Abstract
Phase segregation and domain formation is observed within the protecting monolayer of gold nanoparticles (AuNPs) using ion mobility-mass spectrometry, a two-dimensional gas-phase separation technique. Experimental data is compared to a theoretical model that represents a randomly distributed ligand mixture. Deviations from this model provide evidence for nanophase separation resulting in anisotropic AuNPs.
Keywords: ligand domains, mass spectrometry, mixed-valent compounds, monolayers, self-assembly
One of the prized characteristics of the monolayer-protected gold nanoparticle (AuNP) is the versatility of its surface.[1] The protecting monolayer is comprised of gold-thiolate complexes, whose gold-sulfur backbones are bound to the gold core while the thiolate tails extend into the surrounding media.[2] Most molecules containing a thiol can be integrated into the monolayer, allowing a variety of organic surfaces to be presented on an AuNP scaffold.[1,3] Mixtures of thiols can be utilized to hone chemical functionality and solvation properties,[3a] permitting the exploration of a tremendous amount of chemical space. This chemical space can be further expanded by the formation of ligand domains[4] through “nanophase separation”[5] based on ligand-ligand interactions and entropic energy gains.[6] Surface organization can be harnessed to optimize physical properties and chemical functionalities, from non-destructive membrane transport[7] to controlled assembly[8] and ligand-abundance-dependent solubility.[9] In this context, mixed-ligand AuNPs are somewhat analogous to biomacromolecules, having a versatile nanoarchitecture which can be refined to induce highly specific chemical interactions.[10]
Existing strategies for characterizing AuNPs with ligand domains, such as scanning tunnelling microscopy[4a,11] and other spectroscopic techniques,[4b,12] provide limited tools for establishing the existence of ligand domains. In order to advance application development and scientific understanding of nanophase separation in AuNP monolayers, strategies must be developed for their facile and rapid characterization. This methodology must be able to distinguish between AuNP “isomers” with nominally identical molecular formulas and varying molecular structure. This is particularly true for applications targeted to biological systems, where accurate characterization is necessary to understand biological function.[13]
We have previously established that gold-thiolate complexes can be desorbed from monolayer-protected AuNP surfaces by the matrix-assisted laser desorption/ionization (MALDI) process, revealing structural characteristics of the protecting monolayer.[14] Ion mobility-mass spectrometry (IM-MS), a two-dimensional gas-phase structural separation technique, is particularly effective for the investigation of desorbed gold-thiolate complexes with high sensitivity.[14a,b] Informed by our previous work, we hypothesize that if gold-thiolate complexes are desorbed as discrete portions of the monolayer, nanophase separation in the monolayer will be reflected in the desorbed gold-thiolate ions observed. That is, the relative abundances of homoleptic and heteroleptic gold-thiolate complex ions should reveal the existence and degree of nanophase separation in the monolayer of the parent AuNP.
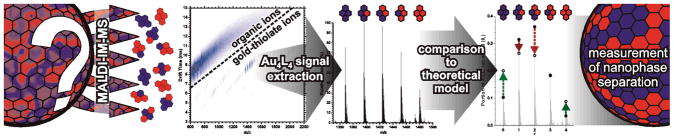
To experimentally test our hypothesis and explore the potential of using MALDI-IM-MS for the identification of ligand domains on AuNP surfaces, we synthesized a number of mixed-ligand AuNPs using both one-step mixed-ligand syntheses[15] and a two-step ligand-exchange process.[16] The formed AuNPs had core diameter averages between 2–4 nm (Figure S1). This size range was predicted to have a greater tendency toward complete phase segregation than larger AuNPs.[6,8b] In a typical experiment (Figure 1), these AuNPs were fragmented by matrix-assisted laser desorption/ionization (MALDI), liberating the gold-thiolate complexes that protect the NP core. The ionized complexes were separated in the gas phase, first by the effective ion surface area and then by the mass-to-charge ratio. A two-dimensional density map was generated, in which dense gold-thiolate ions are clearly separated from organic ions.[14a,b] This gas-phase separation of gold-thiolate complexes from endogenous and exogenous chemical noise allows for the generation of one-dimensional mass spectra which contain only gold-thiolate ions. The Au4L4 ion species was selected as the focus of this work because they were the most abundant gold-thiolate ion species observed.[14a–c,17] These Au4L4 ion species are either directly desorbed from the AuNP surface,[14a,14c] or are products of the rearrangement of “staple” AuxLx+1 species.[2a] In either case, each gold-thiolate complex ion desorbed from the AuNP is expected to contain the ligands present in a given portion of the AuNP surface.
Figure 1.
Typical workflow for experiments presented here. Mixed-ligand AuNPs with unknown levels of nanophase separation are analyzed by MALDI-IM-MS. The MALDI process leads to the fragmentation of protecting gold-thiolate complexes from the AuNP surface. The gold-thiolate ions undergo gas-phase separation from organic ions. The Au4L4 ion species are extracted from the data using software, and their abundances are compared to a theoretical model based on the binomial distribution. Deviations indicate nanophase separation in the AuNP monolayer.
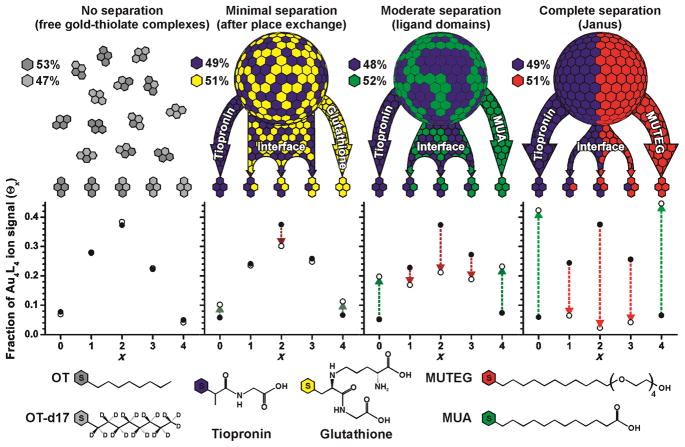
The Au4L4 ions within the resulting spectra were identified and their abundances compared to a theoretical model based on the binomial discrete probability distribution. This model, which represents a random distribution of ligands on the AuNP surface, has been used by Dass and co-workers[18] to study mixed ligand populations on [Au25L18]− molecules. The binomial distribution can also be used as a theoretical model for mixed ligand populations in gold-thiolate ions liberated from AuNPs. Briefly, the binomial distribution describes the probability of x successes for n binary trials based on the probability (p) of success for any one trial. In the context of this work, each trial is considered successful if an alternate ligand, SR′, is found, or unsuccessful if an original ligand, SR, is found. The probability of finding an alternate ligand, SR′, is determined by its relative abundance on the parent AuNP surface (p = % SR′ in monolayer).[14b] Each ligand within the Au4L4 ion is a single trial, yielding four total trials (n = 4), with five possible combinations of SR and SR′ within the Au4L4 ion species. For a mixed-ligand AuNP with a given composition of SR and SR′, the binomial distribution will predict the relative abundances of each Au4L4 ion (Au4(SR)4, Au4(SR)3(SR′)1…). If the ligands are randomly distributed in the protecting monolayer of the AuNP, the binomial distribution will agree with the Au4L4 mass spectral distribution. If nanophase separation is present in the AuNP monolayer, the mass spectral distribution will deviate from the binomial distribution. Homoleptic Au4(SR)4 and Au4(SR′)4 ions will be more abundant than predicted, while heteroleptic Au4(SR)x(SR′)4-x (1 ≥ x ≥ 3) ions will be less abundant. Thus the deviation from the binomial distribution can be correlated to the formation of ligand domains.
As an initial control experiment, we created free gold-thiolate complexes with a mixture of octanethiol (OT) and octanethiol-d17 (OT-d17) ligands. These free complexes are not expected to exhibit any significant phase segregation, and the observed ligand distribution agrees well with the binomial model (Figure 2). The residual sum of squares (r), a measurement of deviation from the binomial model, is 2.6 × 10−4, similar to residual values reported for ligand populations on unfragmented AuNP ions.[18] If the mixed-ligand gold-thiolate complexes are generated by place exchange on an AuNP, the residual is generally higher than the control (r ≈ 10−3). This is true even for ligand mixtures with strong similarities, such as OT:OT-d17 (r = 5.9 × 10−3) or tiopronin:glutathione (Tio:GS, Figure 2). In such cases, the higher residuals may reflect a minimal degree of phase segregation caused by monolayer sites with higher exchange reactivity.[19] If the ligands differ in chemical functionality or length, as with tiopronin:mercaptoundecanoic acid (Tio:MUA), the residual generally increases to >10−2. If the ligand differences are sufficiently strong, the formation of Janus AuNPs with complete nanophase separation can be observed. These AuNPs, such as the tiopronin:mercaptoundecyltetraethylene glycol (Tio:MUTEG) AuNPs, yield residuals above 10−1. For Janus AuNPs with a 50:50 monolayer composition, the amount of ions containing only SR or SR′ should be close to 50% of the total ion signal each. This is the case for Tio:MUTEG, where the homoleptic Au4(Tio)4 and Au4(MUTEG)4 ion species represent 42% and 45% of the Au4L4 ion signal, respectively (Figure 2).
Figure 2.
A comparison of experimental and theoretical ligand distributions for free gold-thiolate complexes and three mixed-ligand AuNPs obtained by ligand-exchange reactions of tiopronin AuNPs with free glutathione, 11-mercaptoundecanoic acid (MUA), or mercaptoundecyltetraethylene glycol (MUTEG). Deviation from the theoretical model indicates the presence of phase-segregated gold-thiolate monolayers on AuNPs. Various ligand mixtures yield different degrees of nanophase separation.
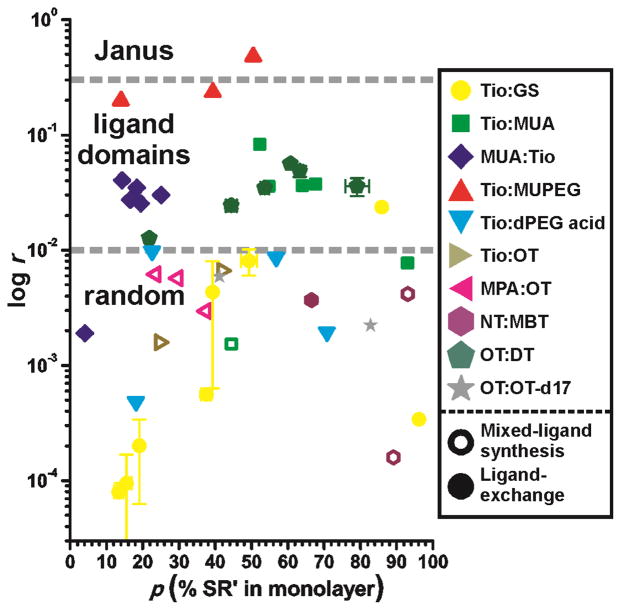
One of the advantages of this strategy is the ability to compare the degree of nanophase separation for multiple AuNP samples. Figure 3 illustrates this ability by comparing the residual sum of squares for various mixed-ligand AuNPs at varying ligand:ligand ratios. In addition to the mixed-ligand AuNPs described in Figure 2, other ligand:ligand ratios and ligand:ligand combinations were tested: MUA:Tio (beginning with MUA AuNPs and adding tiopronin, the inverse order of Tio:MUA shown in the center of Figure 2), Tio:dPEG4 acid® (a mercaptotetraethylene glycol with a carboxylic acid terminus), Tio:OT, mercaptopropionic acid (MPA):OT,[4a] nonanethiol:methylbenzenethiol (NT:MBT),[8a,b] OT:decanethiol (DT),[6] and OT:OT-d17. The lowest residuals are on the order of 10−5, and the pattern illustrated and described in Figure 2 begins to become clear when the residual approaches 10−2. The number of samples scattered between these two values supports the existence of multiple degrees of nanophase separation within a given AuNP sample.[8d,20] Because this technique is averaging with respect to the degree of nanophase separation, the various residuals reflect the relative abundances of AuNPs with phase-segregated or randomly-distributed ligands in the monolayer. For most 50:50 binary ligand mixtures obtained by ligand-exchange reactions, some nanophase segregation is observed (Figure 3). This observation is somewhat surprising, given the strong similarities between ligands such as tiopronin and glutathione. No substantial phase segregation was observed for AuNPs generated by mixed-ligand syntheses, even for ligand mixtures with extreme polarity and length differences. Heating MPA:OT AuNPs at 55 °C for 1h had no apparent effect on the degree of nanophase separation (r = 5.7 and 6.1 × 10−3 before and after heating, respectively).
Figure 3.
A comparison of residual sums of squares (log r, ordinate) for various mixed-ligand AuNPs (above) at different ligand:ligand ratios (SR:SR′, abscissa). Specific ligand mixtures are indicated by color and shape; open and filled symbols indicate mixed-ligand syntheses and ligand exchange reactions, respectively. Abbreviations in the legend are noted in the text. Dashed lines indicate preliminary qualitative assessments of the r values associated with the different types of monolayer structures on AuNPs. The only AuNPs exhibiting phase-segregated monolayers were formed by ligand exchange. Error bars (±1σ, n = 2) are shown for some Tio:GS and OT:DT AuNPs. Deviations are normally within ~3% ligand abundance and ~10−3 for r.
Understanding the lack of phase segregation in mixed-ligand syntheses may require a closer look at the role of gold-thiolate complexes as a precursor to mixed-ligand AuNPs. In solution, the heteroleptic gold-thiolate complexes exhibit no phase segregation (Figure 2). If these complexes are adsorbed to the gold core during the reduction step of the synthesis, the initial state of the AuNP surface will reflect the lack of phase segregation in its component gold-thiolate complexes. Given that nanophase separation requires ligand movement across the AuNP surface post-synthesis, the lack of phase segregation indicates that ligand movement across the surface is minimal. This finding reinforces claims of control over nanophase separation through an interfacial engineering approach,[21] the importance of gold-thiolate complexes to monolayer structure,[14a] and opens the possibility of obtaining diverse AuNP isomers by pursuing multiple synthetic routes.
In this work we have demonstrated the ability to observe and measure phase segregation in the protecting monolayers of AuNPs by characterizing the gold-thiolate complexes which comprise the monolayer. This ability enables a novel strategy for the analysis of nanophase separations on AuNPs which is rapid, semi-quantitative with respect to the degree of phase segregation, and capable of characterizing a wide variety of ligand mixtures. Using this strategy, we were able to compare mixtures of different ligands, ligand:ligand ratios, and synthetic approaches. We found that nanophase separation is often present, though in varying degrees, and that ligand exchange reactions which combine ligands of varying lengths and minimal ligand-ligand interactions maximize the amount of nanophase separation. Though the latter is not a novel aspect of our findings, the observation serves as a partial validation of the results and points to further insights which could be gained using this technique. The strategy described here functions as an excellent characterization technique for mixed-ligand AuNPs, as well as a helpful starting point for future studies of phase-segregated monolayers on AuNPs.
Experimental Section
Nanoparticle synthesis
Tiopronin- and octanethiol-protected AuNPs were synthesized by one-phase and two-phase methods, respectively, as described elsewhere.[14b] Mercaptoundecanoic acid-protected AuNPs were synthesized using a one-phase approach similar to tiopronin with the following modifications: the synthesis was performed in methanol at room temperature using a 1:1:10 Au:MUA:NaBH4 ratio. Some mixed-ligand AuNPs were synthesized using a one-phase approach in ethanol at room temperature.[15b] In each case, Au and the two selected thiols were combined in a 1:0.5:0.5 molar ratio, for a 1:1 Au:thiol ratio overall. Mixed-ligand gold-thiolate complexes were formed by adding AuCl4− to a mixture of OT and OT-d17 in chloroform (1:1.5:1.5 mol ratio). Transmission electron microscopy, thermal gravimetric analysis, and UV-visible spectroscopy were used in addition to IM-MS for the characterization of the AuNP samples.
Ligand exchange reactions
Homoligand AuNPs were dissolved in ~2 mL of a suitable solvent (deionized water, methanol, or dichloromethane). The AuNPs were then combined with various amounts of the chosen free thiol for up to 72 h to allow equilibration. Sample preparation: For hydrophobic samples, a 100 μL solution of dichloromethane containing roughly 0.5 mg of AuNPs was combined with 5 mg of DCTB matrix. A Pasteur pipet was used to deposit roughly 1 μL of the solution on a stainless steel plate. For the remaining samples, a modified sandwich crystallization method[22] was utilized. A 0.5 μL aliquot of a saturated solution of CHCA matrix was deposited on a stainless steel plate. After drying, a 0.5 μL aliquot of the concentrated sample solution was deposited and dried, followed by another 0.5 μL spot of matrix solution. All spectra were obtained on a Waters Synapt G1 or Synapt G2 HDMS in the positive ion mode. Laser intensity was generally set at 20% above threshold, the travelling wave velocity was fixed at 300 m/s while the wave height was ramped from 9 to 16 V. Data were collected for 60 sec for each spectrum.
Data processing and calculations
Mass spectra were extracted from the gold-thiolate region of the ion mobility-mass spectrum using Driftscope v2.1 software (Waters Corp.). Peaks were identified and calibrated using MassLynx 4.1 software (Waters Corp.). The processed spectra were exported to Microsoft Excel, which was used for peak identification and isotopic abundance correction as described previously.[14b] The peak identification cutoffs were placed at 0.5% abundance relative to the base peak and 10 ppm mass accuracy. All ions with an Au4L4 stoichiometry were selected for comparison to a binomial model. For each ligand-ligand combination (i.e., each possible value of x for Au4SRxSR′4-x), the ion abundances were summed and divided by the total abundance of all Au4L4 ions to obtain Θx. The binomial distribution was calculated using Microsoft Excel (function “BINOMDIST”) with the values n = 4, 0 ≤ × ≤ 4, and
where Cx is the sum of ion counts for a given value of x.
Acknowledgments
Thanks to Amanda Agrawal, Tracy Okoli, Prof. Brian Huffman, and Dr. Carrie Simpson for providing samples for analysis, Prof. James McBride and Brian Turner for assistance with TEM measurements, Prof. Richard Caprioli for access to instrumentation, and an anonymous reviewer for insightful and helpful comments. Financial support for this work was provided by the Vanderbilt Chemical Biology Interface (CBI) training program (T32 GM065086), a Vanderbilt Institute of Nanoscale Science and Engineering fellowship, the National Institutes of Health (GM076479), the Vanderbilt College of Arts and Sciences, the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Institute for Integrative Biosystems Research and Education.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author
Contributor Information
Prof. John A. McLean, Email: john.a.mclean@vanderbilt.edu.
Prof. David E. Cliffel, Email: d.cliffel@vanderbilt.edu.
References
- 1.Templeton AC, Wuelfing WP, Murray RW. Acc Chem Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 2.a) Jadzinsky PD, Calero G, Ackerson CJ, Bushnell DA, Kornberg RD. Science. 2007;318:430–433. doi: 10.1126/science.1148624. [DOI] [PubMed] [Google Scholar]; b) Heaven MW, Dass A, White PS, Holt KM, Murray RW. J Am Chem Soc. 2008;130:3754–3755. doi: 10.1021/ja800561b. [DOI] [PubMed] [Google Scholar]; c) Zhu MZ, Eckenhoff WT, Pintauer T, Jin RC. J Phys Chem C. 2008;112:14221–14224. [Google Scholar]; d) Qian H, Eckenhoff WT, Zhu Y, Pintauer T, Jin R. J Am Chem Soc. 2010;132:8280–8281. doi: 10.1021/ja103592z. [DOI] [PubMed] [Google Scholar]; e) Voznyy O, Dubowski JJ, Yates JT, Maksymovych P. J Am Chem Soc. 2009;131:12989–12993. doi: 10.1021/ja902629y. [DOI] [PubMed] [Google Scholar]
- 3.a) Templeton AC, Hostetler MJ, Warmoth EK, Chen S, Hartshorn CM, Krishnamurthy VM, Forbes MDE, Murray RW. J Am Chem Soc. 1998;120:4845–4849. [Google Scholar]; b) Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]; c) Templeton AC, Chen S, Gross SM, Murray RW. Langmuir. 1999;15:66–76. [Google Scholar]
- 4.a) Jackson AM, Myerson JW, Stellacci F. Nat Mater. 2004;3:330–336. doi: 10.1038/nmat1116. [DOI] [PubMed] [Google Scholar]; b) Gentilini C, Franchi P, Mileo E, Polizzi S, Lucarini M, Pasquato L. Angew Chem. 2009;121:3106–3110. doi: 10.1002/anie.200805321. [DOI] [PubMed] [Google Scholar]
- 5.Radzilowski LH, Stupp SI. Macromolecules. 1994;27:7747–7753. [Google Scholar]
- 6.Singh C, Ghorai PK, Horsch MA, Jackson AM, Larson RG, Stellacci F, Glotzer SC. Phys Rev Lett. 2007;99:226106. doi: 10.1103/PhysRevLett.99.226106. [DOI] [PubMed] [Google Scholar]
- 7.Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) DeVries GA, Brunnbauer M, Hu Y, Jackson AM, Long B, Neltner BT, Uzun O, Wunsch BH, Stellacci F. Science. 2007;315:358–361. doi: 10.1126/science.1133162. [DOI] [PubMed] [Google Scholar]; b) Carney RP, DeVries GA, Dubois C, Kim H, Kim JY, Singh C, Ghorai PK, Tracy JB, Stiles RL, Murray RW, Glotzer SC, Stellacci F. J Am Chem Soc. 2007;130:798–799. doi: 10.1021/ja077383m. [DOI] [PubMed] [Google Scholar]; c) Xu Q, Kang X, Bogomolni RA, Chen S. Langmuir. 2010;26:14923–14928. doi: 10.1021/la102540n. [DOI] [PubMed] [Google Scholar]; d) Hu Y, Uzun O, Dubois C, Stellacci F. J Phys Chem C. 2008;112:6279–6284. [Google Scholar]
- 9.Centrone A, Penzo E, Sharma M, Myerson JW, Jackson AM, Marzari N, Stellacci F. Proc Natl Acad Sci U S A. 2008;105:9886–9891. doi: 10.1073/pnas.0803929105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Hung A, Mwenifumbo S, Mager M, Kuna JJ, Stellacci F, Yarovsky I, Stevens MM. J Am Chem Soc. 2011;133:1438–1450. doi: 10.1021/ja108285u. [DOI] [PubMed] [Google Scholar]; b) Liu X, Hu Y, Stellacci F. Small. 2011 doi: 10.1002/smll.201100386. Early View. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AM, Hu Y, Silva PJ, Stellacci F. J Am Chem Soc. 2006;128:11135–11149. doi: 10.1021/ja061545h. [DOI] [PubMed] [Google Scholar]
- 12.a) Centrone A, Hu Y, Jackson Alicia M, Zerbi G, Stellacci F. Small. 2007;3:814–817. doi: 10.1002/smll.200600736. [DOI] [PubMed] [Google Scholar]; b) Pradhan S, Brown L, Konopelski J, Chen S. J Nanopart Res. 2009;11:1895–1903. [Google Scholar]
- 13.Grainger DW, Castner DG. Adv Mater (Weinheim, Ger) 2008;20:867–877. [Google Scholar]
- 14.a) Harkness KM, Fenn LS, Cliffel DE, McLean JA. Anal Chem. 2010;82:3061–3066. doi: 10.1021/ac100251d. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Harkness KM, Hixson BC, Fenn LS, Turner BN, Rape AC, Simpson CA, Huffman BJ, Okoli TC, McLean JA, Cliffel DE. Anal Chem. 2010;82:9268–9274. doi: 10.1021/ac102175z. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gies AP, Hercules DM, Gerdon AE, Cliffel DE. J Am Chem Soc. 2007;129:1095–1104. doi: 10.1021/ja0639057. [DOI] [PubMed] [Google Scholar]; d) Harkness KM, Cliffel DE, McLean JA. Analyst. 2010;135:868–874. doi: 10.1039/b922291j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Chen S, Murray RW. J Phys Chem B. 1999;103:9996–10000. [Google Scholar]; b) Choo H, Cutler E, Shon YS. Langmuir. 2003;19:8555–8559. [Google Scholar]
- 16.Ingram RS, Hostetler MJ, Murray RW. J Am Chem Soc. 1997;119:9175–9178. [Google Scholar]
- 17.a) Fields-Zinna CA, Sampson JS, Crowe MC, Tracy JB, Parker JF, deNey AM, Muddiman DC, Murray RW. J Am Chem Soc. 2009;131:13844–13851. doi: 10.1021/ja905787y. [DOI] [PubMed] [Google Scholar]; b) Fields-Zinna CA, Sardar R, Beasley CA, Murray RW. J Am Chem Soc. 2009;131:16266–16271. doi: 10.1021/ja906976w. [DOI] [PubMed] [Google Scholar]; c) Tang Z, Xu B, Wu B, Germann MW, Wang G. J Am Chem Soc. 2010;132:3367–3374. doi: 10.1021/ja9076149. [DOI] [PubMed] [Google Scholar]
- 18.Dass A, Holt K, Parker JF, Feldberg SW, Murray RW. J Phys Chem C. 2008;112:20276–20283. [Google Scholar]
- 19.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 20.Hu Y, Wunsch BH, Sahni S, Stellacci F. J Scanning Probe Microsc. 2009;4:24–35. [Google Scholar]
- 21.a) Pradhan S, Xu L, Chen S. Adv Funct Mater. 2007;17:2385–2392. [Google Scholar]; b) Kuna JJ, Voitchovsky K, Singh C, Jiang H, Mwenifumbo S, Ghorai PK, Stevens MM, Glotzer SC, Stellacci F. Nat Mater. 2009;8:837–842. doi: 10.1038/nmat2534. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Golding RE, Whittal RM. J Am Chem Soc. 1996;118:11662–11663. [Google Scholar]