Abstract
microRNAs (miRNAs) control a multitude of pathways in human cancers. Differential expression of miRNAs among different histological types of tumors within the same type of tissue offers insight into the mechanism of pathogenesis and may help to direct treatment to improve prognosis. We assessed expression of 667 miRNAs in endometrial endometrioid and serous adenocarcinomas using RNA extracted from benign endometrium as well as from primary endometrial tumors. Quantitative miRNA profiling of endometrial adenocarcinomas revealed four overlapping groups of significantly overexpressed and underexpressed miRNAs. The first group was composed of 20 miRNAs significantly dysregulated in both adenocarcinoma types compared with benign endometrium, two groups were composed of miRNAs significantly dysregulated in either endometrioid adenocarcinomas or in serous adenocarcinomas compared with benign endometrium, and the fourth group was composed of 17 miRNAs that significantly distinguished between endometrioid adenocarcinomas and serous adenocarcinomas themselves. Validation of the expression levels of the selected miRNAs was carried out in a second panel composed of ten endometrioid and five serous tumors. Experimentally validated mRNA targets of these dysregulated miRNAs were identified using published sources, whereas TargetScan was used to predict targets of miRNAs in the first and fourth profile groups. These validated and potential miRNA target lists were filtered using published lists of genes displaying significant overexpression or underexpression in endometrial cancers compared to benign endometrium. Our results revealed a number of dysregulated miRNAs that are commonly found in endometrial (and other) cancers as well as several dysregulated miRNAs not previously identified in endometrial cancers. Understanding these differences may permit the development of both prognostic and diagnostic biomarkers.
Keywords: microRNA, endometrial cancer, endometrioid adenocarcinoma, serous adenocarcinoma
Introduction
Endometrial carcinoma is the fourth most common cancer among women worldwide (1). Endometrioid adenocarcinoma accounts for three-quarters of all endometrial cancers, and serous adenocarcinoma ranks as the second most common type (2). According to the National Cancer Institute, there were 42,160 new cases of endometrial cancer in the US with nearly 8,000 deaths in 2009; the relative 5-year survival has not improved over many decades. In spite of these data, endometrial cancer is perhaps the least studied and understood cancer affecting women. In the past few years, substantial progress has been made in the arenas of clinical trials and translational research, particularly the understanding of the importance of post-transcriptional gene regulation. For example, an area of particular interest is the relative expression patterns of microRNAs (miRNAs) in cancer and how these molecules contribute to oncogenesis (3–7). By understanding how the miRNA expression profile reflects the grade and stage of disease, it may be possible to better design patient treatment regimens.
In the present study we report an analysis of the comparative expression levels of 667 human miRNAs in benign endometrial tissues, endometrial endometrioid adenocarcinomas and endometrial serous adenocarcinomas using a quantitative PCR (qPCR) array platform. Our data demonstrated that the two types of adenocarcinomas (endometrioid and serous) are generally similar with respect to miRNAs that are significantly either overexpressed or underexpressed compared to benign endometrial tissues. However, we also identified numerous miRNAs that differentiate between the two adenocarcinomas. Several of these significantly overexpressed or underexpressed miRNAs correspond to recently identified miRNA expression networks known to be reprogrammed in human cancers (8). We also identified a potential coexpression clique potentially affecting the Wnt signaling pathway in both forms of endometrial adenocarcinoma, signifying the importance of this pathway in the development of all endometrial cancers.
Materials and methods
Endometrial tumor and benign endometrium tissues
Endometrial tumor samples and samples of benign endometrium were obtained under informed consent from patients undergoing surgery at the University of Iowa Hospitals and Clinics. All tumor specimens were from primary sites and were snap-frozen and stored at −80°C following histological examination. Clinicopathologic characteristics of the patients from whom the tissues used in this study were obtained are presented in Table I.
Table I.
Clinical/pathologic characteristics of the tissues used for this study.
| ID | Tumor type | Age | Grade | Stage |
|---|---|---|---|---|
| NE10 | Benign | 58 | NA | NA |
| NE13 | Benign | 48 | NA | NA |
| NE59 | Benign | 48 | NA | NA |
| NE86 | Benign | 63 | NA | NA |
| EA11 | Endometrioid | 72 | 3 | IB |
| EA30 | Endometrioid | 66 | 2 | IC |
| EA38 | Endometrioid | 74 | 1 | IB |
| EA44 | Endometrioid | 53 | 2 | IIA |
| EA46 | Endometrioid | 54 | 1 | IB |
| EA49 | Endometrioid | 64 | 2 | IB |
| EA66 | Endometrioid | 82 | 1 | IB |
| EA68 | Endometrioid | 49 | 2 | IIB |
| EA69 | Endometrioid | 59 | 1 | IB |
| EA71 | Endometrioid | 52 | 3 | IIIC |
| EA73 | Endometrioid | 54 | 1 | IB |
| EA83 | Endometrioid | 85 | 1 | IA |
| EA92 | Endometrioid | 61 | 1 | IC |
| EA96 | Endometrioid | 62 | 1 | IIIC |
| SA12 | Serous | 61 | 3 | IB |
| SA36 | Serous | 59 | 3 | IIIC |
| SA39 | Serous | 64 | 3 | IIIC |
| SA48 | Serous | 60 | 3 | IVB |
| SA50 | Serous | 67 | 3 | IIIC |
| SA55 | Serous | 69 | 3 | IB |
| SA72 | Serous | 83 | 3 | IIIC |
| SA79 | Serous | 87 | 3 | IIIC |
| SA93 | Serous | 85 | 3 | IIIC |
Tissues used for the qPCR arrays are indicated in bold type. NA, not applicable. Age is in years.
RNA extraction
Prior to RNA extraction, tissues were transitioned from −80 to −20°C following perfusion in RNAlater ICE (Ambion) according to the supplier's instructions. Total RNA was extracted using the miRVana miRNA isolation kit (Ambion). RNA quality and concentrations were assessed on a NanoDrop M-1000 and an Agilent 2100 Bioanalyzer. Only RNAs of sufficient quality (RIN ≥7.00) were used for expression assays. RNAs were standardized to 200 ng/μl and these concentrations were reverified on the NanoDrop M-1000.
qPCR arrays
miRNA expression levels were assessed on Applied Biosystems TaqMan Low Density miRNA arrays (TLDA). Both the A-Set TLDA card, containing qPCR assays for 377 human miRNAs and the B-Set TLDA card, containing qPCR assays for 290 additional human miRNAs, were used for expression screening of a panel composed of four benign endometrial tissues, four endometrial endometrioid adenocarcinomas and four endometrial serous adenocarcinomas. A total of 600 ng of tissue RNA was used for each assay. Reverse transcription was carried out using the TaqMan miRNA reverse transcription kit (Applied Biosystems) and either the Human A Pool or Human B Pool Megaplex RT primers (Applied Biosystems).
All 24 TLDA cards were processed on an Applied Biosystems Model 7900 Genetic Analyzer, and the resulting data were analyzed using the Applied Biosystems StatMiner software. Expression of all 667 miRNAs in all samples was normalized against the RNU48 endogenous RNA control prior to statistical analyses. Comparisons between endometrioid adenocarcinoma and benign endometrium, serous adenocarcinoma and benign endometrium, and endometrioid adenocarcinoma and serous adenocarcinoma were carried out using the ΔCt method where fold change was expressed as 2−ΔΔCt (9). Statistical significance of the fold changes was assessed via two-tailed t-tests with unequal variances. We report unadjusted p-values (p≤0.05) as significant. No adjustment of p-values to account for a false discovery rate (FDR) was made as inherent FDR in miRNA studies is insufficient to warrant the type of increased stringency required by other types of arrays (10).
microRNA validation
Following analysis of the TLDA array data, we selected a set of 10 miRNAs with which to validate the array results. miRNA-specific qPCR assays for miR-21, miR-34a, miR-129-3p, miR-135a, miR-135b, miR-137, miR-200c, miR-205, miR-210 and miR-299-3p (Applied Biosystems) were carried out on an RNA panel composed of the original four benign endometrial tissues plus 10 additional endometrial endometrioid adenocarcinomas and five additional endometrial serous adenocarcinomas. For validations, reverse transcriptions and qPCR assays were carried out using miRNA-specific RT primers and qPCR primer/probe sets (Applied Biosystems). These assays were also run on an Applied Biosystems Model 7900 Genetic Analyzer and the resulting data again were analyzed using the Applied Biosystems StatMiner software following normalization against the RNU48 endogenous RNA control.
Results
Twenty microRNAs are significantly dysregulated in both endometrial adenocarcinomas compared with benign endometrium
miRNAs displaying either significant overexpression or underexpression in both endometrioid adenocarcinoma and serous adenocarcinoma as compared to benign endometrium are presented in Table II. A total of seven miRNAs was significantly underexpressed in both adenocarcinomas and a total of 13 miRNAs was significantly overexpressed in both adenocarcinomas. The lowest relatively expressed miRNA in both adenocarcinomas was miR-133b while the highest was miR-205, which is consistent with miRNA studies of endometrial cancers (4–7). Similar to other reports, miR-135b and all five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) were significantly overexpressed in both types of endometrial cancers (4–6).
Table II.
microRNAs significantly underexpressed or overexpressed in both endometrioid and serous adenocarcinomas compared to normal endometrium.
| Endometrioid tumors |
Serous tumors |
|||
|---|---|---|---|---|
| Fold change | p-value | Fold change | p-value | |
| Underexpressed microRNAs | ||||
| miR-1 | −83.3 | 0.001 | −15.9 | 0.048 |
| miR-133a | −200.0 | 0.001 | −83.3 | 0.021 |
| miR-504 | −125.0 | 0.001 | −16.1 | 0.011 |
| miR-133b | −333.3 | 0.002 | −377.2 | 0.022 |
| miR-137 | −200.0 | 0.002 | −30.3 | 0.030 |
| miR-125b-2* | −166.7 | 0.003 | −33.3 | 0.024 |
| miR-129-3p | −47.6 | 0.043 | −37.0 | 0.026 |
| Overexpressed microRNAs | ||||
| miR-15b | 5.7 | 0.005 | 6.9 | 0.020 |
| miR-142-5p | 9.0 | 0.028 | 10.4 | 0.023 |
| miR-142-3p | 13.7 | 0.018 | 15.0 | 0.015 |
| miR-135a | 19.5 | 0.014 | 48.6 | 0.001 |
| miR-519a | 22.9 | 0.027 | 79.6 | 0.037 |
| miR-135b | 41.4 | 0.004 | 90.1 | 0.018 |
| miR-200c | 102.0 | 0.003 | 102.3 | 0.002 |
| miR-141 | 186.8 | 0.001 | 127.0 | 0.003 |
| miR-200a | 194.4 | 0.001 | 85.6 | 0.004 |
| miR-200b | 221.4 | 0.002 | 104.5 | 0.004 |
| miR-429 | 345.0 | 0.003 | 209.2 | 0.006 |
| miR-888 | 400.8 | 0.001 | 74.4 | 0.032 |
| miR-205 | 806.4 | 0.005 | 301.5 | 0.010 |
Endometrioid adenocarcinoma and serous adenocarcinoma each display tumor type-specific dysregulated microRNAs
We next examined how miRNA expression patterns might be used to distinguish between different types of endometrial carcinoma. miRNAs significantly underexpressed or overexpressed in either endometrioid adenocarcinoma or serous adenocarcinoma as compared to benign endometrium are shown in Table III. We identified three times as many miRNAs differentially expressed in endometrioid adenocarcinoma (n=38) than there were in serous adenocarcinoma (n=12) in comparison to the benign endometrium. These included many of the miRNAs commonly observed in other studies such as miR-155, miR-203 and miR-210, the most frequently observed differentially expressed miRNA in endometrial cancers (3–6). For many of these miRNAs, however, the significance of overexpression varied greatly between the two types of adenocarcinomas. For example, miR-155 was significantly overexpressed in serous adenocarcinoma (p=0.049) but was just above statistical significance in endometrioid adenocarcinoma (p=0.053). Similarly, miR-370 was significantly overexpressed in serous adenocarcinoma (p=0.024) but was not statistically significant in endometrioid tumors (p=0.07); miR-210 was significantly overexpressed in endometrioid adenocarcinoma (p=0.029) but, again, fell short of statistical significance in serous tumors (p=0.10). Taken together, these data indicate that, while these individual miRNAs were overexpressed in both types of adenocarcinomas relative to the benign samples, there was a wide range of expression levels within each tumor type.
Table III.
microRNAs significantly underexpressed or overexpressed in either endometrioid and serous adenocarcinomas compared to normal endometrium.
| Fold change | p-value | |
|---|---|---|
| Underexpressed microRNAs | ||
| Endometrioid tumors | ||
| miR-502-3p | −2.2 | 0.003 |
| miR-130a | −3.2 | 0.005 |
| miR-214 | −9.1 | 0.006 |
| miR-218 | −6.7 | 0.010 |
| miR-99a | −7.5 | 0.010 |
| miR-410 | −3.8 | 0.012 |
| miR-100 | −6.6 | 0.014 |
| miR-199a-3p | −4.4 | 0.015 |
| miR-424 | −3.6 | 0.020 |
| miR-199a-5p | −3.4 | 0.021 |
| miR-214* | −5.4 | 0.024 |
| miR-99a* | −15.4 | 0.029 |
| let-7c | −7.8 | 0.030 |
| miR-212 | −2.8 | 0.031 |
| miR-130a* | −32.3 | 0.035 |
| miR-495 | −3.9 | 0.035 |
| miR-100* | −20.4 | 0.036 |
| miR-125b | −6.8 | 0.042 |
| miR-218-2* | −5.2 | 0.043 |
| miR-502-5p | −3.5 | 0.043 |
| miR-532-5p | −1.7 | 0.047 |
| Serous tumors | ||
| miR-370 | −37.0 | 0.024 |
| miR-423-5p | −2.7 | 0.030 |
| Overexpressed microRNAs | ||
| Endometrioid tumors | ||
| let-7g* | 18.1 | 0.002 |
| miR-181c* | 14.7 | 0.007 |
| miR-516a-3p | 8.1 | 0.014 |
| miR-9 | 9.3 | 0.014 |
| miR-203 | 14.1 | 0.017 |
| miR-375 | 333.9 | 0.021 |
| miR-652 | 3.5 | 0.021 |
| miR-146a | 43.1 | 0.022 |
| miR-9* | 5.8 | 0.024 |
| miR-210 | 9.1 | 0.029 |
| miR-32 | 5.9 | 0.030 |
| miR-148a | 4.2 | 0.036 |
| miR-425 | 5.4 | 0.043 |
| miR-592 | 20.0 | 0.047 |
| miR-21 | 6.6 | 0.048 |
| miR-7-1* | 4.8 | 0.049 |
| miR-107 | 9.2 | 0.049 |
| Serous tumors | ||
| miR-31 | 19.0 | 0.006 |
| miR-885-5p | 23.9 | 0.015 |
| miR-490-3p | 20.4 | 0.016 |
| miR-644 | 7.5 | 0.034 |
| miR-522 | 65.1 | 0.037 |
| miR-519d | 37.5 | 0.041 |
| miR-98 | 5.9 | 0.045 |
| miR-425 | 5.4 | 0.049 |
| miR-518e | 27.3 | 0.049 |
| miR-155 | 5.4 | 0.049 |
microRNA-specific expression validations by qPCR
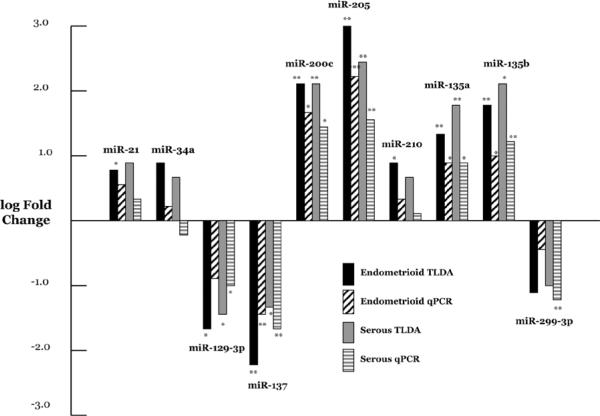
We chose 10 miRNAs for subsequent validation using miRNA-specific qPCR in an expanded panel of patients. These miRNAs (miR-21, miR-34a, miR-129-3p, miR-135a, miR-135b, miR-137, miR-200c, miR-205, miR-210 and miR-299-3p) were selected based in part upon their expression levels as well as reported cancer-related targets. Furthermore, we specifically chose miRNAs with non-significant as well as those with significant fold change compared with benign endometrium. Of the 20 comparisons of endometrial cancer expression vs. benign endometrium, 16 (80.0%) were concordant with the array results (Fig. 1).
Figure 1.
qPCR validations of selected miRNAs. Log fold change is shown for both the TLDA array and the miR-specific qPCR assay for endometrioid adenocarcinoma and serous adenocarcinoma. Significant values (*p<0.05 and **p<0.01) are comparing adenocarcinoma with benign endometrium.
Algorithmically predicted and experimentally validated mRNA targets of dysregulated miRNAs present numerous potential avenues for examining endometrial carcinogenesis
Utilizing both an extensive literature search and the miRNA target prediction algorithm TargetScan, we assembled files composed of algorithmically predicted and experimentally validated mRNA targets for each of the miRNAs significantly dysregulated in both endometrioid and serous adenocarcinomas compared to benign endometrium. We screened the predicted mRNA target files for each miRNA using a filter composed of mRNAs shown to be differentially expressed in endometrial cancers compared to benign endometrium (3,4,11,12). These filtered predicted targets for each of the significantly dysregulated miRNAs are presented in Table IV along with experimentally validated targets for each.
Table IV.
mRNA targets of microRNAs significantly underexpressed or overexpressed in endometrial adenocarcinomas compared to benign endometrium.
| Target mRNAs | |
|---|---|
| Underexpressed microRNAs | |
| miR-133a,b | TAGLN2, BCL2L2, FSCN1, MET, MCL-1, PKM2 |
| miR-137 | MITF, STK38, ZAK, Cdc42 |
| miR-504 | OLFML2A, p53 |
| miR-1 | TRIM2, HDAC4, MET, Pim-1, FOXP1, TAGLN2 |
| miR-125b-2* | CXCL5 |
| miR-129-3p | SOX4, Cdk6 |
| Overexpressed microRNAs | |
| miR-205 | LRP1, HER3, ERBB3, PRKCE |
| miR-888 | PPP1R12B, ZAK, REV3L, PGR |
| miR-135a,b | APC, SFRP4 |
| miR-519a | ZNF238, CXCL12 |
| miR-142-3p | MYLK, ZEB2, BNC2 |
| miR-142-5p | ADAMTS5 |
| miR-15b | BCL2L2 |
| miR-141a | ZEB2, CXCL12, TSHZ3, VEGFA |
| miR-429b | ZEB1, ZEB2, LEPR, PPP1R12B |
The mRNAs shown in bold were experimentally validated. The remaining mRNA targets were identified as underexpressed or overexpressed in the endometrial adenocarcinomas compared to benign endometrium in Boren et al (3), Risinger et al (11) Chung et al (4) or Smid-Koopman et al (12).
includes family members miR-141, miR-200a, miR-200b.
Includes family members miR-429 and miR-200c.
Seventeen microRNAs discriminate between endometrioid and serous adenocarcinomas
In addition to identifying miRNAs that were significantly dysregulated in endometrial cancers compared to benign endometrium, these data also permitted identification of 17 miRNAs that signi ficantly differentiated between endometrioid and serous tumors themselves. These miRNAs are presented in Table V. In 11 of the 17 miRNAs, the fold change for one of the two tumor types compared to benign endometrium was <2, whereas for the remaining six miRNAs there was at least a 2-fold difference in expression compared to benign endometrium in both tumor types. For most of these miRNAs it is clear which of the two tumor types is responsible for the differentiating fold change. These distinctions are important in order to better evaluate the nature of miRNA differentiation between the two tumor types.
Table V.
microRNAs found to significantly differentiate serous tumors from endometnoid tumors.
| microRNA | Fold changea | p-value | EA/BEb | SA/BEb | mRNA targetf |
|---|---|---|---|---|---|
| miR-218 | 5.9 | 0.001 | −6.7d | −1.4 | ZEB2 |
| miR-9 | −9.8 | 0.011 | 9.3c | −1.1 | NEDD4 |
| miR-423-5p | −4.6 | 0.011 | 1.7 | −2.7c | |
| miR-542-3p | 5.8 | 0.018 | −2.5 | 2.3 | BLCAP, PARP12, FIGF |
| miR-490-3p | 11.6 | 0.024 | 1.8 | 20.4 | |
| miR-504 | 8.1 | 0.026 | −131.6e | −16.3d | GPC4, OLFML2A |
| miR-338-3p | 10.9 | 0.030 | −1.9 | 5.9 | |
| miR-9* | −13.6 | 0.030 | −15.4c | −2.3 | |
| miR-146a | −8.9 | 0.031 | 43.1c | 4.9 | ERBB4, ABL2 |
| miR-130a | 5.9 | 0.032 | −3.2d | 1.8 | IGF1, ZAK |
| let-7c | 3.2 | 0.033 | −7.7c | −2.4 | |
| miR-375 | −232.7 | 0.035 | 333.9c | 1.4 | PDK1 |
| miR-675 | 80.2 | 0.037 | −1.5 | 53.2 | RB1 |
| miR-570 | 27.6 | 0.041 | −1.1 | 26.0 | ABL2, XYLT1, IGF1R |
| miR-99a/100 | 4.8 | 0.041 | −7.5 | −1.6 | PPP3CA, IGF1R |
| miR-518e | 19.8 | 0.048 | 1.4 | 27.3c | |
| miR-450a | 5.4 | 0.050 | −2.1 | 2.6 |
Fold change is presented as serous tumors compared to endometrioid tumors.
Fold changes for endometrioid adenocarcinoma (EA) compared to benign endometrium (BE) and for serous adenocarcinoma (SA) compared to benign endometrium (BE) for the same miRNAs.
p<0.05,
p<0.01,
p<0.001
Experimentally validated targets are shown in bold. TargetScan predicted targets were filtered in the same manner as those presented in Table IV.
Discussion
microRNA dysregulation in cancer is a well-established phenomenon (13,14). However, studies of miRNA expression in a wide range of human cancers have shown that the specifics of dysregulation and, by implication, the consequences vary enormously with respect not only to which miRNAs are involved but also to whether specific miRNAs are overexpressed or underexpressed and whether they act as oncogenes or tumor suppressors (15). To date, there have been five surveys of miRNA expression patterns in endometrial cancers (3–7). These studies have employed hybridization array methods and/or qPCR-based methods assessing from 157 to 470 miRNAs in samples ranging from ten to nearly one hundred patients. Cumulative with our data, there are 16 miRNAs in which significant expression dysregulation was observed in at least three studies (data not shown). One of these 16 miRNAs, miR-133b was significantly underexpressed in three of the six studies while the remaining 15 were all significantly overexpressed. miR-133b, and its nearly identical duplicate, miR-133a, target the suspected oncoprotein pyruvate kinase type M2 (PKM2) as well as several other potentially important mRNAs including the oncogenic fascin 1 (FSCN1). In addition, the TargetScan predicted mRNA target transgelin 2 (TAGLN2) has previously been shown in other types of cancers to promote invasiveness of tumorigenic cells (16). The TAGLN2 mRNA is also a predicted target of miR-1 and all three of these miRNAs have recently been linked as a co-regulated clique (8). Significant co-regulated underexpression of miR-133a,b and miR-1 would, therefore, influence overexpression of several tumorigenic proteins. It is important to note in this context that another oncogenic target mRNA of miR-1, MET, is also a target of miR-133a,b.
Among the overexpressed miRNAs, miR-210 was reported in all but one of the extant surveys. Overexpression of miR-210 in these cancers is not unexpected as it is well known as the principle hypoxia-responsive miRNA (17) with mRNA targets involved in DNA repair (BLM), cell cycle (PLK1, E2F3) and metabolism (GPD1L1, LDH-B, ATPSA1) among others. As endometrial adenocarcinomas are solid tumors in which hypoxic microenvironments abound, induction of miR-210 expression through the HIF-1 response mechanism should be an expected outcome.
Similarly represented in the miRNA expression surveys are miR-205 and miR-200a. Recent studies of miR-205 as well as the entire miR-200 family, which is composed of the two polycistrons miR-141/miR-200c and miR-429/miR-200a/miR-220b, show that all of these miRs are coordinately regulated (18) and are involved in mesenchymal transition, tumor invasiveness and metastasis (7,19,20). Expression of these miRNAs is increased in ovarian, bladder and kidney cancers as well but is decreased in prostate and esophageal cancers (7). In addition, miR-182, found to be overexpressed in four of the six surveys, is involved in both tumor invasiveness and anti-apoptosis (21). Overexpression of miR-182, along with miR-210 and miR-200c, was recently reported in ovarian cancers and was linked to tumor progression (22). In addition, miR-182 has been linked to repression of the tumor-suppressor gene FOXO1 in both endometrial (23) and breast cancers (24).
The other miRNA found to be significantly overexpressed in four of the six surveys, miR-203, has been associated with poor survival in pancreatic cancers (25); in ovarian cancers there is evidence that overexpression of this miRNA is due to hypomethylation (26). This latter finding suggests that epigenetic phenomena play an important role in miRNA dysregulation in cancers of the female reproductive tract (27,28).
We also identified a number of significantly dysregulated miRNAs whose expression status in endometrial cancers was either not well known in general or was described for the first time in this study. For example, miR-129-3p was noted here to be significantly underexpressed in both the endometrioid and serous tumors. Whether this was due to the fact that the expression of this miRNA did not achieve statistical significance in other surveys or that it was not one of the miRNAs surveyed is unknown. However, miR-129-3p was found here to be nearly 50-fold underexpressed in the endometrioid tumors and nearly 40-fold underexpressed in the serous tumors compared to benign endometrium. A validated mRNA target of miR-129-3p in endometrial cancers is the SOX4 oncogene. Underexpression of miR-129-3p leads to overexpression of SOX4 and this relationship has been shown to be under epigenetic control through hypomethylation of the miR-129 CpG island (29).
Another important observation in our study is the significant overexpression of miR-135a,b in both endometrioid and serous tumors. This overexpression could have direct consequences for the Wnt signaling pathway, a pathway well known to be important in a variety of tumors (30,31). The adenomatous polyposis coli gene (APC) is a key component of the Wnt pathway (32–34) and its mRNA has recently been shown to be a direct target of miR-135a,b (35). Consistent with the effects of oncogenic mutations arising in various crucial components of the pathway (31), suppression of APC by miR-135a,b overexpression would have the effect of locking the pathway into a ligand-independent state of constitutive activity. The result of leaving the Wnt switch `on' would be accumulation of β-catenin and subsequent overexpression of downstream proteins for which β-catenin is a known transcription factor. One of these downstream targets is c-MYC, which has been shown to be involved in a number of feedback regulatory loops with a host of additional miRNAs that regulate cell progression, angiogenesis and metastasis (36). Thus, overexpression of miR-135a,b in endometrial adenocarcinomas may result in a wide range of downstream miRNA-mediated phenomena.
Another potentially important miRNA dysregulation reported here for the first time is the significant overexpression of miR-888 in both endometrioid and serous tumors compared to benign endometrium. This miRNA lies at Xq27.3 between miR-890 and the two tandem duplication pairs miR-892a,b and miR-891a,b. It has recently been shown that these miRNAs are unique to primate genomes and that they are exclusively expressed in reproductive tissues (37). Consequences of miR-888 overexpression in endometrial cancers are, as yet, unknown. However, one of the TargetScan predicted mRNA targets of miR-888 is the progesterone receptor, PR, which is lost in a substantial proportion of endometrial tumors (38). Moreover, the role of progesterone and progesterone receptors in endometrial carcinogenesis and early treatment is well established (39–41). Thus, investigation of the role of miR-888 in endometrial cancers is warranted.
A number of dysregulated miRNAs are reported here to differentiate endometrioid and serous adenocarcinomas. One of these, miR-675, is particularly notable as it is transcribed as part of the maternally imprinted non-coding RNA H19 (42,43). While its function remains unknown, H19 has been called an `oncofetal' non-coding RNA (44) since it is primarily expressed during fetal development while it has been shown to re-emerge in a variety of cancers including colorectal (45), hepatocellular (46), testicular (47), esophageal (48), bladder (49) and breast (50). The most important observation relative to the potential role of H19 in endometrial cancer is that H19 expression is clearly associated with serous tumors of the ovary (44). This is consistent with our observation that miR-675 expression was greater than 50-fold overexpressed in the serous tumors compared with the benign endometrium but was not different from the benign endometrium in the endometrial tumors. Thus, miR-675 overexpression in serous tumors is the driver of the 80-fold expression differentiation between serous and endometrioid tumors. H19 transcription and miR-675 expression were recently shown to be significantly correlated in colorectal tumors (r=0.86, p<0.001) and cultured cells (r=0.88, p<0.001) (51). Moreover, the one experimentally validated mRNA target of miR-675, retinoblastoma RB1, is a tumor suppressor (51). As H19 and, by extension, miR-675 transcription is maternally imprinted, one mechanism to consider in overexpression in endometrial serous adenocarcinomas is epigenetic. However, it has been shown that the H19 promoter is efficiently repressed by p53 (52) and there is also evidence that H19 expression is negatively regulated by progesterone levels (53). Both p53 mutation and loss of progesterone and its receptor are well characterized phenomena in endometrial cancers so the mechanistic interplay among epigenetics, specific tumor-suppressor mutations and hormones is likely to be complex and warrants future investigation.
We presented evidence of substantial miRNA dysregulation in endometrial endometrioid adenocarcinomas and serous adenocarcinomas compared both to benign endometrium and to each other. Some of these data, such as the consistent overexpression of miR-205, miR-210 and members of the miR-200 family, suggest that these miRNAs are excellent candidates for development as biomarkers for prognosis and possibly early diagnosis. In addition, the opportunity to offer adjuvant chemotherapy after initial hysterectomy for patients at a particularly high risk for recurrence (as identified from predictive miRNA patterns) has the potential to significantly improve survival in this disease. Expression profiles of other miRNAs, notably miR-135a,b, miR-888 and miR-675, point the way to mechanistic studies that may elucidate previously unknown aspects of endometrial carcinogenesis.
Acknowledgements
This study was supported by NIH Grant R01CA99908 to K.K.L. and in part by the Department of Obstetrics and Gynecology Research Development Fund. We thank Kristina W. Thiel for assistance in the manuscript preparation.
References
- 1.Jemal A, Siegal R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Pecorelli S, Pasinetti B, Angioli R, Favalli G, Odicino F. Systematic therapy for gynecological neoplasms ovary, cervix, and endometrium. Cancer Chemother Bio Response Modif. 2005;22:515–544. [PubMed] [Google Scholar]
- 3.Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Chung TK, Cheung TH, Huen NY, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 6.Hiroki E, Akahira JI, Suzuki F, Nagase S, Ito K, Sasano H, Yaegashi N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratner ES, Tuck D, Richter C, et al. MicroRNA signatures differentiate uterine cancer tumor subtypes. Gynecol Oncol. 2010;118:251–257. doi: 10.1016/j.ygyno.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 11.Risinger JI, Maxwell GL, Chandramouli GV, et al. Microarray analysis reveals distinct gene expression profiles among different histologic types of endometrial cancer. Cancer Res. 2003;63:6–11. [PubMed] [Google Scholar]
- 12.Smid-Koopman E, Blok LJ, Chadha-Ajwani S, Helmerhorst TJ, Brinkmann AO, Huikeshoven FJ. Gene expression profiles of human endometrial cancer samples using a cDNA-expression array technique: assessment of an analysis method. Br J Cancer. 2000;83:246–251. doi: 10.1054/bjoc.2000.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davalos V, Esteller M. MicroRNAs and cancer epigenetics a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 14.Garofalo M, Condorelli GL, Croce CM, Condorelli G. microRNAs as regulators of death receptor signaling. Cell Death Differ. 2010;17:200–208. doi: 10.1038/cdd.2009.105. [DOI] [PubMed] [Google Scholar]
- 15.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: microRNAs in cancer. Cell. 2009;137:586.e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 16.Lee E-K, Han GY, Park HW, Song YJ, Kim CW. Transgelin promotes migration and invasion of cancer stem cells. J Proteome Res. 2010;9:5108–5117. doi: 10.1021/pr100378z. [DOI] [PubMed] [Google Scholar]
- 17.Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrba L, Jensen TJ, Garbe JC, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory P, Bert A, Paterson E, et al. The miR-200 family and miR-205 regulate epithelial to msenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 20.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segura M, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalimia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaksman O, Stavnes HT, Kærn J, Trope CG, Davidson B, Reich R. miRNA profiling along tumor progression in ovarian carcinoma. J Cell Mol Med. 2010 Aug 16; doi: 10.1111/j.1582-4934.2010.01148.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myatt SS, Wang J, Monteiro LJ, et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210, and 222 in pancreatic tumors associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 26.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 27.Maldonado L, Hoque MO. Epigenomics and ovarian cancer. Biomark Med. 2010;4:543–570. doi: 10.2217/bmm.10.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiapale J, Beachy PA. The hedgehog and Wnt signaling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 31.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Ann Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 32.Powell SM, Zilz N, Beazer-Barclay Y, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 33.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Sansom OJ, Reed KR, Hayes AJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagel R, Le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 36.Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer. 2010;1:568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Liu Y, Dong D, Zhang Z. Evolution of an X-linked primate-specific microRNA cluster. Mol Biol Evol. 2010;27:671–683. doi: 10.1093/molbev/msp284. [DOI] [PubMed] [Google Scholar]
- 38.Kumar NS, Richer J, Owen G, Litman E, Horwitz KB, Leslie KK. Selective down-regulation of progesterone receptor isoform B in poorly differentiated human endometrial cancer cells implication for unopposed estrogen action. Cancer Res. 1998;58:1860–1865. [PubMed] [Google Scholar]
- 39.Leslie KK, Stein MP, Kumar NS, et al. Progesterone receptor isoform identification and subcellular localization in endometrial cancer. Gynecol Oncol. 2005;96:32–41. doi: 10.1016/j.ygyno.2004.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jongen V, Briët J, De Jong R, et al. Expression of estrogen receptor-alpha and -beta and progesterone receptor-A and -B in a large cohort of patients with endometrioid endometrial cancer. Gynecol Oncol. 2009;112:537–542. doi: 10.1016/j.ygyno.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Kim JJ, Chapman-Davis E. Role of progesterone in endometrial cancer. Sem Reprod Med. 2010;28:81–90. doi: 10.1055/s-0029-1242998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X, Cullen BR. The imprinted H19 non-coding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smits G, Mungall AJ, Griffiths-Jones S, et al. Conservation of the H19 non-coding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 44.Tanos V, Prus D, Ayesh S, et al. Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 1999;85:7–11. doi: 10.1016/s0301-2115(98)00275-9. [DOI] [PubMed] [Google Scholar]
- 45.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 46.Ariel I, Miao HQ, Ji XR, et al. Imprinted H19 oncofetal RNA is a candidate marker for hepatocellular carcinoma. Mol Pathol. 1998;51:21–25. doi: 10.1136/mp.51.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verkerk AJ, Ariel I, Dekker MC, et al. Unique expression patterns of H19 in human testicular cancers of different etiology. Oncogene. 1997;14:95–107. doi: 10.1038/sj.onc.1200802. [DOI] [PubMed] [Google Scholar]
- 48.Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- 49.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 50.Lottin S, Adriaenssens E, Berteaux N, et al. The human H19 gene is frequently overexpressed in myometrium and stroma during pathological endometrial proliferative events. Eur J Cancer. 2005;41:168–177. doi: 10.1016/j.ejca.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Tsang WP, Ng EK, Ng SSM, Jin H, Yu J, Sung JJY, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 52.Dugimont T, Montpellier C, Adriaenssens E, et al. The H19 TATA-les promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene. 1998;16:2395–2401. doi: 10.1038/sj.onc.1201742. [DOI] [PubMed] [Google Scholar]
- 53.Adriaenssens E, Lottin S, Dugimont T, et al. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18:4460–4473. doi: 10.1038/sj.onc.1202819. [DOI] [PubMed] [Google Scholar]