Abstract
The temporal and spatial expression patterns of three 1-aminocyclopropane-1-carboxylate (ACC) synthase genes were investigated in pollinated orchid (Phalaenopsis spp.) flowers. Pollination signals initiate a cascade of development events in multiple floral organs, including the induction of ethylene biosynthesis, which coordinates several postpollination developmental responses. The initiation and propagation of ethylene biosynthesis is regulated by the coordinated expression of three distinct ACC synthase genes in orchid flowers. One ACC synthase gene (Phal-ACS1) is regulated by ethylene and participates in amplification and interorgan transmission of the pollination signal, as we have previously described in a related orchid genus. Two additional ACC synthase genes (Phal-ACS2 and Phal-ACS3) are expressed primarily in the stigma and ovary of pollinated orchid flowers. Phal-ACS2 mRNA accumulated in the stigma within 1 h after pollination, whereas Phal-ACS1 mRNA was not detected until 6 h after pollination. Similar to the expression of Phal-ACS2, the Phal-ACS3 gene was expressed within 2 h after pollination in the ovary. Exogenous application of auxin, but not ACC, mimicked pollination by stimulating a rapid increase in ACC synthase activity in the stigma and ovary and inducing Phal-ACS2 and Phal-ACS3 mRNA accumulation in the stigma and ovary, respectively. These results provide the basis for an expanded model of interorgan regulation of three ACC synthase genes that respond to both primary (Phal-ACS2 and Phal-ACS3) and secondary (Phal-ACS1) pollination signals.
Pollination of flowers is a key regulatory event in plant reproduction. In many flowers pollination causes an initial, dramatic increase in ethylene production in the stigma and style and a subsequent increase in ethylene production by other floral organs. Moreover, ethylene has been shown to play a regulatory role in postpollination developmental events (Larsen et al., 1993; O'Neill et al., 1993; Zhang and O'Neill, 1993; O'Neill, 1997; O'Neill and Nadeau, 1997). In higher plants ethylene is formed via a biosynthetic pathway involving the conversion of AdoMet to ACC and of ACC to ethylene (Adams and Yang, 1979). ACC synthase (EC 4.4.1.14) catalyzes the conversion of AdoMet to ACC, whereas ACC oxidase converts ACC to ethylene. ACC synthase is generally considered to be the rate-limiting step in the ethylene biosynthetic pathway (Yang and Hoffman, 1984). ACC synthase has been purified from several plant tissues, and more than 20 genes and cDNAs encoding ACC synthase have been cloned from numerous plant species (for review, see Zarembinski and Theologis, 1994). It has been shown in several systems that ACC synthase is encoded by a divergent gene family, the members of which are differentially regulated in a tissue-specific manner during plant growth and development, and its expression is regulated at the transcriptional level (Kende, 1993).
In spite of substantial investigation, the precise nature of pollen-borne signals that initiate postpollination development is still uncertain (O'Neill, 1997; O'Neill and Nadeau, 1997). Pollen-borne ACC, which is present at high levels in petunia and tobacco pollen, has been proposed to be the primary pollination signal because it contributes to the early phase of ethylene production in the stigma upon pollination (Whitehead et al., 1983, 1984; Hill et al., 1987), but this conclusion has been contested (Hoekstra and Weges, 1986; Pech et al., 1987; Woltering et al., 1993, 1995).
Alternatively, pollen-borne auxin or the physical stress of pollen-tube growth through the transmitting tract are candidates for the triggering of postpollination developmental events (Abeles and Rubenstein, 1964; Burg and Burg, 1966; Hoekstra and Weges, 1986; Singh et al., 1992; Larsen et al., 1993; Nadeau et al., 1993; Zhang and O'Neill, 1993). In particular, results from several studies demonstrate that auxin can duplicate the postpollination effects of induction of ethylene production and fading of flowers in Phalaenopsis spp. and Vanda spp. orchids (Curtis, 1943; Burg and Dijkman, 1967), as it can induce ovary growth and ovule differentiation in a manner comparable to that triggered by pollination (O'Neill et al., 1993; Zhang and O'Neill, 1993). A substantial quantity of auxin has also been reported to be present in the pollinia of several orchid species (Stead, 1992).
Regardless of the identity of the primary pollination signal, one of its initial effects is the induction of ethylene biosynthesis, which serves to coordinate developmental events in multiple floral organs. We previously described the mechanism of interorgan regulation of ethylene biosynthesis in pollinated flowers of a Phalaenopsis sp. hybrid (cv Doritaenopsis), and identified an ACC synthase gene (Ds-ACS1, formerly called OAS1) that is expressed in the gynoecium and labellum in response to ethylene (O'Neill et al., 1993). The Ds-ACS1 gene was proposed to play a role in amplification and propagation of the pollination signal to distal floral organs but did not account for the initial induction of ethylene biosynthesis that was triggered by the primary pollination signal.
The objectives of the current study were to determine whether additional ACC synthase genes were expressed in the stigma of the pollinated Phalaenopsis spp. flower and whether these were regulated by the primary pollination signal. Two additional ACC synthase cDNAs were isolated from the stigma and ovary of pollinated orchid flowers: a stigma-specific cDNA (Phal-ACS2) and an ovary-specific cDNA (Phal-ACS3) encoding a highly divergent ACC synthase isozyme. The expression of Phal-ACS2 was regulated by auxin and the cDNA accumulated primarily in the stigma, whereas Phal-ACS3 was induced by unknown pollination factor(s) and partially by auxin, and was closely correlated with the pattern of ACC synthase activity found in the ovary of pollinated Phalaenopsis spp. These results indicate a pattern of sequential expression of ACC synthase genes regulated by primary pollination signals (Phal-ACS2 and Phal-ACS3) and by ethylene (Ds-ACS1), which acts as a secondary pollination signal involved in amplification and propagation of the pollination signal.
MATERIALS AND METHODS
Orchid plants of the genus Phalaenopsis (cv SM9108 Silk Moth Wedding Gown; Stewart Orchids, Carpinteria, CA) were maintained under optimal growth conditions, as described by Gordon (1990), in a greenhouse at the University of California, Davis. Flowers were divided into the following floral organ units: stigma (column), ovary (ovary and adjacent pedicel), labellum, and perianth (three sepals and two petals) as previously described by O'Neill et al. (1993).
Experimental Treatments
For the pollination time-course experiments, flowers were pollinated in planta by removing the anther cap and pollinia with blunt forceps and then placing the pollinia on the stigma. At the indicated intervals after pollination, floral parts were collected into liquid N2, pulverized into a fine powder, and stored at −80°C until later use in RNA isolation and enzyme assays. For the 0-h time point, flowers were pollinated and then immediately collected into liquid N2. Six flowers were used for each time point, with the floral parts pooled at each sampling time.
For experiments examining the effect of pollination-associated factors, flowers were treated in planta as follows. ACC treatment was achieved by applying 10 nmol of ACC (Sigma) in a sample volume of 15 μL directly to each stigma. Unless otherwise stated, auxin treatment was carried out by applying 1 μg of NAA (5.4 nmol) in a 20-μL volume at pH 6.5 to 6.8 directly to each stigma. For the treatment with different amounts of NAA solution, 0.1, 1, 5, or 10 μg of NAA was applied in a 20-μL volume directly to each stigma. Control flowers were treated in a similar manner with 20 μL of KPO4 buffer (pH 6.5–6.8).
For experiments determining the effects of NBD on mRNA abundance, control and experimental groups of flowers were treated in parallel as follows. Flowers were harvested by excision at the pedicel/floral stalk junction, placed in tubes containing distilled H2O, and transported to the laboratory for immediate treatments. For NBD treatment, six flowers were incubated in an atmosphere of 2000 μL/L NBD (Aldrich) in a sealed 35-L tank for 26 or 48 h, as previously described (O'Neill et al., 1993). For ACC-plus-NBD treatment, six flowers were pretreated with NBD for 2 h and then removed from the tank. Ten nanomoles of ACC was applied in a sample volume of 15 μL directly to each stigma. ACC-treated flowers were then returned to the sealed tank for 24 h of additional NBD treatment. Six control flowers were treated in a similar manner except that 15 μL of sterile double-distilled H2O was applied to the stigma of each flower.
For NBD-plus-ethylene treatment, six flowers were pretreated with NBD for 24 h and then transferred to another sealed tank supplied with 10 μL/L ethylene for 24 h. For NBD-plus-NAA and NBD-plus-pollination treatments, 12 flowers were pretreated with NBD for 4 h and then pollinated or treated with 1 μg of NAA (pH 6.5–6.8) in a sample volume of 20 μL. These flowers were returned to the sealed tank for 6 h of additional NBD treatment. As controls for the latter treatments, 12 flowers were incubated in a growth chamber at high RH for 4 h. Sterile, double-distilled H2O (20 μL) was applied to each stigma of six flowers designated as AIR. Meanwhile, the identical amount of NAA solution was applied to each stigma of six flowers designated as NAA. These control flowers were returned to the growth chamber for an additional 6 h. After treatments the floral parts were collected into liquid N2, pulverized into a fine powder, and stored at −80°C until later use in RNA isolation and enzyme assays.
Assay of ACC Synthase Activity in Orchid Floral Tissues
ACC synthase activity was assayed in extractions of floral organs at various times after pollination, as described by Yu et al. (1979) with modifications. Two grams of frozen, pulverized tissue was homogenized in 3 mL of 100 mm Hepes-KOH (pH 8.5), 4 mm DTT, 10 μm PLP, and 20% (v/v) glycerol. The homogenate was centrifuged at 25,000g for 20 min at 4°C in a Sorvall SM-24 rotor (DuPont). The supernatant was then passed through a layer of sterile Miracloth (Calbiochem), and the extract was dialyzed at 4°C for 36 h with one change of 10 mm Hepes-KOH (pH 8.5), 10% (v/v) glycerol, and 10 μm PLP. ACC synthase activity was measured by incubating 200 μg of the enzyme extract with 200 μm AdoMet, 10 μm PLP, and 200 mm Hepes-KOH (pH 8.5) in a total sample volume of 600 μL for 60 min at 30°C. The amount of ACC formed was assayed according to the method of Lizada and Yang (1979). To ensure that there was no contamination of enzyme with extracted ACC, a duplicate reaction containing 200 μg of enzyme extract was measured without the substrate AdoMet. The ethylene released to the gas phase was collected and determined by GC using a Carle Analytical GC 211 equipped with a flame-ionization detector and a SP4270 integrator (Spectra-Physics, San Jose, CA). To determine the efficiency of the conversion of ACC to ethylene, 2 nmol of ACC was added as an internal standard to a duplicate reaction mixture and degraded as described above. Protein concentration was determined according to Bradford (1976) using commercially available reagents (Bio-Rad).
For the experiments with ACC, NAA, and pollination, flowers were treated in planta with 10 nmol of ACC, 0.1 or 1 μg of NAA solution, or pollinated for 2 h. For the pollination experiment, flowers were hand pollinated in planta. The stigma tissue was collected at 1, 1.5, 2, 2.5, 4, and 8 h after pollination for RT-PCR analysis and ACC synthase activity assays.
RT-PCR Analysis
Total RNA was isolated from the stigma and ovary tissues of orchid flowers from 1 to 4 h after pollination as previously described. To ensure that the RNA was free of DNA, total RNA (10 μg) was digested with 10 units of RNase-free DNase I (Promega) in a solution containing 40 mm Tris-HCl (pH 7.9), 10 mm NaCl, 6 mm MgCl2, and 40 units of RNasin (Promega) at 37°C for 15 min. The reaction mixture was extracted with RNase-free buffer-saturated phenol and chloroform:isoamyl alcohol (24:1, v/v), followed by ethanol precipitation using standard procedures (Sambrook et al., 1989).
RT-PCR analysis was performed as described by Froh-man et al. (1988) with the following modifications. One microgram of DNA-free RNA was reverse transcribed in a total volume of 20 μL containing 0.5 μg of oligo(dT)17 primer (Pharmacia), 2.5 mm deoxynucleotide triphosphates, and 200 units of SUPERSCRIPT RNase H− reverse transcriptase (GIBCO-BRL) in a RT buffer supplied by the company. RNA and the primer were denatured at 70°C for 10 min in the reaction buffer, then cooled on ice before addition of nucleotides and the enzyme. First-strand cDNA synthesis was carried out at room temperature for 10 min and then at 42°C for 1 h. After incubation, the enzyme was denatured by heating to 90°C for 5 min and then cooled on ice for 10 min. The reaction mixture was treated with 2.4 units of RNase H (Pharmacia) at 37°C for 20 min to degrade RNA. Ten microliters of the cDNA reaction mixture was used for PCR amplification.
PCR amplification was performed using Taq DNA polymerase (Promega) in a reaction volume of 50 μL containing PCR buffer (Promega), 1.25 mm deoxynucleotide triphosphates, and 20 pmol of each PCR primer. A thermocycler (model 480, Perkin-Elmer Cetus) was used to perform the first cycle (94°C, 4 min; 50°C, 2 min; and 72°C, 2 min), followed by 30 cycles (94°C, 1 min; 55°C, 2 min; and 72°C, 1.5 min). The sequences of the oligonucleotide primers corresponding to the conserved regions RDLKW and DEIYS are as follows: upstream, 5′-CAGAATTCAG(A/G)GA(C/T)CT(A/C/G/T)AA(A/G)TGG-3′; downstream, 5′-CAGGATCC(A/C/G/T)GA(A/G)TA(A/G/T)AT(C/T)TC (A/G)TC-3′ (EcoRI and BamHI recognition sites are underlined; these sites were introduced for cloning). The PCR fragments were purified as described below, followed by PCR reamplification using the same set of primers.
For cloning and sequencing of PCR products, 45 μL of the PCR mixtures was digested with EcoRI and BamHI (Promega) and then fractionated on a 1.5% low-melting agarose gel. DNA bands of approximately 300 bp were individually excised from the gel and purified using a standard procedure (Sambrook et al., 1989). The purified PCR products were ligated to pBluescript II (KS+) (Stratagene), and the ligation products were transformed into Escherichia coli DH5α cells. Recombinant bacteria were selected, and plasmid DNA was isolated and subsequently analyzed by restriction digestion for the presence of 300-bp inserts. The complete nucleotide sequence of the cDNA clones was determined for both strands by the dideoxy chain-termination method (Sanger et al., 1977) using a DNA sequencing kit (Sequenase version 2.0, United States Biochemical). Clones encoding ACC synthase isozymes were identified by analyzing DNA sequence data using the BLAST program (Altschul et al., 1990; Gish and States, 1993). Analysis of nucleic acids and amino acid sequences was carried out using the software package by the University of Wisconsin Genetics Computer Group (Devereux et al., 1984).
RNA Isolation and Blot Hybridization Analysis
Total RNA isolation and RNA blot-hybridization analysis were carried out as previously described (O'Neill et al., 1993). RNA blots were hybridized with Ds-ACS1, Phal-ACS2, and Phal-ACS3 cDNA insert probes. Blots were washed twice in a 2× SSC, 0.1% SDS solution at room temperature for 15 min each, and then twice in a 0.2× SSC, 0.1% SDS solution at 55°C for 30 min each. The blots were exposed to Kodak XAR 5 film with one intensifying screen (Cronex Light, DuPont) at −80°C for 4 to 7 d. Genomic DNA gel-blot hybridization analysis with the Ds-ACS1, Phal-ACS2, and Phal-ACS3 cDNA insert probes indicated that each hybridized to a distinct and unique restriction fragment, indicating that each probe hybridized to a unique sequence under the hybridization conditions used in these studies.
RESULTS
Pollination Induces ACC Synthase Activity in Orchid Floral Organs
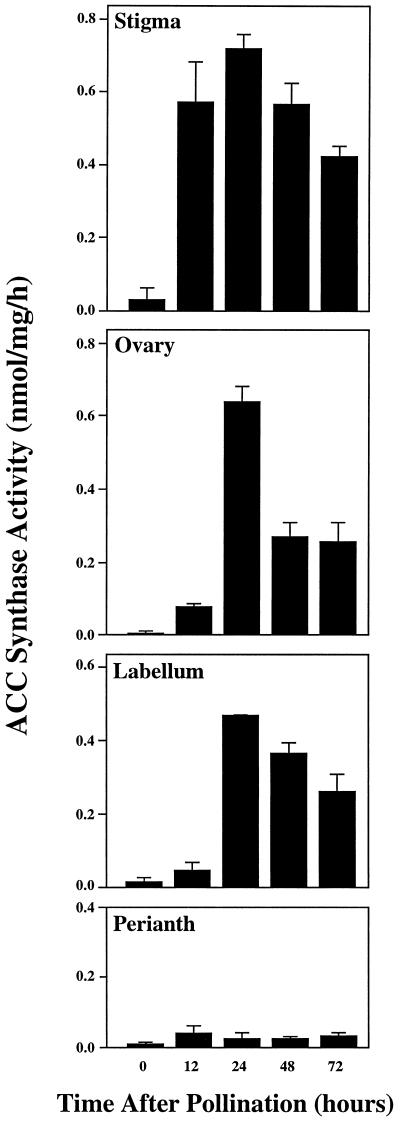
Pollination causes an increase in ethylene production in all floral organs of orchid flowers (O'Neill et al., 1993). To determine if this increase was the result of de novo synthesis of ACC in the four organs or of the translocation of ACC from one organ to the others, crude extracts from all organs were assayed for ACC synthase activity. Figure 1 shows that levels of enzyme activity in the stigma (column), labellum, and ovary were initially very low but increased to peak levels at 24 h after pollination. ACC synthase levels increased first in the stigma, with elevated levels of ACC synthase activity apparent by 12 h after pollination and increasing further until 24 h, at which point the enzyme level was significantly greater than at 12 and 48 h (Student's t test analysis, P < 0.03). In contrast, the perianth had an extremely low level of ACC synthase enzyme activity for a 72-h period after pollination (Fig. 1), consistent with our previous results indicating that ACC synthase mRNA did not accumulate in this floral organ (O'Neill et al., 1993).
Figure 1.
ACC synthase activity in floral organs of pollinated orchid flowers. Enzyme activity was determined in the stigma, ovary, labellum, and perianth. Mean values in nanomoles of ACC per milligrams of protein per hour were obtained from five independent reactions for each floral organ.
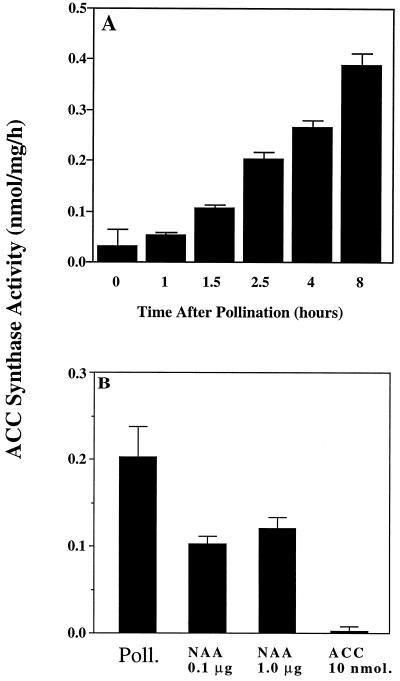
To examine the early induction of ACC synthase in the stigma, a more detailed series of time points after pollination was examined (Fig. 2). The level of enzyme activity in the stigma increased within 1.5 h after pollination. ACC synthase activity in the stigma at 1.5 h after pollination was higher than that observed in the ovary and labellum at 12 h after pollination (see Fig. 1), indicating that ACC synthase activity is induced first in the stigma of pollinated flowers. Exogenous application of auxin (NAA) also mimicked pollination by rapidly inducing ACC synthase activity in the stigma (Fig. 2B). Overall, the above data show that the most rapidly responding tissue with regard to pollination-induced ACC synthase activity is the stigma, and that this response may be mediated by pollen-borne auxin.
Figure 2.
ACC synthase activity in the stigma of orchid flowers. A, Enzyme activity after pollination. B, Enzyme activity in the stigma at 2 h after pollination (as the control), or after 2 h of treatment with 10 nmol of ACC and 0.1 or 1.0 μg of NAA. Mean values were determined from five independent reactions.
Three ACC Synthase cDNAs from the Gynoecium of Pollinated Orchid Flowers
To isolate ACC synthase cDNAs that may contribute to the early induction of ACC synthase enzyme activity, total RNA isolated from stigmas and ovaries of orchid flowers at 2 or 4 h after pollination was subjected to RT-PCR analysis. Several cDNA clones were isolated that corresponded to Phal-ACS2 and Phal-ACS3, from the stigma and ovary, respectively. Figure 3 shows that these cDNA fragments share 63 and 54.1% identity at the nucleotide and amino acid levels, respectively. Additionally, Phal-ACS2 and Phal-ACS3 share 74 and 62% nucleotide identity, and 72.9 and 48.2% amino acid identity, respectively, with the corresponding region of Ds-ACS1, respectively, indicating that these new cDNA fragments represent genes encoding distinct ACC synthase isozymes (Fig. 3). Genomic DNA gel-blot hybridization analysis with the Ds-ACS1, Phal-ACS2, and Phal-ACS3 cDNA insert probes indicated that each hybridized to a distinct and unique restriction fragment (A.Q. Bui and S.D. O'Neill, unpublished data), indicating that each probe hybridized to a unique sequence under the hybridization conditions used in these studies. By analogy to other higher plants, it is likely that ACC synthases in orchids are encoded by a relatively large gene family, but only ACS1, ACS2, and ACS3 have been detected at the level of mRNA in orchid flowers (O'Neill et al., 1993; Zarembinski and Theologis, 1994).
Figure 3.
Comparisons of nucleotide and deduced amino acid sequences of Ds-ACS1, Phal-ACS2, and Phal-ACS3 cDNAs. A, Alignment of nucleotide sequences of ACC synthase cDNAs. B, Clustal analysis of predicted amino acid sequences of cDNAs. Sequences denoting upstream and downstream primers for PCR amplification are underlined. Asterisks represent identical nucleotides and amino acids to counterparts of the Ds-ACS1 cDNA. The five boxed amino acid residues are the invariant amino acids conserved in various aminotransferases (Rottmann et al., 1991).
Ds-ACS1 Encodes Ethylene-Regulated ACC Synthase
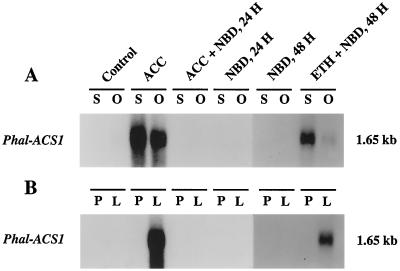
We previously isolated two nearly identical ACC synthase cDNA clones, Ds-ACS1 and Ds-ACS2 (formerly called OAS1 and OAS2; GenBank accession nos. L07882 and L07883; O'Neill et al., 1993). Because of the nearly identical sequences of Ds-ACS1 and Ds-ACS2, we have concluded that they represent transcripts from a single gene (Ds-ACS1). Because Ds-ACS1 mRNA accumulation in the stigma and ovary of pollinated flowers was abolished by pretreatment with aminoethoxyvinylglycine, an inhibitor of ACC synthase, we concluded that the Ds-ACS1 gene was ethylene regulated (O'Neill et al., 1993). To verify this hypothesis in flowers of Phalaenopsis spp., in this study we treated flowers with ACC or ethylene in the presence of NBD, an inhibitor of ethylene action. Figure 4 shows that stigma-applied ACC induced the accumulation of Phal-ACS1 mRNA (homologous to Ds-ACS1) in the stigma, ovary, and labellum; however, this accumulation was inhibited by NBD. In addition, exogenous ethylene overcame the inhibitory effect of NBD on Ds-ACS1 mRNA accumulation in the stigma, ovary, and labellum (Fig. 4). These data confirm that ethylene induces the accumulation of Phal-ACS1 mRNA in orchid flowers.
Figure 4.
Effects of ACC and ethylene on the induction of Ds-ACS1 mRNA in orchid flowers. A, mRNA abundance in the stigma (S) and ovary (O). B, mRNA abundance in the perianth (P) and labellum (L). Treatments were performed on detached flowers as described in Methods. For the stigma and ovary, each lane contained 1 μg of poly(A+) RNA. For the perianth and labellum, each lane contained 2 μg of poly(A+) RNA. Numbers at right indicate the approximate sizes of the detected mRNAs. ETH, Ethylene induced.
Two Distinct ACC Synthase Genes Are Expressed in the Stigma and Ovary of Pollinated Orchids
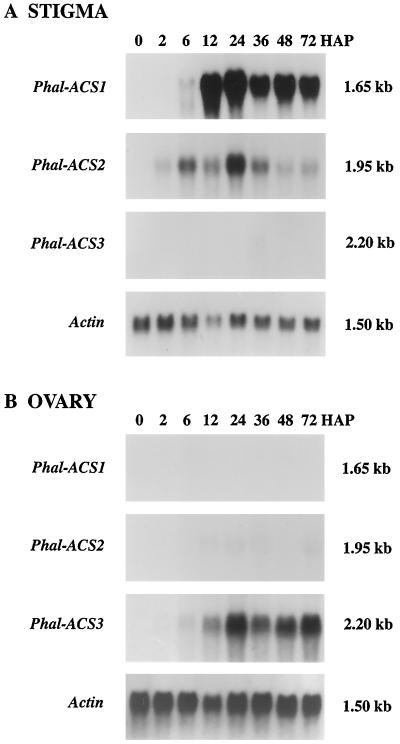
To examine whether expression patterns of Phal-ACS2 and Phal-ACS3 were correlated with patterns of ACC synthase activity in the stigma and ovary of pollinated flowers, we carried out RNA blot-hybridization analysis. Figure 5A shows that the Phal-ACS2 cDNA insert hybridized to a 1.95-kb mRNA in the stigma of pollinated orchid flowers, and that the level of this transcript was detectable at 2 h and increased to a peak at 24 h. In sharp contrast, Phal-ACS3 mRNA was only barely detectable in the stigma of pollinated orchid flowers (Fig. 5A). Phal-ACS2 mRNA was only weakly detected in the ovary at 24 h after pollination (Fig. 5B) and was not detected in the labellum and perianth of pollinated flowers (data not shown). These data indicate that the Phal-ACS2 gene is predominantly expressed in the stigma of the pollinated orchid flowers, and its expression closely parallels the early increase of ACC synthase activity in this tissue after pollination.
Figure 5.
Accumulation of three distinct ACC synthase mRNAs in the orchid gynoecium after pollination. A, Accumulation of Ds-ACS1 and Phal-ACS2 mRNAs in the stigma. B, Accumulation of Phal-ACS3 mRNA in the ovary. Each lane contains 30 μg of total RNA. An actin cDNA probe was used as a control for equal loading of RNA. Numbers at right indicate the approximate sizes of the detected mRNAs. HAP, Hours after pollination.
Figure 5B shows that the Phal-ACS3 cDNA insert hybridized to a 2.2-kb mRNA in the ovary of the pollinated orchid flowers, and that the mRNA accumulation level was relatively low at 6 h but increased to a peak at approximately 24 h. An extremely low level of this mRNA was also detected at 36 h in the stigma (Fig. 5A), but it was not detected in the labellum and perianth of pollinated flowers (data not shown). Overall, these data indicate that the Phal-ACS2 gene is expressed predominantly in the stigma of pollinated orchid flowers and the Phal-ACS3 gene is predominantly expressed in the ovary. Furthermore, the expression of the Phal-ACS3 gene correlates with the pattern of ACC synthase activity in the ovary of pollinated orchids, as shown in Figure 1.
Auxin Induces Phal-ACS2 and Phal-ACS3 Gene Expression in the Orchid Gynoecium
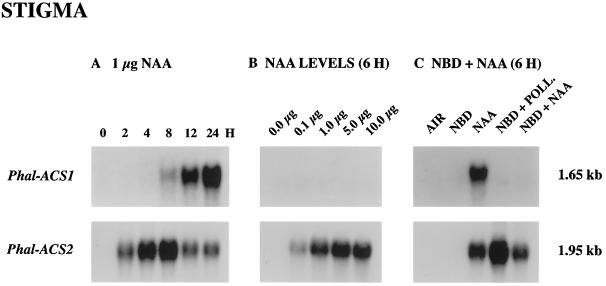
Figure 2B shows that, as in the case of pollination, NAA induced the increase in ACC synthase activity in the stigma within 2 h. By contrast, ACC did not stimulate induced ACC synthase activity in this tissue over a short time period. To determine whether auxin directly induced expression of the Phal-ACS2 gene in the stigma of orchid flowers, we treated the stigma with NAA solution and collected tissue for RNA-blot-hybridization analysis. Figure 6A shows that during 24 h of NAA treatment, Phal-ACS2 mRNA was induced to accumulate in the stigma by 2 h and increased to its peak after 8 h of NAA treatment. By contrast, Phal-ACS1 mRNA was not detected in the stigma until 8 h after NAA treatment. Comparison of the expression patterns of Phal-ACS2 mRNA between NAA treatment and pollination revealed a decline in the level of mRNA after 8 h of NAA treatment; this suggested that a continuous supply of auxin or other pollination signals may be required for the continuous induction of this transcript. As shown in Figure 6B, levels of Phal-ACS2 mRNA increased with increased concentration of the applied NAA solution. This induction was not mediated by ethylene because the level of Phal-ACS2 mRNA accumulation induced by NAA treatment was relatively the same in the presence or absence of NBD (Fig. 6C). Additionally, neither the application of ACC nor emasculation stimulated the accumulation of Phal-ACS2 mRNA in the stigma (data not shown). Thus, all of the above data strongly indicate that auxin induces the expression of the Phal-ACS2 gene, as well as the increase in ACC synthase activity in the stigma tissue. Several other auxin-regulated ACC synthase genes have been identified in other plants (Nakagawa et al., 1991; Botella et al., 1992; Abel et al., 1995).
Figure 6.
Accumulation of Ds-ACS1 and Phal-ACS2 mRNAs induced by auxin (NAA) in the stigma. A, Time course (in hours) of ACC synthase mRNA accumulation after the application of 1 μg of NAA. B, Effects of different levels of NAA on mRNA accumulation in orchid flowers in planta 6 h after the application. C, Induction of ACC synthase mRNA accumulation after the application of 1 μg of NAA for 6 h in the presence or absence of NBD. Each lane contained 30 μg of total RNA. Numbers at right indicate the approximate sizes of mRNAs. POLL., Pollination.
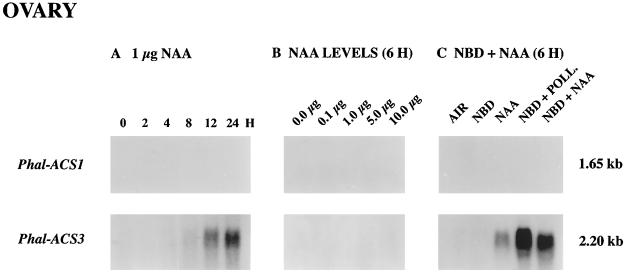
Previously, Zhang and O'Neill (1993) demonstrated that auxin played a crucial role in ovary and ovule development in orchid plants. Therefore, we investigated whether auxin (NAA) induced the accumulation of Phal-ACS3 mRNA in the ovary tissue. Figure 7A shows that levels of Phal-ACS3 mRNA accumulation in the ovary increased during a 24-h period after the application of NAA solution to the stigma tissue. Furthermore, as shown in Figure 7C, this mRNA accumulation was also induced by NAA in the presence of NBD, indicating that auxin and not ethylene induces the expression of the Phal-ACS3 gene. However, the levels of mRNA induced by NAA were much lower and induction occurred later than in the ovary of pollinated flowers, compared at appropriate time points (Figs. 5B and 7A). These results might be attributable to either too small an amount of exogenous NAA being applied or the additional effects of pollination factor(s) other than auxin. Figure 7B demonstrates that the amount of exogenous NAA was not the factor influencing the level of Phal-ACS3 mRNA accumulation, because as much as 10 μg of NAA solution did not result in a detectable level of Phal-ACS3 mRNA at 6 h after the treatment. Thus, it is likely that pollination factor(s) other than auxin also induce the Phal-ACS3 mRNA.
Figure 7.
Accumulation of Ds-ACS1 and Phal-ACS2 mRNAs induced by auxin (NAA) in the ovary. A, Time course (in hours) of ACC synthase mRNA accumulation after the application of 1 μg of NAA. B, Effects of different levels of NAA on mRNA accumulation in flowers in planta 6 h after the application. C, Induction of ACC synthase mRNA accumulation after the application of 1 μg of NAA for 6 h in the presence or absence of NBD. Each lane contained 30 μg of total RNA. Numbers at right indicate the approximate sizes of mRNAs. POLL., Pollination.
DISCUSSION
In many flowers pollination leads to increased production of ethylene, which is involved in developmental changes in floral organs such as floral color change, perianth senescence, ovary growth, and ovule development (Stead, 1992). In carnation, orchid, and petunia flowers, such pollination-induced ethylene production is associated with the expression of ACC synthase and ACC oxidase genes (Woodson et al., 1992; O'Neill et al., 1993; Tang and Woodson, 1996; O'Neill, 1997; O'Neill and Nadeau, 1997). However, the nature of pollen-pistil interactions leading to the expression of ACC synthase genes in floral organs is poorly understood. In this study we further investigated a role of ACC synthase in the postpollination syndrome by cloning additional ACC synthase cDNAs and examining expression patterns of the corresponding genes in pollinated Phalaenopsis spp. orchids. To our knowledge, Phal-ACS2 is the first ACC synthase gene to be induced in the stigma of pollinated flowers. Thus, the expression pattern of Phal-ACS2 may be useful in searching for primary pollination signals that trigger the initial production of ethylene in the stigma of pollinated orchid flowers. The identification of auxin as a regulator of Phal-ACS2 gene expression suggests that auxin may be one such pollination signal.
Unlike Phal-ACS2, the induction of Phal-ACS1 by auxin is mediated by ethylene, because this expression is inhibited when flowers are pretreated with aminoethoxyvinylglycine (O'Neill et al., 1993). In this study we confirmed that, like Ds-ACS1 expression in Doritaenopsis (a Phalaenopsis hybrid cultivar), Phal-ACS1 gene expression is regulated by ethylene in that its expression is inhibited by NBD but can be restored by ethylene treatment. Furthermore, the accumulation of Phal-ACS1 mRNA in the stigma of pollinated flowers was initially detected at 6 h, a time when increased levels of ethylene are first detected in this organ (J.A. Nadeau and S.D. O'Neill, unpublished data). Taken together, the Phal-ACS1 and Phal-ACS2 genes are differentially expressed in the stigma of pollinated orchids, and it is likely that their combined expression is responsible for the de novo synthesis of ACC required for sustained ethylene production in this organ.
In carnation pollination results in a high level of ACC in the ovary, suggesting the activation of ACC synthase gene expression in this organ (Nichols et al., 1983). Two previously isolated ACC synthase cDNAs did not hybridize strongly to an mRNA in the ovary of pollinated carnation, suggesting that a divergent ACC synthase gene may be expressed in this organ (Woodson et al., 1992; Henskens et al., 1994). In contrast to what is seen in carnation, the 2.2-kb Phal-ACS3 mRNA accumulated concomitantly with the increase in ACC synthase activity in the ovary of pollinated orchid flowers, suggesting that the expression of the Phal-ACS3 gene results in ACC synthase enzyme activity in this organ. This mRNA accumulation was also induced by auxin in the ovary; however, its level was severalfold less than that induced by pollination, indicating that auxin is probably not the sole pollination factor stimulating the accumulation of Phal-ACS3 mRNA. We hypothesize that an unknown pollination factor has a synergistic effect with auxin in stimulating Phal-ACS3 gene expression in the ovary.
In this study high levels of ACC synthase activity were detected in the ovary of pollinated orchid flowers. However, very low levels of ethylene and ACC oxidase activity are measured in this organ (Nadeau et al., 1993; O'Neill et al., 1993). These data suggest that only a small amount of ACC synthesized in the ovary is converted into ethylene, which is consistent with the previous report that a small amount of ethylene, together with auxin, regulates ovary growth and ovule differentiation, but that high levels of ethylene result in cell death in the ovary (Zhang and O'Neill, 1993). We hypothesize that a portion of the ACC synthesized in the ovary is available for translocation to other floral organs, especially the perianth. The translocation of ACC to the perianth is plausible because this organ senesces several days after pollination, even though it does not contain ACC synthase mRNA or enzyme activity (Nadeau et al., 1993; O'Neill et al., 1993).
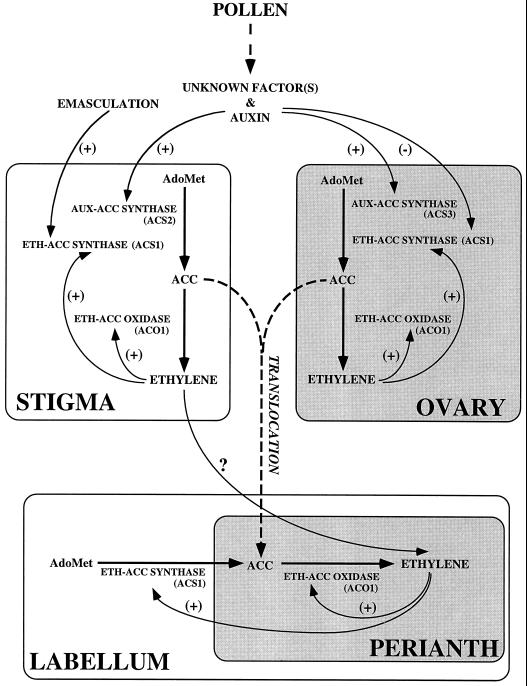
It is now widely appreciated that both ethylene biosynthesis and ethylene perception contribute to the regulation of ethylene responses in plants (Wilkinson et al., 1995). In this study we have focused only on the coordinated regulation of ethylene biosynthesis after pollination, and Figure 8 summarizes the expression patterns of ethylene biosynthetic genes in floral organs of pollinated orchid flowers based on data from studies carried out in our laboratory.
Figure 8.
A model of interorgan regulation of ACC synthase and oxidase gene expression in pollinated orchid flowers. ACO, ACC oxidase; ACS, ACC synthase; AUX, auxin induced; ETH, ethylene induced; and PS, Phalaenopsis species.
Pollen-borne signals, including auxin, induce the expression of Phal-ACS2 in the stigma, leading to increased ACC synthase activity. Newly formed ACC is oxidized by basal constitutive ACC oxidase to ethylene, which then stimulates the expression of Phal-ACS1 and Phal-ACO1 genes for the autocatalytic production of ethylene (Nadeau et al., 1993). ACC or ethylene itself is translocated from the stigma to the labellum and perianth. In the labellum ethylene induces Phal-ACS1 and Phal-ACO1 mRNA accumulation, leading to the autocatalytic production of ethylene, which is responsible for hyponasty and senescence of the labellum. In the perianth ACC translocated from the stigma, ovary, or labellum is oxidized to ethylene by ACC oxidase, which triggers wilting and senescence. In the ovary pollen-borne auxin and unknown pollination factor(s) induce Phal-ACS3 gene expression, resulting in increased ACC synthase activity and ACC synthesis. Based on the low level of ACC oxidase and ethylene production in the ovary, a limited amount of ACC is likely to be converted to ethylene, and this organ may also be a source of ACC for translocation to the perianth. The biochemical tools are now available to extend this model to include the regulation of ethylene perception and ethylene signal transduction that may also participate in regulating postpollination responses in flowers.
Abbreviations:
- AdoMet
S-adenosylmethionine
- NBD
2,5-norbornadiene
- PLP
pyridoxal 5′-phosphate
- RT
reverse transcription
Footnotes
This research was supported by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grant Program-Plant Growth and Development Program (USDA 91-37304-6464 and 93-37304-6464) and the Binational Agriculture Research and Development Fund (US 1867-90R) to S.D.O.
LITERATURE CITED
- Abel S, Nguyen MD, Choe W, Theologis A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- Abeles FB, Rubenstein B. Regulation of ethylene evolution and leaf abscission by auxin. Plant Physiol. 1964;39:963–969. doi: 10.1104/pp.39.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DO, Yang SF. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci USA. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Botella JR, Arteca JM, Schlagnhaufer CD, Arteca RN, Phillips AT. Identification and characterization of a full-length cDNA encoding for an auxin-induced 1-aminocyclopropane-1-carboxylate synthase from etiolated mung bean hypocotyl segments and expression of its mRNA in response to indole-3-acetic acid. Plant Mol Biol. 1992;20:425–436. doi: 10.1007/BF00040602. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burg SP, Burg EA. The interaction between auxin and ethylene and its role in plant growth. Proc Natl Acad Sci USA. 1966;55:262–269. doi: 10.1073/pnas.55.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg SP, Dijkman MJ. Ethylene and auxin participation in pollen-induced fading of Vanda orchid blossoms. Plant Physiol. 1967;42:1648–1650. doi: 10.1104/pp.42.11.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis JT. An unusual pollen reaction in Phalaenopsis. Am Orchid Soc Bull. 1943;11:259–260. [Google Scholar]
- Devereux P, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Gordon B (1990) Culture of the Phalaenopsis Orchid. Laid-Back Publications, CA
- Henskens JAM, Rouwendal GJA, Ten Have A, Woltering EJ. Molecular cloning of two different ACC synthase PCR fragments in carnation flowers and organ-specific expression of the corresponding genes. Plant Mol Biol. 1994;26:453–458. doi: 10.1007/BF00039554. [DOI] [PubMed] [Google Scholar]
- Hill SE, Stead AD, Nichols R. Pollination-induced ethylene and production of 1-aminocyclopropane-1-carboxylic acid by pollen of Nicotiana tabacum cv White Burley. J Plant Growth Regul. 1987;6:1–13. [Google Scholar]
- Hoekstra FA, Weges R. Lack of control by early pistillate ethylene of the accelerated wilting of Petunia hybrida styles. Plant Physiol. 1986;80:4–6. doi: 10.1104/pp.80.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Larsen PB, Woltering EJ, Woodson WR (1993) Ethylene and interorgan signaling in flowers following pollination. In I Raskin, J Schultz, eds, Plant Signals in Interactions with Other Organisms. American Society of Plant Physiologists, Rockville, MD, pp 171–181
- Lizada MCC, Yang SF. A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal Biochem. 1979;100:140–145. doi: 10.1016/0003-2697(79)90123-4. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Zhang XS, Nair H, O'Neill SD. Temporal and spatial regulation of 1-aminocyclopropane-1-carboxylate oxidase in the pollination-induced senescence of orchid flowers. Plant Physiol. 1993;103:31–39. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Mori H, Yamazaki K, Imaseki H. Cloning of a complementary DNA for auxin-induced 1-aminocyclopropane-1-carboxylic acid synthase and differential expression of the gene by auxin and wounding. Plant Cell Physiol. 1991;32:1153–1163. [Google Scholar]
- Nichols R, Bufler G, Mor Y, Fujino DW, Reid MS. Changes in ethylene production and 1-aminocyclopropane-1-carboxylic acid content of pollinated carnation flowers. J Plant Growth Regul. 1983;2:1–8. [Google Scholar]
- O'Neill SD. Postpollination regulation of flower development. Annu Rev Plant Physiol Plant Mol Biol. 1997;58:47–72. doi: 10.1146/annurev.arplant.48.1.547. [DOI] [PubMed] [Google Scholar]
- O'Neill SD, Nadeau JA. Postpollination flower development. Hortic Rev. 1997;19:1–58. [Google Scholar]
- O'Neill SD, Nadeau JA, Zhang XS, Bui AQ, Halevy AH. Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell. 1993;5:419–432. doi: 10.1105/tpc.5.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech JC, Latche A, Larrigaudiere C, Reid MS. Control of early ethylene synthesis in pollinated petunia flowers. Plant Physiol Biochem. 1987;25:431–437. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicken S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Evensen KB, Kao T-H. Ethylene synthesis and floral senescence following compatible and incompatible pollinations in Petunia inflata. Plant Physiol. 1992;99:38–45. doi: 10.1104/pp.99.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead AD. Pollination-induced flower senescence: a review. Plant Growth Regul. 1992;11:13–20. [Google Scholar]
- Tang X, Woodson WR. Temporal and spatial expression of 1-aminocyclopropane-1-carboxylate oxidase mRNA following pollination of immature and mature petunia flowers. Plant Physiol. 1996;112:503–511. doi: 10.1104/pp.112.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead CS, Fujino DW, Reid MS. Identification of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) in pollen. Sci Hortic. 1983;21:291–297. [Google Scholar]
- Whitehead CS, Halevy AH, Reid MS. Roles of ethylene and ACC in pollination and wound-induced senescence of Petunia hybrida flowers. Physiol Plant. 1984;61:643–648. [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee H. An ethylene inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Woltering EJ, Somhorst D, Van der Veer P. The role of ethylene in interorgan signaling during flower senescence. Plant Physiol. 1995;109:1219–1225. doi: 10.1104/pp.109.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering EJ, Van Hout M, Somhorst D, Harren J. Roles of pollination and short-chain saturated fatty acids in flower senescence. Plant Growth Regul. 1993;12:1–10. [Google Scholar]
- Woodson WR, Park KY, Drory A, Larsen PB, Wang H. Expression of ethylene biosynthetic pathway transcripts in senescing carnation flowers. Plant Physiol. 1992;99:526–532. doi: 10.1104/pp.99.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yu Y, Adams DO, Yang SF. 1-Aminocyclopronane-1-carboxylate synthase, a key enzyme in ethylene biosynthesis. Arch Biochem Biophys. 1979;198:280–286. doi: 10.1016/0003-9861(79)90420-x. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case in conservation. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- Zhang XS, O'Neill SD. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell. 1993;5:403–418. doi: 10.1105/tpc.5.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]