SUMMARY
We propose that cell cycle-dependent timing of FEN1 nuclease activity is essential for cell cycle progression and the maintenance of genome stability. After DNA replication is complete at the exit point of the S-phase, removal of excess FEN1 may be crucial. Here, we report a mechanism that controls the programmed degradation of FEN1 via a sequential cascade of post-translational modifications. We found that FEN1 phosphorylation stimulated its SUMOylation, which, in turn, stimulated its ubiquitination and ultimately led to its degradation via the proteasome pathway. Mutations or inhibitors that blocked the modification at any step in this pathway suppressed FEN1 degradation. Critically, the presence of SUMOylation- or ubiquitination- defective, non-degradable FEN1 mutant protein caused accumulation of Cyclin B, delays in the G1 and G2/M phases and polyploidy. These findings may represent a newly identified regulatory mechanism used by cells to ensure precise cell cycle progression and to prevent transformation.
INTRODUCTION
Efficient, accurate processing of up to 50 million Okazaki fragments per cell cycle is required to complete lagging strand DNA synthesis in mammalian cells (Burgers, 2009; Garg and Burgers, 2005). The current model for Okazaki fragment processing involves sequential reactions of gap filling by polymerase δ (Polδ), flap cleavage by flap endonuclease 1 (FEN1) and ligation by ligase I (Lig I), with proliferating cell nuclear antigen (PCNA) functioning as the platform for recruitment and coordination of these enzymes and their activities (Chapados et al., 2004; Liu et al., 2004; Waga and Stillman, 1998). The precise mechanisms that control the sequential switching of enzymes during Okazaki fragment processing to ensure the correct sequence of reactions is still largely unknown. We have recently shown, however, that methylation and phosphorylation are important signals for FEN1 to bind to and dissociate from the PCNA replisome (Henneke et al., 2003). Methylation of FEN1 promotes its association with PCNA, and then, after it removes the RNA primer, FEN1 is phosphorylated, which results in its disassociation from PCNA (Guo et al., 2010).
Because it is a nuclease, the functions of FEN1 must be executed in precise locations and in appropriate protein complexes with perfect timing. Dysregulation of FEN1 activity could result in undesired destruction of the genetic information coded in DNA and disorder to the programmed cell cycle. The fate of soluble phosphorylated FEN1 once it dissociates from the DNA replication fork at the exit of S-phase is unknown. It could be de-phosphorylated and recycled or, alternatively, it may be degraded. Here we report that a cascade of post-translational modifications (PTMs), involving FEN1 phosphorylation, SUMOylation and ubiquitination, mediate the degradation of FEN1 via the proteasome pathway during G2/M phase. Specifically, we have determined that the UBE1/UBE2M/PRP19 complex is responsible for FEN1 protein ubiquitination and its activity is stimulated by conjugation of SUMO3 to FEN1. Disruption of the chain of modifications to FEN1 results in altered levels of Cyclin B, disorder of cell cycle progression and genome instability.
RESULTS
FEN1 is degraded at the exit of S-phase via the ubiquitin-proteasome pathway
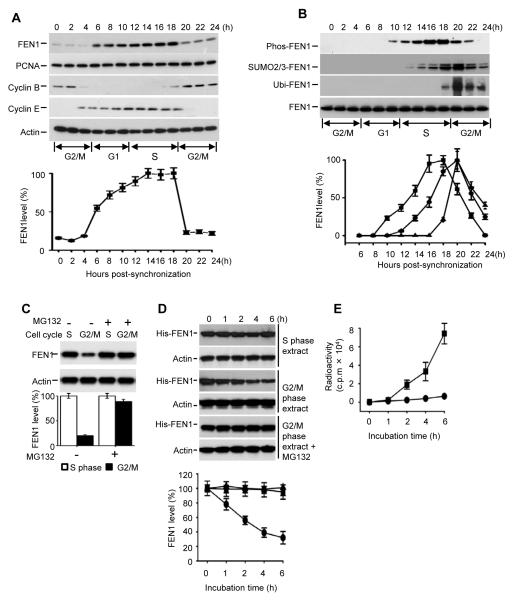
To determine if FEN1 protein levels are differentially regulated at different stages of the cell cycle, we measured the levels of FEN1 protein in HeLa cell extracts at 2-hour intervals for 24 hours after synchronization and release from the G2/M boundary (Xu et al., 2009). The cell cycle phase of each time point was determined by the expression profile of Cyclin B, Cyclin E and FACS (Figure 1A and Figure S1A). Expression of endogenous FEN1 increased during G1, peaked at S phase and then dramatically decreased during the following G2/M phase (Figure 1A). In comparison, we also examined the levels of the PCNA protein, a FEN1 replication partner, at different cell cycle phases but found no cell cycle-dependent expression profile. These data suggest that FEN1 is differentially regulated during cell cycle progression and we confirmed that the fluctuation of FEN1 levels is not due either to differences in FEN1 transcription during different cell cycle phases or to post-transcriptional regulation (shown in Figure S1 B and C).
Figure 1. FEN1 is degraded at G2/M phase via the proteasome pathway.
(A) Endogenous FEN1 levels during the cell cycle. HeLa cells were synchronized at G2/M then released and collected at the indicated time points. FEN1 levels in total cell lysates were determined by Western blotting using an anti-FEN1 antibody. (B) Dynamics of FEN1 modifications during cell cycle. HeLa cells were synchronized and released as shown in Figure 1A. Immunoprecipitates by anti-FEN1 antibody were tested by blotting with different anti-FEN1-modification antibodies as indicated. (C) MG132 inhibits FEN1 degradation in G2/M phase. HeLa cells with or without MG132 treatment were synchronized at the S and G2/M phases. FEN1 levels in total cell lysates were determined by Western blotting and quantified. (D) Degradation of His-tagged FEN1 by cell extracts. Purified recombinant His-FEN1 was incubated with extracts of cells in S or G2/M phase. Aliquots were taken at the indicated times and His-FEN1 detected by Western blotting using an anti-His antibody. Quantitative results are shown in the bottom panel. (E) His-FEN1 was 32P-labeled in vitro and incubated with G2/M cell extract for the indicated times. His-FEN1 was pulled down with Ni-NTA beads and the radioactivity in solution measured by liquid scintillation. Radioactivity from 32P-FEN1 in G2/M cell extracts, ■; Radioactivity from 32P-FEN1 in G2/M cell extracts +MG132, ●. The error bars in all panels represent mean ± SD from three independent experiments. See also Figure S1.
Post-translational modifications (PTMs) are a common mechanism used to efficiently and rapidly regulate protein stability and degradation (Callis, 1995). To test if FEN1 degradation is PTM-mediated, we investigated the dynamics of FEN1 modifications during the cell cycle. In addition to the previously reported phosphorylated form (Guo et al., 2010; Henneke et al., 2003), we discovered two other novel post-translationally modified forms of FEN1: SUMOylated and ubiquitinated (Figure 1B). We observed that all the three modified forms of FEN1 displayed cell cycle-dependent dynamics. We initially observed that phosphorylated FEN1 appeared 2 hours earlier than SUMO2/3 modified FEN1 and SUMO2/3 modified FEN1 appeared 6 hours earlier than ubiquitinated FEN1 (Figure 1B). However, when the Western blotting film is overexposed, we observed that the initial appearance and the maximum levels of SUMOylated and ubiquitinated FEN1 nearly coincide (data not shown). The time interval between these sequential modifications is within 2 hours, which is consistent with what has been reported in other studies (Torres MP et al., 2011; Huang et al., 2003).
The ubiquitin-proteasome pathway is the most common method used by cells to degrade endogenous proteins (Whitfield et al., 2000). Therefore, we tested if it plays a role in FEN1 degradation during G2/M. After treatment with the proteasome inhibitor MG132 (Carbobenzoxyl-leucinyl-leucinyl-leucinal) (Rock et al., 1994), degradation of FEN1 in G2/M phase was blocked (Figure 1C), suggesting FEN1 degradation is regulated by the proteasome pathway. Degradation of recombinant His-tagged FEN1 by G2/M phase cell extracts was also blocked by MG132 treatment (Figures 1D and 1E), further confirming that FEN1 is degraded via the proteasome pathway.
SUMO3 modification mediates FEN1 degradation
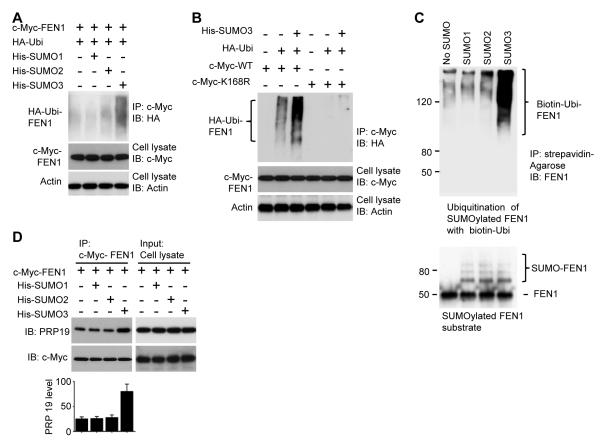
Small ubiquitin-like modifiers (SUMO) can play an important role in protein degradation (Chen et al., 2011) (Chen et al., 2011; Prudden et al., 2007). Mammals have three SUMO paralogues: SUMO 1, 2, and 3, which are attached to various target proteins through the action of two distinct enzymes, a SUMO activating enzyme E1 (SAE1/SAE2 complex) and a SUMO conjugating enzyme E2 (UBC9) (Wilkinson and Henley, 2010). We confirmed that FEN1 can be modified by all three of the SUMO isoforms both in vitro and in cells (Figure S2). We showed that over-expression of SUMO1, 2, or 3 does not affect the cell cycle profile (Figure S3G and H). We then tested if FEN1 degradation is SUMOylation-dependent and if specific SUMO isoforms are involved using three HeLa cell lines which express similar amounts of His-tagged SUMO1, SUMO2 or SUMO3 (Tatham et al., 2009). Levels of FEN1 from the extracts of the three HeLa cell lines were determined by Western blotting. Overexpression of SUMO3 but not SUMO1 or 2, led to a dramatic reduction of FEN1 (Figure 2A, lanes 1, 2, 5 and 8), suggesting that modification with SUMO3 is involved in FEN1 degradation. When UBC9 was co-expressed with SUMO3, FEN1 levels were reduced even further than when SUMO3 was overexpressed alone (Figure 2A, lane 8 vs. lane 9).The increased degradation of FEN1 induced by UBC9 overexpression could be reversed by overexpressing SUMOylation-specific protease 1 (SENP1), ( Ahlmark et al., 1999) (Figure 2A, lane 9 vs. 10). These data together indicated that SUMO3 modification is an important step in the FEN1 degradation pathway. Although FEN1 can be SUMOylated by all three SUMO isoforms both in vitro and in cells (Figure S2), this study focused on the SUMO3 modification because it was the only isoform associated with FEN1 degradation (Figure 2A).
Figure 2. SUMO3 modification mediates FEN1 degradation.
(A) Exogenous expression of SUMO3 in HeLa cells reduces FEN1 levels. Western blotting of FEN1 levels in HeLa cells overexpressing SUMO 1, 2 or 3 with and without UBC 9 and/or SENP1. (B) Validation of SUMO3 modification site in vitro. SUMOylation was conducted in vitro with purified SUMO3 protein and WT or K168R FEN1, then the reactions analyzed by Western blot using an anti-FEN1 antibody. (C) Validation of SUMO3 modification site in cells. HeLa cells were co-transfected with His-tagged SUMO3 and c-Myc-tagged WT or K168R FEN1. Shown is the Western blot of the total cell lysates with or without Ni-NTA purification. (D) Degradation of WT and K168R FEN1 in G2/M cell extracts. WT or K168R His-FEN1 was incubated with G2/M cell extract for the indicated times. His-FEN1 was analyzed by Western blotting using an anti-His antibody. (E) 32P-labeled WT or K168R FEN1 was incubated with G2/M cell extract for the indicated times with or without SENP1 or MG132. His-FEN1 was pulled down by Ni-NTA beads and the radioactivity measured by liquid scintillation. WT FEN1 incubated in G2/M cell extract, ■; K168R FEN1 incubated in G2/M extract,▽; WT FEN1 incubated with G2/M extract and SENP1, ▲; and WT FEN1 incubated with G2/M extract and MG132 proteasome inhibitor, ●. The error bars in the panels D and E represent mean ± SD from three independent experiments. See also Figure S2.
Lysine 168 (K168) was identified by mass spectrometry as the major SUMO3 modification site on FEN1 (Figure S2I). When K168 was mutated to arginine (R), the mutant FEN1 protein (K168R) could no longer be modified by SUMO3 in vitro or in cells (Figure 2B, C), indicating that K168 is the major, if not the only, SUMO3 modification site of FEN1. Furthermore, we determined that K168R FEN1 was resistant to degradation by G2/M cell extracts (Figure 2D and 2E), providing strong evidence that SUMO3 modification of K168 is required for FEN1 degradation.
SUMO3 modification of FEN1 is phosphorylation-dependent
We noticed that not all of the FEN1 protein is degraded in the G2/M phase (Figure 1A & C, Figure S1B), raising the question as to how FEN1 is selected and targeted for degradation. Previously, we showed that FEN1 is phosphorylated in the late S phase, allowing it to dissociate from the DNA replisome after completion of its role in lagging strand DNA synthesis (Guo et al., 2010). However, the fate of phosphorylated FEN1 after dissociation is unknown. Based on previous reports that phosphorylation can play important roles in mediating protein degradation (Welcker et al., 2004; Yada et al., 2004) and SUMOylation may be phosphorylation-dependent (Hietakangas et al., 2006; Mohideen et al., 2009), we speculated that phosphorylation of FEN1 may be required to promote the SUMO3 modification. To test our hypothesis, FEN1 was phosphorylated and then assayed for SUMOylation with SUMO3 in vitro. We observed that the efficiency of SUMOylation was dramatically increased if FEN1 was phosphorylated (Figure 3A). As might be expected, phosphorylation did not induce or enhance the SUMO3 modification of the K168R FEN1 SUMOylation mutant protein (Figure S3A).
Figure 3. SUMO3 modification of FEN1 is phosphorylation-dependent.
(A) Phosphorylation enhances SUMO3 modification of FEN1 in vitro. Wild-type or in vitro phosphorylated FEN1 were in vitro SUMOylated with SUMO3. Shown is the Western blot of the reaction mixtures probed with anti-FEN1 antibody. (B) Elimination of phosphorylation disrupts SUMO3 conjugation. HeLa cells were co-transfected with plasmids encoding His-tagged SUMO3, and c-Myc-tagged WT, S187A or S187D FEN1. Cell lysates were analyzed directly by Western blotting or were pulled-down by Ni-NTA and then analyzed by Western blotting. (C) Olomoucine treatment inhibits FEN1 modification by SUMO3. HeLa cells overexpressing His-SUMO3 and c-Myc tagged FEN1 were treated with or without olomoucine and lysed. Cell lysates were directly analyzed by Western blotting or were pulled-down with Ni-NTA and then analyzed by Western blotting. (D) Olomoucine inhibits SUMO3-induced FEN1 degradation in cells. HeLa cells overexpressing His-SUMO3 were treated with olomoucine or MG132. Cells were lysed and analyzed by Western blotting. Quantitation is shown in the bottom panel. (E) Degradation of WT, S187A or S187D FEN1 in G2/M cell extract. WT or mutant His-FEN1 was incubated with G2/M cell extract for the indicated times. Shown is the Western blotting of His-FEN1 using an anti-His antibody. (F) Interaction of the SUMOylation ligase UBC9 with WT, S187A or S187D FEN1 in cells. HeLa cells overexpressing c-Myc-tagged WT, S187A or S187D FEN1 were lysed. C-Myc-tagged proteins were purified with agarose resin coupled with anti-c-Myc antibody and Western blotted with UBC9 and FEN1. The error bars in the panels D and E represent mean ± SD from three independent experiments. See also Figure S3.
To test the effect of phosphorylation on the SUMOylation of FEN1 in cells, we co-transfected HeLa cells with plasmids expressing His-SUMO3 and either c-Myc-tagged WT FEN1, the phosphorylation-defective S187A mutant or the phosphorylation-mimic S187D mutant (Guo et al., 2010; Henneke et al., 2003).
Lysates were pulled down with Ni-NTA resin then analyzed by Western blotting. In comparison to WT FEN1, the S187A mutation blocked FEN1 SUMOylation, whereas S187D dramatically enhanced it (Figure 3B). Treatment of cells with the kinase inhibitor Olomoucine (Henneke et al., 2003), reduced FEN1 phosphorylation dramatically but did not affect the cell cycle profile using our assay conditions. When we pre-treated the cells with Olomoucine prior to pull-down, the SUMO3 modification of WT FEN1 was significantly inhibited (Figure 3C). Altogether, these results indicate that the SUMO3 modification of FEN1 is regulated or induced by phosphorylation of FEN1 at S187. Consistently, we found that Olomoucine treatment prevented degradation of FEN1 in cells in G2/M phase (Figure 3D). Moreover, the S187A FEN1 mutant protein was resistant to degradation by G2/M cell extract, whereas the S187D mutant showed enhanced degradation compared to WT FEN1 (Figure 3E).
Because SUMOylation requires the interaction of target proteins with the SUMOylation conjugating enzyme UBC9 (Geiss-Friedlander and Melchior, 2007), we speculated that the enhanced SUMOylation of S187D FEN1 may be due to a higher affinity of UBC9 for the S187D mutant protein than WT FEN1. To test this hypothesis, extracts from cells overexpressing c-Myc-tagged WT, S187A or S187D FEN1 were pulled down with anti-c-Myc antibody and analyzed by Western blotting using an anti-UBC9 antibody. The results did indeed indicate that S187D protein had higher binding affinity for the UBC9 ligase than WT FEN1 and S187A (Figure 3F).
Phosphorylation has been reported to reduce FEN1 endonuclease activity (Henneke et al., 2003). However, in our assay the phosphorylation-mimic mutant S187D did not show a significant loss of activity (Figure S5 and Guo et al., 2008). This discrepancy could be due to the different flap substrate that we used in our assay. In the current work, we used the so called “double flap substrate” that contains a 3′ flap and represents an in vivo substrate that can enhance the flap endonuclease activity more than 10 folds. The 3′ flap is an important substrate feature that helps the enzyme to efficiently bind and cleave the substrate (Finger et al., 2009), which minimizes the defects caused by modification or mutation.
FEN1 is ubiquitinated by a UBE1/UBE2M/PRP19 complex
The fact that degradation of FEN1 is inhibited by MG132 (Figure 1C-E) indicated that FEN1 is ubiquitinated so that it can be recognized by the proteasome as a target for degradation. Indeed, we observed that FEN1 was ubiquitinated in vitro (Figure 4A) and in cells (Figure 4B). Moreover, overexpression of ubiquitin in cells led to reduced FEN1 levels (Figure 4C). Mass spectrometry indicated that lysine 354 (K354) of FEN1 was the ubiquitination site (Figure S4) and both in vitro and intracellular experiments showed that the mutation of lysine 354 to arginine (K354R) abolished FEN1 ubiquitination (Figure 4A, B), confirming FEN1 is ubiquitinated at K354. SUMO3-induced degradation of FEN1 in cells was blocked by the ubiquitination inhibitor PYR-41 (Tokumoto et al., 1997) (Figure 4D). In addition, the K354R ubiquitination mutant protein was resistant to degradation by G2/M cell extracts in vitro (Figure 4E). Together, these data strongly suggest that SUMO3-induced FEN1 degradation is mediated by the ubiquitin-proteasome pathway.
Figure 4. Identification of the FEN1 ubiquitin ligase.
(A) In vitro ubiquitination. His-tagged WT or K354R FEN1 and ubiquitin protein were incubated with a HeLa cell fraction that served as a mixture of ubiquitination enzymes (Boston Biochem Inc, K960). The reaction mixture was purified by Ni-NTA resin and analyzed by Western blotting using an anti-ubiquitin antibody. (B) Ubiquitination of FEN1 in cells. Lysates of HeLa cells overexpressing HA-tagged ubiquitin, His-tagged WT or K354R FEN1 were analyzed by directly by Western blot or were purified by Ni-NTA followed by Western blot. (C) Western blot analysis of FEN1 levels in cell lysates from HeLa cells transfected with plasmids encoding ubiquitin. (D) PYR-41 inhibits SUMO3-induced FEN1 degradation in cells. HeLa cells overexpressing His-SUMO3 were treated with the ubiquitination inhibitor PYR-41 or proteasome inhibitor MG132. Cells were lysed and analyzed by Western blotting. (E) 32P-labeled WT or K354R FEN1 was incubated with G2/M cell extract for the indicated time with or without PYR-41 or MG132. His-FEN1 was pulled down by Ni-NTA beads and the radioactivity in solution was measured by liquid scintillation. WT FEN1 incubated with G2/M cell extract, ■; K354R FEN1 incubated with G2/M extract, ▽; WT FEN incubated with G2/M extract and PYR-41, ▲; and WT FEN1 incubated with G2/M extract and MG132, ●. The error bars represent mean ± SD from three independent experiments. (F) Co-immunoprecipitation assay from HeLa cell lysates using a FEN1-specific antibody and then Western blotting using antibodies against UBE1, UBE2M, PRP19 and UBQLN4. (G) Antibody depletion and in vitro ubiquitination assay. His-tagged FEN1 was incubated with ubiquitin and HeLa cell fractions, with or without depletion by the indicated antibodies. The reaction mixture was then analyzed by Western blotting using anti-ubiquitin antibody. (H) In vitro ubiquitination of FEN1 with purified UBE1, UBE2M and PRP19. Shown is the Western blotting of reaction products using an anti-FEN1 antibody. See also Figure S4.
To identify the specific enzymes responsible for FEN1 ubiquitination, FEN1 interacting proteins were pull-down from cell extract and analyzed by MS spectrometry. Four of the FEN1 interacting proteins that we identified (Figure 4F) have been previously implicated as playing a role in the ubiquitination pathway. They are UBE1, UBE2M, PRP19 and UBQLN4. UBE1 has been reported to have E1 ubiquitin-activating activity (Jin et al., 2007) and UBE2M has been reported to have E2 ubiquitin-conjugating activity (Osaka et al., 1998). PRP19 was reported to have E3 ligase activity (Vander Kooi et al., 2006), and UBLNQ4 may play a role in de-ubiquitination (Saharia et al., 2010).
To assess the roles of these proteins in FEN1 ubiquitination, HeLa cell fractions S-100 were pre-depleted with antibodies against each of the four proteins and ubiquitination assays were performed in vitro. Depletion of UBQLN4 had no effect on FEN1 ubiquitination, whereas depletion of UBE1 and UBE2M abolished FEN1 ubiquitination, and depletion of PRP19 showed a substantial reduction of FEN1 ubiquitination (Figure 4G). The partial ubiquitination activity remaining in the PRP19-depleted fraction could be due to the redundant activity of other ubiquitination E3 ligases present in the fraction or an incomplete depletion of PRP19 activity. We also tested purified UBE1, UBE2 and PRP19 and showed they could ubiquitinate FEN1 in vitro (Figure 4H).
SUMO3 modification stimulates FEN1 ubiquitination
The observation that SUMO3-induced degradation of FEN1 was blocked by the ubiquitination inhibitor PYR-41 (Figure 4D) suggested that SUMO3 modification is an upstream event of FEN1 ubiquitination. We hypothesized that SUMO3 modification of FEN1 stimulates ubiquitination and leads to its degradation. Indeed, as shown in Figure 5A, overexpression of SUMO3 resulted in dramatically greater FEN1 ubiquitination than did overexpression of SUMO1 and 2 (Figure 5A). Furthermore, in cells expressing the K168R FEN1 mutant, the stimulatory effect of SUMO3 on FEN1 ubiquitination was blocked (Figure 5B). These results suggest that SUMO3 modification enhances FEN1 ubiquitination. This conclusion was further supported by sequential SUMOylation and ubiquitination assays in vitro (Figure 5C). To explore the mechanism by which SUMO3 stimulates FEN1 ubiquitination, we examined if SUMO3 attachment to FEN1 could stimulate its interaction with PRP19. HeLa cells were co-transfected with c-Myc-FEN1 and SUMO1, 2 or 3. The expressed c-Myc-FEN1 was then pulled down with an anti-c-Myc antibody. Western blotting analysis of PRP19 levels in the pulled-down samples showed that FEN1 bound more PRP19 in cells overexpressing SUMO3 than in cells overexpressing SUMO1 or SUMO2 (Figure 5D), indicating that SUMO3 conjugation to FEN1 strengthens FEN1’s interaction with PRP19.
Figure 5. SUMO3 modification induces FEN1 ubiquitination.
(A) SUMO3 enhances ubiquitination of FEN1 in cells. HeLa cells were co-transfected with plasmids encoding c-Myc FEN1, HA-ubiquitin and/or His-SUMO1/2/3. c-Myc-FEN1 was pulled down by beads pre-coated with anti-c-Myc antibody, followed by Western blotting analysis using an anti-HA antibody. Total cell lysate was also analyzed by Western blotting using anti-c-Myc or anti-actin antibodies. (B) The K168R FEN1 mutation is resistant to SUMO3-stimulated ubiquitination in cells. HeLa cells overexpressing His-SUMO3, HA-ubiquitin or/and c-Myc-tagged WT or K168R FEN1 were lysed and pulled down by anti-c-Myc antibody, followed by Western blotting using an anti-HA antibody. (C) In vitro ubiquitination of SUMOylated FEN1. FEN1 SUMOylated in vitro with SUMO1, 2 or 3 was ubiquitinated with purified UBE1, UBE2M, PRP19 and biotinylated ubiquitin. The ubiquitination mixture was then purified by streptavidin agarose, followed by Western bottling using an anti-FEN1 antibody. (D) PRP19 preferentially binds to SUMO3-FEN1 in cells. Lysates of HeLa cells co-transfected with c-Myc-FEN1 and His-SUMO1/2/3 were pulled down with c-Myc antibody and detected by Western blotting using anti-PRP19 antibody. Quantitative results are shown in the bottom panel. The error bars represent mean ± SD from three independent experiments.
Sequential PTMs modulate cell cycle dependent degradation of FEN1
We speculated that the PTMs that mediate FEN1 degradation occur sequentially and each modification is required for the next reaction to proceed. We overexpressed plasmids encoding mutant versions of FEN1 in the three identified PTM sites in cells, confirmed that none of the mutations affected the FEN1 flap endonuclease activity (Figure S5), and then assessed the FEN1 modification profiles. Although WT FEN1 can be modified by phosphorylation, SUMOylation and ubiquitination (Figure 6A), the phosphorylation mutant S187A blocked all three FEN1 modifications (Figure 6A), indicating that phosphorylation is required upstream of SUMOylation and ubiquitination. The SUMOylation mutant K168R eliminated SUMOylation and ubiquitination, but not phosphorylation, indicating that SUMOylation occurs downstream of phosphorylation but upstream of ubiquitination (Figure 6A). Finally, the ubiquitination mutant K354R only affected the ubiquitination step (Figure 6A), indicating it is downstream of both phosphorylation and SUMOylation. In contrast to the WT protein, the three mutants were not only resistant to their respective PTMs, but also to G2/M degradation (Figure 6B). In agreement with these results, addition of phosphorylation (Olomoucine), SUMOylation (ginkgolic acid) (Fukuda et al., 2009), ubiquitination (PYR-41) or proteasome (MG132) inhibitors all blocked WT FEN1 degradation by G2/M phase cell extracts (Figure 6C).
Figure 6. Sequential PTMs lead to programmed degradation of FEN1.
(A) Phosphorylation, SUMOylation and ubiquitination of FEN1 in sequence. Cells were treated with MG132 to inhibit ubiquitination-mediated protein degradation. Exogenous FEN1 was immunoprecipitated from lysates of cells overexpressing c-Myc-tagged WT, S187A, K168R, and K354R FEN1, followed by Western blotting with indicated antibodies. (B) Mutation of FEN1 phosphorylation, SUMOylation or ubiquitination sites prevents FEN1 degradation. HeLa cells overexpressing exogenous WT, S187A, K168R or K354R FEN1 were synchronized at the S and G2/M phases, lysed and analyzed by Western blotting with anti-FEN1 antibodies. (C) FEN1 degradation is blocked by inhibitors of phosphorylation (olomoucine), SUMOylation (ginkgolic acid), ubiquitination (PYR-41) and the proteasome (MG132). His-tagged FEN1 was incubated with HeLa cell lysates from cells at G2/M phase and different inhibitors for 6 hr. The mixture was then analyzed by Western blotting using anti-His and anti-actin antibodies. (D) His-tagged FEN1 labeled with 32P was subjected to in vitro SUMOylation and ubiquitination. SUMO3-32P-His-FEN1 and ubiquitin-32P-His-FEN1 were purified by Ni-NTA resin, followed by anti-SUMO3 and anti-Ubiquitin antibody resins, respectively. The modified species of FEN1 immobilized on the Ni-NTA or antibody resins were incubated with extracts of cells from G2/M phase in the presence of indicated inhibitors. After 6 hr, the radioactivity in solution was quantified by liquid scintillation. The error bars represent mean ± SD from three independent experiments. (E) A model of sequential modifications to degrade FEN1. In late S phase, FEN1 is phosphorylated, resulting in dissociation from PCNA and the DNA replication fork. Once phosphorylated FEN1 is released from the DNA replication fork, it is then SUMOylated, which triggers ubiquitination by PRP19 and ultimately its degradation. See also Figure S5.
We prepared 32P-His-FEN1, SUMO3-modified 32P-His-FEN1 and ubiquitinated 32P-His-FEN1 in vitro (see Experimental Procedures), immobilized these proteins on Ni-NTA beads and incubated them with extracts from G2/M phase cells in the presence of different PTM inhibitors. The intensity of radioactivity then measured in solution represented the extent of FEN1 degradation. If FEN1 was phosphorylated, its degradation could be blocked by downstream inhibitors of SUMOylation (ginkgolic acid) and ubiquitination (PYR-41). However, if FEN1 was SUMOylated, its degradation could be blocked by the ubiquitination inhibitor PYR-41, but not by the phosphorylation inhibitor olomoucine (Figure 6D). These data further indicate that FEN1 ubiquitination is downstream of SUMOylation, which is downstream of phosphorylation. A model for the sequential modification of FEN1, which leads to its degradation, is illustrated in Figure 6E.
Defects in FEN1 degradation result in cell cycle delay and genome instability
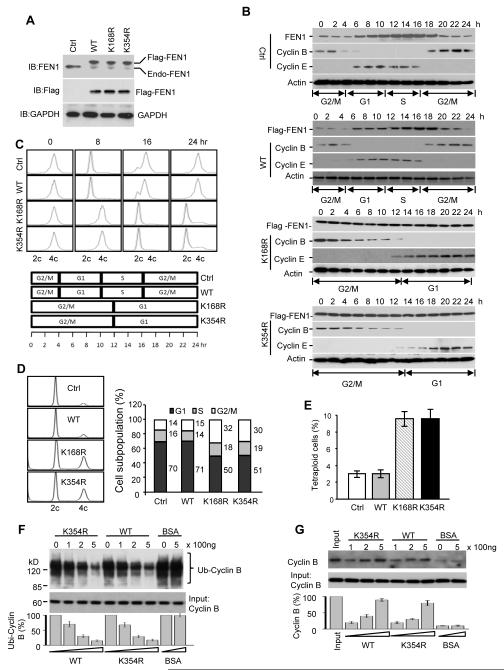
We have proposed that the timely degradation of FEN1 is important for the maintenance of genome integrity and cell cycle progression. To test this hypothesis, we established BJ cell lines, in which flag-tagged WT, K168R or K354R FEN1 mutants were individually overexpressed and then the endogenous FEN1 was depleted by shFEN3 shRNA (Saharia et al., 2008; Saharia et al., 2010) (Figure 7A). Consistent with our previous observations, the endogenous and exogenous WT FEN1 fluctuated throughout the cell cycle but the levels of K168R and K354R FEN1 remained constant. We also noticed that in the cell lines expressing degradation-resistant FEN1 mutant proteins, Cyclin B persists longer than in the parental cell line, raising the possibility that Cylcin B might be stabilized in cells if FEN1 is not degraded. When compared to the parental and WT FEN1 overexpressing cell lines, the K168R and K354R cell lines had an extended G1 and G2/M phase (Figure 7C). Flow cytometric analysis of the non-synchronized cells indicated that cells overexpressing K168R or K354R had a lower G1 population and higher G2/M population. This cell cycle delay had a substantial effect on chromosomal segregation and endoduplication. We found that cell lines overpressing non-degradable FEN1 have higher levels of tetraploidy than WT cell lines.
Figure 7. Defective FEN1 degradation results in cell cycle delay.
(A) Western blot analysis of cell lines with endogenous FEN1 depletion and exogenous FEN1 overexpression. (B) Western blot analysis of FEN1, Cyclin B and Cyclin E expression during the cell cycle. Parental cells (Ctrl) or cells overexpressing WT, K168R or K354R FEN1 were synchronized at G2/M phase and released. Ectopically expressed FEN1 was detected by anti-Flag antibody. (C) FACS profile of cells examined in (B). (D) FACS analysis of cell cycle phase of unsynchronized cells overexpressing WT, K168R or K354R FEN1 is shown in the left panel, while the quantification of FACS analysis is shown in right panel. (E) Chromosome number counting. (F) Inhibition of ubiquitination of Cyclin B by FEN1. Cyclin B was ubiquitinated in vitro using a ubiquitination kit (Boston Biochemical) and biotin-ubiquitin in the presence of gradient amounts of FEN1 or BSA. Ubiquitinated samples were pulled down by Stratavidin beads, followed by Western blotting using anti-Cyclin B antibody. The relative amount of ubiquitinated Cyclin B was quantified in the bottom panel. (G) Inhibition of degradation of Cyclin B by FEN1. Degradation assay of Cyclin B was carried out in vitro using HeLa S100 fraction (Boston Biochemical). Cyclin B was determined by Western blotting and quantified in the bottom panel. The error bars in the panels E, F, and G represent mean ± SD from three independent experiments.
Cyclin B has been reported to be degraded via a ubiquitin-proteasome pathway (Glotzer et al., 1991) and degradation of this protein is critical for completion of the cell cycle (Chang et al., 2003; Tokumoto et al., 1997; Zhang et al., 1998). The prolonged cell cycle period we observed in cell lines overexpressing non-degradable FEN1 and the relative high level of Cyclin B in these cells motivated us to determine if the overexpression of FEN1 could prevent Cyclin B ubiquitination and degradation. Cyclin B was ubiquitinated in vitro in the presence of gradient increases of WT or K354R FEN1. We found that both WT and K354R FEN1 could inhibit Cyclin B ubiquitination in a dose-dependent manner (Figure 7F). Moreover, when WT or K354R FEN1 protein was added to a Cyclin B degradation reaction mixture, the degradation of Cyclin B was reduced compared with BSA controls (Figure 7G). These data suggest that presence of FEN1 blocks Cyclin B degradation by exerting negative effects on its ubiquitination.
DISCUSSION
Because of its critical role in DNA replication, FEN1 is essential for the maintenance of genome stability and integrity. We hypothesized that FEN1 could cause DNA damage and affect the progression of the cell cycle if it was present and active at inappropriate times. Therefore, we were interested in determining the fate of FEN1 in late S phase after DNA replication is complete, when FEN1 is demethylated and phosphorylated, and dissociates from DNA replication fork. We found that FEN1 is degraded after S phase and that the process is regulated by the specific and sequential PTMs, phosphorylation, followed by SUMOylation and then ubiquitination. Soluble phosphorylated FEN1 is degraded by a SUMOylation-dependent, ubiquitin-proteasome pathway. We propose that clearance of soluble FEN1 is a protection mechanism that prevents harmful effects that could be caused if an active RNA/DNA nuclease was present in the wrong location at inappropriate times. We showed that overexpression of a FEN1 degradation mutant resulted in severe cellular phenotypes that included obvious alteration of cell cycle progression time and chromosomal segregation abnormalities (Figure 7). The current work provides evidence not only for a clearance mechanism for FEN1 and, potentially, other nucleases, but also for the programmed regulation of multiple sequential PTMs.
Poly-SUMO chains can exert a signaling function that is in stark contrast to their perceived role as antagonists to the effects of ubiquitin (Ulrich, 2008). Through the action of a newly discovered class of ubiquitin ligases (STUBL), the attachment of poly-SUMO chains to a protein substrate was shown to promote its subsequent ubiquitination and degradation (Tatham et al., 2008) The hallmark of this family of ubiquitin ligases is the presence of multiple SUMO-interacting-motifs (SIMs) (Tatham et al., 2008; Lallemand-breithenbach et al., 2008), which allows the ligases to recognize poly-SUMOylated proteins as ubiquitination targets. STUBLs specifically recognize poly-SUMO chains as a signal to ubiquitinate proteins for degradation (Ulrich, 2008) and recruit the poly-SUMOylated proteins via the tandem SIM repeats. Indeed, UBE1, UBE2 and PRP19, which we propose are responsible for ubiquitinating SUMOylated FEN1, contain multiple SIMs (Figure S6A) and therefore we asked if the PRP19 SIMs are important for interaction of PRP19 with SUMO protein. When the PRP19 SIMs were mutated, the interaction of PRP19 with SUMO3 was substantially reduced (Figure S6B, C).
The mammalian SUMO family includes the SUMO1, SUMO2 and SUMO3 isoforms. SUMO1 has long been believed to have different functions from SUMO2 or 3 due both to its different intracellular localization patterns and its different responses to environmental stimuli (Saitoh and Hinchey, 2000). In contrast, SUMO2 and 3 have always been thought to have identical functions because of their nearly identical sequence (Sekiyama et al., 2008). Recently, however, studies of overexpression, silencing and rescuing SUMO2 and SUMO3 respectively, have shown that these two isoforms are not mutually replaceable (Sang et al., 2011), and suggest that they might be different in terms of substrates and functions. The current study shows that the ubiquitination and subsequent degradation of FEN1 is stimulated by SUMO3 but not SUMO2 (Figure 5), adding more evidence that these two closely related SUMO paralogues have distinct functions. However, it is still possible that there may be competition between the SUMO proteins for substrate availability. We found that co-transfection of SUMO1 with SUMO3 substantially inhibited SUMO3-induced FEN1 degradation in cells (Figure S7). In contrast, co-transfection of SUMO2 rarely inhibited SUMO3-induced FEN1 degradation. The different outcomes observed in our study could be mediated by the N-termini of these isoforms, where the sequences are the most different between the two. The first 5 amino acids of SUMO2 are “MADEK,” whereas in SUMO3 they are “MSEEK”. It has been proposed that SUMO3 contains a phosphorylatable serine (serine 2) at its N-terminus. Phosphorylation of this serine could give SUMO3 different biochemical functions from SUMO2 (Matic et al., 2008). In the current study, we observed that SUMO3-conjugated FEN1 displayed a stronger interaction with the E3 ubiquitin ligase PRP19 compared to WT or SUMO2-conjugated FEN1 (Figure 5D).
Several examples illustrate the biological significance of SUMOylation-induced, ubiquitin-mediated proteasome degradation of target proteins. For example, treatment of mammalian cells with arsenic trioxide, which induces the degradation of the oncogenic protein PML-RAR to cure acute promyelocytic leukemia, promotes SUMOylation of the fusion protein, which, in turn, triggers its proteasomal degradation in an RNF4-dependent manner (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Zhang et al., 2010). Our findings indicate that degradation of FEN1 late in S-phase could be a signal for Cyclin degradation and exit from S phase. Blocking FEN1 degradation by mutating the SUMOylation or ubiquitination sites led to accumulation of Cyclins B and E, and delayed the cell cycle (Figure 7B, C). As a consequence, cells expressing the mutation had abnormal distributions of cell subpopulations in different phases of the cell cycle (Figure 7D). Cells were arrested in G2/M phase, which subsequently induced polyploidization. These observations are consistent with those observed in cancer cells and clinical samples from cancer patients. High FEN1 levels are often associated with cancer cells (Singh et al., 2008), illustrating the potential consequences of impaired FEN1 degradation in cancer etiology. Imbalances in DNA replication and repair enzymes have been implicated in many diseases, including cancer (Canitrot et al., 2006). The study of PTM-mediated regulation of protein functions should be helpful in linking genetic variation with population susceptibility to various cancers, as well as for defining the relationship between environmentally-induced PTM dysfunction and diseases such as cancer.
EXPERIMENTAL PROCEDURES
Cell culture and synchronization and flow cytometry
HeLa cells were synchronized as previously reported (Bernkop-Schnurch et al., 1999; Xu et al., 2009). FACS was performed by flow cytometry core facility in the City of Hope’s Beckman Research Institute.
In vitro phosphorylation and radio-labeling of FEN1 by 32P
In vitro phosphorylation was performed as previously reported (Henneke et al., 2003; Guo et al., 2010).
In vitro SUMOylation
SUMOylation reactions were performed using SUMOylation kits (Boston Biochemicals). The reaction mixtures for SUMOylation contained 5 μg His-tagged FEN1, 1 μg SUMO activating enzyme E1 (SAE1/SAE 2), 1 μg conjugating enzyme E2 (UBC9), 5 μg SUMO 1, 2 or 3 in reaction buffer containing 50 mM Tris (pH 8.0), 5 mM ATP and 5 mM MgCl2 in a total volume of 20 μl. The reaction was incubated at 37 °C for 3 hours. The method for purification of SUMOylated proteins was modified from (Niemi et al., 1999).
In vitro ubiquitination
Ubiquitination assays were conducted either with HeLa cell lysate fractions (Boston Biochemicals, K960) as the enzyme resource or with a mixture of purified enzymes. Recombinant His-FEN1 was incubated with ubiquitin and HeLa cell lysate fractions, or purified UBE1, UBE2M and PRP19 in a reaction buffer provided by the manufacturer (Boston Biochemicals). The reaction was incubated at 37 °C for 3 hours. Ubiquitinated FEN1 was purified as described previously (Laurin et al., 1999).
FEN1 degradation assay by G2/M cell lysate
Recombinant His-tagged FEN1 was incubated with lysate from G2/M cells in the presence of 10 mM ATP, as well as phosphatase inhibitors (Thermo). Aliquots of the mixture were taken at 1, 2, 4 and 6 hour time points. His-FEN1 was identified by Western blotting using an anti-His antibody. In a parallel experiment, 32P-labeld His-FEN1 was immobilized on Ni-NTA resins and incubated with G2/M cell lysate containing 10 mM ATP and phosphatase inhibitors. At the indicated times after incubation, an aliquot of supernatant was taken and its radioactivity measured by liquid scintillation.
Virus production and infection
To establish cell lines with endogenous FEN1 depletion and exogenous FEN1 expression, we requested a lentivirus based vector pResQ-Flag-FEN1-shFEN3 from Sheila Stewart’s laboratory. This vector contains a 3x flag-tagged FEN1 cDNA and lentiviral-expressed RNA interference (RNAi) hairpins targeting FEN1 (shFEN3). Virus production and infection has been reported previously (Saharia et al., 2008; Saharia et al., 2010).
Supplementary Material
HIGHLIGHTS.
FEN1 is sequentially modified by phosphorylation, SUMOylation and ubiquitination
Sequential modification of FEN1 programmed FEN1 degradation after S phase
FEN1 ubiquitination is specifically stimulated by FEN1 SUMO3 modification
Defects in FEN1 degradation delay cell cycle and cause genome instability
ACKNOWLEDGEMENTS
We thank Michael H. Tatham and Ronald T. Hay for HeLa-based cell lines that stably overexpress SUMO1, 2, and 3, and guidance for SUMOylation experiments, the City of Hope Mass Spectrometry Core facility for technical assistance in determining the SUMOylation and ubiquitination sites on FEN1 and the Protein Mass Spectrometry Analysis Center, Institutes of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, for identification of the FEN1 interacting proteins. We thank Drs Margaret Morgan and Keely Walker for editorial assistance. This work was supported by NIH grants R01CA085344 and R01 CA073764 (to B.H.S.), P30 CA033572 to City of Hope Comprehensive Cancer Center from the National Cancer Institute and the Priority Academic Program Development Award for Jiangsu Higher Education Institutions (PAPD) from Jiangsu provincial government to Nanjing Normal University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes supplementary experimental procedures and 7 figures can be found with this article online.
REFERENCES
- Ahlmark M, Vepsalainen J, Taipale H, Niemi R, Jarvinen T. Bisphosphonate prodrugs: synthesis and in vitro evaluation of novel clodronic acid dianhydrides as bioreversible prodrugs of clodronate. J.Med.Chem. 1999;42:1473–1476. doi: 10.1021/jm9810809. [DOI] [PubMed] [Google Scholar]
- Bernkop-Schnurch A, Kirchmayer R, Kratzel M. Synthesis, development and in vitro evaluation of drug delivery systems with protective effect against degradation by pepsin. J. Drug Targeting. 1999;7:55–63. doi: 10.3109/10611869909085492. [DOI] [PubMed] [Google Scholar]
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J. Regulation of Protein Degradation. Plant Cell. 1995;7:845–857. doi: 10.1105/tpc.7.7.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S, Urbe S, Clague MJ. The met receptor degradation pathway: requirement for Lys48-linked polyubiquitin independent of proteasome activity. J. Biol. Chem. 2004;279:52835–52839. doi: 10.1074/jbc.M407769200. [DOI] [PubMed] [Google Scholar]
- Chang DC, Xu N, Luo KQ. Degradation of cyclin B is required for the onset of anaphase in Mammalian cells. J. Biol. Chem. 2003;278:37865–37873. doi: 10.1074/jbc.M306376200. [DOI] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- Chen SC, Chang LY, Wang YW, Chen YC, Weng KF, Shih SR, Shih HM. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J. Biol. Chem. 2011;286:31373–31384. doi: 10.1074/jbc.M111.254896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crinelli R, Bianchi M, Menotta M, Carloni E, Giacomini E, Pennati M, Magnani M. Ubiquitin over-expression promotes E6AP autodegradation and reactivation of the p53/MDM2 pathway in HeLa cells. Mol. Cell. Biol. 2008;318:129–145. doi: 10.1007/s11010-008-9864-8. [DOI] [PubMed] [Google Scholar]
- Finger LD, Blanchard MS, Theimer CA, Sengerová B, Singh P, Chavez V, Liu F, Grasby JA, Shen B. The 3′ Flap Pocket of Human Flap Endonuclease 1 Is Critical for Substrate Binding and Catalysis. J. Biol. Chem. 284(33):22184–94. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Re. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol.Cell.Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nature Chem. Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 2003;22:4301–4313. doi: 10.1038/sj.onc.1206606. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Nat. Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;26(115):565–76. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nature Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Laurin P, Ferroud D, Klich M, Dupuis-Hamelin C, Mauvais P, Lassaigne P, Bonnefoy A, Musicki B. Synthesis and in vitro evaluation of novel highly potent coumarin inhibitors of gyrase B. Bioorg. Medicinal Chem. Lett. 1999;9:2079–2084. doi: 10.1016/s0960-894x(99)00329-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu.Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Matic I, Macek B, Hilger M, Walther TC, Mann M. Phosphorylation of SUMO-1 occurs in vivo and is conserved through evolution. J. Prot. Res. 2008;7:4050–4057. doi: 10.1021/pr800368m. [DOI] [PubMed] [Google Scholar]
- Mohideen F, Capili AD, Bilimoria PM, Yamada T, Bonni A, Lima CD. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi R, Vepsalainen J, Taipale H.a., Jarvinen T. Bisphosphonate prodrugs: synthesis and in vitro evaluation of novel acyloxyalkyl esters of clodronic acid. J. Med. Chem. 1999;42:5053–5058. doi: 10.1021/jm991109o. [DOI] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J. Biol. Chem. 2010;285:27057–27066. doi: 10.1074/jbc.M110.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, Kernstine KH, Lin D, Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nature Protocols. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Torres MP, Clement ST, Cappell SD, Dohlman HG. Cell cycle-dependent phosphorylation and ubiquitination of a G protein alpha subunit. J Biol Chem. 2011;286:20208–16. doi: 10.1074/jbc.M111.239343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto T, Yamashita M, Tokumoto M, Katsu Y, Horiguchi R, Kajiura H, Nagahama Y. Initiation of cyclin B degradation by the 26S proteasome upon egg activation. J. Cell Biol. 1997;138:1313–1322. doi: 10.1083/jcb.138.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. The fast-growing business of SUMO chains. Mol. Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Vander Kooi CW, Ohi MD, Rosenberg JA, Oldham ML, Newcomer ME, Gould KL, Chazin WJ. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochem. 2006;45:121–130. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Liu DX, Pagano M, Ravid K. Ubiquitin-dependent degradation of cyclin B is accelerated in polyploid megakaryocytes. J. Biol. Chem. 1998;273:1387–1392. doi: 10.1074/jbc.273.3.1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.