Abstract
In order to understand the molecular mechanisms of longevity regulation, we recently performed a screen designed to enrich for genes common to several longevity interventions. Using this approach, we identified the Drosophila melanogaster gene takeout. takeout is upregulated in a variety of long-lived flies, and extends life span when overexpressed. Here, we investigate the mechanisms of takeout-dependent longevity.
takeout overexpression specifically in the fat body is sufficient to increase fly longevity and is additive to the longevity effects of dietary restriction. takeout long-lived flies do not show phenotypes often associated with increased longevity, such as enhanced stress resistance or major metabolic abnormalities. However, males exhibit greatly diminished courtship behavior, leading to a reduction in fertility. Interestingly, takeout contains a binding domain for Juvenile Hormone, a fly hormone that plays a role in the regulation of developmental transitions. Importantly, the longevity and courtship phenotypes of takeout overexpressing flies are reversed by treatment with the Juvenile Hormone analog methoprene.
These data suggest that takeout is a key player in the tradeoff-switch between fertility and longevity. takeout may control fertility via modulation of courtship behavior. This regulation may occur through Juvenile Hormone binding to takeout and a subsequent reduction in Juvenile Hormone signaling activity.
Keywords: Drosophila melanogaster, Juvenile Hormone, longevity, fertility, aging
1. Introduction
Numerous genes that play a role in the regulation and modulation of longevity have been discovered since the first demonstration that mutations in single genes affect life span [1]. Progress in the identification of these aging genes has by and large been achieved through methodological dissection of candidate signaling pathways or through the use of high-throughput technologies, such as microarray screening. We have recently combined these two approaches and performed combinatorial microarray experiments in Drosophila melanogaster using longevity interventions related to Dietary Restriction (DR) to identify genes altered under these long-lived conditions. One of the identified genes, takeout (to), extends the life span of flies when overexpressed [2].
takeout is moreover upregulated in a variety of longevity interventions related to DR (Indy or rdp3 downregulation, dSir2 upregulation), but also in interventions not related to DR, such as in chico mutant flies [2]. takeout is a small protein of 249 amino acids, and is produced in and secreted from the fly fat body [3]. takeout mRNA is also present in structures related to feeding, like antennae, the alimentary canal and the crop [4]. takeout is upregulated in response to starvation, and flies lacking takeout are starvation sensitive [4, 5]. Interestingly, takeout null flies consume more food under normal conditions than control flies, but cannot modulate their food intake and foraging behavior, suggesting that takeout plays an important role in feeding related activities [5, 6]. An additional role for takeout in the regulation of male courtship behavior has been suggested [3, 7].
takeout has over 24% identity and 54% similarity to Juvenile Hormone (JH) binding protein (JHBP) from Manduca sexta [4] and contains a putative JH binding domain (JHBD). It may thus belong to a family of JHBP modulating JH function [4, 8]. Flies contain three JHs that control the transition from larval to pupal and adult stages during development [9] and have been implicated in the regulation of life span [10]. However, it is unclear whether the Drosophila takeout JHBD is functional and capable of binding JH. Two possible biological functions could be associated with this binding activity. It has been suggested that JHBP, including takeout, may bind circulating JH in the hemolymph and may thus protect JH from degradation and inactivation by JH esterases. In contrast, reduction of JH levels have been linked to increased longevity. Ablation of the corpora allata, the site of JH production in insects, is associated with increased life span in grasshoppers [11], and flies exposed to methoprene vapors (a synthetic version of JH) have decreased life spans [10]. Therefore, an alternative possible function of takeout may be to bind JH in the hemolymph, thereby reducing JH bioavailability and subsequent JH signaling.
Here, we investigate the mechanisms by which takeout regulates longevity, especially with respects to its role in DR. We show that takeout overexpression specifically in the peri-cerebral fat body increases Drosophila longevity without accompanying changes in stress resistance or gross metabolic abnormalities. However, takeout-dependent long-lived flies have a marked decrease in courtship behavior and reduced fertility. Interestingly, supplementing fly food with the JH analog methoprene reverses the phenotypes of takeout overexpressing flies. These data suggest that takeout-dependent modulation of JH signaling plays an important role in the regulation of the tradeoff between fertility and longevity.
2. Materials and Methods
2.1 Fly culture and strains
All flies were kept in a humidified (50%), temperature-controlled incubator with 12 hour on/off light cycle at 25°C in vials containing standard cornmeal medium [12]. The ELAV-GeneSwitch line was from H. Keshishian (Yale University, New Haven, CT), while lines S1-32 and S1-106 were from M. Tatar (Brown University, Providence, RI), and the GAL4-to, UAS-to and to1 lines were from B. Dauwalder (University of Houston). All other lines were from the Bloomington Drosophila Stockcenter at Indiana University (Bloomington, IN).
2.2 Life span analysis
Flies were collected under light anesthesia, randomly divided into treatment groups and housed at a density of 25 males and 25 females each per vial. At least ten such vials were used per treatment as per [13]. Flies were passed every other day and the number of dead flies recorded.
All life span experiments were performed on regular cornmeal food. For induction with the GeneSwitch system, RU486 (SIGMA) was added directly to the food to the final concentration of 500µM, except where indicated. The same concentration of diluent was added to control food. RU486 was administered from the day of eclosion. For expression with the constitutive to-GAL4 driver, UAS-to was backcrossed to w1118 for 10 generations and isogenic controls were generated from the last backcross.
The Juvenile Hormone analog methoprene (SIGMA) was dissolved in ethanol and directly added to the fly food to a final concentration of 25µg/ml.
2.3 Determination of metabolite levels
Flies were raised on the indicated food at a density of 25 males and 25 females each per vial. At the indicated ages, flies were anesthetized and collected in at least three biological replicates, weighed in groups of maximally 10 flies, transferred to chilled microcentrifuge tubes containing buffer A (20mM HEPES-KOH pH7.5, 10mM KCl, 1.5mM MgCl2, 1mM EDTA, 1mM EGTA, 1mM DTT, protease inhibitors, 0.5mM PMSF) and homogenized using a motorized micropestle. After centrifugation, supernatants were used for determination of metabolite levels. Glucose was determined by using the glucose detection kit (SIGMA). Trehalose and glycogen were converted into glucose through addition of trehalose (SIGMA, 0.2U.ml) or amyloglucosidase (SIGMA, 0.1U/ml), respectively, followed by glucose determination. Assays were measured using a BioTek Synergy2 96-well plate reader in at least triplicate.
2.4 Stress resistance
For assays testing resistance to starvation and H2O2 (SIGMA), at least 5 vials of newly eclosed flies containing 25 males and females each were collected and aged for ten days under the same conditions as for life span analysis. For starvation assays, flies were then shifted to vials containing a 2% agar matrix to avoid desiccation. For H2O2 assays, flies were shifted to vials containing 2% agar with 5% sucrose/5% H2O2. The number of dead flies was counted twice daily.
2.5 Physical activity measurements
For activity measurements, flies were collected and cultured as for life span assays. At the indicated ages, flies were separated by sex, and measurements were taken in a LAM25H-3 Locomotor Activity Monitor (Trikinetics Inc.) over at least a 48hr period using 20 animals per vial. Activity was recorded every 10 minutes.
2.6 Fertility and mating behavior
Fertility was examined using at least 15 individual mating pairs under the same conditions as the longevity analysis. Flies were passed and eggs counted daily over a 20-day period. Mating behavior was determined using at least 6 individual mating pairs of 7-day old naïve virgin male and female flies. Individual mating pairs were observed under a dissection microscope in a mating chamber. First occurrence, frequency and duration of stereotypical male mating behaviors were assessed for 10min directly after placing flies in the observation chamber.
2.7 Gene Set Enrichment Analysis (GSEA)
GSEA was performed as described [14] [15]. Gene expression data were obtained using Drosophila2 Genome Arrays (Affymetrix) and normalized using the GCRMA algorithm. The heatmap in Figure 2C is using gene sets from the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/). Significance cutoff used was a false discovery rate of 0.25, with the false discovery rate calculated by GSEA. Figure 5A shows the enrichment scores calculated by the GSEA algorithm, the rank of each gene in the geneset and the overall distribution of fold changes in the data. The geneset shown represents Juvenile Hormone targets that are usually downregulated, compiled from published data as described in [14].
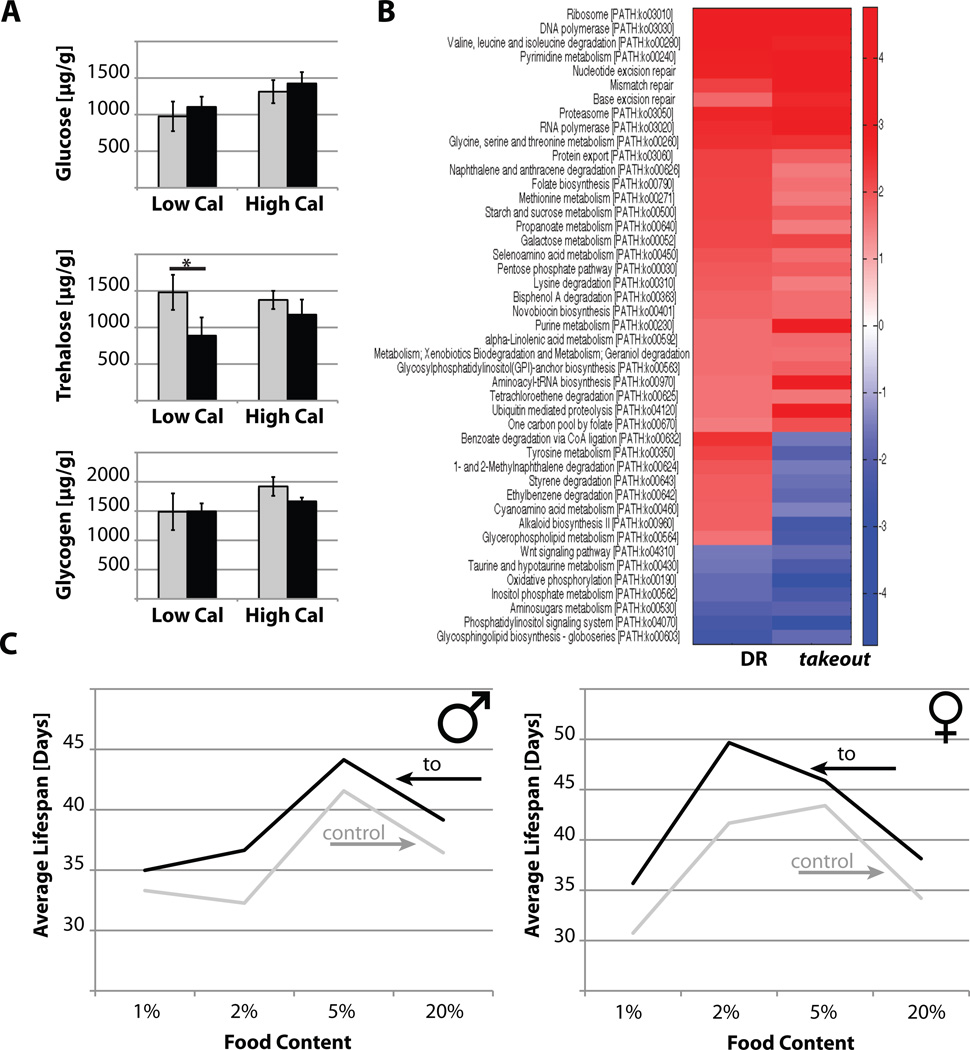
Figure 2. The effects of takeout overexpression on fly physiology.
(A) Flies overexpressing takeout under control of the to-GAL4 driver were raised on high (15%) and low (2%) calorie food. After ten days, female flies were harvested and their whole body levels of glucose, trehalose and glycogen were determined (* p=0.0414). (B) Heat map of KEGG gene sets from DR and long-lived takeout overexpressing female head and thorax (S1-32 driver). Red are gene sets that are statistically significantly upregulated, blue are gene sets that are statistically significantly downregulated. (C) Average life span as a function of dietary content. Flies were raised on food containing the indicated sucrose/yeast extract concentrations (w/V) and their life spans determined. takeout overexpression using a to-GAL4 driver extended life span on all food concentrations (grey: controls, black: takeout; error bars in A represent the standard deviation; shown are representatives of two independent experiments).
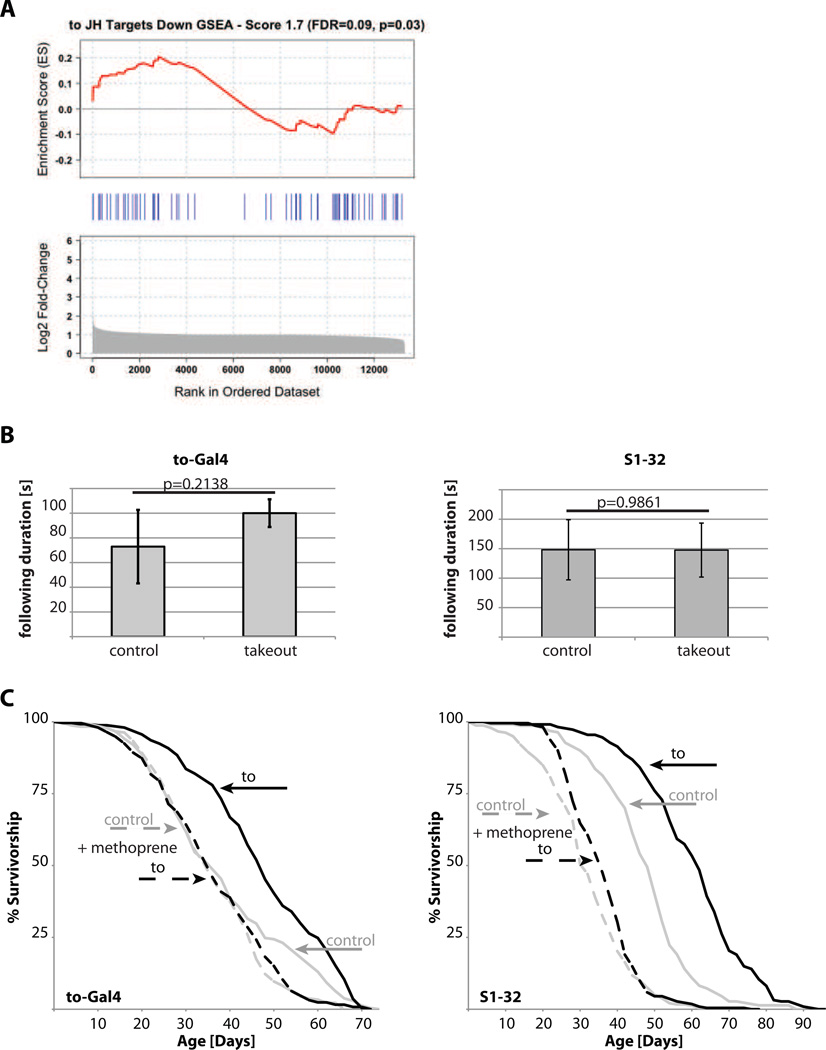
Figure 5. takeout overexpression leads to downregulation of JH signaling.
(A) GSEA analysis from the head and thorax of female takeout overexpressing flies reveals global decreases in JH signaling. A significant and positive ES indicates more genes in the JH signaling pathway rank among the top up-regulated genes than expected by random chance; a significant and negative value reflects more genes in the JH signaling pathway rank among the top down-regulated genes than expected by random chance. The analyzed gene set was compiled from a list of JH down-regulated genes, therefore a positive ES indicates a significant bias for increased expression of genes associated with a reduction of JH signaling. Shown are the enrichment score (ES) from the GSEA algorithm (Top, red curve), the location of the JH down genes in the rank list (Middle, blue lines) and the fold-change in takeout vs. controls of all of the genes on the microarray (Bottom, gray); genes were ranked according to their fold-change. (B) Methoprene inhibited the mating behavior phenotype in takeout long-lived flies. Flies were raised on food containing methoprene and male mating behavior was assessed. No significant differences were observed between control and takeout overexpressing flies. (C). Survivorship curves of takeout overexpressing flies raised on methoprene-containing food. takeout overexpression increased fly longevity when raised on standard food. This life span increase was not observed when flies were raised on food containing methoprene (for statistical analysis, please see Tables 1 and 2; grey: controls, black: takeout; solid lines: control food; dotted lines: methoprene containing food).
2.8 Statistics
Statistical analyses, including log rank tests, were performed using the Prism suit of biostatistical software (GraphPad, San Diego). Maximum life span was calculated as the median age of the last surviving 10% of the population.
3. Results
3.1 takeout overexpression extends life span
We have previously identified Drosophila melanogaster takeout in a screen for mediators of the longevity effects of DR [2]. takeout is upregulated by DR and DR-related genetic interventions, but also in DR-independent long-lived flies. We have subsequently used a variety of inducible Geneswitch drivers to determine the longevity effects of takeout overexpression. This inducible expression system [16] avoids problems often associated with using different genetic background. Furthermore, overexpression can be induced in adult flies only, bypassing possible detrimental effects of overexpression during fly development. Using a variety of Geneswitch drivers, takeout overexpression significantly increases fly longevity [2]. Since takeout is expressed in the peri-cerebral fat body [3], we used the S1-32 Geneswitch driver that directs expression in this tissue. The life span extension observed with this driver was dependent on the dose of the inducer RU486 (Figure 1A and Supplemental Figure 1A). RU486 dose-dependent longevity extension was also observed when the abdominal fat-specific driver S1-106 was used (Figure 1B). Next, we overexpressed takeout using a constitutive takeout driver [3] to determine whether takeout overexpression in its precise endogenous expression pattern would likewise influence fly life span. Similar to overexpression using the S1-32 driver, takeout overexpression using the takeout driver increased fly longevity by ~25% (Figure 1C, and Tables 1 and 2). Therefore, it is sufficient to increase takeout levels in the peri-cerebral fat body to achieve significant longevity increases.
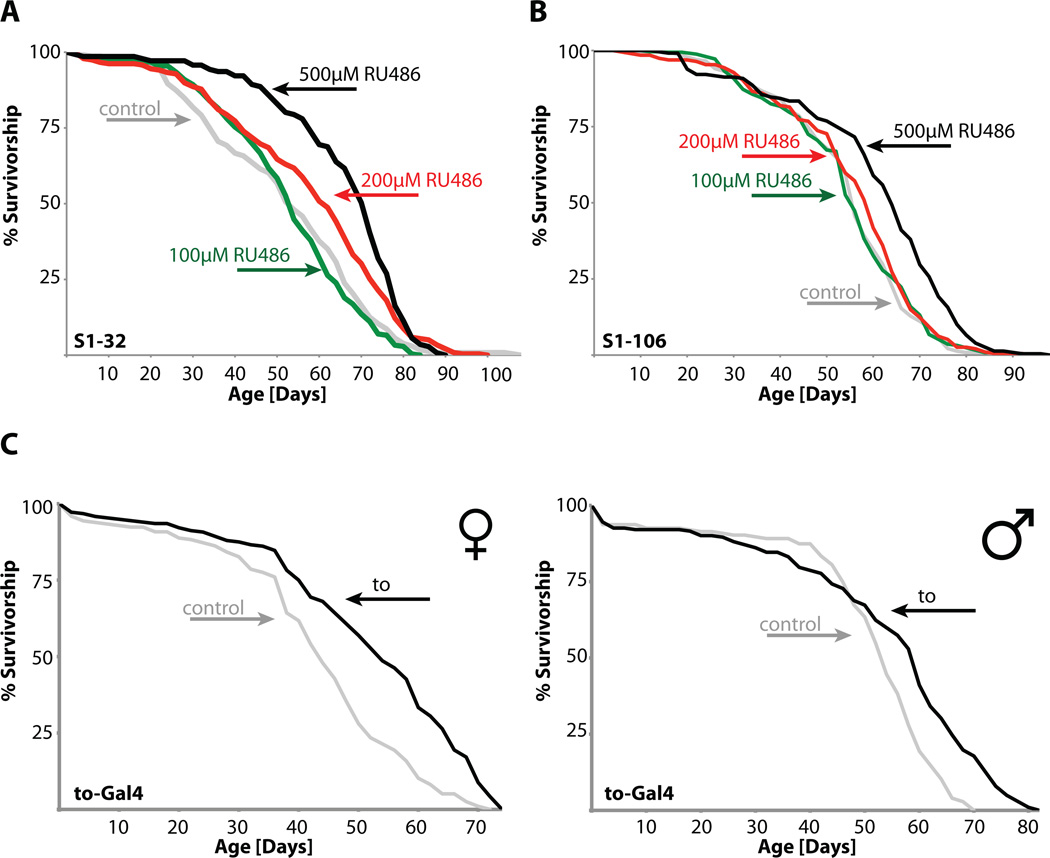
Figure 1. takeout overexpression increases longevity.
Survivorship curves show significant median life span extension of flies overexpressing takeout compared to control flies using either a RU486 inducible peri-cerebral fat body driver (A, females, median life span extension 500µM: 33%), an inducible abdominal fat body driver (B, females, median life span extension 500µM: 14%) or a takeout driver (C, females, median life span extension: 23% and D, males, median life span extension: 11%; for statistical analysis, please see Tables 1 and 2; grey: controls, black: 500µM RU486 or takeout; green: 100µM RU486; red: 200µM RU486).
Table 1.
The effect of to expression on female life span.
| Driver | Mean LS (vs. ctrl) |
Mean LS extension |
Median LS (vs ctrl) |
Median LS extension |
Max LS (vs. ctrl) |
Max LS extension |
Number of flies (control; experimental) |
χ2 | p-value |
|---|---|---|---|---|---|---|---|---|---|
| to-Gal4 | 51/43 | 19% | 54/44 | 23% | 72/66 | 9% | 196 217 |
45.81 | <0.0001 |
| to-Gal4 | 47/39 | 21% | 48/36 | 33% | 68/64 | 6% | 238 250 |
25.89 | <0.0001 |
| to-Gal4 + MP | 36/36 | 0% | 36/36 | 0% | 56/56 | 0% | 269 252 |
0.714 | 0.3981 |
| S1-106, 100µM | 55/55 | 0% | 56/56 | 0% | 74/75 | −1% | 234 251 |
0.1583 | 0.6907 |
| S1-106, 500µM | 61/55 | 11% | 64/56 | 14% | 82/75 | 9% | 234 229 |
41.31 | <0.0001 |
| S1-32, 100µM | 52/52 | 0% | 54/54 | 0% | 80/78 | 3% | 231 243 |
0.9353 | 0.3335 |
| S1-32, 500µM | 66/52 | 27% | 72/54 | 33% | 83/80 | 4% | 231 141 |
39.16 | <0.0001 |
| S1-32, 100µM | 44/41 | 7% | 48/42 | 14% | 66/62 | 7% | 232 227 |
7.14 | 0.0075 |
| S1-32, 500µM | 59/41 | 44% | 64/42 | 52% | 79/62 | 27% | 232 185 |
148.5 | <0.0001 |
| S1-32 | 60/48 | 25% | 62/48 | 29% | 82/68 | 21% | 220 221 |
94.18 | <0.0001 |
| S1-32 + MP | 41/32 | 28% | 36/32 | 12% | 50/51 | −2% | 238 221 |
6.251 | 0.0124 |
Log rank analysis of the survivorship curves of female flies. Mean, median and maximum lifespan, log rank analysis, p-value, percent change in mean, median and maximum lifespan as compared to controls (without RU486 for GeneSwitch experiments), Chi-square and p-values derived from the survivorship curves for each indicated intervention are shown. Maximum life span was calculated as the median life span of the longest surviving 10% of the population.
Table 2.
The effect of to expression on male life span.
| Driver | Mean LS (vs. ctrl) |
Mean LS extension |
Median LS (vs. ctrl) |
Median LS extension |
Max LS (vs. ctrl) |
Max LS extension |
Number of flies (control; experimental) |
χ2 | p-value |
|---|---|---|---|---|---|---|---|---|---|
| to-Gal4 | 53/50 | 6% | 60/54 | 11% | 76/66 | 15% | 175 202 |
30.58 | <0.0001 |
| to-Gal4 | 52/44 | 18% | 54/42 | 29% | 70/70 | 0% | 250 236 |
12.36 | 0.0004 |
| to-Gal4 + MP | 50/51 | −2% | 52/52 | 0% | 70/72 | −3% | 240 241 |
0.4388 | 0.5077 |
| S1-106, 100µM | 66/68 | −3% | 72/68 | 6% | 88/86 | 2% | 224 209 |
0.621 | 0.4307 |
| S1-106, 500µM | 70/68 | 3% | 76/68 | 12% | 96/86 | 12% | 224 215 |
28.78 | <0.0001 |
| S1-32, 100µM | 65/67 | −3% | 70/70 | 0% | 90/88 | 2% | 211 215 |
4.724 | 0.0297 |
| S1-32, 500µM | 61/67 | −9% | 68/70 | −3% | 83/90 | −8% | 211 150 |
22.39 | <0.0001 |
| S1-32, 100µM | 69/67 | 3% | 72/70 | 3% | 93/92 | 1% | 211 200 |
1.361 | 0.2434 |
| S1-32, 500µM | 68/67 | 1% | 70/70 | 0% | 94/92 | 2% | 211 174 |
2.077 | 0.1495 |
| S1-32 | 66/61 | 8% | 66/66 | 0% | 86/84 | 2% | 197 220 |
0.9982 | 0.3178 |
| S1-32 + MP | 70/64 | 9% | 78/70 | 11% | 90/90 | 0% | 224 211 |
3.634 | 0.0566 |
Log rank analysis of the survivorship curves of male flies. Mean, median and maximum lifespan, log rank analysis, p-value, percent change in mean, median and maximum lifespan as compared to controls (without RU486 for GeneSwitch experiments), Chi-square and p-values derived from the survivorship curves for each indicated intervention are shown. Maximum life span was calculated as the median life span of the longest surviving 10% of the population.
3.2 takeout-dependent longevity is not associated with altered feeding regulation
Interestingly, flies carrying the hypomorphic to1 mutation have been reported to have greater feeding frequencies and food uptake. We hypothesized that takeout overexpression may lead to reduced food intake and a state of genetic ‘caloric restriction’, potentially providing a mechanism for the observed longevity phenotype. We thus tested whether takeout overexpressing flies showed feeding defects. No difference in the uptake of food using a dye ingestion assay (as described previously in [5]) was seen in the long-lived takeout overexpressing flies (data not shown).
3.3 takeout long-lived flies show no gross metabolic abnormalities
The dye uptake method to measure food uptake may not be sensitive enough to detect subtle differences in food ingestion and may not be able to discern differences in feeding behavior. In order to investigate more directly whether takeout influences fly metabolism, we measured the weight of flies raised on high and low caloric food over the life span of the animals. takeout overexpressing flies showed no dramatic weight difference to control flies (Supplemental Figure 2A).
Next, we investigated whether takeout long-lived flies had an altered metabolic state by measuring the levels of stored carbohydrates. Interestingly, no difference in the levels of glucose, trehalose or glycogen was observed on high caloric conditions. Under low caloric conditions, however, trehalose levels were reduced in takeout overexpressing flies, while no differences were observed in glucose and glycogen levels (Figure 2A).
3.4 takeout may extend longevity by a pathway largely unrelated to DR
Since takeout was identified in a screen designed to enrich for mediators of DR, we investigated more closely the gene expression profiles of long-lived flies due to DR or takeout overexpression. Recently, we established a signature profile of 83 KEGG pathways altered under DR conditions [14]. This profile was compared to the genetic changes observed in takeout long-lived flies. Interestingly, while there was considerable overlap between the DR signature and the takeout overexpression profile (37 pathways, or 44.6%, Figure 2B), this overlap was not as strong as the overlap observed between DR and resveratrol treated flies (61 pathways, or 73.5%, [14]). Resveratrol in flies has been suggested to employ a longevity mechanism similar to DR [17]. Therefore, these data suggests that takeout may only partially mediate the effects of DR.
Finally, to directly test whether takeout-dependent longevity extension is related to feeding and caloric intake, we exposed takeout overexpressing flies to DR to determine whether the longevity effects of DR and takeout overexpression were additive. As shown in Figure 2C, takeout overexpression extended the life span of flies that had increased longevity due to DR treatment. Moreover, to1 flies, which have severely reduced takeout levels, still responded to DR treatment with extension of life span (Supplemental Figure 2B). Taken together, these data demonstrate that takeout-dependent longevity is independent of DR and does not involve alterations in nutrition-dependent metabolism. Since takeout is upregulated in a variety of longevity interventions, these observations suggest that takeout may mediate only specific phenotypes of altered longevity.
3.5 takeout long-lived flies are not stress resistant
Food intake and metabolic dysregulation could not adequately explain the mechanism of takeout-dependent longevity. Thus, we investigated other phenotypes associated with exceptional longevity. Long-lived flies often show increased stress resistance. Therefore, we exposed takeout overexpressing flies to H2O2 to induce oxidative stress. Surprisingly, takeout long-lived flies did not survive oxidative stress longer than control flies (Figure 3A).
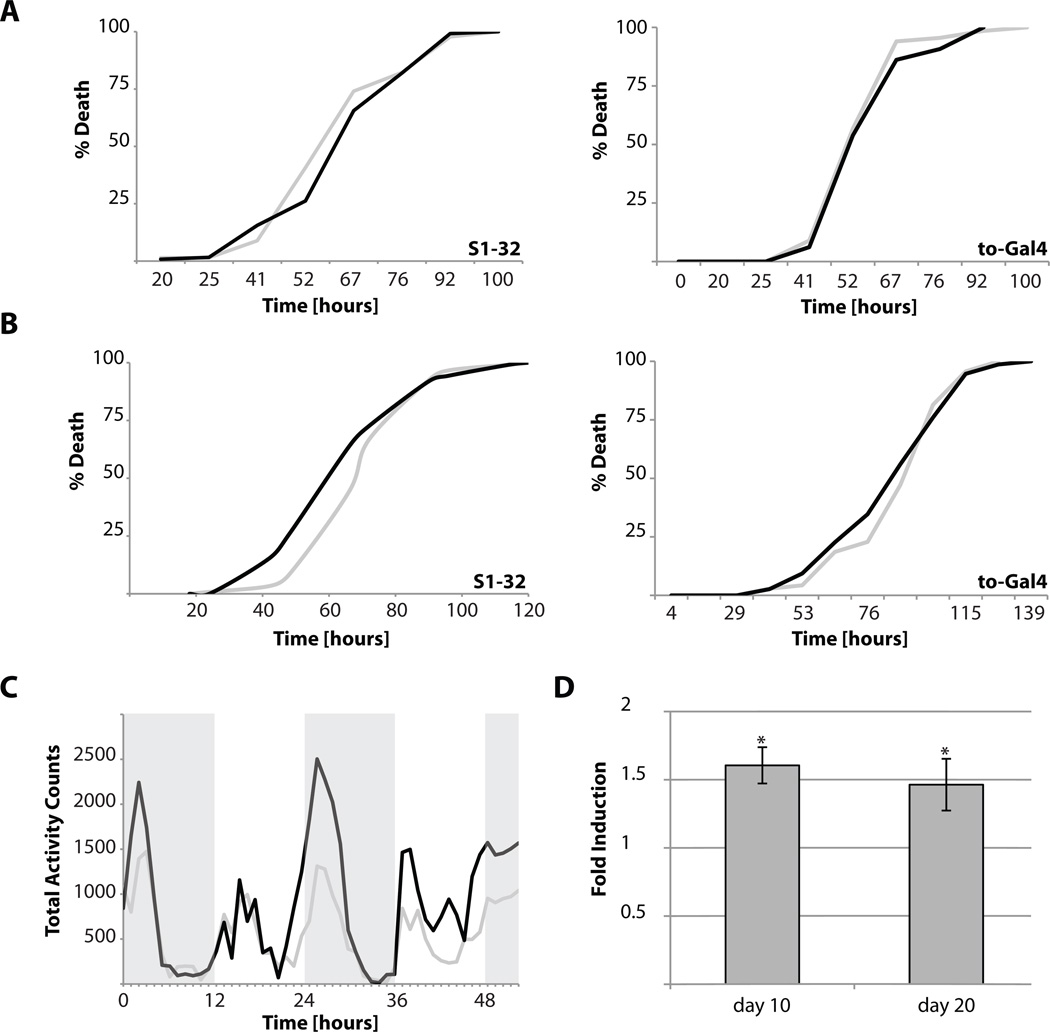
Figure 3. Phenotypes of long-lived takeout overexpressing flies.
10-day old female flies overexpressing takeout under the control of the S1-32 driver or the to-GAL4 driver were exposed to H2O2 (A) or starved (B) and their survival times determined. (C) The spontaneous activity of 10-day old female flies overexpressing takeout under the control of the S1-32 driver was recorded over a 48-hrs period after several hours of acclimatization to the observation chamber. (D) Spontaneous activity of female flies overexpressing takeout under the control of the S1-32 driver was measured at the indicated ages and integrated over the 48-hrs recording interval (grey: controls, black: takeout; shown are averages of at least two independent experiments; asterisks: p<0.05).
Next, we tested whether takeout long-lived flies were more resistant to starvation stress. Similar to the experiments measuring oxidative stress-resistance, takeout overexpressing flies did not survive starvation stress longer than control flies (Figure 3B).
3.6 takeout overexpressing flies are highly active
takeout mRNA is regulated in a circadian fashion [4, 8, 18], and to1 hypomorphs show defects in starvation dependent activity [5]. We thus measured the activity and circadian pattern of takeout long-lived flies. As shown in Figure 3C, takeout overexpressing flies were slightly more active than control flies, especially during normal rest periods. This increased activity of takeout overexpressing flies was maintained as the flies aged (Figure 3D).
3.7 takeout overexpressing flies have defects in mating behavior and fertility
In addition to its role in feeding behavior, takeout has also been implicated in the regulation of male courtship behavior [3, 7]. Flies carrying the to1 allele in conjunction with a mutation in the sex determination gene fru have previously been demonstrated to have a reduction in male courtship behavior. However, no male courtship defects were observed in regular to1 mutant flies [7]. These data suggest that takeout may play a role in regulating male fertility through modulation of courtship behavior.
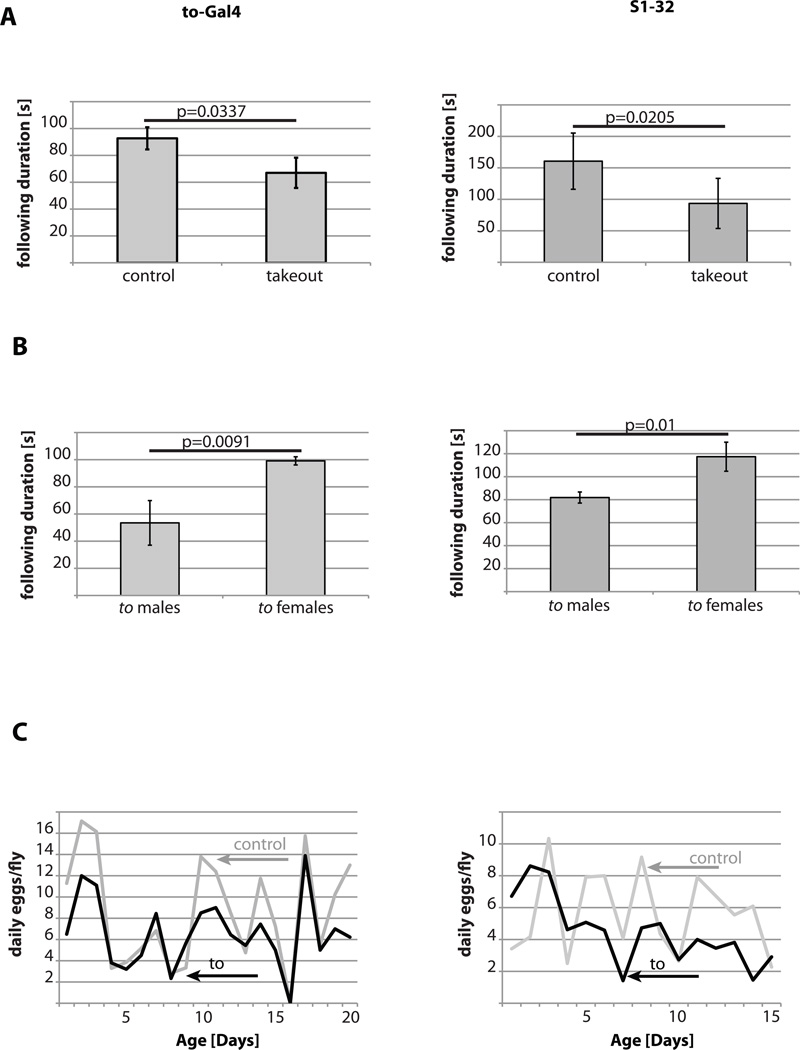
Since longevity is often negatively correlated with fertility [19], we therefore examined male courtship behavior in long-lived takeout overexpressing flies. Interestingly, male flies overexpressing takeout showed a marked reduction in the frequency of stereotypical courtship behaviors. takeout males followed the females less frequently and for shorter periods of time (Figure 4A). In addition, other behaviors, such as wing extension or mounting attempts, were also reduced compared to control males (data not shown). This decrease in courting could not be attributed to lower activity levels, as takeout long-lived flies were more active than control flies (Figure 3C and D). Rather, males did not appear to be interested in the females and began courtship behavior later than control flies (data not shown). Moreover, when takeout overexpressing males were placed with non-takeout overexpressing control females, a similar decrease in male courtship behavior was observed (Figure 4B). In contrast, when control males were placed with takeout overexpressing females, no defects in courtship were observed (Figure 4B).
Figure 4. Long-lived takeout flies show mating behavior and fertility defects.
Mating behavior was assessed by measuring the total time a male followed a female in an observation chamber. (A) Mating pairs, in which both partners overexpressed takeout, had a significant reduction in male following behavior compared to control flies. (B) Mating pairs, in which only the indicated partner overexpressed takeout, were assessed for following behavior. When only the male fly overexpressed takeout, greatly reduced following activity was observed, compared to mating pairs, in which only the female overexpressed takeout. In addition, reduction in following behavior is significant when compared to mating pairs, in which neither partner overexpressed takeout (p=0.0084 for the to-GAL4 driver, p=0.0382 for the S1-32 driver, when compared to control flies from panel A; shown are the averages of at least three independent experiment with at least six individual mating pairs each). (C) Daily egg laying is slightly reduced over the first 20- days of adult life in takeout overexpressing flies (grey: control; black: takeout; shown are representatives of at least two independent experiments).
Next, we investigated whether decreased male mating behavior affected female fertility. We therefore measured the daily egg production of individual mating pairs of takeout overexpressing flies. As shown in Figure 4C, takeout overexpressing females had slightly reduced daily egg production compared to control flies. These data raise the interesting hypothesis that a takeout-dependent decrease of male courtship may influence female longevity by reducing female oviposition and may therefore play a central role in the modulation of the longevity/fertility tradeoff.
3.8 takeout overexpression reduces JH signaling
takeout is a small protein, consisting of a signal peptide and a Juvenile Hormone Binding Domain (JHBD). It is produced in and secreted from the head fat body [3, 4, 8]. The JHBD-containing JH binding proteins (JHBP) bind to the highly lipophilic Juvenile Hormones and thus act as their carriers in insect hemolymph [20, 21]. The effect of these carriers may be two-fold: They could either protect the labile JHs from degradation by JH esterases, or they may function as JH reservoirs, maintaining a proper JH concentration in the hemolymph. Interestingly, modulation of JH signaling has been suggested to regulate insect longevity. Grasshoppers, in which the Corpora Alata (CA), the site of JH production, was surgically removed, exhibited longer life spans, while flies exposed to vapors of the JH analog methoprene had decreased longevity [10, 11].
While binding of JH to Drosophila takeout has not been assessed yet, we hypothesized that takeout may bind JH, leading to a reduction in JH bioavailability and thus JH signaling. In a first test of this hypothesis, we examined the transcriptional profile of takeout overexpressing flies and performed Gene Set Enrichment Analysis (GSEA) [22] for JH regulated genes. Using a list of genes downregulated by JH [14], GSEA showed that some components of JH signaling are diminished leading to an upregulation of the normally downregulated genes. These data suggest a global decrease in normal JH functional activity, or signaling, in the takeout overexpressing flies (Figure 5A). Importantly, a similar comparison for flies exposed to DR showed a non-significant decrease in JH activity [14].
Next, to test directly the hypothesis that takeout may reduce JH signaling activity, we examined whether the phenotypes of takeout overexpressing flies may be blocked by treatment with the JH analog methoprene. First, we examined whether methoprene treatment affected the fertility-related phenotypes of takeout overexpressing flies. Long-lived takeout flies treated with methoprene did not show any difference in courtship behavior to control flies (Figure 5B) and similarly had no reduction in egg laying (data not shown). Importantly, when takeout overexpressing flies were treated with methoprene, the longevity increase was almost completely blocked (Figure 5C). These data demonstrate that methoprene rescues the takeout overexpression phenotype.
The methoprene effects were not simply due to methoprene-dependent inhibition of takeout overexpression, as takeout overexpression levels were only slightly reduced by methoprene treatment (Supplemental Figure 1B). Therefore, takeout may reduce JH signaling activity, leading to increased fly longevity. Taken together, these data indicate that JH signaling is an important modulator of insect longevity.
4. Discussion
We identified takeout in a screen designed to enrich for novel longevity genes [2]. Flies overexpressing takeout in the fat body (using the Geneswitch driver S1-32), or in their endogenous expression pattern (by using a to-GAL4 driver [7]), were long-lived, yet did not display increased resistance to oxidative stress, a phenotype often associated with increased longevity. In addition, takeout long-lived flies did not have any feeding abnormalities or altered starvation survival. These observations are in contrast to what is seen with hypomorphic to1 mutants, which show feeding phenotypes and reduced starvation survival. These data suggest that takeout-associated feeding functions alone cannot account for the increased longevity of takeout overexpressing flies.
Interestingly, even though our original screen utilized genes linked to the longevity effects of DR [2], takeout overexpression was additive to the life span-increasing effects of DR. Moreover, to1 flies still responded to DR with increased longevity. Although to1 flies have residual expression of takeout, taken together, these data suggest that takeout does not play a major role in DR-dependent longevity. However, to1 is a hypomorphic mutation with low basal expression of both takeout mRNA and protein, which may be sufficient to perform residual signaling functions. Additionally, the situation is complicated by the fact that to1 lines have been reported to carry the ry506 mutation as well [7]. This is a mutation in a xanthine dehydrogenase that is involved in the JH biosynthetic pathway. The presence of two mutations that may both affect JH activity levels (ry506 and to1) may lead to compensatory effects on JH signaling [25] and may thus interfere with takeout phenotypes. If takeout regulates longevity via modulation of JH signaling, then the ry506 mutation may counteract these effects. We are currently engineering true takeout null flies in order to directly address the question whether the effects of DR are mediated by takeout.
Our data indicate that increased takeout expression is sufficient, but not required for extended fly longevity. takeout appears to be dispensable for the longevity effects of DR, but it remains to be investigated whether takeout is required for the longevity effects of other interventions with upregulated takeout [2]. It is conceivable that takeout may function as common molecular target mediating specific phenotypes associated with increased longevity. An intriguing role for takeout in this context may be to mediate the tradeoff between fertility and longevity [10], especially as other phenotypes, such as feeding, metabolism and stress resistance, are not affected in takeout long-lived animals.
takeout long-lived flies have a drastic decrease in male courtship display and behavior, which is associated with a decrease in female egg deposition. This courtship defect is not merely due to impaired mobility, as takeout flies are not less active than control flies. The Drosophila fat body has previously been associated with the regulation of male courtship behavior: Expression of the female-specific transformer protein TraF under control of a takeout or lsp2 driver yields male flies with reduced takeout mRNA levels and decreased courtship behavior [3, 7]. However, to1 mutant flies do not display courtship defects. Only when fru alleles are additionally present, does male courtship behavior decrease [7]. Interestingly, supplying those flies with endogenous takeout, via expression in oenocytes or on a genomic rescue construct, reverses the courtship phenotype, suggesting that it is the lowering of takeout levels that lead to courtship defects [3, 7].
It is unclear why takeout overexpression produces similar courtship phenotypes as takeout underexpression. The use of TraF to feminize all takeout expressing cells may affect more biological processes than takeout expression alone, while using to1 mutants could be problematic due to compensatory effects as discussed above [25]. Therefore these models may not adequately represent a takeout loss-of-function scenario. In contrast, overexpression of the JH esterase binding protein DmP29 has been reported to cause hyperactivity, increase male-male courtship behavior, decrease female receptivity and reduce egg counts [26]. These phenotypes are reversed by application of methoprene, suggesting that a decrease in JH signaling may be responsible. Importantly, these observations are similar to what is seen in takeout overexpressing flies, suggesting that reduction in fertility is achieved by decreasing JH signaling.
The mating behavior and longevity phenotypes of takeout overexpressing flies can be reversed by treatment with the JH analog methoprene. Moreover, GSEA analysis indicates that takeout overexpression leads to a reduction of JH signaling activity, as genes negatively regulated by JH are dysinhibited. These data support the hypothesis that takeout may bind JH in the hemolymph, thereby decreasing JH bioavailability and JH signaling capabilities. takeout may thus be the point of convergence for disparate longevity signaling pathways with fertility tradeoffs. takeout is uniquely suited for this role: It is produced in and secreted from the fat body, the major metabolic organ of flies [3]. Manipulation of metabolic pathways, such as insulin or TOR signaling, in the fat body alters fly life span [27–29]. These signaling pathways may generate an inhibitory signal for takeout production, decreased activity of which could raise takeout levels. As a consequence, bio-available JH in the hemolymph would be reduced and likewise JH signaling in target tissues.
During development, JH acts together with 20-Hydroxyecdysone (20E) to control developmental transitions [9], as well as tissue development [30, 31]. Evidence is accumulating that these hormonal signaling pathways are additionally important in the regulation of longevity in adult insects. 20E receptor mutant flies are long-lived [32], while the increased longevity of insulin receptor (InR) mutant flies is reversed by methoprene treatment [33]. In addition, both chico and InR mutants have reduced JH levels [33, 34]. Moreover, exposure of larvae to methoprene has been shown to reduce adult longevity [10], while ablation of the CA, the site of JH production in insects, extends the life span of grasshoppers [11]. Together, these data clearly demonstrate that JH/20E signaling in adult flies is responsible for life span regulation. It is unclear how the JH effects are mediated, as several putative JH receptors exist that interact with each other [30, 35, 36], as well as with 20E receptors [37–40], thus forming a highly complex signaling network with pleiotropic effects in different tissues [30].
Longevity and fertility can be experimentally uncoupled [41], suggesting a molecular longevity signaling pathway with multiple branches for longevity-associated phenotypes. Manipulating those branching pathways directly may alter longevity without affecting other longevity phenotypes. takeout and its modulation of JH signaling may represent such a downstream branch that specifically modulates the fertility aspect of longevity interventions. This modulation may occur via several different mechanisms, such as behavioral changes or alterations in the germline. It will be interesting to dissect the specific components of the JH signaling cascade that affect these pleiotropic outcomes.
5. Conclusions
Here, we investigated the mechanism by which overexpression of Drosophila melanogaster takeout regulates longevity of fruit flies. We found that takeout extended life span at every food condition tested, even on Dietary Restriction (DR) conditions promoting fly longevity. These data indicate that takeout overexpression is additive to the longevity effects of DR and therefore regulates life span by a different mechanism than DR.
In order to understand this mechanism, we examined other phenotypes of takeout long-lived flies. Interestingly, takeout did not impart several of the traits usually associated with increased life span, such as enhanced oxidative stress resistance or metabolic abnormalities. However, takeout long-lived flies showed reduced male mating behavior, and a decrease in female egg laying.
Importantly, the takeout longevity and fertility phenotypes were blocked by administration of a Juvenile Hormone (JH) analog. Since takeout contains a Juvenile Hormone Binding Domain, these observations suggest that takeout negatively regulates JH function by acting as a JH reservoir in the fly hemolymph. Therefore, downregulation of JH activity may be the molecular event that leads to life span extension.
During development, JH regulates the transition between different developmental stages, but our data suggest a novel function for JH in adult flies. JH may act to control fertility and mating behavior, and may thus be central for regulating the tradeoff between fertility and longevity.
Supplementary Material
Highlights.
D. melanogaster takeout extends longevity independent of Dietary Restriction
takeout reduces courtship behavior in males
takeout overexpression is antagonized by a Juvenile Hormone (JH) analog
takeout may act as a reservoir for JH, limiting JH bioavailability and signaling
takeout may be the switch controlling the tradeoff between longevity and fertility
Acknowledgements
The authors would like to thank M. Tatar, B. Dauwalder, and H. Keshishian for the kind gift of fly stocks. We also thank Chengyi Chang, Suzanne Hozier and Chris Thephachanh for technical assistance and Christoph Schorl for help with the microarray studies. This work was supported by NIA grants AG16667, AG24353 and AG25277 to SLH, AG028753 to NN and NIA AG029723 to JHB, and a Hamilton Undergraduate Research Award to KHC. JHB is a recipient of the Ralph. E. Powe Junior Faculty Enhancement Award. SLH is an Ellison Medical Research Foundation Senior Investigator and recipient of a Glenn Award for Research in Biological Mechanisms of Aging.
Abbreviations
- DR
Dietary Restriction
- JH
Juvenile Hormone
- JHBD
JH binding domain
- JHBP
JH binding protein
- CA
Corpora Allata
- MP
methoprene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions
KHC designed and performed all experiments and analyzed the data; SQK performed longevity determinations, MLHN, SNSM and SK performed the metabolic experiments, MA, NN and SLH analyzed the microarray data and JHB designed and performed all experiments, analyzed the data and wrote the manuscript.
References
- 1.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 2.Bauer JH, et al. Comparative transcriptional profiling identifies takeout as a gene that regulates life span. Impact: Aging. 2010;2(5):298–310. doi: 10.18632/aging.100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazareva AA, et al. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3(1):e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarov-Blat L, et al. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101(6):647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 5.Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007;210(Pt 8):1424–1434. doi: 10.1242/jeb.02755. [DOI] [PubMed] [Google Scholar]
- 6.Wong R, et al. Quantification of food intake in Drosophila. PLoS One. 2009;4(6):e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dauwalder B, et al. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16(22):2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.So WV, et al. takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol Cell Biol. 2000;20(18):6935–6944. doi: 10.1128/mcb.20.18.6935-6944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riddiford LM. Hormone receptors and the regulation of insect metamorphosis. Receptor. 1993;3(3):203–209. [PubMed] [Google Scholar]
- 10.Flatt T, Kawecki TJ. Juvenile hormone as a regulator of the trade-off between reproduction and life span in Drosophila melanogaster. Evolution. 2007;61(8):1980–1991. doi: 10.1111/j.1558-5646.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 11.Tatar M, Yin C. Slow aging during insect reproductive diapause: why butterflies, grasshoppers and flies are like worms. Exp Gerontol. 2001;36(4–6):723–738. doi: 10.1016/s0531-5565(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 12.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101(45):15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer JH, et al. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci U S A. 2007;104(33):13355–13360. doi: 10.1073/pnas.0706121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antosh M, et al. Comparative transcriptional pathway bioinformatic analysis of dietary restriction, Sir2, p53 and resveratrol life span extension in Drosophila. Cell Cycle. 2011;10(6):904–911. doi: 10.4161/cc.10.6.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neretti N, et al. Long-lived Indy induces reduced mitochondrial reactive oxygen species production and oxidative damage. Proc Natl Acad Sci U S A. 2009;106(7):2277–2282. doi: 10.1073/pnas.0812484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osterwalder T, et al. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98(22):12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 18.Benito J, et al. The circadian output gene takeout is regulated by Pdp1epsilon. Proc Natl Acad Sci U S A. 2010;107(6):2544–2549. doi: 10.1073/pnas.0906422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc R Soc Lond B Biol Sci. 1996;263(1371):755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- 20.Hagai T, Cohen M, Bloch G. Genes encoding putative Takeout/juvenile hormone binding proteins in the honeybee (Apis mellifera) and modulation by age and juvenile hormone of the takeout-like gene GB19811. Insect Biochem Mol Biol. 2007;37(7):689–701. doi: 10.1016/j.ibmb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Touhara K, Prestwich GD. Binding site mapping of a photoaffinity-labeled juvenile hormone binding protein. Biochem Biophys Res Commun. 1992;182(2):466–473. doi: 10.1016/0006-291x(92)91755-f. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B, et al. Juvenile hormone prevents ecdysteroid-induced expression of broad complex RNAs in the epidermis of the tobacco hornworm, Manduca sexta. Dev Biol. 1998;203(2):233–244. doi: 10.1006/dbio.1998.9059. [DOI] [PubMed] [Google Scholar]
- 24.Minakuchi C, Namiki T, Shinoda T. Kruppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev Biol. 2009;325(2):341–350. doi: 10.1016/j.ydbio.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, Riddiford LM. rosy Function is required for juvenile hormone effects in Drosophila melanogaster. Genetics. 2008;178(1):273–281. doi: 10.1534/genetics.107.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, et al. Overexpression of Drosophila juvenile hormone esterase binding protein results in anti-JH effects and reduced pheromone abundance. Gen Comp Endocrinol. 2008;156(1):164–172. doi: 10.1016/j.ygcen.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305(5682):361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 28.Hwangbo DS, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 29.Kapahi P, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14(10):885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development. 2009;136(12):2015–2025. doi: 10.1242/dev.033712. [DOI] [PubMed] [Google Scholar]
- 31.Riddiford LM, et al. A role for juvenile hormone in the prepupal development of Drosophila melanogaster. Development. 2010;137(7):1117–1126. doi: 10.1242/dev.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon AF, et al. Steroid control of longevity in Drosophila melanogaster. Science. 2003;299(5611):1407–1410. doi: 10.1126/science.1080539. [DOI] [PubMed] [Google Scholar]
- 33.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 34.Tu MP, Yin CM, Tatar M. Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocrinol. 2005;142(3):347–356. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Jones G, et al. Juvenile hormone III-dependent conformational changes of the nuclear receptor ultraspiracle. Insect Biochem Mol Biol. 2001;32(1):33–49. doi: 10.1016/s0965-1748(01)00077-7. [DOI] [PubMed] [Google Scholar]
- 36.Godlewski J, Wang S, Wilson TG. Interaction of bHLH-PAS proteins involved in juvenile hormone reception in Drosophila. Biochem Biophys Res Commun. 2006;342(4):1305–1311. doi: 10.1016/j.bbrc.2006.02.097. [DOI] [PubMed] [Google Scholar]
- 37.Maki A, et al. Juvenile hormones antagonize ecdysone actions through co-repressor recruitment to EcR/USP heterodimers. Biochem Biophys Res Commun. 2004;320(1):262–267. doi: 10.1016/j.bbrc.2004.05.156. [DOI] [PubMed] [Google Scholar]
- 38.Fang F, et al. Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J. 2005;272(7):1577–1589. doi: 10.1111/j.1742-4658.2005.04578.x. [DOI] [PubMed] [Google Scholar]
- 39.Braun S, Azoitei A, Spindler-Barth M. DNA-binding properties of Drosophila ecdysone receptor isoforms and their modification by the heterodimerization partner ultraspiracle. Arch Insect Biochem Physiol. 2009;72(3):172–191. doi: 10.1002/arch.20328. [DOI] [PubMed] [Google Scholar]
- 40.Bitra K, Palli SR. Interaction of proteins involved in ecdysone and juvenile hormone signal transduction. Arch Insect Biochem Physiol. 2009;70(2):90–105. doi: 10.1002/arch.20281. [DOI] [PubMed] [Google Scholar]
- 41.Flatt T, et al. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105(17):6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.