Abstract
The origin of embryos including those created through assisted reproductive technologies (ART) may have profound effects on placental and fetal development, possibly leading to compromised pregnancies associated with poor placental development. To determine the effects of embryo origin on fetal size, and maternal and fetal placental cellular proliferation and global methylation, pregnancies were achieved through natural mating (NAT), or transfer of embryos generated through in vivo (NAT-ET), IVF, or in vitro activation (IVA). On Day 22 of pregnancy, fetuses were measured and placental tissues were collected to immunodetect Ki67 (a marker of proliferating cells) and 5-methyl cytosine (5mC) followed by image analysis, and determination of mRNA expression for three DNA methyltransferases (DNMT). Fetal length and labeling index (proportion of proliferating cells) in maternal caruncles (CAR; maternal placenta) and fetal membranes (FM; fetal placenta) were less (P < 0.001) in NAT-ET, IVF and IVA than in NAT. Expression of 5mC was greater (P < 0.02) in IVF and IVA than in NAT. In CAR, mRNA expression for DNMT1 was greater (P < 0.01) in IVA compared to the other groups, but DNMT3A expression was less (P < 0.04) in NAT-ET and IVA than NAT. In FM, expression of mRNA for DNMT3A was greater (P < 0.01) in IVA compared to the other groups, and was similar in NAT, NAT-ET and IVF groups. Thus, embryo origin may have specific effects on growth and function of ovine utero-placental and fetal tissues through regulation of tissue growth, DNA methylation and likely other mechanisms. These data provide a foundation for determining expression of specific factors regulating placental and fetal tissue growth and function in normal and compromised pregnancies, including those achieved with ART.
Keywords: Growth, Global Methylation, Placenta, Early Pregnancy, Sheep
1. Introduction
Early pregnancy is a critical period, due to the major developmental events that occur, including embryonic organogenesis as well as formation of the placenta, a process known as placentation and manifested by enhanced cell proliferation and vascular development [1–6]. The pattern of placental and fetal growth during early pregnancy after natural breeding has been well established for sheep [1,5–7]. Comparison of the development of placenta in natural pregnancies and pregnancies achieved by various assisted reproductive technologies (ART), such as after transfer of embryos created through IVF, has demonstrated differences in placental and fetal growth in several species [9–13]. For example for cows, sheep and pigs, during early pregnancy fetuses created in vitro and then transferred can be of larger or smaller sizes than fetuses created in vivo [8,14–17]. However, data concerning fetal and placental growth, and especially during early pregnancy after transfer of embryos of different origin are very limited. Factors influencing fetal and placental growth have a dramatic impact on fetal and neonatal survival and development [1–3]. Recent observations indicate that compromised fetal growth impacts not only the neonatal period but also life-long health and productivity in humans and other mammalian species [18–20].
Methylation of DNA, regulated by DNA methyltransferases (DNMT) and other factors, plays a role during fetal and placental development by regulating gene expression, and typically involves methylation of cytosine residues (resulting in formation of 5-methyl cytosine [5mC]) in ‘CG islands’ located throughout the regulatory and coding regions of genes [21–24]. In compromised pregnancies, altered DNA methylation in the placenta may contribute to embryonic/fetal loss or impaired fetal growth [25]. Additionally, origin of the embryo, including those from ART, can alter methylation in the placenta, developing fetus or offspring in several mammalian species [26–30]. Little is known, however, about DNA methylation processes in placental tissues during early stages of normal or compromised pregnancies in any species.
We hypothesize that growth of the fetus and placenta and global methylation during early pregnancy is altered in pregnancies achieved through transfer of embryos generated from various sources, including those from ART compared with natural pregnancies. To test this hypothesis, we established pregnancies using a control group that was naturally mated (NAT), as well as three ART methods, as follows: (i) superovulation induced by multiple injections of follicle stimulating hormone (FSH) combined with natural mating, embryo flushing and transfer to recipients (NAT-ET); (ii) transfer of embryos obtained through IVF of oocytes collected from FSH-treated ewes; and (iii) transfer of embryos obtained through in vitro activation (IVA; i.e., parthenotes, which are clones containing only maternal genes) of oocytes collected from FSH-treated donors. In the NAT-ET group, embryos were only briefly removed from the uterine environment and had maternal and paternal genomes; in the IVF group, embryos were created in culture dishes and possessed both maternal and paternal genomes; and in the IVA group, embryos were created in culture dishes and had only the maternal genome. Parthenogenetic embryos have been used to study the effects of a lack of the paternal genome on embryonic development and expression and role of imprinted genes in several species [15,31–38].
Therefore, in this study we determined fetal size, cell proliferation, and global methylation (measured by expression of 5mC and mRNA for DNMT1, 3A and 3B) in fetal and maternal placenta during early pregnancy in NAT, NAT-ET, IVF, and IVA groups in sheep.
2. Materials and methods
2.1. Animals and Tissue Collection
The North Dakota State University Institutional Animal Care and Use Committee approved all animal procedures used in this study. Estrus was synchronized for ewes (n=67; adult Western range crossbred, primarily Rambouillet, Targhee and Columbia crossbred) randomly assigned to be naturally mated, or to serve as donors or recipients using a CIDR device (MWI, Boise, ID, USA) implanted for 14 d during the breeding season. At 24 h after CIDR removal, NAT ewes were exposed to a fertile ram and naturally mated, but for donor ewes from NAT-ET, IVF and IVA groups, estrus was checked twice daily using a vasectomized ram; 5, 86, and 7% of ewes expressed estrus at 24, 36 and 48 h after CIDR removal, respectively. Beginning on Day 13 of the estrous cycle, donor ewes (n=3) for the NAT-ET group were treated twice daily with FSH (Sioux Biochemical, Sioux Center, IA, USA) for 3 d (Day 13, 5 units/injection; Day 14, 4 units/injection and Day 15, 3 units/injection; unit is equivalent to 3.5 g of NIDDK-oFSH-20), whereas donor ewes (n=22) for the IVF and IVA groups were treated with FSH for 2 d (Days 13 and 14, doses as above) following estrus (d 0) as described before [10,39,40]. On Day 15 of the estrous cycle, ewes from the NAT-ET group were exposed to a fertile ram for 24 to 48 h, but for IVF and IVA groups ovaries were collected, and the oocytes were isolated, matured and then fertilized or activated in vitro as previously described in detail [10,36,40,41]. For breeding of NAT and donor ewes for NAT-ET group two Western range crossbred rams were used. Briefly, cumulus oocyte complexes (COC) were isolated from visible surface antral follicles >3 mm in diameter; the average number of COC collected per ewe was 19.3±1.6. For IVF and IVA procedures, COC (up to 30 COC/0.5 mL in a four-well Nunc culture dish) were incubated overnight in maturation medium (TCM199; Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Sigma), ovine FSH [5 μg/mL; oFSH-RP-1; NIAMDD-NIH, Bethesda, MD, USA], ovine LH [5 μg/mL; oLH-26; NIADDK-NIH], estradiol -17β [1 μg/mL; Sigma], glutamine [2 mM; Sigma], sodium pyruvate [0.25 mM; Sigma], epidermal growth factor [10 ng/mL; Sigma] and penicillin/streptomycin [100 units/mL penicillin and 100 μg/mL streptomycin; Gibco, Grand Island, NY, USA]). After denuding oocytes of cumulus cells [41], half of the oocytes from each ewe were used for IVF, whereas the remainder were used for IVA. For IVF, oocytes were cultured in fertilization medium in the presence of capacitated frozen-thawed sperm (0.5 to 1 × 106 sperm/mL; sperm pooled from five Western range crossbred rams) for 24 h followed by incubation in culture medium until embryo transfer (ET; see below). For IVA, oocytes were incubated for 5 min in TCM199 medium containing 2% FBS and ionomycin (2.5 μM; Sigma) followed by a 3-h incubation with 6-dimethylaminopurine (DMAP; 2 mM; Sigma). In vitro activated oocytes were then transferred to culture medium and incubated until ET (see below).
For the NAT-ET group, on Day 5 post-mating (Day 1 = day of mating), embryos were flushed, evaluated using a stereomicroscope, and then transferred to synchronized recipients (three embryos from the same donor/recipient). For the IVF and IVA groups, in vitro-generated embryos were transferred on Day 5 after fertilization or activation (Day 1 = day of fertilization or activation) to synchronized recipient ewes (three embryos from the same donor/recipient), as described by Grazul-Bilska et al. [10,40]. On Day 22 after mating, fertilization or activation, fetuses and utero-placental tissues were collected from NAT (n=8), NAT-ET (n=7), IVF (n=8), and IVA (n=7) groups. Pregnancy rates for the NAT, NAT-ET, IVF and IVA groups were 80, 78, 80, and 54%, respectively.
As we have previously described [5,6], to maintain specimen morphology for histology/immunohistochemistry, specimen pins were inserted completely through the uterus and FM at the level of the external intercornual bifurcation. Cross sections of the entire gravid uterus (approximately 0.5-cm thick) were the obtained using a Stadie-Riggs microtome blade (Thomas Scientific, Swedesboro, NJ, USA) followed by immersion in formalin (for Ki67 detection) or Carnoy’s solution (for 5mC detection) and embedding in paraffin. For total cellular RNA extraction, caruncular (CAR; maternal placenta) tissues and fetal membranes (FM; fetal placenta [chorioallantois]) were dissected from the area close to the embryo, snap-frozen in isopentane super-cooled in liquid nitrogen, and stored at −70 °C. Fetuses were separated from their fetal membranes, and crown-rump length of each fetus was measured. Day 22 was used for tissue collection, as in our previous experiments, we have demonstrated that on Days 20 to 22, major changes in cell proliferation, vascularization and expression of angiogenic factors occurred in fetal and maternal placenta in pregnancies achieved through natural mating [5,6]. In addition, in sheep by Days 20 to 22 after mating, placentation is already initiated [42].
2.2. Immunohistochemistry
Immunohistochemical procedures were performed as described [5,6]. Briefly, paraffin-embedded uterine tissues containing FM were sectioned at 4 μm and mounted onto glass slides, rinsed several times in PBS containing Triton-X100 (0.3%, v/v) and then treated for 20 min with blocking buffer (PBS containing normal horse serum [2%, vol/vol)]), followed by incubation overnight at 4° C with a specific primary antibody against Ki67 (1:500; mouse monoclonal; Vector Laboratories, Burlingame, CA, USA), an endogenous marker of proliferating cells, or 5mC (1:500; mouse monoclonal; Eurogentec North America, San Diego, CA, USA), a marker of global DNA methylation [43]. Primary antibodies were detected by using an anti-mouse secondary antibody coupled to peroxidase (ImPress Kit; Vector Laboratories) and SG as the peroxidase substrate (Vector Laboratories). Thereafter, sections were counterstained with nuclear fast red (Sigma) to visualize cell nuclei. Control sections were incubated with mouse IgG (4 μg/mL; Vector Laboratories) in place of the primary antibody. Fetal placental cell or tissue (e.g., chorion or allantois) types were not identified due to methodological difficulties, such as a lack of specific markers for these cell/tissue types in sheep or absence of some cell/tissue types in individual tissue sections; thus, the entire fetal placenta was used for immunohistological and other evaluations.
2.3. Image analysis
For each tissue section, images were taken at 400× (Ki67 staining) or 600× (5mC staining) magnification (using a Nikon Eclipse E600 microscope and digital camera) of 5 to 10 randomly-chosen fields (0.025 mm2 per field) from areas containing CAR, inter-CAR (ICAR) and FM, separately. To determine the labeling index (LI; percentage of cellular nuclei stained for Ki67) in CAR, ICAR and FM or the percentage of 5mC-positive area in cell nucleus in FM, an image analysis system (Image-Pro Plus, Media Cybernetics, Bethesda, MD, USA) was used as described [5,6].
2.4. Quantitative Real-Time RT-PCR
All procedures for determining the expression of mRNA for ovine placental genes by quantitative RT-PCR have been reported [6]. Briefly, snap-frozen FM tissues were homogenized in Tri-Reagent (Molecular Research Center; Cincinnati, OH, USA) according to the manufacturer’s specifications. The quality and quantity of total RNA were determined via capillary electrophoresis using the Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE, USA). Real-time RT-PCR reagents, probes, and primers were purchased from and used as recommended by Applied Biosystems (Foster City, CA, USA). For each sample, 30 ng of total RNA was reverse transcribed in triplicate 20-μL reactions using random hexamers. Sequence-specific Taqman probes and primers were designed using the Primer Express Software from Applied Biosystems, and sequences for DNMT1, 3A and 3B have been published [6]. An ABI PRISM 7000 instrument (Applied Biosystems) was used for detection of sequences amplified at 60 ° C, typically for 40 cycles. Quantification was determined relative to a standard curve of dilutions of cDNA generated from total RNA pooled from placental tissues collected on Day 130 of pregnancy. Expression of each gene was normalized to expression of 18S ribosomal RNA (rRNA) in a multiplex reaction using the human 18S pre-developed assay reagent (PDAR) from Applied Biosystems. The PDAR solution, which is primer limited and contains a VIC-labeled probe, was further adjusted by using one-fourth the normal amount, so that it would not interfere with amplification of the FAM-labeled gene of interest. Standard curves were also generated using the multiplex solution, and the quantity of 18S rRNA and the gene of interest were determined using each specific standard curve. The concentrations of mRNA were then normalized to 18S rRNA by dividing each of the mRNA values by their corresponding 18S rRNA value [6].
2.5. Statistical analyses
Data for fetal size, labeling index, and expression of 5mC and mRNA for DNMT1, 3A and 3B were analyzed using the general linear models (GLM) procedure of SAS with the main effect of embryo origin/pregnancy type [44]. When the F-test was significant (P<0.05), differences between specific means were determined by using the least significant differences (LSD) test [45]. Data are presented as means ± SEM.
3. Results
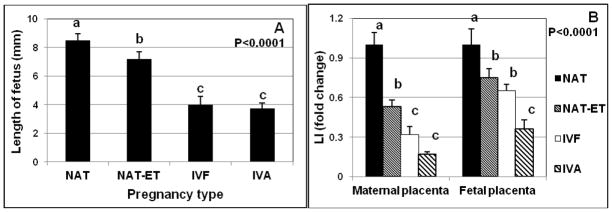
The length of the fetus was the greatest (P < 0.0001) in NAT group, less in NAT-ET, and least in the IVF and IVA groups (Fig. 1A). In the IVF and IVA groups, the length of the fetuses was approximately two-fold less than in the NAT group (Fig. 1A).
Fig. 1.
Mean (± SEM) length of the fetus (A) and labeling index (LI; percentage of proliferating cells) in maternal and fetal placenta (B) in NAT, NAT-ET, IVF and IVA groups on Day 22 of pregnancy in ewes. For LI, data are expressed as fold change compared to the NAT control group, which was arbitrarily set as 1. In the NAT group, the actual LI was 3.5±0.3% in maternal and 24.5±2.9% in fetal placenta.
a–cMeans without a common superscript differed (P < 0.0001).
The Ki67 protein, a marker of proliferating cells, was detected in the nuclei of fetal (FM) and maternal (CAR and ICAR) placental samples from all groups (Fig. 2). Labeling index in CAR and ICAR of the maternal placenta was similar; therefore data were combined for these two uterine compartments within each group. Labeling index was greater (P < 0.001) in FM than in maternal placenta in all groups. In the NAT group, LI was 24.5±2.9% and 3.5±0.3% in FM and maternal placenta, respectively. In maternal placenta, LI was less (P < 0.001) in the NAT-ET group and least in IVF and IVA groups compared to that of the NAT group; in FM, LI was less (P < 0.001) in the NAT-ET and IVF groups and least in IVA group compared to the NAT group (Fig. 1B).
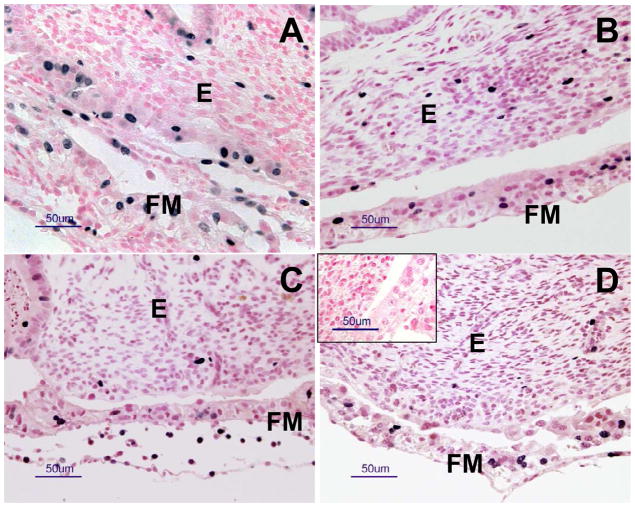
Fig. 2.
Representative photomicrographs of immunohistochemical staining for Ki67 (an endogenous marker of cell proliferation) in maternal and fetal placenta in (A) NAT, (B) NAT-ET, (C) IVF and (D) IVA groups on d 22 of pregnancy. The blackish nuclei represent positive staining and the reddish/pinkish nuclei (nuclear fast red staining) represent unlabeled nuclei. Note nuclear staining of Ki67 in fetal membranes (FM; fetal placenta; chorioallantois) and endometrium (E, maternal placenta). In the inset in (D), note a lack of positive staining in the control sections in which mouse IgG was used in place of the primary antibody.
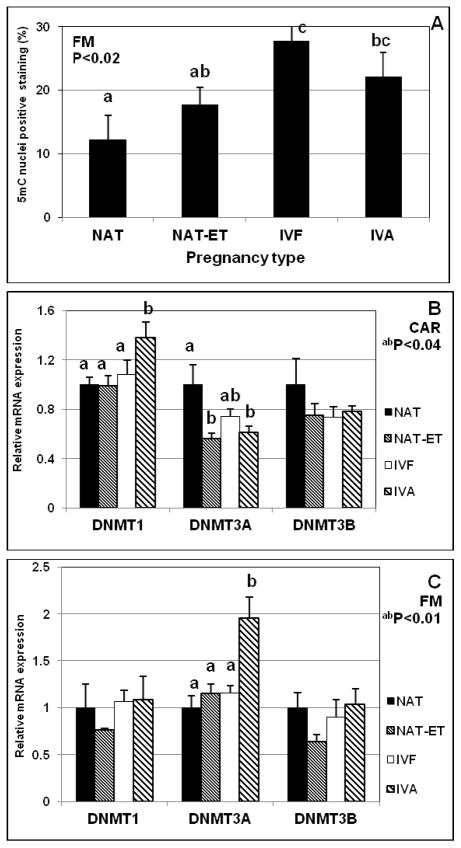
In all groups, 5mC was detected as punctate staining in cell nuclei in FM and maternal placenta (Fig. 3). In FM, expression of 5mC was ~2- to 3-fold greater (P < 0.02) in IVF and IVA compared with the NAT group (Fig. 4A). In maternal placenta, expression of 5mC was similar for all groups (as expected). In CAR, mRNA expression of DNMT1 was greater (P < 0.01) in the IVA group than in the NAT, NAT-ET or IVF groups, whereas expression of DNMT3A was less (P < 0.04) in the NAT-ET and IVA groups than in the NAT group (Fig. 4B). In FM, expression of mRNA for DNMT3A was greater (P < 0.01) in the IVF group that in any other group (Fig. 4C). Expression of mRNA for DNMT3B in CAR and DNMT1 and DNMT3B in FM was similar for all treatment groups (Fig. 4B,C).
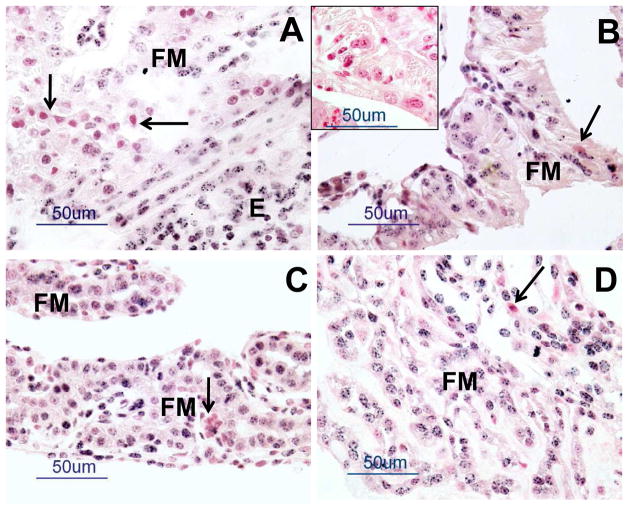
Fig. 3.
Representative photomicrographs of 5mC staining (blackish color in cell nuclei) in fetal membranes (FM; fetal placenta; chorioallantois) and endometrium (E; maternal placenta) in (A) NAT, (B) NAT-ET, (C) IVF and (D) IVA groups on Day 22 of pregnancy. The reddish/pinkish staining represents nuclear fast red counterstaining. Note a very low 5mC expression in some cells (arrows) in each group (A–D). In the inset in (B), note a lack of positive staining in the control sections in which mouse IgG was used in place of the primary antibody.
Fig. 4.
Expression of 5mC (as determined by immunohistochemistry followed by image analysis) in fetal placenta (A), and expression of mRNA for DNMT1, DNMT3A and DNMT3B in maternal placenta (CAR, B) and fetal placenta (FM, C) in NAT, NAT-ET, IVF and IVA groups on Day 22 of pregnancy.
a,bMeans without a common superscript differed (P < 0.0001).
4. Discussion
Embryo origin and/or selected ART may affect embryonic and fetal development, placentation and implantation, placental function and growth, duration of gestation, embryonic loss/survival, pregnancy rate, birth weight, gene expression or other processes, and consequently may lead to compromised pregnancy and subsequent poor pregnancy outcomes [8–13,17,26,46–52].
The present study demonstrated profound effects of embryo origin and selected ART methods on fetal and maternal placental growth, as reflected by smaller fetal size and reduced placental cell proliferation compared with embryos from natural mating. Fetal size was decreased by 15% in the NAT-ET group, where FSH-treatment was combined with natural breeding and ET, and by more than 50% after application of IVF or IVA followed by ET. Reduced fetal size also was observed for ovine and porcine parthenotes at a similar stage of pregnancy as in the present study (Day 26 or 21 of gestation, respectively) compared to naturally-bred controls [15,16]. For cows, shorter crown-rump length of fetuses has been reported for early pregnancy after in vitro compared to in vivo fertilization [8]. In contrast, Farin et al. [17] reported that the length of bovine embryos produced in vitro almost doubled compared to embryos produced in vivo during early pregnancy; and these authors further suggested that this could lead to large offspring syndrome in late pregnancy. Thus, conditions created during FSH treatment followed by natural breeding, IVF or IVA and ET may have negative effects on fetal growth and development very early in pregnancy.
In the present study, for in vivo and in vitro fertilization sperm from several rams with similar genetic background was used. Since it has been demonstrated that semen from individual rams may affect fertilization rates and early embryonic development [53–55], we cannot exclude the possibility that origin of sperm could affect early embryonic development and potentially fetal and placental growth. Furthermore, we transferred three embryos from the same donor to each recipient; this resulted in relatively small variation in various measurements, indicating minimal effects of individual donor ewe on placental development. However, data concerning the effects of sperm origin or oocyte/embryo donor on placental and fetal development are extremely limited; therefore this subject requires further investigation.
In maternal and fetal placenta, a similar pattern of reduced cell proliferation after ET occurred in the present study. In addition, the LI was approximately 10-fold less in maternal than fetal placenta. Placental cell proliferation in pregnancies affected or compromised by application of ART or by other environmental factors (e.g., maternal nutrition, maternal age, maternal genotype, maternal stress, etc.) has received limited attention, and especially during early pregnancy. However, decreased LI was observed in the placenta of adolescent overnourished ewes during mid to late gestation, which were also characterized by impaired fetal and placental growth [56,57]. Moreover, in pregnancies compromised by diabetes, both increased and decreased placental cell proliferation was observed in rats [58,59]. For diabetic mice, decreased cell proliferation was reported in the myometrium during early pregnancy [60]. Conversely, cell proliferation was similar in human term placental from diabetic and non-diabetic mothers [61]. Thus, the high rate of cell proliferation observed in maternal and especially in fetal placenta in natural pregnancy is decreased during early pregnancy after ET and application of ART or in pregnancies compromised by other factors across several mammalian species. This very early defect in placental growth may contribute to impaired fetal and placental growth later in pregnancy, and thereby negatively affect pregnancy outcome and subsequent health and well-being of the offspring.
Tissue growth, a major component of which is cell proliferation, is regulated by growth factors and other regulatory factors in placenta and other tissues [2,3,5–7]. After transfer of embryos of different origin, we have observed reduced expression of several growth factors known to regulate tissue growth and placental function during early pregnancy, including fibroblast growth factor (FGF) 2, FGF receptor, placental growth factor and several others [5,6,62]. We therefore hypothesized that embryo origin and ART would decrease expression of regulatory factors which in turn could contribute to reduced cellular proliferation and fetal size.
In our previous studies, we demonstrated changes in expression of 5mC and mRNA for DMNT1, 3A and 3B in FM during early pregnancy [6]. In the present study, enhanced expression of selected markers of global methylation (5mC or DNMT3A mRNA) was detected in fetal placenta from IVA and IVF groups. In maternal placenta, upregulation of DNMT1 mRNA was observed in the IVA group along with downregulation of DNMT3A mRNA expression in the NAT-ET and IVA groups. Thus, global methylation, as measured by expression of 5mC and mRNA for several DNMTs, was affected in ovine maternal and fetal placenta after transfer of embryos of different origin and application of ART. These epigenetic changes could affect expression of selected genes.
Since there was enhanced expression of 5mC in the IVF and IVA groups, we inferred there was DNA hypermethylation, which is likely associated with silencing of specific genes. Furthermore, differential expression of markers of global methylation in fetal and maternal placenta indicates active methylation/demethylation processes in these tissues. The major role of DNMT1 is to maintain the gene methylation pattern, but DNMT3A and 3B are thought to control de novo methylation [63,64]. Furthermore, it has been suggested that DNMT 3A and 3 B are involved in demethylation process [65,66]. In human placenta, methylation of imprinting control regions of H19 or insulin-like growth factor 2 genes was not affected by IVF and intracytoplasmic sperm injection [67]. Furthermore, the DNA methylation pattern of selected differentially methylated regions in blood samples was similar for ART-conceived and naturally conceived children [29,68]. For other species, including cows and sheep, application of ART (e.g., IVF, IVA or somatic cell nuclear transfer) affected selected gene methylation, global methylation and/or expression of DNMTs in some, but not all studies [22,30,69–71]. Significant variation in global methylation among these reports was likely due to species, time of tissue collection, as well as methodological and other differences, which certainly warrants further investigation in pregnancies compromised by ART. Furthermore, it is currently unclear how changes in any epigenetic mechanism affect placental development and maternal-offspring interactions during pregnancy and postnatally.
The NAT-ET group in the present study underwent two ART manipulations – FSH treatment followed by embryo flushing and transfer. Although several measurements in this and the NAT control group were similar, the NAT-ET group exhibited reduced fetal size, placental cell proliferation and mRNA expression for DNMT3A. Therefore, superovulatory treatments, which are widely used in animal production and human reproductive medicine, may have some negative effects on fetal and placental growth and function during early pregnancy [26]. Conversely, in several experiments in which we used a similar FSH treatment optimized in our laboratory, there were no impaired fertilization or blastocyst formation rates, or impaired development of fetuses or offspring in sheep [10,39–41]. Therefore, we hypothesize that in FSH-treated animals, some compensatory mechanisms may exist during embryonic and/or fetal development to allow for normal fetal and placental growth and function throughout pregnancy.
After transfer of parthenotes in the present experiment, there was decreased fetal and placental growth and enhanced expression of selected markers of global methylation, including increased 5mC and mRNA for DNMT1 in CAR and DNMT3A in FM. In several studies, differences in expression of selected genes, including those involved in epigenetic processes, have been demonstrated in bovine parthenogenetic blastocysts compared to blastocysts created through IVF [37,72–75]. Comparison of the present data with data for blastocysts should be treated with caution, since various stages of embryonic/fetal development (i.e., blastocysts versus 22 d fetus) were evaluated [76]. Nevertheless, these studies indicated epigenetic differences among fetuses of different origin, and that these were likely due to hormonal treatment and/or exposure to in vitro conditions. Because an understanding of the role of epigenetics in the negative effects of ART is incomplete [26,28,77], this area requires further studies.
The major differences in tissue growth or global methylation observed in our study were due to different origins and manipulations of the embryo. Studies of maternal-fetal interactions demonstrate that the embryo affects uterine function and has an active role in initiation of pregnancy, and in turn the uterus affects fetal growth and development [2–4,46,48,52,78–80]. For pregnancies achieved after transfer of embryos created through somatic cell nuclear transfer (SCNT), placental failure is due to abnormal embryo-maternal communication and endometrial remodeling during the peri-implantation period [52,79,81]. Conversely, in pregnancies resulting from the transfer of embryos created through IVF, changes in endometrial remodeling and function were less pronounced than after transfer of embryos from SCNT [52,81]. It has been postulated that endometrial tissues possess mechanisms to adapt to different embryos, which may serve as a biological sensor to meet embryonic demands or adaptation to environmental conditions [48]. These maternal-fetal interactions may have long-term consequences for placental function, and subsequently for offspring outcome and even into adulthood [18–20,82].
In the present study, only placental tissues from Day 22 of pregnancy were evaluated. Therefore, we cannot exclude that the differences in fetal and placental growth and function may be minimized later in pregnancy by compensatory mechanisms, with the exception of pregnancies after transfer of parthenotes which naturally terminate after approximately 4 wk, due to the absence of a paternal genome [15,16]. In our previous study, some compensatory mechanism likely appeared in singleton but not twin pregnancies, since weight of lambs and several placental parameters on Day 140 of gestation (gestational length is ~145 d in sheep) were similar in pregnancies achieved through IVF and natural breeding [10]. Conversely, we and others have reported that in ruminants, pregnancies resulting from various ART including IVF and SCNT exhibit poor placental development and vascularization as well as abnormal/altered fetal growth and development at different stages of gestation [10,17,83–86]. Therefore, the effects observed during early pregnancy in the present study could likely have long-term consequences for pregnancy outcome.
In summary, in this study, transfer of embryos of different origin and application of ART decreased fetal size and placental cell proliferation, and altered expression of selected markers of global methylation in fetal and maternal placenta on Day 22 of pregnancy. Thus, embryo origin may have specific effects on growth and function of the ovine placenta and fetus through regulation of tissue growth and epigenetic processes, as well as other mechanisms such as placental angiogenesis. Since very few studies have focused on evaluation of selected processes in placenta during early gestation without or with application of ART in any species, these data provide novel information concerning fetal and placental tissue growth/cell proliferation and global methylation in relation to embryo origin. Furthermore, these data provide a foundation for determining the expression of specific factors regulating growth of placental and embryonic tissues in pregnancies after application of ART including ET, IVF, IVA and/or cloning. In addition, these data will help us to better understand placental regulatory mechanisms in compromised pregnancies, and to identify strategies for rescuing such pregnancies.
Acknowledgments
The authors acknowledge Dr. Kimberly Vonnahme, Dr. Jerzy Bilski, Ms. Tammi Neville, Mr. James D. Kirsch, Mr. Kim C. Kraft, Mr. Robert Weigl, Mr. Terry Skunberg, and other members of our laboratories and department for their assistance, and Dr. Jodie Haring for critical review of this manuscript. This project was supported by USDA grant (2007-01215) to LPR and ATGB, NIH grant (HL64141) to LPR and DAR, and NSF MRI-R2-ARRA (0959512) grant to ATGB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reynolds LP, Redmer DA. Growth and microvascular development of the uterus during early pregnancy in ewes. Biol Reprod. 1992;47:698–708. doi: 10.1095/biolreprod47.5.698. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds LP, Caton JS, Redmer DA, Grazul-Bilska AT, Vonnahme KA, Borowicz PP, et al. Evidence for altered placental blood flow and vascularity in compromised pregnancies. J Physiol. 2006;572:51–8. doi: 10.1113/jphysiol.2005.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds LP, Borowicz PP, Caton JS, Vonnahme KA, Luther JS, Buchanan DS, et al. Utero placental vascular development and placental function: An update. Int J Develop Biol. 2010;54:355–66. doi: 10.1387/ijdb.082799lr. [DOI] [PubMed] [Google Scholar]
- 4.Spencer TE, Sandra O, Wolf E. Genes involved in conceptus-endometrial interactions in ruminants: insights from reductionism and thoughts on holistic approaches. Reproduction. 2008;135:165–79. doi: 10.1530/REP-07-0327. [DOI] [PubMed] [Google Scholar]
- 5.Grazul-Bilska AT, Borowicz PP, Johnson ML, Minten MA, Bilski JJ, Wroblewski R, et al. Placental development during early pregnancy in sheep: vascular growth and expression of angiogenic factors in maternal placenta. Reproduction. 2010;140:165–74. doi: 10.1530/REP-09-0548. [DOI] [PubMed] [Google Scholar]
- 6.Grazul-Bilska AT, Johnson ML, Borowicz PP, Minten M, Wroblewski R, Coupe LR, et al. Placental development during early pregnancy in sheep: Cell proliferation, methylation and angiogenesis in fetal placenta. Reproduction. 2011;141:529–40. doi: 10.1530/REP-10-0505. [DOI] [PubMed] [Google Scholar]
- 7.Zheng J, Johnson ML, Redmer DA, Reynolds LP. Estrogen and progesterone receptors, cell proliferation, and c-fos expression in the ovine uterus during early pregnancy. Endocrinology. 1996;137:340–8. doi: 10.1210/endo.137.1.8536633. [DOI] [PubMed] [Google Scholar]
- 8.Bertolini M, Mason JB, Beam SW, Carneiro GF, Sween ML, Kominek DJ, et al. Morphology and morphometry of in vivo- and in vitro-produced bovine concepti from early pregnancy to term and association with high birth weights. Theriogenology. 2002;5:973–94. doi: 10.1016/s0093-691x(02)00935-4. [DOI] [PubMed] [Google Scholar]
- 9.Cai LY, Izumi S, Koido S, Uchida N, Suzuki T, Matsubayashi H, et al. Abnormal placental cord insertion may induce intrauterine growth restriction in IVF-twin pregnancies. 2006;21:1285–90. doi: 10.1093/humrep/dei494. [DOI] [PubMed] [Google Scholar]
- 10.Grazul-Bilska AT, Pant D, Luther JS, Choi JT, Borowicz P, Navanukraw C, et al. Pregnancy rates and gravid uterine parameters in single, twin and triplet pregnancies in naturally bred ewes and ewes after transfer of in vitro produced embryos. Anim Reprod Sci. 2006;92:268–83. doi: 10.1016/j.anireprosci.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Collier AC, Miyagi SJ, Yamauchi Y, Ward MA. Assisted reproduction technologies impair placental steroid metabolism. J Steroid Biochem Molec Biol. 2009;116:21–8. doi: 10.1016/j.jsbmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delle Piane L, Lin W, Liu X, Donjacour A, Minasi P, Revelli A, et al. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod. 2010;25:2039–46. doi: 10.1093/humrep/deq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomic V, Tomic J. Neonatal outcome of IVF singletons versus naturally conceived in women aged 35 years and over. Arch Gynecol Obstetr. 2011;284:1411–6. doi: 10.1007/s00404-011-1873-2. [DOI] [PubMed] [Google Scholar]
- 14.Holm P, Walker SK, Seamark RF. Embryo viability, duration of gestation and birth weight in sheep after transfer of in vitro matured and in vitro fertilized zygotes cultured in vitro or in vivo. J Reprod Fertil. 1996;107:175–81. doi: 10.1530/jrf.0.1070175. [DOI] [PubMed] [Google Scholar]
- 15.Loi P, Ledda S, Fulka J, Jr, Cappai P, Moor RM. Development of parthenogenetic and cloned ovine embryos: effect of activation protocols. Biol Reprod. 1998;58:1177–87. doi: 10.1095/biolreprod58.5.1177. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, King T, Dobrinsky J, Harkness L, Ferrier T, Bosma W, et al. In vitro and in vivo developmental competence of ovulated and in vitro matured porcine oocytes activated by electrical activation. Clon Stem Cells. 2003;5:355–65. doi: 10.1089/153623003772032853. [DOI] [PubMed] [Google Scholar]
- 17.Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro produced bovine embryos. Theriogenology. 2006;65:178–91. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Nathanielsz PW. Animal models that elucidate basic principles of the developmental origins of adult diseases. ILAR J. 2006;47:73–82. doi: 10.1093/ilar.47.1.73. [DOI] [PubMed] [Google Scholar]
- 19.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–7. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds LP, Caton JS. Role of the pre- and post-natal environment in developmental programming of health and productivity. Mol Cell Endocrinol. 2012;354:54–9. doi: 10.1016/j.mce.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemann H, Tian XC, King WA, Lee RS. Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning. Reproduction. 2008;135:151–63. doi: 10.1530/REP-07-0397. [DOI] [PubMed] [Google Scholar]
- 23.Miri K, Varmuza S. Imprinting and extraembryonic tissues-mom takes control. International Rev Cell Mol Biol. 2009;276:215–62. doi: 10.1016/S1937-6448(09)76005-8. [DOI] [PubMed] [Google Scholar]
- 24.Szyf M. The early life environment and the epigenome. Biochem Biophys Acta. 2009;1790:878–85. doi: 10.1016/j.bbagen.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 26.Laprise SL. Implications of epigenetics and genomic imprinting in assisted reproductive technologies. Mol Reprod Develop. 2009;76:1006–18. doi: 10.1002/mrd.21058. [DOI] [PubMed] [Google Scholar]
- 27.Díaz-García C, Estella C, Perales-Puchalt A, Simón C. Reproductive medicine and inheritance of infertility by offspring: the role of fetal programming. Fertil Steril. 2011;96:536–45. doi: 10.1016/j.fertnstert.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 28.Iliadou AN, Janson PC, Cnattingius S. Epigenetics and assisted reproductive technology. J Intern Med. 2011;270:414–20. doi: 10.1111/j.1365-2796.2011.02445.x. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Wang L, Le F, Liu X, Yu P, Sheng J, et al. Evaluation of DNA methylation status at differentially methylated regions in IVF-conceived newborn twins. Fertil Steril. 2011;95:1975–9. doi: 10.1016/j.fertnstert.2011.01.173. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Zhao G, Guo R, Zhang D, Li X, Zhou H. DNA methylation status in tissues of sheep clones. Reprod Dom Anim. 2011;47:504–12. doi: 10.1111/j.1439-0531.2011.01911.x. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Yang X. Telomerase activity in early bovine embryos derived from parthenogenetic activation and nuclear transfer. Biol Reprod. 2001;64:770–4. doi: 10.1095/biolreprod64.3.770. [DOI] [PubMed] [Google Scholar]
- 32.Ferrandi B, Cremonesi F, Consiglio AL, Luciano AM, Gandolfi F, Modina S, et al. Microdensitometric assay of enzymatic activities in parthenogenetically activated and in vitro fertilized bovine oocytes. Acta Histochem. 2002;104:193–8. doi: 10.1078/0065-1281-00640. [DOI] [PubMed] [Google Scholar]
- 33.Krivokharchenko A, Popova E, Zaitseva I, Vil’ianovich L, Ganten D, Bader M. Development of parthenogenetic rat embryos. Biol Reprod. 2003;68:829–36. doi: 10.1095/biolreprod.102.006494. [DOI] [PubMed] [Google Scholar]
- 34.Lagutina I, Lazzari G, Duchi R, Galli C. Developmental potential of bovine androgenetic and parthenogenetic embryos: a comparative study. Biol Reprod. 2004;70:400–5. doi: 10.1095/biolreprod.103.021972. [DOI] [PubMed] [Google Scholar]
- 35.Kono T, Obata Y, Yoshimzu T, Nakahara T, Carroll J. Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat Genet. 1996;13:91–4. doi: 10.1038/ng0596-91. [DOI] [PubMed] [Google Scholar]
- 36.Grazul-Bilska AT, Borowicz PP, Redmer DA, Bilski JJ, Reynolds LP. Creation of parthenogenetic sheep embryos: Preliminary study. Western Dakota Sheep and Beef Day. 2008 Report No. 49; http://www.ag.ndsu.edu/HettingerREC/sheep/individual-articles-from-2008-sheep-reserach-report/Creation%20of%20Parthenogenetic%20Sheep%20Embryos.pdf.
- 37.Maalouf WE, Alberio R, Campbell KH. Differential acetylation of histone H4 lysine during development of in vitro fertilized, cloned and parthenogenetically activated bovine embryos. Epigenetics. 2008;3:199–209. doi: 10.4161/epi.3.4.6497. [DOI] [PubMed] [Google Scholar]
- 38.Bebbere D, Bogliolo L, Ariu F, Fois S, Leoni GG, Succu S, et al. Different temporal gene expression patterns for ovine pre-implantation embryos produced by parthenogenesis or in vitro fertilization. Theriogenology. 2010;74:712–23. doi: 10.1016/j.theriogenology.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 39.Stenbak TK, Redmer DA, Berginski HR, Erickson AS, Navanukraw C, Toutges MJ, et al. Effects of follicle stimulating hormone (FSH) on follicular development, oocyte retrieval and in vitro fertilization (IVF) in ewes during breeding season and seasonal anestrous. Theriogenology. 2001;56:51–64. doi: 10.1016/s0093-691x(01)00542-8. [DOI] [PubMed] [Google Scholar]
- 40.Grazul-Bilska AT, Choi JT, Bilski JJ, Weigl RM, Kirsch JD, Kraft KC, et al. Effects of epidermal growth factor on early embryonic development after in vitro fertilization of oocytes collected from ewes treated with follicle stimulating hormone. Theriogenology. 2003;59:1453–61. doi: 10.1016/s0093-691x(02)01192-5. [DOI] [PubMed] [Google Scholar]
- 41.Grazul-Bilska AT, Borowczyk E, Bilski JJ, Reynolds LP, Redmer DA, Caton JS, et al. Overfeeding and underfeeding have detrimental effects on oocyte quality measured by in vitro fertilization and early embryonic development in sheep. Dom Anim Endocrinol. 2012 doi: 10.1016/j.domaniend.2012.05.001. (in press) [DOI] [PubMed] [Google Scholar]
- 42.Igwebuike UM. A review of uterine structural modifications that influence conceptus implantation and development in sheep and goats. Anim Reprod Sci. 2009;112:1–7. doi: 10.1016/j.anireprosci.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Santos F, Dean W. Using immunofluorescence to observe methylation changes in mammalian preimplantation embryos. Methods Mol Biol. 2006;325:129–37. doi: 10.1385/1-59745-005-7:129. [DOI] [PubMed] [Google Scholar]
- 44.SAS Institute. User’s Guide, Statistics. Cary, NC: Statistical Analysis System Inst; 2010. [Google Scholar]
- 45.Kirk RE. Experimental design: procedures for the behavioral sciences. Monterey, CA: Brooks/Cole Publishing Co; 1982. [Google Scholar]
- 46.Barnes FL. The effects of the early uterine environment on the subsequent development of embryo and fetus. Theriogenology. 2000;53:649–58. doi: 10.1016/s0093-691x(99)00264-2. [DOI] [PubMed] [Google Scholar]
- 47.Van Steirteghem A, Bonduelle M, Devroey P, Liebaers I. Follow-up of children born after ICSI. Hum Reprod Update. 2002;8:111–6. doi: 10.1093/humupd/8.2.111. [DOI] [PubMed] [Google Scholar]
- 48.Fleming TP, Kwong WY, Porter R, Ursell E, Fesenko I, Wilkins A, et al. The embryo and its future. Biol Reprod. 2004;71:1046–54. doi: 10.1095/biolreprod.104.030957. [DOI] [PubMed] [Google Scholar]
- 49.Miles JR, Farin CE, Rodriguez KF, Alexander JE, Farin PW. Effects of embryo culture on angiogenesis and morphometry of bovine placentas during early gestation. Biol Reprod. 2005;73:663–71. doi: 10.1095/biolreprod.105.040808. [DOI] [PubMed] [Google Scholar]
- 50.Bukulmez O. Does assisted reproductive technology cause birth defects? Curr Opin Obstet Gynecol. 2009;21:260–464. doi: 10.1097/GCO.0b013e32832924a7. [DOI] [PubMed] [Google Scholar]
- 51.CDC. Centers for Disease Control Assisted Reproductive Technology Reports. National Summary. 2009 http://www.cdc.gov/art/ART2009/index.htm http://apps.nccd.cdc.gov/art/Apps/Marquee.aspx.
- 52.Mansouri-Attia N, Sandra O, Aubert J, Degrelle S, Everts RE, Giraud-Delville C, et al. Endometrium as an early sensor of in vitro embryo manipulation technologies. Proc Nat Acad Sci USA. 2009;106:5687–92. doi: 10.1073/pnas.0812722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukui Y, Glew AM, Gandolfi F, Moor RM. Ram-specific effects on in-vitro fertilization and cleavage of sheep oocytes matured in vitro. J Reprod Fertil. 1988;82:337–40. doi: 10.1530/jrf.0.0820337. [DOI] [PubMed] [Google Scholar]
- 54.Eyestone WH, First NL. Variation in bovine embryo development in vitro due to bulls. Theriogenology. 1989;31:191. [Google Scholar]
- 55.Morris LH, Randall AE, King WA, Johnson WH, Buckrell BC. The contribution of the male to ovine embryogenesis in an in vitro embryo production system. Anim Reprod Sci. 2003;75:9–26. doi: 10.1016/s0378-4320(02)00188-4. [DOI] [PubMed] [Google Scholar]
- 56.Lea RG, Hannah LT, Redmer DA, Aitken RP, Milne JS, Fowler PA, et al. Developmental indices of nutritionally induced placental growth restriction in the adolescent sheep. Pediat Res. 2005;57:599–604. doi: 10.1203/01.PDR.0000155949.08547.66. [DOI] [PubMed] [Google Scholar]
- 57.Redmer DA, Luther JS, Milne JS, Aitken RP, Johnson ML, Borowicz PP, et al. Fetoplacental growth and vascular development in overnourished adolescent sheep at day 50, 90 and 130 of gestation. Reproduction. 2009;137:749–57. doi: 10.1530/REP-08-0516. [DOI] [PubMed] [Google Scholar]
- 58.Caluwaerts S, Pijnenborg R, Luyten C, Van Assche FA. Growth characteristics of diabetic rat ectoplacental cones in vivo and in vitro. Diabetologia. 2000;43:939–45. doi: 10.1007/s001250051473. [DOI] [PubMed] [Google Scholar]
- 59.Zorn TM, Zúñiga M, Madrid E, Tostes R, Fortes Z, Giachini F, et al. Maternal diabetes affects cell proliferation in developing rat placenta. Histol Histopathol. 2011;26:1049–56. doi: 10.14670/HH-26.1049. [DOI] [PubMed] [Google Scholar]
- 60.Favaro RR, Salgado RM, Raspantini PR, Fortes ZB, Zorn TM. Effects of long-term diabetes on the structure and cell proliferation of the myometrium in the early pregnancy of mice. Int J Exp Pathol. 2010;91:426–35. doi: 10.1111/j.1365-2613.2010.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burleigh DW, Stewart K, Grindle KM, Kay HH, Golos TG. Influence of maternal diabetes on placental fibroblast growth factor-2 expression, proliferation, and apoptosis. J Soc Gynecol Invest. 2004;11:36–41. doi: 10.1016/j.jsgi.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Johnson ML, Reynolds LP, Borowicz PP, Redmer DA, Grazul-Bilska AT. Expression of mRNA for factors that influence angiogenesis in ovine utero-placental tissues during early pregnancy: Effects of assisted reproductive technology. Annual meeting of the Society for the Study of Reproduction; Portland, OR. 2011. [Google Scholar]
- 63.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647:30–8. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–8. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Carey N, Marques CJ, Reik W. DNA demethylases: a new epigenetic frontier in drug discovery. Drug Discov Today. 2011;16:683–90. doi: 10.1016/j.drudis.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Wong EC, Hatakeyama C, Robinson WP, Ma S. DNA methylation at H19/IGF2 ICR1 in the placenta of pregnancies conceived by in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril. 2011;95:2524–6. e1–3. doi: 10.1016/j.fertnstert.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 68.Oliver VF, Miles HL, Cutfield WS, Hofman PL, Ludgate JL, Morison IM. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril. 2012;97:147–53.e7. doi: 10.1016/j.fertnstert.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 69.Lan J, Hua S, Zhang H, Song Y, Liu J, Zhang Y. Methylation patterns in 5′ terminal regions of pluripotency-related genes in bovine in vitro fertilized and cloned embryos. J Genet Genom. 2010;37:297–304. doi: 10.1016/S1673-8527(09)60047-3. [DOI] [PubMed] [Google Scholar]
- 70.Golding MC, Williamson GL, Stroud TK, Westhusin ME, Long CR. Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol Reprod Develop. 2011;78:306–17. doi: 10.1002/mrd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su JM, Yang B, Wang YS, Li YY, Xiong XR, Wang LJ, et al. Expression and methylation status of imprinted genes in placentas of deceased and live cloned transgenic calves. Theriogenology. 2011;75:1346–1359. doi: 10.1016/j.theriogenology.2010.11.045. [DOI] [PubMed] [Google Scholar]
- 72.Nowak-Imialek M, Wrenzycki C, Herrmann D, Lucas-Hahn A, Lagutina I, Lemme E, et al. Messenger RNA expression patterns of histone-associated genes in bovine preimplantation embryos derived from different origins. Mol Reprod Dev. 2008;75:731–43. doi: 10.1002/mrd.20816. [DOI] [PubMed] [Google Scholar]
- 73.Gómez E, Caamaño JN, Bermejo-Alvarez P, Díez C, Muñoz M, Martín D, et al. Gene expression in early expanded parthenogenetic and in vitro fertilized bovine blastocysts. J Reprod Dev. 2009;55:607–14. doi: 10.1262/jrd.09-077m. [DOI] [PubMed] [Google Scholar]
- 74.Gómez E, Gutiérrez-Adán A, Díez C, Bermejo-Alvarez P, Muñoz M, Rodriguez A, et al. Biological differences between in vitro produced bovine embryos and parthenotes. Reproduction. 2009;137:285–95. doi: 10.1530/REP-08-0220. [DOI] [PubMed] [Google Scholar]
- 75.Sawai K, Takahashi M, Fujii T, Moriyasu S, Hirayama H, Minamihashi A, et al. DNA methylation status of bovine blastocyst embryos obtained from various procedures. J Reprod Dev. 2011;57:236–41. doi: 10.1262/jrd.10-035a. [DOI] [PubMed] [Google Scholar]
- 76.Thurston A, Taylor J, Gardner J, Sinclair KD, Young LE. Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage. Reproduction. 2008;135:29–40. doi: 10.1530/REP-07-0211. [DOI] [PubMed] [Google Scholar]
- 77.Wilkins-Haug L. Epigenetics and assisted reproduction. Curr Opin Obstetr Gynecol. 2009;21:201–6. doi: 10.1097/GCO.0b013e32832d7b95. [DOI] [PubMed] [Google Scholar]
- 78.Barnea ER. Insight into early pregnancy events: the emerging role of the embryo. Am J Reprod Immunol. 2004;51:319–22. doi: 10.1111/j.1600-0897.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 79.Bauersachs S, Ulbrich SE, Zakhartchenko V, Minten M, Reichenbach M, Reichenbach HD, et al. The endometrium responds differently to cloned versus fertilized embryos. Proc Nat Sci USA. 2009;106:5681–6. doi: 10.1073/pnas.0811841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ostrup E, Hyttel P, Ostrup O. Embryo-maternal communication: signalling before and during placentation in cattle and pig. Reprod, Fertil Develop. 2011;23:964–75. doi: 10.1071/RD11140. [DOI] [PubMed] [Google Scholar]
- 81.Hoffert KA, Batchelder CA, Bertolini M, Moyer AL, Famula TR, Anderson DL, et al. Measures of maternal-fetal interaction in day-30 bovine pregnancies derived from nuclear transfer. Clon Stem Cells. 2005;7:289–305. doi: 10.1089/clo.2005.7.289. [DOI] [PubMed] [Google Scholar]
- 82.Isles AR, Holland AJ. Imprinted genes and mother-offspring interactions. Early Hum Develop. 2005;81:73–7. doi: 10.1016/j.earlhumdev.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 83.Heyman Y, Chavatte-Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod. 2002;66:6–13. doi: 10.1095/biolreprod66.1.6. [DOI] [PubMed] [Google Scholar]
- 84.Fletcher CJ, Roberts CT, Hartwich KM, Walker SK, McMillen IC. Somatic cell nuclear transfer in the sheep induces placental defects that likely precede fetal demise. Reproduction. 2007;133:243–55. doi: 10.1530/rep.1.01203. [DOI] [PubMed] [Google Scholar]
- 85.Loi P, Clinton M, Vackova I, Fulka J, Jr, Feil R, Palmieri C, et al. Placental abnormalities associated with post-natal mortality in sheep somatic cell clones. Theriogenology. 2005;65:1110–21. doi: 10.1016/j.theriogenology.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 86.Palmieri C, Loi P, Reynolds LP, Ptak G, Della Salda L. Placental abnormalities in ovine somatic cell clones at term: A light and electron microscopic investigation. Placenta. 2007;28:577–84. doi: 10.1016/j.placenta.2006.08.003. [DOI] [PubMed] [Google Scholar]