Abstract
Toll-like receptors (TLRs) are critical mediators of the immune response to pathogens. The influence of human TLR6 polymorphisms on susceptibility to infection is only partially understood. Most microbes contain lipopeptides recognized by TLR2/1 or TLR2/6 heterodimers. Our aim was to determine whether single nucleotide polymorphisms (SNPs) in TLR6 are associated with altered immune responses to lipopeptides and whole mycobacteria.
We sequenced the TLR6 coding region in 100 healthy South African adults to assess genetic variation and determined associations between polymorphisms and lipopeptide- and mycobacteria-induced IL-6 production in whole blood. We found 2 polymorphisms, C745T and G1083C that were associated with altered IL-6 secretion. G1083C was associated with altered IL-6 levels in response to lipopeptides, Mycobacterium tuberculosis lysate (Mtb, P = 0.018) and BCG (P = 0.039). The 745T allele was also associated with lower NF-κB signaling in response to di-acylated lipopeptide, PAM2 (P = 0.019) or Mtb (P = 0.026) in a HEK293 cell line reconstitution assay, compared with the 745C allele.
We conclude that TLR6 polymorphisms may be associated with altered lipopeptide-induced cytokine responses and recognition of Mtb. These studies provide new insight into the role of TLR6 variation and the innate immune response to human infection.
Keywords: Toll-like receptor 6, polymorphism, interleukin 6, tuberculosis, immune response
INTRODUCTION
The innate immune system enables the host to recognize invading microbes as foreign. Toll-like receptors (TLRs) are central in this process. These innate immune cell receptors recognize pathogen-associated molecular patterns (PAMPs) present in a wide variety of microbes, via a transmembrane receptor with a leucine-rich repeat (LRR) motif(1). Upon recognition of the ligand by a TLR, a signaling cascade is initiated through activation of a cytoplasmic Toll/IL-1R (TIR) homology domain. This results in activation of transcription factors, including NF-κB, which translocate into the nucleus for induction of pro-inflammatory molecules (2–4). The resulting inflammation is a critical early step in immune responses aimed at controlling the invading microbe.
Genetic variation of human TLRs regulates signaling and inflammatory responses and is associated with susceptibility to infection (5–7). We and others have shown that a polymorphism in TLR1, T1805G, regulates the immune response to tri-acylated lipopeptides, present in cell walls of many microbes (8–10). In contrast, this single nucleotide polymorphism (SNP) was not associated with altered responses to di-acylated ligands (8).
This study’s focus was TLR6. Human TLR6 is a 2 391 base pair gene that encodes a 796-amino acid (aa) type I transmembrane protein with a 630aa extracellular LRR region (including a 31aa signal peptide), a 21aa transmembrane domain and a 145aa intracellular TIR signaling domain. TLR6 forms a heterodimer with TLR2 in a complex incorporating CD14, which preferentially recognizes di-acylated lipopeptides like PAM2 (PAM2CSKKKK, S-[2,3-bis(palmitoyloxy)-propyl]-(R)-cysteinyl-(lysyl)3-lysine). This contrasts with TLR1, which also heterodimerizes with TLR2, in a complex with CD14, but which specifically recognizes tri-acylated lipopeptides like PAM3 (Pam3CSKKKK, N-palmitoyl-S-[2,3-bis- (palmitoyloxy)-propyl]-(R)-cysteinyl-(lysyl)3-lysine)(11–13). Recent studies suggest that ligand preference based on acyl number is partial, and that other structural features may also influence ligand recognition (14, 15). These include length of the fatty acid chain, chirality of the diacyloxypropyl carbon, position of the acyl group, and amino acid composition of the terminal peptides, which have also been shown to affect receptor specificity. The specific role of human TLR6 in recognition of different pathogens is less well understood than that of TLR1 and TLR2. For example, several studies suggest that TLR1 or TLR2 mediate responses to Mycobacterium tuberculosis (Mtb) (16–18), whereas a role of TLR6 in response to this pathogen has only been suggested in a single study (19).
Variation in human cellular responses to di-acylated lipopeptides has been shown in studies that have principally focused on TLR1 polymorphisms (8, 10). Since TLR1 polymorphism did not completely account for the observed variation, it was postulated that this might be due to variation in the TLR6 gene, which mediates differential signaling and cytokine responses; although no specific polymorphism has been defined. Other studies have suggested an association between TLR6 polymorphism and disease susceptibility. For example, the SNP T1932G (A644A) was associated with altered IFN-γ secretion in response to measles virus stimulation of peripheral blood mononuclear cells (PBMC)(20), while C745T was associated with asthma (21, 22) and with invasive aspergillosis(23) after allogeneic stem cell transplantation. It is not known if these polymorphisms alter TLR6 function.
Our aim was to learn about the role of TLR6 polymorphism in recognition and signaling of M. tuberculosis. The current global epidemic of tuberculosis is responsible for 1.7 million deaths annually (24). Chronic inflammation is a hallmark of tuberculosis infection and substantial efforts have been made to identify bacterial components responsible. Mycobacterium tuberculosis (Mtb) is recognized by several TLRs including TLR 1, 2, 4 and 9 (12, 16–18, 25). We hypothesized that TLR6 polymorphisms contribute to differential immune responses, and ultimately differential protection against this disease. We examined whether TLR6 polymorphisms are associated with altered di-acylated lipopeptide- and mycobacteria-induced cytokine responses in humans.
RESULTS
One hundred healthy adults were enrolled, including 56 women and 44 men with an age range from 18 to 57 years, and from a mixture of ethnic backgrounds: 24 Black Africans, 64 participants of mixed ethnicity and 12 Caucasians.
In order to identify common polymorphisms in the TLR6 gene, we sequenced the coding region in 100 healthy adult volunteers. We found 10 polymorphisms, which included 7 non-synonymous and 3 synonymous base pair changes (Table 1). The observed allelic frequencies were consistent with Hardy-Weinberg equilibrium. Nine of these polymorphisms have been reported before in public databases, including HapMap, NCBI and II-PGA (Table 1). One polymorphism (T34A) had not previously been described. All of the remaining nine TLR6 coding region polymorphisms were present in at least one HapMap population.
Table 1.
TLR6 coding region polymorphisms in South Africans, compared with those described in the Hapmap database.
| SNP detail | Current study | MAF data in previously described populations |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | NCBI reference |
Population | AA n (%) |
Aa n (%) |
aa n (%) |
MAF (%) |
HWE | CEU (%) |
YRI (%) |
LWK (%) |
MKK (%) |
| T34A | ss161110012 | Black | 11(50) | 11(50) | 0 (0) | 25 | 0.12 | na | na | na | na |
| (F12I) | Mixed | 39(64) | 22(36) | 0(0) | 18 | 0.08 | |||||
| Caucasian | 9(75) | 3(14) | 0(0) | 12.5 | |||||||
| T359C | rs5743808 | Black | 18(86) | 3(14) | 0(0) | 7.1 | 0.72 | 0 | 8.3 | 1.1 | 9.4 |
| (I120T) | Mixed | 49(82) | 11(18) | 0(0) | 9.1 | 0.43 | |||||
| Caucasian | 12(100) | 0(0) | 0(0) | 0 | |||||||
| T581C | rs5743809 | Black | 21(100) | 0(0) | 0(0) | 0 | 0 | 3.3 | na | na | |
| (L194P) | Mixed | 53(87) | 8(13) | 0(0) | 6.5 | 0.58 | |||||
| Caucasian | 10(100) | 0(0) | 0(0) | 0 | |||||||
| G740A | rs35220466 | Black | 19(86) | 3(14) | 0(0) | 6.8 | 0.73 | 0.8 | 2.5 | na | na |
| (R247K) | Mixed | 59(95) | 3(5) | 0(0) | 2.4 | 0.84 | |||||
| Caucasian | 12(100) | 0(0) | 0(0) | 0 | |||||||
| C745T | rs5743810 | Black | 22(92) | 2(8) | 0(0) | 4.1 | 0.83 | 46.4 | 0 | 1.1 | 7.0 |
| (P249S) | Mixed | 52(81) | 11(17) | 1(2) | 10.1 | 0.64 | |||||
| Caucasian | 3(25) | 7(58) | 2(17) | 45.8 | |||||||
| G979A | rs3796508 | Black | 19(95) | 1(5) | 0(0) | 2.5 | 0.91 | 0 | 0.9 | 2.8 | 2.4 |
| (V327M) | Mixed | 57(93) | 4(7) | 0(0) | 3.2 | 0.79 | |||||
| Caucasian | 11(100) | 0(0) | 0(0) | 0 | |||||||
| G1083C | rs3821985 | Black | 15(65) | 7(30) | 1(4) | 19.5 | 0.87 | 72.4 | 31.4 | na | na |
| (T361T) | Mixed | 27(42) | 25(39) | 12(19) | 38.2 | 0.16 | |||||
| Caucasian | 2(17) | 2(17) | 8(66) | 75 | |||||||
| A1263G | rs3775073 | Black | 15(65) | 7(30) | 1(4) | 15 | 0.87 | 29.2 | 70.8 | 15.7 | 32.5 |
| (K421K) | Mixed | 29(48) | 25(41) | 7(11) | 31.9 | 0.65 | |||||
| Caucasian | 1(9) | 1(9) | 9(82) | 86.3 | |||||||
| T1280C | rs5743815 | Black | 21(100) | 0(0) | 0(0) | 0 | 1.3 | 0 | na | na | |
| (V427A) | Mixed | 61(100) | 0(0) | 0(0) | 0 | ||||||
| Caucasian | 10(91) | 1(9) | 0(0) | 4.5 | |||||||
| T1932G | rs5743818 | Black | 13(62) | 7(33) | 1(5) | 20 | 0.96 | 27 | 31 | 2.2 | 3.1 |
| (A644A) | Mixed | 42(71) | 16(27) | 1(2) | 15.2 | 0.71 | |||||
| Caucasian | 9(75) | 2(17) | 1(8) | 13.6 | |||||||
Genotype and allele frequencies of the ten polymorphisms for South African are listed, along with minor allele frequencies (MAF) described in the Hapmap database, for multiple other populations. CEU–Utah residents with European Ancestry; YRI –Yoruba in Nigeria); LWK – Luhya in Kenya; MKK – Maasai in Kenya. A=common allele, a=minor allele, MAF=minor allele frequency, na = not available. The HWE P-values are calculated for Black and mixed ethnicity populations. There were very few Caucasians and the HWE P-value was not calculated for this population.
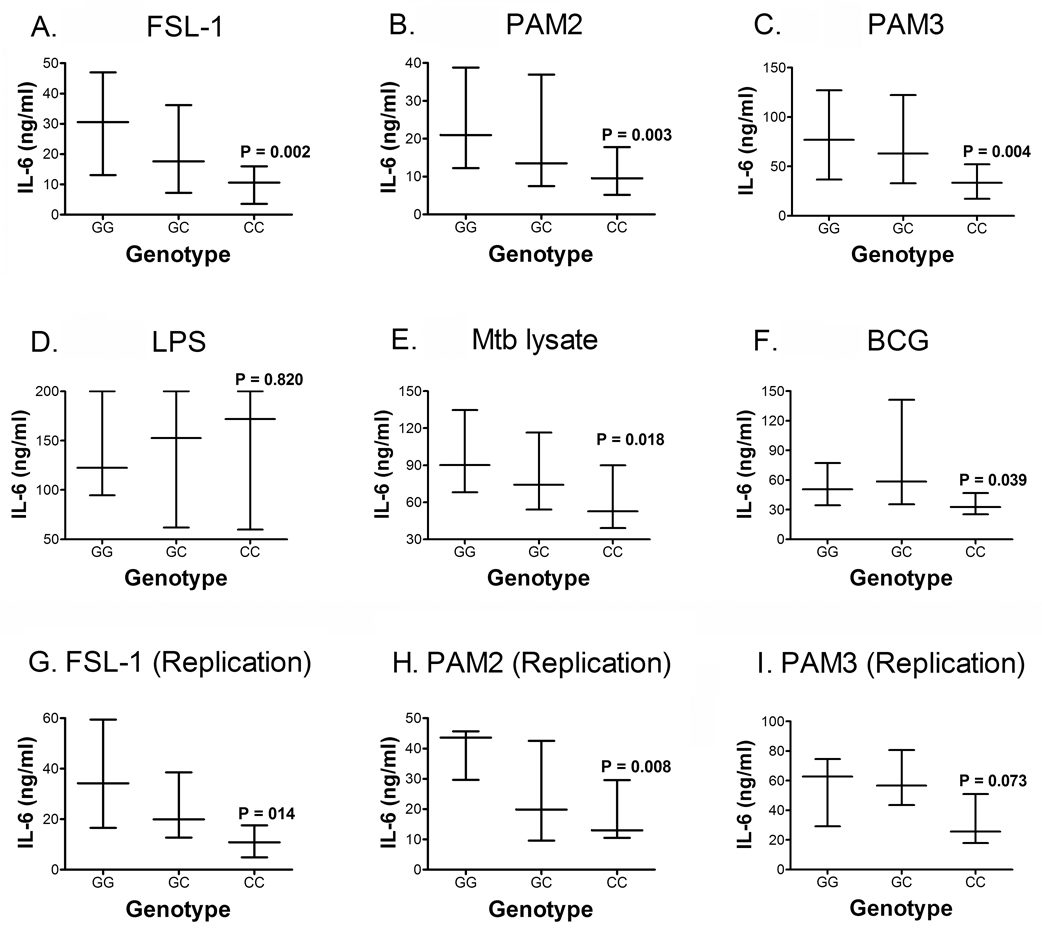
To examine whether any of these polymorphisms were associated with altered cytokine production, we stimulated whole blood from 70 of the 100 participants with di-acylated lipopeptides and several other TLR ligands. We have previously shown that measurement of IL-6 is ideal for assessing innate responses in whole blood stimulated with lipopeptides: IL-6 was secreted at readily detectable levels, whereas IL-12, TNF-α, IL-1β, and IL-10 levels were low (8). Two polymorphisms, C745T and G1083C, were associated with altered IL-6 production. We examined G1083C first: the 1083CC (361T) genotype was associated with lower IL-6 production, compared with 1083GG (361T), in response to FSL-1 (Fig 1A), PAM2 (Fig 1B) and PAM3 (Fig 1C). As a control, stimulation with LPS was not associated with significant differences in the level of IL-6 produced (Fig 1D).
Figure 1. Effect of TLR6 G1083C polymorphism on IL-6 secretion.
Whole blood was stimulated for 20 hours with TLR ligands and whole mycobacteria, followed by measurement of IL-6 levels in plasma, by ELISA. IL-6 levels were stratified by genotypes of G1083C polymorphisms. Ligands used included: (A) FSL-1, a TLR2/6 ligand (B) PAM2, a TLR2/6 ligand, (C) PAM3, a TLR2/1 ligand, (D) Lipopolysaccharide LPS, a TLR4 ligand, (E) H37Rv Mtb lysate, and (F) whole lyophilized live BCG. These figures show results from the following numbers of participants with specific genotypes: GG n=36; GC n=21; and CC n=12. To validate our data, blood from another 26 participants (GG n=8; GC n=12; and CC n=6) was examined: IL-6 levels were determined after stimulation of whole blood with (G) FSL-1, (H) PAM2, and (I) PAM3. Data are shown as medians with interquartile ranges, and a P value representing assessment of differences between the GG and CC genotypes, using a Mann-Whitney test. The GG and GC genotypes were also compared, but no differences were found.
We next examined whether TLR6 SNPs were associated with altered IL-6 response to whole mycobacteria, which have a complex repertoire of lipopeptides and other ligands. SNP G1083C was associated with a decrease in IL-6 secretion after Mtb lysate and BCG stimulation (Fig 1E and F).
To assess the reproducibility of these results, we repeated the experiment in 26 of the 100 individuals, and found the same results for G1083C in response to FSL-1 (Fig 1G) and to PAM2 (Fig 1H), but not to PAM3 (Fig 1I). We also examined the intra- and inter-individual variation of the assay over time by comparing IL-6 levels of individuals drawn at different times. In a paired analysis, we found consistent IL-6 levels within individuals and a similar rank order between individuals suggesting that IL-6 responsiveness is a stable biologic phenotype (P=0.071 for FSL-1; P=0.267 for PAM2; and P=0.480 for PAM3) (Supplementary Figure 1). Mtb lysate and BCG were not used during the repeat experiment.
Together, these results suggest that allele 1083C is associated with decreased IL-6 levels in response to lipopeptide and mycobacteria stimulation.
As a synonymous polymorphism, G1083C may directly regulate lipopeptide- or mycobacteria-induced cytokine secretion through effects on transcriptional regulation of TLR6 or be in linkage disequilibrium with a polymorphism that directly regulates function. To address the latter possibility, we examined whether polymorphisms associated with altered IL-6 levels induced by FSL-1 or PAM-2 were in linkage disequilibrium with G1083C. We first examined the other coding region polymorphisms in TLR6. The highest correlation was observed between the G1083C and the A1263G polymorphisms (R2=0.75, Figure 2, left panel). Although this suggests some LD correlation between 1083 and 1263, the latter SNP was not associated with altered levels of IL-6. Furthermore, A1263 is synonymous and has no known association with function. The remaining TLR6 coding region SNPs had low levels of linkage disequilibrium with 1083 (R2 < 0.12 for all SNPs except 745). We next examined whether non-coding TLR6 SNPs or nearby polymorphisms were associated with IL-6 levels. TLR6 is located on chromosome 4p14 in a region adjacent to TLR1 and TLR10 that spans approximately 54 kb (26). We examined additional non-coding region haplotype-tagging TLR6 polymorphisms (rs1039559, rs7673348, rs7665774) as well as haplotype-tagging polymorphisms (in the CEU and YRI populations; Table 1) in TLR1 (rs17616434, rs3923647, rs3924112, rs4833095, rs5743618) and TLR10 (rs4321646, rs10856837, rs7694115). We also examined seven haplotype-tagging SNPs in TLR2 on chromosome 4q32 (rs3804090, rs5743708, rs11935252, rs1337, rs1339, rs1439166, rs6535946).
Figure 2. Linkage disequilibrium analyses between TLR6 and TLR1 SNPs.
R-squared (R2) values for each SNP combination are shown numerically and by shading, based upon the legend in the middle. TLR6 and TLR1 SNPs were genotyped and analyzed for level of LD with G1083C and C745T polymorphisms. The figure shows R2 values for pairwise comparison of the different polymorphisms. R2 = 0 represents no LD correlation; R2 = 1 represents high (maximum) LD correlation. The program pwld in Stata was used to calculate the values. The minor allele frequency is shown adjacent to each corresponding SNP.
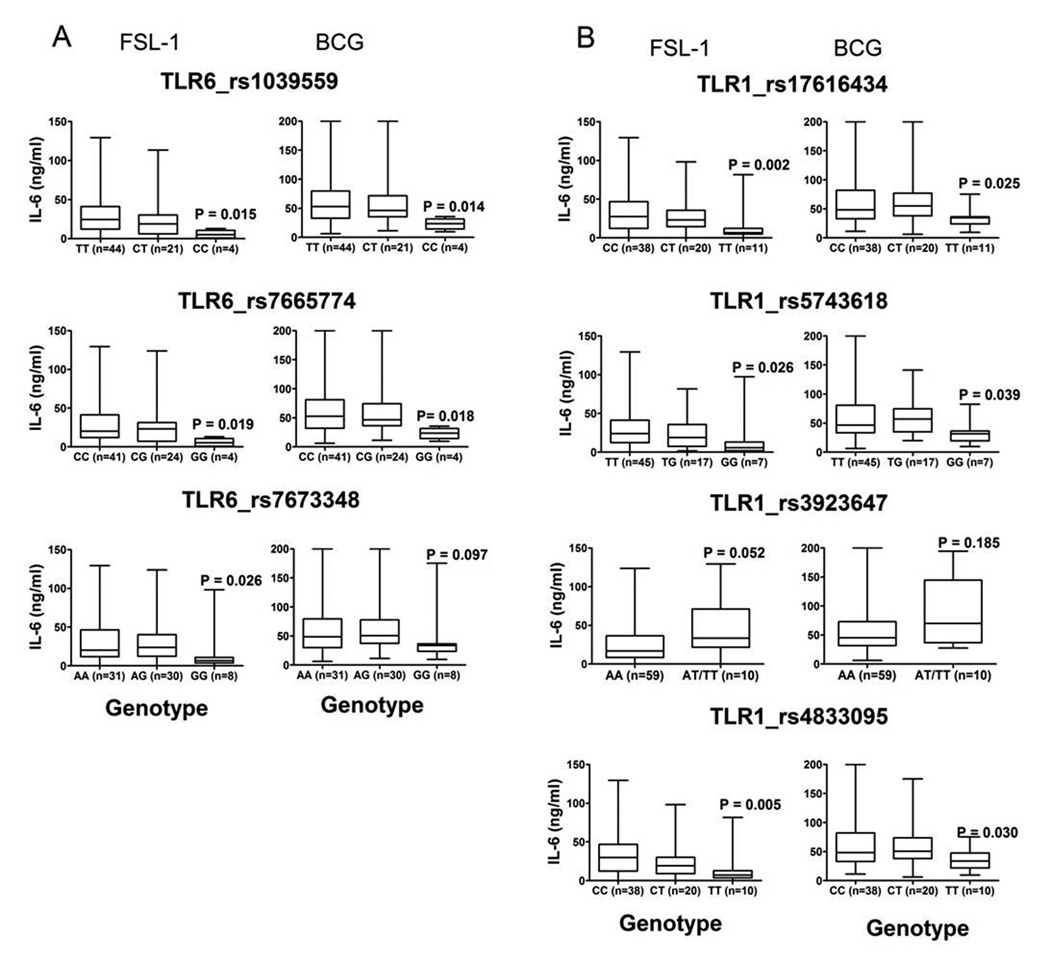
We found that 7 polymorphisms in the TLR6 promoter region and TLR1 were significantly associated with altered IL-6 levels induced by FSL-1, 1 was associated with altered IL-6 induced by Mtb, and none were associated with altered IL-6 induced by PAM-2. The levels of IL-6 secreted in response to FSL-1 and BCG are shown in Figure 2. For the other ligands only the P-values are represented since the trend of IL-6 secretion for these ligands was identical to FSL-1 and BCG (Figure 3 and Supplementary table 1, and data not shown). TLR6 coding region polymorphisms and polymorphisms that were significantly associated with altered levels of IL-6 secretion (Figure 3 and Supplementary table 1) were analyzed for linkage disequilibrium (LD) with the G1083C and C745T polymorphisms for all participants in our study. For the non-coding region polymorphisms, the most prominent correlation with the G1083C polymorphism was observed with the TLR6 promoter region polymorphisms rs1039559 (R2 = 0.42, D’ = 1.0) and rs7665774 (R2 = 0.35, D’ = 0.84), and with the C745T polymorphism (R2 = 0.23, D’ = 1.0) (Figure 2 and Supplementary Figure 2). None of the other TLR6 coding region polymorphisms were associated with altered IL-6 levels (data not shown).
Figure 3. Association of IL-6 secretion in response to FSL-1 and BCG with polymorphisms in the non-coding region of TLR6, and polymorphisms in TLR1.
Polymorphisms in TLR6 and TLR1 were identified from HapMap and genotyped in our participants. Whole blood was stimulated for 20 hours with the TLR ligands and whole mycobacteria. IL-6 levels in plasma were measured by ELISA and responses stratified by genotype of the polymorphism. Data for FSL-1 and BCG are shown as box and whisker plots with medians and interquartile ranges. P values represent differences between the homozygous genotypes with Mann-Whitney test. Panel A: TLR6 tagged polymorphisms, and Panel B: TLR1 tagged polymorphisms. Data for the rest of the ligands is shown in supplementary table 1.
The G1083C polymorphism had very weak LD correlation with the TLR1 polymorphisms with R2 ≤0.18 (Figure 2 and supplementary Figure 2, right panels). One of the associated polymorphisms was TLR1_T1805G (rs5743618), which we and others have previously found to regulate PAM3-induced signaling (8–10). The association of TLR1_T1805G with IL-6 signaling in our dataset was strongest for PAM3 (TT median = 64 920 pg/mL (n=45), GT = 59 980 pg/mL (n=17), GG = 20 970 pg/mL (n=7), P=0.001 for TT vs. GG, Mann-Whitney test). The association was weaker for FSL-1 (TT = 24 118 pg/mL (n=45), GT = 18 968 pg/mL (n=17), GG = 5 545 pg/mL (n=7), P=0.026 for TT vs. GG) and absent for PAM2 (TT = 16 182 pg/mL (n=45), GT = 22 778 pg/mL (n=17), GG = 13 520 pg/mL (n=7), P=0.553 for TT vs. GG). The R2 and D’ values for TLR1_T1805G and TLR6_G1083C were 0.14 and 0.6, respectively, suggesting a low level of linkage disequilibrium. To adjust for a possible effect from this SNP, we stratified the analysis and only examined individuals with the high responding TLR1 genotypes (1805TT/TG). We found the trends for TLR6 G1083C polymorphism were preserved, but the statistical significance was decreased, possibly due to the decreased sample size (FSL-1: GG = 24 750 pg/mL (n=8), CC = 12 061 pg/mL (n=6), P = 0.066; PAM2: GG = 17 010 pg/mL, CC = 11 512 pg/mL, P = 0.074; PAM3: GG = 68 760 pg/mL, CC = 43 756 pg/mL, P = 0.08; LPS: GG = 113 508 pg/mL, CC = 145 040 pg/mL, P = 0.784).
We also examined SNP TLR2_G2258A, which has previously been shown to regulate signaling (6, 27–30). We found that it was not associated with lipopeptide and mycobacteria-induced IL-6 secretion (data not shown) even though the frequency of this SNP in our populations was very rare (97GG, 3GA and no AA) to make any conclusions. These results suggest that these two previously characterized SNPs in TLR1 and TLR2 do not explain the association of G1083C with lipopeptide– and mycobacteria-induced IL-6 secretion.
We next examined possible mechanisms that may explain the observed differences in IL-6 secretion. These factors may include protein and mRNA transcript expression. Since this SNP is synonymous, we did not think it would be associated with differential protein expression. We then examined whether G1083C was associated with altered levels of TLR6 mRNA transcripts. We measured TLR6 mRNA levels in PBMCs from a subgroup of 26 individuals with different G1083C genotypes. No differences were observed in TLR6 mRNA levels when stratified according to genotypes in unstimulated PBMCs or in cells stimulated with PAM2 or LPS (data not shown). We also found that the polymorphisms in Table 2 were not associated with different TLR6 mRNA levels. Together, these results suggest that polymorphism TLR6_G1083C and other SNPs in this region were not associated with altered levels of TLR6 mRNA expression. In summary, G1083C is in linkage disequilibrium with several SNPs that are also associated with altered lipopeptide- or mycobacteria-induced IL-6 secretion. Based on these data, we are unable to determine which of these SNPs is most likely to directly regulate signaling in response to di-acylated lipopeptide stimulation.
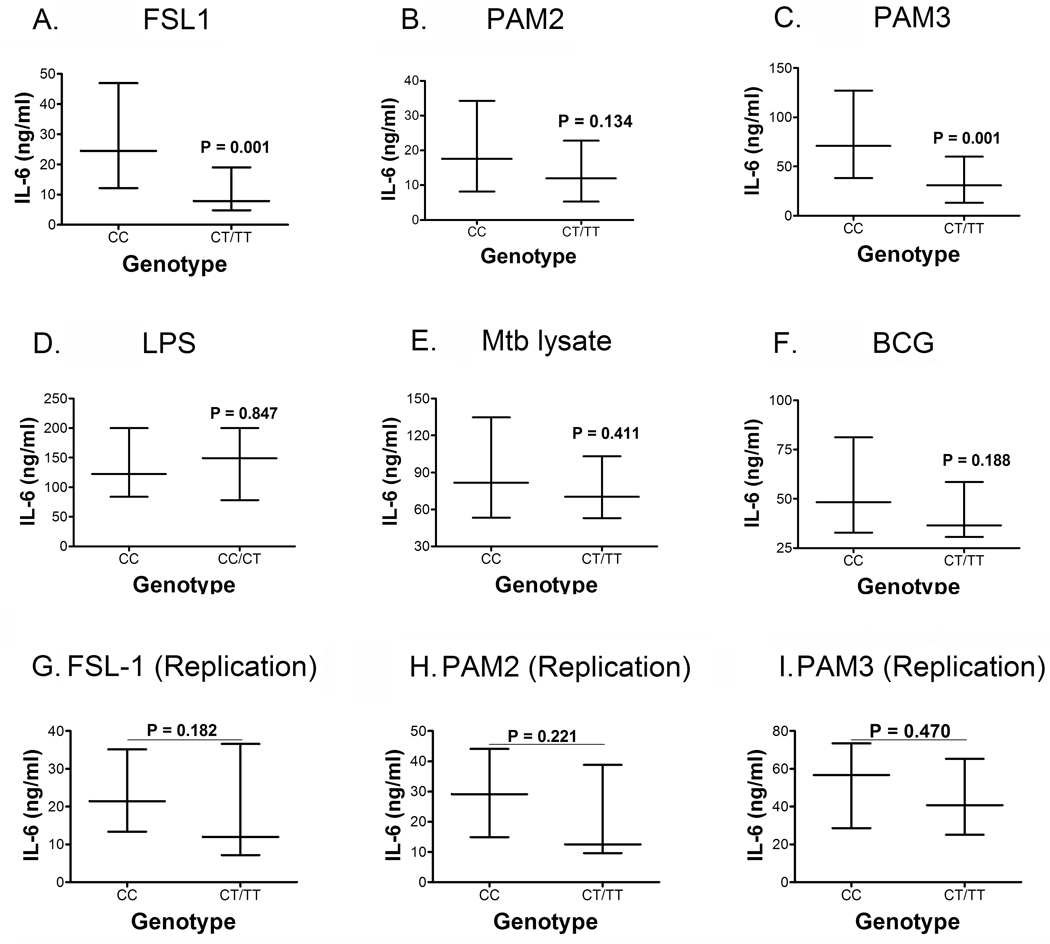
Polymorphism C745T is non-synonymous and encodes a proline to serine (P249S) change at amino acid 249 in the extracellular domain. Due to small numbers of the 745TT genotype (n=2), we were unable to assess its homozygous effect independently. When CC and CT/TT genotypes were compared, individuals with the 745CC genotype (n=55) had increased levels of IL-6 upon stimulation with FSL-1 and PAM3, compared with individuals with the 745CT/TT (n=15) genotypes (Fig 4 A and C, respectively). There were no differences when PAM2, the control LPS, or the mycobacterial antigens Mtb and BCG were used as stimulants (Fig 4B, D, E and F, respectively). We again tested reproducibility of the results by repeating the assay in 26 additional participants (CC, n=18; CT, n=6; TT, n=2). When comparing the CC (n=18) and the CT/TT (n=8) genotypes, similar trends were observed, but the differences were not significant in response to stimulation with FSL-1 (Fig 4G), PAM2 (Fig 4H) or PAM3 (Fig 4I). The lack of a significant difference may have been due to an inadequate sample size. We also observed that this polymorphism had a relatively high LD correlation with TLR6 promoter region and TLR1 polymorphisms (Figure 2). Although the validation results were inconclusive, the data suggests that allele 745T may be associated with decreased IL-6 secretion.
Figure 4. TLR6 C745T and Whole Blood IL-6 Secretion stratified by genotype.
Whole blood IL-6 levels were measured by ELISA and stratified by polymorphism C745T genotypes: Stimuli were (A) Di-acylated lipopeptide FSL-1, a TLR2/6 ligand, (B) Di-acylated lipopeptide PAM2, a TLR2/6 ligand, (C) Tri-acylated lipopeptide PAM3, a TLR2/1 ligand, (D) Lipopolysaccharide LPS, a TLR4 ligand, (E) H37Rv Mtb lysate, and (F) whole lyophilized live BCG. To validate our data, blood was further collected from a subset of 26 participants and IL-6 levels determined after stimulation of whole blood with (G) FSL-1, (H) PAM2, and (I) PAM3. Data depicted as medians and interquartile ranges with P values representing a comparison between the CC and CT/TT genotypes with a Mann-Whitney test. CC n=55; CT n= 13; TT n=2 for A–F, and CC n=18; CT n=6, TT n= 2 for G–I.
To account for the potential confounding effect of the IL-6 promoter polymorphism G-174C, which has been associated with reduced IL-6 levels (31), we genotyped this polymorphism in our study population. We found no association between IL-6 levels and genotype at position -174 of the IL-6 promoter (data not shown).
To further investigate a possible regulatory effect of C745T on lipopeptide signaling, we cloned the 745C (wild type) and 745T variant into the EF6-V5-His-TOPO expression plasmid vector. As a control, we also generated a dominant-negative TLR6 variant with a proline to histidine mutation at amino acid 680 (P680H, C2039A). This mutation is equivalent to the P712H substitution in TLR4, which abrogates LPS signaling in C3H/HeJ mice(32). We examined whether the TLR6 variants were able to mediate NF-κB signalling in HEK293 cells. In response to PAM2 stimulation, allele 745T mediated significantly decreased NF-κB signaling activity in comparison to allele 745C (Fig 5A). The dominant negative variant, 680H, was also associated with decreased NF-κB signaling. As a control, we stimulated the cells with LPS and found no significant signaling above baseline levels in comparison to wells stimulated with media alone (Fig 5B). We next examined whether variant 745T mediated signaling in response to a complex microbial ligand by stimulating with Mtb lysate. We found that allele 745T mediated decreased NF-κB signaling activity, compared with 745C (Fig 5C). As a control, the 2039A dominant negative variant completely abolished signaling to PAM2 and to Mtb lysate (Fig 5A and C).
Figure 5. Regulation of NF-κB signaling by a TLR6 polymorphism.
HEK293 cells were transfected with an NF-κB luciferase reporter, a Renilla luciferase construct to control for transfection efficiency (pRL-TK), and CD14. Additional transfectants varied by condition and included an empty plasmid vector (EV), or, TLR2 with one of 3 TLR6 constructs, 745C, 745T, or 2039A. Polymorphism C2039A (P680H) is a TLR6 variant with a dominant–negative effect on NF-κB signaling. Luciferase activity shown represents the ratio between basal activity (medium only) and that of transfected cells stimulated with (A) PAM2, (B) LPS, or (C) Mtb Lysate. Mean values (+/− standard deviation) are depicted for three independent experiments, each performed in triplicate. P values calculated with Student’s t-test. RLU, relative luciferase units. (D) Expression of TLR6 745C (wild type) and 745T (variant) by immunoblotting using transfected but unstimulated HEK293 cells.
We next examined whether differential localization patterns or expression levels of the 2 variants could mediate observed signaling differences. The TLR6 variants had similar protein levels when assessed by immunoblot (Fig. 5D). Further, TLR6 localization was similar for the variants with expression at or near the plasma membrane and those with expression within the cell (data not shown). The intracellular staining pattern excluded the nucleus and appeared vesicular, suggesting a location within organelles, rather than in the cytoplasm. As a negative control, we transfected the empty backbone pEF6 vector, and did not observe any positive staining (data not shown).
Together, these results suggest that polymorphism C745T directly regulates NF-κB signaling and IL-6 production in response to PAM2 stimulation and Mtb lysates.
DISCUSSION
In this study, we aimed to identify TLR6 polymorphisms occurring in Cape Town, South Africa, and to examine whether TLR6 polymorphisms regulate innate immune responses to lipopeptides and mycobacteria. We identified a novel polymorphism, and also observed that altered IL-6 secretion from stimulated whole blood was associated with the G1083C (T381T) and C745T (P249S) polymorphisms.
The mechanism of altered IL-6 secretion associated with the G1083C polymorphism is not known. G1083C is synonymous and does therefore not alter the amino acid composition of TLR6. Further, individuals with different G1083C genotypes had similar levels of TLR6 mRNA expression, suggesting that transcriptional regulation is an unlikely mechanism. Possible alternative mechanisms include post-transcriptional regulatory mechanisms, or differential stability of the protein or mRNA within the cell. Several studies have shown that mRNA expression and protein levels are not always correlated (33, 34).
Alternatively, SNP G1083C may be in linkage disequilibrium with a polymorphism that acts through a post-transcriptional mechanism. Candidate LD regions include the promoter region of TLR6, or polymorphisms in TLR1 and TLR10, TLRs that are adjacent to TLR6. To examine this possibility, we genotyped haplotype-tagging polymorphisms in TLR1, TLR10, and non-coding regions of TLR6. We found that overall there was a weak LD correlation between several polymorphisms and the G1083C and C745T polymorphisms. This suggests that the G1083C polymorphism may mediate differential IL-6 secretion through association with any of these polymorphisms. The effects of two previously characterized polymorphisms, TLR2_G2258A (R753Q)(6, 27–29) and TLR1_T1805G (I602S)(6, 8, 10), could not explain our results. Together, our data suggest that G1083C is at least a useful genetic marker for regulation of IL-6 responses to di-acylated lipopeptides. Due to our strict selection criteria for the polymorphisms to be genotyped, we might have excluded some polymorphisms which may be associated with mycobacterial and other diseases. Based on current data, we do not know the causative SNP underlying these observations. It is however possible that this polymorphism might be in LD with a yet-to-be described polymorphism either in TLR6 or in genes adjacent to TLR6.
SNP C745T is non-synonymous and was associated with altered NF-κB signaling in a transfected cell line and possibly with differences in IL-6 levels in response to lipopeptide stimulation of primary cells. Although the latter observation was not clearly reproducible, this may have been because we were unable to test sufficient numbers of persons with the rare homozygous 745TT genotype. Based on the NF-κB signaling data in a reconstitution system, this non-synonymous SNP appears to directly alter TLR6 signaling. Given the location of C745T (P249S) in the extracellular domain, we speculate that this polymorphism alters ligand recognition. Alternatively, the substitution could result in a conformational change that affects the assembly of a TLR2/6 signaling complex. A recent study reported no differences in NF-κB signaling between the two C745T variants after PAM2 stimulation of HEK293T cells (35). Compared with our experiments, these investigators used 10-fold more TLR6 DNA for transfection (100ng), and 25-fold less PAM2 (10ng/mL) as TLR6 ligand. These observations suggest that the effect of C745T on NF-κB signaling in a reconstituted system may be partial and results may vary in different in vitro experimental conditions.
Interestingly, the C745T polymorphism was associated with altered IL-6 production in response to stimulation with FSL-1, but not PAM2. Although both of these lipopeptides are di-acylated, they have different peptide moieties, which may affect TLR6 binding (36). Furthermore, PAM3 stimulation induced significantly different IL-6 levels in individuals with the different TLR6 genotypes, an unexpected finding given that PAM3 is predominantly a TLR2/1 ligand. These results may be due to our incomplete understanding of ligand specificity of these receptors. Previous studies indicate that the specificity of lipopeptides for TLR1 and TLR6 is not solely dependent on the number of acylation side groups. Other structural features such as length of the fatty acid chain, chirality of the diacyloxypropyl carbon, position of the acyl group, and amino acid composition of the terminal peptides have also been shown to affect receptor specificity (11, 12, 14, 15, 36, 37). The specificity of our findings was supported by the finding that IL-6 produced in response to LPS, a TLR4 ligand, was not associated with differences when comparing TLR6 genotypes.
Two previous studies have examined the functional role of TLR6 polymorphisms. A recent study demonstrated that rare SNPs in TLR6 were associated with altered NF-κB signaling and an increased risk of TB disease in certain ethnic populations (26). We did not find any of these polymorphisms in our sample of 100 individuals. Recently, three SNPs in TLR6 (rs5743795, rs1039560, and rs3775073) were associated with differential responses to PAM3 (10). PAM2 and FSL-1 were not tested in this study, so it is difficult to directly compare our results. We also examined one of these SNPs, A1263G (rs3775073), but it was not associated with differential responses to PAM3. We did not identify the other 2 SNPs in our cohort. Possible explanations for apparent discrepancies in results may be due to protocol differences including PAM3 dose (10 ng/mL vs. 300 ng/mL), incubation time (6 hours vs. 20 hours), and different population genetic backgrounds (Seattle, USA vs. Cape Town, South Africa). This suggests that other variables including dose of lipoproteins and genetic background may also influence TLR signaling responses.
Although the role of TLR1 in Mtb recognition is well established, the role of TLR6 has been less clear and has not been studied well (19). Our NF-κB signaling data supports a role for TLR6 in Mtb recognition. In addition, we found that TLR6_G1083C genotypes were associated with different levels of IL-6 after stimulation with Mtb lysate and BCG. We were surprised that the response to a combination of ligands may be associated with TLR6 genotypes. Since TLR6 recognizes di-acylated lipopeptides, this suggests the presence of these lipopeptides in Mtb and also that these lipopeptides may play a major role in immunity to Mtb. Previous studies on lipoproteins in TB show that the inactivation of the Mtb LspA by allelic replacement plays an important role in lipoprotein synthesis and the pathogenesis of Mtb(38). The cell wall associated lipoprotein LprA of Mtb was also shown to regulate innate immunity and inhibits antigen presentation in macrophages (39). In addition to Mtb, TLR6 is likely to mediate immune responses to a wide variety of pathogens (40, 41). TLR6 polymorphisms show some evidence of association with IL-6 secretion upon stimulation with lipopeptides and mycobacteria. These results require further replication to confirm the findings. This work thus suggests a role for TLR6 in regulating mycobacterial signaling and offers a strong impetus to evaluate the relationship between TLR6 polymorphisms and susceptibility to mycobacterial diseases including TB.
MATERIALS AND METHODS
Participant recruitment, enrollment, blood collection and processing
Healthy adults were enrolled at the South African Tuberculosis Vaccine Initiative clinical site, near Cape Town in South Africa. Exclusion criteria included HIV and other chronic infections, pregnancy, and active tuberculosis. Heparinized blood was collected, for whole blood incubations (below) and for PBMC isolation by density gradient centrifugation. This study was approved by the Research Ethics Committee of the University of Cape Town, the Western Institutional Review Board (USA), and by the Institutional Review Board of the University of Washington. Written informed consent was obtained from all participants. The study population included individuals from different backgrounds, including Black African (n=24), Caucasian (n=12) and South African Mixed Ethnicity (n=64). The latter is a distinct group that emerged more than 300 years ago and received genetic input from Malaysia, Indonesia, European Caucasoid and Black Africans(42).
Ligands and antigens
Ultrapure lipopolysacharide (LPS, TLR4 ligand, used in whole blood assays at 10ng/mL, concentration in other assays mentioned below) isolated from Salmonella minnesota R595 was obtained from List Biological Labs, Inc. (Campbell, CA, USA). The lipopeptides PAM2 (PAM2CSKKKK, S-[2,3-bis(palmitoyloxy)-propyl]-(R)-cysteinyl-(lysyl)3-lysine, TLR2/6 ligand, 100ng/mL), fibroblast stimulating lipopeptide 1 (FSL-1, TLR2/6 ligand, 300ng/mL) and PAM3 (Pam3CSKKKK, N-palmitoyl-S-[2,3-bis- (palmitoyloxy)-propyl]-(R)-cysteinyl-(lysyl)3-lysine, TLR2/1 ligand, 300ng/mL) were synthetic lipopeptides obtained from EMC Microcollections (Tuebingen, Germany). Lysate from Mtb strain H37Rv (25µg/mL) was obtained from J. Belisle (Colorado State University, Fort Collins, CO, USA; NIAID reagent contract). Lyophilized live Bacille Calmette-Guerin (BCG, 20 × 106CFU/mL) was obtained from Statens Serum Institute (Copenhagen, Denmark).
Whole blood incubation and IL-6 ELISA
Whole blood diluted 1:5 in RPMI medium 1640 (BioWhittaker; Walkersville, MD, USA), was incubated with TLR ligands, BCG or Mtb lysate for 20 hours. Polymixin B (BioChemika, 10µg/mL; Steinheim, Germany) was added to the antigen wells containing the lipopeptides (except to the LPS condition), to minimize any possible effects of LPS contamination. IL-6 secreted into the supernatant was measured by ELISA (Human IL-6 OptEIA, BD Biosciences; San Diego, CA, USA), according to the manufacturer’s instructions.
DNA isolation and sequencing
Genomic DNA was isolated from 5ml blood using the Qiagen Blood Maxi Preparation Kit (Valencia, CA, USA). TLR6 was PCR-amplified using Pfu polymerase (Promega; Madison, WI, USA) in two fragments (1600 and 900 base pairs, respectively) and nucleotide sequencing was performed (commercially by Macrogen, Seoul, South Korea). The following primers were used to amplify the two fragments: TLR6-2 (5’-GTGGAGGTTTGAGAGTAACCATCCG–3’), TLR6-15 (5’- GTGGGCTTCCTCTATAACTTTCTGGG–3’), TLR6-5 (5’-GAGGTCAATAAAAGCAGGGGACAATCC-3’) and TLR6-16 (5’-GGCTAACCTCACCGCCTAGCTCAGTTCCCC – 3’). The sequencing reactions were performed with the following for ward primers: TLR6-3 (5’-CACATGCTGTGTCCTCATGCACCAAGC-3’); TLR6-4 (5’-CACCCAACTAGTTTATTCGCTATCC-3’); TLR6-6 (5’-CCTGCCATCCTATTGTGAGTTTCAGGC-3’) and TLR6-7 (5’-GAGGAACTTTGTCCCTGGCAAGAGC-3’). The sequences were aligned and analyzed using the programs Phred/Phrap and Consed (University of Washington; Seattle, WA, USA) (43, 44).
DNA expression vectors
For functional studies, the single exon coding region of TLR6 was amplified from genomic DNA using Pfu Turbo polymerase (Promega) and primers hTLR6-start (5’- ATGACCAAAGACAAAGAACCTATTG–3’) and hTLR6-nostop (3’-CAGTGACTTTTGTTACTACACTTTAGA-5’). The amplified fragment was then cloned into the pEF6/V5-His-TOPO expression cloning vector with a V5 epitope tag (Invitrogen, Carlsbad, CA, USA). The different polymorphic variants were generated with a whole plasmid PCR mutation strategy with techniques previously described (45). The following primers were used to generate the 745T (249S) variant, hTLR6_745T-F 5’-ACCAGAGGGTCAACCTTACTGAATTTTACC-3’, and hTLR6_745T-R 5’–ATTCAGTAAGGTTGACCCTCTGGTGAGTTCTG–3’. For the dominant-negative control mutant, 2039A (680H), the primer set of hTLR6 2039A-F 5’-GAGGAACTTTGTCCATGGCAAGAGCATTGTGG-3’ and hTLR6 2039A-R 5’-CCACAATGCTCTTGCCATGGACAAAGTTCCTCTCATG-3’ was used. Subsequent digestion with the restriction enzyme DpnI removed the methylated DNA before transformation into JM109 competent E. coli cells (Invitrogen). Human TLR2 was cloned into pEF6/V5-His-TOPO vector after fragment amplification with the following primers, hTLR2-F 5’-ATGCCACATACTTTGTGG-3’ and hTLR2-R 5’ GGACTTTATCGCAGCTCTCAG-3’. All plasmid constructs were verified by sequencing.
HEK293 cell transfections
HEK 293 cells (ATCC #CRL-1573) were grown according to ATCC recommendations. HEK293 cells were cultured in Dulbelco’s modified Eagle medium (DMEM) (BD Biosciences; Lenexa, KS, USA), supplemented with 10% heat inactivated fetal bovine serum, 1% L-Glutamine (Life Technologies) and maintained at 37°C in 5% CO2. HEK293 cells were transfected for 16–20 hours using Polyfect Transfection Reagent (Qiagen) at a cellular concentration of 2 × 104 cells per well in a 96 well plate. Cells were transfected with the following plasmids: 100ng ELAM-luciferase (NF-κB reporter with ELAM promoter and Firefly luciferase), 10 ng pRL-TK luciferase (Thymidine kinase promoter with Renilla luciferase reporter used as a control for transfection efficiency), 40ng CD14 as co-receptor to enhance signaling, 10ng TLR6 or TLR6 variants, and 50ng TLR2. The final concentration of transfected DNA was normalized with empty vector, pEF6. After overnight incubation, the transfected cells were stimulated for 4hrs with PAM2 (250ng/mL), Mtb lysate (25µg/mL) or LPS (100ng/mL). The cells were lysed with cell lysis buffer (Promega) according to the manufacturer’s instructions and luciferase activity was measured with a dual luciferase system (Promega). To control for transfection efficiency, Firefly luciferase activity was normalized to Renilla luciferase activity.
Immunoblotting and immunofluorescence
For immunoblotting, cells were transfected for 16–20 hours and then lysed with 1% Triton X-100 buffer containing a cocktail of protease inhibitors (Sigma Aldrich; Steinheim, Germany). The lysates were blotted on a membrane and probed with a mouse anti-V5 epitope antibody (Serotec; Oxford, UK) that recognizes the V5-His tag on the TOPO cloning vector expressing the protein, followed by HRP-conjugated rabbit anti-mouse IgG (Zymed; San Francisco, CA, USA) and then developed with luminol chemiluminescent reagents (Roche; Basel, Switzerland).
For immunofluorescence, HEK293 cells were transfected for 16–20 hours with TLR6 constructs and plated in a 24 well plate on coverslips. After fixing with 10% neutral-buffered formalin solution (Sigma-Aldrich), the cells were lysed with 0.25% Triton X-100 in PBS, probed with the mouse anti-V5 epitope antibody and then with a FITC-conjugated rabbit anti-mouse conjugated secondary antibody. The coverslips were then washed and mounted for microscopy with mounting medium containing 25% glycerol, 10% polyvinyl alcohol (Sigma-Aldrich), 0.1M Tris-Cl pH 8.5 and 2.5% 1,4-diazabicyclo-[2.2.2)-Octane [DABCO].
mRNA expression
5 × 105 PBMCs were incubated with PAM2 (100ng/mL) and LPS (10ng/mL) or left unstimulated, for 20 hours. RNA was isolated from the stimulated PBMCs using RNeasy Mini Kit (Qiagen) and complementary DNA (cDNA) was synthesized from the extracted RNA using Omniscript RT Kit (Qiagen) with Oligo dT12–18 primers (Invitrogen). Recombinant RNaseOUT Ribonuclease inhibitor (Invitrogen) was used to inhibit ribonuclease activity. Quantitative real time PCR (qRT-PCR) was performed using SensiMix dT (Quantace; Watford, UK) containing 2× SensiMix dT, 50X SYBR Green solution and 50mM MgCl2 Solution with the following primers for TLR6, 5’-CTGTGTCCTCATGCACCAAG-3’ (forward) and 5’– TCAACCCAAGTGCAGTTTC – 3’ (reverse) and Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH): 5’–TTCACCACCATGGAGAGAGGC–3’ (Forward) and 5’– GGCATGGACTGTGGTCATGA – 3’ (Reverse). A standard curve was generated for each run with cloned fragments of TLR6. Results were normalized to relative expression of GAPDH.
Genotyping and linkage disequilibrium
Genotyping was done using Sequenom’s MassARRAY™ technique to identify polymorphisms in the promoter or coding region of TLR6 or in TLRs 2, 1 and 10, as previously described (46). This technique uses allele-specific primer extension reactions to discriminate genotypes. We identified haplotype tagging SNPs from the YRI (Yorubans in Nigeria) and CEU (Utah residents with European ancestry) populations from the International HapMap Project (http://www.hapmap.org) and other public databases with the Genome Variation Server (http://www.ncbi.nlm.nih.gov/SNP/ and www.innateimmunity.net). We searched a region on chromosome 4, 50 kilobases upstream and downstream of genes for TLR1, TLR6, TLR10, as well as TLR2 for tagged SNPs using an R2 cutoff of 0.8 for linkage disequilibrium and a minor allele frequency cut-off of 5%.
Stata/Intercooled v10.0 software program PWLD (StataCorp LP; College Station, TX, USA) was used to calculate R2 and D’ as measurements of linkage disequilibrium between the polymorphisms.
We also genotyped the G-174C polymorphism in the IL-6 promoter region in our population as previously described (31, 47). Briefly, the region flanking position 174 was PCR amplified and the PCR product digested with the restriction enzyme, Hsp92 II (Promega), which cleaves at the C allele, but not the G allele.
Data analysis
The observed allelic frequencies for the SNPs were determined by Hardy-Weinberg equilibrium equation for Black African and mixed ethnicity groups. A Mann-Whitney U test was used for statistical analysis of IL-6 secretion in whole blood while Student’s t-test was used for the luciferase signaling assays. The differences were considered significant if the p value was less than 0.05 using a two-tailed test.
Supplementary Material
Supplementary Figure 1: Reproducible levels of IL-6 in whole blood. Whole blood was stimulated for 20 hours with TLR ligands. Levels of IL-6 are depicted from 26 individuals, collected at two different time points. (A) FSL-1 (B) PAM2 and (C) PAM3. To accommodate values of 0 on the log scale, we replaced these values with 1. P value represents. Wilcoxon rank test comparison between the two bleeds.
Supplementary Figure 2: Linkage disequilibrium analyses between TLR6 and TLR1 SNPs D-prime (D’) values for each SNP combination are shown numerically and by shading, based upon the legend in the middle. The minor allele frequency is shown adjacent to each corresponding SNP. The program pwld in Stata was used to calculate the values.
Acknowledgments
We would like to thank the adult volunteers who participated in the study and also the immunology team at the SATVI research site in Worcester for obtaining informed consent and collecting blood from the participants. We also thank the National Bioinformatics Node (NBN) at the University of Cape Town for providing the PhredPhrap/Consed software. We also thank Rick Wells for excellent technical assistance and Marta Janer and Sarah Li for genotyping work.
This work was supported by the Dana Foundation (T.R.H. and W.A.H.), NIH NO1-AI-70022 (T.R.H. and W.A.H.), and the Burroughs Wellcome Foundation (TRH).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.van Duin D, Medzhitov R, Shaw AC. Triggering TLR signaling in vaccination. Trends in immunology. 2006 Jan;27(1):49–55. doi: 10.1016/j.it.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006 Feb 24;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunological reviews. 2009 Jan;227(1):248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunological reviews. 2009 Jan;227(1):221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 5.Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annual review of genetics. 2006;40:469–486. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- 6.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 2008 Mar;114(5):347–360. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 7.Hill AV. The genomics and genetics of human infectious disease susceptibility. Annual review of genomics and human genetics. 2001;2:373–400. doi: 10.1146/annurev.genom.2.1.373. [DOI] [PubMed] [Google Scholar]
- 8.Hawn TR, Misch EA, Dunstan SJ, Thwaites GE, Lan NT, Quy HT, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. European journal of immunology. 2007 Aug;37(8):2280–2289. doi: 10.1002/eji.200737034. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E, et al. Cutting edge: A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol. 2007 Jun 15;178(12):7520–7524. doi: 10.4049/jimmunol.178.12.7520. [DOI] [PubMed] [Google Scholar]
- 10.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. American journal of respiratory and critical care medicine. 2008 Oct 1;178(7):710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omueti KO, Beyer JM, Johnson CM, Lyle EA, Tapping RI. Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. The Journal of biological chemistry. 2005 Nov 4;280(44):36616–36625. doi: 10.1074/jbc.M504320200. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. International immunology. 2001 Jul;13(7):933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002 Jul 1;169(1):10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 14.Buwitt-Beckmann U, Heine H, Wiesmuller KH, Jung G, Brock R, Akira S, et al. TLR1- and TLR6-independent recognition of bacterial lipopeptides. The Journal of biological chemistry. 2006 Apr 7;281(14):9049–9057. doi: 10.1074/jbc.M512525200. [DOI] [PubMed] [Google Scholar]
- 15.Morr M, Takeuchi O, Akira S, Simon MM, Muhlradt PF. Differential recognition of structural details of bacterial lipopeptides by toll-like receptors. European journal of immunology. 2002 Dec;32(12):3337–3347. doi: 10.1002/1521-4141(200212)32:12<3337::AID-IMMU3337>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Brightbill HD, Modlin RL. Toll-like receptors: molecular mechanisms of the mammalian immune response. Immunology. 2000 Sep;101(1):1–10. doi: 10.1046/j.1365-2567.2000.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. 1999 Oct 1;163(7):3920–3927. [PubMed] [Google Scholar]
- 18.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 1999 Dec 7;96(25):14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulut Y, Faure E, Thomas L, Equils O, Arditi M. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J Immunol. 2001 Jul 15;167(2):987–994. doi: 10.4049/jimmunol.167.2.987. [DOI] [PubMed] [Google Scholar]
- 20.Dhiman N, Ovsyannikova IG, Vierkant RA, Ryan JE, Shane Pankratz V, Jacobson RM, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: Preliminary results. Vaccine. 2008 Mar 25;26(14):1731–1736. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffjan S, Stemmler S, Parwez Q, Petrasch-Parwez E, Arinir U, Rohde G, et al. Evaluation of the toll-like receptor 6 Ser249Pro polymorphism in patients with asthma, atopic dermatitis and chronic obstructive pulmonary disease. BMC medical genetics. 2005 Sep 28;6:34. doi: 10.1186/1471-2350-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tantisira K, Klimecki WT, Lazarus R, Palmer LJ, Raby BA, Kwiatkowski DJ, et al. Toll-like receptor 6 gene (TLR6): single-nucleotide polymorphism frequencies and preliminary association with the diagnosis of asthma. Genes and immunity. 2004 Aug;5(5):343–346. doi: 10.1038/sj.gene.6364096. [DOI] [PubMed] [Google Scholar]
- 23.Kesh S, Mensah NY, Peterlongo P, Jaffe D, Hsu K, M VDB, et al. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Annals of the New York Academy of Sciences. 2005 Dec;1062:95–103. doi: 10.1196/annals.1358.012. [DOI] [PubMed] [Google Scholar]
- 24.WHO. WHO Report 2009. World Health Organisation; 2009. Global Tuberculosis Control: Epidemiology, Strategy, Financing. (WHO/HTM/TB/2009.411) [Google Scholar]
- 25.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. The Journal of experimental medicine. 2005 Dec 19;202(12):1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE. 2007;2(12):e1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infection and immunity. 2000 Nov;68(11):6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroder NW, Diterich I, Zinke A, Eckert J, Draing C, von Baehr V, et al. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005 Aug 15;175(4):2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- 29.Schroder NW, Hermann C, Hamann L, Gobel UB, Hartung T, Schumann RR. High frequency of polymorphism Arg753Gln of the Toll-like receptor-2 gene detected by a novel allele-specific PCR. Journal of molecular medicine (Berlin, Germany) 2003 Jun;81(6):368–372. doi: 10.1007/s00109-003-0443-x. [DOI] [PubMed] [Google Scholar]
- 30.Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. The Lancet infectious diseases. 2005 Mar;5(3):156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 31.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. The Journal of clinical investigation. 1998 Oct 1;102(7):1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (New York, NY. 1998 Dec 11;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002 Apr;1(4):304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome biology. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, Pickrell JK, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009 Jul;5(7) doi: 10.1371/journal.pgen.1000562. e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okusawa T, Fujita M, Nakamura J, Into T, Yasuda M, Yoshimura A, et al. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infection and immunity. 2004 Mar;72(3):1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller SD, Muller MR, Huber M, Esche Uv U, Kirschning CJ, Wagner H, et al. Triacyl-lipopentapeptide adjuvants: TLR2-dependent activation of macrophages and modulation of receptor-mediated cell activation by altering acyl-moieties. International immunopharmacology. 2004 Oct;4(10–11):1287–1300. doi: 10.1016/j.intimp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Rezwan M, Grau T, Tschumi A, Sander P. Lipoprotein synthesis in mycobacteria. Microbiology (Reading, England) 2007 Mar;153(Pt 3):652–658. doi: 10.1099/mic.0.2006/000216-0. [DOI] [PubMed] [Google Scholar]
- 39.Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV. Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol. 2006 Jul 1;177(1):422–429. doi: 10.4049/jimmunol.177.1.422. [DOI] [PubMed] [Google Scholar]
- 40.Henneke P, Dramsi S, Mancuso G, Chraibi K, Pellegrini E, Theilacker C, et al. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J Immunol. 2008 May 1;180(9):6149–6158. doi: 10.4049/jimmunol.180.9.6149. [DOI] [PubMed] [Google Scholar]
- 41.West TE, Ernst RK, Jansson-Hutson MJ, Skerrett SJ. Activation of Toll-like receptors by Burkholderia pseudomallei. BMC immunology. 2008;9:46. doi: 10.1186/1471-2172-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lombard Z, Brune AE, Hoal EG, Babb C, Van Helden PD, Epplen JT, et al. HLA class II disease associations in southern Africa. Tissue Antigens. 2006 Feb;67(2):97–110. doi: 10.1111/j.1399-0039.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 43.Gordon D. Viewing and editing assembled sequences using Consed. Chapter 11. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al. 2003 Aug; doi: 10.1002/0471250953.bi1102s02. Unit11 2. [DOI] [PubMed] [Google Scholar]
- 44.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome research. 1998 Mar;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 45.Bochud PY, Hawn TR, Aderem A. Cutting edge: a Toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J Immunol. 2003 Apr 1;170(7):3451–3454. doi: 10.4049/jimmunol.170.7.3451. [DOI] [PubMed] [Google Scholar]
- 46.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods in molecular biology (Clifton, NJ. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 47.Olomolaiye O, Wood NA, Bidwell JL. A novel NlaIII polymorphism in the human IL-6 promoter. Eur J Immunogenet. 1998 Apr-Jun;25(2–3):267. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Reproducible levels of IL-6 in whole blood. Whole blood was stimulated for 20 hours with TLR ligands. Levels of IL-6 are depicted from 26 individuals, collected at two different time points. (A) FSL-1 (B) PAM2 and (C) PAM3. To accommodate values of 0 on the log scale, we replaced these values with 1. P value represents. Wilcoxon rank test comparison between the two bleeds.
Supplementary Figure 2: Linkage disequilibrium analyses between TLR6 and TLR1 SNPs D-prime (D’) values for each SNP combination are shown numerically and by shading, based upon the legend in the middle. The minor allele frequency is shown adjacent to each corresponding SNP. The program pwld in Stata was used to calculate the values.