Abstract
Macrophages are a critically important component of the innate and adaptive immune systems. They are equipped with oxidative and non-oxidative mechanisms to kill ingested pathogens. Natural Killer Lytic-Associated Molecule (NKLAM) is an E3 ubiquitin ligase expressed in macrophages and natural killer cells. We show that NKLAM expression in macrophages was enhanced by Toll-like receptor agonists and pro-inflammatory cytokines. Using confocal microscopy, we found that NKLAM colocalized with ingested E. coli. In assays using IgG-opsonized latex beads as targets, we demonstrated that NKLAM translocated to the phagosome early during maturation at a time that coincided with elevated levels of ubiquitinated phagosome proteins. In killing assays with bone marrow-derived macrophages from wild type and NKLAM-deficient mice, we found that NKLAM-deficient macrophages demonstrated less killing of Escherichia coli than wild type macrophages. Collectively, our data show that NKLAM is a novel component of macrophage phagosomes and is involved in macrophage bactericidal functions.
Keywords: Macrophage, Innate immunity, Phagocytosis, Phagosome maturation, Killing
1. INTRODUCTION
Macrophages are a critical component of the innate and adaptive immune system and act as a first line of defense against pathogens. Pathogen binding to macrophage cell surface pattern recognition receptors or Fc receptors triggers the engulfment of the pathogen into specialized intracellular compartments termed phagosomes. Newly formed phagosomes possess no killing ability. This characteristic is acquired through a complex and dynamic phagosome maturation process that results in a highly acidic (< pH 5) proteolytic environment [1]. During maturation, the protein composition of the phagosome is continuously remodeled through fusion with early and late endosomes and finally lysosomes. The phagosome proteome contains well over 100 proteins that are involved in phagosome movement along cytoskeletal structures, phagosome acidification and proteolysis [2].
Ubiquitin has been localized to maturing macrophage phagosomes where it is required for the formation of acidic multivesicular bodies [3], but is not required for phagocytosis or phagosome maturation [4]. Ubiquitination is a posttranslational mechanism to target proteins to the proteasome for degradation [5]. However, a novel role for ubiquitin in regulating and facilitating bacterial killing is emerging. Burkholder et al. showed that deubiquitinase inhibition significantly increases the translocation of inducible nitric oxide synthase (iNOS) to the phagosome and enhances the killing of Listeria monocytogenes [6]. Alonso et al. demonstrated that ubiquitin was localized to LAMP1-positive vesicles and cathepsin degradation products of ubiquitin have bactericidal activity against Mycobacterium tuberculosis [7]. Additionally, in vitro studies have shown that the C-terminal fragment of ubiquitin disrupts the membrane of Mycobacterium smegatis and has antifungal properties [8, 9]. Precisely how ubiquitin is trafficked into the lysosomal compartment is under investigation, but it has been suggested that ubiquitinated protein cargo in autophagosomes is a likely source of lysosomal ubiquitin [10].
Natural Killer Lytic-Associated Molecule is an E3 ubiquitin ligase and a member of the RING in between RING (RBR) family of proteins [11]. The N-terminus contains three cysteine-rich domains that comprise the RBR structure and ubiquitin ligase activity [12]. NKLAM also has two predicted transmembrane domains [13]. In resting cells, NKLAM is very weakly expressed; however treatment with cytokines or interleukins such as interferon beta (IFNβ) and IL-2 significantly upregulates NKLAM expression [13]. NKLAM is expressed in mononuclear cells such as monocytes and natural killer (NK) cells and has been colocalized with granzyme B in the cytolytic granules of NK cells [13]. Such a specific subcellular localization implicates NKLAM in the regulation of cytolytic functions and indeed NK cells from mice lacking NKLAM are significantly defective in lysing tumor target cells [14, 15].
In this report, we provide evidence that NKLAM expression is regulated by E. coli lipopolysaccharide (LPS), a Toll-like receptor 4 (TLR4) agonist. We used IgG-opsonized magnetic latex beads to show for the first time that NKLAM is a component of the macrophage phagosome and translocates to the phagosome early in the maturation process. Studies with NKLAM-deficient bone marrow-derived macrophages (BMDM) demonstrate that NKLAM expression in the phagosome coincides with elevated levels of ubiquitinated phagosome proteins. Importantly, we demonstrate that both BMDM and peritoneal macrophages lacking NKLAM have a defective killing response against E. coli.
2. Materials and Methods
2.1 Bacterial strains and macrophage culture
All experiments on mice were approved by the Animal Care and Use Committee at Saint Louis University. Wild type C57BL/6 (WT) and corresponding age-matched NKLAM-deficient knockout (KO) mice were used in all studies. For isolation of bone marrow, femurs and tibias were flushed with DMEM. The collected marrow was resuspended in BM20 media (DMEM supplemented with 20% fetal bovine serum, 20% L929-cell conditioned media, 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, and 1 mM sodium pyruvate). The bone marrow cells were cultured for 7 days in 100 mm non-tissue culture petri dishes with a partial media change on day 3. To isolate peritoneal macrophages, ice-cold PBS plus 3% fetal bovine serum (10 mL) was injected into the peritoneal cavity of euthanized mice. The fluid was aspirated from the peritoneal cavity and the cells were resuspended in DMEM. Escherichia coli (strain JM109) were grown in Luria-Bertani (LB) broth overnight at 37°C with shaking. RAW264.7 and J774A.1 macrophages were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin and cultured at 37°C in 5% CO2.
2.2 Macrophage stimulation
For experiments using stimulated cells, adherent macrophages (RAW264.7, J774A.1 and bone marrow-derived) were incubated with LPS (400 ng/mL) or LPS plus IFNγ (100 U/mL) (LPS/IFNγ) for at least 18 hr at 37°C or for times indicated. For experiments using E. coli, macrophage cultures were infected at a multiplicity of infection (MOI) of 10. At the desired time, the cultures were washed briefly in ice-cold PBS, and then suspended in lysis buffer (65 mM Tris, 0.5% Triton X-100, 137 mM NaCl, 10 % glycerol, 25 mM sodium orthovanadate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 1 mM EDTA). Protein concentrations were determined using bicinchoninic acid (BCA) protein assay reagents (Pierce, Rockford, IL).
2.3 Phagocytosis analysis by flow cytometry
Cultured WT or NKLAM-deficient BMDM (2 × 105) were suspended in DMEM plus 20 mM HEPES and kept on ice. Heat-killed, fluorescently-labeled (FITC) E. coli were added to a final MOI of 20 and the tubes were incubated at 37°C for the times indicated. The cells were then fixed in 2% paraformaldehyde for 10 min on ice. Non-ingested extracellular bacteria were quenched by the addition of 1 mg/mL trypan blue. Macrophage-associated fluorescence was assessed by flow cytometry. To measure changes in phagosomal pH, E. coli labeled with the pH-sensitive dye pHrodo (Life Technologies, Grand Island, NY) were used as targets and the experiments were carried out as described above. Flow cytometric data were analyzed with FlowJo (Treestar, Ashland, OR).
2.4 Macrophage bacteria killing assay
Wild type and NKLAM-deficient BMDM (3 ×105) were suspended in DMEM plus 20 mM HEPES; pH 7.4 and infected with E. coli at an MOI of 10. The cultures were rotated for 20 min at 37°C then washed three times in sterile PBS to remove extracellular bacteria. The infected macrophages were then suspended in DMEM plus 20 mM HEPES and incubated at 37°C for 90 min. At the desired time an aliquot of macrophages was pelleted and lysed in sterile water to release the ingested bacteria. The lysates were serially diluted and plated on LB agar plates. After overnight incubated at 37°C, the colonies were counted to determine colony forming units/mL.
2.5 Bead coating protocol
One micron magnetic beads were obtained from Spherotech, Inc. (Lake Forest, IL). Human IgG was added at a saturating concentration and the beads were incubated at room temperature for 60 min. The coated beads were washed twice in 0.1% BSA/PBS to back-coat the beads and then resuspended at the desired concentration in DMEM plus 20 mM HEPES; pH 7.4.
2.6 Phagosome isolation
Macrophages were incubated with IgG-coated magnetic beads for 10 min on ice to allow bead binding without ingestion. To induce phagocytosis, macrophage suspensions were incubated at 37°C for times indicated. The cell suspensions were then washed twice with ice-cold PBS to stop ingestion. The cells were resuspended in 1 mL homogenization buffer (250 mM sucrose, 20 mM HEPES, 0.5 mM EGTA, 0.1% gelatin, pH 7.0) [7] and disrupted with 40 strokes of a Wheaton dounce homogenizer. The cellular homogenate was centrifuged for 5 min at 150 × g to remove intact cells and nuclei. The post-nuclear supernate was transferred to a new tube and phagosomes were isolated with a magnet. The phagosome pellet was washed once in PBS and then lysed in ice-cold lysis buffer. The protein concentration was determined using BCA protein assay reagents and the lysates were solubilized with the addition of 5X SDS-PAGE sample buffer.
2.7 Immunoblotting
Proteins in lysates were separated using SDS-PAGE then transferred to PVDF membrane. Membranes were incubated with primary antibody overnight at 4°C. The antibodies for LAMP-1 (Cell Signaling Technology, Danvers, MA), ubiquitin, and EEA1 (Santa Cruz Biotechnology, Santa Cruz, CA), were used at 1:1000. Anti β-actin (Sigma Aldrich, St. Louis, MO) was used at 1:4000 and monoclonal anti-NKLAM [11] was used at 1:200. After three washes in TBS-T, the blots were probed with HRP-conjugated secondary antibodies and the proteins were visualized with BioRad Immun-Star Western C chemiluminescence kit. Images were captured and analyzed using a BioRad Chemidoc XRS+ imager (BioRad, Inc, Hercules, CA).
2.8 Confocal microscopy
Wild type BMDM were grown on 18 mm acid-cleaned coverslips until confluent. Heat-killed E. coli labeled with tetramethylrhodamine (Sigma Aldrich, St. Louis, MO) were added to the monolayers at an MOI of 10 and the cultures were incubated for 60 min at 37°C. The monolayers were washed with PBS and fixed in 2% paraformaldehyde. The coverslips were blocked and permeabilized with 5% normal goat serum plus 0.1% saponin for 30 min at room temperature. The coverslips were stained with monoclonal anti-NKLAM at a dilution of 1:50 overnight at 4°C. After washing in PBS, FITC-labeled goat-mouse IgG (1:400) was incubated with the monolayers for 60 min at room temperature. The coverslips were washed in PBS then mounted in polyvinyl alcohol mounting media. Fluorescent photomicrographs were captured using an Olympus FV-1000 MPE. The z-section images were processed using the Colocalization plugin (Pierre Bourdoncle, Institut Jacques Monod) for NIH ImageJ v 1.64.
2.9 Statistical analysis
A two-tailed, unpaired Student’s t-test was used to compare the means of two groups. A p value of 0.05 or lower was considered statistically significant.
3. RESULTS
3.1 NKLAM protein expression is induced by TLR4 agonists
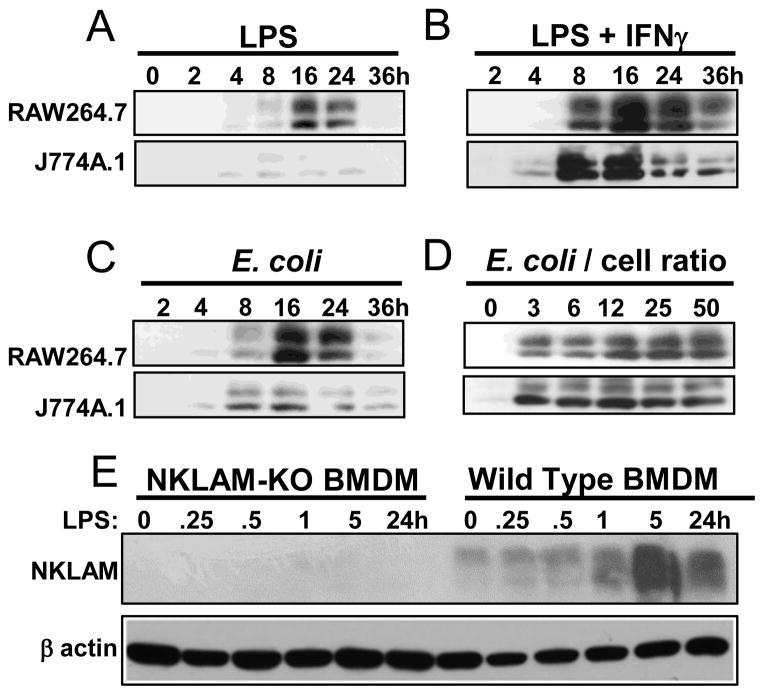
We have previously demonstrated that stimulation of peripheral blood monocytes with IFNβ strongly enhances NKLAM expression [13]. To further study the potential role of NKLAM in macrophage function, we investigated whether treatment with TLR4 agonists would affect NKLAM expression. In Fig. 1A, RAW264.7 or J774A.1 macrophages were treated with 400 ng/mL LPS for 0–36 hours and NKLAM expression was assessed by Western blot. Expression of NKLAM was evident by 4 hours in both cell types, and was maximal at 16 hr. By 24 hr of stimulation, NKLAM expression decreased. The anti-NKLAM monoclonal antibody used was generated in our laboratory [13] and recognizes NKLAM as a doublet. In Fig. 1B, macrophages were stimulated with a combination of LPS (400 ng/mL) plus IFNγ (100U/mL) (LPS/IFNγ). The overall expression pattern was similar to cells treated with LPS alone; however, the expression of NKLAM was prolonged and more robust. We also used heat-killed E. coli (Fig. 1C) and found an NKLAM expression pattern similar to that of LPS. Different E. coli : macrophage ratios were used in Fig. 1D to examine dose dependent effects on NKLAM expression. As the number of E. coli per macrophage was increased, the expression of NKLAM also increased. In Fig. 1E, NKLAM-deficient (KO) and WT BMDM were treated with LPS/IFNγ. In WT BMDM, NKLAM expression was maximal at 5 hr and had decreased by 24 hr (Fig. 1E). There was no NKLAM protein observed in NKLAM-deficient macrophages.
Figure 1. TLR4 stimulation induces NKLAM expression in macrophages.
(A–D) Macrophage cell lines RAW264.7 (top panels) and J774A.1 (bottom panels) were incubated with LPS (400 ng/mL) (A), LPS (400 ng/mL) plus IFNγ (100 U/mL) (B), or E. coli, strain JM109 (C, D). Whole cell lysates were probed for NKLAM expression by Western blot. (E) NKLAM-deficient (KO) and WT BMDM were treated with LPS (400 ng/mL) for times indicated. Cell lysates were probed for NKLAM expression by Western blot. β-actin was run as a loading control.
3.2 NKLAM is a macrophage phagosomal protein
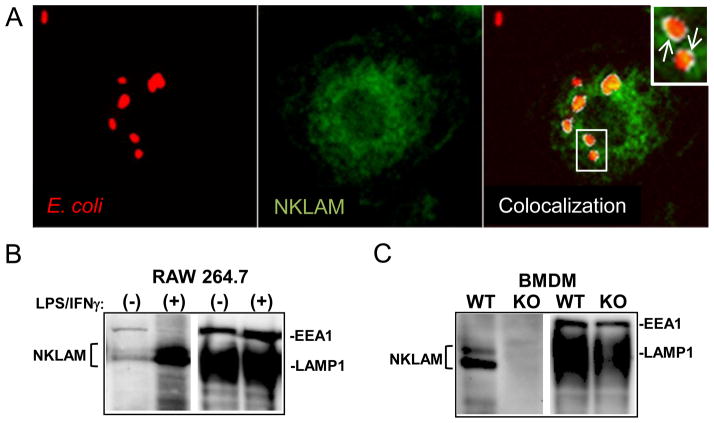
NKLAM is found in the cytotoxic granules of NK cells and is critical for maximal NK anti-tumor activity. [13]. To determine whether NKLAM was also involved in macrophage-mediated killing, we investigated NKLAM expression in the macrophage phagosome. In Fig. 2A, adherent WT BMDM were incubated with red fluorescent E. coli for 60 min (left panel), fixed and then stained for NKLAM. NKLAM had a punctuate, non-nuclear staining pattern (Fig. 2A, middle panel). To determine if NKLAM was localized to the phagosome membrane, we merged the confocal images and selected a z-section midway between the top and bottom surfaces of the cell to ensure we were viewing E. coli that were internalized into phagosomes. We used a colocalization plugin for ImageJ to define areas of specific colocalization (Fig. 2A, right panel). These areas are depicted in white (inset; arrows). We observed significant levels of NKLAM and E. coli colocalization to the phagosome periphery. Biochemical analysis of isolated macrophage phagosomes confirmed the presence of NKLAM in macrophage phagosomes. Untreated and 18h LPS/IFNγ-stimulated RAW264.7 macrophages were incubated with IgG- opsonized magnetic beads (Fig. 2B). Treatment with LPS/IFNγ stimulated a significant increase in phagosome-localized NKLAM. Similar experiments were also performed using LPS/IFNγ-stimulated WT and NKLAM-deficient BMDM (Fig. 2C). NKLAM was present in the phagosomes of WT but not NKLAM-deficient macrophages. The immunoblots (Fig. 2B and C) were stripped and reprobed for phagosome proteins LAMP1 and EEA1 to confirm successful phagosome isolation and to demonstrate that the marked changes in phagosomal NKLAM expression were not due to differences in the amount of protein loaded per well. These results demonstrate that NKLAM is a component of the macrophage phagosome, and is likely localized to the phagosome membrane.
Figure 2. NKLAM is localized to the macrophage phagosome.
(A) Adherent wild type BMDM were incubated with fluorescent red E. coli (left panel) for 60 min at 37°C. The monolayers were fixed in paraformaldehyde and labeled with anti-NKLAM (green, middle panel). Confocal photomicrographs taken at the midpoint of the z series were overlaid (right panel) and processed with the ImageJ Colocalization plugin. Arrows (inset) depict areas of specific colocalization, which are denoted in white. (B) RAW264.7 were untreated (−) or stimulated (+) with LPS/IFNγ for 18hr at 37°C. The cells were then incubated with IgG-opsonized magnetic latex beads for 45 min. The phagosomes were isolated as described in the Materials and Methods section. Equal amounts of phagosome protein were immunoblotted for NKLAM and phagosome markers LAMP1 and EEA1. (C) LPS/IFNγ-stimulated WT and NKLAM-deficient (KO) BMDM were used in experiments that were identical to (B).
3.3 NKLAM expression in the phagosome during maturation
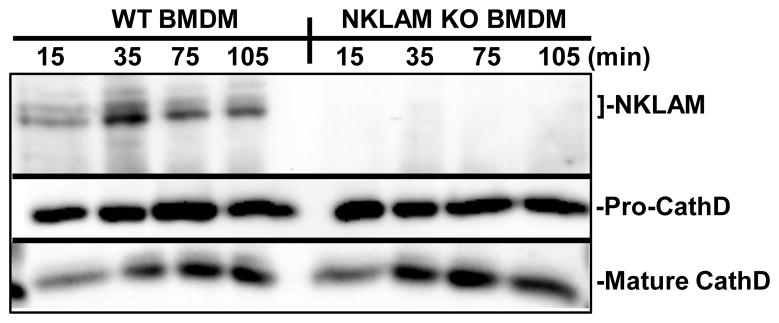
We next examined the levels of NKLAM in the phagosome during the maturation process. In Fig. 3, LPS/IFNγ-stimulated WT or NKLAM-KO BMDM were incubated with IgG-opsonized magnetic beads for 10 min on ice to allow bead binding. Cell suspensions were then switched to 37°C to initiate phagocytosis. Phagosomes were isolated at various times from 15–105 min post-ingestion. NKLAM was transiently expressed in the phagosome, and was observed as early as 15 min post-ingestion. The levels of NKLAM within the phagosome were maximal at 35 min post-ingestion and remained present as late as 105 min post-ingestion. Phagosomes from NKLAM-deficient BMDM did not contain NKLAM at any time point tested. As a control for the kinetics of phagosome maturation, membranes were reprobed for the acid hydrolase cathepsin D. The procathepsin D (pro-CathD) form has been observed in macrophage phagosomes as early as 7 min post-ingestion [16]. As the pH of the phagosome decreases, procathepsin D is proteolytically cleaved to a 48 kDa intermediate form, then cleaved again to a 34 kDa mature form [17]. The expression level of this mature form increases in the phagosome over time [16]. As shown in Fig. 3 (middle panel), the phagosomes of both WT and NKLAM-KO macrophages contained the 48 kDa intermediate form of cathepsin D at all time points. Additionally, the mature 34 kDa form of cathepsin D was present in the phagosomes of both cell types at 15 min and increased during phagosome maturation. These results demonstrate that NKLAM is translocated to the phagosome early in the maturation process.
Figure 3. Phagosomal NKLAM expression decreases during maturation.
WT or NKLAM-KO BMDM were stimulated with LPS/IFNγ for 18 hr. The cells were incubated with IgG-opsonized magnetic beads at 4°C for 10 min to synchronize bead binding. Ingestion was initiated by incubating the cells at 37°C. After incubation for the times indicated, the phagosomes were isolated as described in Materials and Methods. Phagosome lysates were normalized by protein and immunoblotted for NKLAM and phagosome marker cathepsin D.
3.4 Examination of ubiquitinated phagosomal proteins during phagosome maturation
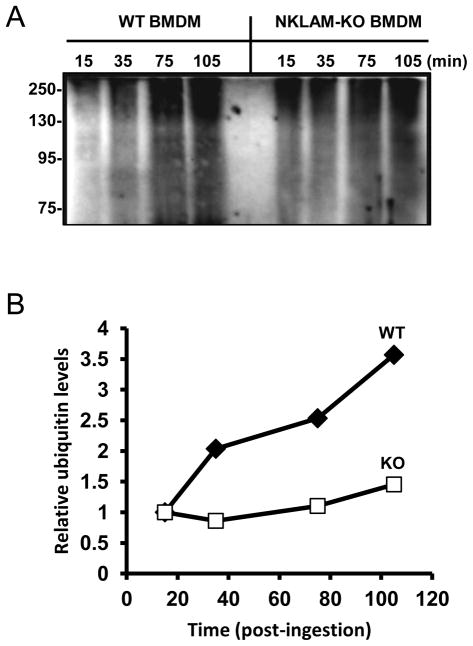
Our data demonstrate that NKLAM, a transmembrane E3 ubiquitin ligase, is localized to the macrophage phagosome. Previous reports have confirmed the presence of mono- and polyubiquitinated proteins in the phagosome membrane [3]. However, studies demonstrating that transmembrane ubiquitin ligases are components of the phagosome and contribute to the ubiquitination of phagosomal proteins are lacking. Our next set of experiments was designed to examine the profile of ubiquitinated phagosomal proteins during phagosome maturation. In Fig. 4, LPS/IFNγ-stimulated WT and NKLAM-deficient BMDM were incubated with IgG-opsonized latex beads. Phagosomes were isolated at various times (15–105 min post-ingestion) and the lysates were immunoblotted with an anti-ubiquitin antibody. A representative blot is shown in Fig. 4A. The phagosomes of both cell types contained significant amounts of ubiquitinated protein. Interestingly, we observed a sharp increase in the amount of ubiquitinated protein between 15 and 35 min post-ingestion in WT phagosomes, which was not observed in the phagosomes isolated from NKLAM-deficient macrophages. In both cell types, the levels of ubiquitinated proteins increased during phagosome maturation. In Fig. 4B, the densitometry values for each lane were determined and were divided by the density value of the 15 min time point. This provided a measure of the change in the levels of ubiquitinated proteins for each time point relative to the earliest time point (15 min) of maturation. t The levels of ubiquitinated proteins in WT phagosomes increase significantly during a time point (35 min post-ingestion) when NKLAM is maximally localized to the phagosome (Fig. 3, top panel). This increase was not observed in phagosomes from NKLAM-deficient macrophages.
Figure 4. Ubiquitination of phagosome proteins during maturation.
(A) WT and NKLAM-KO BMDM were stimulated with LPS/IFNγ for 18 hr. The cells were then incubated with IgG-opsonized magnetic beads at 4°C for 10 min. After incubation at 37°C for the times indicated, the phagosomes were isolated and the phagosome proteins were immunoblotted for ubiquitin. (B) Densitometry was performed on each lane. Graph represents the ratio of each lane relative to the 15 min time point for WT (closed diamonds) and NKLAM-KO (open squares). The immunoblot and graph are representative of five independent experiments.
3.5 NKLAM expression does not affect uptake of E. coli or phagosome acidification
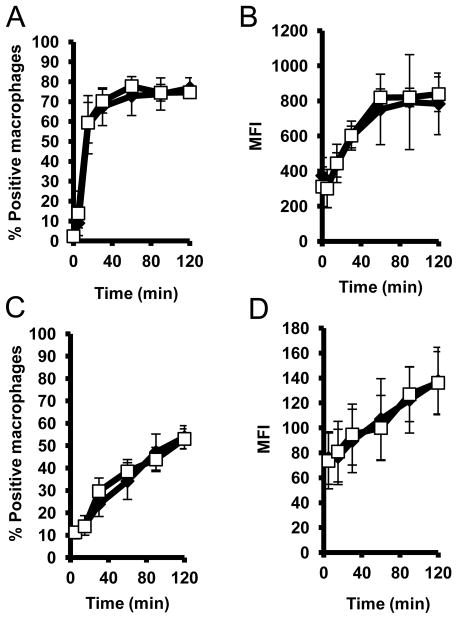
We next determined whether a deficiency in NKLAM protein would influence the kinetics and/or progression of phagocytosis and phagosome acidification. WT or NKLAM-deficient BMDM were incubated with heat-killed fluorescently-labeled E. coli for up to 120 min. At various time points the macrophages were assessed for bacterial uptake by flow cytometry. As shown in Fig. 5A, WT and NKLAM-deficient BMDM ingested fluorescent E. coli with similar kinetics. Ingestion increased rapidly within the first 30 min and then remained constant out to 120 min. The percentage of macrophages (~70%) that ingested E. coli was also equivalent between both cell types. These results suggest that NKLAM does not influence bacteria ingestion. We also determined the mean fluorescence intensity (MFI) for each population as a measure of the quantity of E. coli that was ingested per macrophage. As shown in Fig. 5B, there was no significant difference between WT and NKLAM-KO macrophages with respect to the overall number of bacteria ingested per macrophage.
Figure 5. NKLAM does not affect E. coli ingestion or phagosome acidification.
(A, B) Wild type (closed diamonds) and NKLAM-deficient BMDM (open squares) were incubated with FITC-labeled E. coli for the times indicated, followed by fixation in 1% formalin and analysis by flow cytometry. (A) Graph represents the percentage of macrophages that were positive for fluorescence. (B) Graph represents the mean fluorescence intensity (MFI) of the macrophage populations. Data represent three experiments (± standard deviation). (C–D) Experiments were equivalent to A and B except E. coli were labeled with pH sensitive dye pHrodo to assess changes in phagosomal pH. Data represent six experiments (± standard deviation).
We used E. coli labeled with a pH sensitive dye (pHrodo) to determine whether NKLAM is involved in regulating the decrease in phagosomal pH observed during maturation. Wild type or NKLAM-deficient BMDM were incubated with pHrodo-labeled E. coli and the percentage of fluorescently positive macrophages was assessed by flow cytometry. This population represents the percentage of macrophages that contained acidified phagosomes following bacterial ingestion. As shown in Fig. 5C, the responses of both cell types were similar, suggesting that macrophages of both genotypes have a similar ability to acidify bacteria-containing phagosomes. Additionally, we determined the MFI for these populations. In this assay, the mean fluorescence intensity can be used as a measure of the degree of macrophage phagosome acidification. As shown in Fig. 5D, we found no significant differences between WT and NKLAM-deficient macrophages with respect to the degree of phagosome acidification. Collectively, these results suggest that NKLAM does not play a significant role in either ingestion of bacteria or regulation of phagosome acidification.
3.6 NKLAM-deficient macrophages are defective in killing E. coli
We have shown previously that NKLAM-deficient NK cells are significantly defective in lysing tumor target cells [13, 14]. We next sought to determine whether NKLAM was involved in macrophage bactericidal activity. Bone marrow-derived or peritoneal macrophages were infected with E. coli (strain JM109) at an MOI of 10. After 90 min, macrophages were lysed in sterile water and viable bacteria were enumerated on LB agar plates. As shown in Fig. 6A, NKLAM-deficient BMDM are significantly defective in killing E. coli. At 90 min, WT BMDM killed an average of 77% of the E. coli while NKLAM-KO BMDM killed 57%. To confirm the observed defective killing phenotype in resident macrophages, we isolated peritoneal macrophages from WT and NKLAM-deficient mice and used them as effector cells. As shown in Fig. 6B, WT peritoneal macrophages killed an average of 40% of the E. coli and NKLAM-deficient macrophages killed 20%. Collectively, these data demonstrate that NKLAM is a positive regulator of the macrophage killing response against E. coli.
Figure 6. NKLAM-deficient macrophages have attenuated bactericidal activity.
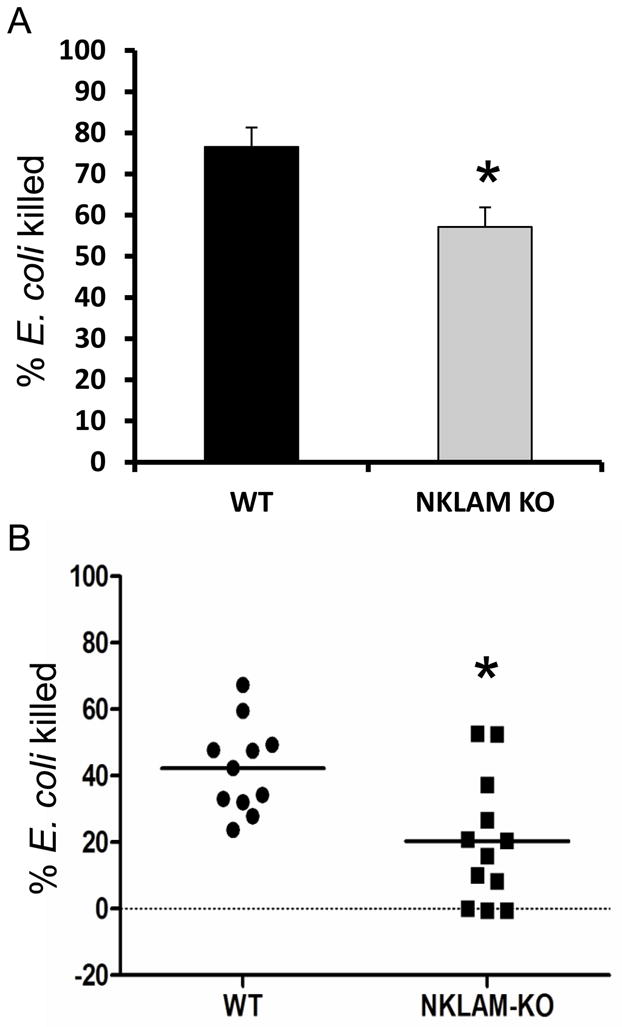
(A) WT (black bar) and NKLAM-deficient (gray bar) BMDM were incubated with live E. coli at an MOI of 10. The cultures were lysed with sterile water after 90 min and viable bacteria were enumerated by colony formation on LB agar plates (*, p = 0.03; ± standard deviation; n = 5). (B) Peritoneal macrophages were isolated from wild type (WT, n = 11) and NKLAM-deficient (NKLAM-KO, n = 12) mice and were assessed for E. coli killing ability as in (A). (*, p < 0.005).
4. DISCUSSION
In the present report, we demonstrate that the E3 ubiquitin ligase NKLAM is a component of the macrophage phagosome proteome. Additionally, our data indicate that NKLAM is a positive modulator of the macrophage killing response against E. coli and functions downstream of bacterial ingestion and phagosome acidification.
NKLAM was originally isolated from and characterized as an NK cell protein that was regulated by cytokines and involved in the NK cell killing response against tumor cell targets [13, 14]. However, cells of the monocyte/macrophage lineage also express NKLAM. Therefore, we tested the effect of LPS, a TLR4 agonist, on NKLAM expression in macrophages. Treatment of RAW264.7 and J774A.1 macrophages with LPS induced a strong and transient increase in NKLAM expression. J774A.1 cells appear significantly less responsive than RAW 264.7 cells to LPS stimulation with respect to NKLAM protein expression. In support of this observation, a study found that RAW264.7 macrophages produced more TNFα in response to LPS than J774A.1 [18]. Wild type BMDM macrophages were the most responsive to LPS and demonstrated elevated NKLAM levels at 5hr. In all cases, NKLAM expression decreases after 24 hr. This expression profile suggests that macrophage NKLAM protein is not preexisting but is rapidly translated upon stimulation. A related RBR ubiquitin ligase, Parkin, is also regulated by LPS [19]. Mutations in the parkin gene (PARK2) are associated with early-onset parkinsonism and recessive juvenile parkinsonism [20, 21]. Interestingly, polymorphisms in PARK2 have been associated with increased susceptibility to infection by Salmonella typhi, Salmonella paratyphi, and Mycobacterium leprae [22–24]. Thus, the cellular functions of RBR ubiquitin ligases are diverse and may be involved in cell signaling pathways that are common to neurological and infectious diseases.
Depending on the method of detection, the number of proteins associated with the phagosome range from over 100 [2] to several thousand [25]. These proteins are involved in phagosome trafficking, protein degradation, phagosome acidification, and antigen presentation [26]. We found that membrane-associated NKLAM was a component of stimulated macrophage phagosomes. Previous research has shown that ubiquitin is translocated to the maturing phagosome where it contributes to the formation of multivesicular bodies [3, 4]. However, studies that examine the potential contribution of ubiquitin ligases to the ubiquitination of phagosome proteins are lacking. After ingestion of opsonized beads, NKLAM expression in the phagosome was maximal at 35 min, suggesting that NKLAM may be translocated to the phagosome via the endosomal pathway. The levels of NKLAM in the phagosome decreased over time in WT BMDM. Whether NKLAM is removed from the phagosome or degraded is currently under investigation. Loss of phagosome proteins can occur through fission of vesicles from the phagosome membrane or through the invagination of the phagosome limiting membrane during the formation of multivesicular bodies, which are then degraded after fusion with lysosomes [27]. NKLAM is also capable of self-ubiquitination, which may target it for degradation by proteasomes at the phagosome surface.
Our studies indicate that NKLAM contributes to the ubiquitination of phagosomal proteins. However, the presence of ubiquitinated proteins in the phagosomes of NKLAM-deficient macrophages suggests that additional ubiquitin ligases are associated with macrophage phagosomes.
There are few studies that examine the role of ubiquitin in phagocytosis. Booth et al. demonstrated that ubiquitin conjugation machinery was not required for phagocytosis of opsonized sheep red blood cells [4]. Our studies with NKLAM-deficient mice support this observation, in that we found no difference between WT and NKLAM-deficient macrophages in the rate of ingestion or the number of E. coli ingested per macrophage. In contrast, Silva et al. demonstrated that a protein named pallbearer acting as an E3 ligase, was required for efficient phagocytosis of apoptotic cells by Drosophila macrophages [28]. Thus, the involvement of ubiquitination in regulating phagocytosis may vary depending upon the type of host macrophage and the target being ingested.
We show that both bone marrow-derived and peritoneal macrophages lacking NKLAM are significantly defective in mounting an effective killing response against E. coli. At present, precisely how ubiquitin ligases function in macrophage bactericidal activity is unknown. Ubiquitination regulates membrane protein sorting [29]; thus one could envision a role for NKLAM in regulating the trafficking of proteins related to bactericidal functions. In support of this concept, recent studies have demonstrated that inhibition of cellular deubiquitinases increased the trafficking of iNOS to phagosomes and significantly augmented macrophage L. monocytogenes killing [6]. Recent studies have also suggested that the ubiquitination machinery normally associated with autophagy is involved in recognizing and degrading bacteria that escape the phagosome into the cytoplasm. In this process, cytoplasmic bacteria become polyubiquitinated, and become associated with autophagosomes [30]. Autophagosomes then fuse with lysosomes and the bacteria cargo are degraded. In this scenario, bacterial ubiquitination would be a critical step towards efficient removal of cytoplasmic pathogens. Further studies are needed to conclusively determine which ubiquitin ligases are involved in this process and which bacterial proteins are targets for ubiquitination.
5. Conclusion
We have demonstrated that RBR family E3 ubiquitin ligase NKLAM is a novel macrophage phagosome protein. Our studies suggest that NKLAM functions downstream of the initial phagocytic event to positively regulate macrophage bactericidal activity. The elucidation of potential NKLAM substrates will be critical for determining the mechanism of action of this ubiquitin ligase. We anticipate that future studies will solidify the role of NKLAM, and possibly other RBR family ligases, as a key component of the leukocyte bactericidal machinery. The role of ubiquitination as part of the leukocyte killing response provides additional therapeutic targets for modulating bacterial killing during infection.
Highlights.
Macrophage NKLAM expression is enhanced by interferon gamma and lipopolysaccharide
NKLAM is localized to the macrophage phagosome
NKLAM is a positive regulator of macrophage bactericidal activity
Acknowledgments
We would like to thank Gail Gullickson for animal care, and expert technical assistance. We also thank Robin Chamberland for many insightful discussions. These studies were supported in part by NIH grant R56AI089758 and by a grant (1 IO1BX000705) from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development.
ABBREVIATIONS
- NKLAM
Natural Killer Lytic-Associated Molecule
- BMDM
Bone marrow-derived macrophage
- TLR
Toll-like receptor
- LPS
Lipopolysaccharide
- NK
Natural Killer
- RBR
RING in between RING
- MFI
Mean fluorescence intensity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 2.Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee WL, Kim MK, Schreiber AD, Grinstein S. Role of ubiquitin and proteasomes in phagosome maturation. Mol Biol Cell. 2005;16:2077–2090. doi: 10.1091/mbc.E04-06-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth JW, Kim MK, Jankowski A, Schreiber AD, Grinstein S. Contrasting requirements for ubiquitylation during Fc receptor-mediated endocytosis and phagocytosis. Embo J. 2002;21:251–258. doi: 10.1093/emboj/21.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 6.Burkholder KM, Perry JW, Wobus CE, Donato NJ, Showalter HD, Kapuria V, O’Riordan MX. A small molecule deubiquitinase inhibitor increases localization of inducible nitric oxide synthase to the macrophage phagosome and enhances bacterial killing. Infect Immun. 2011;79:4850–4857. doi: 10.1128/IAI.05456-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieffer AE, Goumon Y, Ruh O, Chasserot-Golaz S, Nullans G, Gasnier C, Aunis D, Metz-Boutigue MH. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. Faseb J. 2003;17:776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- 9.Purdy GE. Taking Out TB-Lysosomal Trafficking and Mycobactericidal Ubiquitin-Derived Peptides. Front Microbiol. 2011;2:7–16. doi: 10.3389/fmicb.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purdy GE, Russell DG. Ubiquitin trafficking to the lysosome: keeping the house tidy and getting rid of unwanted guests. Autophagy. 2007;3:399–401. doi: 10.4161/auto.4272. [DOI] [PubMed] [Google Scholar]
- 11.Fortier JM, Kornbluth J. NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J Immunol. 2006;176:6454–6463. doi: 10.4049/jimmunol.176.11.6454. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209–218. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlowski M, Schorey J, Portis T, Grigoriev V, Kornbluth J. NK lytic-associated molecule: a novel gene selectively expressed in cells with cytolytic function. J Immunol. 1999;163:1775–1785. [PubMed] [Google Scholar]
- 14.Hoover RG, Gullickson G, Kornbluth J. Impaired NK cytolytic activity and enhanced tumor growth in NK lytic-associated molecule-deficient mice. J Immunol. 2009;183:6913–6921. doi: 10.4049/jimmunol.0901679. [DOI] [PubMed] [Google Scholar]
- 15.Portis T, Anderson J, Esposito A, Kornbluth J. Gene structure of human and mouse NKLAM, a gene associated with cellular cytotoxicity. Immunogenetics. 2000;51:546–555. doi: 10.1007/s002510000182. [DOI] [PubMed] [Google Scholar]
- 16.Ullrich HJ, Beatty WL, Russell DG. Direct delivery of procathepsin D to phagosomes: implications for phagosome biogenesis and parasitism by Mycobacterium. Eur J Cell Biol. 1999;78:739–748. doi: 10.1016/S0171-9335(99)80042-9. [DOI] [PubMed] [Google Scholar]
- 17.Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 2008;376:5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 18.Heming TA, Tuazon DM, Dave SK, Chopra AK, Peterson JW, Bidani A. Post-transcriptional effects of extracellular pH on tumour necrosis factor-alpha production in RAW 246.7 and J774 A.1 cells. Clin Sci (Lond) 2001;100:259–266. [PubMed] [Google Scholar]
- 19.Tran TA, Nguyen AD, Chang J, Goldberg MS, Lee JK, Tansey MG. Lipopolysaccharide and tumor necrosis factor regulate Parkin expression via nuclear factor-kappa B. PLoS ONE. 2011;6:e23660. doi: 10.1371/journal.pone.0023660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cookson MR. The biochemistry of Parkinson’s disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 21.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 22.Ali S, Vollaard AM, Widjaja S, Surjadi C, van de Vosse E, van Dissel JT. PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin Exp Immunol. 2006;144:425–431. doi: 10.1111/j.1365-2249.2006.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra D, Darvishi K, Lohra M, Kumar H, Grover C, Sood S, Reddy BS, Bamezai RN. Association study of major risk single nucleotide polymorphisms in the common regulatory region of PARK2 and PACRG genes with leprosy in an Indian population. Eur J Hum Genet. 2006;14:438–442. doi: 10.1038/sj.ejhg.5201563. [DOI] [PubMed] [Google Scholar]
- 24.Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, Sarno EN, Alter A, Montpetit A, Moraes ME, Moraes JR, Dore C, Gallant CJ, Lepage P, Verner A, Van De Vosse E, Hudson TJ, Abel L, Schurr E. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 25.Trost M, English L, Lemieux S, Courcelles M, Desjardins M, Thibault P. The phagosomal proteome in interferon-gamma-activated macrophages. Immunity. 2009;30:143–154. doi: 10.1016/j.immuni.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33:397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Silva E, Au-Yeung HW, Van Goethem E, Burden J, Franc NC. Requirement for a Drosophila E3-ubiquitin ligase in phagocytosis of apoptotic cells. Immunity. 2007;27:585–596. doi: 10.1016/j.immuni.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 30.Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]