoys must be able to inhibit miRNA activity at single vector copy. To test the efficiency of miRNA inhibition, we stably integrated an mCherry sensor for miR-142-3p into monocytes (142-3p sensor). miR-142-3p is one of the most highly expressed and active miRNAs in monocytes, and thus represents a challenging target for knockdown. We first tested an individual miR-142-3p decoy vector at varying doses, and found that it could effectively interfere with miR-142-3p-mediated suppression at single vector copy (Supplementary Fig. 6b).
Next, we sought to determine whether the miRNA decoy library is effective in a pooled screen. The miR-142-3p decoy vector makes up a small fraction of the vector pool (0.128%, Supplementary Fig. 6c) so identifying cells in which repression is relieved requires a method of selection (Fig. 4d). We transduced the 142-3p sensor cells with the decoy library at low multiplicity of infection, and passaged them for 10 days to reach steady-state. In cells transduced with the library, but not an irrelevant vector, there was a small population of cells with increased mCherry expression (Fig. 4e). To determine whether the miR-142-3p decoy vector was mediating the increased mCherry expression, we isolated the top 1% of mCherryhigh cells by FACS, amplified the region of the vector encoding the target sites, and performed deep sequencing. Strikingly, there was a 37-fold enrichment of the miR-142-3p decoy vector in the mCherryhigh cells over the total population, whereas there was little or no enrichment of the majority of vectors (Fig. 4f). These results validate the effectiveness of the decoy vector for miRNA knockdown, and demonstrate that the decoy library can be employed in a pooled manner for loss-of-function studies.
Discussion
This work provides two valuable resources for studying miRNA biology, and for miRNA-based therapeutics. We also present an assay, Sensor-seq, which can be used to monitor the activity of hundreds of miRNAs within a cell. This technology will be particularly useful for screens aimed at identifying compounds, conditions, or contexts that modulate miRNA activity post-transcriptionally. One could use Sensor-seq to identify miRNAs whose activity, but not concentration, is altered by a particular virus or gene, possibly through the expression of a target mimic29. An important therapeutic application of the target library is for miRNA-based targeting4. For example, Sensor-seq can be used to profile miRNA activity in normal cells and cancer cells to identify target sites that result in sensor suppression only in normal cells. These target sites can then be incorporated into a vector or virus as a means of suppressing expression of a suicide gene or replication of a virus in normal tissue, while permitting expression or replication in cancer cells to selectively kill a tumor8.
We used Sensor-seq to compare the relationship between miRNA concentration and target suppression. Strikingly, we found that only a small number of miRNAs were expressed at a sufficient concentration to mediate sensor regulation. Almost 60% of the miRNAs detected by deep-sequencing had no discernible suppressive activity. This supports our previous assertion that miRNAs expressed below ~100 copies/cell have little regulatory capacity15. Our findings have major implications for interpreting miRNA profiling data since they indicate that a considerable amount of the reported miRNome of a cell may have little biological activity, and highlight the importance of considering absolute expression in addition to differential expression when examining miRNA signatures. It is important to note, however, that profiles of heterogeneous tissues may have low abundant miRNAs that are actually highly expressed in a small fraction of cells, and thus may be functional.
One explanation as to why miRNA concentration must be high for target regulation is that this is necessary to facilitate target interaction. miRNAs must locate their targets in a cell through diffusion and sampling. The rate of interaction between a low abundance miRNA and an mRNA may be slower than the rate of mRNA production and natural decay, and thus have little effect on the mRNA’s expression level. Another factor that may be limiting the activity of a low abundance miRNA is the abundance of its targets in the cell30. That is, if a miRNA is expressed at 10 molecules/cell, and has even as few as 20 different target genes, which are each expressed at 15 transcripts/cell, the miRNA would be titrated to 1 molecule per 30 transcripts. Although a single miRNA molecule can regulate multiple transcripts31,32, this process is inefficient in the non-slicing pathway17, which is how most natural targets are regulated. Thus, a high miRNA concentration is likely required to accelerate target interaction, and overcome a dilution effect mediated by targets and pseudotargets.
So why are there so many low abundance miRNAs that are reliably detected in a cell, but that have little capacity for target regulation? One explanation may be related to the stability of miRNAs. Most of the genome is transcribed to low levels, but since mRNAs have a short half-life16 an aberrantly transcribed gene may not reach a perceptible abundance. However, because miRNAs generally have a long half-life (up to 72 hours)17,33,34, even low-level production can result in a consistent presence.
Sensor-seq also indicated that even highly abundant miRNAs can have weaker activity than similarly or less concentrated miRNAs. Although in some cases this may have been due to intrinsic differences in the miRNA such as a weak SPS24, this did not account for most of the differential activity we observed. Thus, while miRNA activity can be controlled by regulating its concentration, either at the level of production or destruction34, additional factors can clearly affect the miRNA’s activity without affecting its concentration. One mechanism, supported by the studies here, is the concentration of miRNA targets within the cell22–24,30. In addition to target abundance, there is evidence that some miRNAs present in the cell are not in an active state, a phenomenon that has been observed for uridylated miRNAs21. Our results also indicate that subcellular localization of miRNA molecules can result in a lower activity of the miRNA. Some miRNAs are already known to encode sequences that direct their localization to the nucleus33. There may be alternative sequences in the mature or pre-miRNA that direct trafficking to other cellular compartments in the cell, and increase the local concentration of a miRNA with particular targets.
The mechanisms responsible for differential miRNA activity are likely to be varied and there are undoubtedly additional mechanisms to be discovered. Sensor-seq will help to identify miRNAs whose activity is being regulated by some of these mechanisms, and thus to better understand how miRNA regulation is controlled. In parallel, the miRNA decoy library can be used to determine the role and relevance of any of the conserved miRNAs. These new vector collections, along with other recently described large-scale resources for gain and loss of function studies35,36, will help to advance the functional annotation of the genome.
Online Methods
Library design and preparation
Custom oligonucleotide libraries37 were accessed through a collaborative technology program with Agilent Technologies (Santa Clara, CA). For the sensor library, we used a tandem target site configuration. In this design, 5 sequences (21–23 nt each) that are complementary to a specific miRNA are placed in tandem. We separated each target site by an 8 nucleotide spacer. For the decoy library, we utilized the ‘tough decoy’ (TuD) design described by Haraguchi et al.14. The TuD design has only two target sites for a given miRNA, but is designed to make the sites highly accessible and to promote nuclear exit. The oligonucleotides for the tandem library all contained a common 5′ and 3′ primer binding site with AgeI and XbaI restriction sites to enable amplification and cloning. The oligonucleotides for the decoy library also contained a common 5′ and 3′ primer binding site with BsmBI restriction sites on both sides.
The oligonucleotide library was reconstituted in 200 uL of water and PCR amplified using a similar protocol to McManus and colleagues38. Briefly, for a 50 uL reaction, 23.5 uL water, 10 uL 5x Phusion Hot Start II GC Buffer (Finnzymes, Lafayette, CO), 4uL 2.5 mM dNTPs, 5 uL 5 uM 5′ primer, 5 uL 5 uM 3′ primer, 0.025 pmol template, 1 uL DMSO, 0.5 uL Hot StartII Phusion Polymerase. Cycling parameters were 98°C for 40s; 15 cycles of 98°C for 15s, 68°C/63°C for 25s and 72°C for 25s; 72°C for 10 min.
The PCR amplified library was then purified using a PCR purification kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. A restriction digest was then performed as follows: For the sensor library, 200 ng DNA, 6 uL 10x NEB Buffer 4, 0.6 uL of 10 mg/ml BSA and 5 U of AgeI and 5 U of XbaI was prepared in a 60 ul reaction, and incubated at 37°C for 2 hours. For the decoy library, 200 ng DNA, 6 uL 10x NEB Buffer 3, and 10 U BsmBI (NEB, Ipswich, MA) was prepared in a 60 ul reaction and incubated at 55°C for 2 hours. The digested library was then purified by electrophoresis on a 2% Agarose gel with 1X TBE running buffer and purified using a gel extraction kit (Qiagen) following the manufacturer’s protocol.
The sensor library oligonucleotides were cloned into the 3′UTR of GFP in a bidirectional lentiviral vector (BdLV) that also encodes NGFR15,39. The decoy library oligonucleotides were cloned downstream of the human U6 promoter in a lentiviral vector that also contained an eGFP transgene downstream of the human PGK promoter. The vector backbones were digested with either AgeI and XbaI (BdLV) or BsmBI (LV.U6.PGK.GFP), treated with Shrimp Alkaline Phosphatase (NEB), and purified on a 1% agarose gel and recovered using a gel extraction kit (Qiagen).
Ligations were performed in 20 uL reactions containing 50 ng backbone, 3 ng insert, 2 uL 10x T4 DNA Ligase Reaction Buffer (NEB), and 2,000 U T4 DNA Ligase (NEB), and were incubated at 16°C for 14 hours. To prevent loss of library diversity, colonies were collected from ten 10 cm plates after transformation of ligations with TOP10 high efficiency competent cells (Invitrogen, Carlsbad, CA). The pool of plasmids were prepared for transfection using an endotoxin-free Maxi prep kit (Qiagen).
For the studies reported here, the libraries were used as a pool, but individual vectors are being isolated, and will be made available by request and through Addgene.org.
Vector Production and Titration
Lentiviral vectors were produced as previously described17,40. Briefly, 293T cells were seeded 24 hours before Ca3PO4 transfection with third-generation VSV-pseudotyped packaging plasmids and either the sensor or decoy library transfer plasmids. Supernatants were then collected, passed through a 0.22 μm filter, and purified by ultracentrifugation. Viral titer was estimated on 293T cells by limiting dilution.
Cell Culture
Human Embryonic Kidney 293T cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM, Fisher Scientific, Pittsburgh, PA) supplemented with 10% of heat-inactivated fetal bovine serum, 1% of penicillin-streptomycin and 1% glutamine. Human THP-1 monocytic cells were maintained in RPMI medium (Fisher Scientific) supplemented with 10% of heat-inactivated fetal bovine serum, 1% of penicillin-streptomycin and 1% glutamine. 293T and THP-1 cells were transduced as previously described40 with several additional considerations: firstly, to ensure that a majority of transduced cells received only one vector, fewer than 10% of cells were transduced in both sensor and decoy experiments. Secondly, to maintain library diversity, enough cells were plated to ensure at least 100,000 transduction events occurred.
Flow Cytometry
Before FACS analysis, adherent cells were detached with 0.05% trypsin-EDTA, washed and resuspended in sterile PBS. Cells grown in suspension were washed and resuspended in sterile PBS. For analysis of NGFR and GFP expression cells were washed and resuspended in PBS containing 2% FBS. For immune staining cells were blocked in PBS with 2% FBS for 15 minutes at 4°C. After blocking, the cells were incubated with R-phycoerythrin (RPE)-conjugated anti-LNGFR antibody (BD Pharmingen, San Diego, CA) for 25 minutes at 4°C, washed, and resuspended in PBS.
Transduced cells were sorted on a FACS Vantage sorter (Becton-Dickson, Franklin Lakes, NJ). The sorting gates for all cell types were set based on a uniform criteria. The NGFR+ gates were drawn using untransduced, antibody stained cells as a negative comparison. The cells sorted into Bins were restricted to NGFR-positive cells using the same gate as used for NGFR+ cells. For sorting on GFP expression, GFPnegative was gated based on the untransduced (NGFR-negative) cells. To set the gates for GFPpositive and GFPhigh, we used cells transduced with an unregulated BdLV. The gates were drawn to encompass the GFP-positive/NGFR-positive cells from the unregulated reporter. The gate for the GFPlow bin was set so that the mean florescence intensity (MFI) of the population was between the GFPnegative and GFPpositive bin.
Cells transduced with individual sensor vectors were analyzed for NGFR and GFP expression on either the BD Fortessa or BD LSRII (Becton-Dickinson). Analysis was performed using FCSExpress (De Novo Software, Los Angeles, CA) or FlowJo Software (Tree Star, Inc., Ashland, OR).
Target Library Sequencing
Genomic DNA was isolated from FACS sorted cells using a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. The miRNA target sites were amplified from genomic DNA as follows: in a 50 uL reaction — 10 uL 5x Phusion HotStartII HF Buffer, 4 uL 2.5mM dNTPs, 5 uL 5 uM 5′ Primer, 5 uL 5 uM 3′ Primer, 200ng genomic DNA, 1 U Phusion HotStartII Polymerase. Cycling parameters were 98°C 40s; 35 cycles of 98°C 15s, 64°C 25s, 72°C 25s; and 72°C 10 min. The primers used to amplify the target sites contained sequences that hybridize directly to the Illumina flowcell (see below). Barcodes were inserted immediately following the Illumina sequencing primer binding site to allow for multiplexing. PCR amplified libraries were purified on a 2% agarose gel stained with GelStar (Fisher Scientific), visualized using a DarkReader (Clare Chemical Research, Dolores, CO) and purified using a gel extraction kit (Qiagen). Before sequencing all purified library amplification products were analyzed on an Agilent 2100 Bioanalyzer (Agilent). The prepared libraries were multiplexed and sequenced on the Illumina Hi-Seq2000 (Illumina, San Diego, CA). On average we obtained 2,000,000 reads per sample. For each sample there were three replicates (e.g. THP1_bin0_1, THP1_bin0_2, THP1_bin0_3). A Pearson correlation analysis was performed between each sample, and the correlation between replicates was on average 0.91.
The samples were normalized by converting sensor reads to sensor frequency by dividing the number of reads of each sensor by the total number of reads for the sample and multiplying by 100. This is similar to the reads per million normalization, but has the advantage of providing a value that is relatable to the distribution of each sensor within the population of isolated cells. Sensor enrichment was determined by comparing the frequency of a sensor in Bins 1 to 4 to the frequency of the sensor in Bin 0. Enrichment was considered significant if the sensor’s frequency was enriched by ≥2, and P<0.05 by a one-tailed t-test. The validity of the Sensor-seq measurement was confirmed for select miRNAs using individual sensors.
Small RNA Sequencing
For small RNA deep-sequencing, small RNA libraries were prepared using three different protocols to account for biases that can be introduced by library preparation. One protocol was from the Hannon Laboratory41 and another was recently described by us, and uses a panel of different adapters to minimize ligation bias42. The third preparation used Illumina’s kit and followed the manufacture’s recommendations. Each small RNA library was sequenced on an Illumina Genome Analyzer II (Illumina). The sequences were mapped using the computational programs we previously generated43,44, and analyzed on the small RNA dashboard. A subset of this data was reported earlier17. Relative miRNA expression levels were confirmed by real-time PCR using the SABioscience miRNA qPCR whole genome array (Qiagen), and using Taqman miRNA assays for specific miRNAs.
Gene expression analysis
Total RNA was extracted from three different preparation of THP1 monocytic cells (grown in separate flasks) using Tripure Isolation Reagent (Roche Molecular Biochemicals) and Glycogen-blue (Ambion) according to the manufacturers’ instructions. For RNA-Seq, 2 μg of total RNA per sample was fragmented and reverse transcribed and prepared with the Illumina mRNA-seq Sample Prep Kit following the manufacturer’s instructions. Libraries were obtained from paired-end repaired cDNA by PCR with Phusion High-fidelity Taq polymerase (Finnzymes). Each library was prepared with different barcoded primers to permit sample multiplexing. The samples were sequenced on an Illumina Hi-Seq 2000. Sequence reads were mapped and analysed on the small RNA dashboard. The predicted targets for each miRNA were based on TargetScan prediction irrespective of site conservation45. Gene expression levels were converted to reads per kilobase of exon model per million mapped reads (RPKM), as described by Mortazavi et al.46
mRNA Quantification
We measured the number of GFP transcripts expressed in THP1 and 293T cells with a single integrated copy of the BdLV sensor vector per cell, as previously described17. To obtain cells with 1 vector copy per cell, we transduced the cells with the BdLV at <1 MOI. Transduced cells were enriched to 99% purity by two rounds of positive selection using magnetic beads conjugated to an anti-NGFR antibody. Total RNA was extracted from the cells using Tripure Isolation Reagent (Roche Molecular Biochemicals). Reverse transcription was carried out on 1000 ng total RNA using the high capacity RNA to cDNA kit (Applied Biosystems). Real-time PCR analysis of GFP, NGFR, PGK, and Beta Actin expression was performed by Taqman assay. PGK was used to normalize the GFP, NGFR and beta Actin values by the ΔCT method. To determine the absolute concentration of GFP and NGFR transcripts, we extrapolated from a standard curve as previously described17. Samples were analyzed on an ABI Prism 7900HT Real Time PCR System.
Quantification of nuclear and cellular miRNA
Total RNA was extracted from 1,000,000 THP1 monocytic cells using Tripure Isolation Reagent (Roche Molecular Biochemicals), or from the nuclei of 1,075,000 THP1 monocytic cells following a previously described protocol for extraction of nuclear RNA33. Briefly, the cells were centrifuged and the cell pellets were resuspended in 200 μL lysis buffer A [10 mM Tris (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40]. After incubation on ice, the cells were centrifuged, and washed twice to remove the supernatant. The nuclear pellet was lysed, resuspended in Tripure, and total RNA was extracted from the nucleus. The total RNA from the nucleus was consistently 13% (7.525-fold) the amount obtained from the total cell (average of 22,200,000 pg total cell versus 2,950 pg cell nucleus from 1,075,000 cells).
miR-16, miR-21 and U6 RNA were measured by Taqman assay (Life Technologies) according to the manufacture’s instructions. To compare the absolute concentration of miR-16, miR-21, and U6 RNA in the nucleus versus the total cell, we applied a 2.91 threshold cycle (CT) correction (22.91 = 7.525-fold) to the nuclear CT values. We then determined the absolute concentration of the miRNAs by extrapolating from a standard curve, as described17, and compared the fraction of molecules in the nucleus to the total cell.
Hepatitis Infection Assay
Huh-7.5 cells were seeded at 5.0 x 105 cells per well of a 6-well plate and infected the following day with a lentiviral vector vector encoding a miRNA decoy under the control of the U6 promoter and a GFP reporter under the control of the PGK promoter. To allow the cells to recover from any adverse affects of transduction and to stabilize expression, cells were passed into progressively larger plates for 1 to 2 weeks, until a confluent 15-cm-diameter tissue culture plate was derived.
Decoy vector transduced cells were then mixed with untransduced Huh7.5 cells, and infected with a hepatitis C virus that is infectious in cell culture47 (HCVcc) at an MOI of 2, or mock infected, as previously described48. The cells were then passaged for 22 days and analyzed by FACS at day 2, 7, and 22 post infection.
Supplementary Material
Supplementary Figure 1. BdLV-based miRNA Sensor Library.
Supplementary Figure 2. Comparison of miR-9 and miR-99 sensors.
Supplementary Figure 3. Assessing the specificity of Sensor-seq.
Supplementary Figure 4. Assessing the sensitivity of the BdLV sensors.
Supplementary Figure 5. THP1 monocytic cell transcriptome abundance.
Supplementary Figure 6. LV-based microRNA Decoy Library.
Acknowledgments
We thank L. Naldini, B. Gentner, E. Bernstein, E.C. Lai, A. Ventura, and A. Chess for helpful discussions. We thank H. Iba at the University of Tokyo for kindly providing the pmU6-TuD-shuttle plasmid. Oligo libraries were accessed through a collaborative technology program from Agilent Technologies. B.D.B. is supported by a National Institutes of Health Pathfinder Award (DP2DK083052-01) and funding from the Juvenile Diabetes Research Foundation (JDRF-17-2010-770). M.E. is supported by the Pew Charitable Funds and National Institutes of Health (R56AI091792). G.M. is supported by a Helmsley Trust Award.
Footnotes
Author Contributions
G.M., A.B., and A.R. designed and performed research and analyzed data. N.T. and A.D.J. performed research. B.I. and M.J.E. designed and performed research. R.S. designed the project and analyzed data, B.D.B. designed and coordinated the project, and analyzed data.
Competing financial interests
The authors declare no competing financial interests.
Accession codes
RNA sequencing data has been deposited in the Gene Expression Omniubus (GEO): GSE37771.
References
- 1.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 5.Mansfield JH, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 6.Gottwein E, Cai X, Cullen BR. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol. 2006;80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 8.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 9.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentner B, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2:58ra84. doi: 10.1126/scitranslmed.3001522. [DOI] [PubMed] [Google Scholar]
- 11.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 12.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentner B, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 14.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 16.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 17.Baccarini A, et al. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol. 2011;21:369–376. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 21.Jones MR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 26.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 29.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 30.Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 31.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 32.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 33.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 34.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni JQ, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 38.Bassik MC, et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol. 2005;23:108–116. doi: 10.1038/nbt1049. [DOI] [PubMed] [Google Scholar]
- 40.Baccarini A, Brown BD. Monitoring microRNA activity and validating microRNA targets by reporter-based approaches. Methods Mol Biol. 2010;667:215–233. doi: 10.1007/978-1-60761-811-9_15. [DOI] [PubMed] [Google Scholar]
- 41.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 44.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 45.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 48.Narbus CM, et al. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. BdLV-based miRNA Sensor Library.
Supplementary Figure 2. Comparison of miR-9 and miR-99 sensors.
Supplementary Figure 3. Assessing the specificity of Sensor-seq.
Supplementary Figure 4. Assessing the sensitivity of the BdLV sensors.
Supplementary Figure 5. THP1 monocytic cell transcriptome abundance.
Supplementary Figure 6. LV-based microRNA Decoy Library.