Abstract
The muscular dystrophies are a heterogeneous group of genetically caused muscle degenerative disorders. The Kunkel laboratory has had a longstanding research program into the pathogenesis and treatment of these diseases. Starting with our identification of dystrophin as the defective protein in Duchenne muscular dystrophy (DMD), we have continued our work on normal dystrophin function and how it is altered in muscular dystrophy. Our work has led to the identification of the defective genes in three forms of limb girdle muscular dystrophy (LGMD) and a better understanding of how muscle degenerates in many of the different dystrophies. The identification of mutations causing human forms of dystrophy has lead to improved diagnosis for patients with the disease. We are continuing to improve the molecular diagnosis of the dystrophies and have developed a high-throughput sequencing approach for the low-cost rapid diagnosis of all known forms of dystrophy. In addition, we are continuing to work on therapies using available animal models. Currently, there are a number of mouse models of the human dystrophies, the most notable being the mdx mouse with dystrophin deficiency. These mice are being used to test possible therapies, including stem-cell-based approaches. We have been able to systemically deliver human dystrophin to these mice via the arterial circulation and convert 8% of dystrophin-deficient fibers to fibers expressing human dystrophin. We are now expanding our research to identify new forms of LGMD by analyzing zebrafish models of muscular dystrophy. Currently, we have 14 different zebrafish mutants exhibiting various phenotypes of muscular dystrophy, including muscle weakness and inactivity. One of these mutants carries a stop codon mutation in dystrophin, and we have recently identified another carrying a mutation in titin. We are currently positionally cloning the disease-causative mutation in the remaining 12 mutant strains. We hope that one of these new mutant strains of fish will have a mutation in a gene not previously implicated in human muscular dystrophy. This gene would become a candidate gene to be analyzed in patients which do not carry a mutation in any of the known dystrophy-associated genes. By studying both disease pathology and investigating potential therapies, we hope to make a positive difference in the lives of people living with muscular dystrophy.
Keywords: DNA sequencing, Muscle, Muscular dystrophy, Stem cells, Zebrafish
Introduction
The muscular dystrophies represent a genetically heterogeneous group of muscle-wasting disorders characterized by progressive loss of muscle mass, decreased ambulation, and premature death. The most common form of the disease is the X-linked Duchenne muscular dystrophy (DMD), with one in 3,500 males affected worldwide. The second most common form of the disease is X-linked Becker muscular dystrophy (BMD), with one in 20,000 males affected worldwide. For these two dystrophies, comprising nearly 90% of all cases of muscular dystrophy, the underlying cause was identified more than 18 years ago as abnormalities of the gene encoding the protein dystrophin (Burghes 1987; Monaco et al. 1986; Hoffman et al. 1987). The identification of the dystrophin gene was the first example in which a chromosomal map position was used to identify a previously unknown disease gene. From the onset, the gene was recognized to be extremely large and, with the current genome sequence, it is now known to have 79 exons spanning 2,575,000 base pairs of the human X-chromosome (http://www.genome.ucsc.edu/cgi-bin/hgGateway). Large deletions are the most common mutation within the dystrophin locus, most likely because of the large size of the dystrophin gene. The remaining mutations are small insertions/deletions and point mutations in coding sequences and splice sites. The large deletions are readily detected by a PCR-based deletion-screening assay (Chamberlain et al. 1988; Beggs et al. 1990), but the point mutations can be difficult to detect.
Following the cloning of the dystrophin gene, the cDNA was sequenced in its entirety and the encoded amino acids predicted a large rod-like protein of 427 kDa (Koenig et al. 1988). Based on sequence homology, dystrophin was predicted to be a member of the spectrin family of actin binding proteins. Antibodies directed against dystrophin were produced by the immunization of rabbits with different segments of dystrophin expressed as fusion peptides in bacteria. These antibodies detected a low abundance of protein in muscle of a molecular weight of 427 kDa, the molecular weight predicted by the sequence (Hoffman et al. 1987). Antibodies produced by three independent groups localized the dystrophin protein to the inner face of the plasma membrane of muscle cells (Arahata et al. 1988; Bonilla et al. 1988; Zubrzycka-Gaarn et al. 1988). The protein was almost completely absent from the muscle of patients with the severe DMD and of reduced size and abundance in patients with the milder BMD (Hoffman et al. 1988). The detection of dystrophin protein in normal muscle and it's absence from patient muscle led to a protein-antibody-based diagnostic test which could often be prognostic of the disease course. The cloning of the gene responsible for the Duchenne and Becker muscular dystrophies has lead to improved diagnosis of the disorders and the beginnings of an understanding of the molecular mechanisms which cause the disease.
The subsequent biochemical purification of proteins co-purifying with dystrophin led to the identification of a complex of proteins which co-localize and associate with dystrophin at the muscle cell membrane (Ervasti et al. 1990; Yoshida and Ozawa 1990). These co-purifying proteins include: (1) the syntrophins and dystrobrevins, two intracellular proteins which interact directly with the C-terminal domain of dystrophin; (2) the sarcoglycans, a subcomplex of transmembrane proteins; and (3) the dystroglycans, which form a direct link between the extracellular matrix and the intracellular actin cytoskeleton. β-Dystroglycan is a transmembrane protein whose intracellular tail directly binds dystrophin and whose extracellular domain binds α-dystroglycan, which, in turn, binds the extracellular matrix laminins. In the absence of this linkage, the muscle cell membrane is thought to be fragile and more likely to tear under the stress of contraction and relaxation, with the subsequent influx of calcium leading to protein degradation and myofiber degeneration. The identification and characterization of dystrophin and its associated proteins led to new insights into normal muscle function and a better understanding of how abnormalities of dystrophin can lead to myofiber degeneration. As each of the dystrophin-associated proteins was cloned, it was tested for its involvement in the additional autosomal forms of muscular dystrophy that had been described clinically. Interestingly, many of the dystrophin-associated proteins themselves were found to be involved in autosomal forms of dystrophy. Now, more than 20 different genes are known to carry mutations which can give rise to different forms of muscular dystrophy.
Despite this revolution in understanding the pathogenesis and the diagnosis of the muscular dystrophies, there is still no truly effective treatment for patients suffering from the disease. In addition, there are still patients for whom the primary cause of their muscle disease is unknown, either because of inadequate diagnosis or because the mutated gene is still unknown. Here, we will present our strategy to develop an assay to economically and rapidly re-sequence for any patient all genes known to be involved in the various forms of muscular dystrophy. We will also present our current approaches to introduce muscle-derived stem cells into diseased muscle in an attempt to contribute absent gene products, which can contribute to the repair of muscle. We will spend the remainder of the presentation on how one can use zebrafish as a model of human muscle disease and, possibly, identify novel genes responsible for or involved in the muscular dystrophies.
Results and discussions
Improved diagnosis of muscular dystrophy
The current approaches to diagnose muscular dystrophy rely on a PCR-based deletion test (Chamberlain et al. 1988; Beggs et al. 1990) of DNA isolated from patients suspected to have the disease. This is usually followed by protein-antibody-based tissue staining of patient-derived muscle biopsies to assay protein expression and localization. This strategy usually identifies most of the DMD/BMD patients and indicates likely candidate genes for limb girdle muscular dystrophy (LGMD). Based on this diagnostic procedure, only those patients for whom a dystrophin deletion was found will then have an exact molecular diagnosis. Sequencing can be used in some of the LGMD patients for whom protein studies have implicated a particular gene, but, in general, most patients for whom a deletion has not been found will have no specific molecular diagnosis. There have been some recent improvements though, as methods have been developed to scan the dystrophin locus for mutations by SSCP (Hayashi and Yandell 1993) and Wave technology (Bennett et al. 2001). In addition, a group has recently developed an alternative sequencing approach for the dystrophin locus (Flanigan et al. 2003), which is robust and economical, but has not been refined for other loci and has some limitations on its use.
Over the last few years, Applied Biosystems has developed a universal condition PCR strategy for the sequencing of genes. We have been working with them to develop this strategy for sequencing all the exons and promotor regions of all the genes implicated in muscular dystrophy. The goal of this project is to develop a clinical tool to help diagnose patients with the disease (Bennett et al. 2005). The key is to develop primers for PCR amplification which will all work under the same PCR conditions. This will allow the robotic preparation of PCR reactions and their processing in multi-well plates. The genes involved in the dystrophies have been chosen and primers designed to amplify all exons and regulatory regions for each gene. These have been tested for fidelity of amplification and sequencing.
Currently, this sequencing assay has been developed for all exons and areas of interest for ten genes, including dystrophin (Table 1). Some additional genes have only partial coverage, but new primer pairs are currently being tested. The goal will be to have all amplicons for these 12 genes operational and in use by early 2006, and some 20 more genes by early 2007. Our hope is to be able to automate the molecular diagnosis of the dystrophies as schematically outlined in Fig. 1. Our goal in developing this assay is to eventually establish a center that can provide clinical test results for all patients at a reasonable cost.
Table 1.
Table of verified UCDS genes and their amplicons. Tier 1 in row 1 is simply a placeholder to show that 60% of DMD/BMD mutations will be detected by the venerable Beggs–Chamberlain multiplex PCR test, which has been in place for 15 years. All remaining rows are UCDS diagnostic testing tiers and indicate the number of amlicons that have been verified up to 11/22/05. The order of testing will be tier 1 first; if no mutations detected in tier 1, then proceed to tier 2, etc. Costs indicate the minimum cost for reagents and plastic only
| Tier/assay (% detected) | Material cost | Gene name | Protein | Phenotype | No. exons | Approx. no. UCDS amplicons | UCDS amplicons verified as of 11/22/05 | UCDS amplicons not covered as of 11/22/05 |
|---|---|---|---|---|---|---|---|---|
| Tier 1 MUX PCR (60%) | $73 | DMD | Dystrophin | DMD/BMD | 21 | MUX PCR | ||
| Tier 2 (85%) | $118 | DMD | Dystrophin | DMD/BMD | 79 | 118 | 118 | 0 |
| Tier 3 (91%) | $100 | CAPN3 | Calpain 3 | LGMD2A | 24 | 33 | 33 | 0 |
| DYSF | Dysferlin | LGMD2B | 55 | 62 | 55 | 7 | ||
| Tier 4 (95%) | $76 | TRIM32 | Tripatate contain 32 | LGMD2H | 3 | 18 | 18 | 0 |
| CAV3 | Caveolin-3 | LGMD1C | 2 | 12 | 12 | 0 | ||
| FKRP | Fukutin related | LGMD21 (CMD) | 4 | 17 | 17 | 0 | ||
| SGCA | Sarcoglycan alpha | LGMD2D | 10 | 11 | 11 | 0 | ||
| SGCB | Sarcoglycan beta | LGMD2E | 6 | 18 | 18 | 0 | ||
| Tier 5 (98%) | $58 | LMNA | Lamin a/c | LGMD1B (EDMD) | 16 | 28 | ||
| TCAP | Telethonin | LGMD2G | 16 | 8 | ||||
| TTID | Myotilin | LGMD1A | 10 | 22 | ||||
| SGCD | Sarcoglycan delta | LGMD2F | 10 | 13 | 13 | 0 | ||
| SGCG | Sarcoglycan gamma | LGMD2C | 8 | 14 | 14 | 0 | ||
| FCMD | Fukutin | FCMD | 10 | 37 | 37 | 0 | ||
| Tier 6 | NEB | Nebulin | Nemalin | 183 | 169 | 134 | 35 | |
| ACTA1 | Skeletal actin | Nemalin | 7 | 11 |
Fig. 1.
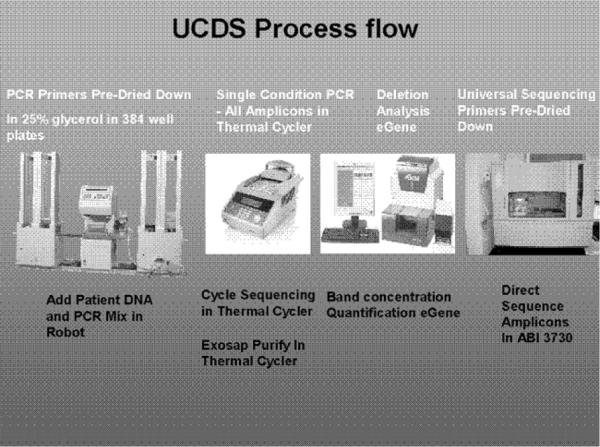
A candidate automation process. A robot, such as a CyBio CyBi-Disk, is used to aliquot primers, DNA, and master mixes to the wells of a 384-well plate, which is then placed in a thermal cycler for amplification of the genetic information. The eGene HDA-GT12 is used to detect deleted genetic regions and to measure the DNA concentration of amplicons to be sequenced. The robot then normalizes the amount of DNA in each well by adding water to some and then prepares cycle sequencing reactions. Sequencing chains are then produced in the thermal cycler, purified on the robot, and sequenced on the ABI 3,730 sequencer
Using a robot, primers will be dispersed into 384 well plates and dried in bulk for future use. Each well will have a different set of primers for a different amplicon of the target gene. Importantly, each primer in the pair will incorporate a universal primer sequence to facilitate future sequencing operations. At later dates, individual plates will be thawed and, with a robot, patient DNA will be added to all 384 wells, along with PCR reagents. Following thermal cycling, the products will be purified using ExoSAP-IT and the products analyzed on an eGene machine for the presence, size, and quantity of the amplicon. In the absence of a deletion, a normalized amount of each sample will be robotically transferred to a different 384-well plate for the addition of sequencing reagents, including the universal sequencing primer. Following thermal cycle sequencing, the reactions will be purified and analyzed on an Applied Biosystems 3730 capillary DNA sequencer. The base calling will be monitored by SeqScape software, which highlights nucleotide differences from the consensus sequence. Base changes will be highlighted and inspected for outcome on the protein sequence. Mutations will be visualized by technicians and compared to known databases for polymorphism status.
The hierarchy of the genes chosen will be based on the incidence of mutant alleles and the fit of the number of amplicons into the 384-well formats. The prioritization of the gene search is outlined in Table 1. We plan to start with the dystrophin locus in tier 1, followed by direct sequencing of dystrophin in tier 2, calpain 3 and dysferlin in tier 3, TRIM 32, caveolin-3, fukutin-related protein, and β-sarcoglycan in tier 4, lamin a/c, telethonin, myotilin, δ-sarcoglycan, γ-sarcoglycan, and fukutin in tier 5, and nebulin and skeletal actin in tier 6. Table 1 shows an estimate of the reagent and plastics costs expected at each tier level. The expectation is to average the costs for one sample over the entire set of costs so that one patient with the rarest form of mutation does not have a cost substantially higher than the others, where the actual test costs were less. The high-throughput nature and the implementation of very small volumes should establish a robust and economical sequencing approach to mutation detection in the various forms of muscular dystrophy. Preliminary results are presented in Fig. 2, where five previously undetected mutations are shown. The new diagnostic approach should really increase the number and accuracy of the molecular diagnosis of muscular dystrophy, something long overdue for patients and their families.
Fig. 2.
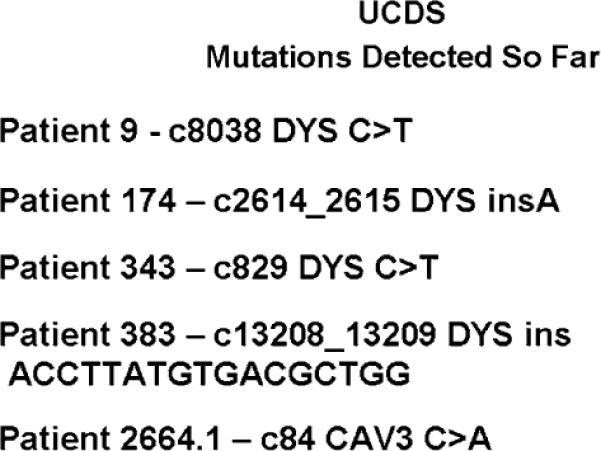
Patient mutations found by UCDS. Patient 9: in the dystrophin gene at cDNA base 8,038, a C in the consensus sequence is replaced by a T in the patient's DNA. This creates a stop codon, resulting in loss of the protein and its function. Patient 174: in the dystrophin gene, the inserted A causes an out-of-frame reading frame, leading eventually to a stop codon and loss of the protein and its function. Patient 343: same as patient 9, but at c829. Patient 383: the inserted sequence causes an out-of-frame reading frame in the dystrophin gene. Patient 2664.1: in the caveolin-3 gene, a C in the consensus sequence is changed to an A. This mis-sense mutation causes an amino acid change from asparagine to glutamate, causing LGMD1C as described by Fischer et al. (2003)
Cell-based therapy of dystrophy
We have learned a lot in the 18 years since the Duchenne gene was cloned, and yet, there is still no long-term effective therapy. It is really a travesty that we have gone that long and still do not have viable therapies for this disease, but there are an enormous number of possibilities coming. To test potential cures, we use animal models of muscle disease, like the mdx mouse. While there are actually five alleles of that model now, the original mdx mouse was first described in 1989 as having a nonsense mutation in exon 23 (Sicinski et al. 1989). This mouse makes no dystrophin and its skeletal muscle shows clear signs of degeneration, yet, for unknown reasons, the animal lives a normal lifespan with minimal muscle weakness. Using muscle degeneration as an assay, Dr. Jeff Chamberlain's group very elegantly showed that, if you transgenically overexpress dystrophin in the mdx mouse, you could completely restore dystrophin expression and actually restore the functional characteristics of muscle (Cox et al. 1993). So, if you put the dystrophin back, you should actually correct the problem. Since then, the goal has been to try to correct the problem by restoring dystrophin expression by some method of gene introduction.
Muscles are a regenerative tissue and our laboratory has felt all along that you should be able to take muscle mononuclear cells called myoblasts from a normal individual, and introduce them into the muscle of a patient with muscular dystrophy. These normal cells should fuse into pre-existing fibers and produce absent gene products, such as dystrophin. Our group has been working on this for many years, along with Dr. Terry Partridge and a number of others. With Dr. Partridge, we were able to convert mdx muscle fibers from dystrophin negative to positive by transplanting fetal mouse muscle progenitor cells (Partridge et al. 1989).
This work led to a series of human experiments using so called myoblast transfer, all of which really did not work well, but caused no additional harm to the patients (Gussoni et al. 1992; Huard et al. 1992; Karpati et al. 1993; Mendell et al. 1995). This led us to go back to the drawing board and ask whether there were any additional ways one could identify cells that had a much higher fusion index. We took advantage of our close proximity to Dr. Richard Mulligan, who was using Hoechst dye to sort bone marrow stem cells. When stained, some bone marrow stem cells are able to efflux the Hoechst dye, leaving a less brightly stained group of cells termed side population (SP) cells. Cells with similar characteristics, when isolated from muscle, were able to restore the hematopoetic compartment of lethally irradiated mice, and could systemically traffic from the circulation, after intravenous injection, into muscle, and contribute to muscle regeneration (Gussoni et al. 1999). We are working to improve this trafficking and, in collaboration with Dr. Jeff Chamberlain, we have transduced muscle SP cells with a self-inactivating lentiviral vector expressing a microdystrophin. Only SP cells, and not the rest of the muscle mononuclear cells (named MP cells), are recruited into damaged muscle, indicating that they were enriched and represented some kind of progenitor cell. Using this intravenous delivery system, we were able to produce human microdystrophin in the non-irradiated mouse muscle, although less than 1% of the fibers expressed the human protein (Bachrach et al. 2004), far below what would be considered therapeutically useful.
A second delivery method developed by Dr. Giulio Cossu (Sampaolesi et al. 2003) is to introduce the cells intra-arterially. Unlike intravenous delivery, where the cells are filtered through various different organs of the animal, the cells go directly to the muscle following arterial delivery. Because of this, arterial delivery gives a much more substantial engraftment of these cells. Indeed, in Dr. Cossu's hands, using a model of α-sarcoglycan muscular dystrophy, he could actually restore function with arterial delivery of stem cells, suggesting that this approach could be very effective. We have performed similar experiments using muscle SP cells in mdx5cv mice (Bachrach et al.2004). The cells were transfected with self-inactivating lentiviral vectors expressing human microdystrophin and, after 20 days in culture, 500,000 cells were transplanted into the right femoral artery of mdx5cv mice. Muscles downstream of the site of injection were harvested at 4 and 12 weeks following transplant. Sections of the right and left tibialis and quadriceps muscles were stained for dystrophin (Fig. 3) using two different antibodies, one specific for human dystrophin, labeled with Texas red, and the other which detects both human and mouse dystrophin, labeled with fluorosein. Mouse-revertant fibers are visualized as green and human-dystrophin-expressing fibers are visualized as orange (Fig. 3). Human dystrophin was only detected in the muscles taken from the right leg and no dystrophin was detected in animals transplanted with the same number of cultured MP cells. By sectioning throughout the leg of one mouse, we noticed variable levels of dystrophin expression in each section, with some sections expressing human dystrophin in 8% of fibers, whereas others had no human dystrophin expression.
Fig. 3.
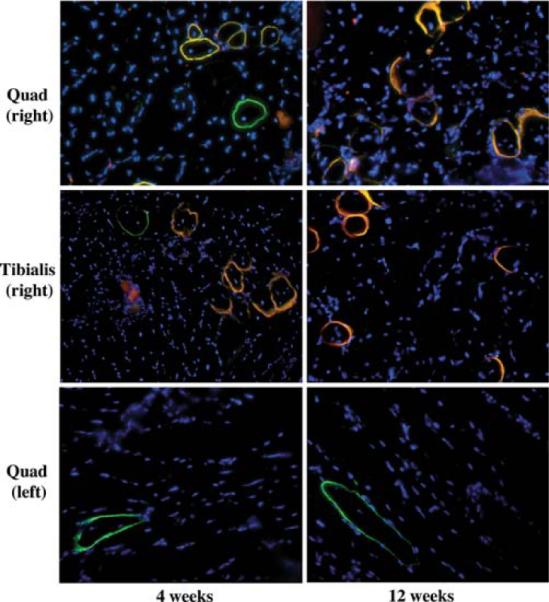
Tissue sections from mdx5cv mice transplanted with human microdystrophin transduced SP cells. Human microdystrophin was detected within the right quadriceps and tibialis anterior muscles from transplanted animals injected with transduced, cultured SP cells, 4 and 12 weeks after transplantation. Both mouse dystrophin positive revertant fibers (FITC, green) and human microdystrophin (FITC and Texas Red, orange–yellow) were detected. No human microdystrophin positive fibers were found in the left quadriceps (control); only murine revertant fibers were observed. 20× magnification. Nuclei stained with DAPI (blue)
Similar experiments were also performed with SP cells transduced with lentiviruses expressing eGFP. Again, in up to 8% of myofibers, the transplanted mice were found to be expressing GFP (Fig. 4). There were clearly some mononuclear cells that expressed GFP (Fig. 4) and, thus, might represent myogenic precursor cells. To test this hypothesis, the muscles of two mice were dissociated into single fibers and the fibers were placed into culture at low density for 15 days. The cultures were followed over time and, even at day 1, there were mononuclear cells surrounding the dissociated fibers and some of these expressed GFP (Fig. 5). Some of these green mononuclear cells also expressed Pax7, an early marker of myogenesis. At later stages in the single-fiber cultures, the mononuclear cells started to express desmin and, at the latest stages, were seen to fuse into new myobers, which expressed GFP. The SP cells that were transplanted did not express Pax7 or desmin at the time of transplant, indicating that these cells, when transplanted into the arterial circulation, could extravasate into muscle and express myogenic markers.
Fig. 4.
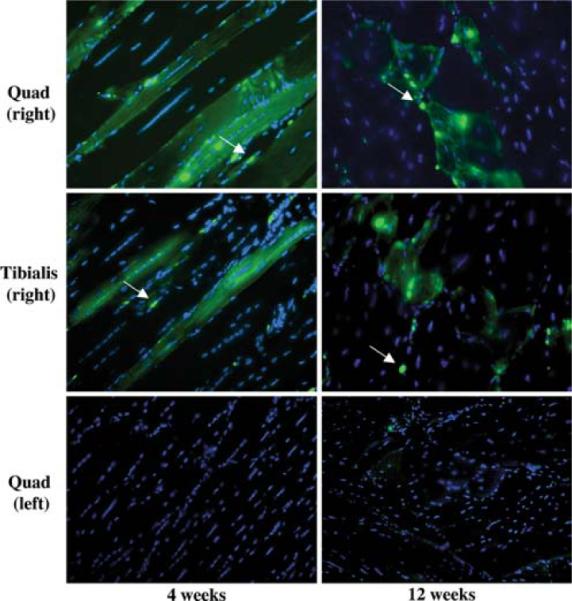
Tissue sections from mdx5cv mice transplanted with eGFP-transduced SP cells. eGFP was detected only in fibers and mononuclear cells (arrows) of transplanted animals injected with SP cells, but not MP cells. Longitudinal (4 weeks) and transverse (12 weeks) muscle sections are presented. eGFP expression was detected in the right quadriceps 4 and 12 weeks after transplantation. Few or no fibers from the left quadriceps (control) express eGFP 4 and 12 weeks after transplantation. Nuclei were stained with DAPI (blue). 20× magnification
Fig. 5.
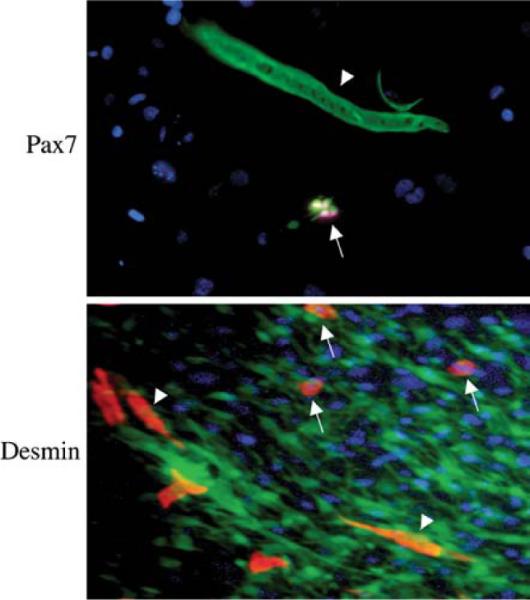
Culture of single myofibers from mdx5cv mouse transplanted with eGFP-transduced SP cells. Fluorescent microscopy of single myofiber culture from mice transplanted with eGFP-positive SP cells. Pax7: at day 4 of culture, one eGFP-positive single myofiber (arrowhead) and four eGFP-derived Pax7-positive mononuclear cells (arrows). Desmin: at day 7 of culture, three eGFP-derived desmin-positive mononuclear cells (arrows) and two eGFP-derived desmin-positive new myotubes (arrowheads). Nuclei were stained with DAPI (blue). 20× magnification
Zebrafish with muscular dystrophy
Several animal models of dystrophin deficiency exist, including cats, dogs, and mice. In the mid 1990s, Dr. Christiane Nusslein-Volhard's group in Tuebingen, Germany, reported a large genetic screen in zebrafish. As part of this screen, they isolated a series of motility mutants that appeared to have muscle degenerative disease (Granato et al. 1996). They reported four different complementation groups, all of which appeared normal at 2 days post-fertilization, but by 5 days post-fertilization, were clearly less mobile than their normal hatchlings. One very striking feature of these mutant fish was the way their muscle refracted polarized light. Using this technique (called birefringence), the muscle appeared torn and disorganized in these particular muscle mutants (Fig. 6).
Fig. 6.
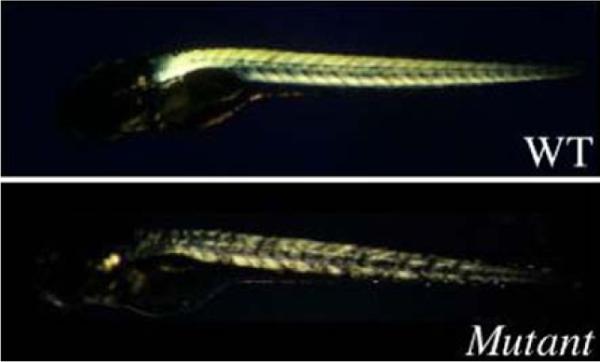
Birefringence pattern of a Tuebingen muscle mutant. Birefringence is assayed by carefully placing a fish in the space between two glass-polarizing filters and rotating the top filter to be perpendicular to the polarizing plane of the bottom filter. Muscle defects can be detected using birefringence by assaying either for an overall decrease in the intensity of refracted light as seen for sarcomeric defects (not shown) or by the presence of a more patchy pattern, suggestive of muscle tearing (see above). WT refers to a wild type fish at 5 days post-fertilization. Mutant refers to a mutant fish at 5 days post-fertilization
The phenotype of the fish in these early genetic studies was partially reproducible by blocking the expression of various proteins known to be involved in mammalian forms of muscular dystrophy (Bassett et al. 2003; Guyon et al. 2003; Guyon et al. 2005). By injecting small oligonucleotides called morpholinos into developing fish embryos, one could block the translation of the dystrophin protein and show that the fish were inactive, bent, and showed disorganized muscle under polarized light to some extent. The absence of dystrophin in these fish led to the destabilization of other members of the dystrophin protein complex, analogous to what is seen in mammals (Guyon et al. 2003). Coincident with these morpholino studies was the work by a group in Australia, who demonstrated that one of the original Tuebingen mutant alleles was caused by a mutation in exon 4 of the zebrafish dystrophin gene (Bassett et al. 2003). This sapje mutant has been sent to us in Boston and we have confirmed the exon 4 mutation in the dystrophin gene. While most of the sapje mutants die within the first month, we have found a subset that can live much longer. Analysis of these mutants confirms that dystrophin is still missing and the muscle is still disorganized (Fig. 7), yet, like the mdx mouse, some sapje mutant fish are still viable. In addition to the sapje fish, we have also obtained six other muscle mutants from the Tuebingin Stock Center, and are currently investigating the molecular cause for the phenotype. Simultaneously, we recently completed our own mutational screen and have identified seven additional mutants with various symptoms of muscle disease. We are currently mapping these different mutant strains in order to identify the underlying mutant gene. The goal will be to identify in zebrafish a gene which has not previously been implicated in mammalian forms of muscular dystrophy. Because seemingly unrelated proteins such as enzymes, structural proteins, and nuclear proteins have all been implicated in various forms of human muscular dystrophy, we believe that there must be additional factors which we have yet to identify that could help us explain the pathogenesis of disease. Novel mutant genes we identify in zebrafish would become prime disease candidates in humans, especially in patients lacking a detectable mutation in any of the human genes already known to be involved in muscle disease.
Fig. 7.
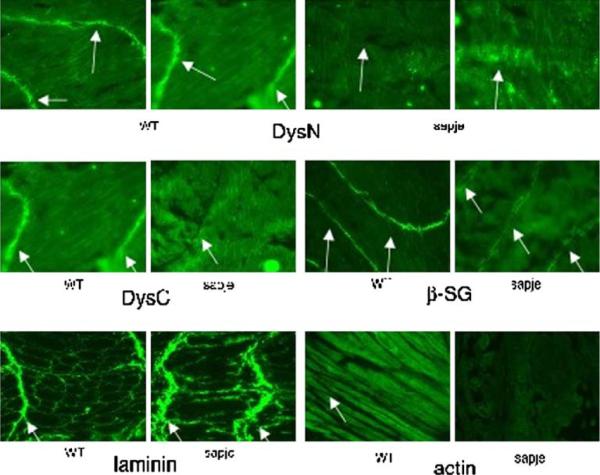
Longitudinal sections of wild type and sapje adult zebrafish were stained against the labeled antibodies. The positions of the transverse myoseptas are indicated with a white arrow. DysN refers to an antibody against the N-terminus of zebrafish dystrophin. DysC refers to an antibody against the C-terminus of zebrafish dystrophin. β-SG refers to an antibody against β-sarcoglycan
The first mutant fish allele we have mapped is the runzel strain isolated from the first Tuebingen screen (Steffen et al. [in preparation]). This mutation maps to the titin locus and, when muscle is stained with antibodies against titin, the level of titin at the sarcomere is markedly reduced. Using a gel system, which allows for the separation of very large protein molecules, we have observed that titin from skeletal muscle seems to switch isoforms during development. One isoform present later in development is either completely missing or markedly reduced in the runzel mutant. Somewhat surprising was the degree of titin isoform switching during skeletal muscle development. Muscle is formed with certain isoforms and, later in development, is replaced with completely different ones. We are currently trying to identify how and from where within the titin gene these isoforms are encoded.
The zebrafish mutants are also great models on which to test different therapies for muscle disease. Development is very rapid and the fish are especially amenable to cell transplantation at different developmental stages (Traver 2004). We have started looking at using stem cells in zebrafish to correct for absent dystrophin in the sapje strain. Zebrafish adult muscle contains mononuclear cells, which can be cultured in vitro, and when the cells reach a high enough density, they fuse to form myotubes. These cells can also be transplanted into a developing embryo and can be shown to contribute nuclei to developing muscle. Cells can be marked by transgenic expression of GFP under the control of either a ubiquitous, such as β-actin, or muscle-specific promoter, such as α-actin. Single cells from these transgenic fish can be prepared and transplanted into developing fish. Shown in Fig. 8 is a cell transplantation experiment in which cells from a GFP positive blastulae embryo (500-cell stage) were transplanted into a GFP negative host blastulae. Within a week, green muscle fibers are readily apparent in the transplanted embryo. This type of assay will allow us to screen for cell populations with the greatest potential to contribute to muscle development.
Fig. 8.
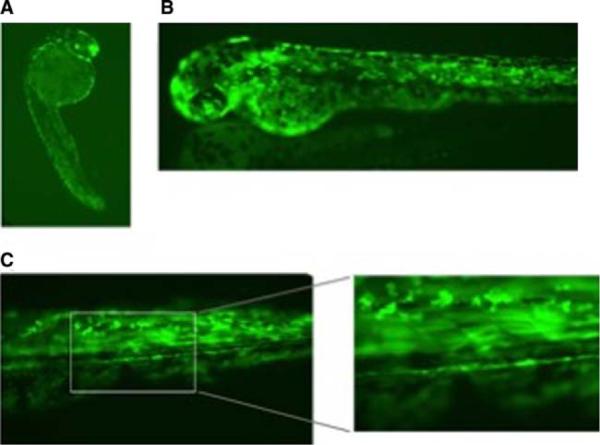
a–c Transfer of cells from a GFP+ zebrafish blastulae to an unlabeled blastulae. Cells from a zebrafish blastulae expressing GFP under the control of the constitutive β-actin promoter were transplanted into unlabeled blastulae. a At 1 dpf, the transplanted cells appear as small dots throughout the embryo. b At 2 dpf, muscle GFP+cells appear to start to elongate. c By 7 dpf, the cells have fully incorporated into the muscle. GFP+muscle fibers are clearly visible and stretch the width of the somite from one myosepta to the next
In mammals, there exists a progenitor cell population resident within adult muscle, which is capable of effluxing the DNA vital dye Hoechst and, thus, stain less brightly than the majority of cells within the muscle. Similar cells exist in adult zebrafish muscle (Fig. 9). They vary in number between different experiments, but generally comprise around 0.5% of the mononuclear cells in muscle. We hope to use the power of zebrafish to rapidly dissect the different developmental stages of muscle differentiation. We also plan to look for different methods of purifying muscle stem cells all in an attempt to identify the earliest progenitor and use it for transplants into zebrafish and, later, into mouse models of dystrophy.
Fig. 9.
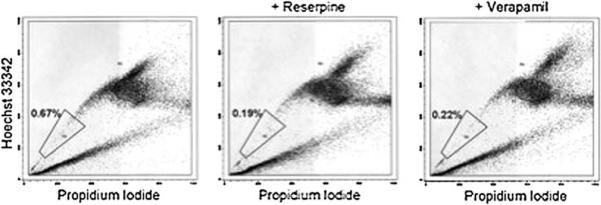
SP profile of adult zebrafish muscle. Adult zebrafish were euthanized and the muscles were partially isolated by removing the head, internal organs, and skin, leaving behind the muscle and bones. Suspended cells were sorted based on their ability to efflux Hoechst dye. The percentage of SP cells relative to the entire population of live mononuclear cells is shown for each gate. Controls using inhibitors of dye transport (reserpine and verapamil) show a decreased number of cells in the SP region
Conclusion
The Kunkel laboratory is attempting to improve the diagnosis of muscle disease through the development of a robust, rapid, and cost-effective sequencing strategy. In an era where therapies are being developed, it is essential that we develop an accurate molecular diagnosis for patients with muscular dystrophy. While diagnosis is useful, it is imperative that we develop a treatment that can help those with the disease. We believe that a cell-based approach offers the best chances for success, and we are investigating this treatment option using both mouse and zebrafish models of muscular dystrophy. Because of the ease of transplanting cells into zebrafish, these animals will be used to identify the cell population with the greatest potential for muscle engraftment. Similar cell populations can then be rapidly tested in mice and might, ultimately, be transferred to humans. The zebrafish experiments might also lead to the identification of novel genes involved in human forms of muscular dystrophy, and these can be added to our sequencing diagnostic services. By studying disease pathology and therapy using both mouse and fish models of disease, we believe we can take the strides necessary to significantly help those afflicted with muscular dystrophy.
Acknowledgements
We would like to thank all members of the Kunkel Laboratory for helpful discussions. L.M.K. is an Investigator of the Howard Hughes Medical Institute. Funding for this work was provided by a Program Project NIH grant awarded to L.M.K. (P01 NS40828-01). J.R.G. is supported by a Resarch Development Grant through the Muscular Dystrophy Association.
References
- Arahata K, Ishiura S, Ishiguro T, Tsukahara T, Suhara Y, Eguchi C, Ishihara T, Nonaka I, Ozawa E, Sugita H. Immuno-staining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature. 1988;333(6176):861–863. doi: 10.1038/333861a0. [DOI] [PubMed] [Google Scholar]
- Bachrach E, Li S, Perez AL, Schienda J, Liadaki K, Volinski J, Flint A, Chamberlain J, Kunkel LM. Systemic delivery of human microdystrophin to regenerating mouse dystrophic muscle by muscle progenitor cells. Proc Natl Acad Sci USA. 2004;101(10):3581–3586. doi: 10.1073/pnas.0400373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Kennan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130(23):5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Koenig M, Boyce FM, Kunkel LM. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86(1):45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- Bennett RR, den Dunnen J, O'Brien KF, Darras BT, Kunkel LM. Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet. 2001;2:17. doi: 10.1186/1471-2156-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RR, Schneider HE, Lakdawalla A, Lai PS, Barrett CE, Lip V, Wu BL, Darras BT, Kunkel LM. Detection of muscular dystrophy genotypes via universal condition direct sequencing (UCDS). Abstracts of the American Society of Human Genetics (ASHG) 55th Annual Meeting; Salt Lake City, Utah. October 2005; 2005. poster 2114. [Google Scholar]
- Bonilla E, Samitt CE, Miranda AF, Hays AP, Salviati G, DiMauro S, Kunkel LM, Hoffman EP, Rowland LP. Duchenne muscular dystrophy: deficiency of dystrophin at the muscle cell surface. Cell. 1988;54(4):447–452. doi: 10.1016/0092-8674(88)90065-7. [DOI] [PubMed] [Google Scholar]
- Burghes AHM, Logan C, Hu X, Belfall B, Worton RG, Ray PN. A cDNA clone from the Duchenne/Becker muscular dystrophy gene. Nature. 1987;328:434–437. doi: 10.1038/328434a0. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16(23):11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GA, Cole NM, Matsumura K, Phelps SF, Hauschka SD, Campbell KP, Faulkner JA, Chamberlain JS. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364(6439):725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345(6273):315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- Fischer D, Schroers A, Blumcke I, Urbach H, Zerres K, Mortier W, Vorgerd M, Schroder R. Consequences of a novel caveolin-3 mutation in a large German family. Ann Neurol. 2003;53(2):233–241. doi: 10.1002/ana.10442. [DOI] [PubMed] [Google Scholar]
- Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72(4):931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- Gussoni E, Pavlath GK, Lanctot AM, Sharma KR, Miller RG, Steinman L, Blau HM. Normal dystrophin transcripts detected in Duchenne muscular dystrophy patients after myoblast transplantation. Nature. 1992;356(6368):435–438. doi: 10.1038/356435a0. [DOI] [PubMed] [Google Scholar]
- Gussoni E, Soneoka Y, Strickland CD, Buzney EA, Khan MK, Flint AF, Kunkel LM, Mulligan RC. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401(6751):390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- Guyon JR, Mosley AN, Jun SJ, Montanaro F, Steffen LS, Zhou Y, Montanaro F, Nigro V, Zon LI, Kunkel LM. Delta-sarcoglycan is required for early zebrafish muscle organization. Exp Cell Res. 2005;304(1):105–115. doi: 10.1016/j.yexcr.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Guyon JR, Mosley AN, O'Brien KF, Volinski JM, Zhou Y, Davidson AJ, Sheng X, Chiang K, Zon LI, Kunkel LM. The dystrophin associated protein complex in zebrafish. Hum Mol Gen. 2003;12(6):601–615. [PubMed] [Google Scholar]
- Hayashi K, Yandell DW. How sensitive is PCR-SSCP? Hum Mutat. 1993;2(5):338–346. doi: 10.1002/humu.1380020503. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, Loike JD, Harris JB, Waterston R, Brooke M, Specht L. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. New Engl J Med. 1988;318(21):1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- Huard J, Bouchard JP, Roy R, Malouin F, Dansereau G, Labecque C, Albert N, Richards CL, Lemieux B, Tremblay JP. Human myoblast transplantation: preliminary results of four cases. Muscle Nerve. 1992;15(5):550–560. doi: 10.1002/mus.880150504. [DOI] [PubMed] [Google Scholar]
- Karpati G, Ajdukovic D, Arnold D, Gledhill RB, Guttmann R, Holland P, Koch PA, Shoubridge E, Spence D, Vanasse M. Myoblast transfer in Duchenne muscular dystrophy. Ann Neurol. 1993;34(1):8–17. doi: 10.1002/ana.410340105. [DOI] [PubMed] [Google Scholar]
- Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53(2):219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Kissel JT, Amato AA, King W, Signore L, Prior TW, Sahenk Z, Benson S, McAndrew PE, Rice R. Myoblast transfer in the treatment of Duchenne's muscular dystrophy. New Engl J Med. 1995;333(13):832–838. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323(6089):646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Partridge TA, Morgan JE, Coulton GR, Hoffman EP, Kunkel LM. Conversion of mdx myofibers from dystrophin-negative to -positive by injection of normal myoblasts. Nature. 1989;337(6203):176–179. doi: 10.1038/337176a0. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D'Antona G, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301(5632):487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244(4912):1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2004;76:127–149. doi: 10.1016/s0091-679x(04)76008-2. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem (Tokyo) 1990;108(5):748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- Zubrzycka-Gaarn EE, Bulman DE, Karpati G, Burghes AH, Bel-fall B, Klamut HJ, Talbot J, Hodges RS, Ray PN, Worton RG. The Duchenne muscular dystrophy gene product is localized in sarcolemma of human skeletal muscle. Nature. 1988;333(6172):466–469. doi: 10.1038/333466a0. [DOI] [PubMed] [Google Scholar]