Summary
The immune system, like all biological systems, operates based on a set of check and balances that strive for homeostasis of the organism. Because of its potent effector mechanisms, the potential for self-destructive immune responses is especially high and, whether to prevent autoreactivity or chronic inflammation, many negative regulatory modalities exist to prevent excessive tissue damage. This Commentary places such regulatory mechanisms in the larger context of system organization at multiple scales. The sometimes counter-intuitive nature of feedback control is discussed and a case is made for greater attention to quantitative spatiotemporal aspects of regulation, rather than limiting ourselves to the qualitative descriptions of pathways that dominate at present.
Newton is well known for his three laws of motion. Law III states that `To every action there is always opposed an equal reaction.'1 This physical law can be paraphrased to better suit an immunological perspective – `To every immune stimulus or pro-inflammatory response, there are one or more opposing control elements'. Such inhibitory activities can be cell intrinsic or extrinsic, they can act immediately or with variable time delay, they can be complete or partial, they can operate at the gene, mRNA, or protein level, they can have varying spatial dimensions. But they all share the common feature of providing the immune system with an ability to regulate the intensity, duration, and scope of inflammatory processes that promote tissue repair or pathogen resistance, but can also damage the host.
The immune system is endowed with a wide diversity of effector modalities, whether soluble or cell-mediated, innate or adaptive. While antibodies can interfere with pathogen spread with only limited impact on host cells or tissue function, though direct neutralization of viruses for example, most effector processes provide anti-infective defense in an indirect manner. They remove infected cells through cytolytic mechanisms to decrease pathogen replication2, 3, sacrificing host elements in the process; they employ proteases that attack microbes but that also damage host constituents;4 they produce reactive oxygen species that are toxic not just for bacteria or some protozoa, but also self components5–7, and so on. Thus, there will always be some level of collateral tissue damage during a response to infection – the trick is to limit this damage to just the amount needed to effectively clear (or maintain stasis of) the pathogen. In this regard, the immune system behaves according to a slightly modified version of the physician's dictum of “Do no harm” that reads “Do as little harm as possible.”8
Unfortunately, the problem facing the immune system is much greater than titrating its response to a clear foreign threat. Because of the nature of T and B cell repertoire generation and the limits of central tolerance mechanisms, there is a constant risk of anti-self responses by cells of the adaptive immune system9, 10. Likewise, for innate effectors, because they have evolved to deal with tissue damage as a source of signals promoting effector activity11–13, there is the risk that the cell disruption occurring during a foreign or self-directed immune response, or even as a result of abnormal tissue turnover14, will drive chronic and debilitating inflammatory responses, absent adequate suppressive controls.
One can read in great detail about the molecular players involved in the (negative) regulation of immune activity in previously published reviews and those in this Focus issue of Nature Immunology - these fine points will not be repeated here. Rather, the purpose of this Commentary is to place such specific information into a broader contextual framework, to move from, for example, which specific cytokines help prevent autoimmunity or inflammatory disease, to principles of design that allow the immune system to develop useful responses, while avoiding excessive or inappropriately directed activity. In other words, how has evolution engineered the immune system to balance its competing interests of potent effector activity with the preservation of the organism? One theme will be the fractal-like use by the immune system of principles of engineering design at various biological scales, from the gene to the molecular, cellular, tissue and organism levels. Another is the rather counter-intuitive nature of negative feedback regulation under many circumstances. Spatiotemporal considerations will also be emphasized, in distinction to the usual classification of particular factors as blandly acting negative controls on an opposing process, without regard for the subtleties of time and space.
A brief systems view of immune regulation
Over the past decade, the emerging discipline of systems biology has included a strong emphasis on network architecture and control mechanisms15. One application of the latter is in the area of synthetic biology, in which investigators seek to create predictable biologic outcomes from molecular or genetic circuits not found in nature16. In attempting to program such pre-defined behaviors, practitioners rely on analogies to electronic circuits and on the seminal work of Alon and colleagues, who described the various ways in which small genetic circuits were organized to achieve specific functional outcomes – suppression of noise, signal amplification, delayed response, entrainment of a sequence of responses and so forth17–21. These genetic motifs can be related to earlier descriptions of regulatory circuits in metabolic processes, some of which are highlighted in this issue22. It is critical, however, to keep in mind that while one can identify such motifs within larger network structures and use our understanding of how these subcircuits each operate to gain insight into the essential design principles of the system, overall biological function involves the integrated activity of these modules. Limitations to our understanding of how the immune system operates in health and disease often come from assuming that a small world view of a few interacting elements will adequately predict the outcome of a perturbation that in reality resonates throughout the organism23.
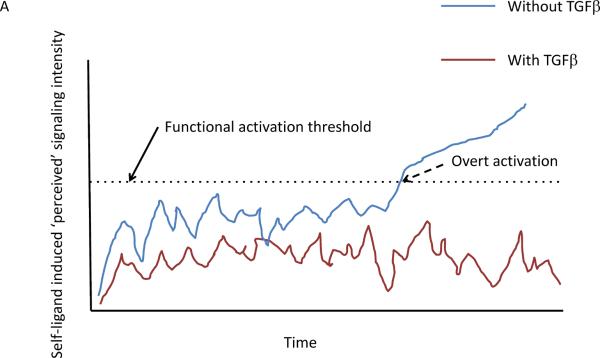
It is nonetheless still valuable to characterize, understand the apparent purpose of, and place in context these limited mini-networks as we seek to develop a more comprehensive understanding of immunity. As a first step in relating emerging insights about subnetwork structure-function to negative immune control, it is important to categorize the general properties of these regulatory pathways. One key insight is the fractal-like nature of regulation – the same strategies operate to provide inhibitory control within a cell, between cells, and at a system-wide level; the specific molecular mediators vary at each level but the design of the regulatory circuits is conserved, so in the discussion that follows, the arguments that are presented apply across these scales even if couched in the specifics of a particular stratum of the system. A second essential point is the fundamental distinction Murray and Smale make so cogently in their Focus Review24 between tonic and reactive regulators-the former set a threshold above which a stimulus must rise to evoke a response; the latter are reactive to the emerging response itself. In the former case, the intent is to prevent an activity from occurring in the first place, whereas the latter seeks to limit the intensity or duration of the response once it begins. These are unquestionably very distinct strategies of regulation, yet they are often not considered by immunologists when describing the function of suppressive factors. But each is closely related to the genetic subcircuits of Alon; the tonic regulators are acting as noise filters suppressing undesired entry into the active state in the presence of non-dangerous self-stimuli25, whereas the reactive regulators fit in with those motifs that control the duration or timing of a response. It is readily apparent that there is less risk of collateral damage if immune activation is prevented in the first place than if the system attempts to squelch an emerging response once it has begun (Fig. 1).
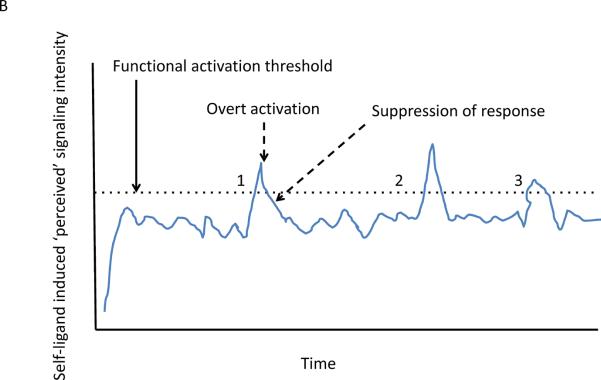
Figure 1.
Graphical depiction of the effect of tonic suppression on lymphocyte activation in response to self-ligand induced signaling input. (a) The blue line shows the behavior of a cell over time in the absence of TFG-β suppression, with occasional traverse (dashed arrow) over the functional activation threshold (dotted line). In contrast, the same T cell in the presence of TFG-β never crosses the activation threshold into a truly activated state. In this scenario, TFG-β prevents cells from responding to self stimuli. (b) The blue line shows the behavior of a cell over time, with occasional traverse (dashed arrow) over the functional activation threshold (dotted line). Here, Tregs act in response to the initial activation to suppress the activated proto-effector back into the resting state after this initial activation response and this occurs repeatedly at times 1, 2, and 3. The potential for escape from control at any of these times is greater than in a, where TFG-β prevents supra-activation responses in the first place.
Did the tree fall in the forest?
To be concrete, let us consider two putatively tonic negative regulators of T cell immune activity, TGF-β and regulatory T (Treg) cells. The former cytokine is present in an active form in many tissues and is considered to act continuously to raise the activation threshold of T cells so as to prevent weak self-stimuli from driving effector development or reactivation26, although in the presence of inflammatory co-stimuli this cytokine can change its role from mediating this tonic suppression to contributing to effector differentiation27. Treg cells are also thought to act in a similar manner, indeed, in large measure through the same TGF-β pathway28–30. But a closer look suggests that there is a substantial difference in the two cases. The continuous production of bioactive TFG-β by non-hematopoietic elements in a tissue will suppress T cell activation and can serve as a threshold device that distinguishes T cell receptor (TCR) engagements involving weak ligands (self and those peptide-major histocompatibility complex (pMHC) from pathogens that are a poor match for particular TCR in the repertoire) from those involving strong interactions. The former will be largely prevented from yielding full activation whereas the latter will still drive the T cell beyond this threshold. TGF-β is thus serving as a high pass filter that eliminates `noise' in the system, that is, undesired response to weak inputs. In this specific case, it prevents responses to prevalent self-ligands such as those involved in positive selection and known to generate tonic signals through the TCR of naïve cells31, while not substantially limiting responses to more avid (foreign) ligands. The trade off is a slight loss in sensitivity to low amounts of antigen for a large gain in avoiding chronic anti-self responses.
What about Treg cells? Don't they do the same? Here the answer is less clear. There are many studies showing that Treg function often acts after initial activation of T cells to suppress effector cell development32 or function33 rather than to prevent initial activation per se, but most such studies examine responses to foreign antigens and not self-ligands. Perhaps with respect to self-reactivity, these inhibitory cells do act as a tonic threshold filter, like tissue TGF-β and there is some evidence supporting this view34. However, close examination suggests a more nuanced picture. Treg cells exist in low and high activity states, in large measure related to exposure to interleukin 2 (IL-2)35. In the steady-state (in the absence of infection), such IL-2 is most likely to come from T cells with the strongest remaining self-reactivity after thymic negative selection trims the repertoire. But these T cells will only make IL-2 once they have reached a level of TCR stimulus beyond the triggering threshold. This means that the Tregs are not tonic suppressors of T cell activation, but rather, feedback regulators whose activity depends on the output of the cells being controlled and whose inhibitory function follows with a time delay the initial activation of the target cells. Considered broadly, the Treg do prevent autoreactive T cell responses, but at a fine-grained level, their true mode of operation is to tamp down a response that has already initiated. Mechanistically, there is a world of difference between true tonic inhibition that prevents a response from exceeding an activation threshold and a reactive negative feedback regulation that must constrain a response already in motion. As the title of this section suggests, if one doesn't listen carefully enough, one may not notice the tree falling, but that doesn't mean it didn't happen. Just because we don't measure the transient activation responses of autoreactive cells doesn't mean these do not regularly occur even with Treg present, with the attendant risks of a control mechanism operating after a response has already begun.
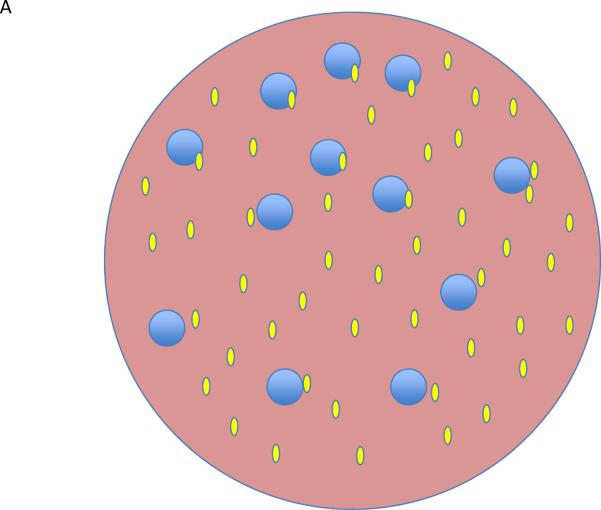
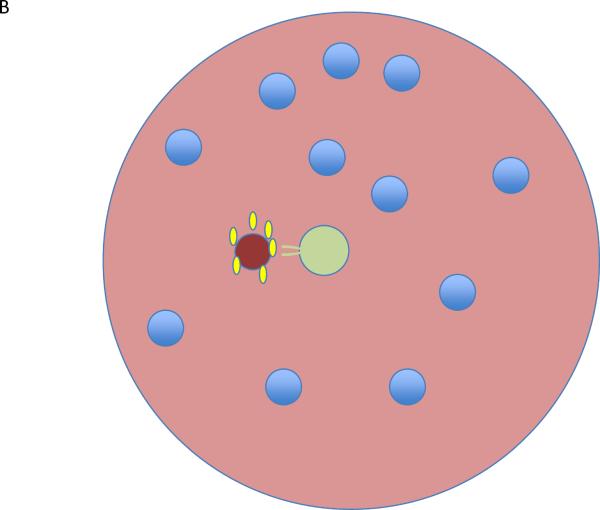
Going one step further, we need to bring spatial rather than just temporal considerations into the mix. Self-ligand activation of T cells will presumably occur rarely and at widely dispersed places in secondary lymphoid tissues, based on a match between self-ligand display (quality and amount) on presenting cells (dendritic cells (DC)) and the TCR specificity of cells probing those DC. IL-2 will be produced locally in response to such engagement, so there will need to be an adequate number of Treg cells in the local vicinity to receive this signal and increase their suppressive capacity and / or outcompete the activated T cells for this cytokine36. The Tregs have an advantage over the newly activated T cells for several hours in accessing and responding to this IL-2, because of their pre-existing CD25 expression, but only if they are present within a small tissue volume near the activated proto-effectors37. The surprising observation of shared transcription factors in effector T cells and Treg cells, driving expression of the same chemokine receptors, is likely to be related to ensuring this co-localization, at least during suppression of effector cells38–42. Furthermore, other data suggest that Tregs are more suppressive if their TCRs are engaged. If they also must react with self-ligands whose distribution is heterogeneous on individual DCs, then how often will there be a match on the same DC between the ligand presented to Tregs and to the self-reactive effector cells? Thus, both the spatial distribution of IL-2 and of TCR ligands will impact the operation of this control system, quite in distinction to one in which a dense network of stromal cells produces TGF-β so that all cells will be within the sphere of influence of this cytokine while in the tissue (Fig. 2).
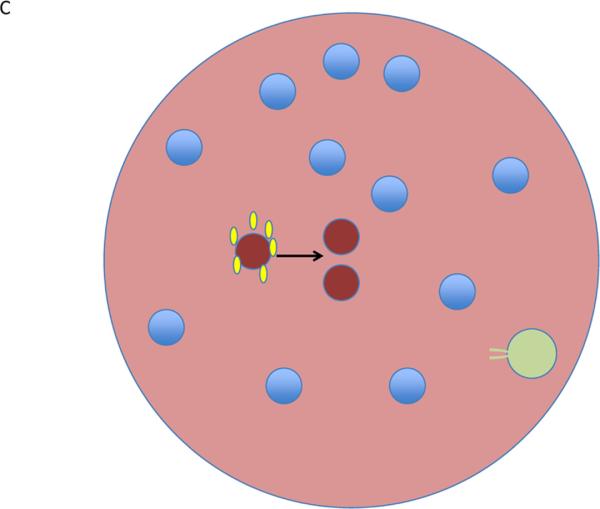
Figure 2.
Lymphocyte activation in tissues. (a) In a tissue, all regions are suffused with TFG-β (green) and all lymphocytes (blue) are subject to suppressive control. (b) A single proto-effector becomes activated (red) and produces IL-2 (yellow). If a Treg (pale green) is nearby, it can bind and remove the IL-2 from the region, becoming activated in the process and further mediating suppressive effects that limit the proto—effector response. (c) If a Treg is not positioned near the proto-effector, then activation and clonal expansion could continue. This contrasts with the volume-encompassing suppressive effect of tissue TGF-β described in a.
Rethinking negative feedback control
If we turn from control of self-reactivity to the avoidance of excessive inflammatory effector function in response to extrinsic stimuli, an important issue is how suppressive regulatory controls operate. It would be counter-productive for the immune system to completely prevent such responses, which are needed for host defense or wound healing. Thus, regulation must be reactive and we typically think in terms of negative feedback responses that are graded in proportion to the input strength, but with a suitable time delay – basically the system we have just outlined for Treg and self-reactive T cells but presumably with a longer time delay between initiation of response and full operation of the inhibitory controls. The latter allows operation of the defense or healing mechanisms for an adequate period and then induces shutdown of these activities to avoid excess tissue damage. Examples of such regulation are detailed in reviews in this issue and include signaling `tolerance' to TLR ligands and the production of suppressive cytokines such as IL-10.
While this is a reasonable description of how such negative control circuits operate, it misses an important point. There is not merely a time delay in operation of the negative regulatory elements, but also a subtle yet critical kinetic aspect to the stimulus-response linkage in such cases. A strong, very sharply rising input will typically evoke a rapid and high potency negative feedback that will truncate the response activity very quickly. The result is a `spiky' output, corresponding to a high peak of short duration43. This contrasts with the behavior seen with a slowly rising input – this evokes slower and less robust feedback, permitting a longer duration of response activity with a lower peak height (Fig. 3). Different biological systems interpret the parameters of peak signal, signal duration above threshold, and total signal in very distinct ways. Consider the fact that many biological circuits operate as concatenations of immediate early, early, and late gene responses. The latter often depend on an initial biochemical activity being sustained until after the immediate early gene products are generated because these products are also substrates of that early enzymatic activity. Only when both activated signaling molecules and immediate early gene products are both present at the same time can the response continue its course44. One example involves MAPK and c-fos; the gene encoding the latter is under control of AP-1, of which c-fos is one component and whose function depends on phosphorylation by MAPK. The new pool of c-fos induced by an initial activation of MAPK only appears after 30 minutes of sequential gene induction, transcription and translation. If MAPK activity is extinguished before this new pool of substrate is produced, the new c-fos cannot contribute to further AP-1 function45, 46. Thus, if strong initial activation of MAPK evokes a rapid shutdown of its own activity through, e.g., DUSP induction44, then the feed-forward potential of the circuit will not be realized. Let us assume that the immune system is geared to generate strong responses to acute insults and further, to show rapid attenuation of these strong responses to protect tissue integrity. In a system structured this way, slower paced stimulation by some infectious agents that do not undergo explosive reproduction, or weak self-reactions that exceed tonic or local feedback controls, could each slip under the radar of these kinetically-tuned regulatory mechanisms. The more prolonged nature of such responses due to the slow imposition of negative feedback control could represent the first step in the establishment of a chronic state of immune activity to the pathogen or self-component.
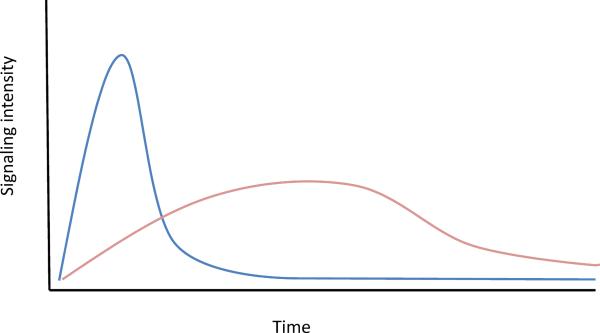
Figure 3.
In many signaling systems, a strong stimulus results is strong and rapid induction of negative feedback pathways that quickly suppress the signaling response (blue). The result is a spiked response, with a high peak but short duration. This contrasts with a weaker stimulus that reaches a lower peak of signaling but where the response is sustained because of a slower and less robust induction of the negative feedback pathway (pink). Because of the way immediate early and early gene transcription programs are kinetically linked to signaling inputs, these two different outcomes of stimulation of the cell can lead to markedly different downstream effects.
The preceding discussion has not made a distinction between controls that operate within a cell and those that operate from the outside, or that are a combination of the two. The immune system uses all these possibilities. Cbl-b is one example of a cell intrinsic regulator and many other ubiquitin-ligases fall into this category47. CTLA-4 and PD-1 on T cells are cell-intrinsic negative regulators whose operation depends on cell extrinsic cues in the form of counter-ligands on other cells48. These counter-ligands concentrate the negative regulatory molecules near to where TCR engagement occurs at the T-APC interface, allowing them to interfere with productive TCR signaling into the cell49. For both CTLA-4 and PD-1, the considerations presented above seem to apply with respect to how the robustness of early signal rise affects the feedback regulation. CTLA-4 relocation from an intracellular pool to the surface membrane at the immunological synapse is controlled by the strength of TCR signaling49, and in a counterintuitive manner, this linkage results in greater suppression of a strong response than a weaker one, apparently serving to preserve polyclonality in the T cell response and avoid easy pathogen escape by mutation50. PD-1 expression is also greatest on the most antigen reactive T cells and reversal of PD-1 inhibition in chronically infected animals tends to allow re-emergence of active effectors from among subdominant responses rather than from among T cells reactive with immunodominant determinants51.
The larger context
This Commentary has only scratched the surface of the complexities inherent in regulation of a network with as many moving parts as the immune system. A smaller array of homeostatic controls may work for tissues with more predictable stresses but prove inadequate for the immune system because of the extremely heterogeneous nature and timing of the challenges facing it. I have concentrated on just a few key issues out of many important ones: (i) the distinction between tonic and induced regulation, and the ease with which the latter can be confused with the former without explicit experiments designed to test whether the regulation is reactive or pre-existing; (ii) the insufficiently examined roles of spatial aspects of immune tissue organization; and (iii) the counterintuitive nature of negative feedback control of strong vs. weak responses. It has been possible only to hint about the emergent properties of systems that are not discoverable from analyses restricted to only very small parts of the larger whole. A deeper understanding of how the immune system achieves a balance between useful activity and a limitation of response-induced host damage will depend on putting the detailed, but isolated observations typically made in individual studies into a more complete framework and developing methods for predicting the behavior of this more complex network when it is perturbed.
Acknowledgements
I wish to thank the many colleagues, both within the Laboratory of Systems Biology, at NIH, and throughout the world, with whom I have discussed aspects of the ideas presented in this Commentary over many years. I am especially grateful to W. Kastenmuller for his careful reading of this manuscript and his insightful suggestions for improvements. This work was supported by the Intramural Research Program of NIAID, NIH.
References
- 1. http://en.wikipedia.org/wiki/Newton's_laws_of_motion.
- 2.Sissons JG, Oldstone MB. Killing of virus-infected cells by cytotoxic lymphocytes. The Journal of infectious diseases. 1980;142:114–119. doi: 10.1093/infdis/142.1.114. [DOI] [PubMed] [Google Scholar]
- 3.Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annual review of immunology. 2000;18:275–308. doi: 10.1146/annurev.immunol.18.1.275. [DOI] [PubMed] [Google Scholar]
- 4.Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell death and differentiation. 2008;15:251–262. doi: 10.1038/sj.cdd.4402244. [DOI] [PubMed] [Google Scholar]
- 5.Bogdan C, Rollinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Current opinion in immunology. 2000;12:64–76. doi: 10.1016/s0952-7915(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 6.Kohchi C, Inagawa H, Nishizawa T, Soma G. ROS and innate immunity. Anticancer research. 2009;29:817–821. [PubMed] [Google Scholar]
- 7.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nature reviews. Immunology. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 8.Segel LA, Bar-Or RL. On the role of feedback in promoting conflicting goals of the adaptive immune system. J Immunol. 1999;163:1342–1349. [PubMed] [Google Scholar]
- 9.Chatenoud L. Protection from autoimmunity: immunological indifference versus T-cell mediated suppression? European journal of immunology. 2006;36:2296–2298. doi: 10.1002/eji.200636591. [DOI] [PubMed] [Google Scholar]
- 10.Joller N, Peters A, Anderson AC, Kuchroo VK. Immune checkpoints in central nervous system autoimmunity. Immunological reviews. 2012;248:122–139. doi: 10.1111/j.1600-065X.2012.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallucci S, Matzinger P. Danger signals: SOS to the immune system. Current opinion in immunology. 2001;13:114–119. doi: 10.1016/s0952-7915(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 12.Matzinger P. Tolerance, danger, and the extended family. Annual review of immunology. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 13.Allam R, Anders HJ. The role of innate immunity in autoimmune tissue injury. Current opinion in rheumatology. 2008;20:538–544. doi: 10.1097/BOR.0b013e3283025ed4. [DOI] [PubMed] [Google Scholar]
- 14.Lleo A, Selmi C, Invernizzi P, Podda M, Gershwin ME. The consequences of apoptosis in autoimmunity. Journal of autoimmunity. 2008;31:257–262. doi: 10.1016/j.jaut.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kholodenko B, Yaffe MB, Kolch W. Computational approaches for analyzing information flow in biological networks. Science signaling. 2012;5:re1. doi: 10.1126/scisignal.2002961. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A, Lu TK. Synthetic Biology: An Emerging Engineering Discipline. Annual review of biomedical engineering. 2012 doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- 17.Alon U. Network motifs: theory and experimental approaches. Nature reviews. Genetics. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 18.Frankenstein Z, Alon U, Cohen IR. The immune-body cytokine network defines a social architecture of cell interactions. Biology direct. 2006;1:32. doi: 10.1186/1745-6150-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashtan N, Itzkovitz S, Milo R, Alon U. Topological generalizations of network motifs. Physical review. E, Statistical, nonlinear, and soft matter physics. 2004;70:031909. doi: 10.1103/PhysRevE.70.031909. [DOI] [PubMed] [Google Scholar]
- 20.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 21.Yeger-Lotem E, et al. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene D. Nature immunology. 2012 [Google Scholar]
- 23.Fraser ID, Germain RN. Navigating the network: signaling cross-talk in hematopoietic cells. Nature immunology. 2009;10:327–331. doi: 10.1038/ni.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray PJ, Smale ST. Nature immunology. 2012 doi: 10.1038/ni.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Germain RN. The art of the probable: system control in the adaptive immune system. Science. 2001;293:240–245. doi: 10.1126/science.1062946. [DOI] [PubMed] [Google Scholar]
- 26.Aoki CA, et al. Transforming growth factor beta (TGF-beta) and autoimmunity. Autoimmunity reviews. 2005;4:450–459. doi: 10.1016/j.autrev.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annual review of immunology. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahl SM, Swisher J, McCartney-Francis N, Chen W. TGF-beta: the perpetrator of immune suppression by regulatory T cells and suicidal T cells. Journal of leukocyte biology. 2004;76:15–24. doi: 10.1189/jlb.1103539. [DOI] [PubMed] [Google Scholar]
- 29.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Pesu M, et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefanova I, Dorfman JR, Germain RN. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature. 2002;420:429–434. doi: 10.1038/nature01146. [DOI] [PubMed] [Google Scholar]
- 32.Kastenmuller W, et al. Regulatory T cells selectively control CD8+ T cell effector pool size via IL-2 restriction. J Immunol. 2011;187:3186–3197. doi: 10.4049/jimmunol.1101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mempel TR, et al. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 2006;25:129–141. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Schildknecht A, et al. FoxP3+ regulatory T cells essentially contribute to peripheral CD8+ T-cell tolerance induced by steady-state dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:199–203. doi: 10.1073/pnas.0910620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuest TY, Willette-Brown J, Durum SK, Hurwitz AA. The influence of IL-2 family cytokines on activation and function of naturally occurring regulatory T cells. Journal of leukocyte biology. 2008;84:973–980. doi: 10.1189/jlb.1107778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nature immunology. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 37.Feinerman O, et al. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Molecular systems biology. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nature medicine. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozaki Y, Sasagawa S, Kuroda S. Dynamic characteristics of transient responses. Journal of biochemistry. 2005;137:659–663. doi: 10.1093/jb/mvi084. [DOI] [PubMed] [Google Scholar]
- 44.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nature reviews. Molecular cell biology. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 45.Glauser DA, Schlegel W. Sequential actions of ERK1/2 on the AP-1 transcription factor allow temporal integration of metabolic signals in pancreatic beta cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:3240–3249. doi: 10.1096/fj.06-7798com. [DOI] [PubMed] [Google Scholar]
- 46.Chalmers CJ, Gilley R, March HN, Balmanno K, Cook SJ. The duration of ERK1/2 activity determines the activation of c-Fos and Fra-1 and the composition and quantitative transcriptional output of AP-1. Cellular signalling. 2007;19:695–704. doi: 10.1016/j.cellsig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Paolino M, Penninger JM. E3 ubiquitin ligases in T-cell tolerance. European journal of immunology. 2009;39:2337–2344. doi: 10.1002/eji.200939662. [DOI] [PubMed] [Google Scholar]
- 48.Bour-Jordan H, et al. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/ B7 family. Immunological reviews. 2011;241:180–205. doi: 10.1111/j.1600-065X.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egen JG, Allison JP. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 50.Chambers CA, Kuhns MS, Egen JG, Allison JP. CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annual review of immunology. 2001;19:565–594. doi: 10.1146/annurev.immunol.19.1.565. [DOI] [PubMed] [Google Scholar]
- 51.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]