Abstract
Cancer chemotherapy inhibits tumor growth, in part, by triggering apoptosis, and anti-apoptotic proteins reduce the effectiveness of chemotherapy. Clusterin, a chaperone-like protein that binds to apoptotic and DNA repair proteins, is induced by chemotherapy and promotes tumor cell survival. Histone deacetylase inhibitors (HDIs) such as sodium butyrate and suberoylanilide hydroxamic acid (SAHA) are pharmacological agents that induce differentiation and apoptosis in cancer cells by altering chromatin structure, and we have found that combinations of chemotherapeutic drugs such as doxorubicin and HDIs efficiently induce apoptosis, even though they paradoxically induce high levels of clusterin. The hyper-expressed form of clusterin localizes to mitochondria, inhibits cytochrome c release, and is inhibited by the proteasome. When HDIs are used as single agents, clusterin suppresses cytochrome c release and apoptosis. However, doxorubicin/HDI-induced apoptosis is not inhibited by clusterin, and clusterin-resistant apoptosis corresponds with markers of the extrinsic/receptor-mediated apoptotic pathway. Thus, chemotherapy-HDI combinations are capable of overcoming an innate anti-apoptotic pathway of tumor cells, suggesting that chemotherapy-HDI combinations have potential for treating advanced stage breast cancer.
Keywords: clusterin, doxorubicin, breast cancer, apoptosis, caspase, PARP, histone deacetylase, calpain, proteasome
1. Introduction
Cancer cells are characterized by increased DNA replication, and many types of cancer chemotherapy target dividing cells by damaging DNA or inhibiting DNA replication. Doxorubicin and etoposide inhibit topoisomerase II, while camptothecins inhibit topoisomerase I [1], and the resulting DNA damage triggers apoptosis. Cancer cells develop resistance to DNA damaging agents, in part, by circumventing apoptotic pathways that are present in non-malignant cells [2]. Histone deacetylase inhibitors (HDIs) are small molecules that preferentially induce apoptosis in cancer cells [3] and also induce differentiation [3, 4]. The binding site for HDIs resembles a pocket which contains a Zinc atom [3], and a broad variety of compounds have HDI activity. Several of these are in clinical trials for cancer [5]. HDIs have also been used in combination with various anti-neoplastic drugs, generally increasing their tumoricidal activity [6–10]. Histone deacetylase inhibitors function, in part, by altering the expression of numerous genes that regulate differentiation [11, 12], apoptosis [13], and components of the proteasome [14].
When exposed to apoptotic stresses, a number of cell types induce clusterin, a pro- or anti-apoptotic protein with chaperone activity [15]. Clusterin, which is also called apolipoprotein J and testosterone repressed prostate message 2 [16], among others, is strongly induced by chemotherapy [17–21], and clusterin up-regulates chemotherapy resistance in tumor cell lines [19, 22, 23]. Clusterin is overexpressed in some tumors [1, 24–28], where it presumably suppresses apoptosis during cellular transformation and metastasis. Clusterin expression decreases in other tumors [18, 28], where it may play a pro-apoptotic role. In some cell types, clusterin is synthesized as a pro-form that is glycosylated, cleaved, and secreted as a heterodimer [16]. Clusterin is also expressed as an intracellular variant [29–31] that can arise through alternate splicing of exons 1 and 3 [32] or as a non-glycosylated full-length protein that is not a splice variant [33]. A number of additional modifications can also alter the electrophoretic mobility of clusterin.
Intracellular clusterin can localize to the membranes of the endoplasmic reticulum or mitochondria [34, 35], where it binds to Bax, a pro-apoptotic member of the Bcl-2 protein family, and suppresses apoptosis [34]. Following cellular damage, Bax and Bak form a membrane pore through which cytochrome c and other mitochondrial proteins are released into the cytoplasm [36]. Cytochrome c then nucleates the formation of the apoptosome, which activates caspase 3 [37]. Clusterin binds directly to Bax and inhibits its oligomerization, but does not alter its conformation or localization [34]. Other clusterin splice variants localize to the nucleus, where they bind to Ku70 [30], a DNA repair protein [38], and promote apoptosis [30]
We found previously that clusterin was induced by doxorubicin in the p53-negative breast cancer cell line MDA-MB-231, but not in p53-positive MCF-7 cells [17]. Furthermore, inhibiting clusterin induction by RNAi sensitized the cells to doxorubicin [17]. Similar results were detected in osteosarcoma cells [19]. In the present study, we demonstrate that clusterin is regulated transcriptionally and post-transcriptionally by histone deacetylases. We also show that clusterin inhibits HDI-induced apoptosis by suppressing the intrinsic/mitochondrial apoptotic pathway, but that the ability of clusterin to suppress apoptosis is overcome by combinations of chemotherapy and HDIs. Our findings suggest that cellular chemoresistance pathways can be circumvented by novel chemotherapy combinations that activate multiple apoptotic pathways.
2. Materials and methods
2.1. Cell growth and treatments
MDA-MB-231 and MDA-MB-435S [39] cells were maintained Dulbecco’s modified Eagle medium containing 10% serum supreme supplemented with penicillin and streptomycin. Doxorubicin (Sigma, St. Louis, MO), camptothecin (Sigma), etoposide (Sigma), sodium butyrate (Alfa Aesar, Ward Hill, MA), and SAHA (Biomol, Plymouth Meeting, PA) were used at doses indicated in the text. For RNAi transfections, cells (500,000/100 mm dish) were transfected with 220 pmoles of RNA oligonucleotide duplexes (clusterin third exon, Ambion ID#146049) diluted in 1 ml of Opti-MEM medium and Oligofectamine (both from Invitrogen, Carlsbad, CA) as described [17]. After an overnight incubation, cells were split in normal medium to a density of 500,000 cells/100 mm dish and treated with the indicated drugs. Cells were then harvested 24–48 hours after drug addition.
2.2. Expression analysis
For western blots, cells were lysed in NP-40 buffer (1% NP-40, 20 mM Tris, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, pH 7.4, and 10 μg/ml of the protease inhibitors aprotinin and leupeptin), separated by SDS-PAGE, transferred to Immobilon P membranes (Millipore, Billerica, MA), and probed as described [40]. The antibodies to clusterin (C-18, #sc-6419), PARP (F-2), pro-caspase 3 (H-227), cytochrome c (A-8), Akt (B-5), Ku70 (A-9), IκBα (H-4) and Bax (2D2) were from Santa Cruz Biotechnologies (Santa Cruz, CA), and were used at a dilution of 1:1000. The clusterin C-18 immunogenic peptide for the C-18 antibody was from Santa Cruz. Antibodies to Fas ligand, RIP, FADD and pro-caspase 8 were from Transduction Labs/BD Biosciences (San Jose, CA). Other antibodies were to tubulin (Labvision/Neomarkers, Inc., Fremont, CA, used at a dilution of 1:2000) and cytochrome c oxidase IV, sub-unit II (COX II, 12C4, Invitrogen).
For RT-PCR, MDA-MB-231 cells were treated with various drugs, and RNA was purified, reverse transcribed with random hexamers, and amplified using Taq polymerase (GenScript, Piscataway, NJ) in an Eppendorf Master Cycler (Eppendorf, Westbury, NY) for 26–36 cycles of 94° C for 1 min., 55° C for 1 min., and 72° C for 1 min. PCR reactions contained primers to clusterin and actin, which served as an internal control for cDNA loading. The primer sequences were hCLU-5′ (ACAGGGTGCCGCTGACC), as described by Leskov [32], and CLU+260R (TGGTCTCATTTAGGGCATCC) for clusterin, BAX+213F (AGTAACATGGAGCTGCAGAGG) and BAX+447R (CCAACAGCCGCTCCCGGAGG) for Bax, and BCL2F (CGACTTCGCCGAGATGTCCAGCCAG) and BCL2R (ACTTGTGGCCCAGATAGGCACCCAG) for Bcl2. The actin primers have been described previously [41]. PCR products were visualized by electrophoresis in 2% agarose 1000 (Invitrogen).
2.3. FACS analysis and caspase 8 assays
FACS analysis was performed by fixing cells after 48 hours treatment in ethanol and resuspending them in phosphate-buffered saline containing 20 μg/ml propidium iodide and 20 μg/ml DNase-free RNase, as described previously [17]. For caspase 8 activity assays, approximately 107 cells were lysed in 100 μl of lysis buffer (50 mM HEPES, pH 7.4, 0.1% CHAPS, 1 mM dithiothreitol, 0.1 mM EDTA, 0.1% NP-40). Caspase activity was measured in triplicate in a reaction containing 20 μl of lysate, 70 μl of reaction buffer (50 mM HEPES, pH 7.4, 0.1% CHAPS, 100 mM NaCl, 10 mM dithiothreitol, and 1 mM EDTA), and 10 μl of 1 mg/ml Ac-IETD-pNA (N-acetyl-Ile-Glu-Thr-Asp p-nitroanilide, Biomol, Inc.). The reaction was incubated at 37°C, and the absorbance at 405 nm was measured after 1, 2, and 3 hours. Protein content was measured by Bradford assay, and the activity was calculated as pmoles pNA cleaved/μg protein/minute.
2.4. Clusterin localization
For mitochondrial fractionation, MDA-MB-231 cells were treated, harvested, and lysed in digitonin lysis buffer (75 mM NaCl, 1 mM NaH2PO4, 8 mM Na2HPO4, 250 mM sucrose, 190 μg/ml digitonin; 250 μl/100 mm dish) on ice for 5 minutes, and then centrifuged at 13,000 g for 5 minutes at 4°C, as described [42]. The pellets were resuspended in radioimmunoprecipitation (RIPA) buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM sodium chloride, and 1 mM NaH2PO4, 8 mM Na2HPO4) on ice for 30 minutes and then centrifuged at 13,000 g for 10 minutes. The supernatant was then sheared by 5–10 passages through a 26 gauge needle.
The procedure for immunofluorescence has been described recently. Briefly, cells were fixed in 3.7% formaldehyde, permeabilized in 0.1% Triton X-100, pre-hybridized with 10% fetal bovine serum, incubated with the anti-clusterin C-18 antibody (Santa Cruz) at a dilution of 1:100, and detected with a Texas Red-labeled anti-goat secondary antibody. COX-II was visualized using the 12C4 monoclonal antibody at a dilution of 1:100 and a FITC-labeled anti-mouse secondary. Cells were detected using a Leica TCS confocal microscope.
3. Results
3.1. Histone deacetylase inhibitors elevate clusterin stability in doxorubicin-treated cells
MDA-MB-231 human breast cancer cells are largely resistant to chemotherapy-induced apoptosis. However, co-administration of 0–1 μM of the topoisomerase II inhibitor doxorubicin (adriamycin) with the histone deacetylase inhibitor (HDI) sodium butyrate for 48 hours caused a marked increase in p112PARP (the full-length form of poly ADP-ribose polymerase) cleavage (Fig. 1A, lanes 3–5) and apoptotic cells (Fig. 1E) compared to doxorubicin or sodium butyrate alone (Fig. 1A, lanes 2 and 6–8, and Fig. 1B–D). Both results are representative of more than three independent experiments. Clusterin was moderately induced by doxorubicin and sodium butyrate (Fig. 2A, lanes 2–3), as reported previously [11, 17] while combined doxorubicin-sodium butyrate treatment caused a 2.4-fold hyper-expression of clusterin (Fig. 2A, lane 4) compared to doxorubicin alone. At this time and dose, p112PARP and pro-caspase 3 were efficiently cleaved (Fig. 2B and C, lanes 4), indicating that the cells were apoptotic. The 60 kDa band detected in Figure 2A was completely blocked by the immunogenic peptide, demonstrating that the antibody was specific for clusterin (Fig. 2D, lanes 3 and 4). Thus, chemotherapy/sodium butyrate combinations synergize to induce apoptosis while paradoxically inducing the anti-apoptotic protein clusterin.
Fig. 1.
Apoptosis is induced by doxorubicin combined with the histone deacetylase inhibitor sodium butyrate (HDI).. A, Western blot for polyADP ribose polymerase (PARP), a marker of apoptosis, or tubulin as a control for protein loading (lower panel). MDA-MB-231 cells were either untreated (lane 1), or treated with 5 mM HDI (sodium butyrate, lane 2), fixed concentrations of HDI with 0.04, 0.2, or 1 μM doxorubicin (lanes 3–5), or the same concentrations of doxorubicin alone (lanes 6–8). The p85 form of PARP is a marker for apoptosis. B–E, Flow cytometry of MDA-MB-231 cells showing the induction of apoptosis by doxorubicin plus HDIs. The sub-G1 DNA is the left-most peak in panel E. Cells were untreated (panel B) or treated with 1 μM doxorubicin (panel C), 5 mM sodium butyrate (panel D), or the same doses of doxorubicin and sodium butyrate in combination (panel E). Samples were analyzed 48 hours after initiating treatment.
Fig. 2.
Clusterin levels are induced by doxorubicin and histone deacetylase inhibitors. For panels A, B, C, and E, MDA-MB-231 cells were untreated (lanes 1), or treated with doxorubicin (lanes 2), a histone deacetylase inhibitor (HDI, sodium butyrate, lanes 3), or doxorubicin plus HDI (lanes 4) for 48 hours. A–C, western blots were analyzed for clusterin (A), polyADP-ribose polymerase, PARP (B), or pro-caspase 3 (C), with tubulin as a control for protein loading. The 60 kDa form of clusterin was hyper-expressed following doxorubicin/HDI treatment (panel A, lane 4), corresponding with a synergistic increase in apoptosis (panels B and C, lane 4). D, Western blots of untreated (lanes 1 and 3) or doxorubicin/HDI-treated samples (lanes 2 and 4) were probed with the clusterin antibody, without (left lanes) or with (right lanes) its immunogenic peptide, which blocked the clusterin band. E, Expression analysis of clusterin (upper band) or actin (lower band) by RT-PCR. As for panels A–C, cells were untreated (lane 1), or treated with doxorubicin (lane 2), HDI (lane 3), or doxorubicin plus HDI (lane 4). The migration of molecular weight markers is indicated to the left of the figure. The results show an additive increase in doxorubicin transcription following doxorubicin/HDI treatment compared to doxorubicin or HDI alone. F, Clusterin expression shown by RT-PCR in which actin primers were not included in the reaction. The reaction was amplified for additional cycles, demonstrating the absence of a 186 bp band representing the exon 1–3 splice variant. Actin was amplified as a loading control in a separate reaction (bottom panel).
Doxorubicin caused a 6-fold induction of clusterin RNA (Fig. 2E, lanes 1 and 2) that increased by 50% with the addition of sodium butyrate (Fig. 2E, lane 4), an effect that is approximately additive. In contrast, the effect of doxorubicin and sodium butyrate on clusterin protein levels is more than additive, suggesting that the hyper-expression of clusterin following doxorubicin/HDI treatment is not due solely to increased transcription. The primers used for this analysis detected exons 1, 2 and 3 as a 313 bp band (Fig. 2E, lanes 2–4), while an exon 1–3 splice variant, which has been reported previously in breast cancer [32], would produce a 186 bp product. Actin was included as an internal control for cDNA loading (Fig. 2E, lanes 1–4), and the shorter 186 bp clusterin fragment was not detectable even when actin primers were not included in the reaction (Fig. 2E, lanes 5 and 6).
The hyper-expression of clusterin following doxorubicin/HDI treatment was also detected in the breast cancer cell line MDA-MB-435S (Fig. 3A, lane 4). Notably, MDA-MB-435S cells were more sensitive to doxorubicin-induced apoptosis than MDA-MB-231 cells when used as a single agent (compare Fig. 3B, lane 2, and Fig. 2B, lane 2). Clusterin hyper-expression was induced by multiple HDIs including SAHA (Fig. 3C, lane 4) and trichostatin A (data not shown). Furthermore, the topoisomerase I and II inhibitors camptothecin and etoposide induced clusterin (Figure 3D, lanes 2–4), as reported previously [17], and led to clusterin hyper-expression when used in combination with sodium butyrate (Fig. 3D, lanes 5–6). We conclude that multiple chemotherapy/HDI combinations lead to hyper-expression of clusterin.
Fig. 3.
Multiple chemotherapeutic drugs and HDI’s induce clusterin hyper-expression. A, Clusterin was induced by doxorubicin/HDI treatment in the p53-negative breast cancer cell line MDA-MB-435S. Cells were untreated (lane 1) or treated with 1 μM doxorubicin (lane 2), HDI (5 mM sodium butyrate, lane 3), or doxorubicin/HDI (lane 4). B, Following doxorubicin/HDI treatment, p112PARP was efficiently cleaved (lane 4). MDA-MB-435S cells also demonstrated increased sensitivity to doxorubicin (lane 2) compared to MDA-MB-231 cells. C, In MDA-MB-231 cells, clusterin was induced by doxorubicin (lane 2) and 50 μM SAHA (lane 3) and hyper-expressed following treatment with doxorubicin plus SAHA (lane 4), indicating that multiple histone deacetylase inhibitors increase clusterin levels. D, Multiple HDI/chemotherapy combinations increase clusterin levels. Clusterin was induced by the HDI sodium butyrate (lane 2), 1 μM camptothecin (cp, lane 3), and 10 μM etoposide (etp, lane 4), and was hyper-expressed following camptothecin/HDI (lane 5) and etoposide/HDI (lane 6) treatment.
3.2. Clusterin localizes to mitochondria and suppresses cytochrome c release
Clusterin co-purified with a heavy membrane fraction containing mitochondria (Fig. 4A, lanes 2–4) after doxorubicin, sodium butyrate, or doxorubicin/sodium butyrate treatment, similar to a recent report which used an exogenous expression system in fibrosarcoma and prostate cancer cells [34]. Clusterin co-fractionated with the mitochondrial marker COXII (Fig. 4B), but not the cytoplasmic marker Akt (Fig. 4C), and tubulin was used as a control for protein loading throughout (Fig. 4D). This result was confirmed by immunofluorescence, which revealed that a fraction of clusterin co-localized with mitochondria (Fig. 4E–G), and there was additional clusterin staining that radiated into the cytoplasm (Fig. 4E).
Fig. 4.
A clusterin fraction localizes to mitochondria. MDA-MB-231 cells that were untreated (lanes 1 and 5) or treated with doxorubicin (lanes 2 and 6), HDI (lanes 3 and 7), or doxorubicin/HDI (lanes 4 and 8) were separated into mitochondrial/heavy membrane (mito/HM, lanes 1–4) and cytoplasmic (lanes 5–8) fractions and analyzed by western blot for clusterin (A), the mitochondrial marker COX II/cytochrome c oxidase IV, sub-unit II (B), the cytoplasmic marker Akt (C), or tubulin (D), as a control for total protein loading. E, Immunofluorescence for clusterin showing a perinuclear and punctuate cytoplasmic staining pattern. F, Immunofluorescence for COXII showing its mitochondrial localization. G, A merged view of panels E and F, showing overlapping staining in yellow. H, Clusterin is inhibited by the proteasome. MDA-MB-231 (lanes 1–4) or MDA-MB-435S (lanes 5–7) cells were untreated (lanes 1 and 5) or treated with 1 μM doxorubicin (lanes 2 and 6), 100 nM epoxomicin (lanes 3 and 7), or 10 μM MG-132 (lane 4). Protein expression was analyzed by western blot, with tubulin as a control for loading.
To test the identity of proteases that might regulate clusterin levels, MDA-MB-231 cells were treated with the proteasome inhibitors epoxomicin [43, 44] or MG132. Clusterin levels increased significantly following treatment with 100 nM epoxomicin (Figure 4H, lane 3) or 10 μM MG132 (Figure 4H, lane 4), both in MDA-MB-231 (Figure 4H, lanes 1–4) and MDA-MB-435S cells (Figure 4H, lanes 5–7). We conclude that clusterin stability is increased by proteasome inhibition in breast cancer cells.
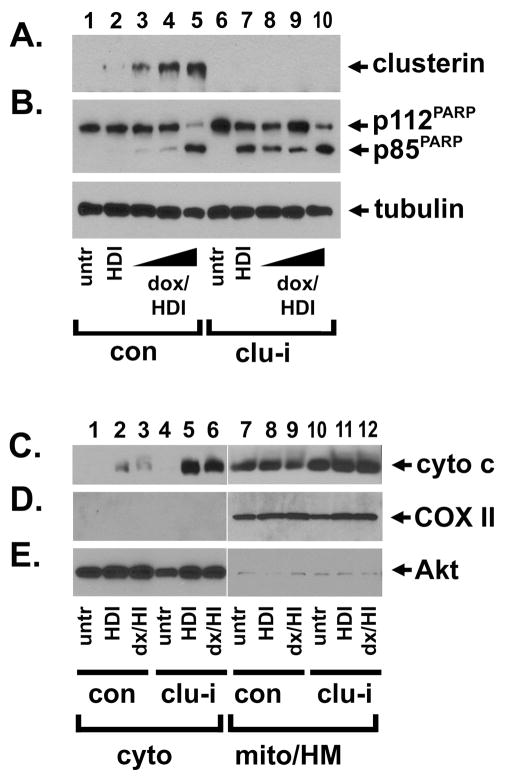
Next, we inhibited clusterin by RNAi and followed with HDI alone or with increasing doses of doxorubicin (Fig. 5A, compare lanes 2–5 with lanes 7–10). Following HDI treatment, clusterin inhibition increased the cleavage of p112PARP (Fig. 5B, lanes 2 and 7), so that the ratio of the p85/p112 forms increased 37-fold when clusterin was absent, suggesting that clusterin suppressed sodium butyrate-induced apoptosis. We detected a similar effect on caspase 3 cleavage, an additional marker of apoptosis (data not shown). In clusterin-inhibited cells, the level of PARP cleavage did not increase further with the addition of 0.04–0.2 μM doxorubicin (Fig. 5B, compare lane 7 with lanes 8–9). At the highest dose of doxorubicin, there was no difference in apoptosis between control and clusterin-inhibited cells (Fig. 5B, lanes 5 and 10).
Fig. 5.
Clusterin inhibits apoptosis and cytochrome c release after histone deacetylase inhibitor treatment. MDA-MB-231 breast cancer cells were transfected with oligonucleotide RNAi duplexes targeting a control sequence (lanes 1–5) or clusterin (lanes 6–10). Cells were left untreated (lanes 1 and 6) or were treated with 5 mM sodium butyrate (lanes 2 and 7), or sodium butyrate in combination with 0.04 μM doxorubicin (lanes 3 and 8), 0.2 μM doxorubicin (lanes 4 and 9), or 1 μM doxorubicin (lanes 5 and 10). A, Clusterin protein levels increased moderately with HDI (lane 2) and increased further with 0.04–1 μM doxorubicin (lanes 3–5), while clusterin expression was effectively inhibited by RNAi (lanes 6–10). B, Treatment with HDI alone did not induce apoptosis in control cells (lane 2) but did where clusterin was inhibited (lane 7). Although high levels of doxorubicin increased apoptosis in combination with HDI (lane 5), there was no further increase in PARP cleavage with the addition of 0.04–1 μM doxorubicin-HDI treatment where clusterin was inhibited (lanes 8–10). Lower panel: tubulin is included as a control for protein loading. C–E, Clusterin inhibits cytochrome c release after HDI treatment. Control (lanes 1–3 and 7–9) or clusterin-inhibited (lanes 4–6 and 10–12) cells were fractionated into cytoplasmic (lanes 1–6) or mitochondrial/heavy membrane (lanes 7–11) fractions and probed for cytochrome c (C), COX II (D), or Akt (E). Cytochrome c release was increased in clusterin-inhibited cells (lanes 5–6) compared to control cells (lanes 2–3). Cytochrome c oxidase (COX II) was a mitochondrial marker, while Akt served as a cytoplasmic marker. We conclude that clusterin suppresses cytochrome c release and apoptosis following treatment with histone deacetylase inhibitors, but that chemotherapy-HDI-induced caspase activation is clusterin-resistant.
Clusterin suppressed the release of cytochrome c into the cytoplasm following HDI treatment (Fig. 5C, compare lanes 2 and 5). In spite of the elevated levels of apoptosis in doxorubicin-sodium butyrate-treated cells (Fig. 2A and E, 3B, and 5B), cytochrome c release was similar to that of sodium butyrate-treated cells (Fig. 5C, lanes 2–3), even in the absence of clusterin (Fig. 5C, lanes 5–6). This suggested that apoptosis induction was not dependent on mitochondrial membrane permeability. For these experiments, the mitochondrial and cytoplasmic markers were the same as described above. We conclude that clusterin localized to mitochondria and suppressed the key event in the extrinsic apoptotic pathway, but that a clusterin-resistant apoptotic mechanism was activated by chemotherapy-HDI treatment.
3.3. The extrinsic apoptotic pathway is activated by doxorubicin-HDI treatment
Our results suggested that apoptotic pathways other than the intrinsic/mitochondrial pathway may be stimulated by doxorubicin/HDI treatment. While clusterin levels increased after doxorubicin-sodium butyrate treatment (Figure 6A, lane 4), pro-caspase 8 was almost completely cleaved (Fig. 6B, lane 4). Pro-caspase 8 proteolysis led to a corresponding increase in its enzymatic activity (Fig. 6G, compare columns 2–3 with column 4). Following clusterin inhibition by RNAi, there was little effect on pro-caspase 8 cleavage, either before or after doxorubicin, sodium butyrate, or doxorubicin/sodium butyrate treatment (Fig. 6H, middle panel, compare lanes 1–4 with lanes 5–8). In addition to pro-caspase 8, IκBα was largely undetectable following doxorubicin/sodium butyrate treatment (Figure 6C, lane 4), while Fas ligand and the Fas binding complex protein FADD (Fas-associated death domain [45, 46]) were increased (Figure 6D and E). In contrast, RIP (receptor interacting protein) increased with doxorubicin and decreased with doxorubicin/HDI treatment (Figure 6F, lanes 2 and 4). We conclude that multiple steps in the extrinsic apoptosis pathway are altered by chemotherapy/HDI combined treatment, but that clusterin does not regulate pro-caspase 8 activation.
Fig. 6.
Chemotherapy-HDI combinations increase pro-caspase 8 activation. A, MDA-MB-231 cells were untreated (lane 1), or treated with 1 μM doxorubicin (lane 2), 5 mM HDI (sodium butyrate, lane 3), or doxorubicin plus HDI (lane 4) for 48 hours. Western blots were analyzed for (A) clusterin, (B) pro-caspase 8, (C) IκBα, (D) FasL, (E) FADD, (F) RIP and tubulin as indicated. Fold changes in protein expression/tubulin ratios, relative to untreated cells, are indicated below each panel. G, Cells were treated as for part A, and caspase 8 activity was measured in the lysates using the chromogenic substrate Ac-IETD-pNA (N-acetyl-Ile-Glu-Thr-Asp p-nitroanilide). The analysis was repeated in triplicate, and fold change in activity relative to untreated cells is shown. H, MDA-MB-231 cells were transfected with oligonucleotide RNAi duplexes targeting a control sequence (lanes 1–4) or clusterin (lanes 5–8). Cells were left untreated (lanes 1 and 5) or were treated with 1 μM doxorubicin (lanes 2 and 6), HDI (5 mM sodium butyrate, lanes 3 and 7), doxorubicin-HDI combined (lanes 4 and 8). Western blots were analyzed for clusterin, pro-caspase 8 and tubulin as indicated. The results show that caspase 8 is activated following doxorubicin-HDI treatment, and that clusterin does not affect caspase 8 cleavage.
4. Discussion
Following DNA damage, we have found that clusterin suppresses the intrinsic apoptosis pathway by limiting cytochrome c release (Figure 7, left), while our data support a model in which histone deacetylases reduce the activity of the extrinsic apoptosis pathway. As a result, apoptosis is largely suppressed, which limits the effectiveness of chemotherapy. In contrast, combining DNA damage with inhibitors of histone deacetylases triggers the extrinsic apoptosis pathway, circumventing the ability of clusterin to suppress apoptosis (Fig. 7, right). The ability of HDIs to suppress the extrinsic apoptosis pathway has been reported previously in leukemia [47], and this report links histone deacetylase-mediated repression of the extrinsic apoptosis pathway to damage-induced apoptosis.
Fig. 7.
Diagram depicting the relationship between clusterin, the intrinsic and extrinsic apoptotic cascades, and histone deacetylases. We have shown that clusterin co-localizes with mitochondria and suppresses cytochrome c release, which triggers apoptosis. Clusterin is regulated by the proteasome, although it is unclear if this regulation is direct. The clusterin-mediated inhibition of the intrinsic apoptotic pathway can be bypassed by activation of the extrinsic pathway, although the precise mechanism through which histone deacetylases perform this function is not known.
It is somewhat paradoxical that histone deacetylase inhibitors induce components of the extrinsic apoptosis pathway while increasing clusterin levels (which inhibits the intrinsic apoptosis pathway). The majority of breast cancers express low levels of clusterin basally [18], and clusterin transcription is induced markedly following treatment with multiple types of chemotherapy [17, 18]. The common regulatory point between clusterin and the extrinsic pathway may be the proteasome [48]. Our results suggest that the proteasome inhibits clusterin levels in multiple cell lines (Figure 4). However, a second proteasome target, IκBα [49], is degraded in doxorubicin/HDI-treated cells under the same conditions that stabilize clusterin. These seemingly contradictory findings can be reconciled if clusterin were inhibited by a second proteolytic pathway, and one candidate is calpain, which inhibits clusterin levels (Figure 4). This multi-step regulation of clusterin may be due to cancer cells degrading clusterin to limit the anti-proliferative effect that it has on some cell lines [50].
There are several notable points involving the induction of apoptosis by chemotherapy/HDI combinations. As a single agent, sodium butyrate treatment triggered modest levels of cytochrome c release and increased levels of IκBα (Figure 6C), as reported previously for other cell types [14, 51]. The addition of a DNA damaging agent induced elevated levels of apoptosis (Figures 1–3, 5, and 6), pro-caspase 8 activation, and induction of components of the extrinsic apoptosis pathway [52], without an increase in cytochrome c release (Figure 5C). Previous work has shown that butyrate induces the transcription of caspase 8 [53], although we did not detect this change in breast cancer cells (data not shown). Butyrate has also been linked to increased expression of death receptors and other apoptosis-related genes in cancer [54–56], and we detected elevated levels of IκBα, Fas ligand, FADD, RIP, and TRAIL in butyrate-treated breast cancer cells (data not shown). However, IκBα and RIP decreased substantially when doxorubicin and HDI were combined, consistent with an anti-apoptotic role for RIP in some cell types [57, 58]. Thus, our results are consistent with a model in which chemotherapy-HDI combinations trigger the extrinsic apoptotic pathway through multiple expression changes in the components of this pathway.
Histone deacetylase inhibitors have been tested in clinical trials for cancer, where they have achieved modest success as single agents. Our results suggest that clusterin reduces the tumoricidal activity of these drugs by inhibiting apoptosis. HDIs induce clusterin transcription [11, 53, 59], and our results suggest that clusterin induction inhibits the effectiveness of the drugs. We chose butyrate for several experiments because it is a natural product of the digestive tract (and is a major component of butter) that was used at concentrations that are lower than physiological levels (which can exceed 100 mM). The effects of butyrate on proliferation and apoptosis are diverse, and include profound effects on differentiation [3, 4, 12, 60], even in breast cancer cells [61]. In MDA-MB-231 cells, sodium butyrate triggered arrest in G2/M, increased sub-G1 content, and G1 arrest (Figure 1). Clusterin can be inhibited clinically by the antisense oligonucleotide OGX-011, which is currently in clinical trials for prostate and lung cancer, and which sensitizes breast cancer cells to several types of chemotherapeutic agents [18, 62]. Our results suggest that OGX-011 combined with histone deacetylase inhibitors may be useful for treating some types of cancer. Furthermore, post-treatment clusterin levels may be a useful biomarker for clinical trials utilizing HDIs, because tumors with high post-treatment levels of clusterin may be more resistant to HDIs clinically than tumors expressing low clusterin levels.
Although this study focuses on cancer chemosensitivity, clusterin expression has been linked to numerous diseases and physiological states, including Alzheimer’s, ischemia, and aging. It is likely that histone deacetylases play an important role in regulating clusterin stability in multiple tissues exposed to various types of stress. Indeed, histone deacetylase inhibitors may be useful in the treatment of inflammatory diseases [63], and clusterin may contribute to the effectiveness of these approaches. Inhibitors of histone deacetylases are undergoing clinical trials for cancer and other diseases, and our results suggest that clusterin may be useful both as a biomarker for the action of these drugs, as well as a target to improve their effectiveness.
Acknowledgments
We are grateful to the advice and encouragement of our colleague, Dr. Steve Zimmer, who passed away in June 2006. We are also grateful to colleagues in the Department of Molecular and Biomedical Pharmacology and to Rachel Chitti and Julia Craven for advice and helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goodman LS, Gilman A, Brunton LL, Lazo JS, Parker KL. Goodman & Gilman’s the pharmacological basis of therapeutics. 11. McGraw-Hill Medical Pub. Division; New York: 2006. [Google Scholar]
- 2.Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW. Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene. 2004;23:2934–2949. doi: 10.1038/sj.onc.1207515. [DOI] [PubMed] [Google Scholar]
- 3.Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27:344–349. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21:351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 5.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 6.Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. In vivo synergy between topoisomerase II and histone deacetylase inhibitors: predictive correlates. Mol Cancer Ther. 2005;4:1993–2000. doi: 10.1158/1535-7163.MCT-05-0194. [DOI] [PubMed] [Google Scholar]
- 7.Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–3822. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- 8.Bevins RL, Zimmer SG. It’s about time: scheduling alters effect of histone deacetylase inhibitors on camptothecin-treated cells. Cancer Res. 2005;65:6957–6966. doi: 10.1158/0008-5472.CAN-05-0836. [DOI] [PubMed] [Google Scholar]
- 9.Marchion DC, Bicaku E, Daud AI, Richon V, Sullivan DM, Munster PN. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–237. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- 10.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 11.Joseph J, Mudduluru G, Antony S, Vashistha S, Ajitkumar P, Somasundaram K. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene. 2004;23:6304–6315. doi: 10.1038/sj.onc.1207852. [DOI] [PubMed] [Google Scholar]
- 12.Davie JR. Inhibition of histone deacetylase activity by butyrate. The Journal of nutrition. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa K, Yasumura S, Atarashi Y, Minemura M, Miyazaki T, Iwamoto M, Higuchi K, Watanabe A. Sodium butyrate enhances Fas-mediated apoptosis of human hepatoma cells. J Hepatol. 2004;40:278–284. doi: 10.1016/j.jhep.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Place RF, Noonan EJ, Giardina C. HDAC inhibition prevents NF-kappa B activation by suppressing proteasome activity: down-regulation of proteasome subunit expression stabilizes I kappa B alpha. Biochem Pharmacol. 2005;70:394–406. doi: 10.1016/j.bcp.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Humphreys DT, Carver JA, Easterbrook-Smith SB, Wilson MR. Clusterin has chaperone-like activity similar to that of small heat shock proteins. J Biol Chem. 1999;274:6875–6881. doi: 10.1074/jbc.274.11.6875. [DOI] [PubMed] [Google Scholar]
- 16.Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34:1430–1448. doi: 10.1016/s1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- 17.Mallory JC, Crudden G, Oliva A, Saunders C, Stromberg A, Craven RJ. A Novel Group of Genes Regulates Susceptibility to Antineoplastic Drugs in Highly Tumorigenic Breast Cancer Cells. Mol Pharmacol. 2005;68:1747–1756. doi: 10.1124/mol.105.016519. [DOI] [PubMed] [Google Scholar]
- 18.So A, Sinnemann S, Huntsman D, Fazli L, Gleave M. Knockdown of the cytoprotective chaperone, clusterin, chemosensitizes human breast cancer cells both in vitro and in vivo. Mol Cancer Ther. 2005;4:1837–1849. doi: 10.1158/1535-7163.MCT-05-0178. [DOI] [PubMed] [Google Scholar]
- 19.Trougakos IP, So A, Jansen B, Gleave ME, Gonos ES. Silencing expression of the clusterin/apolipoprotein j gene in human cancer cells using small interfering RNA induces spontaneous apoptosis, reduced growth ability, and cell sensitization to genotoxic and oxidative stress. Cancer Res. 2004;64:1834–1842. doi: 10.1158/0008-5472.can-03-2664. [DOI] [PubMed] [Google Scholar]
- 20.Miyake H, Hara I, Kamidono S, Gleave ME, Eto H. Resistance to cytotoxic chemotherapy-induced apoptosis in human prostate cancer cells is associated with intracellular clusterin expression. Oncol Rep. 2003;10:469–473. [PubMed] [Google Scholar]
- 21.Miyake H, Nelson C, Rennie PS, Gleave ME. Acquisition of chemoresistant phenotype by overexpression of the antiapoptotic gene testosterone-repressed prostate message-2 in prostate cancer xenograft models. Cancer Res. 2000;60:2547–2554. [PubMed] [Google Scholar]
- 22.Zellweger T, Miyake H, July LV, Akbari M, Kiyama S, Gleave ME. Chemosensitization of human renal cell cancer using antisense oligonucleotides targeting the antiapoptotic gene clusterin. Neoplasia. 2001;3:360–367. doi: 10.1038/sj.neo.7900174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.July LV, Beraldi E, So A, Fazli L, Evans K, English JC, Gleave ME. Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol Cancer Ther. 2004;3:223–232. [PubMed] [Google Scholar]
- 24.Chen X, Halberg RB, Ehrhardt WM, Torrealba J, Dove WF. Clusterin as a biomarker in murine and human intestinal neoplasia. Proc Natl Acad Sci U S A. 2003;100:9530–9535. doi: 10.1073/pnas.1233633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redondo M, Villar E, Torres-Munoz J, Tellez T, Morell M, Petito CK. Overexpression of clusterin in human breast carcinoma. Am J Pathol. 2000;157:393–399. doi: 10.1016/S0002-9440(10)64552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie D, Lau SH, Sham JS, Wu QL, Fang Y, Liang LZ, Che LH, Zeng YX, Guan XY. Up-regulated expression of cytoplasmic clusterin in human ovarian carcinoma. Cancer. 2005;103:277–283. doi: 10.1002/cncr.20765. [DOI] [PubMed] [Google Scholar]
- 27.Lau SH, Sham JS, Xie D, Tzang CH, Tang D, Ma N, Hu L, Wang Y, Wen JM, Xiao G, Zhang WM, Lau GK, Yang M, Guan XY. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- 28.Saffer H, Wahed A, Rassidakis GZ, Medeiros LJ. Clusterin expression in malignant lymphomas: a survey of 266 cases. Mod Pathol. 2002;15:1221–1226. doi: 10.1097/01.MP.0000036386.87517.AA. [DOI] [PubMed] [Google Scholar]
- 29.Humphreys D, Hochgrebe TT, Easterbrook-Smith SB, Tenniswood MP, Wilson MR. Effects of clusterin overexpression on TNFalpha- and TGFbeta-mediated death of L929 cells. Biochemistry. 1997;36:15233–15243. doi: 10.1021/bi9703507. [DOI] [PubMed] [Google Scholar]
- 30.Yang CR, Leskov K, Hosley-Eberlein K, Criswell T, Pink JJ, Kinsella TJ, Boothman DA. Nuclear clusterin/XIP8, an x-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci U S A. 2000;97:5907–5912. doi: 10.1073/pnas.97.11.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy KB, Jin G, Karode MC, Harmony JA, Howe PH. Transforming growth factor beta (TGF beta)-induced nuclear localization of apolipoprotein J/clusterin in epithelial cells. Biochemistry. 1996;35:6157–6163. doi: 10.1021/bi952981b. [DOI] [PubMed] [Google Scholar]
- 32.Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem. 2003;278:11590–11600. doi: 10.1074/jbc.M209233200. [DOI] [PubMed] [Google Scholar]
- 33.O’Sullivan J, Whyte L, Drake J, Tenniswood M. Alterations in the post-translational modification and intracellular trafficking of clusterin in MCF-7 cells during apoptosis. Cell Death Differ. 2003;10:914–927. doi: 10.1038/sj.cdd.4401254. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- 35.Debure L, Vayssiere JL, Rincheval V, Loison F, Le Drean Y, Michel D. Intracellular clusterin causes juxtanuclear aggregate formation and mitochondrial alteration. J Cell Sci. 2003;116:3109–3121. doi: 10.1242/jcs.00619. [DOI] [PubMed] [Google Scholar]
- 36.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell. 2006;10:549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Fisher TS, Zakian VA. Ku: A multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005 doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In vitro. 1978;14:911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 40.Hand RA, Craven RJ. Hpr6.6 protein mediates cell death from oxidative damage in MCF-7 human breast cancer cells. J Cell Biochem. 2003;90:534–547. doi: 10.1002/jcb.10648. [DOI] [PubMed] [Google Scholar]
- 41.Cance WG, Craven RJ, Liu ET. Expression polymerase chain reaction: a sensitive method for analysis of gene expression in human tumours. Surg Oncol. 1992;1:309–314. doi: 10.1016/0960-7404(92)90092-y. [DOI] [PubMed] [Google Scholar]
- 42.Gao M, Fan S, Goldberg ID, Laterra J, Kitsis RN, Rosen EM. Hepatocyte growth factor/scatter factor blocks the mitochondrial pathway of apoptosis signaling in breast cancer cells. J Biol Chem. 2001;276:47257–47265. doi: 10.1074/jbc.M106791200. [DOI] [PubMed] [Google Scholar]
- 43.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bo Kim K, Fonseca FN, Crews CM. Development and characterization of proteasome inhibitors. Methods Enzymol. 2005;399:585–609. doi: 10.1016/S0076-6879(05)99039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 46.Boldin MP, Varfolomeev EE, Pancer Z, Mett IL, Camonis JH, Wallach D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 47.Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, Altucci L, Nervi C, Minucci S, Pelicci PG. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11:71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 48.Roos-Mattjus P, Sistonen L. The ubiquitin-proteasome pathway. Ann Med. 2004;36:285–295. doi: 10.1080/07853890310016324. [DOI] [PubMed] [Google Scholar]
- 49.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 50.Scaltriti M, Santamaria A, Paciucci R, Bettuzzi S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells1. Cancer Res. 2004;64:6174–6182. doi: 10.1158/0008-5472.CAN-04-0920. [DOI] [PubMed] [Google Scholar]
- 51.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Biol Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 52.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 53.Daly K, Shirazi-Beechey SP. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006;25:49–62. doi: 10.1089/dna.2006.25.49. [DOI] [PubMed] [Google Scholar]
- 54.Chopin V, Slomianny C, Hondermarck H, Le Bourhis X. Synergistic induction of apoptosis in breast cancer cells by cotreatment with butyrate and TNF-alpha, TRAIL, or anti-Fas agonist antibody involves enhancement of death receptors’ signaling and requires P21(waf1) Exp Cell Res. 2004;298:560–573. doi: 10.1016/j.yexcr.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 55.Chopin V, Toillon RA, Jouy N, Le Bourhis X. Sodium butyrate induces P53-independent, Fas-mediated apoptosis in MCF-7 human breast cancer cells. Br J Pharmacol. 2002;135:79–86. doi: 10.1038/sj.bjp.0704456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsubaki J, Choi WK, Ingermann AR, Twigg SM, Kim HS, Rosenfeld RG, Oh Y. Effects of sodium butyrate on expression of members of the IGF-binding protein superfamily in human mammary epithelial cells. J Endocrinol. 2001;169:97–110. doi: 10.1677/joe.0.1690097. [DOI] [PubMed] [Google Scholar]
- 57.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 58.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. The EMBO journal. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 59.Lund P, Weisshaupt K, Mikeska T, Jammas D, Chen X, Kuban RJ, Ungethum U, Krapfenbauer U, Herzel HP, Schafer R, Walter J, Sers C. Oncogenic HRAS suppresses clusterin expression through promoter hypermethylation. Oncogene. 2006;25:4890–4903. doi: 10.1038/sj.onc.1209502. [DOI] [PubMed] [Google Scholar]
- 60.Leder A, Leder P. Butyric acid, a potent inducer of erythroid differentiation in cultured erythroleukemic cells. Cell. 1975;5:319–322. doi: 10.1016/0092-8674(75)90107-5. [DOI] [PubMed] [Google Scholar]
- 61.Abe M, Kufe DW. Effect of sodium butyrate on human breast carcinoma (MCF-7) cellular proliferation, morphology, and CEA production. Breast cancer research and treatment. 1984;4:269–274. doi: 10.1007/BF01806038. [DOI] [PubMed] [Google Scholar]
- 62.Biroccio A, D’Angelo C, Jansen B, Gleave ME, Zupi G. Antisense clusterin oligodeoxynucleotides increase the response of HER-2 gene amplified breast cancer cells to Trastuzumab. J Cell Physiol. 2005;204:463–469. doi: 10.1002/jcp.20295. [DOI] [PubMed] [Google Scholar]
- 63.Blanchard F, Chipoy C. Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]