Abstract
The Macrophage Migration Inhibitory Factor (MIF), an inflammatory cytokine, is overexpressed in many solid tumors and is associated with poor prognosis. We previously identified inhibitors of MIF within a class of natural products with demonstrated anti-cancer activities. We therefore sought to determine how MIF contributes to tumor growth and progression. We show here that, in murine tumors including the 4T1 model of aggressive, spontaneously metastatic breast cancer in immunologically intact mice, tumor-derived MIF promotes tumor growth and pulmonary metastasis through control of inflammatory cells within the tumor. Specifically, MIF increases the prevalence of a highly immune suppressive subpopulation of myeloid derived suppressor cells (MDSCs) within the tumor. In vitro, MIF promotes differentiation of myeloid cells into the same population of MDSCs. Pharmacologic inhibition of MIF reduces MDSC accumulation in the tumor similar to MIF depletion, and blocks the MIF-dependent in vitro differentiation of MDSCs. Our results demonstrate that MIF is a therapeutically targetable mechanism for control of tumor growth and metastasis through regulation of the host immune response, and support the potential utility of MIF inhibitors, either alone or in combination with standard tumor-targeting therapeutic or immunotherapy approaches.
Keywords: MIF, microenvironment, MDSC, sulforaphane, 4T1
Introduction
The inflammatory tumor microenvironment contributes to tumor growth and progression through a variety of mechanisms (1). One cellular component of the tumor microenvironment, myeloid derived suppressor cells (MDSCs), strongly enhances tumor growth and metastasis in a murine breast cancer model (2). Circulating MDSCs have been demonstrated in the blood of patients with a variety of tumors, including head and neck (3), lung (3), breast (3), gastric (4), pancreatic (4), and renal cell carcinomas (5). MDSCs utilize a variety of mechanisms to suppress T cell and NK cell functions, thereby inhibiting anti-tumor immunity (6, 7).
In mice, MDSCs are defined as GR1+CD11b+ cells, but this population can be further subdivided into two distinct subsets. The granulocytic subset is characterized by high Ly6G expression and low to intermediate Ly6C expression, while the monocytic subset is characterized by high Ly6C expression and low Ly6G expression (6, 8, 9). These two subtypes of MDSCs also have differing mechanisms of T cell suppression. Both subsets express arginase, which contributes to T cell suppression through consumption of arginine. In addition, the granulocytic subset of MDSCs suppresses T cells by producing reactive oxygen species and the monocytic subset by producing nitric oxide (8). The granulocytic population of MDSCs is usually more increased in tumor models (6), however the monocytic subset is thought to be the more suppressive since this population is able to suppress both antigen-specific and nonspecific T-cell proliferation (9), likely through nitrosylation of either the T-cell receptor (10) or MHC complex (11). Additionally, the monocytic subset of MDSCs has been demonstrated to differentiate into macrophages in vitro (6), possibly linking MDSCs and tumor-supporting macrophages.
The Macrophage Migration Inhibitory Factor (MIF) was identified in the 1960s as a soluble, protein factor secreted by T cells with the ability to inhibit the random migration of macrophages (12, 13). MIF is an inflammatory cytokine that plays a critical role in numerous models of inflammatory disease (14, 15). In addition, MIF is overexpressed in many solid tumors (16–21). In many cases, the degree of MIF overexpression has been correlated with tumor progression and/or metastatic potential, with examples in prostate (22), hepatocellular (23), gastric (24), and lung cancers (25).
We identified MIF as a target for covalent modification and inhibition by a class of natural product compounds with well-established anti-cancer activities (26). MIF has an enzymatic keto-enol tautomerase activity, with the N-terminal proline as its catalytic center (27), and we demonstrated that these compounds covalently modify MIF on this proline and inhibit MIF tautomerase activity (26). No physiological tautomerase substrate has been identified, but the importance of the tautomerase to biological function of MIF is supported by observations of other groups (28, 29) that have also characterized small molecule inhibitors of MIF that inhibit both this enzymatic activity and the biological activities of MIF.
Given the observation that MIF overexpression correlates with tumor growth and progression, several groups have explored the potential contribution of MIF to growth and transformation of tumor cells. As a result, current models proposed to explain the tumorigenic activities of MIF have focused on functional interactions of MIF with cell signaling pathways, tumor suppressors, and oncogenes (30–32). However, as an inflammatory cytokine, we hypothesized that MIF would contribute to the interaction between the tumor and the host immune response. In this study, we provide evidence for a mechanism of MIF action in promoting tumor growth and metastasis that is unrelated to effects within the tumor cells. We demonstrate that the tumor promoting effects of MIF require interaction with a fully competent immune system, and we characterize MIF dependent modulation of monocytic MDSCs within the tumor microenvironment.
Materials and Methods
Cell Lines, Reagents and Antibodies
4T1 and CT26 cells (ATCC) were cultured at 37°C in 5% CO2 in RPMI 1640, supplemented with 10% FBS and 1% Penicillin/Streptomycin. Luciferase-expressing 4T1 lines (Caliper Life Sciences) were cultured with heat-inactivated FBS as recommended. Anti-MIF was from Invitrogen and anti-tubulin from Sigma. L-dopachrome methyl ester and periodate were from Sigma, and sulforaphane from LKT Labs.
Mice
Female 6–12 week old Balb/c mice and 6–8 week old SCID/bg mice were purchased from Charles River Laboratories. All animal experiments were performed with the approval of the UVA ACUC.
Generation of MIF Depleted and Reconstituted Cell Lines
For MIF depleted cell line, the sequences gatccATGCCtATGTTCATCGTgATTCAAGAGATcACGATGAACATaGGCATTTTTTTACG CGTg and aattcACGCGTAAAAAAATGCCtATGTTCATCGTgATCTCTTGAATcACGATGAACATaGG CATg were annealed and ligated into pSiren-RetroQ plasmid (Clontech) and used to make retroviruses. Parental 4T1 cells were infected with either the targeting hairpin or an empty vector and selected with 10μg/mL puromycin. In some experiments, a line expressing a non-targeting RNA directed against the human MIF sequence was used in place of the empty vector control. For preparation of MIF depleted CT26, parental CT26 cells were infected with targeting or non targeting hairpin and selected with 10 ug/ml puromycin.
For the reconstituted cell lines, the coding region for wild type human MIF or P2G MIF in which the codon for the proline at position 2 was mutagenized to glycine (both resistant to the shRNA targeting the murine MIF sequences), was inserted into PQCXI-neo vector and used to produce retroviruses. MIF depleted 4T1 MIF KD cells were infected with empty vector, mutant P2G MIF, or WT MIF expressing viruses and selected with 500μg/mL neomycin.
Tautomerase Assay
Tautomerase activity was measured as previously described (33). Briefly, 4mM dopachrome and 8mM sodium periodate were mixed together in a 1:1 ratio and incubated for 5 minutes. This substrate was diluted 1:9 in tautomerase buffer (25mM Na+PO4, pH6.0, 1mM EDTA) and added to normalized cell lysates. Absorbance was monitored over time at 475nm in a 96 well plate reader (Biorad). The velocity is the rate of decolorization of the colorimetric substrate (measured as a decrease in absorbance at 475nm) per unit time.
Proliferation and Colony Formation Assays
Wild type and MIF depleted 4T1 or CT26 cells (5×104) were plated in triplicate in 6 well plates and incubated at 37°C in 5% CO2 for a total of 4 days. Cells were counted each day using a hemocytometer. For colony formation, 4×104 4T1 cells were suspended in media containing soft agar (Lonza) and incubated at 37°C for two weeks. Photos were taken of 3 independent fields and the number of colonies in each field was enumerated.
Tumor Implantation, Measurement, and Survival Surgery
1×104 4T1 cells were injected into the mammary fat pad of 6–12 week old female Balb/c mice. 2×104 CT26 cells were injected subcutaneously above the flank of 6–12 week old female Balb/c mice. Tumor volumes were estimated using the formula V=0.4 LW2 from 2 perpendicular measurements made with electronic calipers. In experiments with the reconstituted cell lines, tumors were removed at 18 days post implantation. The mice were euthanized 8 days later for enumeration of metastasis.
Sulforaphane (SFN) Treatment of Mice
Mice were treated with 200 μg of SFN (or saline vehicle) by intraperitoneal injection, starting the day of tumor injection and continuing daily until tumor harvest.
Tissue Digestion and Clonogenic Metastasis Assay
Tumors were dissected from the mammary fat pad and digested with 10,000 units of Collagenase I (Worthington Biochemical) for 60 minutes at 37°C. Cell suspensions were strained through a 70μm membrane, and then stained for flow cytometry.
Isolated lungs were digested with 5mg of Collagenase IV (Worthington Biochemical) and 30 units of elastase (Calbiochem) for 75 minutes at 4°C. Strained cell suspensions were plated in IMDM +10% FBS, 1% Penicillin/Streptomycin, containing 60 μM 6-thioguanine. Plates were incubated for at least 10 days at 37°C, fixed with methanol and stained with crystal violet for enumeration of colonies.
Imaging of Lungs Using Luciferase
To image lung metastasis in experiments with the luciferase expressing cell lines, mice were injected intraperitoneally with 100μL/10g body weight of a 15 mg/mL solution of luciferin 5 minutes prior to euthanasia. Lungs were excised and immersed in a 300μg/mL solution of luciferin in PBS for imaging on the Xenogen IVIS System (Caliper Life Sciences).
Flow Cytometry
For flow cytometry analysis, cells were stained with antibodies for CD45 PerCP (BD Biosciences), F4/80 APC-eFluor780 (eBiosciences), Ly6G FITC (Biolegend), Ly6C APC (Biolegend), and CD11b Pacific Blue (Invitrogen), along with LIVE/DEAD Fixable Dead Cell Stain (Invitrogen). Live cells were gated first using LIVE/DEAD Fixable Dead Cells Stain (negative population), then forward and side scatter, followed by CD45pos and, finally CD11bpos. A representative series of gated plots is shown in Supplemental Figure 2A. The cells were analyzed with the Beckman Coulter CyAN ADP LX 9 Color Flow Cytometer.
T Cell Proliferation Assay
CD8+ T cells were purified from a naïve Balb/c mouse using CD8-PE antibody (Biolegend) and the Easy SEP-PE kit (Stem Cell Technologies). MDSCs were purified from tumors by first depleting T cells with CD90.2 magnetic beads (Milltenyi) and subsequently purifying GR1 positive cells with GR1-PE antibody (Biolegend) and the Easy SEP-PE kit. T cells (5×104 per well) were stimulated with anti-CD3 (Cedarlane) and anti-CD28 (eBioscience) at dilutions of 1:1000 each. MDSCs from either a MIF-containing or MIF-deficient tumor were added in 1:1 or 4:1 ratios. After a 72 hour incubation, the culture was pulsed with 3H-thymidine and harvested 18 hours later.
Generation of Conditioned Media and Differentiation Assay
3×105 4T1 WT or KD cells were incubated in RPMI 1640 supplemented with 10% heat-inactivated FBS and 1% Penicillin/Streptomycin for 5 days to generate tumor cell conditioned media. For the differentiation assay, spleens from tumor-bearing mice were harvested and incubated with anti-CD11b beads (Miltenyi) and purified using the Automacs system. 6×106 purified CD11b+ cells were incubated in RPMI 1640 supplemented with 10% heat-inactivated FBS and 1% Penicillin/Streptomycin along with the conditioned media with and without SFN for 5 days before flow staining as described above. All cultures were supplemented with 10ng/mL GM-CSF (R&D Systems) to ensure viability.
MDSC Depletion
Mice were injected intraperitoneally with 500μg of either Rat IgG (Invitrogen) or GR1 Antibody (Clone RB6-8C5) on days 11, 14, and 17 relative to tumor implantation.
Data Analysis and Statistics
All data are graphed as mean ± SEM. Groups were compared by unpaired two-tailed Student’s t test for Figures 1 and 2 and by ANOVA for Figures 3–7.
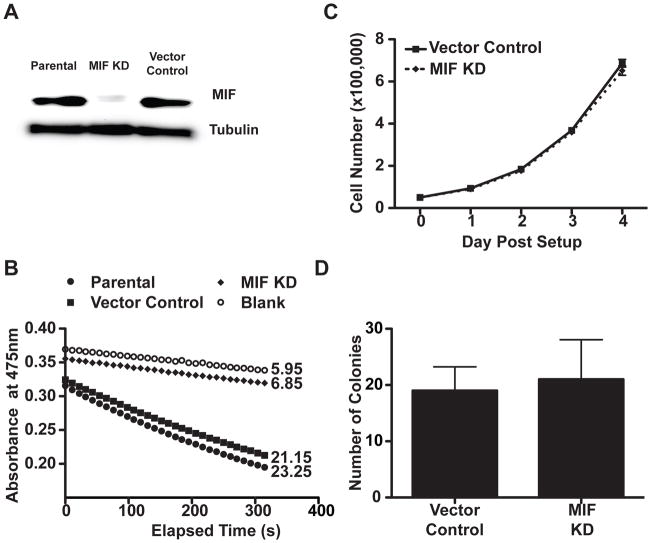
Figure 1. MIF KD cells exhibit similar grow properties in vitro.
A, Immunoblot demonstrates effective MIF depletion in the MIF KD cells compared with the vector control and the parental 4T1 cells. Lysates from cell lines were normalized by Bradford assay for total protein concentration, run on a gel, and the membrane was probed using anti-MIF and anti- tubulin antibodies. B, Effective MIF depletion is confirmed by loss of tautomerase activity. Cell lysates normalized for total protein were assayed for tautomerase activity using a colorimetric substrate. The reaction was monitored for decolorization over 5 minutes at 475nm. Velocities (rate of decolorization/unit time) are indicated to the right of each kinetic plot. C, Depletion of MIF does not alter in vitro proliferation. 5×104 cells per well were plated in 6-well dishes. At the indicated time points, cells were trypsinized and counted. D, Depletion of MIF does not alter anchorage independent growth of 4T1s. 4×104 cells were seeded in soft-agar in a 6 well plates. The plates were incubated at 37°C for 2 weeks, then the colonies counted in three randomly selected fields. For panels C and D, n=3 and each is representative of 3 independent experiments.
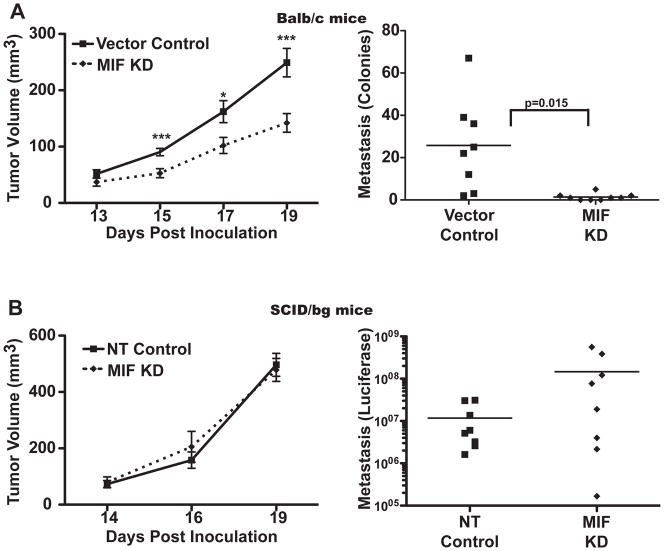
Figure 2. MIF-depleted 4T1 cells form slower growing tumors that are compromised in metastasis to the lung in Balb/c but not SCID/bg mice.
A, MIF depletion leads to compromised tumor growth and metastasis in Balb/c mice. Left, 1×104 4T1 vector control or KD cells were injected into the mammary fat pad of 6–12 week old Balb/c mice. Tumors were measured starting at day 13 and tumor volume was estimated with the formula V=0.4*L*W2. Right, On day 21, lungs were excised, and metastasis detected using the clonogenic assay. n>8 for each group and time point and is representative of three independent experiments. B, MIF depletion does not impact tumor growth and metastasis in immunodeficient mice. Left, 1×104 4T1 luciferase-expressing non-targeting shRNA (NT) or MIF depleted (KD) cells were implanted in the mammary fat pad of 6–8 week old SCID/bg mice, and tumor growth monitored as in A. n=4 for each group and time point and is representative of two independent experiments. Right, On day 21, lungs were excised, immersed in luciferin, and tumor metastasis detected by imaging with the IVIS system. n=8 for each group and includes two independent experiments. * = p<0.05, ***=p<0.005.
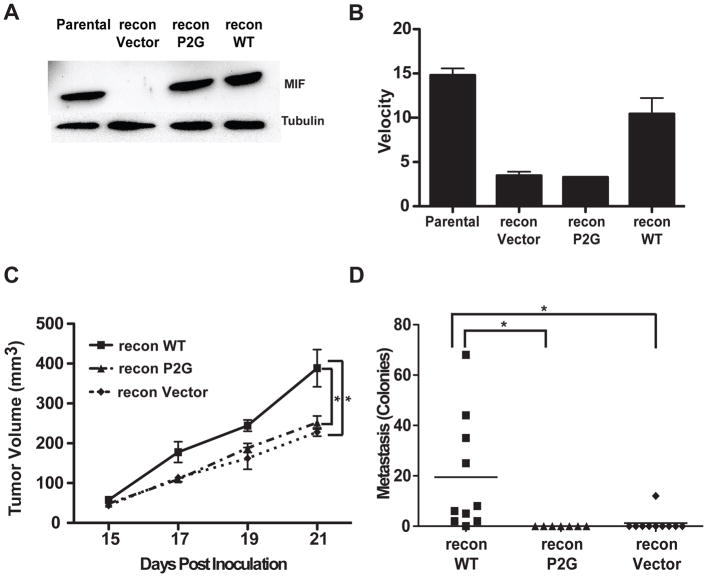
Figure 3. Reconstitution with WT MIF, but not the tautomerase deficient P2G mutant, restores tumor growth and metastasis.
A, Reconstitution of 4T1 KD cells. Immunoblot demonstrates effective reconstitution with mutant P2G MIF, or wild type (WT) MIF. A vector control line was prepared in parallel. B, Tautomerase assay compares the velocities (decolorization/unit time) of the reconstituted cell lines compared with parental 4T1 cells, confirming restoration of tautomerase in the WT reconstitution. C, WT, but not P2G, MIF restores tumor growth. Vector-, P2G-, or WT-reconstituted cells were injected in to the mammary fat pad of 6–12 week old Balb/c mice and tumor monitored as in Figure 2. n>8 for each group and is representative of three independent experiments. D, WT, but not P2G MIF restores pulmonary metastasis. Tumors were implanted as previously described, and surgically removed at day 18. The mice were euthanized eight days later and lungs were excised and assayed for metastasis as in Figure 2A. n>6 for each group and includes results from two independent experiments. * p<0.05 by ANOVA.
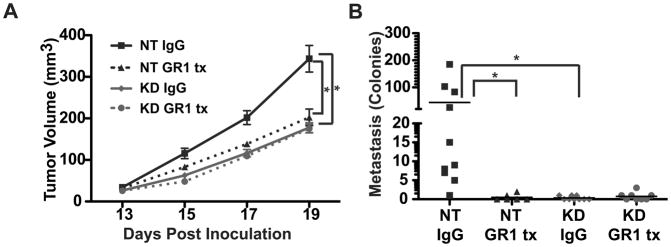
Figure 7. MIF Promotion of Tumor Growth and Metastasis Requires MDSCs.
4T1 non-targeting control (NT) or MIF depleted (KD) cells were implanted as in Figure 2. Mice were injected intraperitoneally with either rat IgG as a control or GR1 antibody on days 11, 14, and 17. A, MDSC depletion in WT tumor-bearing mice decreases tumor growth, while MDSC depletion in KD tumor-bearing mice does not. n>6 for each group and includes 2 independent experiments. B, MDSC depletion in WT tumor-bearing mice decreases metastasis. Lungs were excised at day 20, and metastasis was quantified using the clonogenic assay. n>6 for each group and includes 2 independent experiments. *= p<0.05 by ANOVA.
Results
Depletion of MIF does not alter in vitro growth properties of 4T1 cells
To address the role of MIF in tumor growth and progression, and to test the hypothesis that MIF functions through impact on the immune tumor microenvironment, we utilized the spontaneously metastatic, syngeneic and orthotopic 4T1 model of breast cancer (34).This model mimics stage IV breast cancer, with an organ preference hierarchy for spontaneous metastasis similar to human breast cancer (35). We generated a MIF short hairpin inhibitory RNA (shRNA) knockdown (KD) cell line and a vector control line from the parental 4T1 cell line. Effective MIF depletion was demonstrated by immunoblotting (Figure 1A) and assay of tautomerase activity, measured as the rate of decolorization of a colorimetric substrate per unit time (Figure 1B). Although MIF depletion has been reported to decrease the proliferative capacity of some cell lines (31), and increased expression of MIF to increase colony formation (30), we observed no difference in proliferation between the MIF containing (vector) and MIF depleted (KD) 4T1 cell lines (Figure 1C). We repeated this experiment using a third control cell line expressing a non-targeting shRNA, and again, detected no differences in proliferation (not shown). Because the serum used in the 4T1 cultures likely contains some bovine MIF, we repeated the proliferation assay in low serum conditions, and still the MIF containing and MIF depleted lines exhibited similar proliferation rates (not shown). We compared the ability of the MIF containing and MIF depleted cell lines to grow in soft agar as a measure of anchorage independent growth, and observed no difference in either the number or size of the colonies (Figure 1D). Together, these results demonstrate that MIF depletion does not impact in vitro growth properties of 4T1 cells.
MIF promotes tumor growth and metastasis only in immunocompetent animals
To determine if MIF promotes tumor growth and/or metastasis, we implanted the MIF containing or MIF KD 4T1 cells into the mammary fat pad of Balb/c mice. Both sets of mice developed tumors that were detectable and of similar size at approximately day 13. However, from 15 days post-tumor implantation until harvest, the tumors from MIF depleted KD 4T1 cells grew more slowly than those expressing MIF (Figure 2A, left). Moreover, we detected fewer metastases in the lungs of mice bearing 4T1 MIF KD tumors than MIF containing tumors (Figure 2A, right). This decrease in metastatic burden was not solely due to the difference in size of the primary tumors, as evidenced by comparing the metastasis as a function of tumor mass at the time of harvest. In this analysis, comparing tumors of similar mass, the MIF KD tumor-bearing mice still harbored far fewer metastases (not shown).
To ensure that our observations were not unique to the 4T1 cell line, we generated a MIF depleted CT26 murine colon carcinoma cell line, along with a control line expression a non-targeting shRNA. Effective MIF depletion was confirmed by immunoblot (Supplemental Figure 1A). Similar to the results observed with 4T1 cells, depletion of MIF did not alter the in vitro proliferation of CT26 cells (Supplemental Figure 1B). However, MIF depletion significantly impaired the growth of subcutaneous CT26 tumors in syngeneic Balb/c mice, despite the tumor appearing at similar sizes at early time points (Supplemental Figure 1C). This suggests that, similar to the 4T1 breast tumor model, MIF promotes CT26 tumor growth without influencing the in vitro growth properties of the cells.
Since MIF suppression did not alter cell growth in vitro but caused reduced tumor growth and metastasis in an intact animal, we hypothesized that the difference in tumor growth and metastasis observed in the Balb/c mice was due to an influence of MIF on the immune system. To test this, we implanted MIF-containing and MIF-depleted 4T1 cells expressing luciferase in the mammary fat pad of SCID/bg mice that lack T cells, B cells and NK cells, but have an intact myeloid compartment. In contrast to the results seen in the immunologically intact Balb/c mice, the resulting tumors grew at similar rates in the SCID/bg mice over the duration of the experiment (Figure 2B, left). Moreover, ex vivo imaging of the metastatic burden in the lungs at the time of harvest demonstrated that the MIF containing and MIF depleted tumors were similarly metastatic in immunodeficient SCID/bg animals (Figure 2B, right). These results suggest that the contribution of MIF to tumor growth and metastasis is due to influence over some aspect of the immune system.
Promotion of tumor growth and metastasis is dependent on tautomerase activity of MIF
To confirm that the effects observed in the MIF depleted cells were due to loss of MIF, we reconstituted the KD cells with MIF using an expression plasmid encoding an shRNA resistant version of the MIF gene. In parallel, to test the importance of the tautomerase activity of MIF in promoting tumor growth and metastasis, we reconstituted the MIF KD cell line with a similar plasmid encoding a tautomerase deficient MIF protein containing a proline to glycine substitution at residue 2 (P2G). Expression of the MIF protein in the two reconstituted cell lines was equivalent and similar to the parental cells (Figure 3A). Tautomerase activity was restored in the WT reconstituted line but remained absent in the P2G line since the P2G mutant abolishes the tautomerase activity (Figure 3B). Incipient tumors resulting from either of these cell lines were detected in the mammary glands of injected mice at the same time, but tumors derived from cells reconstituted with WT MIF grew at a faster rate (Figure 3C) when compared to those from the MIF depleted line mock-reconstituted with an empty vector. Importantly, reconstitution with the tautomerase-defective P2G MIF did not restore the rapid tumor growth seen in WT MIF-expressing tumors. This suggests that the tautomerase activity of MIF is required for the tumor growth promoting activity of MIF. Furthermore, the tautomerase activity was also important for promotion of metastasis by MIF, since reconstitution with wild type MIF restored the ability of tumors to produce pulmonary metastases while the P2G mutant MIF did not (Figure 3D).
MIF expression influences the prevalence of monocytic MDSCs in the tumor
The dependence of MIF tumor promoting activity on an intact immune system suggests that MIF does not directly impact an intrinsic property of the tumor cells but rather regulates an aspect of the tumor growth-promoting immune microenvironment. To identify MIF dependent alterations in the immune cell infiltrates within the tumors, we examined the phenotype of cells prepared from the MIF containing and MIF depleted tumors using a panel of markers to detect myeloid derived suppressor cells (MDSCs). Specifically, we examined the two major MDSC populations, defined as Ly6Cint/lowLy6Ghigh granulocytic MDSCs and Ly6ChighLy6Glow monocytic MDSCs (6). The CD11b+ myeloid cells in the WT MIF-reconstituted tumors contained a significantly larger proportion of monocytic MDSCs when compared to the P2G-and vector-reconstituted tumors (Figure 4A). No differences were observed in this subpopulation in either the blood or spleen of these animals (Supplemental Figure 2B). Since MIF is named for its effects on macrophage migration, we also examined F4/80 positive macrophage populations in the 4T1 tumors. Over the same three experiments in which we detected significant differences in the monocytic MDSC population, there was no difference in the GR1 -/lo, F4/80+ macrophage population (Supplemental Figure 2C).Therefore, MIF expression in the tumor cells acts to specifically increase the abundance of the monocytic population of MDSCs within the tumor. A reduction in monocytic MDSC abundance was also observed in the MIF deficient CT26 tumors (Supplemental Figure 1D). These results suggest that MIF may promote tumor growth and metastasis by enhancing the immune suppressive properties of the tumor microenvironment through influence over MDSC populations.
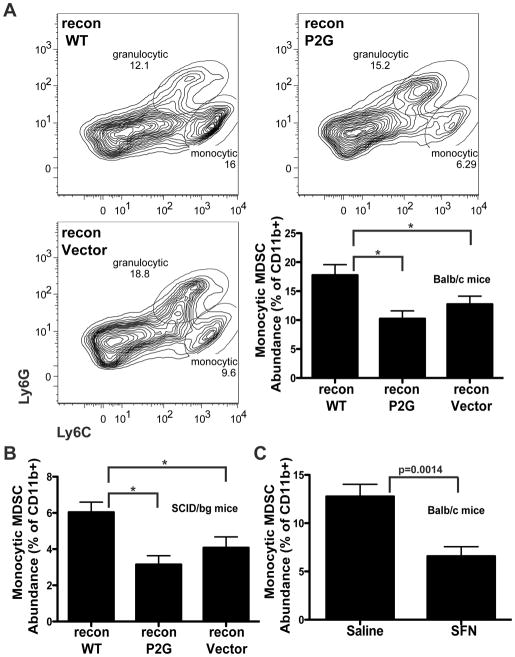
Figure 4. Tumors expressing WT MIF contain a greater proportion of monocytic MDSCs.
A, MIF expression in the tumor correlates with increased monocytic MDSCs. Vector-, P2G-, or WT- reconstituted tumors were implanted as previously described. On day 18, cell suspensions prepared from isolated tumors were stained for analysis by flow cytometry using a viability marker and the cell surface markers CD45, CD11b, Ly6G, and Ly6C. Top, representative plots of Ly6C/Ly6G staining from WT-, P2G-, and vector-reconstituted tumors are shown with prior gating on live, CD45+, CD11b+ cells (complete gating strategy shown in Supplemental Figure 2A). Gated populations indicate Ly6Chigh, Lys6Glow monocytic MDSCs and Ly6Clow/int, Ly6Ghigh granulocytic MDSCs. Bottom right, Quantification of monocytic MDSCs from tumors isolated from Balb/c mice, n=15 for each group and includes 3 independent experiments. B, SCID/bg mice were implanted with reconstituted tumors. On day 22, tumors were processed as described in A. n>8 for each group and includes 3 independent experiments. C, Parental 4T1 tumors were implanted into mice that received either saline or SFN (200μg/day) starting the day of tumor cell implantation. On day 21, tumors were processed as described in A. n>8 for each group and includes 2 independent experiments. * p<0.05 by ANOVA.
The differences in MDSC abundance was not simply the result of differences in tumor size since MDSC populations characterized in WT MIF expressing tumors in SCID/bg mice also displayed a greater fraction of monocytic MDSCs than those from MIF depleted or P2G reconstituted tumors (Figure 4B), despite all of the tumors being of similar size. Significantly, while the MIF dependent differences in monocytic MDSC populations were observed in the immunocompromised mice, tumors that express MIF do not display accelerated tumor growth and metastasis in these mice (Figure 2B). Therefore, while MIF is still able to influence the abundance of monocytic MDSCs in the SCID/beige hosts, in the absence of an anti-tumor immune response mediated by T cells and NK cells, there is no impact of MIF on tumor growth and metastasis.
Parallel to this genetic demonstration of the importance of the tautomerase activity of MIF in regulation of MDSC populations, we used sulforaphane, a natural product inhibitor of MIF (26) to pharmacologically suppress the MIF tautomerase. Treatment of 4T1 tumor bearing mice with SFN reduced the prevalence of monocytic MDSCs in tumors, similar to the effects of MIF depletion from tumors (Figure 4C). Together with the failure of the tautomerase deficient P2G mutant to rescue MDSC recruitment, this suggests that MIF controls the prevalence of monocytic MDSCs through a mechanism that requires its tautomerase activity.
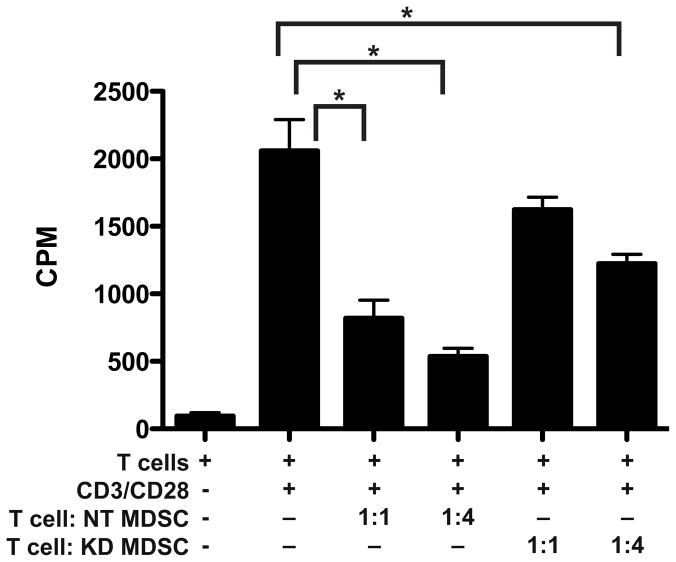
Finally, to confirm that the MIF status of the tumor and the resultant shift in MDSC populations resulted in functional changes within the tumor microenvironment, we isolated GR1 positive cells from MIF-containing and MIF-depleted tumors and evaluated their ability to suppress CD8+ T cell proliferation in vitro. The purified GR1 positive cells isolated from MIF expressing tumors suppressed T cell proliferation significantly more than those from the MIF depleted tumors (Figure 5), when comparing an equivalent number of cells. In fact, at a 1:1 ratio of MDSCs to T cells, the MDSCs isolated from the MIF depleted tumor failed to significantly suppress proliferation, while the cells from the MIF containing tumor were effective. These results demonstrate that the MIF expressing tumors have a more highly suppressive MDSC infiltrate, and therefore likely benefit from a more immunosuppressive microenvironment.
Figure 5. MDSCs isolated from MIF containing 4T1 tumors more effectively suppress CD8+ T cell proliferation than MDSCs from MIF depleted tumors.
4T1 NT or KD tumors were implanted into mice as in Figure 2. At harvest, tumors were digested and GR1+ MDSCs were purified. Purified CD8+ T cells were stimulated with CD3/CD28 in the presence or absence of MDSCs at ratios of 1:1 and 1:4 (T cell:MDSC). The graph shows combined results from two independent experiments * p<0.05 by ANOVA.
MIF promotes the differentiation of CD11b+ cells into monocytic MDSCs in vitro
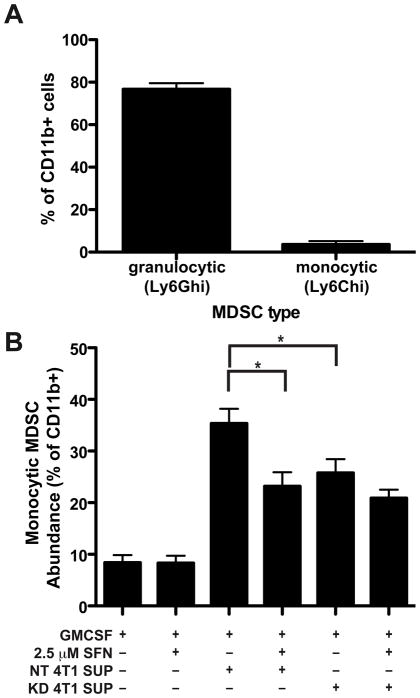
The MIF dependent increase in prevalence of monocytic MDSCs in the 4T1 tumors could result from either increased recruitment of these cells into the tumor or differentiation of the MDSCs towards the monocytic phenotype, perhaps within the tumor. Others have reported that factors such as GM-CSF can control differentiation between the two subsets of MDSCs (36). Using a similar approach (diagrammed in Supplemental Figure 3), we evaluated the effect of MIF on MDSC differentiation in vitro by flow cytometry. After purification of CD11b+ MDSCs from the spleen of a tumor bearing mouse, the majority of cells were of the less suppressive granulocytic phenotype, with the monocytic subset comprising less than 10% of the total population (Figure 6A). Tumor-conditioned media from MIF-containing 4T1 cells induced differentiation of CD11b+ myeloid cells into monocytic MDSCs (Figure 6B). Conditioned medium from MIF depleted (KD) 4T1 cells resulted in a significantly reduced proportion of the CD11b+ cells differentiating into monocytic MDSCs. Cell viability in the cultures at the end of the incubation ranged between 25 and 50% (Supplemental Figure 3B). Notably there was no difference in cell viability between the cultures treated with the conditioned media from the MIF containing vs. the MIF depleted 4T1 cells, suggesting that the differences seen in monocytic MDSCSs cannot be solely attributed to differences in survival. These observations suggest that either MIF itself or some MIF-induced secreted factor is responsible for the accumulation of monocytic MDSCs in this assay..
Figure 6. Conditioned media from MIF expressing 4T1 cells induces differentiation of monocytic MDSCs.
CD11b+ cells were isolated from the spleen of WT tumor-bearing mice and incubated with conditioned media from MIF non-targeting (NT) control cells or MIF KD 4T1 cells with or without SFN as indicated for 5 days (schematic of experiment in Supplemental Figure 3A). A, Phenotype of purified CD11b cells at start of experiment, stained as in Figure 4A n=3. B, Phenotype of cells after 5 days of culture. Cells were stained for analysis by flow cytometry as in Figure 4A. n>12 for each group and includes results of 4 independent experiments. * p<0.05 by ANOVA.
Addition of the MIF inhibitor SFN to the supernatant from the MIF expressing 4T1 cells significantly decreased the induction of monocytic MDSCs (Figure 6B). In contrast, SFN did not alter the induction of monocytic MDSCs in the cultures containing MIF KD supernatants. These results suggest that the differing prevalence of MDSCs in the MIF containing versus MIF depleted 4T1 tumors results from MIF induced differentiation of myeloid cells in the microenvironment into the monocytic MDSC population. Inhibition of this process by SFN suggests that it occurs through a mechanism dependent on MIF tautomerase activity.
MIF promotion of tumor growth and metastasis requires MDSCs
To verify that the contribution of MIF to tumor growth and metastasis was due to the reduction of MDSC abundance in the tumor, we depleted MDSC populations from mice and determined the effect on tumor growth and metastasis. In a pilot experiment, we demonstrated that intraperitoneal delivery of three doses of GR1 antibody on day 11, 14 and 17 post tumor implantation resulted in a near complete depletion of MDSCs from the tumor, when assayed on day 17 (Supplemental Figure 4B). Subsquently, mice bearing either MIF-containing or MIF-deficient tumors were treated with anti-GR1 antibody to deplete MDSCs using the same dosing regimen (Supplemental Figure 4A) with depletion initiated at day 11 post implantation, before the presence of a measurable tumor. The experiment was harvested at day 20. Depletion of MDSCs in mice bearing MIF WT tumors resulted in impaired tumor growth (Figure 7A) and inhibition of metastasis to lung (Figure 7B). In contrast, depletion of MDSC in mice bearing MIF KD tumors had no further impact on tumor growth (Figure 7A) when compared to MIF KD tumors in mice administered an isotype control antibody. Since MIF KD tumors fail to metastasize to lung (Figure 7B) it was not possible to determine if MDSC depletion had any further impact on pulmonary metastasis in the absence of MIF. From this, we conclude that the contribution of MIF to 4T1 tumor growth and metastasis can be attributed to the effects of MIF in controlling MDSCs in the tumor microenvironment.
Discussion
Our results demonstrate a link between the proinflammatory cytokine MIF and the prevalence of monocytic MDSCs in the tumor microenvironment, leading to promotion of tumor growth and metastasis (see the model diagrammed in Figure 8). This differs from previous reports that attribute the tumor growth promoting effects of MIF to activities within the tumor cells (30–32, 37). Our work reveals a new mechanism for MIF as an immune modulator in the tumor microenvironment.
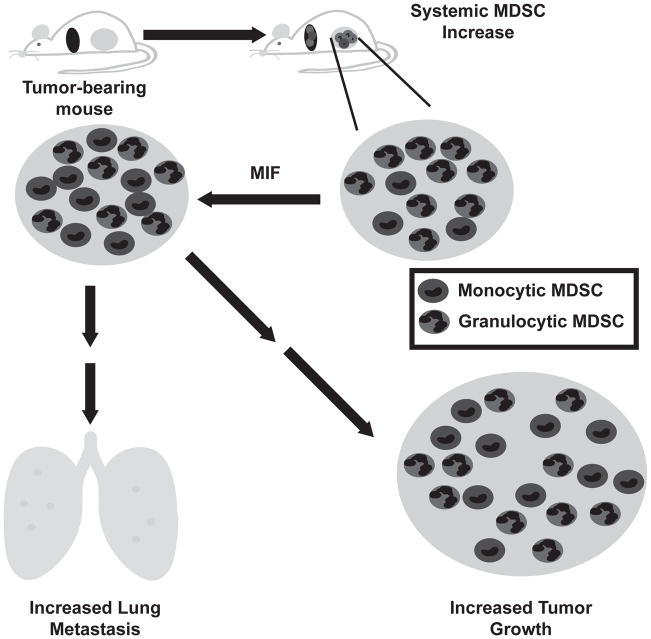
Figure 8. Proposed model of the role of MIF in cancer progression.
Tumor bearing animals demonstrate a systemic increase in myeloid derived suppressor cells, detectable both in the blood and the spleen. The majority of these cells are of the granulocytic phenotype. Within the tumor, high levels of MIF promote the abundance of the more suppressive monocytic phenotype, correlating with an increase in tumor growth and metastasis in the presence of an intact immune system.
Significantly, we demonstrate that the tautomerase activity of MIF is required for promotion of tumor growth and metastasis. Using mutation of the catalytic proline and/or inhibition of MIF tautomerase with the enzyme inhibitor sulforaphane, we further demonstrate that this enzymatic activity is important for inducing monocytic MDSCs both in tumors and in our in vitro differentiation assay. The importance of the MIF tautomerase activity has been debated (28, 38, 39). However, other inhibitors of the MIF tautomerase also influence downstream biological outcomes (28, 29, 40). Our current findings lend support to the conclusion that, while a physiological target has yet to be identified, the tautomerase activity is important for functions of MIF that contribute to its role in tumor growth and metastasis.
In this study, we observed MIF dependent differences in the prevalence of monocytic MDSCs in the tumor microenvironment. As previously described, the monocytic MDSC subset is thought to have a higher capacity to suppress host anti-tumor response (9), ultimately supporting increased tumor growth and metastasis. Therefore, our demonstration that MIF contributes to the prevalence of these cells within the tumor suggests a possible mechanism through which MIF promotes tumor progression by influence over the tumor microenvironment.
Given that human cancers demonstrate increased MIF expression (17–21), and that MDSCs have been observed in the blood of human cancer patients (3–5, 41, 42), MIF is potentially a valuable therapeutic target for efforts aimed at inhibiting the tumor-supportive host immune response. Specifically, inhibition of MIF could block the accumulation of immune suppressive MDSCs in the tumor microenvironment, and restore effective anti-tumor immunity. This strategy, either alone or in combination with immunotherapy approaches designed to boost anti-tumor immune responses, offers an alternative to therapies that directly target the tumor cells. These approaches might be particularly useful in cancers that do not have recognized molecular therapeutic targets but that exhibit a strong inflammatory response or for those that are resistant to currently available targeted therapies. Finally, since tumors can recur and/or reappear as metastatic disease even years after primary tumor resection, our observation that MIF is important for spontaneous pulmonary metastasis suggests that MIF inhibition could represent a key approach not only for preventing primary tumor outgrowth, but also for prevention of metastatic disease.
Our work not only furthers our understanding of the role of MIF in tumor progression, but also suggests a previously unappreciated mechanism of action for cancer chemopreventives. We characterized the isothiocyanate chemopreventives, including sulforaphane, as effective, covalent, natural product inhibitors of MIF (26). Epidemiological evidence and experimental models have long supported the conclusion that isothiocyanates exhibit anti-cancer activities (43–46). However, much of the function of isothiocyanates was thought to be through the detoxification of carcinogens (43–46), preventing the very earliest steps in tumor formation. Alternative models have focused on the ability of isothiocyanates to decrease in proliferation of the cells (47, 48) or induce apoptosis (49, 50). Our study provides a novel molecular mechanism to explain isothiocyanate chemoprevention through inhibition of MIF and the resulting influence over the tumor microenvironment through specific impact on the immunosuppressive immune infiltrate.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance of the Joanne Lannigan and the Flow Cytometry Core Facility as well as Marya Dunlap-Brown and the Molecular Assessment and Preclinical Studies (MAPS) Core. The authors thank Angela Rose for technical assistance and Claudia Rival for advice on flow staining. Finally, the authors thank Sarah Conine, Denise Joplin, Carrie Leonard, Amy Bouton, Michael Gutknecht, and Scott Vande Pol for many helpful discussions and Thomas Braciale, Timothy Bender, Amy Bouton, Dan Conrad, Sheinei Saleem and Scott Vande Pol for critical reading of the manuscript.
Financial Support: This work was supported by NIH grants R01AT004323 (JVC) and R01CA113899 (DJT) as well assupport from the UVa Cancer Center through the Patients and Friends Research Fund and the NCI Cancer Center Support Grant, P30 CA44579 (JVC).
References
- 1.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 2.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–290. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 3.Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, Gabrilovich DI. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 4.Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60:1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, McDermott D, Quiceno D, Youmans A, O’Neill A, Mier J, Ochoa AC. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 6.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietlin TA, Hofman FM, Lund BT, Gilmore W, Stohlman SA, van der Veen RC. Mycobacteria-induced Gr-1+ subsets from distinct myeloid lineages have opposite effects on T cell expansion. J Leukoc Biol. 2007;81:1205–1212. doi: 10.1189/jlb.1006640. [DOI] [PubMed] [Google Scholar]
- 9.Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, De Baetselier P, Van Ginderachter JA. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 10.Nagaraj S, Schrum AG, Cho HI, Celis E, Gabrilovich DI. Mechanism of T cell tolerance induced by myeloid-derived suppressor cells. J Immunol. 2010;184:3106–3116. doi: 10.4049/jimmunol.0902661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, Pisarev V, Sherman S, Sporn MB, Gabrilovich D. Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J Clin Invest. 2011;121:4015–4029. doi: 10.1172/JCI45862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 13.David JR. Delayed hypersensitivity in vitro: its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci U S A. 1966;56:72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, Mizoguchi E, Metz CN, Alsahli M, ten Hove T, Keates AC, Lubetsky JB, Farrell RJ, Michetti P, van Deventer SJ, Lolis E, David JR, Bhan AK, Terhorst C. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–1066. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- 15.Ichiyama H, Onodera S, Nishihira J, Ishibashi T, Nakayama T, Minami A, Yasuda K, Tohyama H. Inhibition of joint inflammation and destruction induced by anti-type II collagen antibody/lipopolysaccharide (LPS)-induced arthritis in mice due to deletion of macrophage migration inhibitory factor (MIF) Cytokine. 2004;26:187–194. doi: 10.1016/j.cyto.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorino M, Sanchez JC, Frutiger S, Hughes G, Pallini V, Hochstrasser DF, Tosi P. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- 17.Meyer-Siegler K, Fattor RA, Hudson PB. Expression of macrophage migration inhibitory factor in the human prostate. Diagn Mol Pathol. 1998;7:44–50. doi: 10.1097/00019606-199802000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu T, Abe R, Nakamura H, Ohkawara A, Suzuki M, Nishihira J. High expression of macrophage migration inhibitory factor in human melanoma cells and its role in tumor cell growth and angiogenesis. Biochem Biophys Res Commun. 1999;264:751–758. doi: 10.1006/bbrc.1999.1584. [DOI] [PubMed] [Google Scholar]
- 19.Bando H, Matsumoto G, Bando M, Muta M, Ogawa T, Funata N, Nishihira J, Koike M, Toi M. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn J Cancer Res. 2002;93:389–396. doi: 10.1111/j.1349-7006.2002.tb01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Y, Tsui HT, Poon RT, Ng IO, Li Z, Chen Y, Jiang G, Lau C, Yu WC, Bacher M, Fan ST. Macrophage migration inhibitory factor: roles in regulating tumor cell migration and expression of angiogenic factors in hepatocellular carcinoma. Int J Cancer. 2003;107:22–29. doi: 10.1002/ijc.11287. [DOI] [PubMed] [Google Scholar]
- 21.He XX, Yang J, Ding YW, Liu W, Shen QY, Xia HH. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 2006;55:797–802. doi: 10.1136/gut.2005.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer-Siegler K, Hudson PB. Enhanced expression of macrophage migration inhibitory factor in prostatic adenocarcinoma metastases. Urology. 1996;48:448–452. doi: 10.1016/S0090-4295(96)00207-5. [DOI] [PubMed] [Google Scholar]
- 23.Hira E, Ono T, Dhar DK, El-Assal ON, Hishikawa Y, Yamanoi A, Nagasue N. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 2005;103:588–598. doi: 10.1002/cncr.20818. [DOI] [PubMed] [Google Scholar]
- 24.Xia HH, Yang Y, Chu KM, Gu Q, Zhang YY, He H, Wong WM, Leung SY, Yuen ST, Yuen MF, Chan AO, Wong BC. Serum macrophage migration-inhibitory factor as a diagnostic and prognostic biomarker for gastric cancer. Cancer. 2009;115:5441–5449. doi: 10.1002/cncr.24609. [DOI] [PubMed] [Google Scholar]
- 25.Kamimura A, Kamachi M, Nishihira J, Ogura S, Isobe H, Dosaka-Akita H, Ogata A, Shindoh M, Ohbuchi T, Kawakami Y. Intracellular distribution of macrophage migration inhibitory factor predicts the prognosis of patients with adenocarcinoma of the lung. Cancer. 2000;89:334–341. [PubMed] [Google Scholar]
- 26.Cross JV, Rady JM, Foss FW, Lyons CE, Macdonald TL, Templeton DJ. Nutrient isothiocyanates covalently modify and inhibit the inflammatory cytokine macrophage migration inhibitory factor (MIF) Biochem J. 2009;423:315–321. doi: 10.1042/BJ20091170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swope M, Sun HW, Blake PR, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lubetsky JB, Dios A, Han J, Aljabari B, Ruzsicska B, Mitchell R, Lolis E, Al-Abed Y. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277:24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 29.Ouertatani-Sakouhi H, El-Turk F, Fauvet B, Roger T, Le Roy D, Karpinar DP, Leng L, Bucala R, Zweckstetter M, Calandra T, Lashuel HA. A new class of isothiocyanate-based irreversible inhibitors of macrophage migration inhibitory factor. Biochemistry. 2009;48:9858–9870. doi: 10.1021/bi900957e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrenko O, Fingerle-Rowson G, Peng T, Mitchell RA, Metz CN. Macrophage migration inhibitory factor deficiency is associated with altered cell growth and reduced susceptibility to Ras-mediated transformation. J Biol Chem. 2003;278:11078–11085. doi: 10.1074/jbc.M211985200. [DOI] [PubMed] [Google Scholar]
- 32.Fingerle-Rowson G, Petrenko O, Metz CN, Forsthuber TG, Mitchell R, Huss R, Moll U, Muller W, Bucala R. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100:9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosengren E, Bucala R, Aman P, Jacobsson L, Odh G, Metz CN, Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- 34.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 35.Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol. 2001;Chapter 20(Unit 20):22. doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 36.Morales JK, Kmieciak M, Knutson KL, Bear HD, Manjili MH. GM-CSF is one of the main breast tumor-derived soluble factors involved in the differentiation of CD11b-Gr1- bone marrow progenitor cells into myeloid-derived suppressor cells. Breast Cancer Res Treat. 2010;123:39–49. doi: 10.1007/s10549-009-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi N, Nishihira J, Sato Y, Kondo M, Ogawa H, Ohshima T, Une Y, Todo S. Involvement of macrophage migration inhibitory factor (MIF) in the mechanism of tumor cell growth. Mol Med. 1998;4:707–714. [PMC free article] [PubMed] [Google Scholar]
- 38.Bendrat K, Al-Abed Y, Callaway DJ, Peng T, Calandra T, Metz CN, Bucala R. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry. 1997;36:15356–15362. doi: 10.1021/bi971153a. [DOI] [PubMed] [Google Scholar]
- 39.Hermanowski-Vosatka A, Mundt SS, Ayala JM, Goyal S, Hanlon WA, Czerwinski RM, Wright SD, Whitman CP. Enzymatically inactive macrophage migration inhibitory factor inhibits monocyte chemotaxis and random migration. Biochemistry. 1999;38:12841–12849. doi: 10.1021/bi991352p. [DOI] [PubMed] [Google Scholar]
- 40.Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc Natl Acad Sci U S A. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wattenberg LW. Chemoprevention of cancer. Cancer Res. 1985;45:1–8. [PubMed] [Google Scholar]
- 44.Prochaska HJ, Talalay P. Regulatory mechanisms of monofunctional and bifunctional anticarcinogenic enzyme inducers in murine liver. Cancer Res. 1988;48:4776–4782. [PubMed] [Google Scholar]
- 45.Morse MA, Reinhardt JC, Amin SG, Hecht SS, Stoner GD, Chung FL. Effect of dietary aromatic isothiocyanates fed subsequent to the administration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone on lung tumorigenicity in mice. Cancer Lett. 1990;49:225–230. doi: 10.1016/0304-3835(90)90163-r. [DOI] [PubMed] [Google Scholar]
- 46.Hecht SS, Kenney PM, Wang M, Upadhyaya P. Benzyl isothiocyanate: an effective inhibitor of polycyclic aromatic hydrocarbon tumorigenesis in A/J mouse lung. Cancer Lett. 2002;187:87–94. doi: 10.1016/s0304-3835(02)00410-x. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava SK, Xiao D, Lew KL, Hershberger P, Kokkinakis DM, Johnson CS, Trump DL, Singh SV. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits growth of PC-3 human prostate cancer xenografts in vivo. Carcinogenesis. 2003;24:1665–1670. doi: 10.1093/carcin/bgg123. [DOI] [PubMed] [Google Scholar]
- 48.Chiao JW, Wu H, Ramaswamy G, Conaway CC, Chung FL, Wang L, Liu D. Ingestion of an isothiocyanate metabolite from cruciferous vegetables inhibits growth of human prostate cancer cell xenografts by apoptosis and cell cycle arrest. Carcinogenesis. 2004;25:1403–1408. doi: 10.1093/carcin/bgh136. [DOI] [PubMed] [Google Scholar]
- 49.Chiao JW, Chung FL, Kancherla R, Ahmed T, Mittelman A, Conaway CC. Sulforaphane and its metabolite mediate growth arrest and apoptosis in human prostate cancer cells. Int J Oncol. 2002;20:631–636. doi: 10.3892/ijo.20.3.631. [DOI] [PubMed] [Google Scholar]
- 50.Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004;25:83–90. doi: 10.1093/carcin/bgg178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.