Abstract
Reading disability (RD) is a complex genetic disorder with unknown etiology. Genes on chromosome 6p22, including DCDC2, KIAA0319, and TTRAP, have been identified as RD associated genes. Imaging studies have shown both functional and structural differences between brains of individuals with and without RD. There are limited association studies performed between RD genes, specifically genes on 6p22, and regional brain activation during reading tasks. Using fourteen variants in DCDC2, KIAA0319, and TTRAP and exhaustive reading measures, we first tested for association with reading performance in 82 parent-offspring families (326 individuals). Next, we determined the association of these variants with activation of sixteen brain regions of interest during four functional magnetic resonance imaging-reading tasks. We nominally replicated associations between reading performance and variants of DCDC2 and KIAA0319. Furthermore, we observed a number of associations with brain activation patterns during imaging-reading tasks with all three genes. The strongest association occurred between activation of the left anterior inferior parietal lobe and complex tandem repeat BV677278 in DCDC2 (uncorrected p=0.00003, q=0.0442). Our results show that activation patterns across regions of interest in the brain are influenced by variants in the DYX2 locus. The combination of genetic and functional imaging data show a link between genes and brain functioning during reading tasks in subjects with RD. This study highlights the many advantages of imaging data as an endophenotype for discerning genetic risk factors for RD and other communication disorders and underscores the importance of integrating neurocognitive, imaging, and genetic data in future investigations.
Keywords: dyslexia, DCDC2, TTRAP, imaging-genetics, neuroimaging
INTRODUCTION
Reading disability (RD) or dyslexia is the most commonly observed learning disability with a prevalence in Western countries estimated to be between 5% and 17% (Interagency Committee on Learning Disabilities 1987; Shaywitz et al. 1994; Pennington and Bishop 2009). RD is marked by difficulty reading despite adequate education, motivation, and intelligence (Shaywitz et al. 1994; Pennington and Bishop 2009). Twin and family studies showed that RD has both environmental and genetic determinants with heritability estimates of 44-75% (DeFries et al. 1987). Our group and others have identified various RD associated genes in the DYX2 locus on chromosome 6p22 including replicated associations of DCDC2, KIAA0319, and a haplotype including KIAA0319/TTRAP (Francks et al. 2004; Cope et al. 2005; Meng et al. 2005; Harold et al. 2006; Schumacher et al. 2006; Luciano et al. 2007; Ludwig et al. 2008; Paracchini et al. 2008; Wilcke et al. 2009; Lind et al. 2010; Scerri et al. 2011; Marino et al. 2012). How these genes function in neuronal circuitry and mechanistically contribute to RD remains unknown, especially since most of the associated variants have not yet been shown to be functional. However, two possible functional variants on DYX2 have been identified. Our group has identified a microdeletion/complex tandem repeat in intron 2 of DCDC2 (GenBank ID: BV677278) that associates with RD, influences DCDC2 gene expression, and binds a putative transcriptional regulatory complex (Meng et al. 2005; Wilcke et al. 2009; Meng et al. 2011; Marino et al. 2012). Paracchini et al. showed that the KIAA0319/TTRAP risk haplotype yielded decreased expression of KIAA0319 but not TTRAP (Paracchini et al. 2006). Additionally, animal studies demonstrated that DCDC2 and KIAA0319 are involved in neuronal migration (Paracchini et al. 2006; Velayos-Baeza et al. 2008; Levecque et al. 2009). The role of DCDC2 and KIAA0319 in neuronal migration suggests variants of these genes may cause abnormalities in brain morphology and functioning.
Functional magnetic resonance imaging (fMRI) studies show that subjects with RD display different patterns of brain activation during reading tasks compared to non-impaired controls (Rumsey et al. 1992; Paulesu et al. 1996; Rumsey et al. 1997; Shaywitz et al. 1998; Horwitz et al. 1998; Brunswick et al. 1999; Paulesu et al. 2001). Subjects with RD also have structural brain differences compared to non-impaired controls, both in grey matter density and in white matter microstructure connectivity (Eliez et al. 2000; Klingberg et al. 2000; Brown et al. 2001; Paulesu et al. 2001; Brambati et al. 2004; Silani et al. 2005; Vickenbosch et al. 2005; Deutsch et al. 2005; Beaulieu et al. 2005; Niogi and McCandliss 2006; Hoeft et al. 2007). These functional and structural differences are most prominent in the superior temporal gyrus, inferior frontal gyrus, parietal-temporal gyrus, and left inferior parietal lobule. In general, subjects with RD utilize different brain regions and neuronal circuits during reading tasks compared to non-impaired controls, suggesting determinants of RD alter brain development and functioning.
Despite the evidence of functional and structural differences between individuals with RD and non-impaired controls, the use of imaging data as an endophenotype to explore mechanisms of neurobehavioral disorders remains under-utilized. Specifically, investigations of the interactions among RD associated genes, behavioral measures, and imaging data are few in number and limited in functional imaging data. One study found a relationship between grey matter volume in subjects with RD and the BV677278 microdeletion (Meda et al. 2008). Recent studies by Wilcke et al. in subjects with RD and Pinel et al. in non-impaired individuals found differences among variants in FOXP2 in regional brain activation during reading tasks including temporo-parietal, inferior frontal, and precentral brain regions (Wilcke et al. 2011, Pinel et al. 2012). Pinel et al. also observed an association between KIAA0319/TTRAP/THEM2 and lower asymmetric activation of the posterior superior temporal sulcus during reading and phonology tasks. These studies suggest that functional brain patterning can be an informative endophenotype for conditioning genetic associations.
The goals of this investigation are to replicate associations of DCDC2, KIAA0319, and TTRAP with behavioral reading measures and to use fMRI to interrogate brain regions involved in reading tasks in the same subjects. This investigation is one of the first comprehensive analyses of behavioral, functional imaging, and genetic data in the same RD cohort. By using both behavioral and imaging-reading tasks, we aim to strengthen associations of DYX2 variants with RD and to evaluate functional consequences of RD associated variants using fMRI data. We hypothesize that associations of DYX2 variants with RD using behavioral data will be replicated and that these variants will influence brain activation patterns during imaging-reading tasks in reading-related regions of interest (ROIs).
MATERIALS AND METHODS
Subjects
Eighty-two unrelated subjects of European American ancestry (50 RD cases / 25 non-impaired controls / 7 with unknown affectation status) from the Yale Center for the Study of Learning and Attention were used in this investigation (Supplementary Table S1). The 50 RD cases were identified by scores below the 25th percentile on either the Word Identification or Word Attack portions of the Woodcock Johnson III Achievement Battery or reading fluency on the Gray Oral Reading Test (GORT) (Woodcock 1987; Woodcock and Johnson 1989; Wiederholt and Bryant 1992). The 25 non-impaired controls were defined as subjects with scores above the 40th percentile on the same tasks. Among subjects, there were 51 males and 31 females, with a mean age of 8.8 years (range 7 – 12 years). Exclusion criteria for subjects included IQ < 85, left-handedness, hearing loss, severe articulation problems, severe emotional disturbance, autism, mental retardation, brain injury, neurologic disorders, and speaking English as a second language. Reading measures were performed on 75 subjects, imaging-reading data were collected on 82 subjects (51 males/31 females), and DNA was collected by buccal swab from subjects and all available family members (n=326). All subjects gave informed consent approved by the Human Investigation Committee of the Yale University School of Medicine, and all studies were performed in accordance with the Declaration of Helsinki.
Behavioral Measures of IQ and Reading Performance
Verbal, performance and full scale IQ were determined by the Wechsler Abbreviated Scale of Intelligence (WASI) (Psychological Corporation 1999). A range of quantitative neurobehavioral tasks measured reading performance of subjects. The Woodcock Johnson III Achievement Battery determined basic reading, letter-word identification, word attack, passage comprehension, and spelling (Woodcock 1987; Woodcock and Johnson 1989). The GORT assessed reading rate, accuracy, fluency, and comprehension (Wiederholt and Bryant 1992). The Test of Word Reading Efficiency (TOWRE) measured sight word efficiency, phonemic decoding efficiency and total word efficiency (Torgesen et al. 1999). The Comprehensive Test of Phonological Processing (CTOPP) measured phonological awareness ability, blending words, memory for digits, rapid digit naming, blending of non-words, and segmenting non-words (Wagner et al. 1999). Behavioral measures are compiled in Supplementary Table S1, and correlations between tests are shown in Supplementary Table S2.
Imaging-Reading Tasks
Subjects completed four reading tasks while being imaged: Word Rhyming (WR)/Non-Word Rhyming (NWR) and Print Categorization (PC)/Auditory Categorization (AC) (Shaywitz et al. 1998; Pugh et al. 2008; Landi et al. 2010). In the WR/NWR tasks, subjects were asked to match a printed target consisting of a word (WR) or pseudoword (NWR) to a pictorially presented cue. A small set of cue pictures was used for both tasks consisting of pictures of either living or nonliving things. Subjects observed the cue picture, while a target stimulus was printed just below. Subjects were instructed to press one button if the cue and target stimulus rhymed, another if they did not. The WR/NWR tasks measured brain activation patterns involved in phonological processing. Phonological processing refers to the detection and separation of specific phonemes or speech sounds within words. Phonological processing is essential to reading and impaired in individuals with RD (Shaywitz et al. 1999; Pugh et al. 2001).
In the PC/AC tasks, subjects were asked to match either a printed (PC) or spoken (AC) target to a pictorially presented cue of living and non-living things. Subjects were instructed to press one button if the two stimuli were in the same category and another if they were not. For instance, “pig” and a picture of a fox are in the same category because both are living. The PC/AC tasks measure the brain activation patterns involved in semantic processing. Semantic processing refers to how one connects written and/or spoken words to actual language meaning. Individuals with RD can have deficits in semantic processing, yielding inaccurate comprehension of written language (Pugh et al. 2001).
Imaging Protocol
Head positioning in the magnet was standardized using the canthomeatal landmarks. In the scanner, cushions inside the head coil reduced head movement, and headphones (RTC technologies) were used to dampen scanner noise, to communicate with participants, and to deliver audio components of the task. Conventional T1-weighted spin-echo sagittal anatomical images were acquired for slice localization using a 1.5T whole body imaging system with a quadrature head coil (Sonata; Siemens AG, Erlangen, Germany). After a 3-plane localizer and a multiple-slice sagittal localizer, fourteen T-1 weighted axial slices (TR=420 ms; TE=11 ms; bandwidth=130 Hz/pixel; FA= 90°; slice thickness=7mm; FOV=200 × 200 mm; matrix=256 × 256) were obtained using flash spin-echo imaging parallel to the anterior and posterior commissure. Eight functional data series (four runs alternating between both category/rhyming tasks and baseline within each run) were acquired with a single-shot gradient-echo echo planar imaging (EPI) sequence (TR=1500 ms; TE=60 ms; bandwidth=1735 Hz/pixel; FA=60°; slice thickness=7mm; FOV=200 × 200 mm; matrix=64 × 64; images per slice=91). Stimuli were projected onto a semi-transparent screen at the head of the bore and viewed by the subject via a mirror mounted on the head coil. At the end of functional imaging, a high resolution 3D Spoiled Gradient Recalled Acquisition in the Steady State (SPGR) T1-weighted sequence (TR=24 ms; TE=4.73 ms; bandwidth=130 Hz/pixel; FA= 45°; slice thickness=1.5mm; FOV=240 × 240 mm; matrix=256 × 256) was used to acquire sagittal images for multi-subject registration.
Imaging Data Analysis
Imaging data were converted from Digital Imaging and Communication in Medicine (DICOM) format to analyze format using XMedCon (Nolf et al. 2003). The first six images at the beginning of each functional series were discarded to enable the signal to achieve steady-state equilibrium between radio frequency pulsing and relaxation, leaving 85 images per slice per trial for analysis. Motion-correction of images was performed with Statistical Parametric Mapping (SPM) (www.fil.ion.ucl.ac.uk/spm) for three translational directions (x, y or z) and three possible rotations (pitch, yaw or roll). Trials with linear motion that had a displacement in excess of 2 mm or rotation in excess of 3° were rejected. Individual subject data was analyzed using a General Linear Model (GLM) on each voxel in the entire brain volume with regressors specific for each task. The resulting functional images were spatially smoothed with a 6 mm Gaussian kernel to account for variations in the location of activation across subjects. Output maps were normalized beta-maps, which were in the acquired space (3.125mm × 3.125mm × 7mm).
Image Registrations
To take these data into a common reference space, three registrations were calculated within the Yale BioImage Suite software package (v2.6.1) (http://www.bioimagesuite.org/) (Evans et al. 1993; Nolf et al. 2003; Duncan et al. 2004). The first registration performs a linear registration between the individual subject raw functional image and that subject’s 2D anatomical image. The 2D anatomical image is then linearly registered to the individual’s 3D anatomical image. The 3D differs from the 2D in that it has a 1×1×1 mm resolution whereas the 2D z-dimension is set by slice-thickness and its x-y dimensions are set by voxel size. Finally, a non-linear registration is computed between the individual 3D anatomical image and a reference 3D image. The reference brain used was a single control child’s high resolution anatomical that was manually stripped to remove all skull and meninges. All three registrations were applied sequentially to the individual normalized beta-maps to bring all data into the common reference space. The 3D anatomical reference brain was non-linearly registered to the Colin27 Brain which is in Montreal Neurological Institute (MNI) space (Evans et al. 1993; Holmes et al. 1998). Talairach coordinates are based on a conversion from MNI space to Talairach coordinates (Talairach and Tournoux 1988; Lacadie et al. 2008).
Regions of Interest
Based on previous reports of altered activation in phonological processing tasks using fMRI, 16 ROIs were chosen for examination with genotype data (Table 1) (Shaywitz et al. 1998). ROIs were defined functionally in a separate sample of 25 control subjects by using activated language areas in the NWR and AC tasks (Shaywitz et al. 1998). Activation values were used as quantitative measures for testing genetic association.
Table 1.
Regions of Interest (ROIs), abbreviations, volumes, and Talairach coordinates
| Region of Interest | ROI Abbreviation | Volume (mm3) | Volume (voxels) | Talairach Coordinates | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Right inferior frontal gyrus, inferior aspect | RIFGI | 1899.51 | 112.66 | 32 | 20 | 0 |
| Left inferior frontal gyrus, inferior aspect | LIFGI | 6203.05 | 367.92 | -42 | 16 | 1 |
| Right temporal lobe | RT | 2834.85 | 168.14 | 49 | -27 | 2 |
| Left temporal lobe | LT | 4832.07 | 286.60 | -52 | -35 | 5 |
| Superior anterior cingulate gyrus | SAC | 2408.83 | 142.87 | -4 | 22 | 41 |
| Left paracentral lobule | LPC | 2580.20 | 153.03 | 0 | -25 | 47 |
| Posterior cingulate gyrus | PC | 10794.90 | 640.27 | 0 | -46 | 32 |
| Inferior anterior cingulate gyrus | IAC | 5431.07 | 322.12 | -1 | 42 | -1 |
| Right posterior inferior parietal lobe (angular gyrus) | RPIPL | 2160.58 | 128.15 | 26 | -64 | 34 |
| Left posterior inferior parietal lobe (angular gyrus) | LPIPL | 1950.76 | 115.70 | -34 | -61 | 37 |
| Right anterior inferior parietal lobe (supramarginal gyrus) | RAIPL | 4215.44 | 250.02 | 52 | -28 | 33 |
| Left anterior inferior parietal lobe (supramarginal gyrus) | LAIPL | 2396.01 | 142.11 | -48 | -28 | 39 |
| Right lateral occipital temporal gyrus | RLOTG | 1370.98 | 81.32 | 51 | -60 | 2 |
| Left medial occipital temporal gyrus | LMOTG | 1905.92 | 113.04 | -39 | -51 | -7 |
| Right inferior frontal gyrus, superior aspect | RIFGS | 3220.84 | 191.03 | 38 | 19 | 20 |
| Left inferior frontal gyrus, superior aspect | LIFGS | 4782.42 | 283.65 | -45 | 17 | 23 |
Genotyping
Fourteen variants spanning DCDC2, KIAA0319, and TTRAP were chosen based on previous association studies and are summarized in Supplementary Table S3 (Deffenbacher et al. 2004; Francks et al. 2004; Cope et al. 2005; Meng et al. 2005; Schumacher et al. 2006;). Genotyping methods by TaqMan Assay-on-Demand (Applied Biosystems, Foster City, CA), TaqMan Assay-by-Design, Pyrosequencing (Biotage, Uppsala), fluorescent dideoxynucleotide DNA sequencing with sequence trace analysis by Mutation Surveyor v2.6 (SoftGenetics, State College, PA), and allele specific PCR for the BV677278 microdeletion/complex tandem repeat (rsdel and rsSTR) are summarized in Supplementary Tables S3-S4 (Meng et al. 2005; Meng et al. 2011). Table 2 presents the multiple alleles (1-10) of the BV677278 complex tandem repeat alleles observed in this sample.
Table 2.
BV677278 complex tandem repeat in DCDC2
| Allele | Repeat Unit 1 | Repeat unit 2 | SNP1 | Repeat Unit 3 | Repeat Unit 4 | Repeat Unit 5 | Frequency in Sample |
|---|---|---|---|---|---|---|---|
| 1 | (GAGAGGAAGGAAA)2 | (GGAA)7 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.650 | |
| 2 | (GAGAGGAAGGAAA)1 | (GGAA)9 | DelGAA A | (GGAA)0 | (GGAA)4 | (GGGA)2 | 0.027 |
| 3 | (GAGAGGAAGGAAA)1 | (GGAA)6 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.012 | |
| 4 | (GAGAGGAAGGAAA)2 | (GGAA)6 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.114 | |
| 5 | (GAGAGGAAGGAAA)2 | (GGAA)8 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.034 | |
| 6 | (GAGAGGAAGGAAA)2 | (GGAA)8 | (GGAA)2 | (GGAA)3 | (GGGA)2 | 0.024 | |
| 8 | (GAGAGGAAGGAAA)2 | (GGAA)7 | DelGAA A | (GGAA)0 | (GGAA)4 | (GGGA)2 | 0.005 |
| 9 | (GAGAGGAAGGAAA)1 | (GGAA)7 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.005 | |
| 10 | (GAGAGGAAGGAAA)2 | (GGAA)4 | (GGAA)2 | (GGAA)4 | (GGGA)2 | 0.051 | |
| deletion | X | X | X | X | X | X | 0.078 |
Statistical Analysis
Statistical Package for the Social Sciences (SPSS) (v18) was used to determine mean activation, standard error, correlations, and principle component analysis (PCA). PCA was performed on 11 highly correlated behavioral measures including subtests of the Woodcock Johnson (basic reading, letter-word identification, word attack, passage comprehension, spelling), GORT (measured reading test, accuracy, fluency), and TOWRE (sight word efficiency, phonemic decoding efficiency, total word efficiency). Mendelian transmission of alleles was assessed using the Genetic Analysis System (GAS; http://users.ox.ac.uk/~ayoung/gas.html). Identity-by-Descent probabilities were estimated using SIMWALK2 (v2.91) (Sobel et al. 2001). Quantitative transmission disequilibrium was assessed with QTDT (v2.6.1) using a total association, variance components model (-at -wega), with behavioral reading measures and brain activation data as quantitative measures. QTDT examines transmission of alleles from heterozygous parent to child in order to determine genetic associations with quantitative traits. QTDT correlates performance on a quantitative trait (in this case, brain activation in ROIs) with the observed number of an allele transmitted to offspring on quantitative measures from parents compared to the number expected following Mendelian transmission. The accumulation of an allele in subjects with similar levels of brain activation during tasks indicates association between the allele and trait analyzed. Haploview (v4.1) was used to determine linkage disequilibrium between markers. False discovery rate (FDR) was used to correct for multiple testing and to establish a threshold for statistical significance (Benjamini and Hochberg 1995).
RESULTS
Linkage Disequilibrium
Linkage disequilibrium (LD) among markers is shown in Figure 1. Two LD blocks were identified: one within DCDC2 and the second within KIAA0319. There was little evidence of LD among the three genes as the TTRAP variant rs2143340 did not fall within either block and there was no significant LD between DCDC2 and KIAA0319. Associations with multiple markers from the same gene with activation of a ROI reflected local LD. All markers were in Hardy-Weinberg equilibrium and followed Mendelian inheritance.
Figure 1.
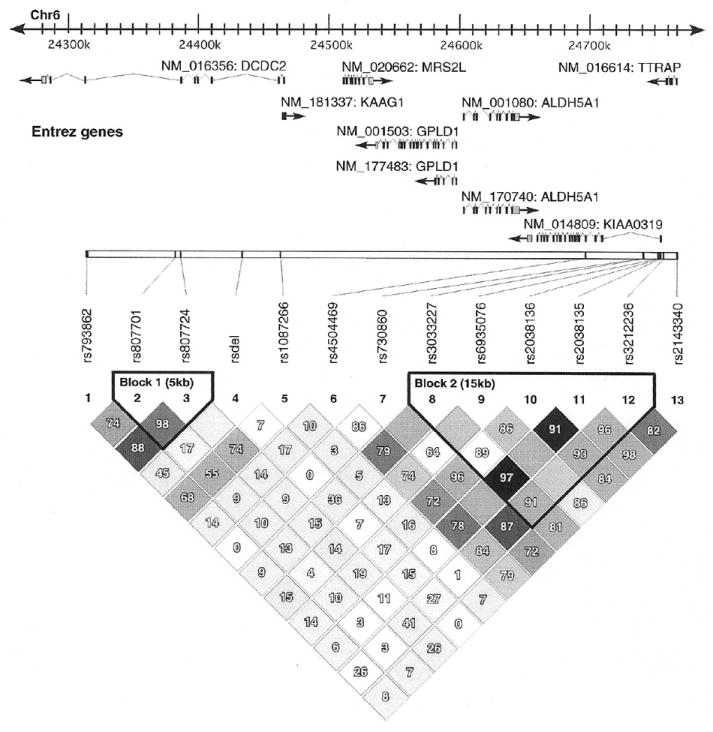
Linkage disequilibrium (LD) structure across DCDC2, KIAA0319, and TTRAP in the sample genotyped. The top portion of the figure depicts the chromosomal region spanning DCDC2 and KIAA0319 with known genes and transcripts. The bottom is a plot of LD calculated as D’ for all possible pairs of markers. rsdel refers to the BV677278 microdeletion. Darker shading indicates increased LD. Two haplotype blocks, block 1 (DCDC2) and block 2 (KIAA0319) were identified. There was no evidence for significant LD between DCDC2, KIAA0319, and TTRAP.
RD Genes and Behavioral Reading Phenotypes
Correlations between most reading measures were highly significant with p ≤ 0.01 (Supplementary Table S2). Due to the consistent high correlations among 11 of the behavioral measures (Woodcock Johnson, GORT, and TOWRE), we performed PCA to reduce them to a single vector that showed strong correlations with the other behavioral measures and accounted for 88.3% of the phenotypic variance (Supplementary Table S2). Nominal associations were observed between DCDC2 markers and reading measures, supporting previous observations that DCDC2 variants contribute to reading performance and RD (Table 3). One KIAA0319 variant (rs6935076) showed nominal association with the blending word subtest of the CTOPP. However, the majority of the associations were observed between reading measures and DCDC2. TTRAP (rs2143340) did not show any associations with reading measures. Although we observed nominal associations between DCDC2 and KIAA0319 variants and reading measures, none of these associations remained significant following FDR correction for multiple testing.
Table 3.
Nominally significant associations with cognitive and behavioral reading and cognitive measures
| Task | Variant | Gene | Direction of Effecta | P-valuec |
|---|---|---|---|---|
| WASIVIQ | rsSTR (allele 5) | DCDC2 | 18.491 | 0.019 |
| WASIVIQ | rsSTR (allele 6) | DCDC2 | -21.092 | 0.0194 |
| WASIPIQ | rs807724 | DCDC2 | 6.264 | 0.0359 |
| WJBAR | rsSTR (allele 1) | DCDC2 | 6.4 | 0.0291 |
| WJLW | rsSTR (allele 1) | DCDC2 | 7.63 | 0.0197 |
| WJPC | rsSTR (allele 1) | DCDC2 | 6.981 | 0.0289 |
| WJPC | rsSTR (allele 10) | DCDC2 | -10.69 | 0.0328 |
| WJSP | rsSTR (allele 1) | DCDC2 | 9.872 | 0.0099 |
| GORTAC | rs807701 | DCDC2 | 1.417 | 0.035 |
| GORTCP | rs807701 | DCDC2 | 1.195 | 0.0411 |
| GORTCP | rs807724 | DCDC2 | 1.642 | 0.0221 |
| GORTCP | rsSTR (allele 1) | DCDC2 | 1.985 | 0.0028 |
| CTOPPBW | rs6935076 | KIAA0319 | -0.881 | 0.049 |
| CTOPPMD | rsSTR (allele 3) | DCDC2 | 3.399 | 0.0308 |
| CTOPPRD | rsdelb | DCDC2 | 1.629 | 0.0263 |
| CTOPPRD | rsSTR (allele10) | DCDC2 | -2.021 | 0.011 |
For SNPs, the direction of effect refers to allele 1 (see Supplementary Table S3), allele 2 has the opposite effect.
Biallelic association (presence/absence of the BV677278 deletion)
No p-value remained significant following correction for multiple testing (437 tests performed).
RD Genes and Region of Interest Activation
Before assessing genetic associations with our ROIs, we assessed the correlations between brain activation patterns and behavioral reading measures (Supplementary Tables S5a–S5d). As shown in previous imaging studies in RD, there were significant correlations between brain activation patterns during reading tasks and performance on reading measures. These correlations suggest that activation of ROIs was related to reading processes in the brain and that genetic associations with these ROIs would be informative as to the etiology of RD.
There were nominal associations between DCDC2 and TTRAP and reading-related ROIs during WR and NWR tasks (Figure 2, Table 4). As with behavioral measures, most associations were observed with DCDC2 and not with KIAA0319 or TTRAP. The most significant associations were seen with alleles of the DCDC2 BV677278 complex tandem repeat and the superior anterior cingulate gyrus (SAC), posterior cingulate gyrus (PC), left paracentral lobule (LPC), and left inferior frontal gyrus, inferior aspect (LIFGI) along with rs2143340 in TTRAP with the right and left anterior inferior parietal lobe (RAIPL, LAIPL). However, none of the associations seen with brain activation during WR and NWR tasks remained significant after correction for multiple testing by FDR.
Figure 2.
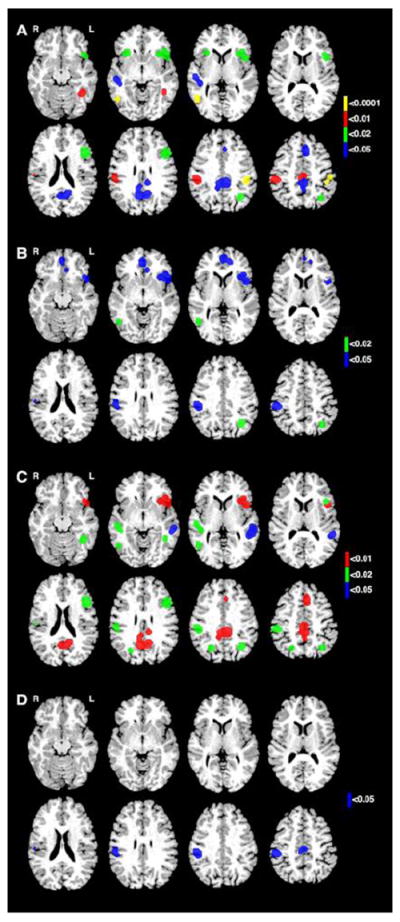
Significant P-values for association projected onto brain slices. A) DCDC2 from AC and PC tasks. P-values for DCDC2 (Tables 3a-3b) mapped onto LAIPL and RLOTG (P<0.0001, yellow); LPC, RAIPL, LMOTG (P<0.01, red); LIFGI, RIFGI, LIPL, LIFGS (P<0.02, green); PC, RT, SAC (P<0.05, blue). B) KIAA0319 from AC and PC tasks. P-values for KIAA0319 (Tables 3a-3b) mapped onto LMOTG and LPIPL (P<0.02, green); RAIPL, LIFGI, IAC (P<0.05, blue). C) DCDC2 from WR and NWR tasks. P-values for DCDC2 (Tables 3c and 3d) mapped onto PC, LIFGI, LPC, SAC (P<0.01, red); RLOTG, LAIPL, LIFGS, LPIPL, RPIPL, LMOTG, RT (P<0.02, green); LT (P<0.05, blue). D) KIAA0319 and TTRAP from WR and NWR tasks. P-values for KIAA0319 and TTRAP (Table 3c and 3d) mapped onto LAIPL and RAIPL (P<0.01, red); LT and LPC (P<0.05, blue).
Table 4.
Genetic associations (p<0.01) with auditory categorization (AC), print categorization (PC), word rhyming (WR), and nonword rhyming (NWR) imaging-reading tasks.
| Task | ROI | Variant | Gene | Direction of Effecta | p-value | q-valued |
|---|---|---|---|---|---|---|
| AC | RLOTG | rsSTR (allele 8) | DCDC2 | -0.319 | 0.0001 | 0.0490 |
| AC | LMOTG | rs1087266 | DCDC2 | 0.033 | 0.0080 | 0.8337 |
| PC | LPC | rsSTR (allele 1) | DCDC2 | -0.026 | 0.0032 | 0.7824 |
| PC | LAIPL | rs793862 | DCDC2 | 0.027 | 0.0065 | 0.9509 |
| PC | LAIPL | rsSTR (allele 4) | DCDC2 | 0.08 | 0.00003 | 0.0442 |
| PC | RLOTG | rsSTR (allele 8) | DCDC2 | -0.263 | 0.00008 | 0.0588 |
| PC | LMOTG | rs793862 | DCDC2 | 0.023 | 0.0074 | 0.9835 |
| PC | LMOTG | rsSTR (allele 4) | DCDC2 | 0.046 | 0.0076 | 0.9253 |
| WR | SAC | rsSTR (allele 5) | DCDC2 | 0.096 | 0.0083 | 0.8068 |
| WR | RAIPL | rs2143340 | TTRAP | -0.029 | 0.0038 | 0.7425 |
| WR | LAIPL | rs2143340 | TTRAP | -0.024 | 0.0029 | 0.8514 |
| NWR | LIFGI | rs1087266 | DCDC2 | 0.02 | 0.0038 | 0.7425 |
| NWR | LPC | rsSTR (deletion)b | DCDC2 | 0.042 | 0.0050 | 0.8133 |
| NWR | LPC | rsdelc | DCDC2 | 0.039 | 0.0079 | 0.8872 |
| NWR | PC | rsSTR (allele 8) | DCDC2 | -0.02 | 0.0016 | 0.5876 |
For SNPs, the direction of effect refers to allele 1 (see Supplementary Table S3), allele 2 has the opposite effect.
multiallelic association (all STR alleles)
biallelic association (presence/absence of the BV677278 deletion)
q-values were calculated by the Benjamini and Hochberg method of False Discovery Rate (FDR) correction for multiple testing with 1472 hypotheses of imaging-genetic associations performed. Those in bold reached or trended toward statistical significance after FDR correction for multiple testing.
Associations were observed between DCDC2 and brain activation in ROIs during AC and PC tasks (Figure 2, Table 4). No nominal associations were seen for TTRAP. The strongest associations occurred between ROIs and DCDC2, specifically with alleles of the BV677278 complex tandem repeat. During the PC task, alleles 4 and 8 of the DCDC2 complex tandem repeat were associated with activation with the LAIPL (uncorrected p=0.00003, q=0.0442) and right lateral occipital temporal gyrus (RLOTG) (uncorrected p=0.00008, q=0.0588), respectively. During the AC task, allele 8 of the complex tandem repeat displayed association with activation of the RLOTG (uncorrected p=0.0001, q=0.0490).
DISCUSSION
In this investigation, we nominally replicated the association of DCDC2 and KIAA0319 with behavioral reading measures and associated markers in the DYX2 locus to brain activation patterns during imaging-reading tasks. Instead of only using diagnostic status and behavioral measures, genetic associations were further conditioned on brain regions activated during reading tasks. These associations not only implicate genes in RD but give functional insight into their effects on brain function. More importantly, our work demonstrates the utility of imaging-genetics in the study of RD and other neurobehavioral disorders and shows the need for future studies to analyze behavioral, imaging, and genetics data while examining these disorders.
Association of DYX2 to RD
DYX2, containing DCDC2, KIAA0319, and TTRAP, is the most replicated RD risk locus (Deffenbacher et al. 2004; Francks et al. 2004; Cope et al. 2005; Meng et al. 2005; Harold et al. 2006; Luciano et al. 2007; Schumacher et al. 2006; Paracchini et al. 2006; Ludwig et al. 2008; Paracchini et al. 2008; Wilcke et al. 2009; Lind et al. 2010; Scerri et al. 2011). In the current investigation, we observed nominal association primarily between DCDC2 and neurocognitive reading measures. There was association between one reading measure (CTOPP-blending words) and KIAA0319 (rs6935076). The lack of substantial associations between reading performance and TTRAP and other variants in KIAA0319 may reflect a lack of statistical power due to sparse SNP coverage of the gene and relative small sample size. Regardless, this study further strengthens the implication of DCDC2 and KIAA0319 in RD and reading performance.
Association of DYX2 to Reading-Related ROIs
Genetic associations were observed between variants in DYX2 and brain activation levels in reading-related ROIs during imaging-reading tasks. In this study, the strongest genetic associations were between the BV677278 complex tandem repeat in DCDC2 and LAIPL and RLOTG (uncorrected p≤0.0001, q≤0.0588). The LAIPL and RLOTG regions of the brain have been implicated in dyslexia in past investigations and are a part of the temporo-parietal and occipito-temporal reading circuits, respectively. (Richlan et al 2009). The LAIPL contains Weirnicke’s area, which is vital for the accurate integration of language stimuli. A recent meta-analysis of functional imaging studies revealed that non-impaired readers have increased activation of the left inferior parietal lobe compared to subjects with dyslexia (Richlan et al. 2009). Our investigation found that having allele 4 of BV677278 within DCDC2 positively influences activation of the LAIPL during imaging-reading tasks. This suggests that allele 4 may improve the integration and activation of reading circuits and have a protective effect against dyslexia. Studies have shown a negative correlation between right occipito-temporal activation and reading ability (Shaywitz et al. 2002, Shaywitz and Shaywitz 2005). Our analyses showed that both allele 4 and 8 of BV677278 were associated with decreased activation of RLOTG, perhaps because they preserve function in temporo-parietal and occipito-temporal circuits and are therefore protective. The BV677278 repeat displays strong associations with RD in multiple studies (Meng et al. 2005; Wilcke et al. 2009; Meng et al. 2011; Marino et al. 2012). Meng et al. investigated possible regulatory functions by showing that different alleles altered DCDC2 gene expression (Meng et al. 2005; Meng et al. 2011). Electrophoretic mobility shift assays showed that BV677278 is a specific binding site for as-yet unidentified transcription regulatory complex(es) found in human brain nuclear lysate (Meng et al. 2011). Considered together, these data suggest that differential, allele-dependent expression of DCDC2 due to BV677278 variants during brain development may influence brain activation patterns in reading-related ROIs.
Pinel et al. recently reported an association between a locus containing KIAA0319, TTRAP, and THEM2 and lower asymmetry of brain activation during imaging-reading tasks in the posterior superior temporal sulcus (Pinel et al. 2012). This locus had previously been identified as part of a risk haplotype for RD in KIAA0319 and TTRAP and includes the rs2143340 variant in TTRAP examined in our study (Francks et al. 2004, Cope et al. 2005, Luciano et al. 2007, Paracchini et al. 2008). In this investigation, we observed an association between rs2143340 and activation in the RAIPL and LAIPL (supramarginal gyri) during the WR task. Although our association did not meet significance following correction for multiple testing, we did observe a nominal association in a brain region anatomically proximal to the posterior superior temporal sulcus. These brain regions have been implicated in reading and language processing and deficits in these regions are associated with RD (Paulesu et al. 2001). Our results, in combination with those of Pinel et al., suggest that TTRAP plays an undefined role in brain functioning in the supramarginal gyri and the posterior superior temporal sulcus that contributes to deficits in reading ability.
Our study was able to detect multiple genetic associations between DYX2 variants and fMRI data during reading tasks. While fMRI studies are state-dependent and influenced by many factors including effort, attention, and motivation, our methodologies were standardized and performed in order to minimize inter-subject variability. Additionally, performance on imaging-reading stimuli and the neurological process being interrogated (e.g. phonology and semantics) will influence the brain activation levels measured by fMRI (Pugh et al. 2008; Landi et al. 2010). The current study cannot distinguish whether activation levels reflect that of disorder state (i.e. RD or non-impaired) as opposed to deficits in performance on each task. However, the correlations between fMRI tasks (data not shown) and between fMRI tasks and behavioral assessments (Supplemental Tables S5a-S5d) support our findings of genotype associations with true physiologic differences in brain activity. These results, however, are limited by statistical power for genetic association due to sparse marker coverage of DYX2 and small sample size. Using data from this analysis, a sample size of 250 trios with severe RD would be needed for imaging-reading tasks to achieve 80% power for detecting associations (alpha = 0.05, total QTL variance = 0.01, cutoff = 2 SDs, additive genetic variance) (Purcell et al. 2003, http://pngu.mgh.harvard.edu/~purcell/gpc/qtdt.html). Despite these limitations, the observation of significant and suggestive genetic associations following correction for multiple testing demonstrates that imaging-genetics data is more sensitive than behavioral data, and the utility of imaging-genetics in further studies with larger sample size and increased coverage of RD associated genes.
Future of Imaging-Genetics
Our data suggest there are complex interactions between genes in DYX2, reading ability, and brain activation of reading-related ROIs. This is one of the first investigations to utilize both behavioral and functional imaging phenotypes of reading in genetic associations in the same sample. The examination of such a multidimensional dataset implies a functional role of these RD associated variants in brain activation. However, the current study cannot distinguish whether DYX2 variants directly influence the activation of reading-related ROIs or whether these variants affect an intermediate process which then alters ROI activation. For instance, variants of DCDC2 have been associated with brain morphological differences in schizophrenic and in control groups (Meda et al. 2008). It is plausible that the genetic associations with brain activation levels are actually a result of brain morphology differences that then alter ROI activation during reading tasks. DCDC2 and KIAA0319 function in neuronal migration during neurodevelopment, which may alter brain structure. Future studies are necessary to further discern the true molecular and gross neurological roles genes in the DYX2 locus play and to determine how these changes in gene and brain function contribute to RD.
Previous association studies conditioned on behavioral reading measures or a categorical RD diagnosis could not reliably distinguish single-gene effects. Our data preliminarily suggest that imaging-genetics is able to make this distinction. This conclusion is supported by the reported low false-positive rate of imaging-genetics studies, and that regardless of the relatively small sample size in this study, there were genetic associations that maintained and approached statistical significance following correction for multiple testing (Meyer-Lindenberg et al. 2008). Additionally, we employed a conservative correction for multiple testing for genetic associations with behavioral and imaging measures. Our corrections ignored the lack of independence among genetic markers due to LD and intra- and inter- correlations amongst behavioral and imaging measures (Figure 1, Supplemental Table S2). Some of the associations that were nominally significant but did not reach our stringent FDR threshold may reflect true positive associations. The lack of imaging-genetics studies reflect the inherent difficulties, technological capabilities, and costs of performing such studies and underscore the need for large, widely available consortia examining neurobehavioral disorders. If the present work can be replicated in a larger sample, our findings make an effective case for using functional imaging data to define informative endophenotypes for genetic studies of RD. The combination of functional imaging and genetic studies of RD could serve as a template for the investigation of other neurobehavioral disorders with unknown etiologies such as specific language impairment, attention deficit hyperactivity disorder, and speech sound disorder.
Conclusions
This study addresses the interaction between reading, regional brain activation, and genetics. We detected associations between a putative functional variant in DCDC2, previously associated with RD, and reading-related ROIs and nominally supported findings of the role of TTRAP in brain activation during imaging-reading tasks. Imaging data provides a viable, valuable endophenotype for RD and other neurobehavioral disorders due to the inherent complexity of brain functioning and the heterogeneous, intricate clinical presentations of these disorders. Further work is needed to validate our results in larger samples and with expanded networks of reading-related genes and ROIs. Additionally, future investigations of RD should examine the influence of various environmental determinants on brain activation of reading related regions.
Supplementary Material
Highlights.
Functional imaging data are an informative endophenotype for reading disability. A functional variant in DCDC2 associated with brain activation during reading tasks. We nominally replicated association of TTRAP with imaging endophenotypes. Future studies should examine cognitive, imaging, and genetic data in studying language deficits.
Acknowledgments
The authors would like to thank all subjects and families in the Yale Reading Disability Sample for their participation in this investigation. The authors also thank Professor Bennett Shaywitz, Professor Sally Shaywitz, Karen Marchione, and Dr. John Holahan for recruiting and testing all the subjects at the Yale Center for the Study of Learning and Attention. Support for JRG is from the National Institute of Health (NIH) R01 NS43530, and support for JDE is from NIH F31 DC012270 and T32 GM007223. Further support for RKF, and CL, is from R01 HD046171. Support for CJG is from a Yale-Rosenberg Genetics Fellowship. Support for RTC is from R01 NS038467 and NS051622. The authors wish to thank Dr. Xenophon Papademetris (XP) for image analysis guidance, Stefan Halvorsen for extracting the ROIs, and to acknowledge the NIH/National Institute of Biomedical Imaging and Bioengineering (NIBIB) under grant 1 R01 EB006494-01 for support of XP and CL.
Abbreviations
- RD
Reading disability
- fMRI
functional magnetic imaging resonance
- ROIs
regions of interest
- GORT
Gray Oral Reading Test
- WASI
Wechsler Abbreviated Scale of Intelligence
- TOWRE
Test of Word Reading Efficiency
- CTOPP
Comprehensive Test of Phonological Processing
- WR
Word Rhyming
- NWR
Non-Word Rhyming
- PC
Print Categorization
- AC
Auditory Categorization
- PCA
principle component analysis
- FDR
false discovery rate
- LD
linkage disequilibrium
- SAC
superior anterior cingulated gyrus
- PC
posterior cingulated gyrus
- LPC
left paracentral lobule
- LIFGI
left inferior frontal gyrus, inferior aspect
- RAIPL
right anterior inferior parietal lobe
- LAIPL
left anterior inferior parietal lobe
- RLOTG
right lateral occipital termporal gyrus
Footnotes
FINANCIAL DISCLOSURES
Yale University has applied for a patent covering the complex tandem repeat and deletion in BV677278 (inventors: JRG and HM), and sublicensed it to JS Genetics, Inc. JRG is a founder and equity holder of JS Genetics, Inc. KH is currently employed by JS Genetics, Inc. NC, JDE, CJG, CL, RKF, RTC, and GPP each reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beaulieu C, Plewes C, Paulson LA, Roy D, Snook L, Concha L, Phillips L. Imaging brain connectivity in children with diverse reading ability. Neuroimage. 2005;25:1266–1271. doi: 10.1016/j.neuroimage.2004.12.053. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statst Soc B. 1995;57(1):289–300. [Google Scholar]
- Brambati SM, Termine C, Ruffino M, Stella G, Fazio F, Cappa SF, Perani D. Regional reductions of gray matter volume in familial dyslexia. Neurology. 2004;63:742–745. doi: 10.1212/01.wnl.0000134673.95020.ee. [DOI] [PubMed] [Google Scholar]
- Brown WE, Eliez S, Menon V, Rumsey JM, White CD, Reiss AL. Preliminary evidence of widespread morphological variations of the brain in dyslexia. Neurology. 2001;56:781–783. doi: 10.1212/wnl.56.6.781. [DOI] [PubMed] [Google Scholar]
- Brunswick N, McCrory E, Price CJ, Frith CD, Frith U. Explicit and implicit processing of words and pseudowords by adult developmental dyslexics: A search for Wernicke’s Wortschatz? Brain. 1999;122(Pt 10):1901–1917. doi: 10.1093/brain/122.10.1901. [DOI] [PubMed] [Google Scholar]
- Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O’Donovan MC, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115:128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Fulker DW, LaBuda MC. Evidence for a genetic aetiology in reading disability of twins. Nature. 1987;329(6139):537–9. doi: 10.1038/329537a0. [DOI] [PubMed] [Google Scholar]
- Deutsch GK, Dougherty RF, Bammer R, Siok WT, Gabrieli JD, Wandell B. Children’s reading performance is correlated with white matter structure measured by diffusion tensor imaging. Cortex. 2005;41:354–363. doi: 10.1016/s0010-9452(08)70272-7. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Papademetris X, Yang J, Jackowski M, Zeng X, Staib LH. Geometric strategies for neuroanatomic analysis from MRI. NeuroImage. 2004;23(Suppl 1):S34–45. doi: 10.1016/j.neuroimage.2004.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliez S, Rumsey JM, Giedd JN, Schmitt JE, Patwardhan AJ, Reiss AL. Morphological alteration of temporal lobe gray matter in dyslexia: an MRI study. J Child Psychol Psychiatry. 2000;41:637–644. doi: 10.1111/1469-7610.00650. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. Proc IEEE Nucl Sci Symp Med Imaging Conf; Nuclear Science Symposium and Medical Imaging Conference; 1993. pp. 1813–1817. [Google Scholar]
- Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie Il, Walter J, Pennington BF, Fisher SE, Olson RK, DeFries JC, Stein JF, Monaco AP. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Paracchini S, Scerri T, Dennis M, Cope N, Hill G, Moskvina V, Walter J, Richardson AJ, Owen MJ, Stein JF, Green ED, O’Donovan MC, Williams J, Monaco AP. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11(12):1085-1091–1061. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- Hoeft F, Meyler A, Hernandez A, Juel C, Taylor-Hill H, Martindale JL, McMillon G, Kolchugina G, Black JM, Faizi A, Deutsch GK, Siok WT, Reiss AL, Whitfield-Gabrieli S, Gabrieli JD. Functional and morphometric brain dissociation between dyslexia and reading ability. Proc Natl Acad Sci USA. 2007;104:4234–4239. doi: 10.1073/pnas.0609399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22(2):324–33. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Horwitz B, Rumsey JM, Donohue BC. Functional connectivity of the angular gyrus in normal reading and dyslexia. Proc Natl Acad Sci USA. 1998;95:8939–8944. doi: 10.1073/pnas.95.15.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interagency Committee on Learning Disabilities. Learning disabilities: a report to the U.S. Congress. Government Printing Office; Washington, D.C: 1987. [Google Scholar]
- Lacadie CM, Fulbright RK, Rajeevan N, Constable RT, Papademetris X. More accurate Talairach coordinates for neuroimaging using non-linear registration. Neuroimage. 2008;42:717–725. doi: 10.1016/j.neuroimage.2008.04.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi N, Mencl WE, Frost SJ, Sandak R, Pugh KR. An fMRI study of multimodal semantic and phonological processing in reading disabled adolescents. Ann Dyslexia. 2010;60(1):102–21. doi: 10.1007/s11881-009-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levecque C, Velayos-Baeza A, Holloway ZG, Monaco AP. The dyslexia-associated protein KIAA0319 interacts with Adaptor Protein 2 and follows the classical clathrin-mediated endocytosis pathway. Am J Physiol Cell Physiol. 2009;297(1):C160–8. doi: 10.1152/ajpcell.00630.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Luciano M, Wright MJ, Montgomery GW, Martin NG, Bates TC. Dyslexia and DCDC2: normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur J Hum Genet. 2010;18:668–673. doi: 10.1038/ejhg.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, Martin NG, Bates TC. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–817. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ludwig KU, Roeske D, Schumacher J, Schulte-Korne G, Konig IR, Warnke A, Plume E, Ziegler A, Remschmidt H, Muller-Myhsok B, Nothen MM, Hoffmann P. Investigation of interaction between DCDC2 and KIAA0319 in a large German dyslexia sample. J Neural Transm. 2008;115:1587–1589. doi: 10.1007/s00702-008-0124-6. [DOI] [PubMed] [Google Scholar]
- Klingberg T, Hedehus M, Temple E, Salz T, Gabrieli JD, Moseley ME, Poldrack RA. Microstructure of temporo-parietal white matter as a basis for reading ability: evidence from diffusion tensor magnetic resonance imaging. Neuron. 2000;25:493–500. doi: 10.1016/s0896-6273(00)80911-3. [DOI] [PubMed] [Google Scholar]
- Marino C, Meng H, Mascherettu S, Rusconi M, Cope N, Giorda, Moleteni M, Gruen JR. DCDC2 genetic variants and susceptibility to developmental dyslexia. Psychiatr Genet. 2012;22(1):25–30. doi: 10.1097/YPG.0b013e32834acdb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Gelernter J, Gruen JR, Calhoun VD, Meng H, Cope NA, Pearlson GD. Polymorphism of DCDC2 reveals differences in cortical morphology of healthy individuals-A preliminary voxel based morphometry study. Brain Imaging Behav. 2008;2(1):21–26. doi: 10.1007/s11682-007-9012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O’Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Powers NR, Tang L, Cope NA, Zhang PX, Fuleihan R, Gibson C, Page GP, Gruen JR. A dyslexia-associated variant in DCDC2 changes gene expression. Behav Genet. 2011;41(1):58–66. doi: 10.1007/s10519-010-9408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;40:655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Nolf E, Voet T, Jacobs F, Dierckx R, Lemahieu I. Annual Congress of the European Association of Nuclear Medicine - EANM’03. European Journal of Nuclear Medicine; Amsterdam, The Netherlands: 2003. p. S246. [Google Scholar]
- Niogi SN, McCandliss BD. Left lateralized white matter microstructure accounts for individual differences in reading ability and disability. Neuropsychologia. 2006;44:2178–2188. doi: 10.1016/j.neuropsychologia.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, LoTurco JJ, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, Monaco AP. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Paulesu E, Frith U, Snowling M, Gallagher A, Morton J, Frackowiak RS, Frith CD. Is developmental dyslexia a disconnection syndrome? Evidence from PET scanning. Brain. 1996;119(Pt 1):143–157. doi: 10.1093/brain/119.1.143. [DOI] [PubMed] [Google Scholar]
- Paulesu E, Demonet JF, Fazio F, McCrory E, Chanoine V, Brunswick N, Cappa SF, COssu G, Habib M, Frith CD, Frith U. Dyslexia: cultural diversity and biological unity. Science. 2001;291:2165–2167. doi: 10.1126/science.1057179. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Bishop DVM. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- Pinel P, Fauchereau F, Moreno A, Barbot A, Lathrop M, Zelenika D, Le Bihan D, Poline JB, Bourgeron T, Dehaene S. Genetics variants of FOXP2 and KIAA0319/TTRAP/THEM2 locus are associated with altered brain activation in distinct language-related regions. J Neurosci. 2012;32(3):817–25. doi: 10.1523/JNEUROSCI.5996-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychological Corporation. Wechsler Abbreviated Scale of Intelligence (WASI) manual. San Antonio, TX: Author; 1999. [Google Scholar]
- Pugh KR, Frost SJ, Sandak R, Landi N, Rueckl JG, Constable RT, Seidenberg MS, Fulbright RK, Katz L, Mencl WE. Effects of stimulus and repetition on printed word identification: an fMRI comparison of nonimpaired and reading-disabled adolescent controls. J Cogn Neurosci. 2008;20(7):1146–60. doi: 10.1162/jocn.2008.20079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh KR, Menci WE, Jenner AR, Katz L, Frost SJ, Lee JR, Shaywitz SE, Shaywitz BA. Neurobiological studies of reading and reading disability. J Commun Disord. 2001;34(6):479–92. doi: 10.1016/s0021-9924(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Richlan F, Kronbichler M, Wimmer H. Functional abnormalities in the dyslexic brain: a quantitative meta-analysis of neuroimaging studies. Hum Brain Mapp. 2009;30:3299–3308. doi: 10.1002/hbm.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsey JM, Andreason P, Zametkin AJ, Aquino T, King AC, Hamburger SD, Pikus A, Rapoport JL, Cohen RM. Failure to activate the left temporoparietal cortex in dyslexia. An oxygen 15 positron emission tomographic study. Arch Neurol. 1992;49:527–534. doi: 10.1001/archneur.1992.00530290115020. [DOI] [PubMed] [Google Scholar]
- Rumsey JM, Nace K, Donohue B, Wise D, Maisog JM, Andreason P. A positron emission tomographic study of impaired word recognition and phonological processing in dyslexic men. Arch Neurol. 1997;54:562–573. doi: 10.1001/archneur.1997.00550170042013. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Morris AP, Buckingham LL, Newbury DF, Miller LL, Monaco AP, Bishop DV, Paracchini S. DCDC2, KIAA0319, and CMIP are associated with reading-related traits. Biol Psychiatry. 2011 doi: 10.1016/j.biopsych. 2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Anthoni H, Dahdouh F, Konig IR, Hillmer AM, Kluck N, Manthey M, Plume E, Warnke A, Remschmidt H, Hulsmann J, Cichon S, Lindgren CM, Propping P, Zucchelli M, Ziegler A, Peyrard-Janvid M, Schulte-Korne G, Nothen MM, Kere J. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78:52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz BA, Shaywitz SE, Pugh KR, Mencl WE, Fulbright RK, Skudlarski P, Constable RT, Marchione KE, Fletcher JM, Lyon GR, Gore JC. Disruption of posterior brain systems for reading in children with developmental dyslexia. Biol Psychiatry. 2002;52(2):101–110. doi: 10.1016/s0006-3223(02)01365-3. [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA. Dyslexia (specific reading disability) Biol Psychiatry. 2005;57(11):1301–9. doi: 10.1016/j.biopsych.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Fletcher JM, Holahan JM, Shneider AE, Marchione KE, Stuebing KK, Francis DJ, Pugh KR, Shaywitz BA. Persistence of dyslexia: the Connecticut Longitudinal Study at adolescence. Pediatrics. 1999;104(6):1351–9. doi: 10.1542/peds.104.6.1351. [DOI] [PubMed] [Google Scholar]
- Shaywitz S, Fletcher J, Shaywitz B. Issues in the definition and classification of attention deficit disorder. Topics in Language Disorders. 1994;14:1–25. [Google Scholar]
- Shaywitz SE, Shaywitz BA, Pugh KR, Fulbright RK, Constable RT, Mencl WE, Shankweiler DP, Liberman AM, Skudlarski P, Fletcher JM, Katz L, Marchione KE, Lacadie C, Gatenby C, Gore JC. Functional disruption in the organization of the brain for reading in dyslexia. Proc Natl Acad Sci USA. 1998;95:2636–2641. doi: 10.1073/pnas.95.5.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silani G, Frith U, Demonet JF, Fazio F, Perani D, Price C, Frith CD, Paulesu E. Brain abnormalities underlying altered activation in dyslexia: a voxel based morphometry study. Brain. 2005;128:2453–2461. doi: 10.1093/brain/awh579. [DOI] [PubMed] [Google Scholar]
- Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–131. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxis atlas of the human brain. Thieme; New York: 1988. [Google Scholar]
- Torgesen JK, Wagner R, Rashotte C. Test of Word Reading Efficiency. Austin, TX: Pro-ed; 1999. [Google Scholar]
- Velayos-Baeza A, Toma C, Paracchini S, Monaco AP. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum Mol Genet. 2008;17:859–871. doi: 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch E, Robichon F, Eliez S. Gray matter alteration in dyslexia: converging evidence from volumetric and voxel-by-voxel MRI analyses. Neuropsychologia. 2005;43:324–331. doi: 10.1016/j.neuropsychologia.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Wagner RK, Torgesen JK, Rashotte CA. Comprehensive Test of Phonological Processing. Austin, TX: Pro-ED, Inc; 1999. [Google Scholar]
- Wiederholt JR, Bryant BR. Gray Oral Reading Test. 3 Pro-Ed; Austin: 1992. [Google Scholar]
- Wilcke A, Weissfuss J, Kirsten H, Wolfram G, Boltze J, Ahnert P. The role of gene DCDC2 in German dyslexics. Ann Dyslexia. 2009;59:1–11. doi: 10.1007/s11881-008-0020-7. [DOI] [PubMed] [Google Scholar]
- Wilcke A, Ligges C, Burkhardt J, Alexander M, Wolf C, Quente E, Ahnert P, Hoffmann P, Becker A, Muller-Myshok B, Cichon S, Boltze J, Kirsten H. Imaging genetics of FOXP2 in dyslexia. Eur J Hum Genet. 2011 doi: 10.1038/ejhg. 2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock RM. Woodcock Reading Mastery Test-Revised. American Guidance Corp; Circle Pines, MN: 1987. [Google Scholar]
- Woodcock RM, Johnson MB. Woodcock-Johnson Psycho-Educational Battery-Revised. Developmental Learning Materials; Allen, TX: 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


