Abstract
The complete genome sequences of over 220 mycobacteriophages reveal them to be highly diverse, with numerous types sharing little or no nucleotide sequence identity with each other. We have determined the preferences of these phages for M. tuberculosis and for other strains of M. smegmatis, and find there is a correlation between genome type (cluster, subcluster, singleton) and host range. For many of the phages, expansion of host range occurs at relatively high frequencies, and we describe several examples in which host constraints occur at early stages of infection (adsorption or DNA injection), and phages have the ability to expand their host range through mutations in tail genes. We present a model in which phage diversity is a function of both the ability of phages to rapidly adapt to new hosts and the richness of the diversity of the bacterial population from which those phages are isolated.
Keywords: Bacteriophage, Mycobacteriophage, Host Range, Genome
Introduction
The bacteriophage population is vast (Hendrix, 2002, 2003; Suttle, 2007), dynamic (Wilhelm et al., 2002), and likely extremely old (Abrescia et al., 2012). The genome sequences of approximately 1,000 phages, isolated from a broad range of bacterial hosts, have been deposited in public databanks, and it is clear that we have only begun to appreciate the enormous sequence diversity of the phage population as a whole. For some bacterial genera, a substantial number of phage genome sequences have been determined, including Escherichia, Lactococcus, Mycobacterium, Pseudomonas, Staphylococcus, and Streptococcus, and these also typically span a broad range of diversity (Adriaenssens et al., 2012; Brussow, 2001; Hatfull, 2012b; Kwan et al., 2005, 2006; Smith et al., 2012). However, for many such collections, the phages were not isolated on the same species or strain, the host ranges are not well-defined, and there are many examples where phages have preferences for different strains of the same bacterial species (Lobocka et al., 2012).
The constraints on infection are not completely understood, but involve many different mechanisms, including restriction-modification (Hyman and Abedon, 2010), receptor availability (Hyman and Abedon, 2010), abortive infection, which can be mediated by a variety of processes (Fineran et al., 2009; Hoskisson and Smith, 2007), Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) (Deveau et al., 2010), and modification of the translational apparatus (Amitsur et al., 1987). In turn, phages have evolved ways to overcome these constraints, such as through spontaneous mutation (Desplats and Krisch, 2003), diversity generating systems (Medhekar and Miller, 2007), site-specific recombination (Plasterk et al., 1984; Sandmeier, 1994), or molecular mimicry (Roberts et al., 2012). The totality of these processes can be viewed in the context of a grand microbial battle, in which the hosts are constantly under attack from the viruses, bacteria evolve effective ways to resist infection, and the viruses co-evolve to either overcome the resistance, or acquire the ability to infect a new host. After perhaps more than three billion years, an equilibrium has been established where both bacterial and phage populations are maintained (Stern and Sorek, 2011).
Mycobacteriophages are viruses that infect mycobacterial hosts and are of interest for two primary reasons. First, the large set of complete mycobacteriophage genome sequences shows them to be highly varied, thus offering insights into phage genome diversity and the evolutionary mechanisms acting to generate such diversity (Hatfull, 2010). Secondly, mycobacteriophages provide a rich set of tools for developing genetic systems for their hosts –including Mycobacterium tuberculosis, the causative agent of human tuberculosis (TB) – as well as fueling novel strategies for diagnostic, preventative, and therapeutic approaches for TB (Hatfull, 2012b). In addition, the discovery of new mycobacteriophages and their genome characterization represents a powerful platform for introducing novice scientists to authentic research experiences (Hanauer et al., 2006; Hatfull et al., 2006).
Currently, there are 221 completely sequenced mycobacteriophage genomes (Hatfull, 2012a; Henry et al., 2010; Pope et al., 2011a; Pope et al., 2011b). All of these [with the sole exception of DS6A (Redmond and Carter, 1960)] were either isolated on M. smegmatis mc2155, or are known to infect this strain. Comparative genomic analysis at both the nucleotide and gene content levels shows that even though these phages enjoy a common host and are thus capable of being in genetic contact, they span a broad range of diversity, albeit in a heterogeneous manner (Pope et al., 2011b). Specifically, there are groups of phages that share substantial levels and extents of DNA similarity, as revealed by either Dotplot analyses or BLAST comparisons. These can be grouped into clusters, where genomes within a cluster have recognizable sequence similarity over a span of more than 50% of their genome lengths (Hatfull et al., 2006). Some phages, however, have no close relatives, and these are designated as singletons. The 221 sequenced genomes fall into 15 clusters, and eight singletons, and there is little or no DNA sequence similarity between these groups (Hatfull, 2012a). The host ranges of only a few of these phages have been reported, suggesting at least a partial correlation between host preference and genetic relationships (Rybniker et al., 2006).
Both the size and the diversity of the different clusters vary enormously. The largest is Cluster A, containing 60 genomes, and the smallest – aside from the eight singletons – are Clusters M, N and O, each with two. Within some clusters, the genomes are extremely similar, and the five Cluster G phages are a good example of this. These all have a similar (although non identical) gene content, and differ mostly by a small number of nucleotide substitutions (for example, Angel and BPs differ by the insertion of a small mobile element MPME1 in BPs, and 138 nucleotide substitutions) (Sampson et al., 2009). Other clusters are more diverse and can be further divided into subclusters, such that genomes within a cluster have similar gene contents and genome organizations, but different subclusters have different degrees of nucleotide similarity. For example, Cluster A can be divided into at least nine subclusters. The total number of non-divided clusters, subclusters and singletons is 44 (Hatfull, 2012a). Mycobacteriophage genomes are also pervasively mosaic such that each genome can be considered as a specific assemblage of individual segments – commonly single genes – joined together in a particular combination (Hatfull, 2010; Pedulla et al., 2003). However, these mosaic relationships are most evident through comparisons of amino acid sequences, and represent distant evolutionary relationships generated by illegitimate recombination over an extended period of evolutionary time (Hatfull, 2010; Hendrix et al., 1999).
We describe here the ability of a large set of mycobacteriophages to infect M. tuberculosis and two strains of M. smegmatis distinct from their known common host (M. smegmatis mc2155). We show that relatively few of these phages infect M. tuberculosis, but that these generally fall within particular clusters/subclusters. In contrast, many of these phages infect other M. smegmatis strains, but there are groups of clusters or subclusters that either don’t infect or only form plaques at low frequencies. There is thus a correlation between cluster/subcluster type and host preference. We also describe two examples, both involving tail fiber mutations, by which mycobacteriophages overcome host barriers, which strongly implicate host recognition or DNA injection as the basis for constrained host range. Finally, we present a model to account for the diversity of mycobacteriophage genome sequences based on host diversity, and their ability to rapidly switch or expand host preferences.
Results
Mycobacteriophage diversity
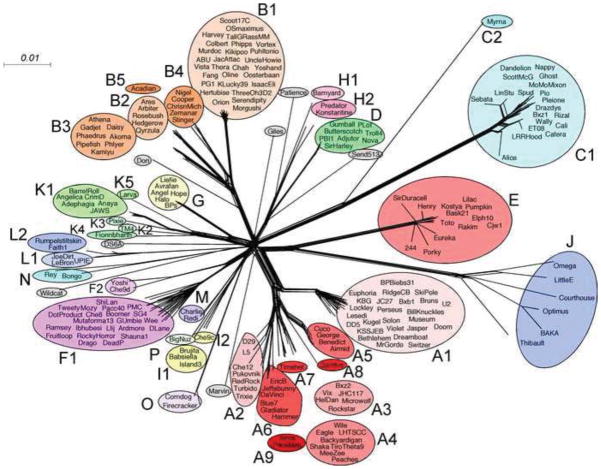
The sequenced mycobacteriophage genomes can be grouped into clusters and subclusters based on sequence identity. However, while this classification is pragmatically useful, it is also inherently untidy, in that the grouping is essentially arbitrary, and the degrees of diversity within and between groups are highly varied. This is illustrated by representation of the phylogenetic relationships based on gene content (i.e. the distribution of homologous genes – phamilies – among different phages) (Fig. 1). For example, some singleton phages – such as Giles and Marvin – appear to be rooted such that they are either unrelated to any other phage, or they are no more closely related to any one group of phages than to the collection as a whole. Both phages actually contain a significant proportion of their genes (21.5% and 23.3%, respectively) that are shared with other mycobacteriophages, but the phages with homologues are broadly distributed across the phylogenetic landscape. Similarly, the Cluster C phages, which have a different virion morphology (i.e. myoviral) than all other mycobacteriophages (siphoviral), share about 10% of the genes with them. Thus these phages presumably have access to the larger pool of mycobacteriophage genes, but with different levels of access and participation in genetic exchange.
Figure 1.
Gene content relationships among mycobacteriophages. The relationships among 220 mycobacteriophages are displayed using the NeighborNet function in Splitstree4 (Huson, 1998). Data were generated from the database Mycobacteriophages_220 in the program Phamerator (Cresawn et al., 2011). Phages within a cluster or subcluster are circled and labeled accordingly. Singletons are shown in grey circles.
We have noted previously that there is little variation in GC% content between phages within clusters, but that the GC% content varies considerably from cluster to cluster (Hatfull, 2012b). There are also GC% content differences, albeit to a lesser degree, between subclusters within a given cluster such as for the subclusters of Cluster A (Table 1; Fig. 2). In this case, several of the subclusters have a narrow range of GC% content that does not overlap with that of other subclusters (e.g. A5, A6, and A1 (Fig. 2). The overall span of GC% content is impressive, ranging from 50.3% in Patience, to 69.1% in Cooper. This contrasts to the 67.4% GC% content of M. smegmatis, fairly typical for the actinomycetes. M. avium has the highest GC% among the mycobacteria (69.0%), and M. leprae has the lowest (57.8%); M. tuberculosis is 65.6% GC. There are several plausible factors that may play a role in the wide range of mycobacteriophage GC% contents. First, amelioration of phage DNA GC% content towards its host may be relatively slow compared to the frequencies with which phages can change or expand their host ranges. Second, M. smegmatis may represent a plausible host, but it may not be the host that was most recently used by the phages within the environment from which they were isolated. As such, the variety of ‘preferred’ hosts, or those most recently infected within their natural history, could be potentially large and varied. Furthermore, although M. smegmatis mc2155 is a common host, the range of host preferences could vary considerably among the phages. We note that the microbial population in soil and compost – from where many of the phages were isolated – is highly diverse (Maron et al., 2011; Schloss and Handelsman, 2008). Furthermore, in this scenario, we would predict that phages isolated from environments of limited bacterial diversity would be much less diverse, and this is what is observed with phages of Propionibacterium acnes isolated from sebaceous follicles of skin, even though about 25% of the genes are homologues to mycobacteriophage genes with weak but detectable amino acid sequence similarity (Marinelli et al., 2012).
Table 1.
Efficiencies of phage plating on mycobacterial strains
| Phage | Cluster | GC% | Jucho1 | MKD81 | Mtb1 |
|---|---|---|---|---|---|
| Bethlehem | A1 | 63.2 | 2.2×10−2 | 1.0×10−3 | <10−7 |
| BillKnuckles | A1 | 63.4 | 3.3×10−3 | <10−8 | <10−8 |
| BpBieps31 | A1 | 63.4 | 9.0×10−2 | <10−8 | <10−8 |
| Bruns | A1 | 63.6 | 1.4×10−1 | <10−4 | <10−4 |
| Bxb1 | A1 | 63.6 | 8.0×10−1 | 1.0×10−4 | 1.0×10−2 |
| DD5 | A1 | 63.4 | 6.6×10−3 | <10−6 | <10−6 |
| Doom | A1 | 63.8 | 4.0×10−2 | <10−8 | <10−8 |
| Dreamboat | A1 | 63.9 | 7.6×10−2 | <10−8 | <10−8 |
| Euphoria | A1 | 63.7 | 2.5 | <10−8 | <10−8 |
| Jasper | A1 | 63.7 | 1.7×10−1 | <10−7 | <10−7 |
| JC27 | A1 | 63.6 | 1.5×10−1 | <10−7 | <10−7 |
| KBG | A1 | 63.6 | 2.0×10−1 | <10−6 | <10−6 |
| KSSJEB | A1 | 63.6 | 3.0×10−1 | <10−9 | <10−9 |
| Kugel | A1 | 63.8 | 1.3×10−1 | <10−10 | 1.0×10−6 |
| Lesedi | A1 | 63.8 | 1.3 | <10−9 | <10−9 |
| Lockley | A1 | 63.4 | 4.3×10−2 | <10−6 | <10−6 |
| McGordo | A1 | 63.8 | 2.2 | <10−8 | <10−8 |
| Museum | A1 | 63.6 | 3.6×10−1 | <10−4 | <10−4 |
| Perseus | A1 | 63.7 | 6.7 | <10−8 | <10−8 |
| RidgeCB | A1 | 64.0 | 3.3 | <10−6 | <10−6 |
| SkiPole | A1 | 63.6 | 1.0×101 | <10−7 | <10−7 |
| Solon | A1 | 63.8 | 1.6×10−1 | <10−4 | <10−4 |
| U2 | A1 | 63.7 | 4.5×10−1 | 4.5×10−6 | 4.5×10−3 |
| Violet | A1 | 63.8 | 1.5×10−1 | <10−7 | <10−7 |
|
| |||||
| Che12 | A2 | 62.9 | 1.7×10−1 | <10−9 | 5.8×10−4 |
| D29 | A2 | 63.5 | 5.9×10−1 | 5.9×10−3 | 5.9×10−1 |
| L5 | A2 | 62.3 | 5.0×10−1 | <10−7 | 7.5 |
| Pukovnik | A2 | 63.3 | 1.0 | 7.0×10−5 | <10−10 |
| RedRock | A2 | 64.5 | 1.2 | <10−9 | <10−9 |
| Trixie | A2 | 64.5 | 3.3 | 3.3×10−1 | <10−6 |
| Turbido | A2 | 63.3 | 7.7×10−2 | 7.7×10−1 | 7.7×10−1 |
|
| |||||
| Bxz2 | A3 | 64.2 | 1.0 | <10−6 | 1.5 |
| HelDan | A3 | 64.0 | 7.7×10−2 | <10−7 | <10−7 |
| JHC117 | A3 | 64.0 | 3.3×10−6 | <10−9 | <10−9 |
| Microwolf | A3 | 64.0 | 1.0 | <10−4 | 2.2 |
| Rockstar | A3 | 64.3 | 1.0 | 1.0 | 5.0 |
| Vix | A3 | 64.0 | 1.0 | <10−6 | 4.5×10−1 |
|
| |||||
| Backyardigan | A4 | 63.7 | 1.0×10−1 | 1.0×10−1 | <10−7 |
| Eagle | A4 | 63.9 | 1.0 | 3.3×10−1 | <10−6 |
| LHTSCC | A4 | 63.9 | 1.0 | <10−8 | <10−8 |
| MeeZee | A4 | 63.9 | 2.0 | 1.0×10−6 | <10−9 |
| Peaches | A4 | 63.9 | 3.7×10−5 | 1.1×10−5 | <10−9 |
| Shaka | A4 | 63.9 | 3.3 | 3.3×10−1 | <10−6 |
| TiroTheta9 | A4 | 63.9 | 1.0×10−1 | <10−9 | <10−9 |
| Wile | A4 | 63.7 | 6.6×10−1 | <10−8 | <10−8 |
|
| |||||
| Airmid | A5 | 60.0 | 7.6×10−1 | 3.9×10−1 | <10−9 |
| Benedict | A5 | 59.8 | 1.0 | 2.3×10−1 | <10−9 |
| Cuco | A5 | 60.9 | 1.0 | <10−8 | <10−8 |
|
| |||||
| Blue7 | A6 | 61.4 | 1.0 | 5.5×10−2 | <10−8 |
| DaVinci | A6 | 61.5 | 3.0×10−1 | 3.0×10−2 | <10−9 |
| Gladiator | A6 | 61.4 | 1.0 | 7.7×10−2 | <10−8 |
| Hammer | A6 | 61.3 | 1.0 | 1.4×10−2 | <10−6 |
| Jeffabunny | A6 | 61.6 | <10−7 | 6.6×10−2 | <10−7 |
|
| |||||
| TimShel | A7 | 63.1 | <10−6 | <10−6 | <10−6 |
|
| |||||
| Saintus | A8 | 61.2 | 1.5×10−1 | <10−5 | <10−5 |
|
| |||||
| Alma | A9 | 62.5 | 1.0×10−1 | <10−7 | <10−7 |
| PackMan | A9 | 62.6 | 1.0×10−2 | <10−6 | 4.3×10−4 |
|
| |||||
| ABU | B1 | 66.5 | <10−9 | 8.1×10−6 | <10−8 |
| Chah | B1 | 66.5 | <10−10 | 3.0×10−6 | <10−10 |
| Colbert | B1 | 66.5 | <10−7 | <10−7 | <10−7 |
| Fang | B1 | 66.5 | <10−9 | 6.5×10−7 | <10−9 |
| Harvey | B1 | 66.5 | <10−7 | <10−7 | <10−7 |
| Hertubise | B1 | 66.4 | <10−9 | 4.3×10−7 | <10−9 |
| IsaacEli | B1 | 66.5 | <10−9 | <10−9 | <10−9 |
| JacAttac | B1 | 66.5 | <10−7 | <10−7 | <10−7 |
| Kikipoo | B1 | 66.5 | <10−9 | 4.8×10−6 | <10−9 |
| Morgushi | B1 | 66.4 | <10−8 | <10−8 | <10−8 |
| Murdoc | B1 | 66.4 | <10−8 | <10−8 | <10−8 |
| Oline | B1 | 66.4 | <10−10 | 3.3×10−6 | 1.3×10−7 |
| Oosterbaan | B1 | 66.5 | <10−7 | <10−7 | <10−7 |
| Orion | B1 | 66.5 | <10−8 | 3.3×10−6 | <10−8 |
| PG1 | B1 | 66.5 | <10−7 | <10−7 | <10−7 |
| Phipps | B1 | 66.5 | <10−4 | 1.0×10−1 | <10−4 |
| Puhltonio | B1 | 66.4 | <10−8 | <10−8 | <10−8 |
| Scoot17c | B1 | 66.5 | 1.8×10−1 | 7.6×10−3 | 5.9×10−3 |
| Serendipity | B1 | 66.5 | <10−8 | <10−8 | <10−8 |
| TallGrassMM | B1 | 66.5 | <10−9 | <10−9 | <10−9 |
| Thora | B1 | 66.5 | <10−9 | 5.4×10−7 | <10−9 |
| ThreeOh3D2 | B1 | 66.5 | <10−9 | <10−9 | <10−9 |
| UncleHowie | B1 | 66.5 | <10−8 | <10−8 | <10−8 |
| Vista | B1 | 66.5 | 1.0×10−4 | 3.0×10−4 | <10−7 |
| Vortex | B1 | 66.5 | <10−10 | <10−10 | <10−10 |
| Yoshand | B1 | 66.5 | 6.7×10−2 | 4.3×10−7 | 2.9×10−6 |
|
| |||||
| Arbiter | B2 | 68.9 | 8.5×10−2 | 5.0×10−2 | 5.0×10−6 |
| Ares | B2 | 69.0 | <10−8 | 1.0×10−2 | 3.0×10−6 |
| Hedgerow | B2 | 69.0 | 1.0 | 2.3×10−3 | 1.0×10−6 |
| Qyrzyla | B2 | 68.9 | 6.7×10−21 | <10−3 | <10−3 |
| Rosebush | B2 | 68.9 | <10−10 | 1.0×10−6 | <10−10 |
|
| |||||
| Athena | B3 | 67.5 | 2.0×10−1 | <10−5 | <10−5 |
| Akoma | B3 | 67.5 | 2.3×10−2 | 2.3×10−2 | <10−9 |
| Daisy | B3 | 67.6 | 1.8×10−1 | 1.5×10−5 | <10−9 |
| Gadjet | B3 | 67.5 | 6.6×10−3 | <10−5 | <10−5 |
| Kamiyu | B3 | 67.5 | 5.9×10−2 | <10−10 | 1.8×10−6 |
| Phaedrus | B3 | 67.6 | 3.0 | 5×10−6 | <10−9 |
| Phlyer | B3 | 67.5 | 7.5×10−1 | <10−8 | <10−8 |
| Pipefish | B3 | 67.3 | 1.1×10−1 | 5.0×10−6 | <10−9 |
|
| |||||
| ChrisnMich | B4 | 69.1 | <10−6 | <10−6 | <10−6 |
| Cooper | B4 | 69.1 | 5.0×10−1 | <10−6 | <10−6 |
| Nigel | B4 | 68.3 | <10−9 | 1.0×10−4 | <10−9 |
| Stinger | B4 | 68.6 | 3.0×10−1 | 1.0×10−2 | <10−5 |
| Zemanar | B4 | 68.9 | <10−9 | <10−9 | <10−9 |
|
| |||||
| Acadian | B5 | 68.4 | 1.0×10−3 | <10−9 | <10−9 |
|
| |||||
| Bxz1 | C1 | 64.8 | 2.6×10−2 | 1.0 | <10−8 |
| Cali | C1 | 64.7 | 1.0×10−2 | 1.0 | <10−8 |
| Catera | C1 | 64.7 | 1.0×103 | 1.0×102 | <10−6 |
| Dandelion | C1 | 64.7 | 3.0×10−3 | 1.0×10−1 | <10−9 |
| Drazdys | C1 | 64.7 | 1.0×10−2 | 3.3 | <10−6 |
| ET08 | C1 | 64.6 | 3.3×10−3 | 3.3 | <10−6 |
| LinStu | C1 | 64.8 | 4.4×10−6 | 4.4×10−6 | <10−9 |
| LRRHood | C1 | 64.5 | 1.1×10−5 | 4.8×10−2 | <10−9 |
| MoMoMixon | C1 | 64.8 | <10−6 | 1.0×10−3 | <10−6 |
| Nappy | C1 | 64.7 | 1.3×10−5 | 1.1×10−1 | <10−9 |
| Pio | C1 | 64.8 | 3.0 | 3.0×10−5 | <10−9 |
| Pleione | C1 | 64.7 | <10−5 | 5.0×10−2 | <10−5 |
| Rizal | C1 | 64.7 | <10−8 | 2.3×10−1 | <10−8 |
| ScottMcG | C1 | 64.8 | <10−7 | 3.0×10−1 | <10−7 |
| Sebata | C1 | 64.8 | 6.0×10−2 | 6.0×10−2 | <10−5 |
| Spud | C1 | 64.8 | <10−6 | 1.0×10−2 | <10−6 |
| Wally | C1 | 64.7 | 1.0 | 4.5 | <10−10 |
|
| |||||
| Adjutor | D | 59.9 | <10−8 | <10−8 | <10−8 |
| Gumball | D | 59.6 | 1.0×10−2 | <10−8 | <10−8 |
| Nova | D | 59.7 | 5.4×10−1 | <10−10 | <10−10 |
| PBI1 | D | 59.8 | 1.1×10−5 | <10−8 | <10−8 |
| P-Lot | D | 59.8 | 3.4×10−4 | <10−8 | <10−8 |
| SirHarley | D | 59.6 | 7.5×10−2 | 7.5×10−6 | <10−9 |
| Troll4 | D | 59.6 | <10−8 | <10−8 | <10−8 |
|
| |||||
| Bask21 | E | 62.9 | 3.3 | <10−5 | <10−5 |
| CJW1 | E | 63.7 | 5.6×10−1 | <10−5 | <10−5 |
| Elph10 | E | 63.0 | 3.3 | <10−6 | <10−6 |
| Eureka | E | 62.9 | 5.0×10−2 | <10−7 | <10−7 |
| Henry | E | 63.0 | 1.7 | <10−5 | <10−5 |
| Kostya | E | 63.5 | 5.0×10−1 | <10−6 | <10−6 |
| Lilac | E | 63.0 | 2.5×10−1 | 3.5×10−2 | <10−8 |
| Porky | E | 63.5 | 6.6×10−1 | <10−8 | <10−8 |
| Pumpkin | E | 63.0 | 1.3×10−2 | 1.0×10−6 | <10−10 |
| Rakim | E | 62.9 | 1.0 | <10−6 | <10−6 |
| Toto | E | 63.0 | 2.0 | <10−7 | <10−7 |
|
| |||||
| Boomer | F1 | 61.1 | 9.5×10−1 | 4.2×10−7 | <10−9 |
| Che8 | F1 | 61.3 | 9.0×10−4 | 2.7×10−4 | <10−10 |
| DeadP | F1 | 61.6 | 4.2×10−4 | <10−7 | <10−7 |
| DLane | F1 | 61.9 | 2.3×10−3 | <10−7 | <10−7 |
| DotProduct | F1 | 61.8 | 1.3×101 | <10−8 | <10−8 |
| Drago | F1 | 61.2 | 8.0×10−1 | <10−10 | <10−10 |
| Fruitloop | F1 | 61.8 | 3.3×10−2 | <10−9 | <10−9 |
| GUmbie | F1 | 61.4 | 4.5×10−1 | <10−6 | <10−6 |
| Ibhubesi | F1 | 61.2 | 4.4×10−1 | <10−8 | <10−8 |
| Llij | F1 | 61.5 | 1.3 | <10−9 | <10−9 |
| Mozy | F1 | 61.1 | 3.0×10−3 | <10−9 | <10−9 |
| Mutaform13 | F1 | 61.3 | 2.0×10−1 | <10−9 | <10−9 |
| Pacc40 | F1 | 61.3 | 4.0×10−1 | <10−10 | <10−10 |
| PMC | F1 | 61.4 | 7.6×10−1 | 1.0×10−6 | <10−9 |
| Ramsey | F1 | 61.2 | 6.5×10−1 | <10−8 | <10−8 |
| RockyHorror | F1 | 61.1 | 6.6 | <10−8 | <10−8 |
| SG4 | F1 | 61.9 | <10−7 | <10−7 | <10−7 |
| Shilan | F1 | 61.4 | 6.6×10−1 | 5.6×10−7 | <10−9 |
| Spartacus | F1 | 61.7 | 8.7×10−1 | 1.3×10−6 | <10−10 |
| Tweety | F1 | 61.7 | 4.3×10−1 | <10−9 | <10−9 |
| Wee | F1 | 61.8 | 2.3×10−4 | 5.1×10−7 | <10−9 |
|
| |||||
| Che9d | F2 | 60.9 | 3.7×10−4 | <10−9 | <10−9 |
| Yoshi | F2 | 61.0 | 1.2×10−1 | <10−10 | <10−10 |
|
| |||||
| Angel | G | 66.7 | 5.4 | 1.1×10−2 | 9.0×10−1 |
| Avrafan | G | 66.6 | 5.0×10−1 | 1.0×10−3 | 1.0×10−5 |
| BPs | G | 66.6 | 7.6×10−1 | 1.0×10−3 | 1.5×10−5 |
| Halo | G | 66.7 | 1.7 | 6.7×10−3 | 6.0×10−4 |
| Hope | G | 66.6 | 7.5×10−1 | 7.5×10−2 | <10−7 |
| Liefie | G | 66.8 | 1.7×10−1 | 5.8×10−3 | 2.3×10−2 |
|
| |||||
| Konstantine | H1 | 57.4 | 7.5×10−5 | 7.5×10−6 | <10−9 |
| Predator | H1 | 56.4 | <10−6 | <10−6 | <10−6 |
|
| |||||
| Barnyard | H2 | 57.5 | <10−4 | <10−4 | <10−4 |
|
| |||||
| Babsiella | I1 | 67.1 | 3.0×10−3 | <10−9 | <10−9 |
| Island3 | I1 | 66.8 | <10−8 | 4.4×10−5 | <10−8 |
|
| |||||
| Che9c | I2 | 65.4 | 1.5×10−3 | 1.5×10−3 | <10−9 |
|
| |||||
| Baka | J | 60.7 | 2.9×10−3 | <10−8 | <10−8 |
| Courthouse | J | 60.9 | 1.1×10−4 | <10−10 | <10−10 |
| LittleE | J | 61.3 | <10−7 | <10−7 | <10−7 |
| Omega | J | 61.4 | <10−5 | <10−5 | <10−5 |
| Optimus | J | 60.8 | 3.3×10−2 | <10−6 | <10−6 |
| Thibault | J | 60.8 | 7.0×10−2 | <10−9 | <10−9 |
|
| |||||
| Adephagia | K1 | 66.6 | 1.0×10−2 | <10−8 | 3.3 |
| CrimD | K1 | 66.9 | 3.0 | <10−6 | 1.0 |
| Angelica | K1 | 66.4 | <10−9 | <10−9 | <10−9 |
| Jaws | K1 | 66.6 | 4.3×10−1 | 1.2×10−2 | 1.3 |
|
| |||||
| TM4 | K2 | 68.1 | 1.5 | 2.3×10−5 | 7.7×10−1 |
|
| |||||
| Pixie | K3 | 67.3 | 3.3×10−1 | 2.0×10−2 | 4.6×10−1 |
|
| |||||
| Fionnbharth | K4 | 68.0 | 1.3×10−1 | <10−8 | 3.0×101 |
|
| |||||
| Larva | K5 | 65.3 | 1.0 | <10−8 | <10−8 |
|
| |||||
| JoeDirt | L1 | 58.8 | 3.3×10−5 | 2.3×10−1 | <10−8 |
| LeBron | L1 | 58.8 | <10−9 | 1.0×10−1 | <10−9 |
| UPIE | L1 | 58.8 | 3.0×10−5 | 4.0×10−2 | <10−9 |
|
| |||||
| Faith1 | L2 | 58.9 | 1.0 | 1×10−9 | 3.3×10−6 |
| Rumpelstiltskin | L2 | 58.9 | <10−9 | <10−9 | 3.3×10−6 |
|
| |||||
| Bongo | M | 61.6 | 5.2×10−1 | 5.4×10−3 | 7.7×10−3 |
| Rey | M | 60.9 | 2.2 | <10−8 | 3.3×10−5 |
|
| |||||
| Charlie | N | 66.3 | 1.3×10−1 | 1.0×10−4 | <10−8 |
| Redi | N | 66.1 | 1.3×10−1 | <10−9 | <10−9 |
|
| |||||
| Corndog | O | 65.4 | <10−8 | 4.4×10−5 | <10−8 |
| Firecracker | O | 65.5 | <10−8 | 4.3×10−1 | <10−8 |
|
| |||||
| BigNuz | Sin | 66.7 | 2.9×10−1 | 1.4×10−5 | <10−8 |
|
| |||||
| Dori | Sin | 66.0 | 2.3 | <10−9 | 4.0×10−2 |
|
| |||||
| Giles | Sin | 67.3 | 2.9×10−3 | 2.5×10−5 | <10−8 |
|
| |||||
| Marvin | Sin | 64.7 | <10−10 | 2.5×10−4 | <10−10 |
|
| |||||
| Patience | Sin | 50.3 | <10−6 | 4.4×10−3 | <10−6 |
|
| |||||
| Send513 | Sin | 56.0 | <10−8 | 2.3×10−1 | 3.3×10−6 |
|
| |||||
| Wildcat | Sin | 57.2 | <10−10 | 1.7×10−3 | 1.0×10−4 |
Efficiencies of plating are relative to infection of M. smegmatis mc2155
Figure 2.
Gene content relationships among the subclusters of Cluster A mycobacteriophages. An expanded representation of the Cluster A phages from Figure 1 is shown with each of the subclusters circled and the GC% range for each subcluster indicated.
Mycobacteriophage infection of M. tuberculosis
Although a subset of the sequenced mycobacteriophages previously have been tested for infection of M. tuberculosis, we have extended this to the larger collection of phages (Table 1; Fig. S1). Interestingly, there is a strong correlation between cluster or subcluster designation and the ability to efficiently infect M. tuberculosis mc27000 (using a cutoff value for infection at an e.o.p. of 0.1) in a chi squared test (x2 = 97.85, df =26, P = 2.9×10−10). The infectivity across the clusters of phages is restricted primarily to phages in Cluster K, and two subclusters (A2 and A3) of Cluster A, although some plaques were also observed with Bxb1 and U2 from Subcluster A1 (Table 1, Fig. S1). The phages in other subclusters of cluster A do not form plaques on M. tuberculosis, even those most closely related phylogenetically, such as in subclusters A6/A9 and A4/A7, respectively (Fig. 2). However, Cluster A phages span considerable diversity (Fig. 2), and genome alignments do not provide obvious clues as to the genetic basis of this discrimination for M. tuberculosis. As reported previously for BPs and Halo (Sampson et al., 2009), all of the Cluster G phages are able to form plaques on M. tuberculosis but at a reduced efficiency of plating (Table 1, Fig. S1). We also observed plaques when plating large numbers of particles on M. tuberculosis for several other phages including some Cluster B phages.
Mycobacteriophage infection of M. smegmatis strains
M. smegmatis and M. tuberculosis are distantly related mycobacterial species with vastly different growth rates (doubling times of 3 hours and 24 hours, respectively) and genome sizes (7.0 Mbp and 4.4 Mbp, respectively). It is therefore perhaps not surprising that relatively few mycobacteriophages efficiently infect both strains. To further explore mycobacteriophage host preferences on more closely related bacteria, we tested these phages for their ability to infect two other strains of M. smegmatis, Jucho, and MKD8. M. smegmatis Jucho (Mizuguchi and Tokunaga, 1971) and MKD8 (Parsons et al., 1998) – a derivative of M. smegmatis PM5 (Tokunaga et al., 1973) – have been described previously, and have distinct patterns of conjugation with M. smegmatis mc2155, a derivative of M. smegmatis ATCC607 (Parsons et al., 1998; Snapper et al., 1990; Wang et al., 2003). We reasoned that since these strains have independent origins, some of the phages would discriminate between them, and exhibit patterns of infection distinct from those observed on M. tuberculosis. We also tested these phages on Mycobacterium aichiense – a fast grower and distant relative of M. smegmatis – but none of the tested phages are able to infect this bacterium (data not shown).
The plating properties of over 200 phages on the various M. smegmatis strains are shown in Table 1 and Fig. S1, and there are three main patterns that emerge. First, it is clear that the phages exhibit distinct patterns of infection on various M. smegmatis strains. Second, there are correlations between the abilities of phages to infect individual strains and their cluster/subcluster designation, although not as strongly as for M. tuberculosis (Jucho: x2 = 40.00, df = 26, p = .039; MKD8: x2 = 67.03, df = 26, p = 1.8×10−5; cutoff e.o.p. of 0.1) (Table 1, Fig. S1). A third observation is that although there are many examples where no plaques are observed at all on a particular strain, there are numerous examples where the e.o.p. is merely reduced as compared to mc2155, presumably either through escape of a restriction mechanism, or through phage mutations. Overall, these data are consistent with the idea that there is a correlation between cluster/subcluster designation and host range.
There are many potential mechanisms for resistance and host range determination, but their contributions to mycobacteriophage host preferences are poorly understood. One potential mechanism is superinfection immunity conferred by the presence of a prophage to which escape is typically very infrequent, and in the phage lambda prototype, multiple mutations (at least three) are required to confer a virulent phenotype (Hopkins and Ptashne, 1971). This could account for example for the inability of subcluster A1 phages to infect MKD8, although prophages in this strain have yet to be reported. However, for M. tuberculosis mc27000 – an avirulent derivative of strain H37Rv – the genome sequence is known and there are no full-length prophages [there are two small prophage-like elements, but these do not encode any recognizable repressor proteins (Cole et al., 1998; Hendrix et al., 1999)]. Thus the inability for phages to infect M. tuberculosis is unlikely to result from phage immunity.
Expanding the host range of Cluster G phages
The Cluster G phages do not efficiently infect M. tuberculosis but can form plaques on this bacterium at a reduced frequency, relative to M. smegmatis mc2155 (Sampson et al., 2009)(Fig. 3A). This pattern of infection might arise due to the escape from restriction or from the generation of mutations with expanded or altered host range. To explore these possibilities, 12 independently isolated plaques of either BPs or Halo were recovered from M. tuberculosis lawns; all were re-plated and found to infect M. smegmatis and M. tuberculosis at equivalent e.o.p’s (Fig. 3B). These were then plaque purified again on M. smegmatis, and the resulting phages were also shown to infect M. tuberculosis with an e.o.p. of one, relative to M. smegmatis (Fig. 3B). These properties strongly suggest that the enhanced efficiency of plating of the Cluster G phage isolates on M. tuberculosis results from mutational and heritable changes, rather than phenotypic survival, as would be expected if these phages were escaping from host restriction/modification systems. In this latter case, we predict that plating on M. smegmatis would have resulted in loss of the enhanced e.o.p. phenotype.
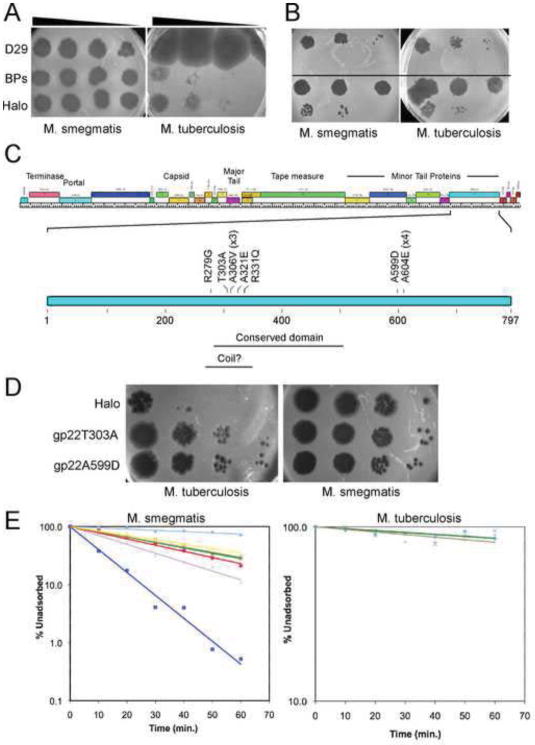
Figure 3.
Host Range Expansion of Cluster G mycobacteriophages. (A) Mycobacteriophages BPs and Halo form plaques on M. tuberculosis mc27000 at an efficiency of 10−4 – 10−5, relative to M. smegmatis mc2155. A control phage, D29, infects both bacteria with equal efficiency. (B) A single Halo plaque picked from an M. tuberculosis and re-plated on M. tuberculosis and M. smegmatis infects both bacteria with equal efficiency (top part). A single plaque picked from the M. smegmatis plate in the top part of the figure retains the ability to efficiently infect M. tuberculosis (bottom part), showing it has acquired a heritable mutation conferring host range expansion. (C) The left arms of the BPs and Halo genomes contain the virion structure and assembly genes (1 – 26). The positions of a conserved domain and a weakly predicted coiled domain are shown. (D) Two of the mutations conferring host range expansion – T303A and A599D – were engineered back into mycobacteriophage Halo using BRED mutagenesis. Both mutant phage derivatives efficiently infect M. tuberculosis. (E) Halo and BPs host range mutants [A604E (blue squares); R331Q (purples crosses); T303A (brown squares); A306V (yellow triangles); A321E (green crosses); A599D (red circles) display increased adsorption rates on M. smegmatis relative to wild-type phage (light blue diamonds); adsorption to M. tuberculosis is unaffected.
To map the mutations responsible for the ability to efficiently infect M. tuberculosis, we first determined the complete genome sequence of one of the phage isolates, Halo enhanced host range 1 (ehr1). A single point mutation was identified at position 22,253 within gene 22, which changes an alanine residue at position 604 to a glutamic acid (A604E) (Fig. 3C). Halo gene 22 is located within a group of putative minor tail protein genes (Fig. 3C) and encodes a product of 797 residues; BPs encodes a nearly identical product that differs in only a single residue at position 12. The structure of BPs/Halo gp22 is not known, but it is rich in glycine (12.5%) and alanine (11.8%), characteristic of phage tail fiber proteins. There is a weakly predicted coiled domain between residues 270 and 330, but no significant matches were found using HHPred (Fig. 3C).
To map the locations of the mutations in the other host range mutants, segments containing gene 22 were amplified by PCR and sequenced. A single base substitution was found in each, corresponding to a total of seven different amino acid changes (Fig. 3C). Seven of the 12 mutants map to just two residues, at positions 306 and 604, and in general, the mutations cluster to two regions, one spanning residues 279 – 331 near the weakly predicted coiled domain, and the other around position 600 (Fig. 3C). The roles of these substitutions in gene 22 in host range was confirmed by using BRED mutagenesis (Marinelli et al., 2008) to separately engineer two of the substitutions (T303A and A599D) into a wild-type Halo genome. These engineered mutants recapitulated the phenotypes of the mutants isolated directly on M. tuberculosis (Fig. 3D). The mapping of host range mutations to tail fibers is not unexpected, and suggests strongly that the inability of Halo and BPs to efficiently infect M. tuberculosis results from surface interactions rather than defects in expression, replication, assembly, or lysis. In support of this interpretation, when DNA from either wild-type Halo or from a host range mutant is electroporated into M. tuberculosis and the transformed cells are plated on a lawn of permissive M. smegmatis, approximately equivalent numbers of plaques (~130 per 250 ng DNA) are observed.
To determine if these expanded host range mutants have altered surface interactions, we measured adsorption rates for M. smegmatis and M. tuberculosis (Fig. 3E). Although M. tuberculosis adsorption assays are technically challenging because of cell clumping, we observed no differences in the binding of the two mutants tested as compared to wild-type phage. Surprisingly, all of the mutants we tested have increased adsorption rates to M. smegmatis with A604E showing the strongest phenotype (Fig. 3E). This is a surprise because the mutants were isolated solely for their ability to infect M. tuberculosis, and although we might have expected to see phenotypic changes that compromise the ability of the mutants to infect M. smegmatis, dramatic improvements in adsorption were not expected. One plausible explanation for this phenomenon is that these mutants are not altered in receptor recognition at the initial stage of infection, but differ in a subsequent event that renders binding irreversible, such as DNA injection across the cell membrane, and which presumably requires dynamic events within the receptor-tail fiber complex. It should be noted, however, that these experiments were conducted with planktonically growing cells under laboratory conditions, and it is possible that these mutants could be impaired in infection of M. smegmatis growing environmentally [for example as biofilms (Ojha et al., 2005)], as noted for high adsorption rate mutants of phage λ (Gallet et al., 2009). This might explain why phages such as Halo have evolved to have rather poor adsorption rates.
Expanded host range of Subcluster B2 phages
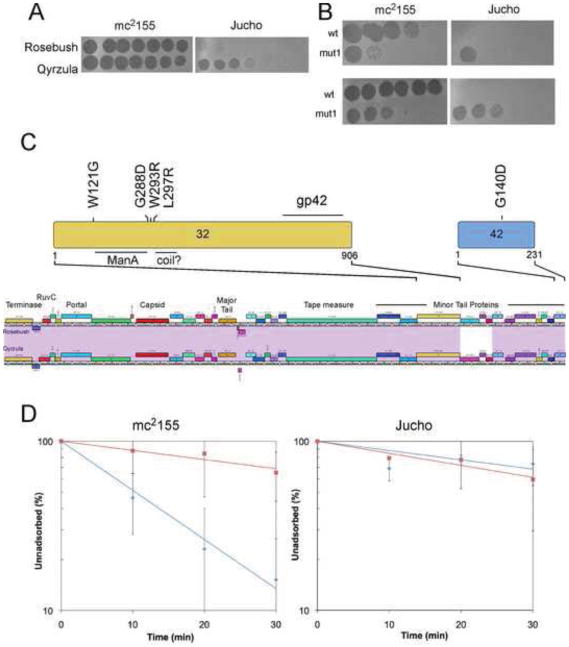
To further explore the basis of host range determination in the mycobacteriophages, we examined the ability of the subcluster B2 phage Rosebush against the host strain M. smegmatis Jucho. As noted above, Rosebush does not efficiently infect Jucho, although plaques with a normal appearance are observed at a frequency of ~10−5 (Fig. 4A). We isolated and purified five independent mutants, all of which were shown to infect mc2155 and Jucho at equivalent efficiencies of plating (Fig. 4B). We determined the sequence of one of these mutants and identified a single base substitution at position 30,977 within Rosebush gene 32 that introduces a L297R substitution in gp32 (Fig. 4C). Gene 32 is located within a group of putative minor tail proteins, and – like BPs/Halo gp22 – Rosebush gp32 has a relatively high glycine (9.0%) and alanine (11.4%) content, consistent with its role as a phage tail fiber. Furthermore, like BPs gp22, Rosebush gp32 has a weakly predicted coiled domain (Fig. 4C). Comparative analysis suggests a modular structure to the protein. The N-terminal 400 residues are similar to a large group of putative tail proteins of phages from other clusters such as H, I, M, N, and other subclusters of B, and the C-terminal 500 residues have similarity to putative tail proteins of Cluster L phages, with much weaker similarity in the N-terminal domain. Interestingly, HHPred reveals a highly probable match (98.93%) of the segment containing residues 125-284 to the 147 aa carbohydrate-binding module of Thermoanaerobacterium polysaccharolyticum ManA (Bae et al., 2008), suggesting that Rosebush binds to a receptor with a carbohydrate moiety. However, the L297R substitution lies outside of this region.
Figure 4.
Isolation and characterization of Rosebush expanded host range mutants. (A) Plating of serial dilutions Rosebush on M. smegmatis mc2155 and M. smegmatis Jucho shows the reduced efficiency of plating; the control phage Qyrzula (which, like Rosebush, is in Subcluster B2). (B) A Rosebush plaque picked from the Jucho plate (mut1) plates on mc2155 and Jucho with equivalent efficiencies of plating (upper). A mut1 plaque recovered from M. smegmatis mc2155 plates with equivalent efficiencies on both strains, in contrast to wild-type Rosebush (wt; lower). (C) Locations of amino acid substitutions in gp32 and gp42 of Rosebush in the expanded host range mutants. Positions of the ManA domain and a weakly predicted coiled domain are shown, as well as a region at the C-terminus of gp32 that is homologous with gp42. (D) Adsorption assays (n = 2) of wild-type Rosebush (red squares) and expanded host range mutant 1 (blue diamonds) on M. smegmatis mc2155 and M. smegmatis Jucho as indicated.
The remaining four mutants were mapped by PCR amplification and sequencing of gene 32. Three of the mutants each have a single base substitution in 32, conferring the substitutions W121G, G288D and W293R. The identification of three substitutions within a span of fewer than 10 residues suggests that this is a particularly important region for host range determination; all of the substitutions lie outside of the ManA carbohydrate binding domain. The fourth mutant has a wild-type gene 32, and the complete genome sequence was determined to identify the mutation, a single base change at position 39,077 conferring a G140D substitution in gp42 (Fig. 4C). This suggests that gp42 is also important for host range determination. Gene 42 lies at the extreme end of the structural gene operon (the genes to its immediate right are transcribed leftwards) and encodes a 231 aa product. BLASTP comparison reveals that gp42 has sequence similarity albeit weakly (24% identity) to the C-terminal portion of Rosebush gp32 (Fig. 4C). HHPred analysis shows that Rosebush gp42 (residues 35 – 209) has highly probable matches (85.8 – 96.6%) to nine glycosyl hydrolases for which structures have been determined, with the closest being to a mouse Galactocerebrosidase (Deane et al., 2011). Curiously, HHPred also shows similarity (residues 35-188) to the baseplate protein gp10 of phage T4 (Leiman et al., 2006), although with a much weaker probability (37.1%). These data are consistent with the interpretation that gp42 is a tail component that assists in infection through hydrolytic degradation of sugar-containing molecules on the cell surface.
Collectively, these observations suggest that the inability of wild-type Rosebush to infect M. smegmatis Jucho is because of poor receptor recognition, or – as proposed for BPs – downstream events such as DNA injection. To confirm that there was no defect in replication, expression, assembly, or lysis, we performed a transfection experiment in which Rosebush genomic DNA was electroporated into competent Jucho cells, and following a period of recovery, plated with permissive M. smegmatis mc2155 cells (Fig. 5A). An equivalent number of plaques were recovered as when using M. smegmatis mc2155 competent cells, or when using Giles genomic DNA (as a control for cell competency) with both strains.
Figure 5.
Transfection and replication in non-permissive hosts.
(A) Transfection of electrocompetent M. smegmatis Jucho (top row) and mc2155 (bottom row) with 150 ng of either Rosebush genomic DNA (left column) or Giles genomic DNA (right column) followed by recovery and plating with M. smegmatis mc2155 cells. (B) Transfection of electrocompetent M. tuberculosis mc27000 with 150 ng of either Giles genomic DNA (left plate) or D29 genomic DNA (right plate) followed by recovery and plating with M. smegmatis mc2155 cells. (C) Transfection of electrocompetent M. smegmatis mc2155 with either 200 ng (left plate) or 400 ng (right plate) of Streptomyces phage Zemlya followed by recovery and plating with S. lividans. Individual plaques were picked and tested for infection of S. lividans and M. smegmatis; all infected S. lividans but failed to infect M. smegmatis, showing that these are not mutants or contaminants.
Adsorption assays performed with the gp32 L297R mutant show no apparent difference in binding to Jucho cells as compared to wild-type Rosebush, reminiscent of the behavior of the BPs/Halo mutants in adsorption to M. tuberculosis (Fig. 4D). In addition, we observed that – like the BPs/Halo mutants – the Rosebush mutant adsorbs better to M. smegmatis mc2155 than wild-type Rosebush (Fig. 4D). As described above for the BPs/Halo host range expansion, the basis for this unexpected phenotype is not clear.
Cross-species and cross-genus transfection
The transfection assay described above for Rosebush is a powerful approach for determining whether a host range constraint derives from a surface or DNA injection blockage, or from subsequent events post-DNA entry into the cell. The observation that wild-type Rosebush can propagate in Jucho following DNA transfection is perhaps not surprising given that the bacterial hosts are strains within the same species. However, we have explored the possibility that this approach could be applied more broadly including both hosts of different species, and host from different genera. First, we tested the ability of M. tuberculosis to support the propagation of Giles DNA (Giles is normally unable to infect M. tuberculosis), by transfecting Giles genomic DNA into electrocompetent M. tuberculosis mc27000 cells, allowing recovery, and plating with M. smegmatis cells (Fig. 5B). Equivalent numbers of plaques were recovered as when using electrocompetent M. smegmatis cells, indicating that surface recognition and DNA injection represent the barrier to Giles infection of M. tuberculosis. As described above, we performed similar transfection assays with wild-type Halo and one of the host range mutants and found that both produce similar numbers of plaques when equivalent amounts are electroporated into M. tuberculosis mc27000 and plated on a lawn of M. smegmatis (data not shown).
To our knowledge, phages infecting either mycobacterial hosts or Streptomyces hosts do not cross the genus barrier between Mycobacterium and Streptomyces. In order to determine if surface interactions are responsible (at least in part) for this barrier, we performed a similar experiment to that described above, using DNA of Streptomyces phage Zemlya, which was electroporated into M. smegmatis, and plated with S. lividans cells (Fig. 5C). Although recovery was not efficient, using 200 ng and 400 ng of DNA, we recovered approximately 18 and 30 plaques, respectively, indicating that M. smegmatis can support the post-DNA injection events of Zemlya’s growth. In order to rule out host range mutants or contamination, 20 random plaques were picked and tested for their ability to infect both M. smegmatis and S. lividans. All 20 were able to infect S. lividans and none were able to infect M. smegmatis, indicating that these phages were not contaminants and were not host range mutants.
Discussion
Analysis of the host preferences for a large collection of mycobacteriophages indicates correlations between host range and genome type, with varying strengths of correlation depending on the strain. Although only a few hosts were tested, we strongly predict that these patterns will extend to other potential mycobacterial hosts. However, in general, these data provide good support for the idea that mycobacteriophages within different cluster or subcluster designations have distinct host preferences, and that these host preferences confer barriers to genetic exchange, even though under laboratory conditions they can all infect the same host. However, because the designation of phages into clusters and subclusters is predicated upon essentially arbitrary parameters, and genetic mosaicism from horizontal genetic exchanges contributes to a considerable variability within these groups, the correlation between cluster/subcluster type is a relatively crude one. Presumably the molecular basis of host preference depends on the presence and behavior of specific genes, raising the possibility of searching for a correlation between gene phamilies and host preferences. However, this is further complicated by the finding that host preference is also allele specific, as demonstrated here for phage BPs and Rosebush.
Although there are no similarly large collections of phages with sequenced genomes that are known to infect a common host that have been examined for host preferences, the patterns observed for the mycobacteriophages are not atypical. For example, phages rarely infect hosts from other genera, and have preferences for particular species and sometimes strains or serovars within a species, such as the Listeria phages (Loessner and Busse, 1990; Loessner and Calendar, 2006). However, there are abundant examples of phages that can expand their host range within a species and to a different species as observed here, either by adapting to a new receptor or overcoming restriction, immunity, CRISPRs or Toxin-antitoxin systems (Deveau et al., 2010; Fineran et al., 2009; Hopkins and Ptashne, 1971). There are also examples of phages that can ‘jump’ to new genera (Bielke et al., 2007; Jensen et al., 1998). To our knowledge, no phages have been described that traverse the barrier between gram-negative and gram-positive bacteria, or from which mutants can be isolated that permit jumping of the gram-barrier.
There are a multitude of mechanisms that can determine host range, including receptor availability, restriction/modification, CRISPRs, toxin-antitoxin systems and immunity; infection is also influenced by conditions such as the growth state of the cells and metal ion availability (Fullner and Hatfull, 1997). The number and variety of such systems is no surprise given the highly dynamic nature of host-phage interactions that have dominated the microbial world for perhaps more than three billion years. Interestingly, we note that not only is infection of M. tuberculosis restricted to specific genomic types – Cluster K, and subclusters A2, A3 – but of the others, only the Cluster G phages appear to be able to generate mutants that can overcome the constraint at a moderate frequency (10−4 – 10−5). This is different than the behaviors on other strains of M. smegmatis, where there are relatively few examples of clusters or subclusters failing to produce plaques at all, and it is more common to see reduced efficiencies of plating. Nonetheless, the mechanisms of mutant escape that we’ve described here both involve single residue substitutions in tail proteins.
The experiments reported here provide further insights into the nature of mycobacteriophage tails and their host interactions. The gp42 protein of Rosebush is a strong candidate for providing an enzymatic activity that allows for the phage tail tip to ‘bore’ its way through the complex outer surface of the mycobacterial cell for it to reach the membrane and complete the infection process, similar to the endorhamnosidase activity of the P22 tail spike (Goldenberg and King, 1982) or the lysozyme function of T4 gp5 (Arisaka et al., 2003). It is unclear how the mutation in Rosebush 42 generates an expanded host range phenotype, but it is plausible that the enzyme acquires a change in substrate specificity that permits processing of cell wall components in M. smegmatis Jucho that otherwise prevent infection. Such enzymes are of some interest as possible agents for anti-mycobacterial therapies.
M. tuberculosis contains two or three CRISPR loci and a total of 30 – 40 spacers, depending on the strain. It is unclear whether these are functional or not, but it is notable that there are no identifiable protospacer sequences corresponding to any of the 83 spacers in the entire phage collection (He et al., 2012). The CRISPRs are thus unlikely to contribute to the failure of any of the mycobacteriophages to infect M. tuberculosis. M. smegmatis mc2155 does not contain any CRISPRs; however, it is not known if any are found in either Jucho or MKD8 and therefore, whether they could contribute to the observed resistance patterns in these strains. We also note that M. tuberculosis encodes a large number of toxin-antitoxin systems, and although these are also not known to play any role in phage resistance, this has been demonstrated for a type III TA in Erwinia carotovora (Fineran et al., 2009) and is worthy of further exploration. Finally, although there is no evidence for restriction-modification systems in either M. smegmatis mc2155 or M. tuberculosis these could play a role in Jucho or MKD8 infection, and restriction systems have been reported in other strains such as M. chelonei and M. gordonae (Jones and Greenberg, 1977; Shankar and Tyagi, 1993a, b).
Based on these data, we present a model to account for the diversity of the mycobacteriophage population in which the diversity of the bacterial population in the environments where these microbes exist plays a central role (Fig. 6). In essence, the model recognizes that bacteriophages have the capacity to switch or expand their host ranges at frequencies that are vastly greater than the overall rates of genetic exchange. The changes in host preference are more likely to occur between hosts that are more similar – such as different strains of the same species (e.g. M. smegmatis mc2155 and M. smegmatis Jucho) – but can also occur with different species within the same genus (e.g. between M. smegmatis mc2155 and M. tuberculosis). Presumably, this switch can also happen between hosts of different genera, but this is likely to be a much less frequent event. For example, we have tested more than 60 mycobacteriophages for infection of S. lividans, and none formed plaques (data not shown), and no Streptomyces phages are known to infect M. smegmatis. Nonetheless, Streptomyces phage Zemlya can clearly replicate within and lyse M. smegmatis. Because the soil microbial diversity is large – especially given the substantial proportion of non-cultivatable bacteria (Rondon et al., 2000; Rondon et al., 1999) – and the genus barrier is not an insurmountable one, it is possible that a series of host range switches and expansions between closely related hosts collectively enable a random-walk across large swaths of the bacterial landscape.
Figure 6.
A model accounting for mycobacteriophage diversity. The large number of different types of mycobacteriophages isolated on M. smegmatis mc2155 can be explained by a model in which a) phages can readily infect a new bacterial host – either by a switch or an expansion of host range – and b) a highly diverse bacterial population, including many closely related strains, in the environments from which the phages are isolated. As such, phages with distinctly different genome sequences and GC% contents infecting distantly related bacterial hosts – such as those to the left (red) or right (blue) extremes of a spectrum of hosts – can migrate across a microbial landscape through multiple steps. Each host switch occurs at a relatively high frequency (~1 in 105 particles, or an average of about one every 103 bursts of lytic growth), and much faster than either amelioration of phage GC% to its new host, or genetic recombination. Two phages (such as those shown in red and blue) can thus ‘arrive’ at a common host (M. smegmatis mc2155) but be of distinctly different types (clusters, subclusters, and singletons). The variety of hosts is shown two dimensionally for simplicity, and the actual relationships among bacteria in environments such as soil and compost is likely to be considerably more complicated. Because host range switching or expansion is a common feature of bacteriophages, the model predicts that a high degree of phage diversity will be seen for any particular host if the microbial population from which the phages are isolated from is also highly diverse and rich in closely related strains. Because none of the phages isolated on M. smegmatis mc2155 also infect M. aichiense, we assume that this strain and its close relatives are absent from the soil and compost environments where most of these phages were isolated from [M. aichiense was isolated from soil in Japan (Ichiyama et al., 1988)].
Such diverse environments are known to exist in compost and soil, but other environments – such as in the human sebaceous follicles from which P. acnes phages can be isolated – are much less diverse, and this is reflected in the restricted diversity of P. acnes phages (Marinelli et al., 2012). Thus in a highly diverse bacterial context, phages can ‘arrive’ at a common host – such as M. smegmatis mc2155 – having ‘traveled’ from numerous phylogenetically distinct (from the host perspective) origins, with distinctive GC% contents, and particular genetic neighborhoods. The effects of this are expected to broadly impact the phage population, such that collections of phages isolated on any bacterial host from this environment will reflect the diversity of the underlying bacterial population. Furthermore, although it is often assumed that phage GC% will reflect that of its host (even though the ‘natural’ host may not be known), we predict that no such correlation will be seen for phages isolated from diverse microbial environments. We note that a broad range of GC% contents is also seen with both Pseudomonas and Staphylococcus phages (Ceyssens and Lavigne, 2010; Kwan et al., 2005, 2006; Marinelli et al., 2012). This model also underscores the potential for phages to readily act as vehicles of genetic exchange between different bacteria in a complex environment like the soil, as these phages readily move in and out of different hosts. Future studies aimed at characterizing phages of various hosts in both restricted and diversity rich environments will be important to determine the extent to which this model applies to a broad range of environmental niches, and ultimately, to better understand the complex ways in which bacteria and their phages interact in the context of the natural world.
Materials and Methods
Bacterial Strains and Media
M. smegmatis Jucho, M. smegmatis, MKD8, M. smegmatis mc2155 – a high efficiency transformation strain (Snapper et al., 1990) were grown in Middlebrook 7H10 supplemented with 0.5% glycerol, 1 mM CaCl2 10% and 10% Albumin Dextrose Complex (ADC), and the avirulent strain M. tuberculosis mc27000 (Ojha et al., 2005; Sambandamurthy et al., 2006), was grow in Middlebrook 7H11 agar (Difco) supplemented with 0.5% glycerol, 1 mM CaCl2 10% and 10% Oleic Acid Albumin Dextrose Complex (OADC). Liquid cultures were grown in 7H9 liquid media (Difco), supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% ADC for M. smegmatis and 10% OADC for M. tuberculosis; Tween 80 was omitted, and CaCl2 was added at a final concentration of 1 mM to cultures used for phage infections. M. smegmatis was grown shaking at 250 rpm, and M. tuberculosis cultures were grown either on a roller or stationary; both were incubated at 37°C. Unless otherwise specified, carbenicillin (CB) (50 μg/ml) and cyclohexamide (CHX) (10 μg/ml) were routinely added to mycobacterial media. S. lividans (ATCC 69441) was cultured at 30°C in YEME (yeast extract, malt extract), on MS agar and on Nutrient agar containing 8mM Ca(NO3)2, 10mM MgSO4, and 0.5% glucose (Kieser et al., 2000).
Propagation of Bacteriophages
All of the mycobacteriophages used here have been described previously (Hatfull, 2012a; Pope et al., 2011b). Streptomyces phage Zemlya is newly isolated, has a siphoviral morphology with a 51 kbp genome and will be described elsewhere. High-titer bacteriophage stocks and plate lysates were prepared as described previously (Sarkis and Hatfull, 1998). In some cases, phage were concentrated by ultracentrifugation in Ti45 rotor tubes at 20,000 μg for 1.5 hours and further purified by CsCl density gradient if necessary. Top agar overlays, seeded with ~300 μl of M. smegmatis or 1–2 ml of M. tuberculosis, were prepared in 0.35% mycobacterial top agar (MBTA) with 1 mM CaCl2 and incubated at 37°C overnight or 1–2 weeks, respectively. Phage lysates were diluted in phage buffer (10 mM Tris-HCl, pH 7.5; 10 mM MgSO4; 68.5 mM NaCl; 1 mM CaCl2).
Host range tests
Lysates of phages were serially diluted in phage buffer and 3 μl of one hundred-fold dilutions were spotted onto fresh lawns of M.smegmatis mc2155, Jucho, MKD8, M. aichiense and M. tuberculosis mc2 7000. The plates were incubated at 37°C and plaque formation was assayed after 24–48 hours for M. smegmatis and M. aichiense strains and 6 days for M. tuberculosis.
Isolation of expanded host range mutants
Independent host range mutants of BPs and Halo were isolated by preparing multiple lysates on M. smegmatis mc2155 – each derived from a single plaque – and using these to infect M. tuberculosis mc27000. Individual plaques were picked into phage buffer and re-plated on both M. smegmatis mc2155 and M. tuberculosis mc27000. Individual plaques were picked from the M. smegmatis mc2155 plate, plaque-purified, and shown to infect both strains at equal efficiencies of plating. Halo ehr1 was identified by 454 sequencing, and the remaining mutants were identified by Sanger sequencing of BPs/Halo gene 22.
Rosebush host range mutants were isolated by plating independently prepared lysates onto M. smegmatis Jucho, and individual plaques were picked and purified. Following re-plating onto both strains, phage mutants recovered from M. smegmatis mc2155 were shown to infect both strains at equivalent efficiencies. A clone library of Rosebush mut1 DNA was prepared in pBluescript IIKS+ vector and sequenced to 92% coverage using an ABI 3730 sequencer. A mutation was identified at coordinate 30,977 in gene 32; no additional mutations were identified. Sequence coverage was subsequently extended to 99.9% using IonTorrent sequencing. Gene 32 of the remaining four mutants was amplified via PCR from phage lysates using primers RB_gp32_F and RB_gp32_R and sequenced using an ABI 3730 Sanger sequencer form the primers RB_gp32_F, RB_gp32_R, gp32seqF1-4, and gp32seqR1-4, identifying three additional mutations residing in 32. The final mutant (mut4) was sequenced using phenol-extracted genomic DNA via 454 to 99.8% coverage, identifying a point mutation in gene 42; no additional mutations were identified.
Transfection
Transfection was typically performed using phage genomic DNA (150–400 ng) and electrocompetent bacteria, and after electroporation allowed to recover for 1 hour in 7H9/ADC/CaCl2 and then plated with addition of a permissive host. Plates were assayed for plaque formation after 48hr of growth at 37°C. For transfection of Zemlya into M. smegmatis, recovery was completed as above, but plated with soft nutrient agar and S. lividans pre-germinated spores (50°C for 10min in YEME) onto Nutrient agar plates containing Chloramphenicol (2ug/ml), which S. lividans is naturally resistant to.
BRED Mutagenesis
Point mutations in Halo gene 22 were introduced using BRED mutagenesis, as described previously (Marinelli et al., 2008). Briefly, for each mutation, induced electrocompetent cells of the recombineering strain mc2155:pJV53, prepared as described previously (van Kessel and Hatfull, 2007), were transfected with 100–300 ng phage DNA and 50 ng each of two complementary 71-mer recombineering oligonucleotides (oligos) containing the mutation to be incorporated centrally located. Reactions were recovered for approximately 1 hour in 7H9 with 10% ADC and 1 mM CaCl2 and plated prior to lysis as top agar lawns with ~300 μl M. smegmatis mc2155 cells. Plaques were picked into 100 μl phage buffer, and 1 μl of these were screened by PCR using the highly sensitive mismatch amplification mutation assay (MAMA)-PCR (Swaminathan et al., 2001) and Platinum Taq High Fidelity DNA Polymerase (Invitrogen). Primary plaques containing a mixture of mutant and wild-type alleles were serially diluted, and re-plated on M. smegmatis. Individual secondary plaques were then screened for the mutation by MAMA-PCR, and mutants were confirmed by Sanger sequencing.
All oligonucleotides were purchased from IDT Inc; the 71-mer recombineering oligos were gel purified. PCR products were processed using QIAquick PCR-Purification (QIAGEN) or MinElute PCR Purification Kits (QIAGEN), eluting DNA in a minimal volume of sterile water.
Adsorption Assays
M. smegmatis mc2155, M. smegmatis Jucho, or M. tuberculosis mc27000 cells in mid-log phase were diluted to an OD600 of ~0.2 and infected with phage at an multiplicity of infection (M.O.I.) of 0.00005 – 0.01. At 10-minute intervals, aliquots were removed, the bacterial cells along with any adsorbed phage were pelleted by centrifugation, and the supernatant was serially diluted and tittered on a fresh lawn of M. smegmatis mc2155. The percentage of total infectious particles remaining in the supernatant was calculated and plotted.
Supplementary Material
Table S1. Oligonucleotides used in this study.
Mycobacteriophage host preference correlates with genome type
Mycobacteriophage expanded host range results from tail gene mutations
A diverse host population and easy host range expansion promotes phage diversity
Acknowledgments
We thank Christina Ferreira for excellent technical assistance and Ching-Chung Ko, Max Garber and Dan Russell for expert assistance with DNA sequencing. We are grateful to statistical assistance from Dr. Jeffrey Lawrence and advice from Dr. William Jacobs, Jr. We would also like to thank all the students, faculty, and teaching assistants in the Howard Hughes Medical Institute Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Biology ((HHMI SEA-PHAGES) program for the isolation and characterization of mycobacteriophages. We would like to thank Julian Baptiste, Alexandra Cathcart, Zane Foster, Forrest Guilfoile, Abby McPherson, Matthew Olm, Terrence Parker, Kathleen Pulice, Kate Rockenbach, Emilee Shine, Enoch Tse and Philip Williams for their helpful preparation of phage lysates. This work was supported in part by a grant to the University of Pittsburgh by the Howard Hughes Medical Institute in support of GFH under HHMI’s Professorship program, by National Institutes of Health grants GM093901 to GFH, and R21AR060382 to RLM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- Adriaenssens EM, Mattheus W, Cornelissen A, Shaburova O, Krylov VN, Kropinski AM, Lavigne R. Complete genome sequence of the giant Pseudomonas phage Lu11. J Virol. 2012;86:6369–6370. doi: 10.1128/JVI.00641-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Kanamaru S, Leiman P, Rossmann MG. The tail lysozyme complex of bacteriophage T4. Int J Biochem Cell Biol. 2003;35:16–21. doi: 10.1016/s1357-2725(02)00098-5. [DOI] [PubMed] [Google Scholar]
- Bae B, Ohene-Adjei S, Kocherginskaya S, Mackie RI, Spies MA, Cann IK, Nair SK. Molecular basis for the selectivity and specificity of ligand recognition by the family 16 carbohydrate-binding modules from Thermoanaerobacterium polysaccharolyticum ManA. J Biol Chem. 2008;283:12415–12425. doi: 10.1074/jbc.M706513200. [DOI] [PubMed] [Google Scholar]
- Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci. 2007;86:2536–2540. doi: 10.3382/ps.2007-00250. [DOI] [PubMed] [Google Scholar]
- Brussow H. Phages of dairy bacteria. Annu Rev Microbiol. 2001;55:283–303. doi: 10.1146/annurev.micro.55.1.283. [DOI] [PubMed] [Google Scholar]
- Ceyssens PJ, Lavigne R. Bacteriophages of Pseudomonas. Future Microbiol. 2010;5:1041–1055. doi: 10.2217/fmb.10.66. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JE, Graham SC, Kim NN, Stein PE, McNair R, Cachon-Gonzalez MB, Cox TM, Read RJ. Insights into Krabbe disease from structures of galactocerebrosidase. Proc Natl Acad Sci U S A. 2011;108:15169–15173. doi: 10.1073/pnas.1105639108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C, Krisch HM. The diversity and evolution of the T4-type bacteriophages. Res Microbiol. 2003;154:259–267. doi: 10.1016/S0923-2508(03)00069-X. [DOI] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner KJ, Hatfull GF. Mycobacteriophage L5 infection of Mycobacterium bovis BCG: implications for phage genetics in the slow-growing mycobacteria. Mol Microbiol. 1997;26:755–766. doi: 10.1046/j.1365-2958.1997.6111984.x. [DOI] [PubMed] [Google Scholar]
- Gallet R, Shao Y, Wang IN. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evol Biol. 2009;9:241. doi: 10.1186/1471-2148-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg D, King J. Trimeric intermediate in the in vivo folding and subunit assembly of the tail spike endorhamnosidase of bacteriophage P22. Proc Natl Acad Sci U S A. 1982;79:3403–3407. doi: 10.1073/pnas.79.11.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. Inquiry learning. Teaching scientific inquiry. Science. 2006;314:1880–1881. doi: 10.1126/science.1136796. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Mycobacteriophages: genes and genomes. Annu Rev Microbiol. 2010;64:331–356. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Complete Genome Sequences of 138 Mycobacteriophages. J Virol. 2012a;86:2382–2384. doi: 10.1128/JVI.06870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF. The secret lives of mycobacteriophages. Adv Virus Res. 2012b;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WR, Jr, Lawrence JG, Hendrix RW. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Fan X, Xie J. Comparative genomic structures of Mycobacterium CRISPR-Cas. J Cell Biochem. 2012;113:2464–2473. doi: 10.1002/jcb.24121. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M, O’Sullivan O, Sleator RD, Coffey A, Ross RP, McAuliffe O, O’Mahony JM. In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene. 2010;453:9–23. doi: 10.1016/j.gene.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hopkins N, Ptashne M. Genetics of virulence. In: Hershey AD, editor. The Bacteriophage Lambda. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1971. pp. 571–574. [Google Scholar]
- Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage ‘resistome’. Curr Opin Microbiol. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217–248. doi: 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- Ichiyama S, Shimokata K, Tsukamura M. Relationship between mycobacterial species and their carotenoid pigments. Microbiol Immunol. 1988;32:473–479. doi: 10.1111/j.1348-0421.1988.tb01407.x. [DOI] [PubMed] [Google Scholar]
- Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, Kokjohn TA. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol. 1998;64:575–580. doi: 10.1128/aem.64.2.575-580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Jr, Greenberg J. Host modification and restriction with a mycobacteriophage isolated from a pseudolysogenic Mycobacterium chelonei. J Gen Microbiol. 1977;99:389–395. doi: 10.1099/00221287-99-2-389. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. Evolution of bacteriophage tails: Structure of T4 gene product 10. J Mol Biol. 2006;358:912–921. doi: 10.1016/j.jmb.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Lobocka M, Hejnowicz MS, Dabrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dabrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Glowacka A. Genomics of Staphylococcal Twort-like Phages - Potential Therapeutics of the Post-Antibiotic Era. Adv Virus Res. 2012;83:143–216. doi: 10.1016/B978-0-12-394438-2.00005-0. [DOI] [PubMed] [Google Scholar]
- Loessner MJ, Busse M. Bacteriophage typing of Listeria species. Appl Environ Microbiol. 1990;56:1912–1918. doi: 10.1128/aem.56.6.1912-1918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ, Calendar R. The Listeria Bacteriophages. In: Calendar R, editor. The Bacteriophages. Oxford University Press; New York, NY: 2006. pp. 593–601. [Google Scholar]
- Marinelli LJ, Fitz-Gibbons S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, Kim J, Miller JF, Hatfull GF, Modlin RL. Propionibacterium acnes bacteriophages display limited genetic diveristy ajd broad killing activity against bacterial skin isolates. mBio. 2012 doi: 10.1128/mBio.00279-12. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron PA, Mougel C, Ranjard L. Soil microbial diversity: Methodological strategy, spatial overview and functional interest. C R Biol. 2011;334:403–411. doi: 10.1016/j.crvi.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi Y, Tokunaga T. Recombination between Mycobacterium smegmatis strains Jucho and Lacticola. Jpn J Microbiol. 1971;15:359–366. doi: 10.1111/j.1348-0421.1971.tb00592.x. [DOI] [PubMed] [Google Scholar]
- Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Parsons LM, Jankowski CS, Derbyshire KM. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Microbiol. 1998;28:571–582. doi: 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Kanaar R, van de Putte P. A genetic switch in vitro: DNA inversion by Gin protein of phage Mu. Proc Natl Acad Sci U S A. 1984;81:2689–2692. doi: 10.1073/pnas.81.9.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, DeJong RJ, Elgin SCR, Guilfoile FR, Forsyth MH, Harris AD, Harvey SE, Hughes LE, Hynes PM, Jackson AS, Jalal MD, MacMurray EA, Manley CM, McDonough MJ, Mosier JL, Osterbann LJ, Rabinowitz HS, Rhyan CN, Russell DA, Saha MS, Shaffer CD, Simon SE, Sims EF, Tovar IG, Weisser EG, Wertz JT, Weston-Hafer KA, Williamson KE, Zhang B, Cresawn SG, Jain P, Piuri M, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Cluster K Mycobacteriophages: Insights into the Evolutionary Origins of Mycobacteriophage TM4. PLoS ONE. 2011a;6:e26750. doi: 10.1371/journal.pone.0026750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD, Anderson AG, Ang AAS, Ares M, Jr, Barber AJ, Barker LP, Barrett JM, Barshop WD, Bauerle CM, Bayles IM, Belfield KL, Best AA, Borjon A, Jr, Bowman CA, Boyer CA, Bradley KW, Bradley VA, Broadway LN, Budwal K, Busby KN, Campbell IW, Campbell AM, Carey A, Caruso SM, Chew RD, Cockburn CL, Cohen LB, Corajod JM, Cresawn SG, Davis KR, Deng L, Denver DR, Dixon BR, Ekram S, Elgin SCR, Engelsen AE, English BEV, Erb ML, Estrada C, Filliger LZ, Findley AM, Forbes L, Forsyth MH, Fox TM, Fritz MJ, Garcia R, George ZD, Georges AE, Gissendanner CR, Goff S, Goldstein R, Gordon KC, Green RD, Guerra SL, Guiney-Olsen KR, Guiza BG, Haghighat L, Hagopian GV, Harmon CJ, Harmson JS, Hartzog GA, Harvey SE, He S, He KJ, Healy KE, Higinbotham ER, Hildebrandt EN, Ho JH, Hogan GM, Hohenstein VG, Holz NA, Huang VJ, Hufford EL, Hynes PM, Jackson AS, Jansen EC, Jarvik J, Jasinto PG, Jordan TC, Kasza T, Katelyn MA, Kelsey JS, Kerrigan LA, Khaw D, Kim J, Knutter JZ, Ko CC, Larkin GV, Laroche JR, Latif A, Leuba KD, Leuba SI, Lewis LO, Loesser-Casey KE, Long CA, Lopez AJ, Lowery N, Lu TQ, Mac V, Masters IR, McCloud JJ, McDonough MJ, Medenbach AJ, Menon A, Miller R, Morgan BK, Ng PC, Nguyen E, Nguyen KT, Nguyen ET, Nicholson KM, Parnell LA, Peirce CE, Perz AM, Peterson LJ, Pferdehirt RE, Philip SV, Pogliano K, Pogliano J, Polley T, Puopolo EJ, Rabinowitz HS, Resiss MJ, Rhyan CN, Robinson YM, Rodriguez LL, Rose AC, Rubin JD, Ruby JA, Saha MS, Sandoz JW, Savitskaya J, Schipper DJ, Schnitzler CE, Schott AR, Segal JB, Shaffer CD, Sheldon KE, Shepard EM, Shepardson JW, Shroff MK, Simmons JM, Simms EF, Simpson BM, Sinclair KM, Sjoholm RL, Slette IJ, Spaulding BC, Straub CL, Stukey J, Sughrue T, Tang TY, Tatyana LM, Taylor SB, Taylor BJ, Temple LM, Thompson JV, Tokarz MP, Trapani SE, Troum AP, Tsay J, Tubbs AT, Walton JM, Wang DH, Wang H, Warner JR, Weisser EG, Wendler SC, Weston-Hafer KA, Whelan HM, Williamson KE, Willis AN, Wirtshafter HS, Wong TW, Wu P, Yang Yj, Yee BC, Zaidins DA, Zhang B, Zúniga MY, Hendrix RW, Hatfull GF. Expanding the Diversity of Mycobacteriophages: Insights into Genome Architecture and Evolution. PLoS ONE. 2011b;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond WB, Carter JC. A bacteriophage specific to Mycobacterium tuberculosis varieties hominis and bovis. Am Rv Respir Dis. 1960;82:781–786. doi: 10.1164/arrd.1960.82.6.781. [DOI] [PubMed] [Google Scholar]
- Roberts GA, Stephanou AS, Kanwar N, Dawson A, Cooper LP, Chen K, Nutley M, Cooper A, Blakely GW, Dryden DT. Exploring the DNA mimicry of the Ocr protein of phage T7. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, Goodman RM, Handelsman J. The Earth’s bounty: assessing and accessing soil microbial diversity. Trends Biotechnol. 1999;17:403–409. doi: 10.1016/s0167-7799(99)01352-9. [DOI] [PubMed] [Google Scholar]
- Rybniker J, Kramme S, Small PL. Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis--application for identification and susceptibility testing. J Med Microbiol. 2006;55:37–42. doi: 10.1099/jmm.0.46238-0. [DOI] [PubMed] [Google Scholar]
- Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR., Jr Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine. 2006;24:6309–6320. doi: 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, Russell D, Hendrix RW, Hatfull GF. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology. 2009;155:2962–2977. doi: 10.1099/mic.0.030486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol Microbiol. 1994;12:343–350. doi: 10.1111/j.1365-2958.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Sarkis GJ, Hatfull GF. Mycobacteriophages. Methods Mol Biol. 1998;101:145–173. doi: 10.1385/0-89603-471-2:145. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9:34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S, Tyagi AK. MchAI and MchAII, two class-II restriction endonucleases from Mycobacterium chelonei. Gene. 1993a;132:119–123. doi: 10.1016/0378-1119(93)90523-6. [DOI] [PubMed] [Google Scholar]
- Shankar S, Tyagi AK. Purification and characterization of restriction endonuclease MgoI from Mycobacterium gordonae. Gene. 1993b;131:153–154. doi: 10.1016/0378-1119(93)90686-w. [DOI] [PubMed] [Google Scholar]
- Smith DL, Rooks DJ, Fogg PC, Darby AC, Thomson NR, McCarthy AJ, Allison HE. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics. 2012;13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Ellis HM, Waters LS, Yu D, Lee EC, Court DL, Sharan SK. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Tokunaga T, Mizuguchi Y, Suga K. Genetic recombination in mycobacteria. J Bacteriol. 1973;113:1104–1111. doi: 10.1128/jb.113.3.1104-1111.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nature Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- Wang J, Parsons LM, Derbyshire KM. Unconventional conjugal DNA transfer in mycobacteria. Nat Genet. 2003;34:80–84. doi: 10.1038/ng1139. [DOI] [PubMed] [Google Scholar]
- Wilhelm SW, Brigden SM, Suttle CA. A dilution technique for the direct measurement of viral production: a comparison in stratified and tidally mixed coastal waters. Microb Ecol. 2002;43:168–173. doi: 10.1007/s00248-001-1021-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotides used in this study.