Abstract
The trophectoderm epithelium is the first differentiated cell layer to arise during mammalian development. Blastocyst formation requires the proper expression and localization of tight junction, polarity, ion gradient and H2O channel proteins in the outer cell membranes. However, the underlying transcriptional mechanisms that control their expression are largely unknown. Here, we report that transcription factor AP-2γ (Tcfap2c) is a core regulator of blastocyst formation in mice. Bioinformatics, chromatin immunoprecipitation and transcriptional analysis revealed that Tcfap2c binds and regulates a diverse group of genes expressed during blastocyst formation. RNA interference experiments demonstrated that Tcfap2c regulates genes important for tight junctions, cell polarity and fluid accumulation. Functional and ultrastructural studies revealed that Tcfap2c is necessary for tight junction assembly and paracellular sealing in trophectoderm epithelium. Aggregation of control eight-cell embryos with Tcfap2c knockdown embryos rescued blastocyst formation via direct contribution to the trophectoderm epithelium. Finally, we found that Tcfap2c promotes cellular proliferation via direct repression of p21 transcription during the morula-to-blastocyst transition. We propose a model in which Tcfap2c acts in a hierarchy to facilitate blastocyst formation through transcriptional regulation of core genes involved in tight junction assembly, fluid accumulation and cellular proliferation.
Keywords: Blastocyst formation, Tight junction, Tcfap2c, Tfap2c
INTRODUCTION
The blastocyst trophectoderm (TE) epithelium is the first differentiated cell layer to emerge during mammalian development. A key function of the developing TE layer is to act as a barrier and to regulate the exchange and accumulation of small molecules and fluid during blastocoel formation (Cockburn and Rossant, 2010). This property is mediated primarily by the action of tight junction (TJ) complexes, ion gradient pumps and H2O channels that assemble on the apical and basolateral membranes of the outer cell layer. Members of several gene families are important for blastocyst formation, including claudins, zonal occludins (TJ proteins), occludins, jam proteins, cell polarity proteins, Na/K-ATPases and aquaporins. The proper expression and localization of these gene products is crucial for blastocyst development (Watson and Barcroft, 2001; Eckert and Fleming, 2008). However, the underlying transcriptional mechanisms that control the spatial and temporal expression of these genes are largely unknown.
Recent studies established a role for developmental transcription factors (TFs) such as Oct4 (Pou5f1 – Mouse Genome Informatics), Nanog, Sox2, Tead4, Gata3, Cdx2, Eomes and Elf5 in cell fate specification, pluripotency and trophoblast development (Nichols et al., 1998; Russ et al., 2000; Mitsui et al., 2003; Niwa et al., 2005; Strumpf et al., 2005; Dietrich and Hiiragi, 2007; Yagi et al., 2007; Ng et al., 2008; Nishioka et al., 2008; Parfitt and Zernicka-Goetz, 2010; Ralston et al., 2010). Interestingly, many of these TFs are not essential for blastocyst formation per se, but are necessary for implantation or postimplantation development. Functional studies uncovered a potential dual role for a subset of these TFs in blastocyst formation and cell fate specification. For example, ablation of Tead4, Gata3 or Sox2 causes developmental arrest at the morula stage and defects in cell fate specification (Nishioka et al., 2008; Home et al., 2009; Keramari et al., 2010). Nonetheless, their precise role in the cellular processes associated with blastocyst formation is unknown. Thus, a fundamental question persists: which developmental TFs directly govern the cellular processes associated with blastocyst formation in mammals (i.e. TJ and fluid accumulation)?
One such TF is AP-2γ (Tcfap2c; Tfap2c – Mouse Genome Informatics), a DNA-binding protein that acts as both an activator and repressor of gene transcription (Eckert et al., 2005). Tcfap2c is expressed in both the oocyte and preimplantation embryo (Winger et al., 2006). During blastocyst formation, Tcfap2c is enriched in the TE lineage (Kuckenberg et al., 2010). Loss of zygotic Tcfap2c results in abnormal placental development and embryonic lethality between days 7.5 and 8.5 (Auman et al., 2002; Winger et al., 2006). To test whether maternally derived Tcfap2c was required for preimplantation development, Winger et al. (Winger et al., 2006) crossed mice with a ZP3-Cre transgene and floxed Tcfap2c allele to generate female mice lacking Tcfap2c in their oocytes. Subsequently, two of these females were mated with Tcfap2c-null males to generate embryos that lacked both maternal and zygotic Tcfap2c and no preimplantation phenotype was observed. However, the results of this experiment are inconclusive because only seven preimplantation embryos from a total of two mice were evaluated and studies to confirm that Tcfap2c protein was effectively ablated using this approach were not performed. Therefore, we further investigated whether Tcfap2c is required for preimplantation development.
Here, we report that Tcfap2c is essential for the morula-to-blastocyst transition in mouse preimplantation embryos. An RNAi approach was employed to effectively ablate maternally and zygotically derived Tcfap2c protein. Using a combination of bioinformatics and transcriptional analysis, we found that Tcfap2c is required for the proper expression of a diverse family of genes involved in cell polarity, TJ biogenesis, fluid accumulation and cellular proliferation. Furthermore, functional and ultrastructural analyses revealed that Tcfap2c is obligatory for the formation of TJ complexes and paracellular sealing in the TE epithelium crucial to blastocyst formation. To our knowledge, this is the first study to describe the function of a single TF in controlling TJ assembly, fluid accumulation and cell proliferation in early mammalian embryogenesis.
MATERIALS AND METHODS
Embryo collection and microinjection
Mouse embryos were derived from superovulated B6D2/F1 females mated with B6D2/F1 males (Charles River Laboratories, Wilmington, MA, USA). Fertilized one-cell embryos were collected at 16-17 hours post-human chorionic gonadotropin treatment (hph) and then cultured in modified KSOM medium (EMD Millipore, Billerica, MA, USA) under mineral oil at 37°C in a humidified atmosphere of 5% O2, 5% CO2, 90% N2. Microinjection was carried out as described previously (Wang et al., 2010). Briefly, 5-10 pl 100 μM Tcfap2c siRNA (siGenome and On-target plus; Dharmacon, Lafayette, CO, USA), 100 μM p21 siRNA (Dharmacon), 100 μM control siRNA (Dharmacon) or 0.7 μg/μl Tcfap2c mRNA was injected into the cytoplasm of one-cell zygote embryos using a PL100 picoinjector (Harvard Apparatus, Hollistan, MA, USA) at 19-21 hph. Animal care was in accordance with the institutional guidelines of Michigan State University.
siRNA sequences
siRNA sequences (5′-3′) were as follows. Tcfap2c (pool 1): AAGCUGAGUUCCCUAGUAA, GCACGGGACUUCGCCUAUG, AGCGGUGGCUGACUAUUUA and CCGCAGUGCAGAAUUAUAU. Tcfap2c (pool 2): UGAAAGGUGCUACGAGUUU, CAGAUAAAGGGAUCGAUCA, GCAAAGGACCCAUUUCGAU and UGAGAAAUGGGAUUCGAUU. p21: CGAGAACGGUGGAACUUUG, GAACAUCUCAGGGCCGAAA, GGAGCAAAGUGUGCCGUUG and GGUGAUGUCCGACCUGUUC. Control: UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA and UGGUUUACAUGUCGACUAA.
Embryonic stem (ES) cell culture and differentiation
A Cdx2-inducible ES cell line provided by Coriell Cell Repositories (Camden, NJ, USA) was cultured on mitomycin-treated puromycin-resistant mouse embryonic fibroblasts in ES cell medium supplemented with 0.2 mg/ml doxycycline and 1.0 mg/ml puromycin (Nishiyama et al., 2009). Prior to Cdx2 induction, cells were switched onto gelatin and cultured in the presence of 1.5 mg/ml puromycin for 3 days. Transgene expression was induced by removal of doxycycline and was verified by immunocytochemistry and quantitative (q) RT-PCR analysis (Wang et al., 2010).
Microarray and bioinformatics analysis
Microarray analysis was carried out utilizing Affymetrix 430v2 gene chips as described (Kidder et al., 2009). Blastocyst array data were compared with eight-cell embryo array data using GeneSpring software (Agilent Technologies, Santa Clara, CA, USA). Genes that were upregulated ≥2-fold (P<0.05) in blastocysts were cross-referenced with a published Tcfap2c ChIP-Chip dataset in trophoblast stem (TS) cells (Kidder and Palmer, 2010). Genes that were both upregulated in blastocysts and bound by Tcfap2c in TS cells were then subjected to Tcfap2c binding motif analysis using TRANSFAC and ExPlain 3.0 (BIOBASE, Wolfenbüttel, Germany). Functional annotation and gene ontology analysis were conducted using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) and GOrilla (http://cbl-gorilla.cs.technion.ac.il/). Microarray data are available at GEO under accession number GSE41925.
Gene expression analysis by qRT-PCR
Total RNA was isolated using the PicoPure RNA isolation kit (Arcturus, Mountain View, CA, USA) or an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA synthesis was carried out using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). qRT-PCR analysis was conducted utilizing TaqMan probes (Applied Biosystems, Foster City, CA, USA) or gene-specific designed primers (SYBR Green detection) and a StepOnePlus real-time PCR system (Applied Biosystems). Ubtf and Eef1a1 were used as endogenous controls for embryos and Cdx2-inducible ES cells, respectively. To determine the developmental expression of Tcfap2c transcripts in metaphase II (MII) eggs to blastocysts, samples were normalized to exogenous GFP that was spiked into each sample prior to RNA isolation. SYBR Green PCR primer sequences are listed in supplementary material Table S1.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed as previously described (Wang et al., 2010). Briefly, at 0, 48 and 96 hours after Cdx2 induction, samples were collected for ChIP and gene expression analysis. ES cells were sonicated and chromatin extracts were frozen at –80°C. ChIP was carried out using a Tcfap2c antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a rabbit IgG (Millipore) and chromatin extracts equivalent to 2×106 cells. ChIP samples were quantified by qPCR (SYBR Green Master Mix; Applied Biosystems) and ChIP-qPCR data were normalized using the percent input method.
Immunocytochemistry
Preimplantation embryos were fixed with 3.7% paraformaldehyde for 20 minutes, permeabilized with PBS containing 0.1% Tween 20 for 15 minutes, blocked with PBS containing 0.1% BSA for 1 hour at room temperature, and incubated with primary antibodies in blocking solution overnight at 4°C, followed by incubation with Alexa Fluor 488 and 594 (Molecular Probes, Eugene, OR, USA) secondary antibodies. Embryos were then mounted in Vectashield containing DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA, USA) and imaged using a spinning disc confocal module (CARV; Atto Bioscience, Rockville, MD, USA) with MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). Validated primary antibodies for Tcfap2c, Oct4, p21, and Pard6b were obtained from Santa Cruz. Antibodies for Cdh1, Cldn4 and Tjp2 were from Invitrogen. Aqp3 and Krt18 antibodies were purchased from Alpha Diagnostic (San Antonio, TX, USA) and Abcam (Cambridge, MA, USA), respectively. Cdx2 antibody was obtained from Biogenex (San Ramon, CA, USA). F-actin was stained using Alexa Fluor 586 phalloidin (Molecular Probes).
TJ permeability assay by FITC-dextran uptake
To investigate the effects of Tcfap2c depletion on TJ permeability, control and Tcfap2c knockdown (KD) embryos were cultured until 120 hph, and then the blastocysts were incubated in modified KSOM medium containing 1 mg/ml 4 kDa FITC-dextran (Sigma-Aldrich, St Louis, MO, USA) for 10 minutes. Following the incubation, the blastocysts were immediately washed and visualized under an inverted fluorescence microscope.
Transmission electron microscopy (TEM)
Control and Tcfap2c KD morulae at 90 hph were washed with 0.1 M cacodylate buffer (pH 7.4) and fixed with 2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 M cacodylate buffer for 2 hours at room temperature. The fixed embryos were embedded in 1% agarose. The agarose blocks were postfixed in 1% (w/v) osmium tetraoxide in 0.1 M cacodylate buffer for 30 minutes. The postfixed specimens were dehydrated through a graded ethanol series in the cacodylate buffer, transferred into mixtures of Spurr’s resin and ethanol at room temperature and polymerized for 2 days at 60°C, followed by preparation of ultrathin sections using a Power Tome XL ultramicrotome (RMC, Boekeler Instruments, Tucson, AZ, USA) and examination by a JEOL100 CXII transmission electron microscope (Japan Electron Optics Laboratories, Tokyo, Japan).
Generation of chimeras by aggregation
The zona pellucida of control and Tcfap2c KD embryos at the compacted eight-cell stage was removed by acid Tyrode solution (Sigma) and two zona pellucida-free embryos were paired in microwells to generate chimeric embryos that consisted of: control-control, control-Tcfap2c KD, or Tcfap2c KD-KD embryos. To track the fate of Tcfap2c KD blastomeres, GFP mRNA and Tcfap2c siRNA were co-injected at the one-cell stage. Chimeric embryos were visualized using an inverted microscope equipped with UV light and an FITC filter.
Statistical analysis
Data were analyzed by analysis of variance (ANOVA) or χ2 test using INSTAT3 (GraphPad Software, San Diego, CA, USA). The data are presented as mean ± s.e.m. P<0.05 was considered statistically significant unless otherwise stated.
RESULTS
Combined depletion of maternal and zygotic Tcfap2c blocks blastocyst formation
A recent study demonstrated that forced expression of Tcfap2c in mouse ES cells could induce a TE cell fate, indicating that Tcfap2c may promote development of the TE epithelium (Kuckenberg et al., 2010). However, the biological role of Tcfap2c during blastocyst formation has not been clarified. To address this, we first examined the expression of Tcfap2 during preimplantation development. Consistent with previous studies (Auman et al., 2002; Winger et al., 2006), Tcfap2c transcripts were detected at all stages of preimplantation development (Fig. 1A). Notably, after fertilization, the levels of maternal transcripts were significantly reduced. Furthermore, from the four-cell stage onward there was a dramatic increase in zygotic Tcfap2c mRNA, peaking at the morula stage. Likewise, Tcfap2c protein was detected at all stages of preimplantation development, and at the blastocyst stage the levels of Tcfap2c protein were much greater in the TE than in the Oct4-positive inner cell mass (ICM) (Fig. 1B). Interestingly, the localization of Tcfap2c was heterogeneous in the TE; Tcfap2c was expressed in the mural TE, but was not detectable in the polar TE (supplementary material Fig. S1).
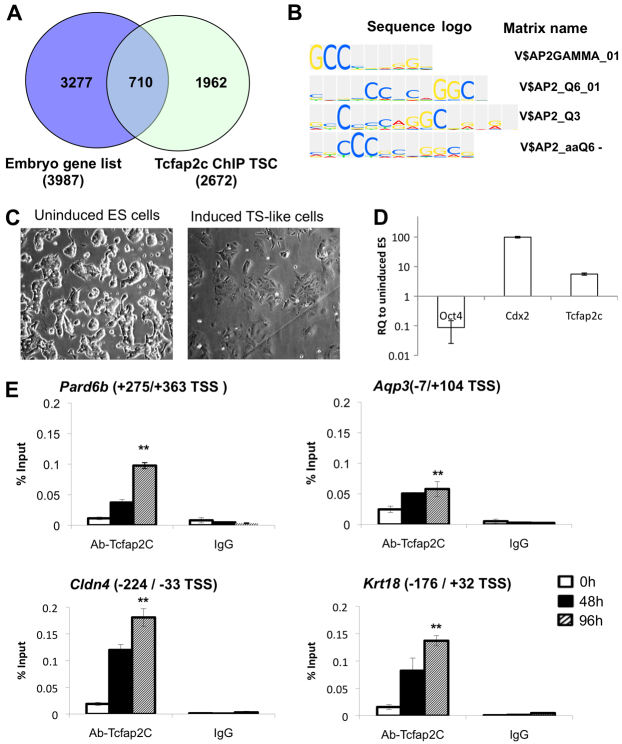
Fig. 1.
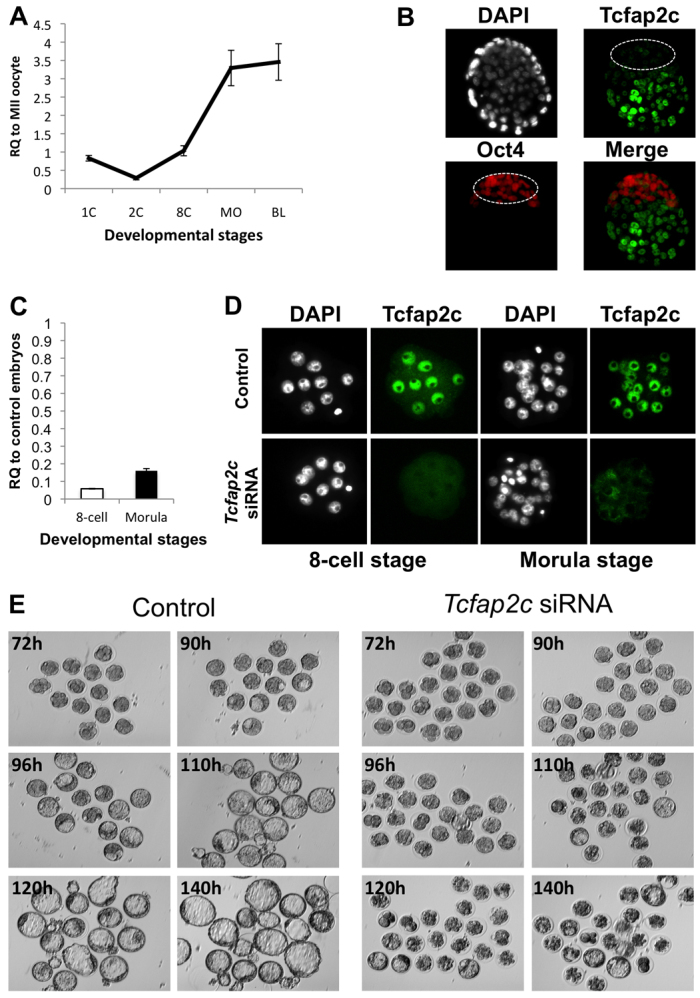
Developmental expression and RNAi-mediated ablation of Tcfap2c in mouse preimplantation embryos. (A) qRT-PCR analysis of Tcfap2c transcripts in mouse MII oocytes and preimplantation embryos. Expression data from each stage were normalized to exogenous GFP and are relative to MII oocytes. 1C, one-cell zygote; 2C, two-cell; 8C, eight-cell; MO, morula; BL, blastocyst. (B) Immunocytochemistry (ICC) analysis revealed that Tcfap2c protein is enriched in the blastocyst mural trophectoderm (TE). Blastocysts were double stained for Oct4 to label the inner cell mass (ICM; outlined). Nuclei were counterstained with DAPI. (C) Validation of siRNA-mediated knockdown (KD) of Tcfap2c transcripts in eight-cell and morula stage embryos by qRT-PCR. (D) Confirmation of Tcfap2c protein ablation in eight-cell and morula stage embryos by ICC. (E) Depletion of Tcfap2c blocks blastocyst formation. Representative images of Tcfap2c KD and control embryos cultured for 72 to 140 hph. Error bars indicate mean ± s.e.m. RQ, relative quantification.
Next, we examined the function of Tcfap2c during preimplantation development. Previously, it was demonstrated that knockout of zygotic Tcfap2c resulted in embryonic lethality at around day 7.5 of development (Auman et al., 2002; Winger et al., 2006). However, in these studies expression of maternal Tcfap2c protein might have masked any potential phenotype during preimplantation development. To circumvent this, we used an RNAi approach to deplete both maternally and zygotically derived Tcfap2c. One-cell embryos were injected with either 100 μM Tcfap2c or control siRNA and cultured for 2 to 4 days. Microinjection of Tcfap2c siRNA resulted in a 96% and 80% reduction in Tcfap2c mRNA and in the complete loss of protein as assayed by immunocytochemistry (ICC) at the eight-cell and morula stages, respectively (Fig. 1C,D). Importantly, Tcfap2c siRNA did not target Tcfap2a transcripts, demonstrating that the siRNAs were specific to Tcfap2c mRNA (supplementary material Fig. S2). Remarkably, Tcfap2c knockdown (KD) embryos underwent compaction and developed normally to the morula stage (83.71% for KD versus 93.64% for control; P>0.05). However, only a small percentage of Tcfap2c KD morulae formed blastocysts compared with control embryos injected with scrambled siRNA (Fig. 1E; 13.73% versus 92.64%; P<0.001). Moreover, Tcfap2c KD embryos that developed to the blastocyst stage were not able to fully expand and hatch after extended culture (data not shown).
To rule out any potential siRNA off-targeting effects, several experiments were carried out. First, we showed by qRT-PCR that the control siRNA did not target Tcfap2c transcripts in injected embryos (supplementary material Fig. S2). Second, different concentrations of Tcfap2c siRNA were injected into one-cell embryos and KD efficiency and effects on blastocyst formation were assayed (supplementary material Fig. S2). These experiments revealed that 100 μM was the most effective concentration for depleting Tcfap2c and blocking blastocyst formation. Third, rescue experiments were performed by co-injection of Tcfap2c mRNA and Tcfap2c siRNA (pool 1) into one-cell embryos (supplementary material Fig. S3). These experiments revealed that a higher percentage of one-cell embryos co-injected with Tcfap2c siRNA and mRNA developed to the blastocyst stage compared with embryos injected with Tcfap2c siRNA alone (57.69% versus 13.73%; P<0.05). Fourth, a second pool of Tcfap2c siRNA was injected into one-cell embryos (supplementary material Fig. S3). Targeting of Tcfap2c via this second pool also reduced blastocyst formation.
As stated above, a major difference between Tcfap2c KD and control embryos was the ability to form a blastocoel cavity during the morula-to-blastocyst transition. Therefore, we monitored the timing of cavity formation in Tcfap2c KD and control morulae from 76 to 120 hph. As shown in supplementary material Fig. S3, ∼25% of control embryos formed cavities at 90 hph, and by 120 hph over 90% of embryos formed a blastocoel cavity. By contrast, only 3% of Tcfap2c KD embryos formed a cavity at 90 hph, and by 120 hours only 36% of embryos formed a small cavity. To better understand the etiology of this phenotype, we used a time-lapse camera to record Tcfap2c KD and control embryos undergoing cavity formation in real-time (supplementary material Movie 1). This revealed that Tcfap2c KD morulae repeatedly initiated cavity formation but failed to form an expanded blastocoel cavity. Altogether, these data demonstrate that Tcfap2c is required for blastocyst formation.
Tcfap2c target genes in the preimplantation embryo
Because Tcfap2c KD embryos arrest at the morula stage, we hypothesized that Tcfap2c is required for transcriptional regulation of key genes involved in the morula-to-blastocyst transition. To address this, we (1) conducted transcriptome analysis on normal embryos to identify genes upregulated between the eight-cell and blastocyst stage and (2) cross-referenced this gene list with a published Tcfap2c ChIP-Chip dataset (n=2673 genes) in TS cells (Kidder and Palmer, 2010). We identified a total of 3987 genes that were upregulated ≥2-fold between the eight-cell and blastocyst stages. When cross-referenced with the Tcfap2c ChIP-Chip dataset we found a total of 710 common genes that were both bound by Tcfap2c in TS cells and upregulated in blastocysts (Fig. 2A; supplementary material Table S2). To confirm that these genes were enriched with Tcfap2c binding motifs, we used TRANSFAC and ExPlain 3.0 (Fig. 2B). This analysis confirmed that Tcfap2c binding motifs were enriched in this gene list.
Fig. 2.
Tcfap2c binding is enriched on genes that are upregulated during the morula-to-blastocyst transition. (A) Venn diagram illustrating genes that are both upregulated during blastocyst formation (embryo gene list) and enriched for Tcfap2c binding in trophoblast stem cells (TSC). (B) Tcfap2c motif analysis of putative Tcfap2c target genes using ExPlain 3.0. (C) Representative images of Cdx2-inducible ES cells (0 hours) differentiating into TE-like cells (96 hours post-induction). (D) qRT-PCR analysis of Cdx2-inducible ES cells at 0 and 96 hours post-induction. Oct4 (ICM marker) was downregulated, but Cdx2 (TE marker) and Tcfap2c transcripts were upregulated in TE-like cells at 96 hours post-induction. (E) ChIP analysis in Cdx2-inducible ES cells demonstrated that Tcfap2c is recruited to target gene promoters during differentiation into TE-like cells at 48 to 96 hours. Error bars indicate mean ± s.e.m. **P<0.01. TSS, transcription start site.
Next we used a web-based application tool called GOrilla (Gene Ontology Enrichment Analysis and Visualization) to obtain meaningful and systematic biological information from the upregulated/Tcfap2c-bound gene list. This analysis revealed that epithelial cell differentiation, cellular response to cytokine stimulus, and cytokine-mediated signaling pathway presented a unique set of enriched GO terms (P<10–3), suggesting that Tcfap2c might regulate genes involved in epithelial differentiation (supplementary material Table S3). Moreover, we utilized the DAVID (Database for Annotation, Visualization and Integrated Discovery) annotation pathway analysis tool to identify possible functions or molecular interactions of the listed genes. According to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database, we found that these genes are highly involved in cell communication, including TJs, focal adhesion and adherens junctions (supplementary material Table S4). This analysis identified putative Tcfap2c target genes involved in TJ assembly and paracellular sealing (supplementary material Fig. S4). Finally, in line with this systematic data mining and morphological observation, such as the delayed or retarded cavity formation, we selected 19 genes that are responsible for cell polarity, paracellular sealing and fluid accumulation. This list contains genes that are important for adherens junctions (Cdh1, also known as E-cadherin), cellular polarization (Pard6b, Rock2), cytoskeleton organization (Krt18, Fn1), TJ assembly (Cldn4, Cldn6, Cldn7, Tjp1, Tjp2, Ocln, Inadl, Jam2, F11r), ion gradients (Atp1b1, Atp1b3, Atp1a1) and the formation of membrane H2O channels (Aqp3, Aqp9). This list of selected genes with a brief description of their known function/phenotype is presented in supplementary material Table S5.
To test whether Tcfap2c can directly bind to target gene promoters during TE formation, ChIP analysis was conducted in ES cells differentiating into TE-like cells. To accomplish this, we utilized a doxycycline-controllable, Cdx2-inducible ES cell line as a model system for the developing TE epithelium (Nishiyama et al., 2009; Wang et al., 2010). ChIP primers flanking Tcfap2c binding motifs were designed for a subset of genes. As a negative control, primers were designed for an intergenic region that does not contain binding motifs. At 0, 48 and 96 hours after Cdx2 induction, cells were isolated for ChIP and gene expression analysis (Fig. 2C,D). We found that at 48 and 96 hours post-induction, Tcfap2c was largely recruited to the promoters of Tjp2, Pard6b, Cldn4, Krt18 and Aqp3 (P<0.01), whereas Tcfap2c enrichment was not observed at an intergenic region (Fig. 2E; supplementary material Fig. S5). Moreover, at these same time points there was a significant increase in mRNA expression for these genes (P<0.05) (supplementary material Fig. S6). Furthermore, examination of the developmental expression of Tjp2, Pard6b, Cldn4, Krt18 and Aqp3 in preimplantation embryos revealed that these genes were upregulated between the eight-cell and blastocyst stage (supplementary material Fig. S6). Altogether, these data suggest that Tcfap2c might play an important role in the transcriptional regulation of genes that potentiate blastocyst formation.
Tcfap2c is required for transcriptional regulation of genes important for cell polarity, TJ assembly and fluid accumulation
As described above, a significant proportion of Tcfap2c KD embryos arrested at the morula stage and failed to form and/or maintain a blastocoel cavity. Importantly, the phenotype of Tcfap2c KD embryos resembles that observed in several other loss-of-function studies in mice and other mammalian species, implying that Tcfap2c might function as a core regulator of blastocyst formation genes. To test this, we utilized qRT-PCR analysis to determine the expression levels of 20 putative Tcfap2c target genes in Tcfap2c KD embryos (Fig. 3A). We first evaluated the expression of Pard6b, Pard3, Fn1, Rock2 and Krt18. These genes are implicated in cell polarity and cytoskeleton organization and are important for blastocyst formation in mouse and/or bovine embryos (Kawagishi et al., 2004; Goossens et al., 2007; Alarcon, 2010; Goossens et al., 2010). The expression of Pard6b, Rock2, Fn1 and Krt18 mRNA was significantly reduced in Tcfap2c KD embryos (P<0.05), suggesting that Tcfap2c is required for the correct expression of genes during the morula-to-blastocyst transition. By contrast, the levels of Pard3 transcripts were unchanged in Tcfap2c KD embryos.
Fig. 3.
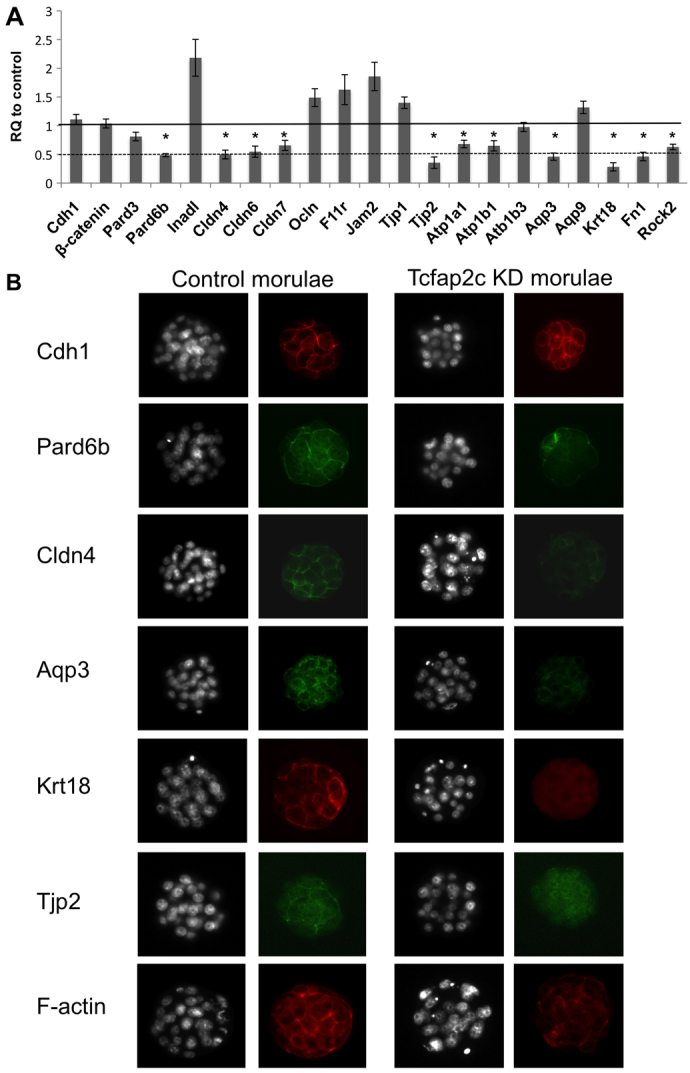
Tcfap2c regulates genes that are crucial for cell polarity, TJ assembly and ion gradients. (A) qRT-PCR analysis of Tcfap2c target genes in Tcfap2c KD and control morulae (90 hph). Genes important for cell polarity, tight junction (TJ) assembly and fluid gradient were significantly downregulated in Tcfap2c KD embryos. Error bars indicate mean ± s.e.m. *P<0.05. (B) Protein expression and localization of Tcfap2c target genes in Tcfap2c KD and control morulae. Depletion of Tcfap2c disrupted the expression and subcellular localization of Pard6b, Cldn4, Aqp3, Krt18, Tjp2 and F-actin. Embryos were counterstained with DAPI.
To further examine the molecular basis of the Tcfap2c KD phenotype, we examined the expression of genes necessary for TJ assembly and fluid accumulation (Fig. 3A). The assembly of TJ complexes is crucial for paracellular sealing of the TE epithelium and formation of a fluid-filled cavity (Saitou et al., 2000; Kim et al., 2004; Thomas et al., 2004; Moriwaki et al., 2007; Katsuno et al., 2008). We observed a significant reduction in the expression levels of Cldn4, Cldn6, Cldn7 and Tjp2 in Tcfap2c KD morulae (P<0.05). By contrast, the levels of Tjp1, Ocln and Jam2 mRNA were increased in Tcfap2c KD morulae (P<0.05). The expression of F11r (previously known as Jam1) did not differ between Tcfap2c KD and control morulae (P=0.07). In addition to the barrier function of TJs, the establishment of a trans-TE ionic gradient by Na/K-ATPase and H2O transporters is essential for the accumulation of fluid within the blastocoel cavity. We found that Atp1a1 and Atp1b1 transcripts were reduced (P<0.05), whereas the expression of Atp1b3 was unaffected (P>0.05), in Tcfap2c KD embryos. Likewise, the expression of Aqp3 was significantly reduced (P<0.05), whereas Aqp9 expression was unchanged (P>0.05), in Tcfap2c KD embryos. Finally, we examined the expression of Cdh1 and Ctnnb1 (β-catenin), two genes required for adherens junctions. These genes were unchanged in Tcfap2c KD morulae.
Subsequently, we analyzed the expression and localization of Tcfap2c target genes by ICC analysis. We selected genes that were downregulated at the transcript level by ≥50% in Tcfap2c KD embryos. Consistent with the reduction in target gene mRNA there was a dramatic reduction in the expression of Pard6b, Tjp2, Cldn4, Krt18 and Aqp3 proteins (Fig. 3B). For example, in control morulae Pard6b expression was observed as a continuous band, and at the apical pole of the outer cells Pard6b accumulated at the cell-to-cell contact sites. By contrast, in Tcfap2c KD morulae the overall expression of Pard6b was reduced and lost altogether at the cell-to-cell contacts. Likewise, in control morulae the TJ proteins Cldn4 and Tjp2 were distributed as a continuous belt around each blastomere and were concentrated at the cell-to-cell contacts in the apical region. By contrast, in Tcfap2c KD morulae the expression of Cldn4 and Tjp2 was severely reduced and not visible in the apical region at the cell-to-cell contacts. The intermediate filament protein Krt18 was highly expressed and localized to the cell membrane in control morulae, whereas in Tcfap2c KD embryos it was lost completely. Expression of Aqp3 was localized to the basolateral region in control morulae, whereas in Tcfap2c KD morulae Aqp3 expression was diminished. Finally, we stained control and Tcfap2c KD morulae for Cdh1 and F-actin (Fig. 3B). Consistent with the qRT-PCR and morphological data, the expression and localization of Cdh1 were unimpaired in Tcfap2c KD embryos. However, F-actin was highly disorganized in Tcfap2c KD embryos compared with control embryos. This is consistent with observations made in several other loss-of-function studies on TJ and cell polarity-related proteins (Madan et al., 2007; Sheth et al., 2008; Alarcon, 2010). Altogether, these results demonstrate that Tcfap2c is required for the proper expression and localization of key TJ, cell polarity, cytoskeletal and H2O channel proteins during the morula-to-blastocyst transition.
Functional evidence that Tcfap2c is required for TJ assembly and paracellular sealing during blastocyst formation
During blastocyst formation TJ complexes are essential for paracellular sealing, which excludes the transport of higher molecular weight molecules from the apical region into the basolateral region and allows the formation of a fluid-filled cavity (Watson and Barcroft, 2001). These complexes are assembled during the morula-to-blastocyst transition and become functional at the blastocyst stage (Eckert and Fleming, 2008). Because key TJ and cytoskeletal proteins are downregulated in Tcfap2c KD morulae we hypothesized that the function and/or integrity of these complexes would be compromised. To test this we analyzed the permeability of TJ complexes using a 4 kDa FITC-dextran assay (Moriwaki et al., 2007; Giannatselis et al., 2011). Since most Tcfap2c KD embryos do not form blastocysts, we confined our analysis to partially recovered blastocysts from Tcfap2c KD embryos; despite developing to the blastocyst stage this subset of embryos contained a reduced complement of Tcfap2c, Pard6b, Krt18 and Cldn4 (supplementary material Fig. S7). A higher percentage of Tcfap2c KD blastocysts were permeable to FITC-dextran than control blastocysts (55±4% versus 25±7%; Fig. 4A,B), suggesting that the function of TJ complexes was impaired in Tcfap2c KD embryos. In addition, we analyzed a subset of Tcfap2c KD embryos that had been rescued by Tcfap2c mRNA injection. Importantly, Tcfap2c KD embryos that were rescued by mRNA injection exhibited functional paracellular sealing in the TE epithelium, with normal expression and localization of Cldn4 and F-actin (supplementary material Fig. S8). The average total cell number of the Tcfap2c mRNA-rescued group was lower than that of control blastocysts (63.20 versus 71.38; P<0.05).
Fig. 4.
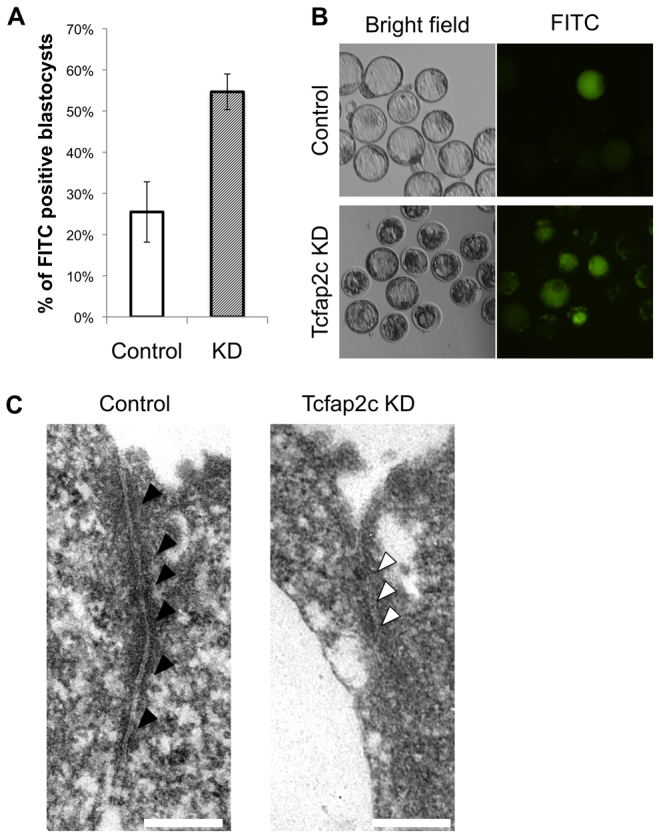
Disruption of TJ complexes and paracellular sealing in Tcfap2c KD embryos. (A) A 4 kDa FITC-conjugated dextran was used to test the permeability of TJs in Tcfap2c KD and control blastocysts. A difference in permeability was observed between Tcfap2c KD and control blastocysts. Error bars indicate mean ± s.e.m. (B) Representative brightfield and fluorescence images of Tcfap2c KD and control blastocysts subjected to the FITC-dextran assay. (C) TEM analysis of TJ complexes in Tcfap2c KD and control morulae. At 90 hph, several electron-dense plaques (black arrowheads) were observed at apical cell-to-cell contact sites in control embryos. By contrast, in Tcfap2c KD embryos the electron-dense regions (white arrowheads) at the apical cell-to-cell contact site were absent. Scale bars: 200 nm.
To examine the ultrastructural integrity of TJ complexes in Tcfap2c KD morulae, we performed TEM in Tcfap2c KD and control embryos at the morula stage. These experiments revealed that Tcfap2c KD morulae failed to form TJ complexes at the apical cell boundaries, consistent with the downregulation and/or mislocalization of TJ complex proteins in these embryos (Fig. 4C). Collectively, these results demonstrate that Tcfap2c is required for the correct expression and assembly of TJ complex proteins during blastocyst formation and that dysregulation of these genes results in a failure to establish paracellular sealing.
Aggregation of control with Tcfap2c KD eight-cell embryos rescues blastocyst formation
Since Tcfap2c is required for the proper expression of TJ proteins and assembly of functional TJ complexes, we hypothesized that the aggregation of control eight-cell embryos (Tcfap2c+) with Tcfap2c KD eight-cell embryos would restore blastocoel formation in Tcfap2c KD embryos. We produced three types of chimeras by aggregation: (1) control-control (C:C) embryos; (2) control-Tcfap2c KD (C:K) embryos; and (3) Tcfap2c KD-Tcfap2c KD (K:K) embryos. Regardless of the type of chimera, over 90% of the paired embryos successfully aggregated and formed chimeras (Fig. 5A). More importantly, over 90% of C:K paired chimeras overcame the morula-to-blastocyst transition block and developed to the blastocyst stage at a rate that was similar to that of C:C chimeras (P>0.05) (Fig. 5A,B). By contrast, only 15% of K:K paired chimeras formed blastocyst-like structures and the majority of chimeras arrested at the morula stage (P<0.05). To confirm whether features of the TE epithelium were restored in rescued blastocysts, C:K and C:C chimeras were stained for F-actin, Cldn4 and Cdx2 (Fig. 5C,D). This revealed that in C:K blastocysts the expression and localization of Cldn4 and F-actin were restored in the Cdx2-positive TE epithelium.
Fig. 5.
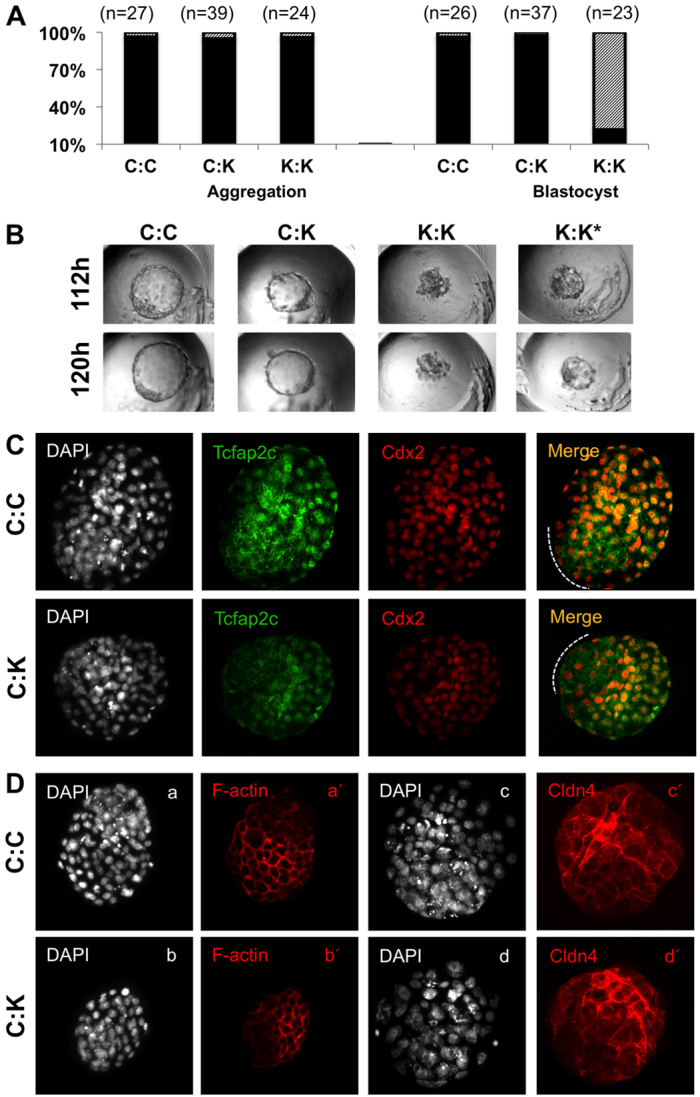
Aggregation of control embryos with Tcfap2c KD embryos rescues blastocyst formation. (A) Developmental rates of control eight-cell embryos aggregated with Tcfap2c KD eight-cell embryos. To the left is shown the percentage of aggregated (black) or unaggregated (hatched) embryos and to the right is shown the percentage of chimeric embryos that developed to blastocyst stage (black) or failed to reach blastocyst (hatched). (B) Representative brightfield images of chimeric embryos produced by aggregation of C:C, C:K and K:K embryos at 112 and 120 hph. Asterisk denotes blastocyst-like vesicle in K:K chimeras. (C) ICC analysis of the subcellular localization of Tcfap2c and Cdx2. Orange indicates merge of Tcfap2c and Cdx2. Dotted line indicates the polar TE. (D) ICC analysis of F-actin and Cldn4 in C:C and C:K embryos (lowercase letters denote different groups of C:C and C:K aggregated embryos). C:C, aggregation of two control embryos; C:K, aggregation of control and Tcfap2c KD embryo; K:K, aggregation of two Tcfap2c KD embryos.
To determine whether control blastomeres contribute to the TE epithelium of C:K blastocysts two experiments were carried out. First, ICC analysis revealed that Tcfap2c was localized mainly in the TE of C:C and C:K chimeras (Fig. 5C). Second, GFP mRNA was co-injected with Tcfap2c siRNA into embryos prior to aggregation with control eight-cell embryos (C:K-GFP labeled). In C:K blastocysts, GFP expression was predominantly localized to the Oct4-positive ICM and not the TE, demonstrating that the control cells contributed predominantly to the TE lineage (supplementary material Fig. S9). Collectively, these data demonstrate that developmental arrest of Tcfap2c KD morulae can be reversed through cell-to-cell aggregation.
Tcfap2c negatively regulates p21 transcription to promote cellular proliferation during the morula-to-blastocyst transition
In addition to TJ assembly and establishment of trans-TE ion gradients, blastocyst formation requires an increase in cellular proliferation for blastocoel expansion (Copp, 1978). The observed developmental delay or arrest in Tcfap2c KD embryos suggested that cell cycle regulation was altered. Moreover, in blastocysts derived from C:K chimeras there was a reduction in total cell number (106.31±3.42 for C:K versus 132.98±7.79 for C:C; P<0.05). To more accurately assess this phenotype we calculated the total cell number in Tcfap2c KD and control embryos at the morula stage, which differed significantly (26.52±0.64 for control versus 21.21±0.49 for KD; P<0.01). To identify potential target genes involved in proliferation, we focused on genes that negatively regulate proliferation, such as p21, p27 and p57 (Cdkn1a, Cdkn1b and Cdkn1c – Mouse Genome Informatics), which are important in the regulation of cell cycle progression (Sherr and Roberts, 1995; Sherr and Roberts, 1999). A previous study demonstrated that increased levels of p21 expression during preimplantation development was associated with arrest around the morula stage (Adiga et al., 2007). Using TRANSFAC and ExPlain and ChIP assays, we identified a Tcfap2c binding site in the p21 promoter (Fig. 6A). Remarkably, qRT-PCR analysis revealed that p21 transcripts were upregulated more than 8-fold in Tcfap2c KD morulae compared with control embryos (Fig. 6B), whereas expression of p27 and p57 was unaffected (data not shown). ICC revealed increased expression and nuclear localization of p21 in Tcfap2c KD morulae versus controls (Fig. 6C). Because p21 expression can be regulated by p53-dependent and -independent mechanisms (Gartel and Tyner, 1999), we ascertained whether upregulation of p21 expression was mediated by p53 (Trp53 – Mouse Genome Informatics). qRT-PCR analysis revealed that p53 transcript levels did not differ between Tcfap2c KD and control morulae. Furthermore, we evaluated the expression of Hdac1, a negative regulator of p21 expression (Ma and Schultz, 2008; Zupkovitz et al., 2010). The levels of Hdac1 did not differ between Tcfap2c KD and control embryos (Fig. 6B), suggesting that the increased expression of p21 is mediated via Tcfap2c and not by other mechanisms.
Fig. 6.
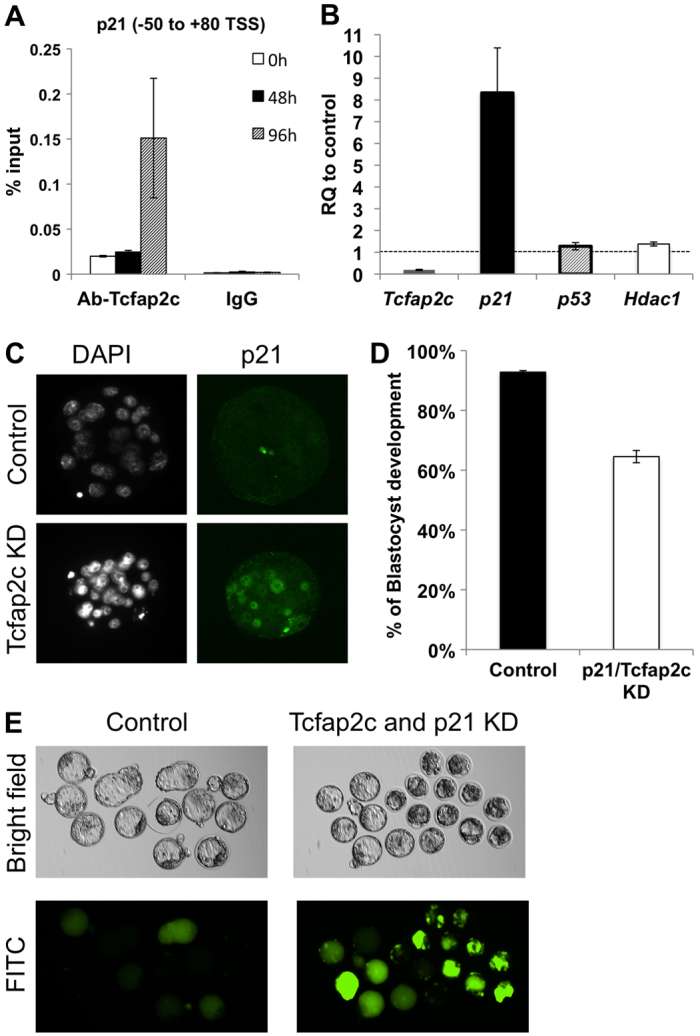
Tcfap2c regulates cellular proliferation via direct repression of p21. (A) ChIP analysis of Tcfap2c binding to the p21 proximal promoter. (B) p21 is upregulated in Tcfap2c KD morulae. Upregulation of p21 is independent of p53 and Hdac1 expression. (C) Expression and subcellular localization of p21 protein in Tcfap2c KD and control morulae. (D) Combined KD of p21 and Tcfap2c partially rescues blastocyst formation. (E) Representative brightfield and fluorescence images of p21/Tcfap2c KD and control blastocysts subjected to the FITC-dextran assay. Error bars indicate s.e.m.
To test whether inhibiting the increase in p21 expression could rescue the preimplantation arrest phenotype of Tcfap2c KD embryos, we co-injected p21 siRNA and Tcfap2c siRNA at the one-cell stage and cultured them for 4 days. Importantly, p21 siRNA efficiently reduced the levels of p21 mRNA without compromising the efficacy of the Tcfap2c siRNA (data not shown). A higher percentage of p21/Tcfap2c KD embryos developed to the blastocyst stage (65%, versus 93% in control embryos; Fig. 6D). However, after extended culture these embryos failed to fully expand and exhibited increased uptake of FITC-dextran into the blastocoel cavity (Fig. 6E), suggesting that the barrier function in these blastocysts was still impaired. Collectively, these results demonstrate that, in addition to regulating the expression of TJ and cell polarity genes, Tcfap2c negatively regulates p21 transcription to promote blastocyst formation in mice.
DISCUSSION
Results presented here reveal a previously unknown role of Tcfap2c in early embryonic development. Our results in preimplantation mouse embryos demonstrate that: (1) Tcfap2c is required for blastocyst formation; (2) Tcfap2c binds to and regulates a diverse set of genes involved in cell polarity, TJs and fluid accumulation; (3) Tcfap2c is necessary for the assembly of functional TJ complexes in TE apical cell membranes; and (4) Tcfap2c controls cellular proliferation via negative regulation of p21. We propose a model in which Tcfap2c acts in a hierarchy to facilitate blastocyst formation via transcriptional regulation of core genes involved in TJ assembly, fluid accumulation and cellular proliferation (Fig. 7).
Fig. 7.
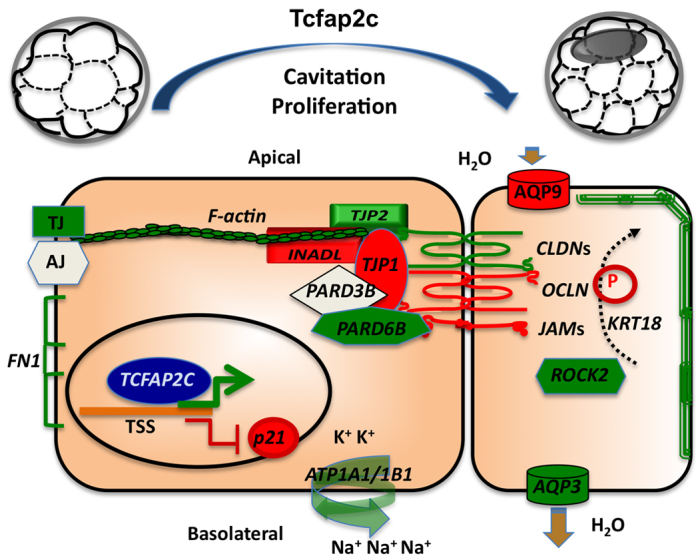
Model for Tcfap2c-mediated transcriptional regulation of blastocyst formation. Tcfap2c regulates a diverse group of genes during the morula-to-blastocyst transition. Genes shown in green and red are positively and negatively regulated by Tcfap2c, respectively; genes in white are not regulated by Tcfap2c. TJ, tight junction; AJ, adherens junction; P, phosphorylation.
In the present study we identified a diverse group of Tcfap2c-regulated genes with established roles in blastocyst formation. These include genes that are important for TJ assembly (Cldn4, Cldn6, Tjp2, Tjp1), cell polarity (Pard6b) and fluid accumulation (Atp1b1, Aqp3). Claudin family members encode tetraspanin membrane proteins that serve crucial roles in TJ assembly and epithelial cell barrier function (Krause et al., 2008). In preimplantation embryos, disruption of Cldn4 and Cldn6 function via an inhibitory peptide impairs blastocyst development (Moriwaki et al., 2007). The TJ proteins Tjp1 and Tjp2 play an important role in connecting the actin skeleton with TJ complexes at the apical membrane (Schneeberger and Lynch, 2004). Embryos that lack Tjp1 or Tjp2 exhibit defects in blastocyst formation and/or undergo early embryonic lethality (Katsuno et al., 2008; Sheth et al., 2008). Likewise, in mouse preimplantation embryos the cell polarity regulator Pard6b is essential for blastocyst formation and proper localization of Tjp1 in the apical cell membrane (Alarcon, 2010). Atp1b1 and Aqp3 play major roles in fluid accumulation. For example, RNAi-mediated KD of Atp1b1 and pharmacological inhibition of aquaporins blocks blastocyst formation and expansion, respectively, in mouse preimplantation embryos (Barcroft et al., 2003; Madan et al., 2007). These results in combination with our findings demonstrate that the proper expression of these genes is required for blastocyst formation.
Consistent with the altered expression of TJ, cell polarity and ion gradient-related genes, the barrier function of the TE epithelium was impaired in Tcfap2c KD embryos. Previous studies showed that TJ complexes are assembled at the morula stage and become functional near the time of blastocyst formation (Eckert and Fleming, 2008). In the present study, ultrastructural and functional analyses revealed the absence of TJ complexes and defective paracellular sealing in Tcfap2c KD embryos. In line with these findings, inhibition of Cldn4 and Cldn6 has been shown to disrupt paracellular sealing in mouse preimplantation embryos (Moriwaki et al., 2007). Altogether, our data demonstrate that Tcfap2c is a core regulator of TJ assembly and paracellular sealing in the TE epithelium.
To address whether blastocyst formation could be rescued in Tcfap2c KD embryos, chimera experiments were performed. We found that aggregation of control eight-cell embryos with Tcfap2c KD embryos could fully restore blastocyst formation. Interestingly, in chimeric blastocysts, Tcfap2c KD blastomeres (GFP+) contributed predominantly to the ICM epithelium, whereas control blastomeres contributed to the TE epithelium. This finding, in combination with the observed enrichment of Tcfap2c in the TE epithelium (this study) (Kuckenberg et al., 2010), suggest that Tcfap2c might have a potential role in cell fate specification. In support of this idea, forced expression of Tcfap2c in mouse ES cells triggers differentiation into TE-like cells by Cdx2-dependent and -independent mechanisms (Kuckenberg et al., 2010). In Tcfap2c KD embryos we observed a significant decrease in Cdx2 expression, suggesting that Tcfap2c might play an important role in regulating Cdx2 expression during preimplantation development (our unpublished data). Additional studies are necessary to establish a regulatory relationship between Tcfap2c and Cdx2 in the preimplantation embryo.
Our data also provide some tantalizing clues as to the role of Tcfap2c in cellular proliferation. Molecular analysis of Tcfap2c KD embryos revealed that p21 expression was upregulated greater than 8-fold. Several lines of evidence implicate p21 in preimplantation embryo arrest. First, mouse embryos fertilized with γ-irradiated sperm exhibit an increase in p21 expression and developmental arrest around the morula stage, whereas, p21-null embryos fertilized with irradiated sperm develop normally to the blastocyst stage, demonstrating that p21 is required for DNA damage-induced arrest at the morula stage (Adiga et al., 2007). Second, depletion of Hdac1, a negative regulator of p21 transcription, resulted in increased levels of p21 expression and lower rates of blastocyst formation (Ma and Schultz, 2008). Third, in growth-arrested human preimplantation embryos, the p21-related family member P27 is upregulated (Civico et al., 2002). To address the relationship between Tcfap2c and p21 we performed double KDs and found that ablation of p21 in Tcfap2c KD embryos could improve blastocyst formation. Interestingly, in other cellular contexts, p21 expression is regulated by Tcfap2c. For example, in MCF-7 breast cancer cells Tcfap2c promotes cellular proliferation via direct repression of p21 transcription (Williams et al., 2009). Because Tcfap2c expression is dysregulated in breast cancer it is tempting to speculate that Tcfap2c confers a dual function in controlling both cellular proliferation and TJ assembly. Collectively, our data demonstrate that Tcfap2c plays a crucial role in negatively regulating p21 expression during the morula-to-blastocyst transition in mouse preimplantation embryos.
In summary, results reported here provide strong evidence that Tcfap2c is a core regulator of blastocyst formation in mouse embryos. These results have significant implications in understanding preimplantation embryo failure in humans, where 50-70% of embryos produced via assisted reproductive technologies fail to develop to the blastocyst stage (Gardner et al., 2004; Blake et al., 2007). From a broader perspective, our findings might have an impact on understanding the etiology of more invasive forms of cancer in which TJ complexes are lost (Brennan et al., 2010).
Supplementary Material
Acknowledgments
We thank Dr George Smith for critically reading the manuscript.
Footnotes
Funding
This research was supported by a grant from the National Institute of General Medical Sciences [R01GM095347 to J.G.K.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.086645/-/DC1
References
- Adiga S. K., Toyoshima M., Shiraishi K., Shimura T., Takeda J., Taga M., Nagai H., Kumar P., Niwa O. (2007). p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene 26, 6141–6149 [DOI] [PubMed] [Google Scholar]
- Adjaye J., Huntriss J., Herwig R., BenKahla A., Brink T. C., Wierling C., Hultschig C., Groth D., Yaspo M.-L., Picton H. M., et al. (2005). Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23, 1514–1525 [DOI] [PubMed] [Google Scholar]
- Alarcon V. B. (2010). Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman H. J., Nottoli T., Lakiza O., Winger Q., Donaldson S., Williams T. (2002). Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 129, 2733–2747 [DOI] [PubMed] [Google Scholar]
- Barcroft L. C., Offenberg H., Thomsen P., Watson A. J. (2003). Aquaporin proteins in murine trophectoderm mediate transepithelial water movements during cavitation. Dev. Biol. 256, 342–354 [DOI] [PubMed] [Google Scholar]
- Barcroft L. C., Moseley A. E., Lingrel J. B., Watson A. J. (2004). Deletion of the Na/K-ATPase alpha1-subunit gene (Atp1a1) does not prevent cavitation of the preimplantation mouse embryo. Mech. Dev. 121, 417–426 [DOI] [PubMed] [Google Scholar]
- Blake D., Farquhar C., Johnson N., Proctor M. (2007). Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst. Rev. doi: 10.1002/14651858.CD002118.pub3 [DOI] [PubMed]
- Brennan K., Offiah G., McSherry E. A., Hopkins A. M. (2010). Tight junctions: a barrier to the initiation and progression of breast cancer? J. Biomed. Biotechnol. 2010, 460607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civico S., Agell N., Bachs O., Vanrell J. A., Balasch J. (2002). Increased expression of the cyclin-dependent kinase inhibitor p27 in cleavage-stage human embryos exhibiting developmental arrest. Mol. Hum. Reprod. 8, 919–922 [DOI] [PubMed] [Google Scholar]
- Cockburn K., Rossant J. (2010). Making the blastocyst: lessons from the mouse. J. Clin. Invest. 120, 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp A. J. (1978). Interaction between inner cell mass and trophectoderm of the mouse blastocyst. I. A study of cellular proliferation. J. Embryol. Exp. Morphol. 48, 109–125 [PubMed] [Google Scholar]
- de Vries W. N., Evsikov A. V., Haac B. E., Fancher K. S., Holbrook A. E., Kemler R., Solter D., Knowles B. B. (2004). Maternal beta-catenin and E-cadherin in mouse development. Development 131, 4435–4445 [DOI] [PubMed] [Google Scholar]
- Dietrich J.-E., Hiiragi T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231 [DOI] [PubMed] [Google Scholar]
- Ebnet K., Aurrand-Lions M., Kuhn A., Kiefer F., Butz S., Zander K., Meyer zu Brickwedde M.-K., Suzuki A., Imhof B. A., Vestweber D. (2003). The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J. Cell Sci. 116, 3879–3891 [DOI] [PubMed] [Google Scholar]
- Eckert D., Buhl S., Weber S., Jäger R., Schorle H. (2005). The AP-2 family of transcription factors. Genome Biol. 6, 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert J. J., Fleming T. P. (2008). Tight junction biogenesis during early development. Biochim. Biophys. Acta 1778, 717–728 [DOI] [PubMed] [Google Scholar]
- Gardner D. K., Surrey E., Minjarez D., Leitz A., Stevens J., Schoolcraft W. B. (2004). Single blastocyst transfer: a prospective randomized trial. Fertil. Steril. 81, 551–555 [DOI] [PubMed] [Google Scholar]
- Gartel A. L., Tyner A. L. (1999). Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp. Cell Res. 246, 280–289 [DOI] [PubMed] [Google Scholar]
- Giannatselis H., Calder M., Watson A. J. (2011). Ouabain stimulates a Na+/K+-ATPase-mediated SFK-activated signalling pathway that regulates tight junction function in the mouse blastocyst. PLoS ONE 6, e23704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens K., Van Soom A., Van Poucke M., Vandaele L., Vandesompele J., Van Zeveren A., Peelman L. J. (2007). Identification and expression analysis of genes associated with bovine blastocyst formation. BMC Dev. Biol. 7, 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens K., Tesfaye D., Rings F., Schellander K., Hölker M., Van Poucke M., Van Zeveren A., Lemahieu I., Van Soom A., Peelman L. J. (2010). Suppression of keratin 18 gene expression in bovine blastocysts by RNA interference. Reprod. Fertil. Dev. 22, 395–404 [DOI] [PubMed] [Google Scholar]
- Home P., Ray S., Dutta D., Bronshteyn I., Larson M., Paul S. (2009). GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno T., Umeda K., Matsui T., Hata M., Tamura A., Itoh M., Takeuchi K., Fujimori T., Nabeshima Y.-i., Noda T., et al. (2008). Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol. Biol. Cell 19, 2465–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi R., Tahara M., Sawada K., Ikebuchi Y., Morishige K., Sakata M., Tasaka K., Murata Y. (2004). Rho-kinase is involved in mouse blastocyst cavity formation. Biochem. Biophys. Res. Commun. 319, 643–648 [DOI] [PubMed] [Google Scholar]
- Keramari M., Razavi J., Ingman K. A., Patsch C., Edenhofer F., Ward C. M., Kimber S. J. (2010). Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS ONE 5, e13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S. (2010). Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 20, 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S., Knott J. G. (2009). SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317–328 [DOI] [PubMed] [Google Scholar]
- Kim J., Gye M. C., Kim M. K. (2004). Role of occludin, a tight junction protein, in blastocoel formation, and in the paracellular permeability and differentiation of trophectoderm in preimplantation mouse embryos. Mol. Cells 17, 248–254 [PubMed] [Google Scholar]
- Krause G., Winkler L., Mueller S. L., Haseloff R. F., Piontek J., Blasig I. E. (2008). Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 [DOI] [PubMed] [Google Scholar]
- Kuckenberg P., Buhl S., Woynecki T., van Fürden B., Tolkunova E., Seiffe F., Moser M., Tomilin A., Winterhager E., Schorle H. (2010). The transcription factor TCFAP2C/AP-2γ cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell. Biol. 30, 3310–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L., Ohsugi M., Hirchenhain J., Kemler R. (1994). E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA 91, 8263–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Schultz R. M. (2008). Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev. Biol. 319, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan P., Rose K., Watson A. J. (2007). Na/K-ATPase β1 subunit expression is required for blastocyst formation and normal assembly of trophectoderm tight junction-associated proteins. J. Biol. Chem. 282, 12127–12134 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- Moriwaki K., Tsukita S., Furuse M. (2007). Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev. Biol. 312, 509–522 [DOI] [PubMed] [Google Scholar]
- Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. (2008). Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. (2008). Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283 [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Xin L., Sharov A. A., Thomas M., Mowrer G., Meyers E., Piao Y., Mehta S., Yee S., Nakatake Y., et al. (2009). Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5, 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- Parfitt D.-E., Zernicka-Goetz M. (2010). Epigenetic modification affecting expression of cell polarity and cell fate genes to regulate lineage specification in the early mouse embryo. Mol. Biol. Cell 21, 2649–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S., Rossant J. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 [DOI] [PubMed] [Google Scholar]
- Riethmacher D., Brinkmann V., Birchmeier C. (1995). A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. USA 92, 855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ A. P., Wattler S., Colledge W. H., Aparicio S. A., Carlton M. B. L., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C., et al. (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 [DOI] [PubMed] [Google Scholar]
- Saitou M., Furuse M., Sasaki H., Schulzke J.-D., Fromm M., Takano H., Noda T., Tsukita S. (2000). Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell 11, 4131–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger E. E., Lynch R. D. (2004). The tight junction: a multifunctional complex. Am. J. Physiol. 286, C1213–C1228 [DOI] [PubMed] [Google Scholar]
- Sherr C. J., Roberts J. M. (1995). Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9, 1149–1163 [DOI] [PubMed] [Google Scholar]
- Sherr C. J., Roberts J. M. (1999). CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- Sheth B., Nowak R. L., Anderson R., Kwong W. Y., Papenbrock T., Fleming T. P. (2008). Tight junction protein ZO-2 expression and relative function of ZO-1 and ZO-2 during mouse blastocyst formation. Exp. Cell Res. 314, 3356–3368 [DOI] [PubMed] [Google Scholar]
- Shin K., Straight S., Margolis B. (2005). PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J. Cell Biol. 168, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C.-A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 [DOI] [PubMed] [Google Scholar]
- Thomas F. C., Sheth B., Eckert J. J., Bazzoni G., Dejana E., Fleming T. P. (2004). Contribution of JAM-1 to epithelial differentiation and tight-junction biogenesis in the mouse preimplantation embryo. J. Cell Sci. 117, 5599–5608 [DOI] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C. A., Henry R. W., Knott J. G. (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS ONE 5, e10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A. J., Barcroft L. C. (2001). Regulation of blastocyst formation. Front. Biosci. 6, D708–D730 [DOI] [PubMed] [Google Scholar]
- Williams C. M. J., Scibetta A. G., Friedrich J. K., Canosa M., Berlato C., Moss C. H., Hurst H. C. (2009). AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J. 28, 3591–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger Q., Huang J., Auman H. J., Lewandoski M., Williams T. (2006). Analysis of transcription factor AP-2 expression and function during mouse preimplantation development. Biol. Reprod. 75, 324–333 [DOI] [PubMed] [Google Scholar]
- Xu J., Kausalya P. J., Phua D. C. Y., Ali S. M., Hossain Z., Hunziker W. (2008). Early embryonic lethality of mice lacking ZO-2, but not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol. Cell. Biol. 28, 1669–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., DePamphilis M. L., Buonanno A. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836 [DOI] [PubMed] [Google Scholar]
- Zupkovitz G., Grausenburger R., Brunmeir R., Senese S., Tischler J., Jurkin J., Rembold M., Meunier D., Egger G., Lagger S., et al. (2010). The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol. Cell. Biol. 30, 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.