Abstract
Dragline silk from orb-weaving spiders is a copolymer of two large proteins, major ampullate spidroin 1 (MaSp1) and 2 (MaSp2). The ratio of these proteins is known to have a large variation across different species of orb-weaving spiders. NMR results from gland material of two different species of spiders, N. clavipes and A. aurantia, indicates that MaSp1 proteins are more easily formed into β-sheet nanostructures, while MaSp2 proteins form random coil and helical structures. To test if this behavior of natural silk proteins could be reproduced by recombinantly produced spider silk mimic protein, recombinant MaSp1/MaSp2 mixed fibers as well as chimeric silk fibers from MaSp1 and MaSp2 sequences in a single protein were produced based on the variable ratio and conserved motifs of MaSp1 and MaSp2 in native silk fiber. Mechanical properties, solid-state NMR, and XRD results of tested synthetic fibers indicate the differing roles of MaSp1 and MaSp2 in the fiber and verify the importance of postspin stretching treatment in helping the fiber to form the proper spatial structure.
INTRODUCTION
Dragline silks from orb-weaving spiders display both high tensile strength and elasticity making them tougher than almost all natural and synthetic materials.1 The extraordinary mechanical properties of spider silks have been noted since ancient time. However, only in the late 20th century have researchers started to unravel the reasons for the unique properties that spider silk possesses.2,3 Unlike silkworms, the territorial nature of spiders makes them difficult to raise for silk farming. Thus, understanding the molecular nature of spider silk and creating synthetic fibers to mimic native spider silk using modern bioengineering techniques is the most viable way to use this ancient material and is a major focus of current spider silk research.
Typical orb-weaving spiders have seven types of silk gland; each type of gland produces a single silk fiber with distinct properties and functions.4 Major ampullate gland silk (also known as dragline silk) is the strongest of all. It is more than three times tougher than the man-made fiber, Kevlar, and five times stronger than steel wire by weight.5,6 Spiders use dragline silk to build the main framework of their web and also use it as a lifeline and walking thread to quickly move between places safely. Two main proteins comprise the dragline silk: major ampullate Spidroin 1 (MaSp1)7 and major ampullate Spidroin 2 (MaSp2).8 Previous studies on dragline silk revealed that the amino acids are organized as several basic motifs repeated hundreds of times in the silk protein.9–11 Those highly conserved motifs have been retained with little change for more than 150 million years in the Araneidae spider family.12,13 MaSp1 is composed of two distinct motifs: (i) a polyA region which forms tightly knit crystalline β-sheet structures and contribute to the tensile strength of silk10 and (ii) a GGX motif (X = L, Y, Q, A), which based on recent NMR results, forms a glycine II helix.14–16 The function of this motif is still unclear, but it is believed to have effects on fiber formation by providing a stabilizing energy/force between proteins.17,18 The MaSp2 sequence can be divided into two motifs as well: (i) a GPGXX repetitive region (X = G, Q, Y), which forms a β-spiral and is responsible for the elasticity of the silk19,20 and (ii) a polyA region similar to MaSp1.10 These motifs control various structural and functional aspects of the silk that help balance the strength and elasticity of the fibers. By manipulating the amount and ratio of different motifs, synthetic fibers with different mechanical properties could be created.
Several groups have produced recombinant spider silk proteins based on MaSp1 or MaSp2 consensus sequences in a great array of organisms,21–26 including E. coli, yeast, tobacco, silk worm, mammalian cell, or even from goat’s milk. However, MaSp1 and MaSp2 in native spider silk always come together, with a ratio that varies between species.27,28 Using amino acid and cDNA analyses, the ratio of MaSp1 to MaSp2 in dragline silks have been determined from two spider species: N. clavipes and A. aurantia. For N. clavipes, an average of 80% of its major ampullate silk is formed by MaSp1,29 while only around 40% of the dragline silk protein in A. aurantia is MaSp1.30 MaSp1 and MaSp2 coexist in the silk gland and later spun into a single fiber.6 Several concepts of fiber structural model has been proposed,31,32 however, how MaSp1 and MaSp2 interact during fiber assembly and their precise roles in the fiber mechanical properties are still unknown.
In this study, we produced four types of synthetic spider silk fibers. Two were MaSp1 and MaSp2 protein mixtures with the ratios of two proteins close to those of native dragline silk in N. clavipes and A. aurantia. The other two types of fibers were single chimeric proteins with amino acids sequences from both MaSp1 and MaSp2 in different ratios. The mechanical properties of all four types of fibers were tested. Solid-state NMR and XRD were used to analyze both natural spider silk protein from dissected gland as well as dragline silk fibers and then compared closely with our synthetic mimics to investigate the differing roles of MaSp1, MaSp2 and the effect of postspin stretching in fiber spinning.
There are three major reasons to design, express, test, and compare MaSp1/MaSp2 mixtures and chimeric fibers in this study: First reason is to assess the role MaSp1 and MaSp2 play in synthetic fiber, orb web spiders have been evolving for over 150 million years, so there must be critical reason(s) for dragline silk to contain two distinct proteins. Second reason is to assess how different MaSp1 and MaSp2 ratios would affect fiber properties, so synthetic spider silk biomaterials could be better tuned for their specific applications. For example, bullet-proof vests or artificial bone replacements need high tensile strength with little extensibility, however, good artificial ligament or airbag materials need a substantial elasticity. With current knowledge of silk protein motifs, we believe the mechanical properties of silk can be controlled by altering the ratios of each motif.21,33 For example, if we want more elasticity in a specific fiber, add more GPGXX motif. Third reason is to determine whether chimeric protein fibers could replace the mixed protein fibers by providing better and more consistent mechanical properties. Consistency is definitely one important factor in fiber production. By mixing two proteins together, we cannot control the distribution of MaSp1 and MaSp2 in the fiber during fiber spinning. This could potentially cause consistency problems. A chimeric fiber, on the other hand, is a single repetitive protein, with evenly distributed protein motifs from both proteins throughout the whole fiber, which could eliminate this problem.
MATERIALS AND METHODS
Natural Glands Protein and Silk Collection
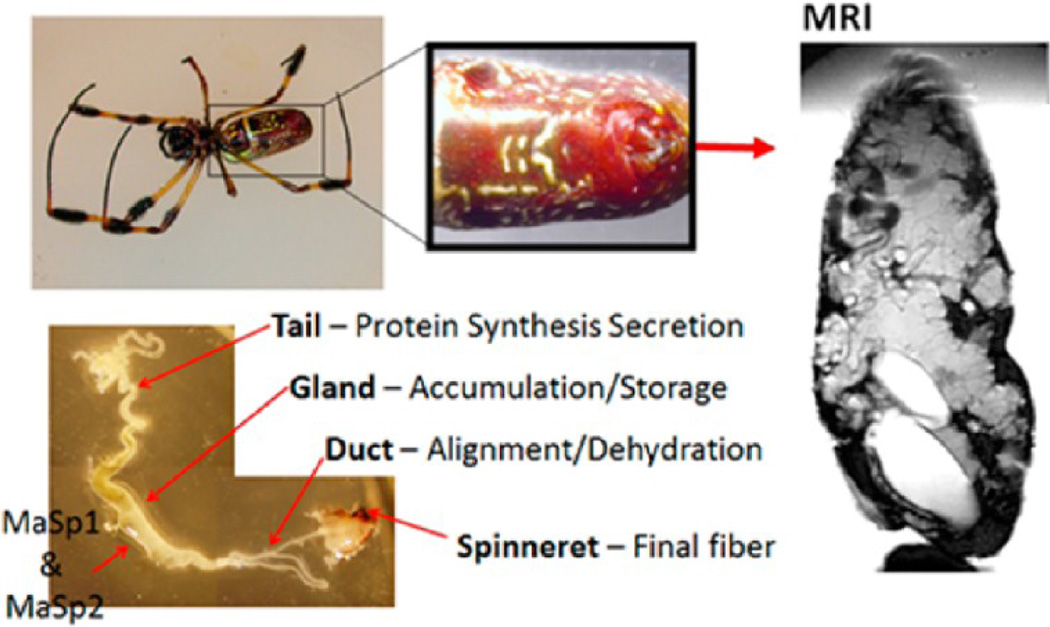
Adult female N. clavipes and A. aurantia were fed a 10% w/v solution of uniformly 13C/15N-enriched alanine (U-13C, 15N-Ala) in water every other day to isotopically label the protein solution in the glands. The metabolic distribution of isotopes is complex but, in general, the U-13C/15N-alanine labeling protocol enriches 13C for alanine, glycine, glutamine, and serine in spider silk.17 The spiders were forcibly silked34 at a rate of 2 cm/s for 1 to 1.5 h during each feeding session to remove unlabeled protein from the major glands and to ensure that the new protein solution in the gland would incorporate the U-13C15N alanine. Major ampullate silk was collected during silking with a dissection microscope to ensure other types of silk were not combined with the major samples. On average, spiders drank between 20 and 100 µL of the U-13C15N alanine solutions per feeding, depending on the size of the spider. Spiders were not silked on the feeding prior to dissection to ensure that the protein solution inside the major glands was not depleted. All spiders were anesthetized with carbon dioxide for approximately 60 s prior to dissection. Spiders were dissected using microscissors and blades under a dissection microscope. The major ampullate glands were identified by their size, shape, and location. During dissection, DI water was utilized to irrigate the abdomen to prevent dehydration. The major glands were removed immediately from the spider abdomen and placed on glass slides. These glands were then placed in a covered beaker to prevent particles from settling on the gland while it dried. To shear the glands, a gland was placed between two slides and the top slide was pressed unilaterally against the bottom slide to produce a protein film approximately 3 times the original length of the gland. The slides were held in place until the film had completely dried to produce a sheared film. The glands were dried for at least a week prior to experiments. Figure 1 illustrates the spider abdomen as well as the structure of major ampullate silk gland. For the X-ray measurements, N. clavipes major ampullate gland was retrieved as mentioned above, except without feeding the spiders with U-13C15N isotopes. Once dried, the glands were carefully removed from the glass slide with a razor blade to ensure that the gland remained in a single piece. The glands were then mounted on metal washers. To mount the gland, the main section of the gland was centered over the hole in the washer and small amounts of super glue were utilized to tack the gland ends onto the washer. Care was taken to prevent any glue from contaminating the detection area of the gland.
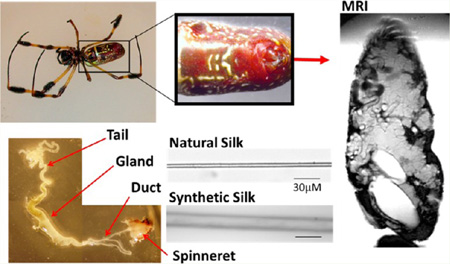
Figure 1.
Nephila clavipes spider picture and associated glands. Also, a magnetic resonance image (MRI) is shown of the abdomen of the same spider. The major ampullate glands can easily be discerned from this image.
Synthetic Silk Gene Cloning
Synthetic spider silk genes were made based on the N. clavipes MaSp1 and MaSp2 conserved amino acids sequence (Table 1). The last four constructs in Table 1 are the genes expressed in this study. The basic MaSp1 repeating unit contains multiple GGX motifs followed by a hexa-polyalanine tail. The MaSp2 repeating unit contains two GPGXX motifs (GPGGYGPGQQ) followed by 8-alanine strength motif (row 2 in Table 1). MaSp1 was duplicated 24 times, while MaSp2 was duplicated 16 times in order to have similar protein molecular weights. The repeating units of the chimeric silk genes in this study were made using one MaSp1 repeating unit followed by either four or eight MaSp2 elasticity cassettes, and then a strength 8-alanine cassette. The repeating units were duplicated 8 or 6 times to make a silk gene around 2.4 kb. These two constructs were named (148)8 or (188)6, respectively. Duplication of the monomers was achieved by a compatible, but nonregenerable, cloning strategy.35 All monomers were flanked by 5′-XmaI and 3′-BspEI. Gene manipulation was completed in pBluescript II SK+ (Ampr) vector in E. coli strain GM2163 (Cmr). The final constructs were ligated into pET19K (a modified pET-19b vector where the ampicillin resistance gene was replaced with the kanamycin resistance gene from the pET-26b vector)35 through 5′-NdeI and 3′-BamHI, then transformed into E. coli strain BL21(DE3) for protein expression. A deca-histidine-tag was included at the N-terminal of the silk protein for purification purposes. Restriction enzymes used in the gene cloning were purchased from New England Biolabs (Ipswich, MA). DNA electrophoresis was performed on each step of cloning using an 8% agarose gel at 100 V in TAE buffer, gels were stained by ethidium bromide and visualized under UV light. Chemicals used in all studies were purchased from Sigma Aldrich unless otherwise indicated. Plasmid sequencing was performed in University of Wyoming, Nucleic Acid Exploration Facility (NAEF).
Table 1.
Synthetic Spider Silk Protein Sequences with Their Gene and Protein Sizes Listeda
| protein sequence | DNA size (bp) | protein Mw (kDa) | |
|---|---|---|---|
| elasticity cassette (E) | GPGGYGPGQQ | 30 | |
| strength 8 cassette (S8) | GPGGPSGPGSA8 | 54 | |
| (MaSp1)24 | (GGAGQGGYGGLGSQGAGRGGLGGQGAGA6)24 | 2376 | 66 (62) |
| (MaSp2)16 | (GPGGYGPGQQGPGGYGPGQQGPSGPGSA8)16 | 1680 | 48 (47) |
| chimeric (148)8 | (PGG + MaSp1 + LAGPGQQG + E4 + S8)8 | 2448 | 70 (68) |
| chimeric (188)6 | (PGG + MaSp1 + LAGPGQQG + E8 + S8)6 | 2556 | 75 (73) |
For protein molecular weight, numbers in the parentheses are calculated Mw. His-tag sequence at N-terminal of the proteins is not shown and calculated.
Recombinant Protein Expression
Positive colonies were tested by small scale (5–20 mL) protein expression in LB media before large-scale protein production. This was done by growing 12 selected colonies in LB media in small culture tubes (10–50 mL) until an OD600 of 0.8. Follow-up IPTG induction, cell collection and silk protein detection processes were similar to the large-scale expression method described below. E. coli colonies with highest silk protein yields were selected for large-scale protein production. Selected colonies were then grown in a 19.5L BioFlo 415 Fermentor (New Brunswick Scientific, Edison, NJ) with fermentor recommended media (provided in the New Brunswick fermentor standard protocols) overnight till it reached an OD600 of 15–20, then 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside, Biosynth AG, Switzerland) was added to induce synthesis of synthetic silk protein from lac promoter. Induced cells were allowed to grow for 4 h before harvesting. The resultant bacteria culture was collected and centrifuged at 3500 rpm for 20 min (Allegra 6KR, Beckman, Brea, CA). Cell pellets were resuspended in lysis buffer (0.25 mg/mL lysozyme, 5 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 8.0) at a weight to volume ratio of 1:3. The resuspended cell mixtures were immediately frozen in the −80 °C freezer for at least 24 h.
Protein Purification and Analysis
Approximately 800 mL of cell suspension was thawed and sonicated for two 4 min bursts at 90W to ensure complete lysis. Cell debris was separated by centrifugation at 13000g for 25 min (Beckman J2–21M) and discarded. Supernatants were heat treated at 80 °C in a water bath for 15 min to precipitate heat unstable E. coli proteins. Spider silk proteins of interest remained soluble with this treatment. The supernatant containing synthetic silk protein after heat treatment was collected by 25 min centrifuge at 13000g, and the silk protein with a 10X N-terminal His-tag was captured and purified by immobilized metal ion(nickel) affinity chromatography (IMAC) using an ÄKTAExplorer (GE Healthcare, Piscataway, NJ). The protein binding column was washed with wash buffer (100 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 7.9), except MaSp2, which was washed with 50 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 7.9. Silk protein was eluted with buffer at an imidazole concentration of 250 mM and the same concentration of other salts. Silk protein eluates were concentrated to 150–200 mL and then dialyzed against ddH2O in a stirred cell with a 30 kDa membrane (Millipore Amicon, Billerica, MA) with at least 5 vol of doubly distilled H2O. Dry protein powders were produced by lyophilization of dialyzed silk protein solution. Protein electrophoresis was done on precast 4%–20% SDS-PAGE gel (Precise™ Protein Gel, Pierce Biotechnology, Rockford, IL) at 90 V with the Tris-Hepes-SDS buffer. Silk proteins of interest were detected by either His-tag In-gel stain (InVision His-Tag In-gel Stain, Invitrogen, Carlsbad, CA) or traditional Western blot with His-tag antibody (Novagen) following a standard protocol.33 Coomassie staining with Bio-Safe Coomassie (Bio-Rad, Hercules, CA) was used to visualize all proteins on the gel. Amino acids analysis was performed at University of Wyoming Macromolecular Core Equipment Facility (MCEF) with their standard protocols. MALDI-TOF mass spectrometry were performed at Tufts Chemistry Department on a Microflex LT system (Bruker Corporation, Billerica, MA) with 75% laser intensity using standard LP (linear positive) 60 kDa method provided by the software, sinapic acid (Sigma) was used as MALDI matrix.
Fiber Spinning and Postspin Treatment
Spinning dopes were made by dissolving lyophilized silk protein powders into 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP; TCI America, Portland, OR). For MaSp1/MaSp2 mixed fibers, (MaSp1)24 and (MaSp2)16 proteins were mixed at a molar ratio of 4:1 or 1:1 and dissolved in HFIP. The protein dopes in glass vials were put on a mixing rotor for 2 days and vortexed regularly to achieve full dissolution and maximum mixing. Protein concentration in the spinning dope was about 30% (w/v). Fiber spinning was performed on a spinning apparatus custom designed at DACA Instruments (Santa Barbara, CA). Spinning dopes were loaded into 1 mL Hamilton syringes (Hamilton, Reno, NV) and extruded through 0.005 in. PEEK tubing (SUPELCO, Bellefonte, PA) at a syringe plunger speed of 0.7 mm/min into a 100% isopropanol coagulation bath. Fibers formed in the coagulation bath were lifted out carefully with tweezers, threaded onto two sets of spinning godets (speed 0.5–1 m/min) and then collected on a winder spool. Usually, at this setting, 2–3 m of fibers could be collected without breakage. As-spun fibers were examined by eye for visible large defects. Defect-free fibers were cut into 2 cm pieces. Fibers were soaked in 75% isopropanol/water for 1 min to achieve maximum water penetration. Then, the fibers were stretched from one end slowly by tweezers till it reaching a total length of 6 cm (3× stretch).
Supercontraction Test
Postspin stretched fibers were cut into 3 cm pieces, and pictures of individual fibers were taken under microscope at 10× ocular lens plus 40× objective lens (Nikon Eclipse E200 microscope). Fiber diameter was measured at 9 different places using ImageJ 1.42q (National Institute of Health, USA). The average was calculated to determine the diameter of each fiber sample. Fibers were soaked in pure water for one minute. The lengths and diameters of the contracted fibers were measured again in the same way. No mechanical tests were done on supercontracted fibers in this study.
Mechanical Testing
Collected natural fibers or stretch treated synthetic fibers were air-dried, cut, and mounted onto a 30 × 20 mm rectangular testing card with a 15 mm square gap in the middle of the card. Microscope pictures of each fiber were taken and diameters were calculated, as described above. Mechanical tests were done at 25 °C with approximately 18% humidity on an MTS Synergie 100 (MTS corporation, Eden Prairie, MN) using a 10 g custom-built load cell (Transducer Techniques, Temecula, CA) at a pulling rate of 10 mm/min, with data collection at 30 Hz frequency. The data was plotted in Matlab 7.6.0 (R2008a) Mac and a polynomial regression curve was fit to the data points with a seventh degree polynomial.
Solid State Nuclear Magnetic Resonance Studies
Solid state nuclear magnetic resonance studies of all samples (5–10 mg each) were done in Arizona State University’s Magnetic Resonance Research Center (MRRC). All 13C solid-state NMR (ssNMR) data was collected on a 400 MHz Varian VNMRS wide-bore spectrometer utilizing a 3.2 mm triple resonance MAS probe operating in 1H/13C mode. The 13C isotropic chemical shift was referenced to the downfield peak of adamantane (38.56 ppm). Samples were all center-packed in a 3.2 mm zirconia rotor. 1H → 13C cross-polarization (CP) was performed at 10 kHz magic angle spinning (MAS), with the 1H → 13C CP condition matched to the −1 spinning sideband (ssb) of the Hartmann–Hahn (HH) profile. A 1 ms contact pulse was utilized with two-pulse phase-modulated (TPPM) applied during acquisition with an rf-field strength of 100 kHz. A 4 s recycle delay was utilized in all 1H → 13C CP experiments.
X-ray Fiber Diffraction
X-ray fiber diffraction (XRFD) was performed on the BioCars 14BM-C beamline at the Advanced Photon Source at Argonne National Laboratory, Argonne, IL, U.S.A. The wavelength of the X-ray beam was 0.9 Å, with a flux of 6 × 1011 photons/sec, and the beam size on the sample was 150 × 200 µm. Data were recorded using an ADSC Quantum-315 detector. A bundle of 20 individual fibers of 4:1 synthetic silk or N. clavipes major ampullate silk were held taut (but unstretched) in a metal frame, with the fiber axis normal to the X-ray beam. The N. clavipes major ampullate gland material was prepared on a metal frame, as described in Materials and Methods. The sample to detector distance was 200 mm for the N. clavipes major ampullate dehydrated gland material and silk and 150 mm for 4:1 synthetic silk. Data collection times were 30 s for one image for the N. clavipes major ampullate silk, 180 s for the 4:1 synthetic silk, and 120 s for the gland material. Background measurement was performed with the sample displaced from the beam and the image was recorded under the same conditions, as used with the sample in the beam. Multiple images were taken to get better statistics and improve on the signal/background ratio. CeO2 powder was used for instrument calibration.
RESULTS
For gene cloning, each step of cloning was confirmed by restriction enzyme digest and DNA sequencing. SDS-PAGE and MALDI-TOF MS confirmed the correct size of the protein produced by fermentation (Figure S1). The average yield of recombinant silk protein from the soluble fraction of bacteria lysate is approximately 120 mg/L of media. Small amounts of each type of protein were analyzed for amino acid content to confirm the correct amino acid composition. As-spun fibers were postspin stretched in a 75% isopropanol/water solution, as these synthetic as-spun fibers are soluble or semisoluble in pure water. The malleability of different types of fibers actually differs in the solution, for example, (188)6 fiber could easily be stretched 10×, while the 4:1 fiber barely made 3×. For comparison purposes, all fibers were stretched 3× for mechanical testing. Postspin stretched fibers were no longer soluble in pure water, except (188)6 fibers, which became very weak and gel-like. They easily broke when trying to pick them up by tweezers, presumably because of the low amount of hydrophobic strength cassettes (polyA) in the protein.
It was noticed during postspin stretching that the chimeric fibers supercontracted like the native dragline silk in humid conditions.36 No significant water contraction was seen in MaSp1 only37 or MaSp1/MaSp2 mixed fibers. A preliminary supercontraction study was carried out to determine the length and diameter change of those synthetic chimeric fibers. A total of 10 (148)8 fibers were tested with an original length of 3 cm; after contraction, the average length was 1.23 ± 0.13 cm, which was a 59 ± 4% shrink. The average diameters were 34.70 ± 1.35 µm before contraction and 57.89 ± 5.51 µm after, a 40 ± 6% increase in diameter. Individual fiber length and diameter changes are listed in Table S1. Besides diameter change, the fiber morphology also noticeably changed. Figure S2 shows the microscopic images of (148)8 fibers before and after supercontraction. The original straight fiber became slightly wavy with increased surface roughness after supercontraction, likely due to the contraction ratio difference between the water-resistant polyA regions and the supercontraction sensitive GPGXX regions. Changes on the optical refraction pattern of the fiber may also indicate regions with different levels of water penetration. Although (188)6 fibers also supercontracted drastically in water, it also became weak and easily broken. No tests were done on (188)6 because we could not lift the contracted fibers out of water. Noteworthy, (188)6 fibers also contracted in aqueous isopropanol solutions with decreased contraction ratio, however, no change in length was observed when soaked in 100% isopropanol.
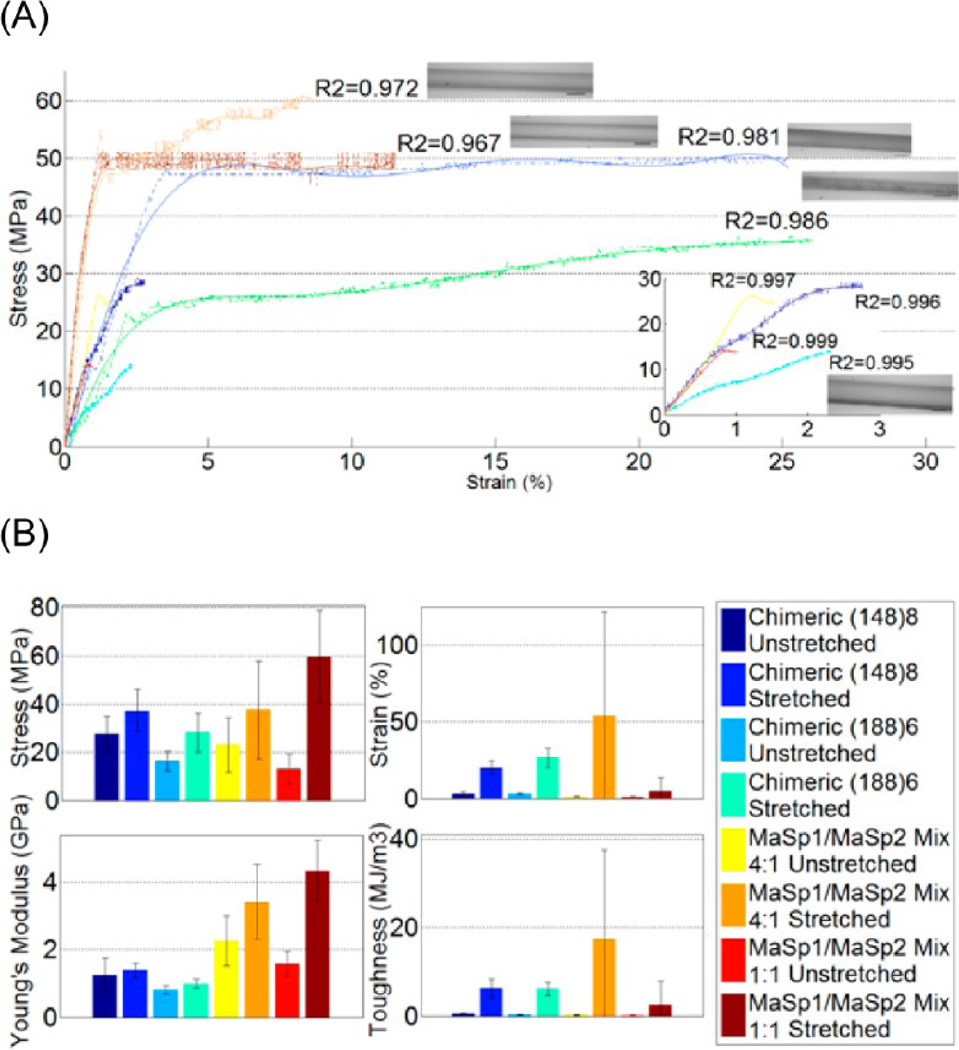
Mechanical properties of both as-spun and postspin stretch treated synthetic silk fibers were measured and are summarized in Table 2 and shown graphically in Figure 2. Table 4 shows selected better performers from each fiber group to represent the best single fiber produced in this study and compare them with best natural dragline silks from N. clavipes and A. aurantia. A total of 10 or more fibers in each group were tested. The average diameters of as-spun fibers were 58.13 ± 4.93 µm, while 29.12 ± 5.44 µm were the average diameters for postspin stretched fibers.
Table 2.
Summary of the Average Mechanical Test Performance of All the Fibers Tested in the Study
| materials | strength; σmax(MPa) | extension; εmax (%) | Young’s modulus; Einitial (GPa) | toughness (MJ/m3) | |
|---|---|---|---|---|---|
| chimeric (148)8 | as-spun | 27.54 | 3.00 ± 1.38 | 1.26 ± 0.49 | 0.45 ± 0.19 |
| stretched | 37.26 ± 8.70 | 20.07 ± 4.53 | 1.39 ± 0.21 | 6.18 ± 2.10 | |
| chimeric (188)6 | as-spun | 16.26 ± 3.94 | 3.13 ± 0.55 | 0.81 ± 0.12 | 0.24 ± 0.05 |
| stretched | 28.07 ± 8.08 | 26.55 ± 6.52 | 0.99 ± 0.13 | 6.09 ± 1.48 | |
| MaSp1/MaSp2 4:1 mix | as-spun | 23.15 ± 11.33 | 1.16 ± 0.36 | 2.26 ± 0.74 | 0.18 ± 0.13 |
| stretched | 37.63 ± 20.35 | 53.87 ± 67.95 | 3.42 ± 1.10 | 17.36 ± 20.10 | |
| MaSp1/MaSp2 1:1 mix | as-spun | 13.08 ± 6.24 | 0.88 ± 0.42 | 1.58 ± 0.37 | 0.07 ± 0.07 |
| stretched | 59.61 ± 19.23 | 4.83 ± 8.64 | 4.33 ± 0.91 | 2.52 ± 5.43 | |
Figure 2.
Mechanical testing data analysis of synthetic silk fibers (A) Typical stress–strain curves (dash dot line) of chimeric (148)8 as-spun (dark blue)/stretched (blue), chimeric (188)6 as-spun (cyan)/stretched (green), MaSp1/MaSp2 = 4:1 As-spun (yellow)/stretched (orange), MaSp1/MaSp2 = 1:1 as-spun (red)/stretched (maroon), with trendline (solid line with corresponding colors) fitted to an order of 7th degree polynomial. Inserted chart shows as-spun fiber only. Microscopic pictures next to the curve represent each type of fiber. Scale bar = 30 µm. Only one picture is shown for as-spun fiber. (B) Column charts present the average performance in stress, strain, Young’s modulus, and toughness of all the fibers tested in each group. All error bars represent standard deviation, with n ≥ 10.
Table 4.
Selected Better Performers from Each Synthetic Fiber Group Compared with Native Dragline Silks
| materials | strength; σmax (MPa) |
extension; εmax (%) |
Young’s modulus; Einitial (GPa) |
toughness (MJ/m3) |
|---|---|---|---|---|
| chimeric (148)8 | 50.08 | 25.21 | 1.44 | 11.4 |
| chimeric (188)6 | 37.81 | 33.45 | 1.13 | 7.52 |
| MaSp1/MaSp2 4:1 mix | 73.73 | 104.43 | 2.79 | 47.05 |
| 21.99 | 200.25 | 1.62 | 34.13 | |
| MaSp1/MaSp2 1:1 mix | 69.02 | 34.65 | 4.5 | 21.63 |
| 100.20 | 1.84 | 6.17 | 1.1 | |
| N. clavipes dragline | 1854 | 20 | 10.2 | 223 |
| A. aurantia dragline | 1702 | 34 | 5.6 | 180 |
Stress–strain curves of all fibers showed good polynomial fits at an order of 7, as reflected by R2 value of the polynomial regression in each trendline (R2 > 0.97). Generally, postspin stretched fibers clearly showed superior mechanical properties with improved fiber strength (tenacity) and extensibility. The rupture sites of the postspin stretched fibers are close to the middle of the fibers, while as-spun fibers are more brittle and tend to break at the end point in contact with the glue and clamp. The average increase in tenacity is 12 MPa, with a greater than 15% increase in extensibility. (148)8 does show better tenacity, while extending less than (188)6. This result supports our hypothesis of the roles of the strength and elasticity motifs, as (148)8 has more strength cassettes and less elasticity cassettes than (188)6. When compared, the chimeric fibers with 4:1 and 1:1 mixture fibers, no significant mechanical property differences were observed. However, our chimeric fiber does show much better consistency in all mechanical properties. For the 4:1 mixture fibers, the extensibility of individual fibers varied substantially, resulting in large error bars in both extension and toughness. To better present the mechanical properties of MaSp1/MaSp2 mixture fibers, the fibers have been further divided into three subgroups based on their extensibility (Table 3). In the case of the 4:1 mixture fiber, four fibers extended more than 80%, with the highest extending 200% and a second fiber from this group having a 104% elongation. Four fibers extended close to 15%. The other three fibers extended less than 2%. The 1:1 mixture fiber data, compared with 4:1 fiber, was more uniform, but most of them did not extend much before breaking (~5%).
Table 3.
Further Categorization of Stretched MaSp1/MaSp2 Mixture Fibers Based on Fiber Extension (in Bold)
| fiber group | number of fibers in subgroups |
strength; σmax (MPa) |
extension; εmax (%) |
Young’s modulus; Einitial (GPa) |
toughness (MJ/ m3) |
|---|---|---|---|---|---|
| MaSp1/MaSp2 4:1 mix stretched | 4 | 47.20 ± 22.33 | 132.01 ± 49.54 | 3.21 ± 1.26 | 41.90 ± 7.51 |
| 4 | 41.83 ± 18.89 | 15.45 ± 5.04 | 4.01 ± 1.16 | 5.76 ± 3.45 | |
| 3 | 19.27 ± 7.97 | 0.92 ± 0.21 | 2.92 ± 0.73 | 0.13 ± 0.06 | |
| MaSp1/MaSp2 1:1 mix stretched | 2 | 60 ± 12.76 | 23.09 ± 16.36 | 4.4 ± 0.15 | 13.52 ± 11.47 |
| 8 | 59.55 ± 20.44 | 2.02 ± 0.84 | 4.32 ± 0.98 | 0.83 ± 0.49 |
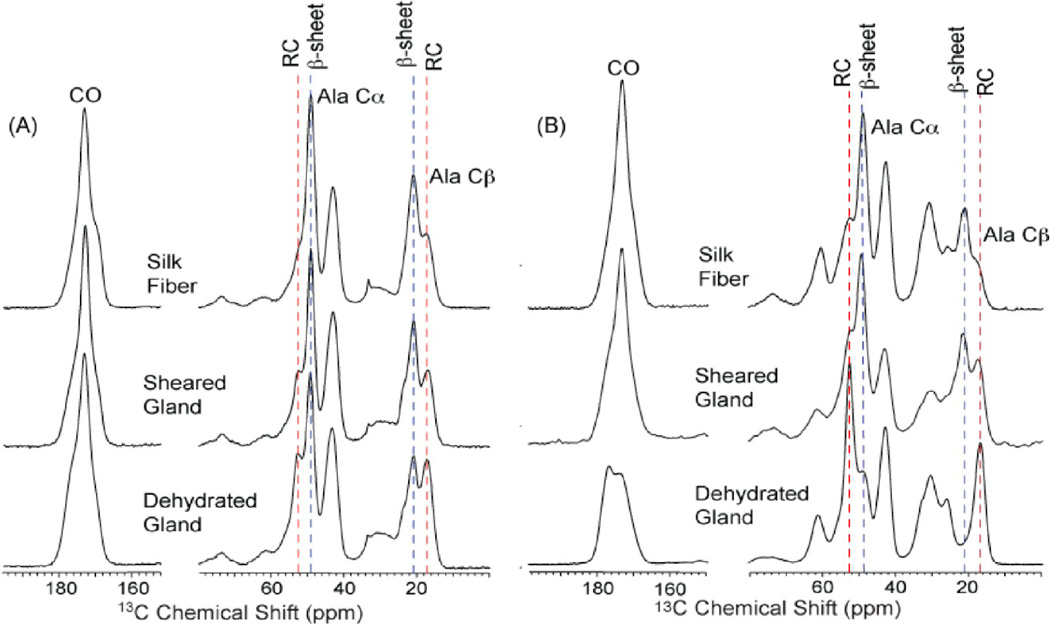
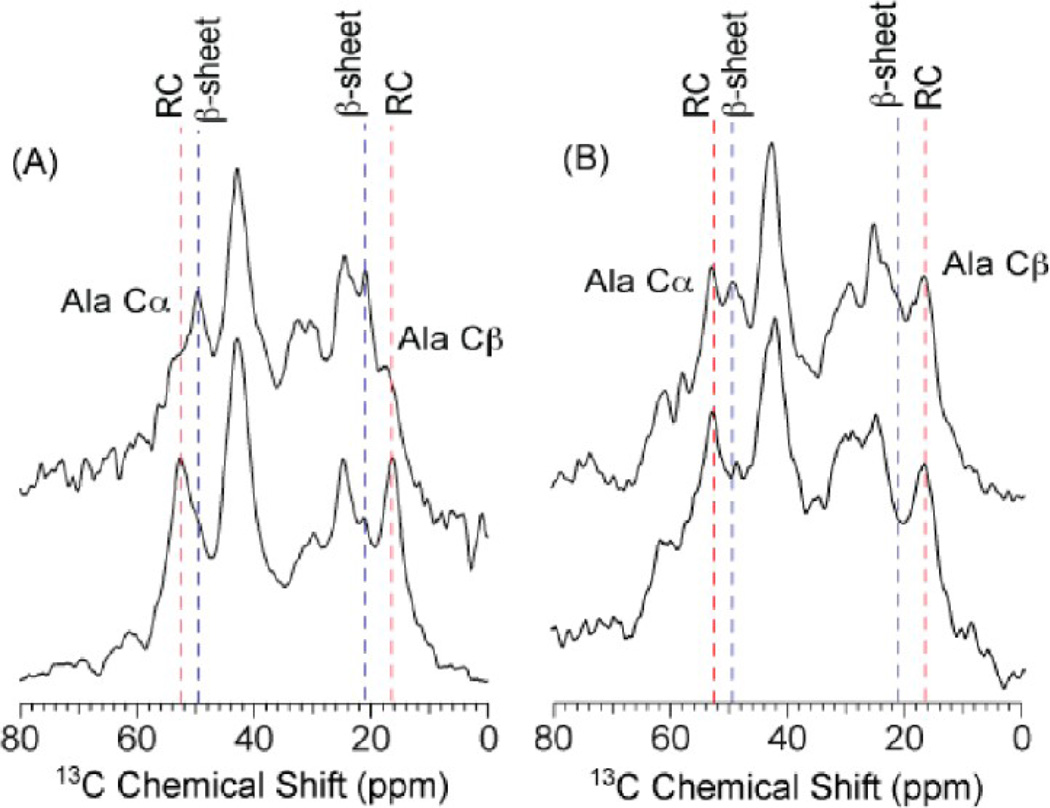
The 1H → 13C cross-polarization magic angle spinning (CP-MAS) solid-state nuclear magnetic resonance (ssNMR) spectra of the major ampullate gland and forcibly pulled spider dragline silk is presented in Figure 3. The spectra were collected on glandular materials extracted from A. aurantia and N. clavipes adult female spiders and dragline silk (fiber) was forcibly collected at 2 cm/s. In Figure 3, the focus is on the 13C alanine chemical shifts, which are sensitive to the protein secondary structure.38 Specifically seen is the amount of β-sheet formation for alanine, which is known to be the dominant structure in the polyalanine region of natural spider dragline silk. This is shown for both A. aurantia and N. clavipes spider spiders, with their forcibly dragline silk fibers showing primarily β-sheet structure in the CP-MAS ssNMR spectra at the top of Figure 3. Conversely, the dehydrated gland material for A. aurantia spiders has almost no β-sheet structure and is instead found as a helical and random coil structure. This in contrast with the same results from the N. clavipes spider dehydrated gland material where almost half the material is in a β-sheet conformation and half is in a helical or random coil conformation. The major difference between A. aurantia and N. clavipes major ampullate gland and fiber material is the ratio of the two proteins, MaSp1 and MaSp2. Hence, the observations of this indicate that MaSp1 has a high propensity toward spontaneous β-sheet formation upon removal of water (dehydration). Furthermore, it can be crudely shown that shearing aids in the formation of β-sheet and does so for both MaSp1- and MaSp2-rich silk materials.
Figure 3.
1H → 13C CP-MAS ssNMR data of U-13C/15N alanine-labeled (A) N. clavipes and (B) A. aurantia major ampullate silk fibers, sheared glands, and dehydrated glands. The conversion of alanine from a random coil (RC) conformation (blue dashed line) in the dehydrated gland into a β-sheet (red dashed lines) secondary structure in the fiber is shown.
The 1H → 13C CP-MAS ssNMR data of two types of synthetic fibers in the as-spun and stretched form are seen in Figure 4. The stacked plot of the MaSp1/MaSp2 4:1 fibers in the as-spun and stretched form (Figure 4A) highlights the difference in chemical shifts of the alanine resonances, which show the changes that occur to the secondary protein structure during the stretching process. After postspin stretching process, the intensity of the Ala Cβ chemical shift is indicative of a random coil or helical conformation decreases in intensity, while the Ala Cβ chemical shift increases for the β-sheet chemical shift. The same trend is observed for the Ala Cα, with the higher ppm shift, which indicates a helical-type conformation, decreasing in intensity with stretching and the lower ppm Ala Cα chemical shift increasing in intensity with stretching. The same trends are seen in the MaSp1/MaSp2 1:1 data (Figure 4B), with the intensity of the Ala Cα and Ala Cβ helical components decreasing, while the intensity of the β-sheet chemical shifts increases. However, the extent of β-sheet formation is to a lesser degree in the MaSp1/MaSp2 1:1 stretched fiber than the MaSp1/MaSp2 4:1 stretched fiber.
Figure 4.
1H → 13C CP-MAS ssNMR data of synthetic fibers with MaSp1/MaSp2 ratios of (A) 4:1 and (B) 1:1 in the as-spun (bottom) and postspin stretched forms (top). The formation of alanine β-sheet (blue dashed lines) is seen in the stretched fibers.
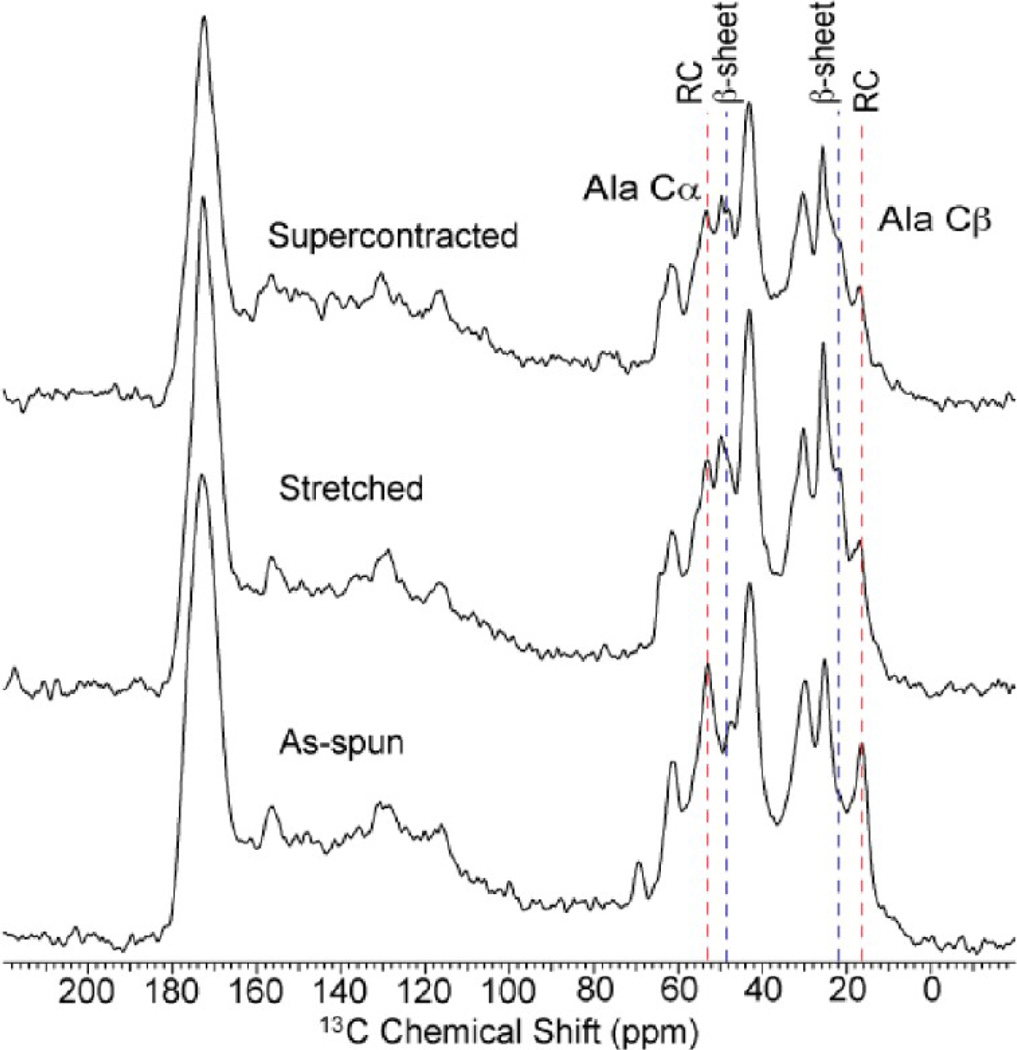
The (148)8 fiber displayed significant supercontraction in water similar to native dragline silk.36 To investigate how supercontraction affects synthetic fiber structure, 13C CP-MAS spectra were collected for the as-spun, stretched, and supercontracted (148)8 fibers (Figure 5). The stretching converts a substantial fraction of the alanine to β-sheet structure compared to the as-spun fibers similar to the other synthetic spider silks discussed above. Interestingly, the β-sheet structures still persist following the supercontraction process indicating that the water does not penetrate the β-sheet domains. This result is similar to previous reports on native dragline silk where the β-sheets remained intact following supercontraction process.39–41 This shows that the supercontraction property can be incorporated into synthetic spider silks and the behavior is reminiscent of that observed in native spider dragline silk.
Figure 5.
1H → 13C CP-MAS ssNMR spectra for (148)8 synthetic chimeric fibers as-spun, stretched, and following supercontraction. The formation of alanine β-sheet structures (blue dashed lines) is seen in the stretched fibers. These β-sheet structures are still present following supercontraction in water.
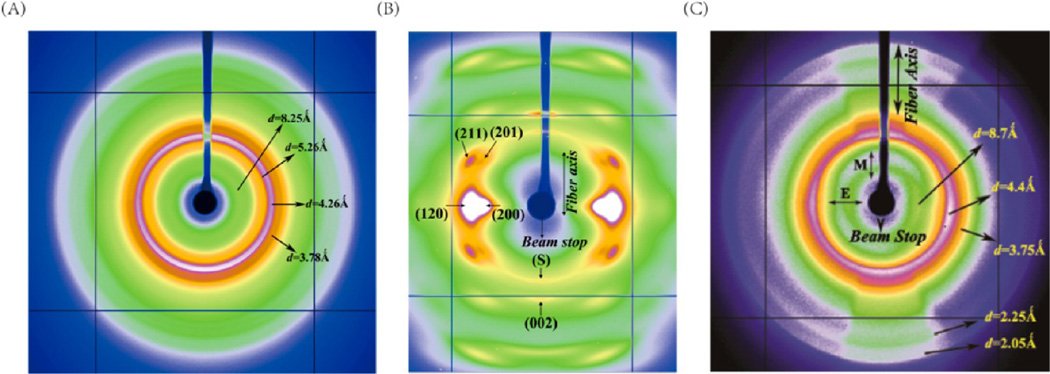
X-ray 2D diffraction patterns of the dehydrated N. clavipes major ampullate gland material, native N. clavipes major ampullate silk, and 4:1 synthetic silk are shown in Figure 6. The 2D diffraction patterns were analyzed using the software package FIT2D.42 Concentric broad diffuse diffraction rings are seen in the 2D diffraction patterns of the N. clavipes major ampullate gland material and the 4:1 synthetic silk. The d-spacings at which these diffraction rings occur are shown. The 2D X-ray diffraction pattern of N. clavipes major ampullate silk depicts a semicrystalline structure characterized by crystalline-Bragg reflections and an amorphous fraction.43 Intense crystalline reflections (210)/(200) are observed along the equator (perpendicular to the fiber axis). Also observed are higher order layer-line reflections, parallel to the equator. Along the meridian (parallel to the fiber axis), the main (002) reflection is strong, while subsequent higher order reflections are weaker. The reflections comprising the crystalline fraction correspond to the pattern of β-poly(l-alanine) structure.43–45 The reflections were indexed based on an orthogonal unit cell with the c-axis as the fiber axis.46–48
Figure 6.
XRFD pattern of (A) dehydrated N. clavipes major ampullate gland material. (B) N. clavipes major ampullate silk (C) 4:1 synthetic silk single fiber.
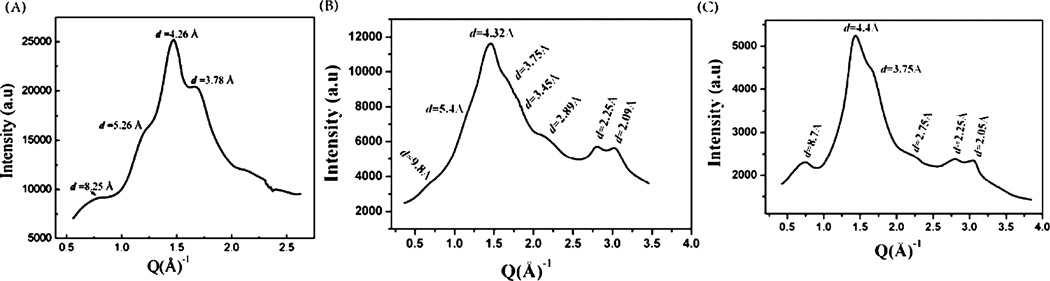
Radial and azimuthal 1D profiles were obtained from the deconvolution of 2D diffraction images using FIT2D. Radial profiles are intensity as a function of radius integrated azimuthally for the whole 2D pattern, while azimuthal profiles are integrated intensity as a function of all azimuthal angles over a thin annular ring centered at the peak maximum of the desired diffraction ring.43–45,49 Microcal Origin was used for the analysis of the deconvoluted 1D X-ray data. Figure 7 shows the 1D radial intensity profiles as a function of all azimuthal angles, for dehydrated N. clavipes major ampullate gland material, native N. clavipes major ampullate silk and 4:1 synthetic silk, respectively. The peaks in the dehydrated gland and 4:1 synthetic silk correspond to the d-spacings of the equatorial and layer-line reflections (perpendicular to the fiber axis) in the native N. clavipes major ampullate silk, but the meridional reflections (parallel to the fiber axis) are totally absent, which is interpreted as arising from a secondary structure, which has a certain degree of spatial order but with little or no preferred orientation, in the gland and 4:1 synthetic silk.
Figure 7.
1D radial integration profile of the whole 2D pattern of (A) gland material of N. clavipes major ampullate gland. (B) N. clavipes major ampullate silk. (C) Synthetic silk (4:1).
Structural features corresponding to certain d-spacings are present in all the three samples. The peak at d ~ 4.4 Å with the highest intensity in all three samples corresponds to the (210) crystalline reflection in the native N. clavipes major ampullate silk. This peak denotes the interchain distance. The peak at d ~ 5.4 Å, which is present in the gland, and the native silk is absent in the 4:1 synthetic silk. This peak corresponds to the (200) crystalline reflection in the native N. clavipes major ampullate silk and denotes the distance between β-sheets, which are stacked antiparallel in the nanocrystallites. NMR results in this work show the presence of β-sheets in both the gland and the 4:1 silk, hence, presence of this peak, the X-ray data in the gland material, could be interpreted as coming from β-sheets that are correlated spatially to some degree, giving rise to a diffraction peak, but with no preferred orientation about any direction. Absence of this peak in the 4:1 synthetic silk indicates that in the synthetic silk the β-sheets are not spatially correlated but could be dispersed through the fiber. For the native N. clavipes major ampullate silk, the average crystallite size was found to be 2.7, 4.0, and 7.0 nm along the a, b, and c directions, where a, b, and c are calculated from the (200), (210), and the (002) reflections, respectively. The details of the crystallinity and orientation parameters calculation for the N. clavipes major ampullate silk has been described in our previously published work.43
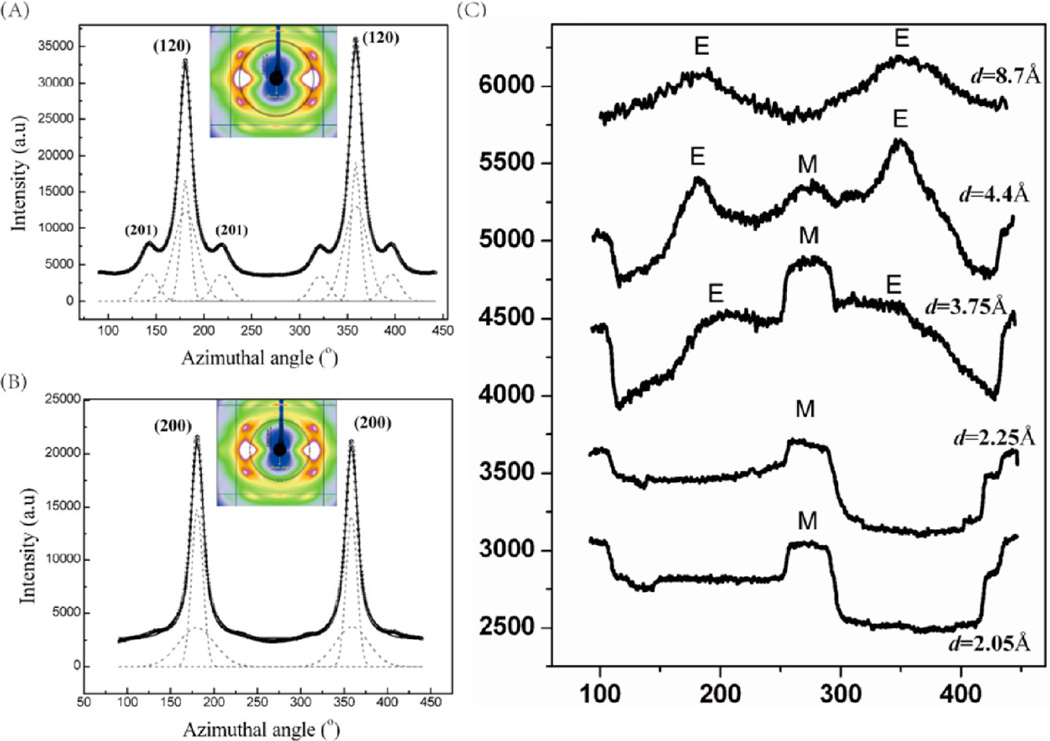
The axial orientation distribution of the crystallites about the fiber axis is determined from azimuthal broadening (fwhm) of the (200)/(120) equatorial reflections in the native spider silks.43–45 Figure 8A,B show the azimuthal intensity profiles as a function of angle measured from the equator at the radial position of the strong (120) and (200) peaks, respectively (the region used for integration is shown in the insets). In the dehydrated gland, it is seen from the XRFD pattern that the intensity distribution in the diffraction rings is almost uniform, which means that the protein chains although could be forming spatially correlated β-sheets, they do not have a preferred orientation. In the 4:1 synthetic silk however, the intensity distribution in the diffraction rings is not uniform. The intensity is higher along the meridian (parallel to the fiber axis) for the lower d-spacing rings (d = 2.05 and 2.25 Å), intensity along the equator (perpendicular to the fiber axis) starts to appear for the d = 3.75 Å ring, increases for the d = 4.4 Å ring, and finally for the d = 8.7 Å, almost all the intensity is concentrated along the equator, as shown in Figure 8C (E and M denote equatorial and meridional positions, respectively). The axial ordering gets better as the size of the secondary structure increases, as seen in the azimuthal profiles at the different d-spacings (order increasing from d = 2.05 to 8.7 Å).
Figure 8.
1D azimuthal intensity profile of (A) the radially integrated (120) peak with Gaussian fits for N. clavipes major ampullate silk. (B) Radially integrated (200) peak with Gaussian fits for N. clavipes major ampullate silk. Inset in (A) and (B) shows the annular section chosen for the integration. (C) Theradially integrated diffraction rings centered at different d-spacings (E and M denote equatorial and meridional positions, respectively).
DISCUSSION
The overall mechanical properties of synthetic fiber under-performed the natural fibers in all regards, likely due to the much smaller sizes of the recombinant silk proteins as well as limitations in fiber spinning to exactly replicate the spinning condition natural fiber undergoes in spider’s spinneret. However, it is comparable to several similar-sized synthetic silk protein fibers being produced before.33,50 On the other hand, regardless of how well synthetic fiber performs, we have observed the trend of mechanical property changes among different types of fibers, which may correlate to the primary sequence as well as the ratio of protein motifs in the fibers.
Data from the chimeric fibers support our hypothesis about spider silk protein motifs and their roles in controlling mechanical properties. The (148)8 protein has a larger molar ratio of the polyA motif than (188)6, thus, its fibers have a higher tenacity and Young’s modulus (about 10 MPa higher in tenacity and 0.4 GPa higher in Young’s modulus for both stretched and as-spun). The (188)6 protein contains more GPGXX motifs that make it ~6% more extensible. During our supercontraction test, we found post-spin-stretched (188)6 fibers are partially soluble in water. The fibers became weak, easy to break, and hard to lift by tweezers. It is likely the result of too few hydrophobic polyA motifs present in the fiber to help prevent water penetration. In contrast, stretched (148)8 fiber, which has a higher ratio of polyA motifs, were insoluble in water. This phenomenon may indicate that a certain level of β-sheet motif must be maintained in all silks in order to prevent easy water dissolution of the fiber.
Meanwhile, as predicted, chimeric fibers were more consistent in their mechanical properties than the two protein mixed fibers, especially in fiber elongation. All fibers in this study were prepared under the same conditions, thus, an uneven distribution of MaSp1 and MaSp2 in the mixed fiber could be the main factor causing this inconsistency. Based on our hypothesis about protein motifs and their relation to fiber mechanical properties, we were expecting the 4:1 mixed fibers to have a higher tenacity but lower elongation than the 1:1 mixed fibers. However, the average mechanical data showed a contradictory result to our prediction. Despite the large variability, several 4:1 mixed fibers extended more than 100%, but most 1:1 mixed fibers only elongated 5%. The average tenacity of 4:1 fibers is also 20 MPa lower than 1:1 fibers. Some plausible explanations for these results are: First, even after mixing two days on the rotor, MaSp1 and MaSp2 proteins in the fibers are still not well-blended in solution, suggesting that the two protein could not be simply mixed together to achieve a fully homogeneous fiber with our current wet spinning techniques. Second, the postspin stretching process could be uneven, as it was performed by hand, like other reported studies.51–53 We tried our best to minimize the variability of hand pulling, but ideally, postspin stretching should be performed on automatic controlled mechanical rotors with constant stretching force and speed.54,55 Regrettably, the requirement of post-spin stretching silk fiber in an aqueous environment prevented us from using our current automatic spinning system. Third, the β-sheets in some fibers might fold back on themselves and form a stack. If the crystalline β-sheet structures are not spread well enough or are not oriented parallel to the fiber axis, it may not provide extra strength to the fiber or may even have some weak spots in specific areas leading to premature breakage. Interestingly, compare to MaSp1 only fiber, higher breaking strength and Young’s modulus have also been observed on pure MaSp2 fiber previously through synthetic fiber testing as well as computational molecular dynamic simulation.50 However, the fibers tested in that study do not involve post-spin stretching, a treatment that is now believed to significantly affect fiber quality.
Almost all commercially available synthetic fibers need some degree of post-spin treatment. Post-spin stretching is involved in the production of nylon as well as many types of polyester fibers.56,57 Improved mechanical properties of various types of protein-based fibers have been observed by post-spin drawing.37,58,59 It has also been adapted as a standard procedure for synthetic spider silk production.51,52,60 Without stretching in a proper solution during spinning, many critical spatial structures of the protein may fail to form properly, thus, affecting the final quality of the fiber.61
Data from as-spun and post-spin stretched fibers in this study once again showed that the overall mechanical performance of the fiber improves after post-spin stretching. Solid-state NMR results support a clear shift from helix structure into β-sheet in the polyA regions of the fiber. Comparing the 1H → 13C CP-MAS ssNMR data of synthetic fibers to the natural glands with similar protein ratios show that the synthetic fibers and glands with a higher percentage of MaSp1, in this case, N. clavipes and MaSp1/MaSp2 4:1 more readily for alanine β-sheets with shearing or stretching. While the A. aurantia glands and the MaSp1/MaSp2 1:1 fibers do not as readily form alanine β-sheets secondary structure. The propensity for fibers and glands with a higher MaSp1 content to form alanine β-sheet structure may indicate that MaSp1 is a templating agent for β-sheet formation. Unlike MaSp1, MaSp2 has a high proline content, which is known to form β-turns in proteins. In the MaSp2-rich fibers (MaSp1/MaSp2 1:1) and A. aurantia glands, the higher proline content may cause turns in the protein backbone that inhibit β-sheet formation. Although stretching or shearing does cause β-sheet formation in the MaSp2-rich materials, the extent of β-sheet formation is greater in the MaSp1-rich materials, indicating that the ratio of MaSp1 to MaSp2 can be utilized to tune β-sheet formation in synthetic fibers.
X-ray fiber diffraction results show that crystalline reflections that occur in the native N. clavipes major ampullate silk are absent in the N. clavipes major ampullate gland material and the 4:1 synthetic silk. While sharp diffraction rings in an X-ray diffraction pattern indicate a polycrystalline structure with no preferred orientation, presence of broad diffuse diffraction rings in the N. clavipes major ampullate gland and the 4:1 synthetic silk indicate that both of these samples are comprised of secondary structures that have not formed crystallites. Moreover, these secondary structures do not have a preferred axial orientation seen in the native N. clavipes major ampullate silks. From the azimuthal intensity profiles, it is seen that, while there is no orientational ordering of the protein chains in the gland, there is partial orientation in the 4:1 synthetic silk. In the native N. clavipes major ampullate silk, the nanocrystallites comprising of β-sheets are almost fully oriented with respect to the fiber axis (Hermann’s orientation fc ≈ 0.985). Details of orientation function calculation are described in our already published work.43
CONCLUSION
From the current point of view, recombinant expression of synthetic spider silk in other organisms seems to be the most viable way for us to produce spider silk protein in large scale. Understanding the molecular nature of spider silk and replicating fibers comparable to native silk will continue to be the main focus for spider silk researchers. The amino acid sequences of the orb weaving spider silk proteins have remained almost unchanged for more than 100 million years.12,13 Interestingly, MaSp1 and MaSp2 are the major two components in all studied dragline silk fibers from these spiders. The mechanical properties of native dragline silk vary between species, which is believed to relate to the ratio of MaSp1 to MaSp2 in the silk.27 Both of these facts clearly indicate the importance of these two proteins to the mechanical properties of the fiber.
The results presented here confirm that altering protein motif composition could modify the mechanical properties of synthetic silk fibers. More consistent mechanical data from our MaSp1/MaSp2 chimeric fibers indicate a better way to produce uniform fiber with consistent mechanical properties. Meanwhile, as revealed by our post-spin stretch, NMR and XRD study, many elements other than silk protein sequences also have significant effects on the mechanical properties of synthetic silk. Those elements will continue to be investigated to improve the silk fiber property we currently produce.
Supplementary Material
ACKNOWLEDGMENTS
This material is based on work by the Lewis laboratory supported by NIH (EB000490) the AFOSR (FA9550-09-1-0717) and DOE (DE-SC0004791) and by Jeff Yarger and Greg Holland by the AFOSR (FA9550-10-1-0275) and the NSF (DMR-0805197 and CHE-1011937) for the NMR component of this project. The authors would also like to thank Dr. Florence Teule for suggestions on post-spin stretching, Dr. Brian Cherry for help with NMR instrumentation and student training, and Dr. David Wilbur and the Tufts Chemistry Department for allowing us to use the MALDI-TOF MS equipment. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357. Use of the BioCARS Sector 14 was also supported by grants from the National Center for Research Resources (5P41RR007707) and the National Institute of General Medical Sciences (8P41GM103543) from the National Institutes of Health. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the AFOSR, NSF, or NIH.
Footnotes
Supporting Information
SDS-PAGE and MALDI-TOF MS spectra of recombinant silk proteins expressed in this study. Microscopic images and a list of diameter and length changes of chimeric (148)8 protein fiber before and after water supercontraction. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Gosline JM, DeMont ME, Denny MW. Endeavour. 1986;10:37–43. [Google Scholar]
- 2.Lewis RV. Chem. Rev. 2006;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 3.Lewis RV. Acc. Chem. Res. 1992;25:392–298. [Google Scholar]
- 4.Vollrath F. Sci. Am. 1992;266:70–76. [Google Scholar]
- 5.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. J. Exp. Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 6.Vollrath F, Knight DP. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 7.Xu M, Lewis R. Proc. Natl. Acad. Sci. U.S.A. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinman M, Lewis RV. J. Biol. Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 9.Mita K, Ichimura S, James TC. J. Mol. Evol. 1994;38:583–592. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi CY, Shipley NH, Lewis RV. Int. J. Biol. Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 11.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. PLoS ONE. 2007;2:e514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 13.Garb JE, Dimauro T, Vo V, Hayashi CY. Science. 2006;312:1762. doi: 10.1126/science.1127946. [DOI] [PubMed] [Google Scholar]
- 14.Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Chem. Commun. (Cambridge, U.K.) 2008;43:5568–5570. doi: 10.1039/b812928b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhe AD, Seeley SK, Gardner K, Thompson L, Lewis RV. J. Mol. Recognit. 1997;10:1–6. doi: 10.1002/(SICI)1099-1352(199701/02)10:1<1::AID-JMR338>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Fraser RDB, MacRae TP. Conformation in Fibrous Proteins and Related Synthetic Polypeptides. New York: Academic Press; 1973. Poly(glycine) II, poly(L-proline) I, and related conformations. [Google Scholar]
- 17.Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. J. Am. Chem. Soc. 2008;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- 18.van Beek JD, Hess S, Vollrath F, Meier BH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi CY, Lewis RV. Science. 2000;287:1477–1479. doi: 10.1126/science.287.5457.1477. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins J, Creager M, Butler E, Lewis R, Yarger J, Holland G. Chem. Commun. (Cambridge, U.K.) 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- 21.Teulé F, Furin WA, Cooper AR, Duncan JR, Lewis RV. J. Mater. Sci. 2007;42:8974–8985. [Google Scholar]
- 22.Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 23.Scheller J, Guhrs KH, Grosse F, Conrad U. Nat. Biotechnol. 2001;19:573–577. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- 24.Menassa R, Zhu H, Karatzas CN, Lazaris A, Richman A, Brandle J. Plant Biotechnol. J. 2004;2:431–438. doi: 10.1111/j.1467-7652.2004.00087.x. [DOI] [PubMed] [Google Scholar]
- 25.Fahnestock SR, Bedzyk LA. Appl. Microbiol. Biotechnol. 1997;47:33–39. doi: 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- 26.Teulé F, Miao YG, Sohn BH, Kim YS, Hull JJ, Fraser MJJ, Lewis RV, Jarvis DL. Proc. Natl. Acad. Sci. U.S.A. 2012;109:923–928. doi: 10.1073/pnas.1109420109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. Biomacromolecules. 2005;6:3095–3099. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- 28.Swanson BO, Blackledge TA, Hayashi CY. J. Exp. Zool., Part A: Ecol. Genet. Physiol. 2007;307:654–666. doi: 10.1002/jez.420. [DOI] [PubMed] [Google Scholar]
- 29.Lombardi S, Kaplan D. J. Arachnol. 1990:297–306. [Google Scholar]
- 30.Tillinghast EK. Insect Biochem. 1984;14:115–120. [Google Scholar]
- 31.Gosline JM, Denny MW, DeMont ME. Nature. 1984;309:551–552. [Google Scholar]
- 32.Work RW. Trans. Am. Microsc. Soc. 1984;103:113–121. [Google Scholar]
- 33.Brooks AE, Stricker SM, Joshi SB, Kamerzell TJ, Middaugh CR, Lewis RV. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- 34.Work RW, Emerson PD. J. Arachnol. 1982;10:1–10. [Google Scholar]
- 35.Teulé F, Cooper AR, Furin WA, Bittencourt D, Rech EL, Brooks A, Lewis RV. Nat. Protoc. 2009;4:341–355. doi: 10.1038/nprot.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell FI, McEwen IJ, Viney C. Nature. 2002;416:37. doi: 10.1038/416037a. [DOI] [PubMed] [Google Scholar]
- 37.An B, Hinman MB, Holland GP, Yarger JL, Lewis RV. Biomacromolecules. 2011;12:2375–2381. doi: 10.1021/bm200463e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitô H. Magn. Reson. Chem. 1986;24:835–852. [Google Scholar]
- 39.Yang Z, Liivak O, Seidel A, LaVerde G, Zax DB, Jelinski LW. J. Am. Chem. Soc. 2000;122:9019–9025. [Google Scholar]
- 40.Holland GP, Lewis RV, Yarger JL. J. Am. Chem. Soc. 2004;126:5867–5872. doi: 10.1021/ja031930w. [DOI] [PubMed] [Google Scholar]
- 41.Fu C, Porter D, Chen X, Vollrath F, Shao Z. Adv. Funct. Mater. 2011;21:729–737. [Google Scholar]
- 42.Hammersley, FIT2D website. http://www.esrf.fr/computing/expg.
- 43.Sampath S, Isdebski T, Jenkins JE, Ayon J, Henning RW, Orgel J, Antipoa O, Yarger JL. Soft Matter. 2012;8:6713–6722. doi: 10.1039/C2SM25373A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grubb DT, Jelinski LW. Macromolecules. 1997;30:2860–2867. [Google Scholar]
- 45.Riekel C, Branden C, Craig C, Ferrero C, Heidelbach F, Muller M. Int. J. Biol. Macromol. 1999;24:179–186. doi: 10.1016/s0141-8130(98)00084-1. [DOI] [PubMed] [Google Scholar]
- 46.Warwicker JO. J. Mol. Biol. 1960;2:350–362. doi: 10.1016/s0022-2836(60)80046-0. [DOI] [PubMed] [Google Scholar]
- 47.Marsh R, Corey R, Pauling L. Biochim. Biophys. Acta. 1955;16:1–34. doi: 10.1016/0006-3002(55)90178-5. [DOI] [PubMed] [Google Scholar]
- 48.Arnott S, Dover SD, Elliott A. J. Mol. Biol. 1967;30:201–208. doi: 10.1016/0022-2836(67)90252-5. [DOI] [PubMed] [Google Scholar]
- 49.Zussman E, Burman M, Yarin AL, Khalfin R, Cohen Y. J. Polym. Sci., Part B: Polym. Phys. 2006;44:1482–1489. [Google Scholar]
- 50.Brooks AE, Nelson SR, Jones JA, Koenig C, Hinman MB, Stricker S, Lewis RV. Nanotechnol. Sci. Appl. 2008;8:9–16. doi: 10.2147/nsa.s3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teulé F, Addison B, Cooper AR, Ayon J, Henning RW, Benmore CJ, Holland GP, Yarger JL, Lewis RV. Biopolymers. 2012;97:418–431. doi: 10.1002/bip.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia X, Qian Z, Ki C, Park Y, Kaplan DL, Lee S. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14059–14063. doi: 10.1073/pnas.1003366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks AE, Creager MS, Lewis RV. Biomed. Sci. Instrum. 2005;41:1–6. [PubMed] [Google Scholar]
- 54.Corsini P, Perez-Rigueiro J, Guinea GV, Plaza GR, Elices M, Marsano E, Carnasciali MM, Freddi G. J. Polym. Sci., Part B: Polym. Phys. 2007;45:2568–2579. [Google Scholar]
- 55.Zhou G, Shao Z, Knight DP, Yan J, Chen X. Adv. Mater. (Weinheim, Ger.) 2009;21:366–370. [Google Scholar]
- 56.Sakuma Y, Rebenfeld L. J. Appl. Polym. Sci. 1966;10:637–652. [Google Scholar]
- 57.LeBourvellec G, Monnerie L, Jarry JP. Polymer. 1986;27:856–860. [Google Scholar]
- 58.Yin J, Chen E, Porter D, Shao Z. Biomacromolecules. 2010;11:2890–2895. doi: 10.1021/bm100643q. [DOI] [PubMed] [Google Scholar]
- 59.Fudge DS, Hillis S, Levy N, Gosline JM. Bioinspiration Biomimetics. 2010;5 doi: 10.1088/1748-3182/5/3/035002. 035002. [DOI] [PubMed] [Google Scholar]
- 60.Gnesa E, Hsia Y, JL Y, Weber W, Lin-Cereghino G, Tang S, Agari K, Vierra C. Biomacromolecules. 2012;13:304–312. doi: 10.1021/bm201262n. [DOI] [PubMed] [Google Scholar]
- 61.Holland C, Terry AE, Porter D, Vollrath F. Polymer. 2007;48:3388–3392. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.