Abstract
Antigen-specific immune responses against peptides derived from missense gene mutations have been identified in multiple cancers. The application of personalized peptide vaccines based on the tumor mutation repertoire of each cancer patient is a near-term clinical reality. These peptides can be identified for pre-validation by leveraging the results of massive gene sequencing efforts in cancer. In this study, we utilized NetMHC 3.2 to predict nanomolar peptide binding affinity to 57 human HLA-A and B alleles. All peptides were derived from 5,685 missense mutations in 312 genes annotated as functionally relevant in the Cancer Genome Project. Of the 26,672,189 potential 8–11 mer peptide-HLA pairs evaluated, 0.4% (127,800) display binding affinities < 50 nM, predicting high affinity interactions. These peptides can be segregated into two groups based on the binding affinity to HLA proteins relative to germline-encoded sequences: peptides for which both the mutant and wild-type forms are high affinity binders, and peptides for which only the mutant form is a high affinity binder. Current evidence directs the attention to mutations that increase HLA binding affinity, as compared with cognate wild-type peptide sequences, as these potentially are more relevant for vaccine development from a clinical perspective. Our analysis generated a database including all predicted HLA binding peptides and the corresponding change in binding affinity as a result of point mutations. Our study constitutes a broad foundation for the development of personalized peptide vaccines that hone-in on functionally relevant targets in multiple cancers in individuals with diverse HLA haplotypes.
Keywords: cancer vaccines, computational biology, immunomics, immunotherapy, missense mutation, protein database, T cell therapy
Introduction
Missense mutations in cancer cells can generate unique T cell epitopes.1 Natural antigen-specific T-cell responses against missense mutant epitopes in acute myeloid leukemia (AML), melanoma, renal and lung cancer have been discovered using traditional discovery methods based on CD8+ tumor infiltrating lymphocytes (TILs).2-6 Such foreign epitopes have been shown is to facilitate immunosurveillance and cancer control in chemically-induced murine models of carcinogens.7 Furthermore, sequencing efforts have identified 50 mutated peptides that can serve as rejection antigens in transplanted B16 murine melanoma.8 The marked increase in tumor sequencing efforts has provided an opportunity to discover mutant epitopes that strongly bind to human HLA, compared with wild-type peptides, and may thus be effective mediators of T-cell responses. Thus, knowledge of a patient’s HLA type and missense mutation profile provides the opportunity to develop personalized peptide vaccines. Evidence supporting the efficacy of vaccination strategies based on mutant epitopes has already been generated. For instance, peptides derived from codon 3 mutations in RAS family members can readily induce immunity in patients with pancreatic, lung, and colon cancer patients.1,9-14
However, RAS mutations do not generate optimal antigens in most cancer patients. Indeed, RAS is a relatively commonly mutated gene, yet is present only in a minority of patients. Models of peptide-HLA binding affinity can facilitate the identification of novel and “personal” targets for cancer vaccines. To the best of our knowledge, computational methods to discover mutant epitopes and the differential binding affinity to HLA were first applied by Segal et al. to a data set from breast and colorectal tumors.15 In this study, 1,152 peptides were interrogated for HLA-A*02:01 binding in silico. Similarly, Warren et al. subjected a general survey of mutations to multiple in silico HLA-binding algorithms in an effort to identify a polyvalent peptide vaccine optimized for prophylactic use.16 HLA allelic frequency in the United States, mutation frequency and tumor subtype frequency were given equal consideration to generate the proposed vaccine formulation.
Since the binding affinity of a peptide for HLA proteins is associated with the immunogenicity of the peptide, computational prediction is a convenient and practical first step toward the identification of optimal vaccine targets for cancer therapy.17 The ongoing development of peptide-MHC binding algorithms has benefitted from progressively more empirical binding data, allowing for increased prediction accuracy.18,19 By means of the artificial neural network-based prediction algorithm NetMHC3.2, developed by Lundegaard et al., highly accurate predictions of peptide binding to large numbers of HLA alleles are now possible.18 The prediction of peptide sequence-dependent antigen presentation, which entails multiple steps including proteosomal cleavage and TAP binding, is by far more complex (and hence less developed) than that of peptide-HLA interactions. Moreover, peptide processing is highly variable across multiple types of cancer cells and inflammatory states.
The analysis performed in this study yields a potential functional class of immunotherapeutic targets that possess increased HLA binding affinity upon mutation. In addition, we present a set of peptides predicted to bind HLA with similar affinity before and after a cancer-associated mutation. Thus, we offer a foundational database to support research of hypotheses related to the immunogenicity of peptides derived from missense cancer-associated mutations.
Results
Execution of MHC Class I binding peptide prediction with NetMHC 3.2
In order to identify peptides that may serve as tumor rejection antigens based on the predicted ability to bind human HLA, a database of missense mutation-derived peptides was assembled from the Catalogue of Somatic Mutations in Cancer (COSMIC) database. Mutations (n = 5,590) from 312 genes (Table S1) that are represented in the COSMIC database were used as a resource for the amino acid substitution affecting the wild-type sequence. Short peptide strings of 8, 9, 10 and 11 amino acid lengths were generated from every mutation in the primary database. Each mutation resulted in 76 unique strings. In total, 26,672,189 peptides were generated for analysis. Each short peptide string was submitted to the NetMHC 3.2 HLA-binding algorithm. The results of this analysis were nanomolar (nM) binding affinity prediction scores for human HLA-A alleles and HLA-B alleles as indicated in the methods section.
From the 26,672,189 mutated peptides and 57 distinct human HLA alleles interrogated, 127,801 (0.3%) unique peptide-HLA pairs achieved a binding score of 50 nM of less (i.e., tight binding). This set of peptides was subjected to further analysis.
Mutation-mediated alteration in HLA-binding affinity
For the purpose of comparison, a delta table was generated to display the change in predicted HLA affinity that occurred as a result of each missense mutation, compared with the wild-type sequence. This table is organized with a FASTA-formatted identifier for each mutation and HLA pair, reference to the position of the mutation in the peptide string, the peptide length, and whether the peptide referenced is the wild-type or mutant sequence. Following the FASTA-formatted identifier is the peptide sequence and the nM binding affinity score for the peptide and indicated HLA allele. This order is repeated for the mutated peptide sequence and each row ends with the calculated difference in nM affinity between the wildtype and mutated peptides for an individual HLA allele (Table S2).
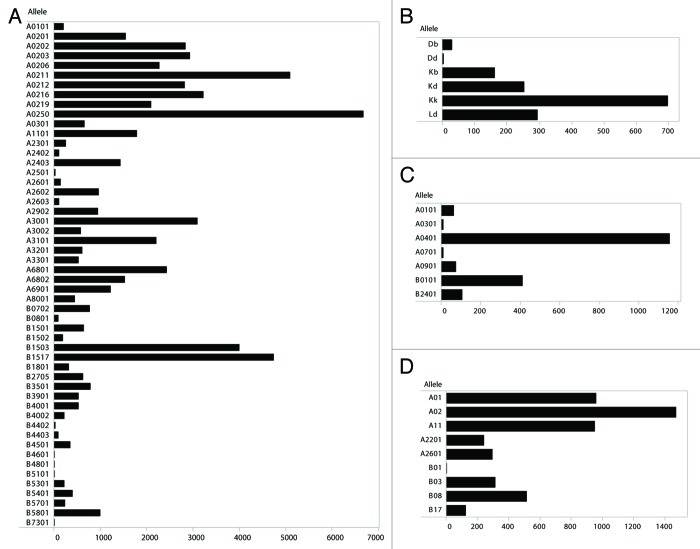
Some human HLA alleles present a broader repertoire of mutated peptide than others (Fig. 1A). The frequently studied human allele HLA-A0201 ranks 15th among 57 alleles in the breadth of its repertoire. The mean number of mutated peptides that human alleles may bind is 1,767 with a standard deviation of 2,377. The number of mutated peptides that any human allele may bind with high affinity range from 0–11,005; the quartiles are bound by 0, 131, 768, 2,692 and 11,005. Analysis of mutated peptide binding to a limited number of MHC class I alleles from mouse, rhesus macaque and chimpanzees demonstrates that these species may also be capable of presenting this set of antigens (Figure 1B-D).
Figure 1. Allelic distribution of tight binding mutated peptide-HLA pairs. Allelic distribution of predicted tight binding (affinity score < 50 nM) mutated peptides from the Cancer Gene Census among MHC class I alleles. (A) Human HLA (n = 52), null data not shown for n = 5 HLA alleles (n = 5). (B) Murine H-2 alleles (n = 6). (C) Rhesus macaque Mamu alleles (n = 7). (D) Chimpanzee Patr alleles (n = 9).
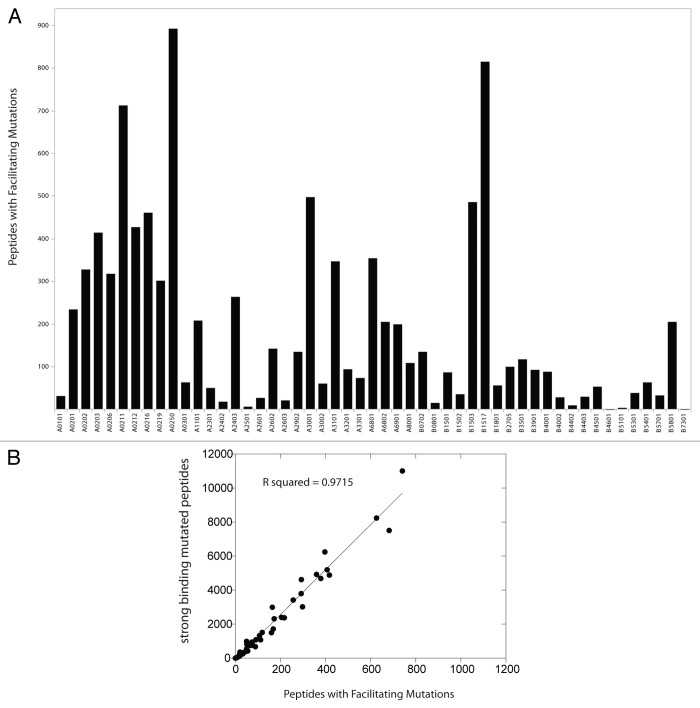
Analysis of the number of tight-binding mutated peptides for which the corresponding wild-type peptide has a nonbinding score (greater than 500 nM) is reported in Figure 2. These peptide-MHC pairs numbered 7,982 (8% of the total tight-binding mutated peptides). The mutations leading to peptides that may exhibit greater binding to HLA than wild-type counterparts are referred to as facilitating mutations. A partial set of facilitating mutations for HLA-A02:01 is collected in Table 1 (n = 15).
Figure 2. Tight binding mutated peptides derived from facilitating mutations and human HLA alleles. (A) Allelic distribution of predicted tight-binding peptides derived from facilitating mutations (mutated peptide affinity score < 50 nM AND wild-type peptide affinity score > 500 nM) from the Cancer Gene Census among HLA alleles (n = 52), null data not shown for n = 5 HLA alleles. (B) Plot of tight-binding mutated peptides and peptides derived from faciliating mutations for each cognate HLA allele. Linear regression, y = 13.22(x)-82.942, R2 as indicated.
Table 1. Facilitating, mutated strong binding HLA-A 02:01 peptides.
| FASTA wt | wt Peptide | nM (wt) | FASTA mt | mt Peptide | nM (mt) | Delta nM |
|---|---|---|---|---|---|---|
| > ALK-R401Q-wildtype-sequence-1-HLA-A0201 |
FRVALEYI |
15009 |
> ALK-R401Q-mutant-sequence-1-HLA-A0201 |
FQVALEYI |
29 |
14980 |
| > BAP1-H169Q-wildtype-sequence-1-HLA-A0201 |
FHFVSYVPI |
13143 |
> BAP1-H169Q-mutant-sequence-1-HLA-A0201 |
FQFVSYVPI |
21 |
13122 |
| > BRAF-K475M-wildtype-sequence-1-HLA-A0201 |
GKWHGDVAV |
15489 |
> BRAF-K475M-mutant-sequence-1-HLA-A0201 |
GMWHGDVAV |
13 |
15476 |
| > CDK6-P199L-wildtype-sequence-1-HLA-A0201 |
TPVDLWSV |
14601 |
> CDK6-P199L-mutant-sequence-1-HLA-A0201 |
TLVDLWSV |
12 |
14589 |
| > CHEK2-P536L-wildtype-sequence-1-HLA-A0201 |
RPAVCAAV |
20392 |
> CHEK2-P536L-mutant-sequence-1-HLA-A0201 |
RLAVCAAV |
25 |
20367 |
| > EGFR-H773L-wildtype-sequence-8-HLA-A0201 |
VMASVDNPH |
22464 |
> EGFR-H773L-mutant-sequence-8-HLA-A0201 |
VMASVDNPL |
48 |
22416 |
| > FANCF-P185L-wildtype-sequence-1-HLA-A0201 |
RPARFLSSL |
22304 |
> FANCF-P185L-mutant-sequence-1-HLA-A0201 |
RLARFLSSL |
38 |
22266 |
| > GNAS-D141V-wildtype-sequence-8-HLA-A0201 |
SVMNVPDFD |
20809 |
> GNAS-D141V-mutant-sequence-8-HLA-A0201 |
SVMNVPDFV |
24 |
20785 |
| > ITK-G372V-wildtype-sequence-7-HLA-A0201 |
FVQEIGSG |
19247 |
> ITK-G372V-mutant-sequence-7-HLA-A0201 |
FVQEIGSV |
48 |
19199 |
| > JAK1-E966V-wildtype-sequence-8-HLA-A0201 |
FLPSGSLKE |
17955 |
> JAK1-E966V-mutant-sequence-8-HLA-A0201 |
FLPSGSLKV |
11 |
17944 |
| > JAK2-K539L-wildtype-sequence-8-HLA-A0201 |
HMNQMVFHK |
18253 |
> JAK2-K539L-mutant-sequence-8-HLA-A0201 |
HMNQMVFHL |
35 |
18218 |
| > KRAS-Q61L-wildtype-sequence-10-HLA-A0201 |
CLLDILDTAGQ |
6354 |
> KRAS-Q61L-mutant-sequence-10-HLA-A0201 |
CLLDILDTAGL |
26 |
6328 |
| > NOTCH1-R1634L-wildtype-sequence-1-HLA-A0201 |
KRAAEGWAA |
21734 |
> NOTCH1-R1634L-mutant-sequence-1-HLA-A0201 |
KLAAEGWAA |
24 |
21710 |
| > RB1-P515L-wildtype-sequence-1-HLA-A0201 |
FPWILNVL |
10736 |
> RB1-P515L-mutant-sequence-1-HLA-A0201 |
FLWILNVL |
13 |
10723 |
| > TP53-P47L-wildtype-sequence-8-HLA-A0201 | AMDDLMLSP | 10776 | > TP53-P47L-mutant-sequence-8-HLA-A0201 | AMDDLMLSL | 11 | 10765 |
Table of representative mutated peptides from the Cancer Gene Census predicted to be tight binders (affinity < 50 nM) to HLA-A02:01, for which cognate wild-type peptides are predicted to be non-binders (affinity > 500nM). n = 15.
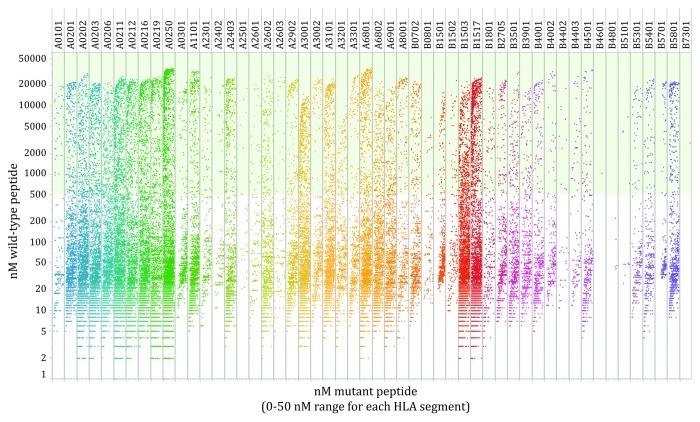
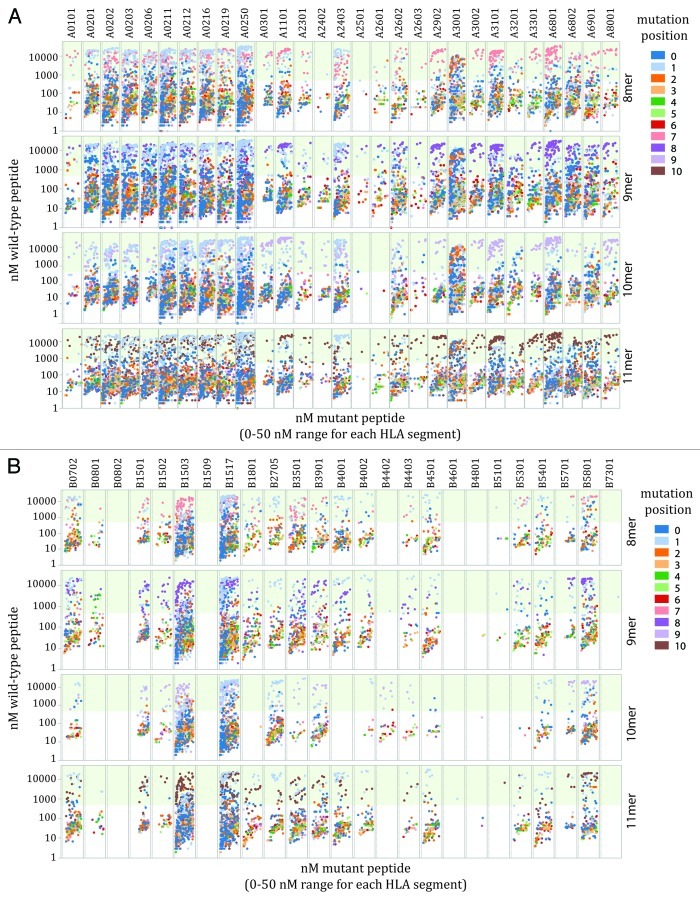
Neither the number of peptides binding each HLA molecule, nor facilitating mutations are equally distributed among the HLA alleles analyzed. However, the number of facilitating mutations was highly associated with the total number of tight binding mutated peptides analyzed R = 0.97 (n = 57) (Fig. 2). There is no enrichment or depletion of facilitating mutations among peptid-HLA pairs as represented in the Cancer Gene Census database, a collection of the observable functional missense mutations in human cancer (Fig. 3). The identified facilitating mutations are largely composed of peptides in which the mutated residue is located in a peripheral residue position (Fig. 4A and B).
Figure 3. Facilitating mutation distribution for human HLA-A and HLA-B alleles. Plot of tight binding mutated peptides (< 50 nM mutated peptide affinity score) from the Cancer Gene Census and corresponding wild-type peptides affinity score for each cognate HLA allele. Light green background indicates the threshold (500 nM) of predicted non-binding wild-type peptides.
Figure 4. Facilitating mutations utilize peripheral anchor residues. Plot of tight binding mutated 8, 9, 10 and 11 mer peptides (mutated peptide affinity score < 50 nM) and corresponding wild-type peptides affinity score for each cognate HLA-A (A) and HLA-B (B) allele. Light green background indicates the threshold (500 nM) of predicted non-binding wild-type peptides. Coloring indicates the position of each mutation in the peptide string (starting from the C terminus): dark blue (1), light blue (2), orange (3), light orange (4), dark green (5), light green (6), red (7), pink (8), purple (9), light purple (10), brown (11).
In the large majority of peptide-MHC pairs that remain in the total data set, mutations exert limited effect upon the HLA-binding scores. In this set of 79,733 neutral mutations, 79% of the tight-binding peptides retain < 50 nM binding scores with and without the point mutation numbers (Fig. 3). A partial set of neutral mutations for HLA-A02:01 is collected in Table 2 (n = 15).
Table 2. Neutral, mutated strong binding HLA-A 02:01 peptides.
| FASTA wt | wt Peptide | nM (wt) | FASTA mt | mt Peptide | nM (mt) | Delta nM |
|---|---|---|---|---|---|---|
| > ALK-R1275Q-wildtype-sequence-3-HLA-A0201 |
GMARDIYRA |
43 |
> ALK-R1275Q-mutant-sequence-3-HLA-A0201 |
GMAQDIYRA |
33 |
10 |
| > BAP1-H169Q-wildtype-sequence-6-HLA-A0201 |
RTMEAFHFV |
9 |
> BAP1-H169Q-mutant-sequence-6-HLA-A0201 |
RTMEAFQFV |
15 |
6 |
| > BRAF-L618W-wildtype-sequence-2-HLA-A0201 |
SILWMAPEV |
20 |
> BRAF-L618W-mutant-sequence-2-HLA-A0201 |
SIWWMAPEV |
10 |
10 |
| > CDK6-P199L-wildtype-sequence-9-HLA-A0201 |
VLLQSSYATPV |
12 |
> CDK6-P199L-mutant-sequence-9-HLA-A0201 |
VLLQSSYATLV |
13 |
1 |
| > CRLF2-F232C-wildtype-sequence-4-HLA-A0201 |
KLSKFILI |
25 |
> CRLF2-F232C-mutant-sequence-4-HLA-A0201 |
KLSKCILI |
24 |
1 |
| > EGFR-D761G-wildtype-sequence-2-HLA-A0201 |
ILDEAYVMASV |
11 |
> EGFR-D761G-mutant-sequence-2-HLA-A0201 |
ILGEAYVMASV |
20 |
9 |
| > FBXW7-I563T-wildtype-sequence-5-HLA-A0201 |
SLDTSIRV |
23 |
> FBXW7-I563T-mutant-sequence-5-HLA-A0201 |
SLDTSTRV |
44 |
21 |
| > GNA11-V223M-wildtype-sequence-1-HLA-A0201 |
NVTSIMFLV |
48 |
> GNA11-V223M-mutant-sequence-1-HLA-A0201 |
NMTSIMFLV |
9 |
39 |
| > IDH2-V294M-wildtype-sequence-5-HLA-A0201 |
LIDDMVAQV |
18 |
> IDH2-V294M-mutant-sequence-5-HLA-A0201 |
LIDDMMAQV |
23 |
5 |
| > JAK1-D660H-wildtype-sequence-8-HLA-A0201 |
YLYGVCVRDV |
23 |
> JAK1-D660H-mutant-sequence-8-HLA-A0201 |
YLYGVCVRHV |
15 |
8 |
| > JAK2-K607N-wildtype-sequence-4-HLA-A0201 |
KLSHKHLV |
44 |
> JAK2-K607N-mutant-sequence-4-HLA-A0201 |
KLSHNHLV |
29 |
15 |
| > KRAS-Q22K-wildtype-sequence-0-HLA-A0201 |
QLIQNHFV |
42 |
> KRAS-Q22K-mutant-sequence-0-HLA-A0201 |
KLIQNHFV |
11 |
31 |
| > NOTCH1-S1598I-wildtype-sequence-5-HLA-A0201 |
FLRELSRV |
40 |
> NOTCH1-S1598I-mutant-sequence-5-HLA-A0201 |
FLRELIRV |
20 |
20 |
| > RET-D925H-wildtype-sequence-3-HLA-A0201 |
SLFDHIYTT |
13 |
> RET-D925H-mutant-sequence-3-HLA-A0201 |
SLFHHIYTT |
32 |
19 |
| > TCL1A-K23N-wildtype-sequence-6-HLA-A0201 | RLWAWEKFV | 11 | > TCL1A-K23N-mutant-sequence-6-HLA-A0201 | RLWAWENFV | 8 | 3 |
Table of representative mutated peptides from the Cancer Gene Census predicted to be tight binders (affinity < 50 nM) to HLA-A02:01, for which cognate wild-type peptides are also predicted to be tight binders (affinity < 50 nM). n = 15.
Discussion
In this work, we have analyzed peptides that contain mutated amino acids from 5685 mutations in 312 genes for binding to HLA alleles in silico. All genes and mutations were selected from the Cancer Gene Census and annotated in the COSMIC database to have known functional roles in cancer.20 This curated data set of functional mutations was selected for analysis to facilitate vaccine efforts that may be able to exert immunological pressure on genes required for tumor cell survival. In addition to focusing our search on genes and mutations with important survival properties, we have identified peptides that we predict will demonstrate enrich surface presentation as a result of enhanced HLA-binding affinity.21
Mutated peptides are a class of antigens with superior properties as immunological targets for cancer therapy. Self-protein derived peptides must overcome multiple and redundant mechanisms of T-cell suppression due to peripheral tolerance.22 Additionally, the frequency of autoreactive T cells is attenuated due to central tolerance. Mutated peptides are absent in the thymus during development and are not endogenously expressed in normal tissues. Moreover, altered protein folding of mutated proteins generally reduces their stability. Reduced stability in some cases is observed in the presence of stabilizing the heat-shock 90 KDa protein (HSP90) and in other cases by the increased proteosomal degradation accounted for by peptide and protein decay experiments.23 Loss-of-function of tumor suppressor genes can be facilitated by a rapid protein turnover due to destabilization. The increased source of peptides resulting from decreased stability can enrich these peptides as potential HLA binders as compared with their wild-type counterparts, as in the case of neutral effects on HLA-binding affinity.24 A simplistic mass action mechanism for increased peptide presentation may underlie the enhanced surface presentation of these antigens.
However, if the HLA-binding affinity of a peptide is increased due to a mutation, this may translate into increased presentation of the mutant peptide. During the loading of peptides on HLA in the ER and endosomes, competition can occur. Thus, facilitating mutations are subject to this advantage in loading and presentation.24 Once exposed on the cell surface, stable HLA/B2M/peptide complexes are presented for a relatively long time and hence have a higher chance to activate T cells.21,24 Interestingly, in a study that determined the antigen specificity of T cells in an immunosurveillance model of murine sarcoma, the mutated peptide from spectrin β2 was shown to contain a facilitating mutation. Identified in silico as a MHC class I (H-2-Db) binder after exome sequencing, the VAVVNQIAL peptide possesses a binding score of 9 nM based on the methods used in this study. The cognate wildtype peptide VAVVNQIAR achieves only a score of 5304 nM.7 In part, the principle of HLA-specific peptide anchor residues motivates the search for these target peptides. However, the artificial neural network-trained binding approach of NetMHC 3.2 functions independently of the recognized dominant influence that some residues acquire in peripheral positions. Therefore it was not surprising to observe the high prevalence of peptides with mutations in the anchor position within the population of facilitating mutations. This result indicates that the machine-learning model embedded in NetMHC 3.2 for predicting peptide-HLA binding confirms experimentally informed theoretical expectations.
This study provides an analysis of the Cancer Gene Census database revealing a subset of mutated peptides with superior HLA-binding affinity for the development of personalized peptide vaccines that target functionally relevant targets in diverse cancer and diverse HLA haplotypes. For this approach, substantial rationale exists for the continued use of immunostimulatory treatments such as anti-CTLA4 or anti-PD-1 monoclonal antibodies in combination with peptide vaccines.22 Still, in the clinical and experimental use of self peptides as tumor antigens, autoimmune responses are a source of significant toxicity when combined with anti-CTLA4 therapy22,25. For this reason, as we and others previously stated, the discovery and characterization of tumor-specific epitopes is still a priority in immunotherapy. Exerting pressure on cancer cells with functional mutations that facilitate tumor survival may provide an additional level of protection, as the loss of such antigens may be detrimental to the tumorigenic phenotype. The generation of this evolutionary double bind is predicted to have superior effects in a mutating target cell population.26 This class of peptides displays conserved molecular homology and exerts lethal pressure on cells in relation to oncogenic properties, which underlies their theoretical value as clinical targets for T-cell therapy and as ligands for TCR-like antibodies.27-29 The results of this study are critical to the high-throughput experimental validation of mutated tumor epitopes in anticipation of the broad implementation of personalized mutation-specific cancer vaccines.
Materials and Methods
Mutated peptide library generation
Point substitutions observed in cancer cell lines and tumor specimens were collected from the Catalogue of Somatic Mutations in Cancer (COSMIC) database in February 2011 and February 2012.20 Mutant genes were taken from the Cancer Genes of the Cancer Gene Census. This expanding collection of validated cancer mutations is curated to maintain a collection of mutations extending functional properties to cancer cells.20 Gene mutations resulting in altered peptide sequences were used to generate a library of 8, 9, 10 and 11 amino acid long peptides for further in silico screening. Reference sequences for each gene were similarly acquired from the COSMIC database, and 8, 9, 10, and 11 amino acid wild-type peptides corresponding to each mutated peptide were also generated.
In silico prediction of antigen presentation
Peptides were submitted to NetMHC 3.2 for prediction of binding affinity for all MHC class I alleles available.18,30 Human HLA-A alleles: 01:01, 02:01, 02:02, 02:03, 02:06, 02:11, 02:12, 02:16, 02:19, 02:50, 03:01, 11:01, 23:01, 24:02, 24:03, 25:01, 26:01, 26:02, 26:03, 29:02, 30:01, 30:02, 31:01, 32:01, 33:01, 68:01, 68:02, 69:01, 80:01; HLA-B alleles; 07:02, 08:01, 08:02, 08:03, 15:01, 15:02, 15:03, 15:09, 15:17, 18:01, 27:03, 27:05, 35:01, 38:01, 39:01, 40:01, 40:02, 44:02, 44:03, 45:01, 46:01, 48:01, 51:01, 53:01, 54:01, 57:01, 58:01, and 73:01 were included in the analysis. Murine alleles: H-2-Db, H-2-Dd, H-2-Kb, H-2-Kd, H-2Kk, H-2Ld. Rhesus macaque alleles: Mamu-A01, Mamu-A02, Mamu-A11, Mamu-A2:201, Mamu-A26:01, Mamu-B01, Mamu-B03, Mamu-B08, Mamu-B17. Chimpanzees alleles: Patr-A01:01, Patr-A03:01, Patr-A04:01, Patr-A07:01, Patr-A09:01, Patr-B01:01, Patr-B24:01. Nanomolar affinity values for wild-type reference and mutation-derived peptides were ranked, with those less that 50 nM referred to “tight binders” and those less that 500 nM as “loose binders.” These thresholds were previously adopted by Istail et al.31
Data Analysis
Linear regression was analyzed with Graphpad Prism (GraphPad Software Inc.). Figures were generated with Graphpad Prism and Tableau Professional (Tableau Software).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Z.S. is supported by the Breast Cancer Research Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/21511
References
- 1.Tuttle TM. Oncogene products represent potential targets of tumor vaccines. Cancer Immunol Immunother. 1996;43:135–41. doi: 10.1007/s002620050314. [DOI] [PubMed] [Google Scholar]
- 2.Gaudin C, Kremer F, Angevin E, Scott V, Triebel F. A hsp70-2 mutation recognized by CTL on a human renal cell carcinoma. J Immunol. 1999;162:1730–8. [PubMed] [Google Scholar]
- 3.Echchakir H, Mami-Chouaib F, Vergnon I, Baurain JF, Karanikas V, Chouaib S, et al. A point mutation in the alpha-actinin-4 gene generates an antigenic peptide recognized by autologous cytolytic T lymphocytes on a human lung carcinoma. Cancer Res. 2001;61:4078–83. [PubMed] [Google Scholar]
- 4.Corbière V, Chapiro J, Stroobant V, Ma W, Lurquin C, Lethé B, et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res. 2011;71:1253–62. doi: 10.1158/0008-5472.CAN-10-2693. [DOI] [PubMed] [Google Scholar]
- 5.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–92. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greiner J, Ono Y, Hofmann S, Schmitt A, Mehring E, Götz M, et al. Mutated regions of nucleophosmin 1 (NPM1) elicit both CD4+ and CD8+ T cell responses in patients with acute myeloid leukemia. Blood. 2012 doi: 10.1182/blood-2011-11-394395. Epub ahead of print; [DOI] [PubMed] [Google Scholar]
- 7.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 9.Toubaji A, Achtar M, Provenzano M, Herrin VE, Behrens R, Hamilton M, et al. Pilot study of mutant ras peptide-based vaccine as an adjuvant treatment in pancreatic and colorectal cancers. Cancer Immunol Immunother. 2008;57:1413–20. doi: 10.1007/s00262-008-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer RG, Korn S, Micke P, Becker K, Huber C, Wölfel T, et al. An open-label, prospective phase I/II study evaluating the immunogenicity and safety of a ras peptide vaccine plus GM-CSF in patients with non-small cell lung cancer. Lung Cancer. 2007;58:88–94. doi: 10.1016/j.lungcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Fenton RG, Keller CJ, Hanna N, Taub DD. Induction of T-cell immunity against Ras oncoproteins by soluble protein or Ras-expressing Escherichia coli. J Natl Cancer Inst. 1995;87:1853–61. doi: 10.1093/jnci/87.24.1853. [DOI] [PubMed] [Google Scholar]
- 12.Gjertsen MK, Bakka A, Breivik J, Saeterdal I, Gedde-Dahl T, 3rd, Stokke KT, et al. Ex vivo ras peptide vaccination in patients with advanced pancreatic cancer: results of a phase I/II study. Int J Cancer. 1996;65:450–3. doi: 10.1002/(SICI)1097-0215(19960208)65:4<450::AID-IJC10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Gjertsen MK, Bakka A, Breivik J, Saeterdal I, Solheim BG, Søreide O, et al. Vaccination with mutant ras peptides and induction of T-cell responsiveness in pancreatic carcinoma patients carrying the corresponding RAS mutation. Lancet. 1995;346:1399–400. doi: 10.1016/S0140-6736(95)92408-6. [DOI] [PubMed] [Google Scholar]
- 14.Gjertsen MK, Saeterdal I, Thorsby E, Gaudernack G. Characterisation of immune responses in pancreatic carcinoma patients after mutant p21 ras peptide vaccination. Br J Cancer. 1996;74:1828–33. doi: 10.1038/bjc.1996.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 16.Warren RL, Holt RA. A census of predicted mutational epitopes suitable for immunologic cancer control. Hum Immunol. 2010;71:245–54. doi: 10.1016/j.humimm.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Keogh E, Fikes J, Southwood S, Celis E, Chesnut R, Sette A. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167:787–96. doi: 10.4049/jimmunol.167.2.787. [DOI] [PubMed] [Google Scholar]
- 18.Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods. 2011;374:26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang GL, Ansari HR, Bradley P, Cawley GC, Hertz T, Hu X, et al. Machine learning competition in immunology - Prediction of HLA class I binding peptides. J Immunol Methods. 2011;374:1–4. doi: 10.1016/j.jim.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Burg SH, Visseren MJ, Brandt RM, Kast WM, Melief CJ. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J Immunol. 1996;156:3308–14. [PubMed] [Google Scholar]
- 22.Sanderson K, Scotland R, Lee P, Liu D, Groshen S, Snively J, et al. Autoimmunity in a phase I trial of a fully human anti-cytotoxic T-lymphocyte antigen-4 monoclonal antibody with multiple melanoma peptides and Montanide ISA 51 for patients with resected stages III and IV melanoma. J Clin Oncol. 2005;23:741–50. doi: 10.1200/JCO.2005.01.128. [DOI] [PubMed] [Google Scholar]
- 23.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busch DH, Pamer EG. MHC class I/peptide stability: implications for immunodominance, in vitro proliferation, and diversity of responding CTL. J Immunol. 1998;160:4441–8. [PubMed] [Google Scholar]
- 25.Amos SM, Duong CP, Westwood JA, Ritchie DS, Junghans RP, Darcy PK, et al. Autoimmunity associated with immunotherapy of cancer. Blood. 2011;118:499–509. doi: 10.1182/blood-2011-01-325266. [DOI] [PubMed] [Google Scholar]
- 26.Gatenby RA, Brown J, Vincent T. Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res. 2009;69:7499–502. doi: 10.1158/0008-5472.CAN-09-1354. [DOI] [PubMed] [Google Scholar]
- 27.Klechevsky E, Gallegos M, Denkberg G, Palucka K, Banchereau J, Cohen C, et al. Antitumor activity of immunotoxins with T-cell receptor-like specificity against human melanoma xenografts. Cancer Res. 2008;68:6360–7. doi: 10.1158/0008-5472.CAN-08-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaeli Y, Denkberg G, Sinik K, Lantzy L, Chih-Sheng C, Beauverd C, et al. Expression hierarchy of T cell epitopes from melanoma differentiation antigens: unexpected high level presentation of tyrosinase-HLA-A2 Complexes revealed by peptide-specific, MHC-restricted, TCR-like antibodies. J Immunol. 2009;182:6328–41. doi: 10.4049/jimmunol.0801898. [DOI] [PubMed] [Google Scholar]
- 29.Denkberg G, Lev A, Eisenbach L, Benhar I, Reiter Y. Selective targeting of melanoma and APCs using a recombinant antibody with TCR-like specificity directed toward a melanoma differentiation antigen. J Immunol. 2003;171:2197–207. doi: 10.4049/jimmunol.171.5.2197. [DOI] [PubMed] [Google Scholar]
- 30.Lundegaard C, Hoof I, Lund O, Nielsen M. State of the art and challenges in sequence based T-cell epitope prediction. Immunome Res. 2010;6(Suppl 2):S3. doi: 10.1186/1745-7580-6-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Istrail S, Florea L, Halldórsson BV, Kohlbacher O, Schwartz RS, Yap VB, et al. Comparative immunopeptidomics of humans and their pathogens. Proc Natl Acad Sci U S A. 2004;101:13268–72. doi: 10.1073/pnas.0404740101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.