SUMMARY
Dynamic histone H3K4 methylation is an important epigenetic component of transcriptional regulation. However, most of our current understanding of this histone mark is confined to regulation of transcriptional initiation. We now show that human LSD2/KDM1b/AOF1, the human homolog of LSD1, is an H3K4me1/2 demethylase that specifically regulates histone H3K4 methylation within intragenic regions of its target genes. Genome-wide mapping reveals that LSD2 associates predominantly with the gene bodies of actively transcribed genes, but is markedly absent from promoters. Depletion of endogenous LSD2 results in an increase of H3K4me2 as well as a decrease of H3K9me2 at LSD2 binding sites, and a consequent dysregulation of target gene transcription. Furthermore, characterization of LSD2 complex revealed that LSD2 forms active complexes with euchromatic histone methyltransferases EHMT1/2 and NSD3 as well as cellular factors involved in active transcription elongation. These data provide a possible molecular mechanism linking LSD2 to transcriptional regulation post initiation.
INTRODUCTION
The interplay and dynamics of histone modifications govern the structural diversity of chromatin and the accessibility of DNA, thus representing an important epigenetic mechanism underlying regulation of eukaryotic gene transcription (Goldberg et al., 2007; Jenuwein and Allis, 2001; Strahl and Allis, 2000). Many studies have addressed how histone modifications are established and/or maintained at gene promoters to regulate transcription initiation. However, once transcription initiation events commence, RNA polymerase II (Pol II) can either continue forward with productive elongation, as is the case for constitutively active genes, or demonstrate promoter-proximal pausing, depending on gene context, cellular demands, or environmental stimuli (Core and Lis, 2008; Sims et al., 2004). Several regulatory mechanisms converge at this stage, including the activity of negative and positive elongation factors. While the negative elongation factor complex (NELF) functions by effectively stalling Pol II, positive transcription elongation factor b (P-TEFb), comprised of CDK9 and cyclin T1, phosphorylates the C-terminal domain of Pol II, NELF, DSIF and other targets thereby promoting productive elongation (Bres et al., 2008; Peterlin and Price, 2006).
Although the functional role and dynamic regulation of histone modification during active elongation remain largely unknown, increasing evidence indicates a distinct epigenetic contribution to post-initiation transcriptional regulation. Both (negative) NELF and (positive) P-TEFb have been reported to play important roles in coordinating cotranscriptional histone modifications, such as histone acetylation, H3K4 and H3K36 methylation, and H2B ubiquitination during Pol II elongation (Pirngruber et al., 2009). Further, studies ranging from yeast to humans have suggested that several histone modifiying enzymes are associated with elongating Pol II, functioning to control proper histone methylation within coding regions for productive transcription or mRNA processing. For example, in yeast, SET2 associates with elongating Pol II and mediates co-transcriptional H3K36 methylation within the coding regions of actively transcribed genes (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). The Eaf3/Rpd3 deacetylase complex can recognize this H3K36 methylation mark and remove histone acetylation immediately subsequent to Pol II transcription, thus maintaining a repressive chromatin structure and preventing unwanted intragenic transcription initiation or cryptic transcription. In mammalian cells H3K9 methylation and its associated HP1γ have been shown to be dynamically regulated within actively transcribed regions, and it has been speculated that this mark, in coordination with modifications at H3K4, H3K36, and H3K79, comprises a unique histone modification pattern characteristic of actively transcribed chromatin (Brinkman et al., 2006; Vakoc et al., 2005). Collectively, these findings not only illustrated the interplay and cooperative nature of various histone modifications during elongation but also led to the unexpected finding that maintenance of a `repressive' environment may be critical for optimal transcription (Berger, 2007).
The functions and mechanisms of several LSD1/KDM1a family histone demethylases have been intensively studied in multiple model organisms including human, mouse, Drosophila, C. elegans and S. pombe. Yet, our knowledge of the precise roles of LSD1 family members remains mostly limited to promoter association and regulation. Specifically, one key function of LSD1 is balancing promoter H3K4/H3K9 methylation for activation or repression of LSD1 target genes (reviewed in (Chosed and Dent, 2007)).
LSD2/KDM1b/AOF1 is the only mammalian homolog of LSD1. Here we present evidence for an important function of LSD2 in active gene transcription. LSD2 specifically associates with the coding region of its target genes. Removal of endogenous LSD2 promotes an increase in H3K4me2 levels and concurrent decrease in H3K9me2 levels, with a consequent down-regulation of targeted gene transcription. We propose a model where LSD2, possibly through association with elongation factors and phosphorylated Pol II, is required for coordinating the dynamics of H3K4 and H3K9 methylation within elongating regions of genes, maintaining repressive chromatin structure, and thus contributes to the fine-tuning of gene expression.
RESULTS
LSD2 is an H3K4-specific demethylase with molecular and functional properties distinct from LSD1
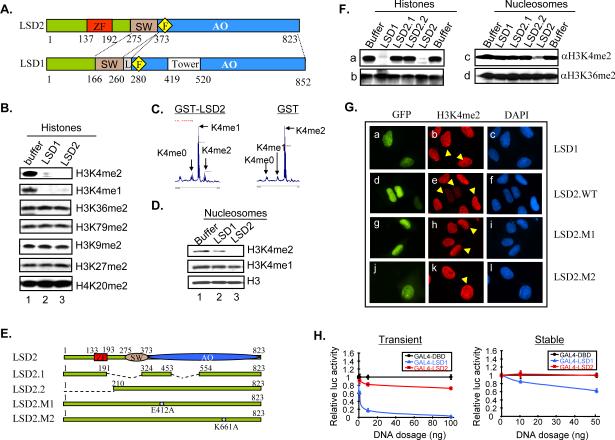
LSD2/KDM1b/AOF1, the only homolog of LSD1/KDM1a/AOF2 in the human genome, shares a similar domain homology with LSD1, but exhibits an overall sequence identity of less than 31%. Both LSD1 and LSD2 contain a SWIRM domain, an FAD coenzyme binding motif, and a C-terminal amine oxidase domain, all of which are integral to the enzymatic activity of LSD family members (Figure 1A). In contrast to LSD1, LSD2 contains a CW-type zinc-finger domain in its N-terminal region; the corresponding region in LSD1 is unstructured. Additionally, LSD2 lacks the “tower domain” present in LSD1. Thus, we predicted that the unique structure of LSD2 may confer biochemical and biological properties distinct from those of LSD1.
Figure 1. LSD2 is an H3K4me2 specific demethylase with distinct properties from LSD1.
A. Schematic representation of LSD2 and LSD1. ZF, zinc finger domain; SW, SWIRM domain; AO, amine oxidase domain; F, FAD binding motif; L, linker.
B. LSD2 specifically demethylates mono- and di-methylated H3K4. Bulk histones were incubated with purified recombinant LSD2 and LSD1, and subject to immunoblot analysis with methylation specific anti-histone antibodies as indicated.
C. MALDI-TOF mass spectrometry analysis of LSD2 demethylation of H3K4me2 histone peptides. Peptide masses corresponding to un-methylated (K4me0), mono-methylated (K4me1) and di-methylated (K4me2) peptides are denoted.
D. In vitro demethylation of nucleosomes.
E. Schematic representation of LSD2 variants and mutants.
F. Demethylation analysis of LSD2 variants using bulk histone (left) and nucleosome (right) substrates.
G. Immunofluorescence analysis of in vivo demethylation activities. Arrows denote cells transfected with GFP fusion proteins. Green, GFP fusion protein; red, H3K4me2; blue, DAPI counterstain of DNA.
H. LSD2 does not repress reporter gene expression when artificially tethered to promoters. Luciferase activity relative to GAL4-DBD control is presented. Transient, transient transfected; stable, stably integrated reporters.
Using a series of in vitro and in vivo assays, we confirmed that LSD2, like its human homolog LSD1, possesses H3K4me1/2 specific histone demethylase activity. As shown in Figure 1B, LSD2 can remove mono- and di-methyl H3K4 modifications from histone substrates, without affecting other marks including dimethyl-H3K36, -H3K79, -H3K9, -H3K27, and -H4K20. Robust H3K4me2 demethylase activities were observed on synthetic H3 peptides (Figure 1C) and nucleosomes (Figure 1D), but only weak or negligible activity towards H3K4me1 under these conditions, suggesting that H3K4me2 is the preferred substrate. While it is possible that optimal demethylase activity of LSD2 may still require additional cofactor(s), LSD2 alone could demonstrate significant H3K4 demethylase activity on nucleosomal substrates—a clear distinction from LSD1.
We next examined a series of mutations to further dissect the critical residues and domains involved in demethylation activity. The panel of proteins includes two point mutations that ablate the predicted FAD binding site (LSD2.M1) and catalytic residue (LSD2.M2), as well as deletions that remove part of the amine oxidase and SWIRM domain (LSD2.1, a splicing isoform, NM_153042.3) or the upstream zinc finger (LSD2.2) (Figure 1E). In vitro demethylase activity assays of these LSD2 mutants demonstrate the requirement of the SWIRM, amine oxidase, and zinc finger regions, as well as critical residues, for robust enzymatic activity of LSD2 protein (Figure 1F and Figure S1A). We also examined LSD2 demethylase activity in vivo, by staining for global di-methylated H3K4 levels after GFP-LSD2 overexpression. We observed a significant reduction in total cellular H3K4me2 levels in cells expressing GFP-LSD2, whereas GFP-LSD1 was unable to do so as previously observed (Shi et al., 2005). H3K4me1 demethylase activity was undetectable under identical experimental conditions (compare Figure S1B and 1C). As expected, LSD2.M1 and LSD2.M2 did not demonstrate demethylase activity in vivo (Figure 1G).
Human LSD1 was first characterized as a transcriptional repressor and has well-studied functions in promoter regulation. To explore any potential corepressor activity of LSD2, we utilized a Gal4-TK-luciferase reporter assay where LSD2 was tethered to an active promoter via a Gal4-DBD. In both transiently transfected and stably integrated reporter systems, LSD2 displays little impact on luciferase reporter expression. Experiments confirmed the proper expression, nuclear localization and enzymatic activity of Gal4-LSD2 in vivo (Figure S1, D–E), hence excluding the possibility that the lack of repressive activity of the Gal4 fusion LSD2 is caused by instability or inactivation of LSD2 protein. These results suggest that LSD2 may act differently from LSD1 in epigenetic gene regulation.
LSD2 is absent from gene promoters but preferentially associates with gene bodies downstream of the promoters
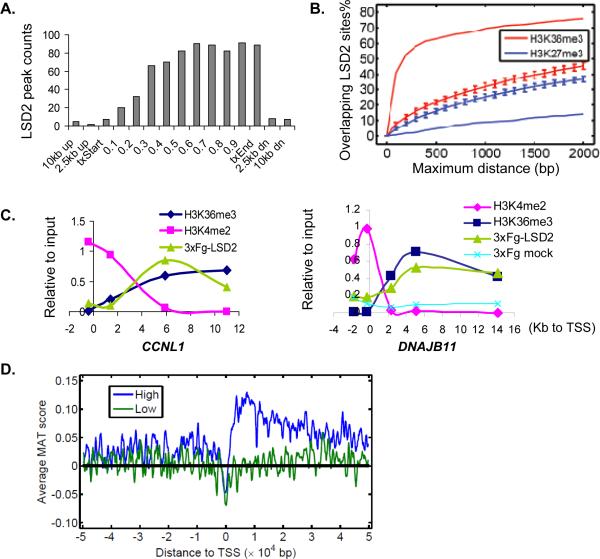
LSD2/KDM1b has been reported to regulate the establishment of maternal imprinting in mouse oocytes (Ciccone et al., 2009). However, little is known about how LSD2 functions in non-oocyte cells, although we found that human LSD2 is expressed in multiple tissues and cell lines (Figure S2A and B). To address this question, we employed a ChIP-chip tiling array strategy to identify LSD2 binding loci on a genome-wide scale in HeLa cells. To ensure chromatin immunoprecipiation (ChIP) efficacy and specificity, we incorporated native ChIP with tandem affinity purification (TAP-ChIP) from HeLa cells stably expressing Flag-HA tagged LSD2. TAP-ChIP DNA was processed for tiling array analysis using an Affymetrix Human Tiling 2.0R Array chip that covers chromosomes 3, 21, 22, X and Y (representing roughly 1/7 of the human genome). Utilizing the Model-based Analysis Tiling-arrays (MAT) algorithm, we identified >800 LSD2 binding sites (p<0.0001) scattered along the arms of these chromosomes (Figure 2A). Remarkably, as exemplified by DNAJB11 and CCNL1 (Figure 2B), the vast majority of LSD2 binding signal was found to be enriched at introns or exons (herein defined as intragenic regions or gene bodies) but absent from promoters. Statistical analysis showed that more than 80% of LSD2 targets were located either at the exons (33.9%) or introns (50.5%) of ~450 genes, while 0.45% of LSD2 was found at promoters, and 15.1% at intergenic regions (Figure 2C). It is noted that 1.1–1.4% of human genome is spanned by exons; 24.4–36.4% by introns; 0.9–1.4% by gene promoters (Venter et al., 2001). Clearly LSD2 does not show any enrichment at promoters, which is strikingly different from the predominant promoter association of all other LSD1 family of histone demethylases characterized so far (Lan et al., 2007; Wang et al., 2007). To rule out any potential artifacts due to ChIP-chip technology or native ChIP conditions, we performed conventional ChIP and validated the specific association of LSD2 with coding regions versus gene promoters on all of the targets examined (Figure 2D).
Figure 2. CHIP-chip analysis indicates enrichment of LSD2 binding sites within the coding regions but not at promoters of genes.
A. Whole genome tiling array analysis of LSD2 binding across human chromosome 3. Vertical bars mark positions of LSD2 target sites (P<0.0001). ChIP-chip signals (MAT score) shown in log 2 scale.
B. ChIP-chip signals of LSD2 binding across CCNL1 and DNAJB11. Arrow denotes promoter orientation.
C. Distribution of LSD2 binding sites.
D. Confirmation of the preferential LSD2 binding to gene bodies. Cross-linked ChIP using anti-Flag antibody was performed from 3×Flag-LSD2 expressing HeLa (purple bars, 3×Fg-LSD2) and empty vector transduced cells (open bars, mock). Fold of enrichment relative to mock was shown. PCR primers specific to coding region or the promoter (-pro, boxed) of each gene were denoted in X-axis. Error bars, s.e.m. of triplicates from representative experiment. LSD2 did not shown significant binding to the promoter (GAPDH-pro) or an intragenic region (GAPDH-intra) of GAPDH.
LSD2 binds to highly transcribed coding regions enriched in H3K36me3
To explore the nature of LSD2 association with genes, we further analyzed the intragenic distribution of LSD2 binding sites. Each target gene (defined in Ensembl 56 gene database GRCh37; http://www.ensembl.org) was divided into ten equal units, and the number of LSD2 binding sites within each segment was counted and plotted versus the distance from the transcription start site (TSS) (Figure 3A). We found that the majority of LSD2 binding occurs downstream of promoter regions, peaking towards the 3' end of genes. Negligible binding was observed beyond the up- or down-stream gene boundaries.
Figure 3. LSD2 binding correlates with high levels of tri-methylated H3K36 within the coding regions of actively transcribed genes.
A. Distribution of LSD2 binding sites across genes. The open reading frame of each LSD2 target gene was divided into 10 equal segments; regions corresponding to 0–2.5kb, 2.5–10kb up or down-stream each gene were also studied. The number of LSD2 binding sites within each defined segment is shown. txStart, transcription start site; txEnd, transcription end.
B. Correlation of LSD2 binding to histone modifications. Enrichment of LSD2 binding sites, presented as the fraction of total peaks, was plotted against the relative distance to known H3K36me3 (red line) and H3K27me3 (blue line) sites. Permuted distribution models are shown as hatched lines.
C. LSD2 binding at CCNL1 and DNAJB11 loci overlaps with H3K36me3 and inversely correlates with H3K4me2. Enrichment relative to input is presented in arbitrary units. Distance of qPCR primers relative to the transcription start site (TSS) is shown.
D. LSD2 preferentially associates with the gene bodies of actively transcribed genes. Genes present on the tiling array were divided into high expressing (top 50%, blue line) and low expressing (low 50%, green line) groups. Average LSD2 binding signals CHIP-chip (or MAT scores) are plotted relative to TSS within each group.
The distinct distribution of LSD2, showing intragenic binding preferentially at the 3' end, is reminiscent of the pattern of H3K36me3 deposition (Barski et al., 2007; Hampsey and Reinberg, 2003). We therefore examined the correlation of LSD2 binding with H3K36me3 modifications. As shown in Figure 3B, >60% of LSD2 binding sites are located within 0.5 kb of an H3K36me3 site, demonstrating a strong correlation between LSD2 localization and H3K36me3 enrichment (p<10−72). In contrast, no apparent correlation of LSD2 loci with the repressive mark H3K27me3 was found (p≈1) (Figure 3B). To illustrate this finding on individual genes, we examined LSD2 binding, H3K36me3 and H3K4me2 levels across LSD2 target genes CCNL1 and DNAJB11 by conventional ChIP (Figure 3C). Typical for actively transcribed genes, H3K4me2 peaks at the TSS and rapidly decreases further downstream; H3K36me3 is low at promoters and peaks in the 3' region. LSD2 binding profile at these loci mirrors that of H3K36me3, but inversely correlates with H3K4me2 along the gene body, consistent with the H3K4me2 histone demethylase activity of LSD2.
The correlation with H3K36me3 strongly suggests an association of LSD2 with active gene transcription. To test this hypothesis, we divided the annotated genes present on the tiling array into high and low expression groups, and examined LSD2 CHIP-chip signals relative to the TSS within each group. As shown in Figure 3D, a significant enrichment of LSD2 binding signals was observed downstream from the TSS of highly expressed genes (blue line within 0~30kb), whereas no obvious enrichment was observed either up- or downstream the TSS in the low expressing group (green line). These observations together indicate that LSD2 preferentially associates with the coding regions but not the promoters of actively transcribed genes, further suggesting a positive role of LSD2 in active gene transcription.
LSD2 is required for maintenance of H3K4 and H3K9 methylation status within transcribing regions of active genes
To explore the mechanism(s) of action of LSD2 in active gene transcription, we next examined the role of LSD2 in regulating histone marks within coding regions of its target genes. We depleted endogenous LSD2 using shRNA and first examined the changes in H3K4 methylation levels on the identified LSD2 targets. LSD2 shRNA significantly depleted LSD2 expression at both mRNA and protein levels, but had no effect on the expression of LSD1 (Figure 4A). Changes to the H3K4me2 profile across CCNL1 and DNAJB11 were examined by quantitative ChIP subsequent to LSD2 depletion. In agreement with the lack of LSD2 association with gene promoters, no significant increases of H3K4me2 levels were noted at these active promoters; coinciding with LSD2 binding within gene bodies, we consistently observed an increase in H3K4me2 levels across the coding regions of these genes (Figure 4B). Examination of additional targets revealed a similar pattern of H3K4me2 changes (Figure 4C). Significantly, no obvious changes of H3K4me2 levels were found within the coding region of SCG10, a repressed gene that does not associate with LSD2 in HeLa cells. The specific H3K4me2 increases at coding regions were confirmed by a second LSD2 shRNA (Figure S3A). Thus, these data suggest that reduction of LSD2 within coding regions elevates the otherwise depressed H3K4me2 levels of these regions.
Figure 4. LSD2 maintains the balance of H3K4 and H3K9 methylation within intragenic regions.
A. Specific LSD2 depletion by shRNA. qRT-PCR result normalized with β-actin (upper) and immunoblot of whole cell lysates (lower) are presented. shCntr, control shRNA; shLSD2, LSD2-specific shRNA. Error bars, s.e.m. of triplicates from representative experiment.
B. LSD2 depletion causes an increase of H3K4me2 within gene bodies but not at promoters of CCNL1 and DNAJB11. Data presented as relative enrichment compared to input. Distance of qPCR primers from the TSS is denoted. Error bars, s.e.m. of triplicates from representative experiment.
C. Effect of LSD2 depletion on H3K4me2 levels at additional LSD2 binding sites. Data presented as fold of change relative to input, comparing shLSD2 HeLa to shCntr HeLa. Primers targeting the 3' regions or promoters (-pro) of the genes are denoted on the X-axis. Error bars, s.e.m. of triplicates from representative experiment.
D. LSD2 depletion results in an increase of intragenic H3K9me2 levels of LSD2 binding targets. Data presented as in C. Negative controls, SCN2A and SCG10, are two repressed genes not associated with LSD2.
We next examined changes of other histone modifications at the same regions to further explore the consequences of LSD2 removal on chromatin structure. Of particular interest, it has been reported that H3K9 methylation, typically a mark of repressive chromatin, is associated with actively transcribed regions and required for active transcription (Brinkman et al., 2006; Vakoc et al., 2005). In contrast to the increase of H3K4me2, a consistent decrease in H3K9me2 (Figure 4D) was detected at LSD2 binding sites. Despite a strong co-alignment with LSD2 binding, no significant change of the H3K36me3 profile was observed upon LSD2 depletion (Figure S3B). In addition, we did not observe significant changes in the levels of acetylated H3 (Figure S3C) or tri-methylated H3K4 (data not shown).
Depletion of LSD2 or inhibiting its demethylase activity results in downregulation of a subset of LSD2 target genes
To interrogate the role of LSD2 in gene transcription, we examined genome-wide mRNA expression profile changes upon LSD2 depletion using microarray analysis. We identified 461 genes that are differentially expressed after LSD2 depletion in HeLa cells, using a criteria of >1.5 fold changes in expression values. Interestingly, the majority (77%) of differentially expressed genes were down-regulated after LSD2 depletion. Similar results were observed using more stringent criteria (>2 fold change), where 89 of the 107 target genes were down-regulated (Figure 5A). 37 out of the 461 differentially expressed (DE) genes identified by microarray are located on the four chromosomes examined by LSD2 ChIP-chip (3, 21, 22 and X). 11 of these 37 DE genes (~30%) are direct LSD2 targets as determined by ChIP-chip analysis, 9 of which are down-regulated after LSD2 depletion comparing to 2 that are up-regulated. Microarray results were confirmed by quantitative RT-PCR (Figure 5B, blue or purple bars show down- and up-regulated genes, respectively). The LSD2-specific knockdown effects on down-regulation of these genes were further validated by a second LSD2 shRNA (Figure S4A). Together, these data indicate that LSD2 is important for active gene transcription.
Figure 5. Depletion of LSD2 or inhibition of its demethylase activity results in downregulation of a subset of actively transcribed genes.
A. The majority of differentially expressed (DE) genes after LSD2 depletion are down-regulated. The number of DE genes filling the criteria of 1.5–2.0 fold (light grey) or ≥2 fold (dark grey) changes is shown. Up, up-regulated; down, down-regulated.
B. Quantitative RT-PCR confirms the down-regulation of a large subset of LSD2 targets after LSD2 depletion. Error bars, s.e.m. of triplicate independent experiments.
C. Inhibition of demethylase activity by tranylcypromine causes down-regulation of LSD2 target genes. T-72, HeLa treated with tranylcypromine for 72 hours; cntr, 72 hour treatment with vehicle only. Error bars, s.e.m. of biological triplicates.
D. LSD2 is required for optimal induction of immediate-early genes. Control and LSD2 shRNA-transduced HeLa were treated with 50ng/mL of EGF for up to 40 minutes. Data is presented as mRNA abundance relative to time 0. Error bars, s.e.m. of triplicates.
To ascertain whether the enzymatic activity of LSD2 is important for its role in gene transcription, we studied the effect of a monoamine oxidase inhibitor, tranylcypromine, on the expression of LSD2 regulated genes. Since tranylcypromine inhibits the demethylase activity of both LSD1 and LSD2 (Karytinos et al., 2009; Lee et al., 2006b), we decided to isolate the effect of tranylcypromine inhibition on LSD2 by focusing on genes that are specifically regulated by LSD2, but not LSD1 (Figure S4B, blue bars). Similar to the effect of LSD2 depletion, the majority of LSD2 targets examined were down-regulated after tranylcypromine treatment (Figure 5C). This result links LSD2 demethylase activity to active gene transcription.
In addition to the constitutively active genes studied, we examined the effect of LSD2 depletion on inducible gene expression. We chose to investigate a set of immediate-early (IE) genes, since without stimulation these genes are “pre-initiated” but blocked at an early stage of elongation, with RNA polymerase II paused proximal to transcription start sites (Wang et al., 2005). Upon EGF treatment, the transcription of a number of IE genes is greatly induced within minutes independent of promoter initiation and de novo protein synthesis. HeLa cells were treated with control and two independent shLSD2 constructs, followed by EGF treatment. Induction efficiency of CTGF, DUSP1, and EGR1 were significantly reduced after LSD2 depletion with the most obvious effect occurring at 30 and 40 minutes post-treatment (Figure 5D). Endogenous LSD2 binding within the gene bodies of these genes was detected by ChIP (Figure S4C). These data together strongly support the finding that LSD2 likely functions to modulate chromatin structure of coding regions to facilitate active elongation.
Isolation and characterization of LSD2-associated complex physically and functionally links LSD2 to active transcription elongation
To investigate how LSD2 is physically and functionally linked to the transcription machinery, we isolated and characterized the LSD2-containing cellular complex using tandem affinity purification (TAP) (Shi et al., 2005; Tahiliani et al., 2007). The LSD2 complex eluted from the second affinity purification step (anti-HA affinity column) is highly specific (Figure 6A, compare lanes 3 and 4). LSD2-associated proteins were identified by MS/MS proteomic analysis.
Figure 6. The LSD2 complex contains proteins involved in active gene transcription and functional links LSD2 to intragenic gene regions.
A. Tandem affinity purification (TAP) and mass spectrometric analysis of LSD2 complex. Silver staining of Flag elutes (lane 1–2) and HA elutes (lane 3–4) of FgHA-LSD2 and mock purification was shown. A complete list of associated polypeptides identified by MS/MS is shown in Figure S5.
B. Reciprocal immunoprecipitation. Nuclear extracts from FgHA-LSD2 HeLa were immunoprecipiated with either IgG control or indicated antibodies. Input and precipitated proteins were analyzed by immunoblot with the indicated antibodies.
C. LSD2 complex demethylates H3K4me2 but lacks deacetylase activity. Bulk histones were incubated with buffer alone (lane 3–4), LSD1 complex (lane 6) or two independent preparations of LSD2 complex (lane 5, 7) and immunobloted with the indicated antibodies. Inputs, nuclear extract of mock (lane 1) and FgHA-LSD2 HeLa (lane 2).
D. LSD2 complex possesses H3K9 methyltransferase activity. Wild-type GST-histone H3 and a panel of arginine substitutions were used. Autoradiography of methylation signal is shown (upper); histone inputs are stained by Coomassie blue (lower).
E. G9a regulates LSD2 target genes. qRT-PCR data was normalized with beta-2 microglobulin (B2M) and is expressed as relative abundance comparing G9a shRNA (shG9a) treated HeLa with control (shCntr). Effects of shG9a on the expression of housekeeping genes HPRT and GAPHD are shown. Blue bars, genes down-regulated after LSD2 depletion shown in Figure 5B; purple bars, up-regulated genes. Error bars, s.e.m. of triplicates.
F. Model of gene transcription regulation by LSD2 complex. LSD2 localizes to the 3' end of actively transcribed genes, where it can interact with phosphorylated RNA polymerase II and elongation factors. The LSD2 complex coordinates intragenic H3K4me2, H3K9 methylation and potentially also H3K36me2 levels via its own intrinsic demethylase activity, and through interaction with histone methyltransferases G9a and NSD3.
The composition of the LSD2 complex is notably distinct from the LSD1 complex (Shi et al., 2005). LSD2-associated proteins fall into several functional groups, including DNA replication and damage repair, nucleosome remodeling and histone modification, and active gene transcription (summarized in Figure S5), suggesting that LSD2 may be involved in multiple cellular functions. The presence of a number of proteins from each functional category was confirmed by immunoblot (Figure S6A).
A number of protein factors in the LSD2 complex are involved in the regulation of active gene transcription and Pol II elongation, and may therefore provide physical links for LSD2 to the regulation of the transcription machinery. Of particular interest, the ser-2 phosphorylated RNA polymerase II, the active form of elongating polymerase, is detected in LSD2 complex. We also identified co-transcriptional factors important for regulation of elongating Pol II activity and chromatin structure of the transcribing coding regions, including the NSD3 H3K36 methyltransferase (Li et al., 2009), euchromatin H3K9 methyltrasferase EHMT1/2, SWI/SNF-like nucleosome remodeling factor SNF2H and components of positive elongating factor b (P-TEFb), cyclin T1 and CDK9. Association of G9a/EHMT2 and SMCI, as well as NSD3 and cyclin T1 with LSD2 was validated by reciprocal co-immunoprecipitation (Figure 6B). Glycerol gradient ultracentrifugation analysis showed that TAP-purified LSD2 complex separates into multiple peaks (marked A–E in Figure S6B), suggesting the existence of more than one sub-complexes of LSD2. Importantly, G9a and NSD3 co-migrate with LSD2 in high density fractions (peak E in Figure S6B), indicating that all these proteins are components of the same complex.
To further characterize the LSD2 associated complex, we examined the enzymatic activities of the purified complex. As expected, LSD2 complex readily demethylates H3K4me2 in vitro using bulk histone as substrates (Figure 6C, panel d). Contrary to the LSD1 complex, no histone deacethylase activity was observed for LSD2 complex (Figure 6D, panel c). The absence of HDAC activity is consistent with MS/MS proteomic analysis and confirmed by immunoblot (Figure 6C, panel b).
The histone methyltransferase (HMTase) activity of the complex was assayed in vitro using recombinant GST-histone H3 proteins. LSD2 complex demonstrated robust HMTase activity on recombinant wild type GST-H3 (Figure 6D, lane 2). This signal is completely abolished when residue lysine 9 is replaced with arginine (H3 K9R, Figure 6D, lanes 4, 6, 7, and 9), while other mutations (K4R, K27R, lane 3 and 5 respectively) do not adversely affect HMTase activity, demonstrating that LSD2 complex indeed possesses H3K9 HMTase activity. Recent studies on the nuclear receptor SET domain-containing (NSD) family of methyltransferses suggested that a nucleosomal substrate (or octamers in conjunction with dsDNA) is required for their H3K36-specific HMTase activities (Li et al., 2009). Taking this into account, we modified the histone methylation assays by using reconstituted nucleosomes. Under these conditions, we again detected methylation activity specific to the LSD2 complex (Figure S6C, lanes 1 and 2). Importantly this signal, although reduced, persisted when MLA nucleosomes carrying H3Kc9me3, an analog of H3K9me3 nucleosomes (Simon et al., 2007), were used (Figure S6C, lane 3). We propose that this activity could be attributed to NSD3 acting on H3K36.
The physical association of G9a with LSD2 and the H3K9 HMTase activity of LSD2 complex are particularly interesting given the concomitant H3K9 methylation changes at LSD2 binding loci observed upon LSD2 depletion. We next asked if G9a might coordinate with LSD2 to regulate active gene transcription. As expected, we detected G9a binding at the coding region of a subset of LSD2 target genes by ChIP (Figure S6D). Importantly, upon G9a depletion, a large majority of LSD2 targets tested showed similar transcription changes as LSD2 knockdown, with most of the LSD2 target genes down regulated (Figure 6E).
Taken together, we conclude that LSD2, in association with elongating Pol II and elongation factors such as cyclin T1 (p-TEFb complex), and additional LSD2 complex-associated histone modifying activities, may be coordinated to set the proper histone modifications for the dynamic chromatin structures of the coding regions, thus facilitating Pol II-mediated transcription elongation (Figure 6F).
DISCUSSION
Histone methylation and the recently identified histone demethylases play important roles in gene transcription regulation. The first identified histone demethylase LSD1/AOF2/KDM1a was originally characterized as a transcriptional repressor, functioning in part by removing active H3K4me2 marks from promoter regions. Here we describe an important function of human LSD2/AOF1/KDM1b in active gene transcription. LSD2 shares similar substrate specificity with LSD1 and demethylates mono- and di-methylated H3K4. Instead of functioning as a co-repressor, LSD2 is important for optimal gene transcription. LSD2 is unique in associating specifically with the gene bodies of actively transcribed genes, but not at promoters. A specific function of LSD2 is to maintain low levels of H3K4 methylation within elongation regions. Furthermore, LSD2 forms complexes with H3K9 and H3K36 methyltransferases. These histone modification enzymes together orchestrate appropriate histone modifications in order to maintain a repressive chromatin structure at elongation regions, which may be important for optimal transcription elongation.
During the preparation of this manuscript, the enzymatic activity and functional studies of LSD2/AOF1 were reported by others (Ciccone et al., 2009; Karytinos et al., 2009; Yang et al., 2010). Work by Ciccone et al has shown that LSD2 knockout mice fail to establish a subset of maternal imprints. These results likely reflect the functional relationship between DNA methylation and H3K4 methylation, although the mechanism underlining how LSD2 directly regulates DNA methylation has yet been discovered. It is noted that this finding is confined to a particular stage of oocyte maturation and a small subset of imprinted genes, while global DNA methylation is unchanged. To explore the function of LSD2 outside of oocytes, we examined more than 90 genes in HeLa cells after LSD2 depletion (including LSD2 targets and a collection of cancer-related genes) and found no significant alterations in DNA methylation on the CpG islands of corresponding promoters (data not shown). These findings indicate multiple functions of LSD2 at different stages of development.
What is the function of LSD2 in somatic cells, given that LSD2 expression persists post-development? A study by Yang et al suggests a functional role of LSD2 in direct regulation of gene transcription, mainly towards transcriptional repression, albeit independent of its own H3K4 demethylase or HDAC activity (Yang et al., 2010). However, it is worth noting that this study failed to demonstrate dosage-dependent effects on reporter gene activity, or identify any endogenous targets of LSD2. On the other hand, we observed a predominant down-regulation of transcription upon depletion of endogenous LSD2, consistent with a positive role for LSD2 in transcription.
The positive role of LSD2 in transcription regulation may at first seem to be at odds with its innate enzymatic activity, which removes H3K4me2/1 marks of active genes. Nonetheless, the incorporation of canonical `repressive' marks into regions of active transcription does represent a newly appreciated and intriguing development. In yeast, Eaf3 of Rdp3 complex binds to tri-methylated H3K36 and recruits HDAC activity to the loci (Carrozza et al., 2005; Joshi and Struhl, 2005). It is proposed that this HDAC activity counteracts the spreading of acetylation from promoters and is required to maintain repressive state of chromatin after elongating Pol II. It is tempting to speculate that an LSD2-H3K4me2 relationship within gene bodies at least in part resonates with the aforementioned role of HDAC-acetylation on active transcription. Interestingly, we did not detect any HDAC activity from the LSD2 complex; nor did we detect any changes in intragenic acetyl-H3 levels upon LSD2 depletion. It is therefore probable that LSD2 function in active gene transcription in mammals is independent of HDAC activity. Instead, it may provide a complementary epigenetic mechanism for maintaining co-transcriptional `repressive' chromatin state.
In addition to H3K4 demethylase activity, LSD2 also contributes to maintaining an optimal `repressive' environment of actively transcribed regions through its interaction with H3K9 methyltransferase G9a. We show that G9a forms stable complexes with LSD2 and regulates H3K9 methylation at LSD2 binding sites within coding regions, and is required for active gene transcription. Consistent with our findings, it has also been reported that G9a binds within actively transcribed genes and can function as a coactivator of nuclear receptors (Lee et al., 2006a), in addition to its well known role as a repressor (Roopra et al., 2004). It is plausible that in mammals, beside HDACs, LSD2 and G9a provide additional layers of control of repressive chromatin structure of elongating chromatin that could be important for efficient and faithful transcription.
Additional factors present in the LSD2 complex may also shed light on the mechanism by which LSD2 positively regulates transcription. We observed and validated the presence of pTEFb in purified LSD2 complex, as well as Ser-2 phosphorylated RNA polymerase. The functional link between elongating Pol II and LSD2 is further supported by our observation that LSD2 depletion impedes the full induction of immediately early responsive genes, such as EGR-1, which are known to be regulated post-initiation, at the level of elongation (Wang et al., 2005).
We note that not all of the LSD2 associated genes identified by ChIP-chip were dramatically affected by LSD2 depletion, despite robust increases of H3K4me2 levels of at the LSD2 binding regions. We believe that this finding is not unexpected. In fact, inactivation of other transcriptional elongation-associated histone modifying enzymes, such as the deacetylase Rpd3 or H3K36 methyltransferase SETD2, also cause only moderate transcriptional changes in a subset of its associated genes in both yeast and C. elegans (Edmunds et al., 2008; Sharma et al., 2007; Wang et al., 2002). This could be explained in part by the understanding that transcription elongation is regulated at multiple layers, by post-translational modification of the CTD of the Pol II controlled by both positive (p-TEFb) or negative (NELF) elongation factors, and finally chromatin remodeling and modifying enzymes (Core and Lis, 2008; Sims et al., 2004). Each layer of regulatory factors likely differentially and perhaps cooperatively contributes to the transcriptional outcomes of specific genes, providing flexibility and fidelity to ensure proper programs of gene expression essential for cellular and biological processes.
It is of great interest to find several PWWP-domain containing proteins in association with LSD2 complex, including the H3K36 methyltransferase NSD3; MSH6, a component of DNA mismatch repair machinery; and an uncharacterized protein NPAC. The PWWP domain is a loosely conserved protein module found in eukaryotic nuclear proteins, which are often associated with chromatin (Maurer-Stroh et al., 2003). Recently, specific recognition of histone methylation marks by PWWP domains has been reported (Vezzoli et al., 2010; Wang et al., 2009). NSD3 is a major component of the LSD2 complex, and demonstrates a strong biochemical intereaction (Figure 6B and Figure S6B). We detected NSD3 binding to a small subset of LSD2 targets by ChIP (Figure S6E) and observed a moderate effect of NSD3 depletion on the expression of LSD2 target genes (Figure S6F). It is possible that NSD3 and LSD2 form complexes in vivo and coordinate the dynamics of H3K4 and H3K36 methylation for transcription elongation of certain genes; however further investigation is required to fully define the nature of this interaction. We are tempted to speculate that PWWP domain-containing factors in LSD2 complex may function in LSD2 targeting through binding to specific histone methylation marks. Future investigation into these possibilities is warranted to further our understanding of the role PWWP- containing proteins in the recruitment of LSD2 and their potential in regulating LSD2 functions.
Human LSD2 is a candidate cancer-related gene located at chromosome 6p22, a genomic region with high incidence of chromosomal translocations, deletions or amplifications in multiple cancer types (Heidenblad et al., 2008; Orlic et al., 2006). Furthermore, data from the ONCOMINE cancer database shows that human LSD2 levels are significantly lower in some leukemias, seminomas, and a few classes of ER-negative breast cancers. These correlations suggest that the biological function of LSD2 and its exact role in human cancer are likely tissue or cell type dependent. The function of LSD2 in transcription regulation may provide insight to the epigenetic mechanism involving gene regulation in tumorigenesis, therefore offering a molecular basis for future development of demethylase inhibitor-based anti-cancer drugs.
EXPERIMENTAL PROCEDURES
Plasmids
Full length LSD2/KDM1b/AOF1 was amplified from HeLa cDNA, cloned into XhoI and NotI sites of pOZ-N, and expressed as N-terminal Flag-HA double-tagged fusion proteins. LSD2 was subcloned into pBlueBac (Invitrogen) for protein expression and purification from Sf9 insect cells as described (Tahiliani et al., 2007).
Histone Demethylase Assays
In vitro and in vivo histone demethylase assays were performed as described previously (Tahiliani et al., 2007).
HMTase assays
HMTase activity assays were performed as described (Shi et al., 2003). In brief, samples were incubated for 60 min at 30°C in total of 40μl of methylation assay buffer (50mM Tris-HCl, pH 8.5, 20mM KCl, 10mM MgCl2, 10% glycerol, 10mM 2-mercaptoethanol, 1mM PMSF, 8pmol S-adenosyl-[methyl-3H]methionine). Samples were resolved on SDS-PAGE and analyzed by autoradiography or scintillation counting.
ChIP and tiling array analysis
Native chromatin of HeLa-S stably expressing FgHA-LSD2 was prepared by micrococcal nuclease digestion as previously described (Umlauf et al., 2004), and was primarily composed of mono-, di-, and tri-nucleosomes. Native chromatin fragments were sequential immunoprecipited using anti-flag and anti-HA agarose beads (Sigma). After extensive washing, Flag or HA peptides (Sigma) were used for elution after each immunoprecipitation. DNA from the final step was purified and processed for tiling array hybridization to Human Genome tiling 2.0 Array (Affymetrix) per manufacture's instructions. Input DNA was used as control. Regions enriched by TAP N-ChIP were identified using algorithm Model-Based Analysis of Tiling arrays (MAT) (Johnson et al., 2006), and displayed by Integrated Genome Browser (IGB). CEAS (cis-element annotation system) was used for statistic analysis of the distribution pattern of LSD2 binding sites on promoter and non-promoter regions (Ji et al., 2006). To estimate the statistical significance of colocalization between LSD2 and histone modification marks previously identified (Barski et al., 2007), we estimated the null distribution of overlap by generating 100 randomly permutated sample of the histone modification loci and the overlap is calculated for each permutated sample.
Conventional X-ChIP was performed as previously described (Shi et al., 2003) using formaldehyde-crosslinked chromatin. Anti-H3K4me2 (Upstate, 07-030), anti-H3K9me2 (Upstate, 05-768), anti-H3K36me3 (Abcam, ab9050), anti-G9a (Abcam, ab40542) and anti-NSD3 (GeneTex) antibodies were used for ChIP. Enrichment of each target was determined by qPCR. Sequences of primers are available upon request.
RNAi and microarray gene expression profile analysis
Retroviral shRNA targeting human LSD2 (5'-GTGGGACCACAATGAATTCTT -3') and control shRNA was used to infect HeLa. RNA was purified 70 hours post infection and processed for Human Genome U133 Plus 2.0 Array (Affymetrix) hybridization per manufacture's instructions. Biological duplicates were analyzed. A common set of 461 differentially expressed genes with at least 1.5 fold changes were obtained by normalization with either Quantile or Invariant Set method using dChip. Microarray result was confirmed by quantitative RT-PCR. Sequences of qRT-PCR primers are available upon request. An independent LSD2 shRNA construct (5'-GCCGTGTTCTATGACATGGATCTCGAGATCCATGTCATAGAACACGGC-3') was used to confirm the specific effects of LSD2 depletion. G9a and NSD3 shRNAs were purchased from ThermoScientific.
Luciferase reporter assay
Luciferase reporter assays were performed 36 hours after transfection and normalized with the activity of cotransfected β-galatosidase as described (Shi et al., 2004).
Complex purification and proteomics analysis by mass spectrometry
LSD2 associated protein complexes were purified from HeLa-S stably expressing Flag-HA-LSD2 by sequential immunoprecipitation using anti-Flag and anti-HA Affinity Gel (Sigma A2220) as previously described (Shi et al., 2003). HeLa-S transformed with pOZ-N vector was used to purify mock complex using identical procedures. LSD2 complex was sequenced by MS/MS at the Harvard Medical School Taplin Biological Mass Spectrometry Facility.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants to Y.S. and NIH NRSA Postdoctoral Fellowships T32HL007609 to R.F and F32GM86021 to A.B. Y.S. is a PEW scholar. We thank Chun-Xiang Liu (SABioscience) for technical support with DNA methylation studies, and Matthew Simon for providing MLA nucleosomes. We also thank Stephen Sugrue and Stephen Buratowski for helpful discussion and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Roelofsen T, Pennings SW, Martens JH, Jenuwein T, Stunnenberg HG. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chosed R, Dent SY. A two-way street: LSD1 regulates chromatin boundary formation in S. pombe and Drosophila. Mol Cell. 2007;26:160–162. doi: 10.1016/j.molcel.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. Embo J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Heidenblad M, Lindgren D, Jonson T, Liedberg F, Veerla S, Chebil G, Gudjonsson S, Borg A, Mansson W, Hoglund M. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA, Shi Y. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006a;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006b;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain `Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009;8:3636–3642. doi: 10.4161/cc.8.22.9890. [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Sharma VM, Tomar RS, Dempsey AE, Reese JC. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol. 2007;27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP corepressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 Histone Demethylase Activity by Its Associated Factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, Zhang Y, Wong J. AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 2010;20:276–287. doi: 10.1038/cr.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.