Abstract
Protein post-translational modifications increase the functional diversity of the proteome by covalently adding chemical moieties onto proteins thereby changing their activation state, cellular localization, interacting partners, and life cycle. Lipidation is one such modification that enables membrane association of naturally cytosolic proteins. Protein prenyltransferases irreversibly install isoprenoid units of varying length via a thioether linkage onto proteins that exert their cellular activity at membranes. Substrates of prenyltransferases are involved in countless signaling pathways and processes within the cell. Identification of new prenylation substrates, prenylation pathway regulators, and dynamic trafficking of prenylated proteins are all avenues of intense, ongoing research that are challenging, exciting, and have the potential to significantly advance the field in the near future.
Introduction
Protein prenyltransferases catalyze attachment of lipid moieties onto the cysteine residue of the C-terminus of a protein substrate to enable normally hydrophilic proteins to localize to cell membranes via a hydrophobic lipid modification [1]. Protein farnesyltransferase (FTase) catalyzes the transfer of a 15-carbon farnesyl group from farnesyl diphosphate (FPP) to a substrate protein, and protein geranylgeranyltransferase (GGTase-I) performs the same reaction using a 20-carbon geranylgeranyl diphosphate (GGPP) donor group. The substrate proteins are proposed to contain a “Ca1a2X” motif, where `C' is the cysteine residue, `a1' and `a2' are aliphatic amino acids, and `X' determines whether a protein is modified by FTase or GGTase-I. This lipidation step is followed by cleavage of the -aaX residues catalyzed by an endoplasmic reticulum (ER)-bound protease (Rce1 or ZMPSTE24) and C-terminal carboxymethylation catalyzed by the integral membrane enzyme, isoprenylcysteine carboxylmethyltransferase (Icmt) [2].
Although the “CaaX” prenylation paradigm has served fairly well in describing many of the prenyltransferase substrates, recent studies have shown that this motif is too narrowly defined and does not adequately predict all potential FTase and GGTase-I substrates. Advances in understanding prenyltransferase substrate recognition have significantly expanded the pool of potential substrates and thus enabled elucidation of new roles for lipidated proteins in cell signaling pathways. Furthermore, several bacteria were recently discovered to use the mammalian prenylation machinery to lipidate bacterial proteins to enhance membrane localization. Although prenylation is irreversible, new modes of regulation have been identified in mammalian cells that modulate progression of proteins through the prenylation pathway. This review will focus on the most significant findings in the field of prenylation published since May 2010.
Substrate Identification
Identification of the substrates of FTase and GGTase-I has been an important goal for understanding the biological function of these enzymes. Traditional radiological methods where cells are treated with a radiolabeled analog of FPP, GGPP or a precursor of these molecules have not been generally useful for identifying prenyltransferase substrates mainly due to the low signal of the radiolabel [3]. Despite these challenges, great progress has been made in identifying potential and substantiated in vivo substrates of the prenyltransferases using peptide library studies, computational techniques, and lipid analogs.
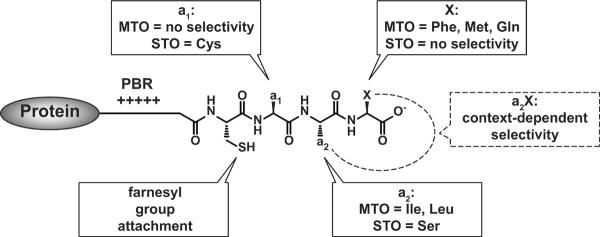
Analysis of the reactivity of prenyltransferases with diverse libraries of peptides has provided significant insight into the substrate selectivity of FTase and GGTase-I, as these enzymes react efficiently with short peptides. Using this approach, Krzysiak and colleagues demonstrated that FTase readily catalyzes farnesylation of a large number of peptides terminating in Leu (-CaaL), contrary to the CaaX paradigm which describes these as canonical GGTase-I substrate sequences [4]. This result expands the pool of FTase substrates and demonstrates additional cross-reactivity with GGTase-I. Hougland and colleagues showed that the identity of the amino acid at the X position affects the selectivity of the amino acid at the a2 position [5], suggesting that FTase recognizes the entire Ca1a2X sequence cooperatively rather than each amino acid individually. Building on this result, a large scale study of the reactivity of a library of 300 dansyl-TKCxxx peptide sequences based on the human proteome [6] revealed two classes of peptide reactivity with FTase: 1) peptides farnesylated under multiple turnover (MTO) conditions; and 2) peptides farnesylated only under single turnover (STO) (excess enzyme) conditions. For the STO substrates, FTase binds the substrate and catalyzes farnesylation but the product dissociates from the enzyme very slowly. Statistical analysis of these two classes of substrates revealed significantly different sequence preferences (Figure 1). The MTO peptides typically display canonical Ca1a2X sequences, including Ile and Leu at the a2 position and Phe, Met, and Gln at the X position. STO FTase substrates are more diverse in sequence, often containing a Ser at the a2 position and little sequence preference at the X position. The biological role of the STO substrates is not yet clear. Together, these studies show that FTase catalyzes farnesylation of a wide range of substrates, suggesting that a large cohort of proteins is farnesylated in vivo.
Figure 1.
The substrate selectivity of FTase for multiple turnover (MTO) and single turnover (STO) substrates. The region upstream of the CaaX sequence may contain a polybasic region (PBR). The selectivity at the a2 and X positions is context-dependent.
Computational techniques have also been developed to probe and predict the molecular recognition of the prenyltransferases. London and coworkers developed FlexPepBind to predict FTase substrates based on a calculated binding energy using the Hougland peptide library as a training set [7]. Analysis of the binding energy of all 8,000 possible CaaX peptide sequences identified potential novel FTase substrates. In a test of the validity of these predictions, FTase catalyzed prenylation of 26 out of 29 of the proposed peptides, including a subgroup containing Asp or Glu at the X residue. These data confirm that FTase catalyzes prenylation of a variety of diverse sequences, a significant portion of which do not fit into the traditional “CaaX” motif. Finally, Yang and colleagues used quantum mechanical molecular mechanical studies (QM/MM) to demonstrate a correlation between peptide structure and transition state structure for FTase [8]. These computational studies indicated that the transition state for farnesylation catalyzed by FTase changes from SN2-like for CVLS to SN1-like for CVIM, consistent with previously determined alterations in secondary kinetic isotope effects [9]. This change in transition state structure is likely due to steric interactions between the peptide and FTase.
Lipid donor analogs with reactive moieties have been used to identify in vivo prenyltransferase substrates. Upon addition of these analogs to cells, the prenyltransferases incorporate the modified lipids into substrate proteins. The modified lipid is used to enrich the labeled proteins which are then identified using mass spectrometric methods. One approach uses geranylgeranyl-azide analogs that are labeled with alkyne tetra-methylrhodamine using Cu(I)-catalyzed “click” reaction. Using this technique, many known GGTase-I substrates were identified, including members of the Rab protein family, as well as Rap2c, a novel member of the Ras family [10]. Using a similar method, a C10-alkyne analog was used to identify seven in vivo FTase substrates, including GNBP, Lamin B1, Rab 1B, Rab 2A, Rab 6A, Rab 7, and Annexin X3 [11]. Alternatively, an immunogenic analog, anilinogeraniol, was incorporated into cellular proteins to identify a number of substrate proteins, including Ras, Rho, and Rac proteins [12]. Finally, a biotin-GGPP analog was used to identify prenylatable proteins in cell lysates using engineered FTase and GGTase-I enzymes [13]. Although these various analogs have successfully identified prenylated proteins in vivo, each analog identifies a somewhat different set of substrates, suggesting that the analog structure may alter the prenyltransferase substrate selectivity. This is not surprising as in vitro and structural studies demonstrate that FPP and GGPP directly contact the peptide, contributing to molecular recognition of the peptide substrates [14,15].
Regulation of prenylation pathway
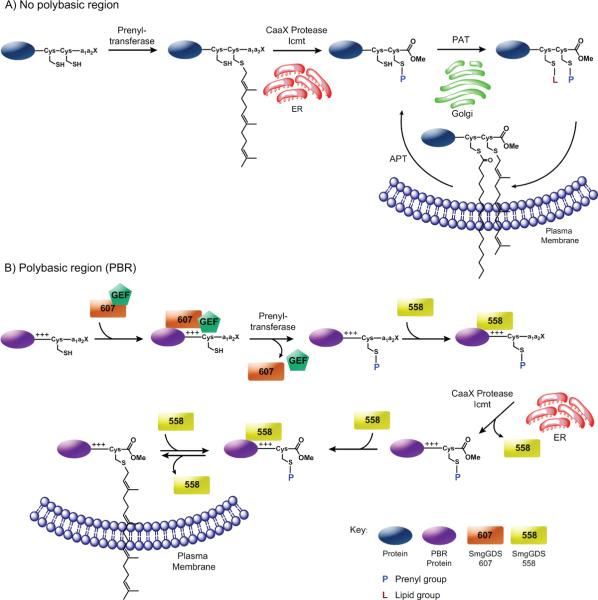
Although the importance of prenylation for biological activity of many proteins has been extensively studied, regulatory mechanisms of this pathway are only now being identified. Presence of a polybasic region (PBR) upstream of the CaaX sequence dictates that a prenylated protein is trafficked directly from the endoplasmic reticulum (ER) to the plasma membrane (PM); absence of such a region dictates that the protein undergoes additional processing via the Golgi compartment (GC) such as attachment of palmitoyl groups (Figure 2A) [16].
Figure 2.
Models for the trafficking and regulation of prenylated proteins. The farnesyl group is depicted here to represent prenylation. A) The prenylation of a Caax-containing protein is catalyzed by FTase or GGTase-I in the cytsosol. Next it is trafficked to the ER where it is proteoylzed by a CaaX protease such as Rce1 and carboxymethylated by isoprenylcysteine methyltransferase (ICMT). If the protein contains another cysteine near the C-terminus, a protein acyltransferase (PAT) can catalyze the palmitoylation of the protein at the Golgi. Then, the modified protein is trafficked to the plasma membrane. A protein acylthioesterase (APT) can catalyze the removal of the palmitoyl group, allowing for regulation of proteins between different cellular compartments. B) The SmgGDS-607 isoform and possibly a GEF-like protein may interact with an unmodified CaaX protein containing a polybasic region (PBR), promote GDP/GTP exchange, and allow the CaaX protein to enter the prenylation pathway. Once prenylated by GGTase-I or FTase, SmgGDS-558 may facilitate the release of the protein from the prenyltransferase and/or traffic the protein to the ER where it is subsequently proteolyzed and methylated. SmgGDS-558 may also transport the fully processed prenylated protein to and from the plasma membrane.
Williams and coworkers demonstrated that prenylation and trafficking of PBR-containing small GTPases is regulated by their interactions with SmgGDS and by GDP/GTP exchange [17] (Figure 2B). The longer SmgGDS-607 variant interacts with non-prenylated small GTPases and regulates their entry into the prenylation pathway, while a shorter SmgGDS-558 splice variant specifically associates with prenylated small GTPases and regulates their trafficking to the PM. SmgGDS-607 captures newly synthesized small GTPases and prevents prenylation until GDP/GTP exchange occurs leading to dissociation of the SmgGDS-607 complex. For Rho GTPases, SmgGDS-607 may function as a guanine nucleotide exchange factor (GEF), while other GTPases require a separate GEF where SmgGDS-607 may act as a scaffold [17,18]. An electronegative surface patch and substrate binding groove in SmgGDS-607 interact with the C-terminal PBR of RhoA and impart its biological activity.
Once prenylated, Rab and Rho proteins are shuttled between different membrane compartments by guanine nucleotide dissociation inhibitors (GDIs) that sequester and bury the hydrophobic lipid moiety. When the lipidated cargo molecules reach their destination, GDI displacement factors (GDFs) unload these proteins from GDIs. The search for analogous chaperones for the Ras protein family identified several candidates, including PDEδ, PRA1, and galectin [19]. PDEδ solubilizing factor was shown to modulate the dynamic shuttling of H-Ras and K-Ras, and it is required for the membrane localization of Ras family proteins by facilitating their diffusion in the cytoplasm [20]. In the structure of PDEδ complexed with farnesylated Rheb-GDP, the prenyl group is buried deeply in a hydrophobic pocket of PDEδ [21]. This interaction occurs primarily with the farnesylated and carboxymethylated C-terminus of Rheb and helps to explain the observed lack of specificity of PDEδ for GTPases or their guanine-nucleotide binding states. Dissociation of the Rheb-PDEδ complex is accelerated by formation of a transient ternary complex with Arl2/3-GTP, and since binding of farnesylated Rheb and Arl2/3-GTP to PDEδ is mutually exclusive, Arl2/3 is presumed to be a GDF.
Palmitoylation of prenylated proteins provides additional regulation of membrane localization [22]. While palmitoylation occurs exclusively in the Golgi, depalmitoylation has been observed in all cellular compartments, thus providing directionality to the membrane targeting process. Depalmitoylation can either be non-enzymatic modulated by prolyl isomerase [23] or catalyzed by enzymes such as acyl protein thioesterase 1 (APT1) [24]. Depalmitoylation weakens the interaction of Ras with the PM, allowing its diffusion into the cytoplasm.
Upon farnesylation, the –aaX group of the C-terminus is proteolyzed and the terminal carboxyl group is methylated, catalyzed by Rce1 or ZMPSTE24 and Icmt enzymes, respectively. These modifications are required for proper function and/or localization of many farnesylated proteins such as Rheb proteins [25], but can be circumvented with geranylgeranylated proteins, such as Rho GTPases [26]. Rce1 deficiency has been implicated in hematological malignancies and is essential for photoreceptor cell survival [27]. Icmt inhibition induces tumor cell death and reduces tumor growth in vivo [28]; thus, Rce1 and Icmt inhibitors are being investigated as potential cancer therapeutics [29–31].
Role of prenylation in bacterial infection
Introduction of macromolecules, such as DNA and protein, into host cells via a secretion system is a common mechanism used by intracellular pathogenic bacteria. Some of these secreted proteins are effectors that modulate host cell processes to favor bacterial growth and survival. Furthermore, recent experiments demonstrate that effector proteins from Salmonella enterica typhimurium and Legionella pneumophila can be post-translationally modified by eukaryotic enzymes, including prenyltransferases.
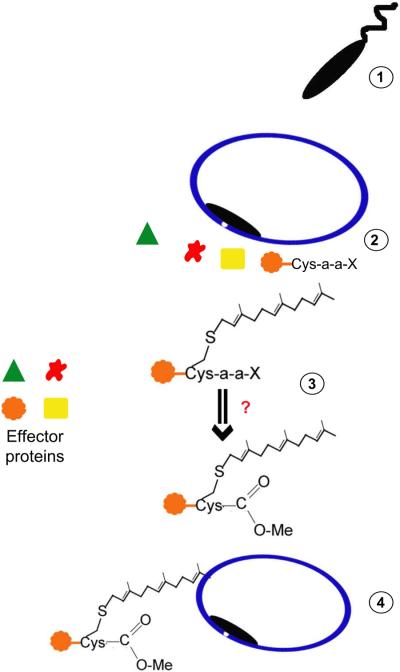
Secreted effector proteins of Legionella pneumophila, an aquatic gram-negative bacterium that can cause pneumonia, are prenylated [32,33]. Several Legionella effector proteins contain C-terminal CaaX motifs and a number of these effectors exhibit localization to membranes when overexpressed in mammalian cells. Mutation of the CaaX cysteine or FTI treatment disrupts localization, suggesting that these bacterial effectors are modified by host protein prenyltransferases. Ankyrin B, a Legionella effector, both interacts with the host ubiquitin machinery and contains a C-terminal CaaX motif (CVLC). The host FTase catalyzes farnesylation of ankyrin B which anchors this protein to the Legionella-containing vacuole during infection (Figure 3). This modification is required for sustained infection; L. pneumophila expressing ankyrin B with a Cys to Ala mutation in the CaaX box cannot sustain infection in mice [32]. This is the first example where host prenylation of a bacterial effector is required for survival of an intracellular pathogen.
Figure 3.
Bacteria hijack host prenylation machinery during infection. Upon bacterial entry via phagocytosis (1), the infectious bacterium is surrounded by a vacuole and immediately secretes effectors into the host cytosol (2). Effectors containing a CaaX box may be prenylated and further modified by other enzymes implicated in the prenylation pathway (3). These modifications may help target bacterial effectors to the cytosolic face of the bacteria-containing vacuole or elsewhere in the host cell (4).
Recently, the FTase inhibitor, FTI-277, has been investigated as treatment of bacterial infection. In septic mice, treatment with FTI-277 significantly increased phagocytosis by peritoneal macrophages in comparison to controls leading to enhanced bacterial clearance and survival [34]. Additionally, FTI-277 treatment of LPS-injected mice increased survival rates and decreased processes associated with LPS-induced apoptosis, such as caspase-3 cleavage and JNK phosphorylation [35]. These data suggest that FTI treatment may be a viable approach to treating some bacterial infections and septicemia.
Numerous infectious bacteria, such as Mycobacterium tuberculosis and Francisella tularensis, are predicted to express secreted effectors containing CaaX motifs that may be prenylated by host protein prenyltransferases. Further exploration of this modification would be beneficial in two ways: 1) FTIs and GGTIs currently approved for clinical use could be added to treatment regimens for bacterial infections, and 2) substrate specificity of FTase and GGTase-I can be further defined, as many of the CaaX sequences observed in bacterial genomes are not observed in the human genome. Further delineation of the human and bacterial prenylome will provide insight into the exploitation of prenylation during bacterial infection.
Clinical Investigations
Inhibition of protein prenylation has been pursued as a strategy for cancer therapy since oncogenic Ras proteins require farnesylation for their biological function [36]. Although FTIs have low toxicity, achieving clinical efficacy has been challenging and the most advanced candidates, tipifarnib and lonafarnib, failed to demonstrate efficacy in Phase III clinical trials. One potential reason for the lack of clinical efficacy of FTIs is that GGTase-I can compensate for the inhibited FTase and catalyze prenylation of key proteins, thereby allowing cancer cells to proliferate. Genetic knockouts of both FTase and GGTase-I in a mouse model of K-Ras-induced lung cancer significantly reduced tumor sizes and improved overall survival, suggesting that dual FTase/GGTase-I inhibitors may prove to be a more effective therapeutic approach [37]. This strategy is further supported by two studies: (1) GGTase-I deficiency decreased the severity of K-Ras-induced myeloid leukemia in a mouse model [38] and (2) a dual FTase/GGTase-I inhibitor was more effective than an FTI at inducing apoptosis of myeloid leukemia cells via inhibition of K-Ras prenylation [39].
A second complication in prenyltransferase-targeted therapeutics is the lack of validated biomarkers for patient selection. Currently the most promising cancer indications for FTIs are in hematological malignancies, specifically for patients with leukemia, lymphoma and myelodysplastic syndrome [40–42], and gene and protein expression signatures of tumors from these patients are being studied to help identify response predictors. A high gene expression ratio of Ras guanyl releasing protein 1 and DNA excision repair protein aprataxin (RASGRP1/APTX) correlated with a positive response to tipifarnib therapy in patients with leukemia [43] and lymphoma [44] and could potentially be used to predict responsiveness to FTIs. Pro-apoptotic Bim and Bcl-2 proteins could also serve as potential FTI sensitivity determinants in patients with lymphoma; tipifarnib inhibits the Raf/MEK/ERK signaling pathway through abrogation of prenylation of H-Ras or N-Ras GTPases, which leads to Bim and Bcl-2 up-regulation [45]. Given this mechanism of FTI-induced apoptosis, combinations of MEK and Akt kinase inhibitors with FTIs showed synergistic effects and may show clinical benefit [46,47].
Glioblastoma tumors have highly overexpressed epidermal growth factor receptor (EGFR) that leads to increased Ras activity. Kieran and coworkers showed that inhibition of H-Ras farnesylation by lonafarnib potentiated both temozolomide, a DNA alkylating agent currently approved for the treatment of astrocytoma, and radiation treatments in a murine model of glioblastoma [48]. Subsequent clinical evaluation of combinations of lonafarnib with temozolomide [49] and tipifarnib with radiation [50] showed encouraging results.
FTIs are also being explored for treatment of other diseases. Hutchinson-Gilford progeria syndrome (HGPS), a premature aging disease, is caused by the accumulation of prelamin A due to lack of proteolysis of the farnesylated C-terminus in the nucleus [51]. FTIs have been shown to improve this disease in a mouse model [52], and based on these data, lonafarnib entered clinical evaluation for HGPS and is currently being evaluated in Phase II clinical trials in combination with zoledronic acid and pravastatin [53]. Additionally, recent studies have provided evidence that alterations in the lamin A processing pathway may play a role in the aging process in the general population [54,55], and could thus ameliorate some of the age-related vascular degeneration. Farnesylation of ubiquitin C-terminal hydrolase-L1 has been linked to progression of neurodegenerative disorders leading FTIs to be evaluated in patients with Alzheimer's disease [56].
Conclusions
Investigation of protein lipidation remains an exciting field that has progressed significantly in the last few years. In vitro peptide library studies with FTase along with computational studies have demonstrated that the CaaX paradigm of substrate recognition does not sufficiently describe the molecular recognition of this enzyme. Expansion of the long-held “CaaX” paradigm heralds a new era of identification of noncanonical prenylated proteins. These studies in combination with multiple novel approaches to identify proteins that are prenylated in vivo suggest that complete identification of the human “prenylome” could be realized within a decade.
Furthermore, the assumption that newly synthesized small GTPases enter the prenylation pathway without regulation is no longer plausible. SmgGDS binds to newly synthesized small GTPases and regulates their entry into the prenylation pathway; this protein sequesters unprenylated small GTPases and releases them for prenylation upon receipt of a specific cellular signal. So far SmgGDS has been demonstrated to interact with only a select group of small GTPases, but it is reasonable to speculate that other prenylated proteins associate with SmgGDS-like chaperones that regulate their prenylation status.
Finally, although FTIs have not yet translated into the clinic as a solid tumor cancer therapeutic, there is still potential for using them to treat hematological malignancies and potentially other diseases as well. Excitingly, lonafarnib has shown promise in treating children with HGPS, a devastating premature aging disease. Treatment with FTIs may both halt disease progression and reverse clinical features of the disease in humans, as phenotypic improvement was observed both on cellular and organismal levels in animal models. FTIs also have potential as antibacterial agents as mammalian FTase was found to be involved in intracellular proliferation of Legionella bacterium.
Highlights
Expansion of the CaaX paradigm by biochemical and computational methodologies
Improvement of chemical biology tools to define mammalian prenylome
Identification of proteins that regulate entry into prenylation pathway
Involvement of mammalian prenyltransferases in bacterial pathogenesis
Clinical evaluation of inhibitors for cancer and premature aging diseases
Acknowledgements
We thank members of the Fierke laboratory for helpful comments and suggestions in preparation of this manuscript. This work was supported by the NIGMS grant GM 40602 (C. A. F.) and PSTP training grant GM07767 (E. A. Z.). We apologize to the many authors whose work could not be included due to space constraints of this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nguyen UT, Goody RS, Alexandrov K. Understanding and exploiting protein prenyltransferases. Chembiochem. 2010;11:1194–1201. doi: 10.1002/cbic.200900727. [DOI] [PubMed] [Google Scholar]
- 2.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 4.Krzysiak AJ, Aditya AV, Hougland JL, Fierke CA, Gibbs RA. Synthesis and screening of a CaaL peptide library versus FTase reveals a surprising number of substrates. Bioorg Med Chem Lett. 2010;20:767–770. doi: 10.1016/j.bmcl.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hougland JL, Lamphear CL, Scott SA, Gibbs RA, Fierke CA. Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry. 2009;48:1691–1701. doi: 10.1021/bi801710g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hougland JL, Hicks KA, Hartman HL, Kelly RA, Watt TJ, Fierke CA. Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities. J Mol Biol. 2010;395:176–190. doi: 10.1016/j.jmb.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This article identifies two FTase substrate classes, those with multiple turnover and single turnover activity, and defined their specificity requirements.
- 7.London N, Lamphear CL, Hougland JL, Fierke CA, Schueler-Furman O. Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. PLoS Comput Biol. 2011;7:e1002170. doi: 10.1371/journal.pcbi.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Wang B, Ucisik MN, Cui G, Fierke CA, Merz KM., Jr. Insights into the mechanistic dichotomy of the protein farnesyltransferase peptide substrates CVIM and CVLS. J Am Chem Soc. 2012;134:820–823. doi: 10.1021/ja209650h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pais JE, Bowers KE, Fierke CA. Measurement of the alpha-secondary kinetic isotope effect for the reaction catalyzed by mammalian protein farnesyltransferase. J Am Chem Soc. 2006;128:15086–15087. doi: 10.1021/ja065838m. [DOI] [PubMed] [Google Scholar]
- 10.Chan LN, Hart C, Guo L, Nyberg T, Davies BS, Fong LG, Young SG, Agnew BJ, Tamanoi F. A novel approach to tag and identify geranylgeranylated proteins. Electrophoresis. 2009;30:3598–3606. doi: 10.1002/elps.200900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Evaluation of alkyne-modified isoprenoids as chemical reporters of protein prenylation. Chem Biol Drug Des. 2010;76:460–471. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This article reports on using alkyne-modified isoprenoids followed by click chemistry with an azide-modified fluorophore. This allows direct visualization of prenylated proteins as well as 2D-gel analysis followed up mass spectrometry in order to identify in vivo prenylated proteins.
- 12.Onono FO, Morgan MA, Spielmann HP, Andres DA, Subramanian T, Ganser A, Reuter CW. A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol Cell Proteomics. 2010;9:742–751. doi: 10.1074/mcp.M900597-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Franzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, et al. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 14.Krzysiak AJ, Rawat DS, Scott SA, Pais JE, Handley M, Harrison ML, Fierke CA, Gibbs RA. Combinatorial modulation of protein prenylation. ACS Chem Biol. 2007;2:385–389. doi: 10.1021/cb700062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 16.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nature Reviews Molecular Cell Biology. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, Williams CL. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem. 2010;285:35255–35266. doi: 10.1074/jbc.M110.129916. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This is the first study to identify a regulatory mechanism of prenylation. Prenylation has long been considered a constitutive process, this study identifies SmgGDS-607 as a protein that binds unprenylated small GTPases and inhibits their prenylation where GDP/GTP exchange regulates their subsequent release into the prenylation pathway.
- 18.Hamel B, Monaghan-Benson E, Rojas RJ, Temple BR, Marston DJ, Burridge K, Sondek J. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem. 2011;286:12141–12148. doi: 10.1074/jbc.M110.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhagatji P, Leventis R, Rich R, Lin CJ, Silvius JR. Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys J. 2010;99:3327–3335. doi: 10.1016/j.bpj.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2012;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 21.Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]; •Co-crystal structure of farnesylated Rheb with PDEδ shows that the prenyl-binding pocket of PDEδ is allosterically regulated by GTP-bound Arl2/3.
- 22.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]; •This study shows that palmitoylation of Ras proteins occurs only in the Golgi, while depalmitoylation can occur anywhere in the cell.
- 23.Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, Philips MR. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41:173–185. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 25.Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene. 2010;29:380–391. doi: 10.1038/onc.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P, Der CJ. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christiansen JR, Kolandaivelu S, Bergo MO, Ramamurthy V. RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc Natl Acad Sci U S A. 2011;108:8862–8866. doi: 10.1073/pnas.1103627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Hossain MS, Tan W, Coolman B, Zhou J, Liu S, Casey PJ. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene. 2010;29:4959–4970. doi: 10.1038/onc.2010.247. [DOI] [PubMed] [Google Scholar]
- 29.Majmudar JD, Hodges-Loaiza HB, Hahne K, Donelson JL, Song J, Shrestha L, Harrison ML, Hrycyna CA, Gibbs RA. Amide-modified prenylcysteine based Icmt inhibitors: Structure-activity relationships, kinetic analysis and cellular characterization. Bioorg Med Chem. 2012;20:283–295. doi: 10.1016/j.bmc.2011.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Go ML, Leow JL, Gorla SK, Schuller AP, Wang M, Casey PJ. Amino derivatives of indole as potent inhibitors of isoprenylcysteine carboxyl methyltransferase. J Med Chem. 2010;53:6838–6850. doi: 10.1021/jm1002843. [DOI] [PubMed] [Google Scholar]
- 31.Judd WR, Slattum PM, Hoang KC, Bhoite L, Valppu L, Alberts G, Brown B, Roth B, Ostanin K, Huang L, et al. Discovery and SAR of methylated tetrahydropyranyl derivatives as inhibitors of isoprenylcysteine carboxyl methyltransferase (ICMT) J Med Chem. 2011;54:5031–5047. doi: 10.1021/jm200249a. [DOI] [PubMed] [Google Scholar]
- 32.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study shows that ankyrin B is farnesylated by host FTase enzyme and targeted to Legionella-containing vacuoles. It shows that this modification of ankyrin B is essential to its biological function, and that it plays a role in Legionella pathogen survival and replication in the host organism.
- 33.Ivanov SS, Charron G, Hang HC, Roy CR. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem. 2010;285:34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W, Yamada M, Tamura Y, Chang K, Mao J, Zou L, Feng Y, Kida K, Scherrer-Crosbie M, Chao W, et al. Farnesyltransferase inhibitor FTI-277 reduces mortality of septic mice along with improved bacterial clearance. J Pharmacol Exp Ther. 2011;339:832–841. doi: 10.1124/jpet.111.183558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinozaki S, Inoue Y, Yang W, Fukaya M, Carter EA, Yu YM, Fischman A, Tompkins R, Kaneki M. Farnesyltransferase inhibitor improved survival following endotoxin challenge in mice. Biochem Biophys Res Commun. 2010;391:1459–1464. doi: 10.1016/j.bbrc.2009.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, Wahlstrom AM, Dalin M, Yu H, Chen Z, et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci U S A. 2010;107:6471–6476. doi: 10.1073/pnas.0908396107. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This article demonstrates that concomitant conditional loss of both FTase and GGTase-I in mice effectively reduces K-Ras-induced lung carcinogenesis, and extends the lifespan of these mice considerably more than FTase or GGTase-I deficiency alone, suggesting that the simultaneous inhibition of FTase and GGTase-I may be therapeutically beneficial in cancer patients.
- 38.Sjogren AK, Andersson KM, Khan O, Olofsson FJ, Karlsson C, Bergo MO. Inactivating GGTase-I reduces disease phenotypes in a mouse model of K-RAS-induced myeloproliferative disease. Leukemia. 2011;25:186–189. doi: 10.1038/leu.2010.242. [DOI] [PubMed] [Google Scholar]
- 39.Morgan MA, Onono FO, Spielmann HP, Subramanian T, Scherr M, Venturini L, Dallmann I, Ganser A, Reuter CW. Modulation of anthracycline-induced cytotoxicity by targeting the prenylated proteome in myeloid leukemia cells. J Mol Med (Berl) 2012;90:149–161. doi: 10.1007/s00109-011-0814-7. [DOI] [PubMed] [Google Scholar]
- 40.Kirschbaum MH, Synold T, Stein AS, Tuscano J, Zain JM, Popplewell L, Karanes C, O'Donnell MR, Pulone B, Rincon A, et al. A phase 1 trial dose-escalation study of tipifarnib on a week-on, week-off schedule in relapsed, refractory or high-risk myeloid leukemia. Leukemia. 2011;25:1543–1547. doi: 10.1038/leu.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witzig TE, Tang H, Micallef IN, Ansell SM, Link BK, Inwards DJ, Porrata LF, Johnston PB, Colgan JP, Markovic SN, et al. Multi-institutional phase 2 study of the farnesyltransferase inhibitor tipifarnib (R115777) in patients with relapsed and refractory lymphomas. Blood. 2011;118:4882–4889. doi: 10.1182/blood-2011-02-334904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karp JE, Vener TI, Raponi M, Ritchie EK, Smith BD, Gore SD, Morris LE, Feldman EJ, Greer JM, Malek S, et al. Multi-institutional phase 2 clinical and pharmacogenomic trial of tipifarnib plus etoposide for elderly adults with newly diagnosed acute myelogenous leukemia. Blood. 2012;119:55–63. doi: 10.1182/blood-2011-08-370825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raponi M, Lancet JE, Fan H, Dossey L, Lee G, Gojo I, Feldman EJ, Gotlib J, Morris LE, Greenberg PL, et al. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood. 2008;111:2589–2596. doi: 10.1182/blood-2007-09-112730. [DOI] [PubMed] [Google Scholar]
- 44.Rolland D, Ribrag V, Haioun C, Ghesquieres H, Jardin F, Bouabdallah R, Franchi P, Briere J, De Kerviler E, Chassagne-Clement C, et al. Phase II trial and prediction of response of single agent tipifarnib in patients with relapsed/refractory mantle cell lymphoma: a Groupe d'Etude des Lymphomes de l'Adulte trial. Cancer Chemother Pharmacol. 2010;65:781–790. doi: 10.1007/s00280-009-1185-4. [DOI] [PubMed] [Google Scholar]
- 45.Ding H, Hackbarth J, Schneider PA, Peterson KL, Meng XW, Dai H, Witzig TE, Kaufmann SH. Cytotoxicity of farnesyltransferase inhibitors in lymphoid cells mediated by MAPK pathway inhibition and Bim up-regulation. Blood. 2011;118:4872–4881. doi: 10.1182/blood-2011-02-334870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellicano F, Simara P, Sinclair A, Helgason GV, Copland M, Grant S, Holyoake TL. The MEK inhibitor PD184352 enhances BMS-214662-induced apoptosis in CD34+ CML stem/progenitor cells. Leukemia. 2011;25:1159–1167. doi: 10.1038/leu.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balasis ME, Forinash KD, Chen YA, Fulp WJ, Coppola D, Hamilton AD, Cheng JQ, Sebti SM. Combination of farnesyltransferase and Akt inhibitors is synergistic in breast cancer cells and causes significant breast tumor regression in ErbB2 transgenic mice. Clin Cancer Res. 2011;17:2852–2862. doi: 10.1158/1078-0432.CCR-10-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaponis D, Barnes JW, Dellagatta JL, Kesari S, Fast E, Sauvageot C, Panagrahy D, Greene ER, Ramakrishna N, Wen PY, et al. Lonafarnib (SCH66336) improves the activity of temozolomide and radiation for orthotopic malignant gliomas. J Neurooncol. 2011;104:179–189. doi: 10.1007/s11060-010-0502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desjardins A, Reardon DA, Peters KB, Threatt S, Coan AD, Herndon JE, 2nd, Friedman AH, Friedman HS, Vredenburgh JJ. A phase I trial of the farnesyl transferase inhibitor, SCH 66336, with temozolomide for patients with malignant glioma. J Neurooncol. 2011;105:601–606. doi: 10.1007/s11060-011-0627-0. [DOI] [PubMed] [Google Scholar]
- 50.Haas-Kogan DA, Banerjee A, Poussaint TY, Kocak M, Prados MD, Geyer JR, Fouladi M, Broniscer A, Minturn JE, Pollack IF, et al. Phase II trial of tipifarnib and radiation in children with newly diagnosed diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13:298–306. doi: 10.1093/neuonc/noq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrowman J, Hamblet C, George CM, Michaelis S. Analysis of prelamin A biogenesis reveals the nucleus to be a CaaX processing compartment. Mol Biol Cell. 2008;19:5398–5408. doi: 10.1091/mbc.E08-07-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang SH, Chang SY, Andres DA, Spielmann HP, Young SG, Fong LG. Assessing the efficacy of protein farnesyltransferase inhibitors in mouse models of progeria. J Lipid Res. 2010;51:400–405. doi: 10.1194/jlr.M002808. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study verified the mode of action of FTI in a mouse model of progeria. FTI was only beneficial to mice that expressed farnesylated version of prelamin A, and was inactive in those mice that expressed a nonfarnesylated prelamin A protein.
- 53.Kieran MW, Gordon L, Kleinman M. New approaches to progeria. Pediatrics. 2007;120:834–841. doi: 10.1542/peds.2007-1356. [DOI] [PubMed] [Google Scholar]
- 54.Olive M, Harten I, Mitchell R, Beers JK, Djabali K, Cao K, Erdos MR, Blair C, Funke B, Smoot L, et al. Cardiovascular pathology in Hutchinson-Gilford progeria: Correlation with the vascular pathology of aging. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30:2301–2309. doi: 10.1161/ATVBAHA.110.209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, Figg N, Shroff R, Skepper J, Shanahan CM. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation. 2010;121:2200–2210. doi: 10.1161/CIRCULATIONAHA.109.902056. [DOI] [PubMed] [Google Scholar]
- 56.Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT., Jr. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson's disease. Proc Natl Acad Sci U S A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]