Abstract
Transglutaminase 2 (TG2) is a widely expressed and multifunctional protein that modulates cell death/survival processes. We have previously shown that TG2 binds to hypoxia inducible factor 1β (HIF1β) and decreases the upregulation of HIF responsive genes; however, the relationship between these observation was not investigated. In this study, we investigated whether endogenous TG2 is sufficient to suppress HIF activity and whether the interaction between TG2 and HIF1β is required for this suppression. shRNA-mediated silencing of TG2 significantly enhanced HIF activation in response to hypoxia. In addition, nuclear localization of TG2 is required for its suppressive effect on HIF activity, with TG2 being recruited to HIF responsive promoters in hypoxic conditions. These observations suggest that TG2 directly regulates hypoxic transcriptional machinery; however, its interaction with HIF1β was not required for this regulation. We also examined whether TG2’s effect on cell death/survival processes in ischemia is due to its effects on HIF signaling. Our results indicate that TG2 mediated HIF suppression can be separated from TG2’s effect on cell survival in hypoxic/hypoglycemic conditions. Lastly, here we show that nuclear TG2 in the closed conformation and non-nuclear TG2 in the open conformation have opposing effects on hypoxic/hypoglycemic cell death, which could explain previous controversial results. Overall, our results further clarify the role of TG2 in mediating the cellular response to ischemia and suggest that manipulating the conformation of TG2 might be of pharmacological interest as a therapeutic strategy for the treatment of ischemia-related pathologies.
Keywords: ischemia, hypoxia, transglutaminase 2, cell death, transcription
1. INTRODUCTION
Transglutaminase 2 (TG2, EC 2.3.2.13) is a multifunctional protein [1] which, in addition to its role in catalyzing calcium-dependent transamidation reactions [2], exhibits GTPase [3], protein disulfide isomerase [4] and possibly even a protein kinase activity [5]. Further, TG2 can regulate cellular function by binding to other proteins resulting in a change in their activity and/or localization [6–8]. The human transglutaminase (TG) family consists of 9 members, and TG2 is the most ubiquitously expressed and most studied member of the family [2]. Although TG2 plays a role in numerous cellular processes, its function in regulating cell survival/death has been an area of particular interest [9].
TG2 was originally considered to be a pro-cell death protein [10, 11], which certainly can be the case when its transamidating activity is robustly increased [12, 13]. However, it is increasingly apparent that TG2 often plays a pro-survival role [8, 12, 14–16, 17]. Interestingly, in many cases the pro-survival effects of TG2 seem to be independent of its transamidating activity [8, 18–20]. There is also a growing awareness that the subcellular localization plays a role in determining the role of TG2 in cell survival processes [8, 19]. Furthermore, the conformation of TG2 could be important for its role in cell survival/death processes. TG2 is proposed to have open and closed conformers, with notably distinct features [21, 22]. The closed form is used as an equivalent to GTP/GDP bound form, which is thought to be more compact than the open conformer. In a previous study, we found that the open conformer was toxic to immortalized mouse striatal neurons [23].
TG2 is primarily cytosolic, however the presence of TG2 in the nucleus is well-documented [24, 25]. More interestingly, a pattern is emerging where TG2 exhibits increased nuclear localization in response to cell stress and this may be a protective response. Earlier reports suggested that treatment with sphingosine [26], glutamate [27] or maitotoxin [24] caused nuclear accumulation of TG2. More recently Tatsukawa et. al. demonstrated that the ethanol-induced stress in hepatic cells resulted in significant TG2 accumulation in the nucleus [28]. We [7, 29] and others [30] have shown that hypoxic/ischemic stress also significantly increases nuclear TG2 levels. Therefore, it is reasonable to expect a functional outcome from this stress-induced nuclear accumulation, and it is becoming evident that this outcome is most likely transcriptional regulation [7, 28–32].
Numerous studies have provided evidence that TG2 does indeed modulate transcriptional processes. For example, TG2 attenuates AP-1 activation by binding to c-Jun which decreases its interaction with c-fos [31], and in mutant huntingtin-expressing cells TG2 repressed the expression of PGC-1α and cytochrome c [32]. TG2 also has been reported to increase NF-κB activation in cancer cells [30, 31, 33]. We have previously shown that TG2 accumulates in the nucleus in response to hypoxic and ischemic stress in neurons; and it can interact with hypoxia inducible factor 1β (HIF1β) and suppress HIF-dependent transcription. HIFs are heterodimeric transcription factors which are induced by hypoxia. Both subunits, α and β, are members of the PAS (PER-ARNT-SIM) protein family which contain a basic helix-loop-helix (bHLH) DNA binding domain [34–39]. In these ischemia models increased TG2 expression was protective and specifically targeting TG2 to the nucleus enhanced this effect [7, 19, 29]. Recently, an exciting study from our lab showed that, in contrast to TG2 expression in neurons, astrocytic TG2 expression is detrimental to the survival of astrocytes and negatively impacts their ability to protect neurons from ischemic insult [40]. The reasons for the cell type specific effects of TG2 on survival in ischemic conditions are currently unknown and an area of investigation.
In this study, we focused on dissecting the role of TG2 in hypoxic signaling. We now demonstrate that knocking down endogenous TG2 results in increased HIF signaling in neurons and in a human neuroblastoma cell line, SH-SY5Y. Using nuclear localization signal (NLS) and nuclear export signal (NES) tagging of TG2 to manipulate the levels of TG2 in the nucleus, we demonstrate that nuclear localization, but not the transamidating activity, of TG2 is required for suppression of HIF signaling. Furthermore, using chromatin immunoprecipitation (ChIP) assay, we demonstrate that TG2 is recruited to the hypoxia response element (HRE) bearing portion of the Enolase 1 promoter in hypoxic, but not normoxic conditions. This finding strongly suggests that TG2 is in a position to control hypoxia responsive transcription directly. We also show that the interaction between TG2 and HIF1β is not required for TG2 to repress HIF dependent transcription. Very intriguingly, the expression of the catalytic core domain of TG2, which lacks transamidase activity, was enough to enhance oxygen and glucose deprivation (OGD)-induced toxicity. This observation suggests that the conformation, not the activity, of TG2 may be more decisive in determining whether TG2 is pro-survival or pro-death in ischemia. Overall, this study strongly suggests that the role of TG2 in the hypoxic response likely includes regulating hypoxia responsive transcription by directly modulating the preinitiation complex responsible for the transcription of HIF target genes. However, our data suggest that its role in HIF signaling is unlikely to be sufficient to explain it role in ischemic cell death.
2. MATERIALS and METHODS
2.1 Constructs
The NES vector was created by cloning the nuclear export signal from PKI-α [41] into pcDNA 3.1 (+) (Invitrogen). Briefly, the sequence: 5’-atcgctcgagcccaacagcaatgaattagccttgaaattagcaggtcttgatatcaacaagacagaataggggcccatcg-3’ was commercially synthesized, cloned into pcDNA3.1(+) vector using the restriction enzyme sites XhoI and ApaI. The resulting NES-pcDNA3.1(+) construct was verified by sequencing. This vector was used previously in our lab to exclude another protein from the nucleus [42]. The remaining constructs that were created are described in Supplemental Table 1 and the primers used in this procedure are described in Supplemental Table 2.
2.2 Cell Culture
The preparation and treatment of primary cortical neurons is explained in the Appendix A. HEK 293A cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Irvine Scientific) supplemented with 5% fetal bovine serum (HyClone); MCF-7 cells were cultured in RPMI (Irvine Scientific) medium supplemented with 5% fetal bovine serum; SH-SY5Y cells were cultured in RPMI medium supplemented with 10% fetal bovine serum. All media were supplemented with 2 mM L-glutamine (Invitrogen, Life Technologies, Inc.), 100 µg/ml streptomycin (Invitrogen, Life Technologies, Inc.) and 100 units/ml penicillin (Invitrogen, Life Technologies, Inc.). Cells were grown in a humidified atmosphere containing 5% CO2 at 37°C. Transient transfections were carried out using FuGENE 6 reagent (Roche Applied Science) or Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. The SH-SY5Y cells which overexpress TG2 were created as described previously [43].
2.3 Cell Treatment Paradigms
24 h after transfection, the cells were transferred to serum-free media and incubated in a humidified atmosphere at 37°C containing 5% CO2 at the specified oxygen concentrations for the indicated lengths of time. For the cell survival/death studies, cells were maintained at 0.1% oxygen and no glucose (complete OGD); mild OGD refers to treatment in 0.2 % oxygen and 2.8 mM glucose. Control cells were maintained in serum free control media (25 mM glucose) and ambient oxygen. For the measurement of HIF activity, cells were incubated in serum free control media at 0.1% oxygen for the indicated lengths of time. Control cells were maintained in serum free control media and ambient oxygen.
For XRE luciferase assay, cells were transferred to serum free control media and treated with 10 µM 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD, dioxin) for 12 h in a humidified atmosphere at 37°C containing 5% CO2 and ambient oxygen. Immediately after the incubation the activity of the reporter was measured. Control cells were treated with vehicle alone, dimethyl sulfoxide (DMSO).
2.4 Dual luciferase assay
The HRE luciferase assay was carried out as described previously [19]. For the XRE luciferase assay, the MCF-7 cells were plated in a 24-well plate and transiently transfected with the TG2 constructs together with a luciferase vector under control of an XRE-bearing fragment from the CYP1A1 promoter (a generous gift from Dr. S. Kato) and a Renilla luciferase vector control [7] using FuGENE 6 reagent as described above. 24 h post-transfection, cells were transferred to serum free media and treated with TCDD or DMSO as described above. Luciferase activity was measured in cellular lysates using the Dual-Luciferase Reporter Assay System Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol and a TD-20/20 Luminometer. For each sample, the luciferase reporter data were normalized to the Renilla luciferase internal control.
2.5 Real time PCR
Following total RNA isolation, cDNA was synthesized using Superscript III first strand synthesis kit (Invitrogen) with oligo-dT primers according to manufacturer’s instructions. Real time PCR was conducted using Taqman gene expression system (Applied Biosciences) and Taqman primers for TG2 (Hs00190278_m1), BNIP3 (Hs00969291_m1), ENO1 (Hs00361415_m1), NOXA (Hs00560402_m1), EPO (Hs00171267_m1), VEGFA (Hs00173626_m1) and β-actin (Hs99999903_m1). Data was analyzed using the ΔΔCt method. Data were normalized to β-actin and expressed relative to normoxic values.
2.6 Cell survival and Cell death assays
The LDH release assay was carried out as described previously [19]. The calcein AM assay was modified from Ruan et. al. [44]. Calcein AM was dissolved in DMSO and was used at a final concentration of 2 µM in the cell media. It was incubated with the cells for 1 h and the fluorescent signal recorded using 485 nm excitation wavelength and 530 nm emission wavelength. The results are expressed relative to normoxic values.
2.7 Nuclear Fractionation
Nuclear fractionation studies were carried out as previously described [19]. The only modification was the final step, in which the crude nuclei were overlaid on the top of 0.5 – 0.7 M sucrose with protease inhibitors, and spun at 1200 × g for 10 min at 4 °C for HEK-293A cells and 2000 × g for 20 min at 4 °C for MCF cells.
Please see the supplementary information section for the remainder of the Materials and Methods.
3. RESULTS
3.1 TG2 regulates HRE activity in rat primary cortical neurons
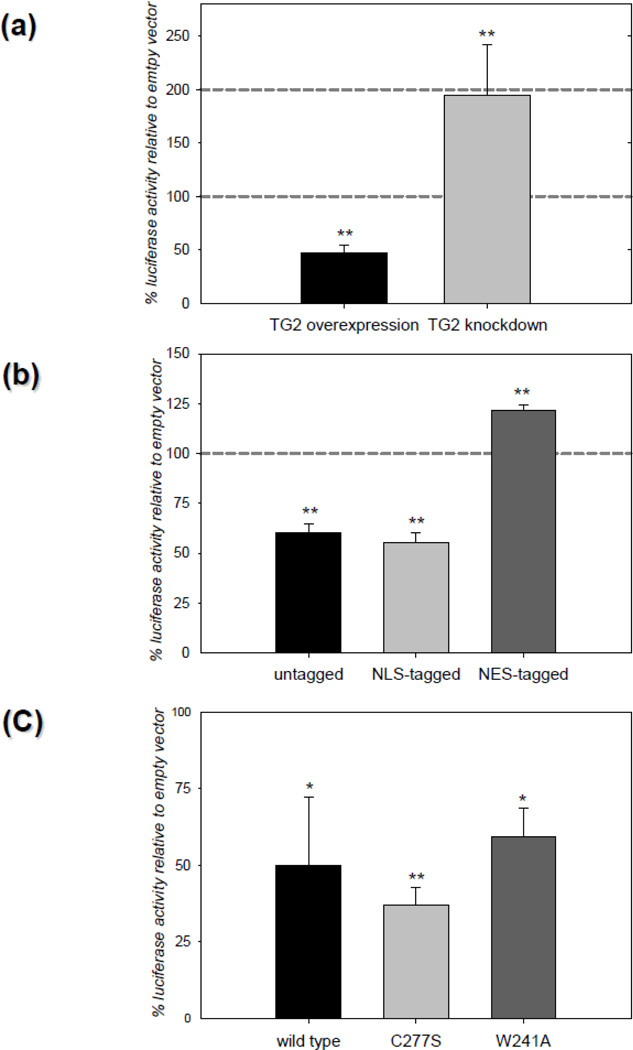
In our previous studies we showed that TG2 overexpression in neurons significantly decreased ischemic cell death in situ [7] and infarct volumes in vivo [29]. Therefore we examined the effects of TG2 on HIF signaling in primary neurons. Rat primary neurons were infected with either a lentivirus encoding human TG2 or the shRNA for TG2. As a control, neurons were transduced with control viral particles containing empty lentiviral vector. All the transduced neurons were subsequently transfected with HRE luciferase and renilla luciferase (internal control). 24 h after transfection, neurons were treated with 100 µM desferrioxamine (DFO) to stabilize HIF1α and increase HIF signaling. DFO had no significant effect on neuronal survival at this concentration (data not shown). DFO treatment and oxygen deprivation result in similar increases in the expression of HIF responsive genes [45, 46]. Data is presented as percent HRE reporter activity relative to control. TG2 overexpression in rat primary neurons reduced the HRE reporter activity ~50% compared to its own control and knocking down endogenous TG2 potentiated HRE reporter activity approximately 2 fold over the control (Figure 1a). When neurons were transfected with untagged TG2 or NLS-tagged TG2 along with HRE luciferase and renilla luciferase vectors and subsequently treated with DFO, HRE activity was decreased in a manner similar to what was observed when the neurons were infected with TG2. Also, as expected, the suppression of HRE activity was lost when the exogenously expressed TG2 was tagged with an NES (Figure 1b). To determine if transamidation activity is required for the suppression of HRE activity in rat primary neurons, the C277S-TG2 and W241A-TG2 constructs were used, both of which lack transamidating activity. These studies demonstrated that C277S-TG2 and W241A-TG2 suppressed HRE reporter activity significantly relative to empty vector (Figure 1c). These results show that in neurons, endogenous TG2, as well as exogenously expressed TG2, suppress HRE activation and that TG2 must localize to the nucleus to downregulate HRE activity. Further, the transamidation activity of TG2 is not required for the suppression of HRE activity in rat primary neurons. These data are in agreement with the results that we have obtained with cell lines as discussed below.
Figure 1. TG2 regulates HIF activity in rat primary cortical neurons.
(a) Endogenous TG2 is sufficient to suppress HIF activity in neurons. Primary neurons were transduced with hTG2 or TG2-shRNA viral particles prior to treatment with DFO for 24 h (chemical hypoxia) and measurement of HRE luciferase reporter activity. Increased TG2 expression significantly suppressed HRE activity (~50%) whereas knocking down endogenous TG2 significantly increased HRE activity (N=3). (b) Nuclear localization of TG2 is required to suppress HIF activity in neurons. Primary neurons were transiently transfected with control, untagged, NLS-tagged or NES-tagged TG2 constructs and the HRE and Renilla luciferase reporter constructs prior to chemical hypoxia and HRE activity measures as in (a). The untagged and NLS-tagged TG2 significantly suppressed HRE activity ~40%. NES-tagged TG2 significantly increased HRE activity ~20% (N=5). (c) Transamidase activity of TG2 is not involved in HIF suppression in neurons. Primary neurons were transiently transfected with control, wild type TG2, C277S-TG2 or W241A-TG2 constructs and the HRE and Renilla luciferase reporter constructs prior to chemical hypoxia and HRE activity measures as in (a). Both the C277S and W241A mutations abolish the transamidating activity of TG2. All three TG2 constructs significantly suppressed chemical hypoxia induced HIF activity relative to the control (N=4). Results are shown as mean ± SEM *p<0.05, **p<0.01.
3.2 TG2 regulates HIF activity and OGD-induced cell death in human neuroblastoma cells
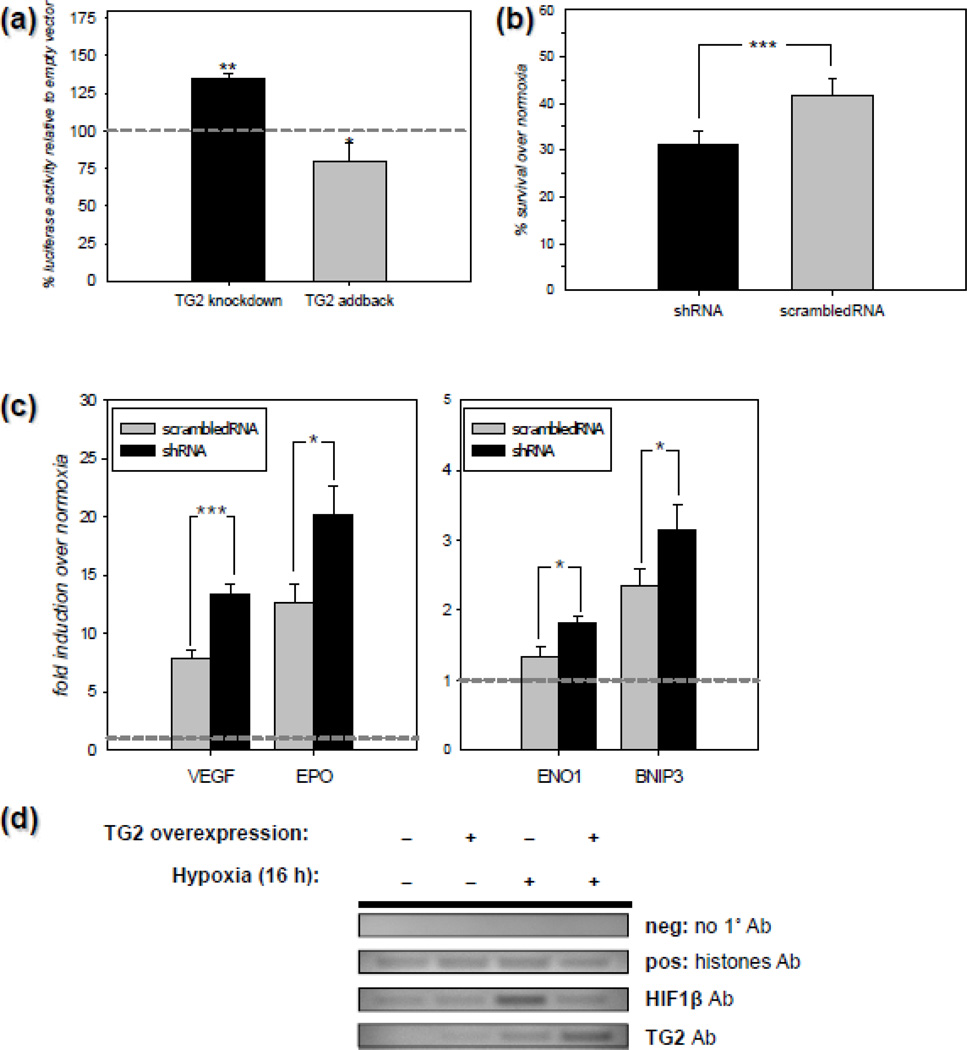
Data shown in Figure 1 clearly demonstrates that endogenous TG2 attenuates HRE activity in rat primary neurons in response to DFO, which is often used to increase HIF activity [45, 46]. However, in primary rat neurons we were unable to detect any increase in HRE activity using HRE luciferase reporters in response to oxygen deprivation, even though we tried numerous different paradigms (data not shown). Therefore, we used another cell type to determine whether endogenous TG2 can modulate HIF signaling in response to oxygen deprivation (i.e., true hypoxia). To this end, we used a human cell line of neuronal origin in order to analyze the ability of TG2 to modulate HIF signaling. In the first series of experiments, we knocked down endogenous TG2 by lentiviral delivery of a shTG2 construct to human neuroblastoma SH-SY5Y cells. In initial studies, the efficiency of TG2 knockdown with the lentiviral constructs was determined by transducting naïve SH-SY5Y human cells and SH-SY5Y cells stably overexpressing human TG2 with shRNA for TG2 [47] at two different dilutions (1:2 and 1:10 as described in Materials and Methods), or with a scrambled shRNA (scrRNA) [47] at one dilution (1:2) and subsequently collected and immunoblotted. TG2 immunoblots from naïve cells were exposed for a longer period of time to visualize the endogenous TG2 compared to the TG2 overexpressing cells. The data demonstrate that TG2-shRNA efficiently knocked down both endogenous and exogenous TG2. Tubulin was used as a loading control (Figure C1). The data was quantified by histogram analysis and it was found that the TG2 expression was decreased by ~51% at 1:10 dilution and ~ 85% at 1:2 dilution (Fig C1). After confirming that the knock down was efficient, naïve human SH-SY5Y neuroblastoma cells transduced with TG2-shRNA or scrRNA were transfected with an HRE luciferase reporter and a Renilla control construct and 24 h later, the cells were incubated for 20 h at 0.1% oxygen. Following incubation, the transcriptional activity was monitored by an HRE luciferase assay. The results shown in Figure 2a demonstrate that knocking down TG2 levels by shRNA significantly increased HRE-driven transcription in response to hypoxia as determined by the luciferase assay. When we transfected a human TG2 (addback) construct together with the HRE luciferase construct before hypoxic treatment, the increase in luciferase activity was attenuated (Figure 2a). To determine if endogenous TG2 positively or negatively affects cell survival in hypoxic/hypoglycemic conditions in addition to its negative effect on HIF activity, naïve SH-SY5Y cells were transduced as described above prior to OGD treatment. For these studies we used 0.2% oxygen and 2.8 mM glucose (~10% of the amount in regular media), which we refer to as ‘mild OGD’ for 20 h. Cell survival/death was monitored by calcein AM assay. As shown in Figure 2b, depleting endogenous TG2 by shRNA significantly reduced cell survival in SH-SY5Y cells in response to mild OGD. These results demonstrate that endogenous TG2 is sufficient to suppress HIF signaling and protect the neuroblastoma cells against OGD-induced cell death. HIF can induce both pro-survival and pro-death genes; therefore it is difficult to predict the overall effect of HIF suppression on cell survival. A selective suppression of pro-death HIF responsive genes might explain why TG2 is protective in this cell line. To test this possibility, we screened a small subset of HIF responsive genes. For this purpose, naïve human SH-SY5Y neuroblastoma cells infected with TG2-shRNA or scrRNA were incubated for at 0.1% oxygen for 6–12 h. Following incubation, the transcriptional activity was monitored by Q-RT-PCR analysis of several HIF responsive genes. Depleting endogenous TG2 by shRNA significantly increased the expression of EPO, ENO1, VEGF and BNIP3 (Figure 2c) indicating a lack of selectivity in the regulation of HIF responsive gene expression. We also checked the expression of Noxa but we did not detect a hypoxic upregulation of this gene (data not shown). To further elucidate the mechanisms by which TG2 suppresses HIF-dependent transcription, we next examined if TG2 is recruited into the complex that forms on HRE-bearing promoters in response to hypoxia. To this end, we used ChIP with primers that amplify ~150–200 bp portions from the promoters of human Eno1, BNIP3 and Noxa genes that bear functional HREs. As shown in Figure 2d, TG2 was recruited to the promoter of human Eno1 gene in response to 16 h of 0.1% oxygen treatment, while only trace amounts of TG2 could be detected in normoxic samples. Immunoprecipitation of histones was used as a positive control (Figure 2d). TG2 was also found on the human Bnip3 promoter after 6 h of 0.1% oxygen, however we did not detect TG2 at the HRE bearing portion of the Noxa promoter in any of the conditions that were used (data not shown).
Figure 2. TG2 regulates HIF activity and ischemic cell death in human neuroblastoma cells.
(a) Endogenous TG2 is sufficient to suppress HIF activity in SH-SY5Y cells. Relative HRE luciferase activity in SH-SY5Y cells after 20 h of 0.1% oxygen treatment compared to normoxia. Data is presented as percent of scrambled RNA or empty vector infection controls. Depleting endogenous TG2 by shRNA (TG2 knockdown) significantly increased HRE-driven luciferase activity and this activation was reversed by heterologous expression of human TG2 (TG2 addback) (N=5). (b) Depleting endogenous TG2 decreases survival after mild OGD in SH-SY5Y cells. Percent survival compared to normoxia/normoglycemia after 20 h of 2.8 mM glucose and 0.2% oxygen treatment as determined by the calcein-AM assay in SH-SY5Y cells. (N=7). (c) Depleting endogenous TG2 upregulates HIF target genes in SH-SY5Y cells. Fold induction in the expression of certain HIF target genes at the mRNA level following 6–12 h of 0.1% oxygen treatment determined by qRT-PCR. Depleting endogenous TG2 by shRNA significantly increased the expression of EPO, ENO1, VEGF and BNIP3 (N=3). (d) TG2 is recruited to the HRE spanning portions of the Eno1 promoter in SH-SY5Y cells in response to hypoxia. Representative agarose gel electrophoresis results from chromatin immunoprecipitation (ChIP) experiments (N=3). TG2 is recruited to the promoter of human Eno1 gene in response to 16 h of 0.1% oxygen treatment. Results are shown as mean ± SEM *p<0.05, **p<0.01.
3.3 Nuclear localization, but not transamidase activity, of TG2 is required to suppress HIF activity in different human cell lines
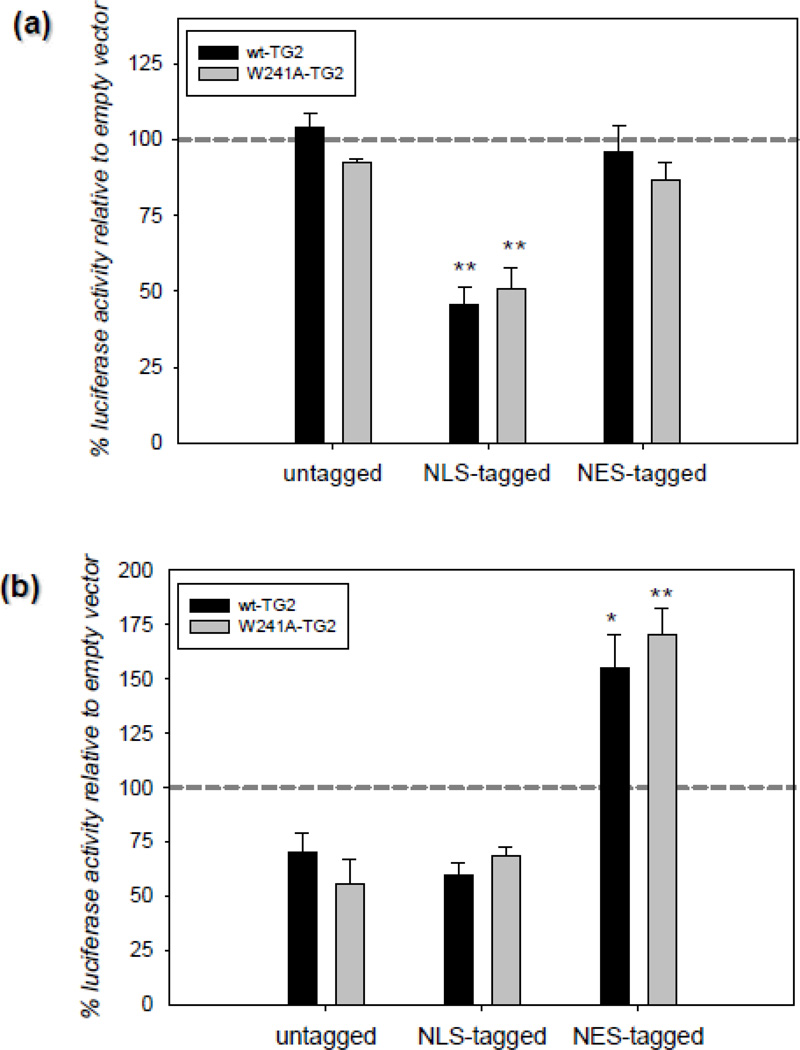
The fact that TG2 is physically present on HRE bearing promoters in hypoxia clearly suggest that it might be directly affecting the HIF-dependent transcriptional machinery. In order to further delineate this possibility, we designed an experiment in which we manipulated the nuclear TG2 amount by expressing it with an NLS or an NES tag. According to our rationale, if TG2 is suppressing HIF activity at the transcriptional machinery, there should be an inverse correlation between the nuclear TG2 amounts and the HIF activity. We carried out these studies in two human cell lines; HEK-293A (Figure 3a) and MCF-7 (Figure 3b) cells. These cell types, unlike SH-SY5Y cells, have low endogenous TG2 levels and therefore are better models for overexpression studies. HEK-293A and MCF-7 cells were transiently transfected with TG2 and luciferase constructs and 24 h post-transfection the cells were treated with 15 h of 0.1% oxygen. In HEK-293A cells, there were only trace amounts of TG2 in the nucleus in both normoxic and hypoxic conditions unless the TG2 that was expressed was specifically localized to the nucleus with an NLS-tag (data not shown). In line with this observation and our previous studies [19], NLS-tagged TG2 significantly suppressed HRE activity; whereas untagged and NES-tagged TG2 had no effect (Figure 3a). The same treatment paradigm was used in MCF-7 cells. In contrast to HEK-293A cells, untagged TG2 showed significant nuclear localization even under normoxic conditions (Figure C2). In MCF-7 cells both untagged and NLS-tagged TG2 tended to suppress HRE activity without reaching statistical significance. However, NES-tagged TG2 not only reversed this suppression, but resulted in a significant increase in HRE activity relative to vector controls (Figure 3b) (NES tag was confirmed to successfully export TG2 out of the nucleus by cellular fractionation studies; see Figure C2). The effects of TG2 on HRE activity were not dependent on transamidating activity, as a transamidating inactive form of TG2 (W241A) was as effective as wild type in suppressing HRE activity (Figure 3a & 3b). These results clearly show that there is an inverse correlation between the nuclear TG2 levels and HIF activity.
Figure 3. Nuclear localization, but not transamidase activity, of TG2 is required to suppress HIF activity in different human cell lines.
(a) Nuclear localization of TG2 is required to suppress HIF activity in HEK cells. Relative HRE luciferase activity in HEK-293A cells transiently transfected with TG2 constructs after 15 h of 0.1% oxygen treatment. Data is presented as percent of empty vector controls. NLS-tagged TG2 significantly suppressed HRE activity; whereas untagged and NES-tagged TG2 had no effect. The effect of the transamidating inactive TG2 (W241A-TG2) was the same as wild type TG2 (N=4). (b) Nuclear localization of TG2 is required to suppress HIF activity in MCF-7 cells. Relative HRE luciferase activity in MCF-7 cells transiently transfected with TG2 constructs after 15 h of 0.1% oxygen treatment. Data is presented as percent of empty vectors. NES-tagged TG2 significantly induced HRE activity. The effect of the transamidating inactive TG2 (W241A-TG2) was the same as wild type TG2 (N=4). Results are shown as mean ± SEM *p<0.05, **p<0.01.
3.4 All domains of TG2 are required for maximal transamidase activity
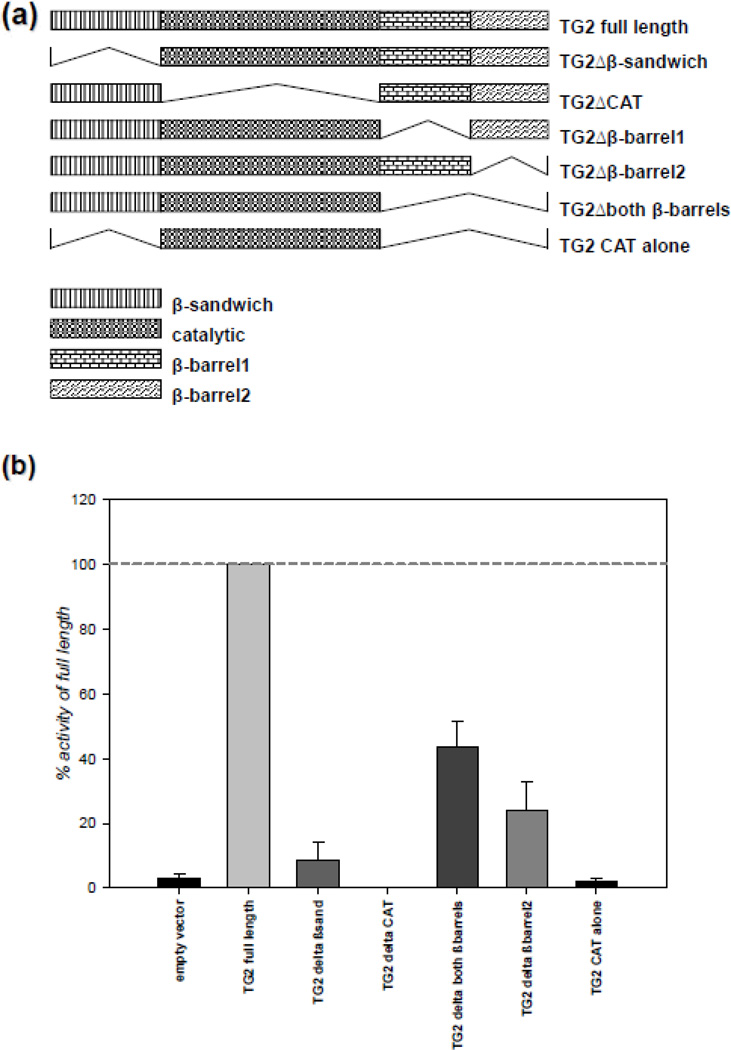
We previously demonstrated that TG2 interacts with HIF1β and suppresses HIF transcriptional activity [7, 19]. However, it was not clear whether there is a link between these two observations. To determine whether the TG2-HIF1β interaction is required for the TG2-dependent suppression of HIF signaling, we wanted to determine the interacting domain of TG2 with HIF1β. To this end, we made several TG2 constructs that lack certain domains and measured their in vitro transamidase activities prior to using them in our assays. The TG2 deletion constructs were made based on well defined domains within the protein [48]. Diagrams of these constructs are shown in Figure 4a, and their maximal in vitro transamidase activities are shown in Figure 4b. As expected, catalytic core domain deletion causes complete loss of transamidase activity of TG2. Also, N-terminal β sandwich domain was proved to be indispensible for the activity; however constructs lacking C-terminal β barrels retained some activity. Nevertheless, it should be noted that the deletion of β barrel domains also impairs guanine nucleotide binding; therefore, the in situ activity profiles of the C-terminal deletion construct could be different than the in vitro profiles.
Figure 4. All domains of TG2 are required for maximal transamidase activity.
(a) The schematic representation of TG2 deletions used. The TG2 deletions were based on the well-defined domains of TG2 which were determined by its crystal structure [48]. (b) N-terminal deletion causes complete and C-terminal deletion causes partial loss of transamidase activity of TG2. HEK-293A cells were transfected with V5-tagged TG2 deletion constructs and the cells were lysed after 48 h. Transamidase activity in the lysates was detected by a ELISA based assay and the results are presented as percent of full length TG2. (N=4). Results are shown as mean ± SEM.
3.5 Deletion of the catalytic domain of TG2 abolishes the interaction between TG2 and HIF1β
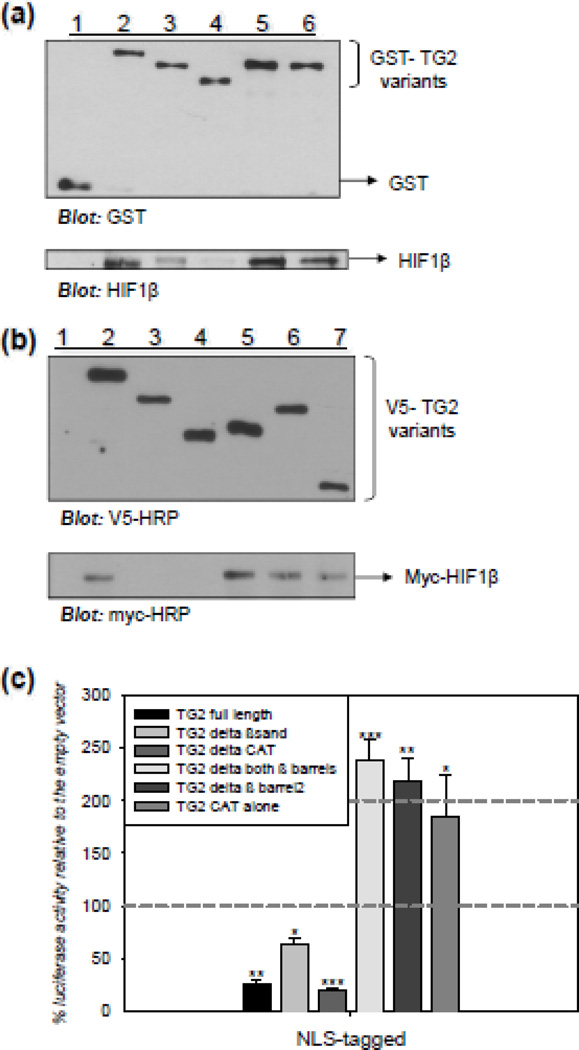
GST pull-down assays were used to determine the interactions between the constructs in vitro and co-immunoprecipitation studies were conducted to confirm the results. For the GST pull-down experiment, GST-HIF1β was expressed and used in the pull down assay after removal of the GST tag by PreScission Protease treatment as shown in Figure C3b. GST-pull-down assays were carried out using the GST-TG2 constructs (Fig C3a) as bait and HIF1β (Fig C3b) as prey. The results shown in Figure 5a demonstrate that HIF1β was pulled down by GST-TG2 (lane 2), GST-TG2Δβ-barrel1 (lane 5), and GST-TG2Δβ-barrel2 (lane 6); but not by GST alone (lane 1) or GST-TG2ΔCAT (lane 4). HIF1β was pulled down by GST-TG2Δβ-sandwich, but to a lesser extent (lane 3). These data indicate that the catalytic domain of TG2 is required for HIF1β and TG2 interactions. In addition, the deletion of the β sandwich domain from TG2 impaired the interaction with HIF1β, possibly because of the steric hindrance caused by the β barrel domains in the absence of the β sandwich domain. For the co-immunoprecipitation assay, the V5-TG2 variants were used as bait and the myc-HIF1β as prey. HEK-293A cells were transfected with V5-tagged TG2 deletion constructs and the myc-HIF1β construct and the assay was conducted 48 h post-transfection. The results shown in Figure 5b demonstrate that HIF1β interacts with full length V5-TG2 (lane 2), V5-TG2Δboth-β-barrels (lane 5), V5-TG2Δβ-barrel2 (lane 6), and V5-TG2-CAT alone (lane 7); but not with V5 tag alone (lane 1), V5-TG2Δβ-sandwich (lane 3), and V5-TG2ΔCAT (lane 4). The interaction between the catalytic domain alone with HIF1β (lane 7) is direct evidence which suggests that the catalytic core domain is the interacting domain.
Figure 5. Interacting with HIF1β is not required for TG2 to suppress HIF-dependent transcription.
(a) TG2 and HIF1β interact in vitro through the catalytic core domain of TG2. Representative immunoblot from a GST pull-down assay with GST-TG2 variants as bait and HIF1β as prey. Immunoblot analysis for HIF1β confirms that HIF1β was pulled down by full length GST-TG2 (lane 2), GST-TG2Δβ-barrel1 (lane 5), and GST-TG2Δβ-barrel2 (lane 6); but not by GST alone (lane 1) and GST-TG2ΔCAT (lane 4). HIF1β was pulled down by GST-TG2Δβ-sandwich to a lesser extent (lane 3). (b) TG2 and HIF1β interact through the catalytic core domain of TG2 in human cells. Representative immunoblot from a co-immunoprecipitation assay with V5-TG2 variants as bait and myc-HIF1β as prey. HEK-293A cells were transfected with V5-tagged TG2 deletion constructs and the myc-HIF1β construct. The cells were lysed after 48 h. Immunoblot analysis for myc confirms that HIF1β interacts with full length V5-TG2 (lane 2), V5-TG2Δboth-β-barrels (lane 5), V5-TG2Δβ-barrel2 (lane 6), and V5-TG2-CAT alone (lane 7); but not by V5 tag alone (lane 1), V5-TG2Δβ-sandwich (lane 3), and V5-TG2ΔCAT (lane 4). (c) Interacting with HIF1β is not required for TG2 to suppress HIF-dependent transcription. Relative HRE luciferase activity in MCF-7 cells transiently transfected with NLS-TG2 constructs after 15 h of 0.1% oxygen treatment in the presence of 2.5 µM NC9. Data is presented as percent of empty vectors. NLS-tagged TG2 full length, TG2Δβ-sandwich and TG2ΔCAT constructs significantly suppressed HRE activity; however, NLS-tagged TG2Δboth-β-barrels, TG2Δβ-barrel2 and TG2-CAT alone constructs significantly induced HRE activity (N=4). Results are shown as mean ± SEM *p<0.05, **p<0.01, ***p<0.005.
3.6 Interacting with HIF1β is not required for TG2 to suppress HIF-dependent transcription
After identifying the domain of TG2 required for interaction with HIF1β, we used this information to determine whether the interaction between TG2 and HIF1β is required for TG2 to modulate HIF-dependent transcription. To examine the effects of the TG2-HIF1β interactions on TG2-modulated transcription and to avoid confounding variables such as differential nuclear localization, we constructed the TG2 deletion variants as NLS-tagged proteins in mammalian expression vectors for these studies. MCF-7 cells were transiently transfected with NLS-TG2 full-length and deletion constructs. Again, in order to avoid other confounding variables, such as differential activities and the conformations of the deletion constructs, we used the irreversible TG inhibitor NC-9, which inhibits all activity and locks the protein in the open conformation [23]. Cells were transfected with TG2 and luciferase constructs and 24 h post-transfection they were treated with 15 h of 0.1% oxygen in the presence of 2.5 µM NC-9. The results are shown in Figure 5c. NLS-tagged TG2 full length, TG2Δβ-sandwich and TG2ΔCAT, which cannot interact with HIF1β, significantly suppressed HRE activity (Figure 5c). Very intriguingly, NLS-tagged TG2Δboth-β-barrels, TG2Δβ-barrel2 and TG2-CAT alone constructs significantly activated HRE (Figure 5c). Overall, this result suggests that, interacting with HIF1β is not required for TG2 to suppress HIF-dependent transcription.
3.7 Conformation and localization of TG2 is important in determining its role in OGD-induced cell death
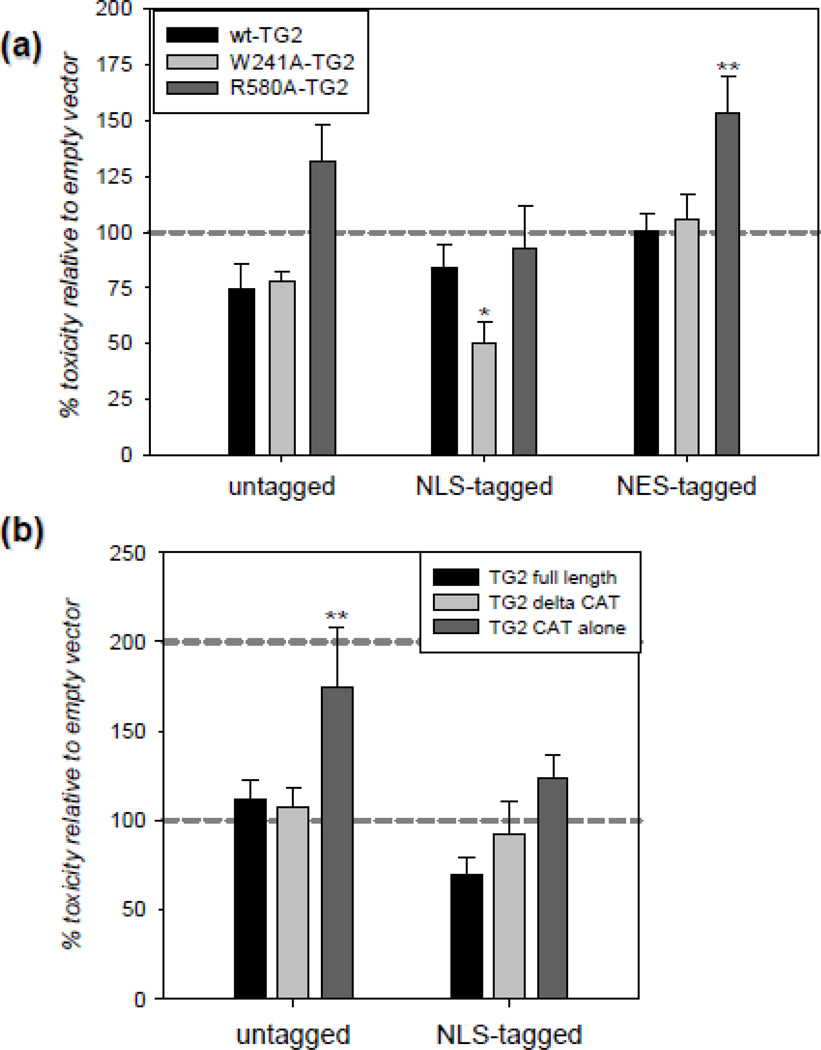
HEK-293A cells were transfected with untagged, NLS-tagged and NES-tagged TG2 constructs and 24 h post-transfection they were treated with 20 h of 2.8 mM g/L glucose and 0.2% oxygen. OGD-induced toxicity was measured with the LDH release assay and the data are presented as a percent of control (empty vector). For the first part of the cell death study, we used wild type TG2, a transamidating inactive mutant (W241A) which is in the closed conformation [19, 21] and a mutant that does not bind GTP and thus is more prone to attain open conformation and exhibit increased transamidating activity in situ (R580A) [19, 21]. LDH release was significantly decreased in cells which express NLS-tagged W241A-TG2; whereas, NES-tagged R580A [49] significantly increased ODG-induced toxicity (Figure 6a). Untagged-R580A tended to increase toxicity without reaching statistical significance (Figure 6a). For the second part of this study, we used full length V5-TG2, V5-TG2ΔCAT and V5-TG2-CAT alone constructs with or without an NLS tag. Only the V5-TG2-CAT alone construct without an NLS tag significantly increased OGD-induced toxicity; none of the other constructs significantly changed LDH release.
Figure 6. Effect of conformation and localization of TG2 on survival under OGD-induced stress in HEK-293A cells.
(a) Nuclear closed TG2 and non-nuclear open TG2 have opposing effects on survival in OGD. LDH release in HEK-293A after 20 h of mild OGD conditions. Data is normalized to normoxic/normoglycemic group and presented as percent of empty vector. LDH release is significantly decreased in HEK-293A cells which express NLS-tagged W241A-TG2; whereas, NES-tagged R580A-TG2 significantly increased LDH release. (N=6). (b) Non-nuclear expression of the catalytic domain of TG2 significantly enhances OGD-induced toxicity. LDH release in HEK-293A after 20 h of mild OGD conditions. Data is normalized to normoxic/normoglycemic group and presented as percent of empty vector. LDH release is significantly in increased in HEK-293A cells which express V5-TG2 CAT alone. (N=6). Results are shown as mean ± SEM *p<0.05, **p<0.01, ***p<0.005.
3.8 TG2 has no effect on xenobiotic responsive transcription
In addition to the HIF pathway, HIF1β is an important player in xenobiotic responsive transcription [50]. Given that TG2 binds HIF1β, TG2 could also attenuate xenobiotic response element (XRE) signaling; therefore we measured the effects of TG2 on XRE activity in MCF-7 cells. Treatment with 10 nM of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) for 12 h [51] resulted in a ~3 fold induction in XRE luciferase activity compared to vehicle alone (data not shown). However, none of the TG2 constructs had any significant effect on XRE activity (Figure C4).
4. DISCUSSION
Although we had previously shown that exogenous TG2 decreases HIF activity in human cell lines, in this study we demonstrate for the first time that endogenous TG2 also suppresses HIF signaling in rat primary neurons and in a human neuroblastoma cell line giving strong physiological relevance to our findings. We also show that TG2 must localize to the nucleus to suppress HIF activity and that TG2 is recruited to the HRE preinitiation complex. Further, nuclear localization, but not activity, of TG2 is essential for protection against OGD-induced cell death. In addition to these findings, we demonstrate that the catalytic domain of TG2 is required for its interaction with HIF1β; however this domain is neither necessary nor sufficient for TG2 to suppress HIF activity. On the other hand, the C-terminal β-barrel 2 domain does appear to be essential for TG2 to suppress HIF activity. These data suggest that the binding of HIF1β and TG2 and the ability of TG2 to suppress HIF activity are independent events.
HIF is composed of two subunits: a constitutively expressed β subunit (HIF1β), and the oxygen sensing HIFα subunit [52]. The HIF1α and HIF2α are structurally similar in their DNA binding and dimerization domains but differ in their transactivation domains and regulate unique target genes [53, 54]. In our previous report we demonstrated that overexpressed TG2 suppressed both HIF1 and HIF2 dependent transcription [7]. In this study, we examined the effects of TG2 on expression levels of both HIF1 and HIF2 targets in SH-SY5Y cells. ENO1 [53], and BNIP3 [55] are primarily targets for HIF1α, while EPO is preferentially regulated by HIF2α [56]. VEGF can be regulated by both HIF1α and HIF2α [53]. EPO and VEGF were highly upregulated in response to hypoxia, whereas ENO1 and BNIP3 were upregulated to a lesser extent. These results suggest that HIF2α may play a more prominent role in HIF signaling in SH-SY5Y cells, which is a neuroblastoma cell line. This is in line with a previous report which demonstrated the relevance of HIF2α in neuroblastoma models [57]. Nonetheless the hypoxiainduced expression of all HIF responsive genes we have examined thus far significantly increased when TG2 is knocked down. Overall, these results clearly show that endogenous TG2 modulates the transcription of HIF target genes. We examined both prodeath (BNIP3) and prosurvival (EPO and VEGF) genes in this study in order to see if TG2 has a selective effect. From our limited set of genes, we could not detect any selective suppression which could tip the balance of survival and death in one direction; therefore the biological significance of TG2 mediated HIF suppression is not clear in the context of ischemic cell death.
The most straightforward mechanism for this suppression would be direct regulation of the transcriptional machinery forming on the HRE bearing promoters. The results of the ChIP assay (Figure 2d) clearly show that endogenous TG2 is recruited to the HRE spanning portions of HIF target genes in response to hypoxia in SH-SY5Y cells. The recruitment was only observed in hypoxic conditions, which strongly indicates that this recruitment has functional outcomes. Furthermore, we failed to detect any TG2 on the Noxa promoter in either hypoxic or normoxic conditions, which argues for specificity and functionality. An important question is whether the conformation of TG2 is important for the recruitment to promoters. Although we lack direct evidence, the likely answer is no; the deletion constructs (except the ones that lacked the β-barrel2 domain), which without question lack the native open or closed conformation of the full length protein, still suppressed HIF activity. If the recruitment to the promoter is necessary for HRE suppression, this observation clearly shows that β-barrel2, not the conformation of the protein is crucial in this phenomenon.
The presence of TG2 in preinitiation complexes has also been shown in other models. For example, in the presence of mutant huntingtin, TG2 localizes to the promoters for cytochrome c and PGC-1α [32]. Although TG2 is at HRE containing promoters, the exact mechanism by which TG2 is repressing HIF activity is still being investigated. At this point we speculate that TG2 is either mediating the recruitment of certain corepressors or preventing the recruitment of certain coactivators. If TG2 is suppressing HIF activity at the promoter of HIF responsive genes, an inverse correlation between the nuclear TG2 amounts and the HIF activity should be expected. The results shown in Figure 3 strongly indicate that there is indeed an inverse correlation. Whether there is a relationship between the TG2 - HIF1β interaction and TG2-mediated HIF suppression has been a fundamental question for our studies. To this end, we have demonstrated that the HIF1β interacting domain of TG2 is within its catalytic core domain (Figure 5a & 5b). Further, our results suggest that the interaction between HIF1β and TG2 is not required for TG2 to suppress HIF-dependent transcription (Figure 5c). The failure of TG2 to influence another HIF1β dependent pathway, namely xenobiotic responsive transcription (Figure C4), supports the conclusion that the TG2 - HIF1β interaction is not essential per se for transcriptional repression. However, this interaction might have other physiological roles. For example we have preliminary evidence suggesting that this interaction might be important for the nuclear accumulation of TG2 in hypoxia (data not shown). The question whether HIF1β binding affects in situ transamidating activity of TG2 is an interesting one. However, given that the transamidating activity of TG2 is not crucial for HRE suppression, this question was not central to our studies and we did not examine this possibility.
TG2 can differentially affect a signaling process in a cell type specific manner. Therefore it is crucial to exercise caution when interpreting data obtained through the use of more than one cell line. In this study we have used several cell models and the amount of nuclear TG2 varies greatly between the cell lines used. However if we manipulate nuclear TG2 amounts using genetic approaches, the outcome is very similar. This observation suggests that the mechanism of HRE suppression by TG2 is conserved among cell types and the amount of TG2 in the nucleus is the variable that determines whether TG2 suppresses HIF signaling or not. This common response among various cell types is informative in this regard. Therefore, using more than one cell line has advantages, as well as drawbacks. In a similar trade-off situation, we had to choose whether to manipulate the nuclear TG2 amounts through mutating the proposed endogenous NLS residues in the protein or by adding exogenous NLS and NES signals. Both approaches have its merits and drawbacks; however, mutating the endogenous NLS sites may have secondary unforeseeable effects on HIF signaling other than changing the localization of the protein. Furthermore, the presence of endogenous NLS sites in TG2 have not been unequivocally confirmed experimentally [32]. Therefore, the nuclear TG2 amounts were manipulated by exogenous tags in this study. It should be noted that this approach does not allow us to titrate the TG2 amounts in the nucleus; rather the use of NLS and NES tagged constructs allow us to relatively increase or decrease the nuclear TG2 amounts compared to untagged constructs. Since we lack the tools to quantitatively manipulate nuclear TG2 amounts, we are unable to test whether there is a linear relationship between nuclear TG2 amounts and survival in OGD.
The depletion of endogenous TG2 not only upregulated HIF target genes, but also decreased survival of SH-SY5Y cells in response to OGD. However it still needs to be established whether or not there is a causal relationship between the TG2-mediated HIF suppression and TG2-mediated improvement in survival. Considering that TG2 modulates many aspects of cell survival/death processes [19], it would not be surprising if they were not directly linked. More work is required to clarify the mechanisms by which TG2 protects cells against ischemic cell death. However the data we obtained using HEK-293A cells suggest that the protection conferred by TG2 in OGD and the suppression of HIF activity are separate phenomena. Both wild type and W241A-TG2 with NLS-tags were equally effective in suppressing HRE activity in HEK-293A cells (Figure 3a), although, the W241A mutant version, and not the wild type, protected the cells in OGD (Figure 6a). More importantly, NLS-V5-TG2ΔCAT and NLS-V5-TG2-CAT alone failed to diminish or facilitate OGD-induced cell death in HEK-293A cells (Figure 6b). However, their effect on HIF signaling was dramatically different: while NLS-V5-TG2ΔCAT strongly suppressed HIF activity, NLS-V5-TG2-CAT alone activated HIF (Figure 5c). This result indicates that the two events are not likely to be directly linked. Nonetheless, the extent of the present data does not allow us to rule out the possibility that TG2-dependent suppression of HIF is partly contributing to TG2-mediated protection in OGD.
Although some TG2 constructs protected HEK-293A cells against OGD-induced cell death, two of them, namely non-nuclear R580A (Figure 6a) and non-nuclear V5-TG2 CAT (Figure 6b), facilitated it. The R580 residue is located at the center of the guanine nucleotide binding site and has been shown to be indispensable for binding [19, 49]. Mutation of this residue, therefore, has two important outcomes: the mutant is more likely to exist in an open conformation (and hence certain domains which are buried in the closed conformation are exposed) and it exhibits higher transamidating activity inside the cell [19, 21, 49]. Therefore, the facilitation of OGD-induced cell death by R580A (Figure 6a) could be attributed either to this mutant’s conformation and/or its increased intracellular transamidase activity. Interestingly, the expression of the catalytic core domain without the NLS tag also facilitated OGD-induced cell death (Figure 6b). The catalytic core domain alone has no transamidating activity (Figure 4b), but it might present a potentially “toxic” region of the molecule due to the deletion of the N-terminal and C-terminal domains. Unless R580A-TG2 and the catalytic core domain exert their toxicities through different mechanisms; these data suggest that the conformation, not the disinhibited transamidating activity, of TG2 is facilitating the ODG-induced cell death. This conclusion is also supported by our previous findings which showed that the open conformation of TG2 exacerbated OGD-induced cell death in an immortalized striatal cell model [23].
5. CONCLUSION
HIF-dependent transcription has great relevance to at least two major pathologies: cancer [58] and stroke [59, 60]. Although initial studies indicated that the HIF responsive genes were largely pro-survival, it is now evident that the role of HIF activity in mediating cell death/survival pathways is complex and dependent on many variables such as cell type and duration and severity of the hypoxic episode [59–61]. Further, there are other important pathways which are activated in hypoxia such as NFκB and AP-1 [62–65] and modulated by TG2 [30, 31, 33]. Therefore it needs to be considered that TG2- mediated modulation of these or other pathways may also contribute to the protective role of nuclear TG2 in ischemia and further investigations are required to delineate their possible involvement.
In summary, these studies have provided significant insights into the TG2-mediated protection against, or facilitation of, ischemic cell death. These findings clearly demonstrate the dual nature of the role TG2 plays in modulating cell death processes depending on its localization and conformation. We also provide data supporting the physiological relevance of TG2 in mediating the cell’s response to hypoxia, as endogenous TG2 mediates HIF activity. Our data suggest that nuclear localization of TG2 is required for suppression of HIF activity and that this suppression is likely due to the direct regulation of hypoxia responsive transcription as TG2 localizes to HRE containing promoters. Overall, it is becoming clear that an important function of TG2 is the modulation of the transcription of hypoxic responsive genes, and that TG2 clearly plays an important role in the complicated survival or death decisions that occur in response to ischemic stress.
Supplementary Material
Highlights.
Endogenous TG2 suppresses HIF activity.
Nuclear localization of TG2 is necessary for suppression of HIF activity.
Interaction between TG2 and HIF1β is not required for HIF activity suppression.
TG2 is found on HIF target promoters in hypoxia.
Conformation of TG2 is crucial in determining the fate of the cell in ischemia.
Acknowledgements
The authors thank Dr. S. Kato for the XRE-luciferase reporter and Dr. R.S. Freeman for providing the HRE luciferase reporter. This work was supported by NIH grant NS065825.
Abbreviations
- TG2
transglutaminase 2
- HIF
hypoxia inducible factor
- HRE
hypoxia response element
- XRE
xenobiotic response element
- OGD
oxygen glucose deprivation
- NLS
nuclear localization signal
- NES
nuclear export signal
- CAT
catalytic
- ChIP
chromatin immunoprecipitation
- HEK
human embryonic kidney
- shRNA
short hairpin RNA
- scrRNA
scrambled RNA
- DFO
desferrioxamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gundemir S, Colak G, Tucholski J, Johnson GV. Transglutaminase 2: a molecular Swiss army knife. Biochimica et biophysica acta. 2012;1823:406–419. doi: 10.1016/j.bbamcr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 3.Nakaoka H, Perez DM, Baek KJ, Das T, Husain A, Misono K, Im MJ, Graham RM. Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y. A novel function of tissue-type transglutaminase: protein disulphide isomerase. Biochem J. 2003;373:793–803. doi: 10.1042/BJ20021084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra S, Melino G, Murphy LJ. Transglutaminase 2 kinase activity facilitates protein kinase A-induced phosphorylation of retinoblastoma protein. J Biol Chem. 2007;282:18108–18115. doi: 10.1074/jbc.M607413200. [DOI] [PubMed] [Google Scholar]
- 6.Akimov SS, Belkin AM. Cell-surface transglutaminase promotes fibronectin assembly via interaction with the gelatin-binding domain of fibronectin: a role in TGFbeta-dependent matrix deposition. J Cell Sci. 2001;114:2989–3000. doi: 10.1242/jcs.114.16.2989. [DOI] [PubMed] [Google Scholar]
- 7.Filiano AJ, Bailey CD, Tucholski J, Gundemir S, Johnson GV. Transglutaminase 2 protects against ischemic insult, interacts with HIF1beta, and attenuates HIF1 signaling. FASEB J. 2008;22:2662–2675. doi: 10.1096/fj.07-097709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milakovic T, Tucholski J, McCoy E, Johnson GV. Intracellular localization and activity state of tissue transglutaminase differentially impacts cell death. J Biol Chem. 2004;279:8715–8722. doi: 10.1074/jbc.M308479200. [DOI] [PubMed] [Google Scholar]
- 9.Mastroberardino PG, Piacentini M. Type 2 transglutaminase in Huntington's disease: a double-edged sword with clinical potential. Journal of internal medicine. 2010;268:419–431. doi: 10.1111/j.1365-2796.2010.02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliverio S, Amendola A, Rodolfo C, Spinedi A, Piacentini M. Inhibition of "tissue" transglutaminase increases cell survival by preventing apoptosis. J Biol Chem. 1999;274:34123–34128. doi: 10.1074/jbc.274.48.34123. [DOI] [PubMed] [Google Scholar]
- 11.Fesus L, Thomazy V, Falus A. Induction and activation of tissue transglutaminase during programmed cell death. FEBS letters. 1987;224:104–108. doi: 10.1016/0014-5793(87)80430-1. [DOI] [PubMed] [Google Scholar]
- 12.Tucholski J, Johnson GV. Tissue transglutaminase differentially modulates apoptosis in a stimuli-dependent manner. J Neurochem. 2002;81:780–791. doi: 10.1046/j.1471-4159.2002.00859.x. [DOI] [PubMed] [Google Scholar]
- 13.Tucholski J, Roth KA, Johnson GV. Tissue transglutaminase overexpression in the brain potentiates calcium-induced hippocampal damage. J Neurochem. 2006;97:582–594. doi: 10.1111/j.1471-4159.2006.03780.x. [DOI] [PubMed] [Google Scholar]
- 14.Mangala LS, Fok JY, Zorrilla-Calancha IR, Verma A, Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2007;26:2459–2470. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 15.Antonyak MA, Singh US, Lee DA, Boehm JE, Combs C, Zgola MM, Page RL, Cerione RA. Effects of tissue transglutaminase on retinoic acid-induced cellular differentiation and protection against apoptosis. J Biol Chem. 2001;276:33582–33587. doi: 10.1074/jbc.M105318200. [DOI] [PubMed] [Google Scholar]
- 16.Antonyak MA, Jansen JM, Miller AM, Ly TK, Endo M, Cerione RA. Two isoforms of tissue transglutaminase mediate opposing cellular fates. Proc Natl Acad Sci U S A. 2006;103:18609–18614. doi: 10.1073/pnas.0604844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang JY, Mangala LS, Fok JY, Lin YG, Merritt WM, Spannuth WA, Nick AM, Fiterman DJ, Vivas-Mejia PE, Deavers MT, Coleman RL, Lopez-Berestein G, Mehta K, Sood AK. Clinical and biological significance of tissue transglutaminase in ovarian carcinoma. Cancer Res. 2008;68:5849–5858. doi: 10.1158/0008-5472.CAN-07-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyer LM, Schooler KP, Ai L, Klop C, Qiu J, Robertson KD, Brown KD. The transglutaminase 2 gene is aberrantly hypermethylated in glioma. J Neurooncol. 2010 doi: 10.1007/s11060-010-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gundemir S, Johnson GV. Intracellular localization and conformational state of transglutaminase 2: implications for cell death. PLoS ONE. 2009;4:e6123. doi: 10.1371/journal.pone.0006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datta S, Antonyak MA, Cerione RA. GTP-binding-defective forms of tissue transglutaminase trigger cell death. Biochemistry. 2007;46:14819–14829. doi: 10.1021/bi701422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begg GE, Carrington L, Stokes PH, Matthews JM, Wouters MA, Husain A, Lorand L, Iismaa SE, Graham RM. Mechanism of allosteric regulation of transglutaminase 2 by GTP. Proc Natl Acad Sci U S A. 2006;103:19683–19688. doi: 10.1073/pnas.0609283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinkas DM, Strop P, Brunger AT, Khosla C. Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 2007;5:e327. doi: 10.1371/journal.pbio.0050327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colak G, Keillor JW, Johnson GV. Cytosolic guanine nucledotide binding deficient form of transglutaminase 2 (R580a) potentiates cell death in oxygen glucose deprivation. PLoS ONE. 2011;6:e16665. doi: 10.1371/journal.pone.0016665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesort M, Attanavanich K, Zhang J, Johnson GV. Distinct nuclear localization and activity of tissue transglutaminase. J Biol Chem. 1998;273:11991–11994. doi: 10.1074/jbc.273.20.11991. [DOI] [PubMed] [Google Scholar]
- 25.Singh US, Erickson JW, Cerione RA. Identification and biochemical characterization of an 80 kilodalton GTP-binding/transglutaminase from rabbit liver nuclei. Biochemistry. 1995;34:15863–15871. doi: 10.1021/bi00048a032. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi Y, Ohashi H, Birckbichler PJ, Ikejima T. Nuclear translocation of tissue type transglutaminase during sphingosine-induced cell death: a novel aspect of the enzyme with DNA hydrolytic activity. Z Naturforsch C. 1998;53:352–358. doi: 10.1515/znc-1998-5-609. [DOI] [PubMed] [Google Scholar]
- 27.Campisi A, Caccamo D, Li Volti G, Curro M, Parisi G, Avola R, Vanella A, Ientile R. Glutamate-evoked redox state alterations are involved in tissue transglutaminase upregulation in primary astrocyte cultures. FEBS letters. 2004;578:80–84. doi: 10.1016/j.febslet.2004.10.074. [DOI] [PubMed] [Google Scholar]
- 28.Tatsukawa H, Fukaya Y, Frampton G, Martinez-Fuentes A, Suzuki K, Kuo TF, Nagatsuma K, Shimokado K, Okuno M, Wu J, Iismaa S, Matsuura T, Tsukamoto H, Zern MA, Graham RM, Kojima S. Role of transglutaminase 2 in liver injury via cross-linking and silencing of transcription factor Sp1. Gastroenterology. 2009;136:1783–1795. doi: 10.1053/j.gastro.2009.01.007. e1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filiano AJ, Tucholski J, Dolan PJ, Colak G, Johnson GV. Transglutaminase 2 protects against ischemic stroke. Neurobiol Dis. 2010;39:334–343. doi: 10.1016/j.nbd.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang GY, Jeon JH, Cho SY, Shin DM, Kim CW, Jeong EM, Bae HC, Kim TW, Lee SH, Choi Y, Lee DS, Park SC, Kim IG. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene. 2010;29:356–367. doi: 10.1038/onc.2009.342. [DOI] [PubMed] [Google Scholar]
- 31.Ahn JS, Kim MK, Hahn JH, Park JH, Park KH, Cho BR, Park SB, Kim DJ. Tissue transglutaminase-induced down-regulation of matrix metalloproteinase-9. Biochem Biophys Res Commun. 2008;376:743–747. doi: 10.1016/j.bbrc.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 32.McConoughey SJ, Basso M, Niatsetskaya ZV, Sleiman SF, Smirnova NA, Langley BC, Mahishi L, Cooper AJ, Antonyak MA, Cerione RA, Li B, Starkov A, Chaturvedi RK, Beal MF, Coppola G, Geschwind DH, Ryu H, Xia L, Iismaa SE, Pallos J, Pasternack R, Hils M, Fan J, Raymond LA, Marsh JL, Thompson LM, Ratan RR. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol Med. 2010;2:349–370. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, Kunnumakkara AB, Kumar R, Aggarwal BB, Mehta K. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 34.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basichelix- loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 36.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- 37.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 38.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 39.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 40.Colak G, Johnson GV. Complete transglutaminase 2 ablation results in reduced stroke volumes and astrocytes that exhibit increased survival in response to ischemia. Neurobiol Dis. 2012;45:1042–1050. doi: 10.1016/j.nbd.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 42.Beagle B, Johnson GV. Differential modulation of TCF/LEF-1 activity by the soluble LRP6-ICD. PLoS ONE. 2010;5:e11821. doi: 10.1371/journal.pone.0011821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucholski J, Lesort M, Johnson GV. Tissue transglutaminase is essential for neurite outgrowth in human neuroblastoma SH-SY5Y cells. Neuroscience. 2001;102:481–491. doi: 10.1016/s0306-4522(00)00482-6. [DOI] [PubMed] [Google Scholar]
- 44.Ruan Q, Lesort M, MacDonald ME, Johnson GV. Striatal cells from mutant huntingtin knock-in mice are selectively vulnerable to mitochondrial complex II inhibitor-induced cell death through a non-apoptotic pathway. Hum Mol Genet. 2004;13:669–681. doi: 10.1093/hmg/ddh082. [DOI] [PubMed] [Google Scholar]
- 45.Chavez JC, Baranova O, Lin J, Pichiule P. The transcriptional activator hypoxia inducible factor 2 (HIF-2/EPAS-1) regulates the oxygen-dependent expression of erythropoietin in cortical astrocytes. J Neurosci. 2006;26:9471–9481. doi: 10.1523/JNEUROSCI.2838-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ran R, Xu H, Lu A, Bernaudin M, Sharp FR. Hypoxia preconditioning in the brain. Developmental neuroscience. 2005;27:87–92. doi: 10.1159/000085979. [DOI] [PubMed] [Google Scholar]
- 47.Robitaille K, Daviau A, Lachance G, Couture JP, Blouin R. Calphostin C-induced apoptosis is mediated by a tissue transglutaminase-dependent mechanism involving the DLK/JNK signaling pathway. Cell Death Differ. 2008;15:1522–1531. doi: 10.1038/cdd.2008.77. [DOI] [PubMed] [Google Scholar]
- 48.Liu S, Cerione RA, Clardy J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc Natl Acad Sci U S A. 2002;99:2743–2747. doi: 10.1073/pnas.042454899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Begg GE, Holman SR, Stokes PH, Matthews JM, Graham RM, Iismaa SE. Mutation of a critical arginine in the GTP-binding site of transglutaminase 2 disinhibits intracellular cross-linking activity. J Biol Chem. 2006;281:12603–12609. doi: 10.1074/jbc.M600146200. [DOI] [PubMed] [Google Scholar]
- 50.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 51.Klinge CM, Bowers JL, Kulakosky PC, Kamboj KK, Swanson HI. The aryl hydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT) heterodimer interacts with naturally occurring estrogen response elements. Mol Cell Endocrinol. 1999;157:105–119. doi: 10.1016/s0303-7207(99)00165-3. [DOI] [PubMed] [Google Scholar]
- 52.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 53.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol Cells. 2010;29:435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- 55.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rankin EB, Biju MP, Liu Q, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, Poellinger L, Pahlman S. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 58.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baranova O, Miranda LF, Pichiule P, Dragatsis I, Johnson RS, Chavez JC. Neuron-specific inactivation of the hypoxia inducible factor 1 alpha increases brain injury in a mouse model of transient focal cerebral ischemia. J Neurosci. 2007;27:6320–6332. doi: 10.1523/JNEUROSCI.0449-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helton R, Cui J, Scheel JR, Ellison JA, Ames C, Gibson C, Blouw B, Ouyang L, Dragatsis I, Zeitlin S, Johnson RS, Lipton SA, Barlow C. Brain-specific knock-out of hypoxia-inducible factor-1alpha reduces rather than increases hypoxic-ischemic damage. J Neurosci. 2005;25:4099–4107. doi: 10.1523/JNEUROSCI.4555-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor CT, Cummins EP. The role of NF-kappaB in hypoxia-induced gene expression. Ann N Y Acad Sci. 2009;1177:178–184. doi: 10.1111/j.1749-6632.2009.05024.x. [DOI] [PubMed] [Google Scholar]
- 63.Faller DV. Endothelial cell responses to hypoxic stress. Clin Exp Pharmacol Physiol. 1999;26:74–84. doi: 10.1046/j.1440-1681.1999.02992.x. [DOI] [PubMed] [Google Scholar]
- 64.Ruas JL, Poellinger L, Pereira T. Role of CBP in regulating HIF-1-mediated activation of transcription. J Cell Sci. 2005;118:301–311. doi: 10.1242/jcs.01617. [DOI] [PubMed] [Google Scholar]
- 65.Piret JP, Cosse JP, Ninane N, Raes M, Michiels C. Hypoxia protects HepG2 cells against etoposide-induced apoptosis via a HIF-1-independent pathway. Exp Cell Res. 2006;312:2908–2920. doi: 10.1016/j.yexcr.2006.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.