Abstract
The revolution in virus genome sequencing promises to effectively map the extant biological universe and reveal fundamental relationships between viral biology, genome structure, and evolution. Indeed, microbial virus genomes include large numbers of conserved coding sequences of unknown function as well as unique gene combinations, implying that that these viruses will be a significant source of novel protein biochemistry and genome architecture. Yet, making sense of the approaching phalanx of A’s, G’s, T’s, and C’s stretching across the genome sequencing horizon will require innovation and an unprecedented coordination of annotation efforts among stakeholders.
Keywords: viral genomics, genome annotation, biological sequence databases
Introduction
Typically there is a chronological disconnect between the publication of genome sequences and follow up experiments which reveal the biochemistry encoded within these sequences. Hence it is important to treat sequence records deposited in GenBank and other public databases as dynamic documents which are routinely updated to maintain an accurate representation of latest experimental evidence. Though this is a fundamental departure from the typical, incremental approach to scientific publication, treating sequence records essentially as eDocuments provides authors - and whole communities - the ability to maintain “living” records of current knowledge - a genome blog of sorts - where a wealth of information is aggregated.
With possibility comes responsibility, and maintenance of genome sequence records as living documents requires periodic updating to capture evolving experimental data as well as sequence record data model innovations. This necessity for constant vigilance is effectively at odds with the sheer number of virus genome sequence records, and whereas previous curation models emphasized the role of expert curators within public databases, the emergent annotation landscape is complex with a number of additional stakeholders, including sequencing centers and community databases. In this view only comprehensive efforts which embrace and aggregate annotation from a variety of resources can bridge the growing gap between sequencing efforts and the need for accurate genome annotation.
Results and Discussion
What is genome sequence annotation?
Annotation is the process by which a raw genome sequence assembly is transformed into a documented record of biological features. Sequence records submitted to the International Nucleotide Sequence Database Collaboration (INSDC) member databases - DDBJ, ENA, and GenBank - include several discrete types of annotation activity (Karsch-Mizrachi, Nakamura, and Cochrane, 2012). First, the span of a given feature - gene, mRNA, coding sequence, ect. - must be predicted along the genome sequence. Second, each feature must be assigned a set of descriptors - the most basic of which is a name. Finally, the sequence source must be described and placed within a biological context using a set of metadata.
Feature prediction generally relies on computer algorithms which rely either on information intrinsic to the sequence itself, extrinsic information such as homologous coding sequences, or some combination of the two (Besemer and Borodovsky, 2005). Obviously, the accuracy of gene prediction can be greatly enhanced when driven by experimentally validated reference protein sequences, and in this way, annotation of one protein can inform the annotation of many others. Current INSDC sequence record data structure allows the use of evidence codes, allowing one to indicate when a particular gene model has been experimentally validated, predicted based on homology to other sequences, or simply predicted ab initio (Klimke et al., 2011).
Descriptors used in INSDC sequence records are also used as labels in other resources. A prime example is “protein name.” This descriptor is included in the definition lines of results returned from protein BLAST searches. When conducting such searches, one often one finds a multitude of names referencing nearly identical peptides with the same function. To prevent such inconsistencies, there has been an international effort to harmonize protein names across databases using a standard functional name format initially developed by UniProtKB/Swiss-Prot (Brister et al., 2010; Klimke et al., 2011).
While it is likely that source descriptors - metadata - will remain stable over time, other feature annotations are apt to change in response to experimental progression. So while the initial INSDC sequence record may include the current state of science at the time of submission, the same record may be out of date within a few years. This natural progression of knowledge necessitates an equally dynamic approach to genome sequence annotation.
Virus genome annotation efforts at NCBI
The primary source of annotation for most genome sequence records is the actual sequence submitter. While some data structure elements are validated by GenBank indexers, most annotations are not reviewed extensively. This approach is consistent with the archival role of GenBank, and while this policy creates a relatively low burden for submission, it also makes it difficult to enforce annotation standards. Moreover, the submitter is not required to update sequence records as the current state of knowledge evolves, so even initially well annotated records can erode with the passage of time.
The curated RefSeq database was introduced to help mitigate the problems caused by inconsistent, outdated, or simply incorrect annotations (Pruitt, Tatusova, and Maglott, 2005). The idea is to create a single representative reference genome for each group of related viruses using submitted GenBank records as templates. Reference genomes are then curated by biologists using in house annotation tools and the scientific literature as guides. A panel of Viral Genome Advisors from outside NCBI bolsters these curation efforts by offering expert guidance or taking responsibility for specific RefSeq records themselves.
Typically, one virus RefSeq record is made for each virus species as defined by the International Committee on the Taxonomy of Viruses (International Committee on Taxonomy of Viruses., King, and International Union of Microbiological Societies. Virology Division., 2012). However, it is sometimes difficult to capture the genetic content of a particular species within a single genome sequence, and multiple RefSeq records must be created. This is often done for bacterial viruses where horizontal gene transfer contributes to genetic diversity, but multiple references are also maintained for a number of other species – most notable double-stranded DNA viruses - as dictated by genetic diversity. RefSeq also maintains multiple reference records in cases where there are more than one well studied isolates.
While an “expert curation” approach to all RefSeq records was arguably feasible a decade ago, it could not keep pace with the rate of virus genome sequencing (Figure 1) and a somewhat ad hoc curation model has followed in its wake. Accordingly, the curation of most virus RefSeq records is limited to taxonomic and data structure considerations. This light hand approach is reflected in the RefSeq status key where these records are marked “provisional.” Intense curation efforts are limited to a small subset of virus genome records, typically denoted by their “reviewed” RefSeq status (Pruitt et al., 2002; Pruitt et al., 2009).
Figure 1. Growth of virus RefSeq records.
A. Cumulative number of virus nucleotide sequence records deposited in the RefSeq database from 1999 to 20121,2. B. Cumulative number of protein records deposited in the RefSeq database from 1999 to 20122. 1Individual viral segments are included in tabulations, not complete constellations. 2Number of records calculated on September 11, 2012.
Protein Clusters
The current RefSeq approach to genome curation reflects two realities: One, there are simply too many genome sequence records to manually curate and keep up to date. Two, only a small number of viruses or viral gene products will be investigated in the laboratory. Without detailed experimental evidence from individual genomes, one must infer annotation from other genome or protein homologs. Hence, successful annotation requires the ability to transfer annotation from the genome sequence records of well-studied viruses to those of less studied ones.
The NCBI Protein Clusters resource was created to help bridge the gap between experimentally characterized proteins and those less well studied (http://www.ncbi.nlm.nih.gov/proteinclusters) (Klimke et al., 2009). The resource groups nearly identical proteins into clusters and aggregates any data associated with individual protein sequences into a single shared space. While each cluster may contain dozens of nearly identical protein sequences, often only a few - maybe just one - have actually been characterized in the laboratory. Yet, based on these experiments, it is possible to infer functionality of other, uncharacterized proteins within the same cluster. In this way data from one protein can inform the aggregate.
Currently the Protein Clusters database only includes RefSeq proteins. While this reference protein approach excludes other GenBank sequences including metagenomic sequences, it reflects the goal of seeding the database with well annotated proteins and transferring this annotation to other RefSeq records. Yet, even within this restricted data set there is evidence for a large annotation gap since 43,434 of the 108,988 current viral RefSeq proteins include “hypothetical protein” in their name. This gap appears even larger among bacterial virus RefSeq proteins where 18,694 out of a total of 38,364 proteins are named “hypothetical protein.”
The Protein Clusters curation approach underscores the importance of well-studied genomes, or more accurately, experimentally validated and well curated genome sequence records. Data associated with these so called “gold standard” records - such as gene models, protein names, and ontology terms - can be used to guide annotation of homologous sequences which are part of the same cluster. The net effect is that one well annotated genome can inform the annotation of dozens if not hundreds of proteins - resulting in standardized protein names and other annotations across all constituent proteins within a given cluster.
Experimentally derived data is critical to evaluating protein functionality. The Protein Clusters resource includes a multi-sequence alignment viewer where the relationships between clustered sequences can be directly visualized – allowing one to appraise the conservation of functional domains and discrete residues across all proteins in a cluster. Given sequences annotated as deficient in a specific biochemical activity or other data regarding required residues, it is possible to manually predict the functional potential of a given sequence within an alignment. It is also possible to use the functional data aggregated in Protein Clusters to build gene models for use in automated sequence annotation pipelines, improving the accuracy of these tools.
NCBI virus genome annotation does not occur in a vacuum, and other databases and community groups are actively involved in various aspects of annotation. This global network has the potential to greatly enhance the availability of well annotated genomes and proteins but also presents a significant challenge with regard to data association and aggregation. Protein Clusters solves this problem by capturing data linked to a particular protein from a variety of internal and external sources like Entrez Gene (Maglott et al., 2011), UniProtKB/Swiss-Prot (2012), and ACLAME (Leplae, Lima-Mendez, and Toussaint, 2010) and aggregating it within a common cluster space where it can be assessed by hand or machine and ultimately, assigned to all member proteins within the cluster.
Protein annotation efforts at UniProtKB/SwissProt
UniProtKB is a comprehensive protein sequence knowledgebase that consists of two sections: UniProtKB/Swiss-Prot (Boutet et al., 2007), which contains manually annotated entries, and UniProtKB/TrEMBL, which contains computer-annotated entries. Until recently, SwissProt curation activities were focused on annotating all available protein sequences. Yet, manual protein annotation using SwissProt standards is an involved process, which requires reading relevant publications and annotating function, sequence features, locations, protein interactions, post translational modifications, and other descriptors. This process requires several hours for each protein and extending it across a single viral genome can be very time consuming. Given that thousands of new genome sequences are submitted for viruses like influenza, there is no question that the time of manual annotation of all available records has come to an end.
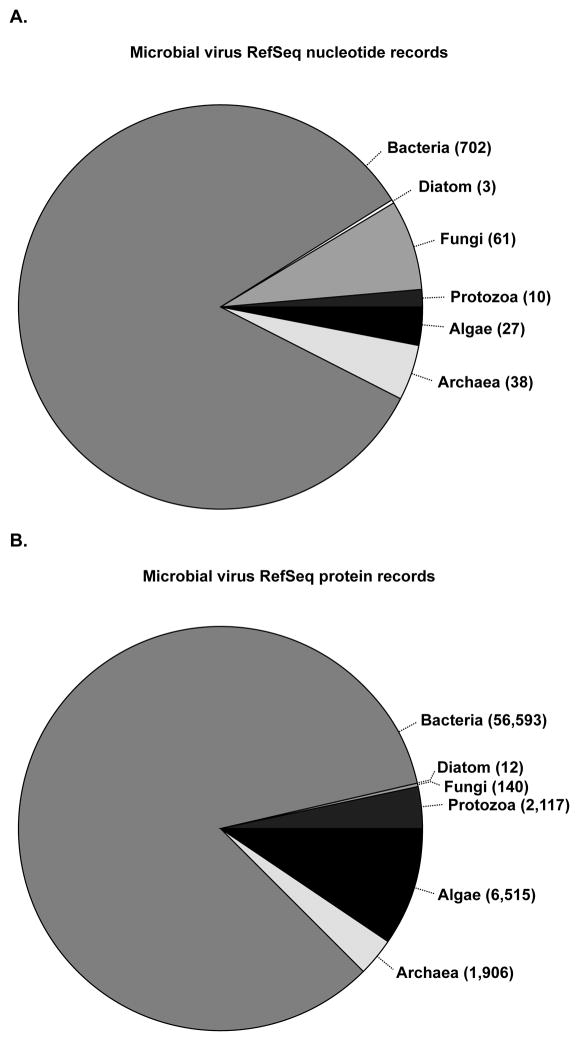
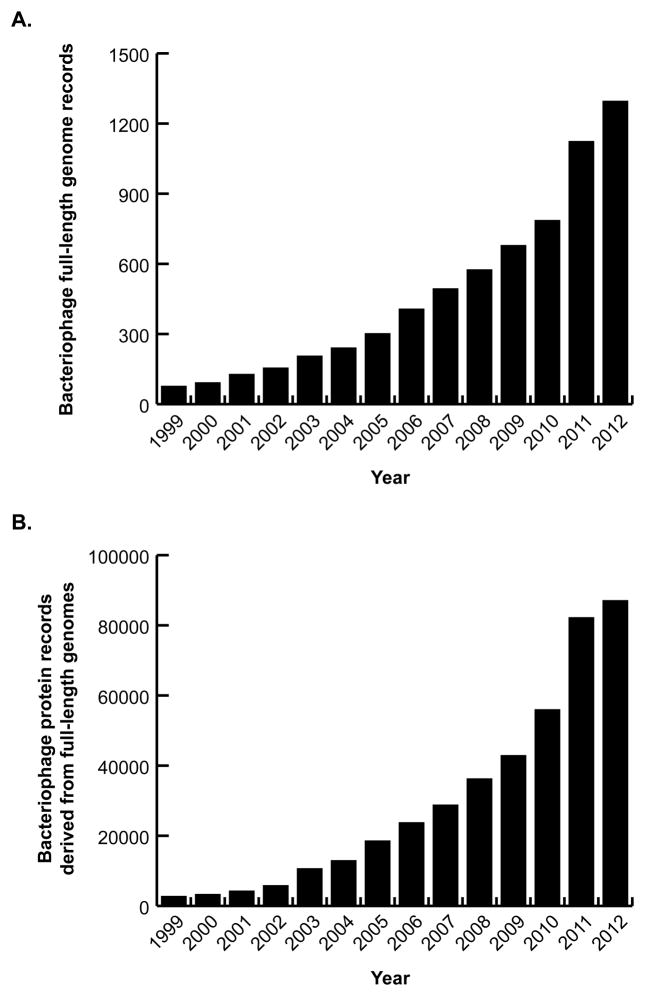
NCBI has attempted to ameliorate problems related to the rapid increase in virus genome sequencing using reference genomes (RefSeq), typically one for each virus species. This approach limits the number of manually curated genomes, but maintains comprehensive coverage of virus diversity. There are currently 4,218 viral RefSeq genome records publicly available from NCBI. Most of the 841 microbial virus RefSeq records are bacteriophages (Figure 2), which is not surprising given the current rate of bacteriophage genome sequencing (Figure 3). Although this reference genome approach offers a significant reduction from the 35,410 validated virus genome sequence records in GenBank (a figure which includes 1126 validated microbial virus genome records but does not include influenza sequences), there are still too many individual RefSeq protein records to manually annotate.
Figure 2. Distribution of microbial virus RefSeq records.
A. Current number1 of microbial virus nucleotide sequence records deposited in the RefSeq database broken down by host – algae, archaea, bacteria, diatom, fungi, protozoa. B. Current number1 of microbial virus protein sequence records deposited in the RefSeq database broken down by host – algae, archaea, bacteria, diatom, fungi, protozoa. 1Number of records calculated on September 11, 2012.
Figure 3. “Complete genome” sequences of virus RefSeq records.
A. Cumulative number of bacteriophage genome sequence records indexed by GenBank as “complete genomes” by year, 1999 to 20121. Note some of these sequences have yet to be validated by RefSeq as full-length genomes. B. Cumulative number of bacteriophage protein sequence records derived from genome sequences in (A). 1Number of records calculated on September 11, 2012.
Reference proteomes
The current SwissProt curation model is based on the manual annotation of one complete proteome per taxonomic genus. These “reference proteomes” are chosen from existing NCBI RefSeq genomes, so each reference has manually curated genome and proteome components and can be used as a gold standard (The UniProt Consortium, 2012). In UniProt release 07_12, there are 353 viral reference proteomes: 188 from animal, 87 from plant and 78 from microbial viruses. Reference proteomes comprise 12,758 individual protein entries - a sum which can be manually annotated and updated with reasonable effort. This number is expected to grow slowly as new viral genera are described – unlike the expected rapid increase in viral sequences – so reference proteomes should continue to provide a stable representative list of well curated viral proteins which can be used to access and/or propagate the annotation.
Unfortunately the diversity of microbial viruses complicates the establishment of references for proteomic annotation. Indeed, many bacterial viruses are subject to cross-species recombination, challenging their taxonomic classification (Krupovic et al., 2011). Moreover, the microbial virus gene diversity within a taxonomic family is high compared to vertebrate viruses, and many predicted proteins are named “uncharacterized” because there is no experimental information, and nothing of their function can be predicted. For example, the Mimivirus proteome comprises 909 proteins, 560 of which are uncharacterized in SwissProt (Suzan-Monti, La Scola, and Raoult, 2006). Therefore, SwissProt reference proteomes are useful standards for “core” microbial virus proteins, i.e. the proteins that all viruses of a genus have in common, but may not include many important proteins.
Virus ontology
Information published in free-text or transcribed in protein annotation is extremely valuable for users, but it is a complicated task for computers to extract and search this data. To deal with large scale annotation, a common set of concepts is needed to structure the protein knowledge within databases. This is especially true for important fields like protein function, which is written in free-text in UniProtKB. To ameliorate this problem, an ontology of 135 concepts has been created for eukaryotic viral proteins. This ontology is comprised of five parts - virion, entry, gene expression/replication, exit, and host-defense modulation - and has been used to annotate viral proteins in SwissProt/UniProtKB and in the ViralZone (viralzone.expasy.org) (Hulo et al., 2011) web resource where concepts are detailed and linked to viruses and proteins. For example the page “Viral penetration into host nucleus” (http://viralzone.expasy.org/all_by_protein/989.html) displays a picture of the different strategies to cross nuclear membranes during viral entry, a text detailing the various molecular processes involved, and a list of the 17 viral families involved in this process with references to PubMed articles. All proteins playing an active role in this process are also listed; that is the 572 viral proteins having received the keyword in SwissProt/UniProtKB. Though the 135 concepts developed at UniProt are currently focused on the eukaryotic virus replication cycle, in the future additional concepts will describe the prokaryotic virus counterpart. The UniProtKB viral ontology is currently in the process of being integrated into Gene Ontology (GO) (Gene Ontology Consortium, 2012) in coordination with GO Consortium efforts to cover both eukaryotic and prokaryotic viruses.
The advantage of ontology annotation versus free-text annotation is scalability. Controlled vocabulary provides annotation that can be propagated efficiently and allows exhaustive search in databases using one or several concepts. The ontology approach also facilitates the annotation process since it’s easier to assign function from a limited list of 135 keywords than writing a text from scratch to explain a protein/gene function.
Community annotation
Although the importance of bacteriophage in the molecular understanding of gene function goes back to the earliest days of molecular biology, annotation of viruses in general and phage in particular has lagged compared to other model systems. This may be attributed in part to the lack of stably funded dedicated model organism databases for viruses, which means that professional curation of viral genomes is being done only as part of larger, more general curation efforts. To our knowledge, the PortEco project is the only data resource where improving the databases for bacteriophage is an explicit part of a model organism resource’s mission, and even for PortEco, the mandate was only for the phage, plasmids, and mobile elements using laboratory E. coli as a host. This situation is unlikely to change, so alternative models for increasing annotation of viral genomes will require more involvement of the broader scientific community.
Enlisting the broader community to participate in annotation has obvious benefits in terms of scale, prioritization of areas of interest, and cost. However, community annotation has met with limited success for a variety of reasons (Bunt et al., 2012). FlyBase (Tweedie et al., 2009) and the Arabidopsis Information Resource (TAIR; (Swarbreck et al., 2008)) have had some success with recruiting authors of recent papers to participate in annotation (Bunt et al., 2012); the TAIR effort is aided by cooperation from several leading plant biology journals who notify authors of accepted papers that there is a website where they can contribute annotations to TAIR (Berardini et al., 2012). We hope that Virology and other journals relevant to bacteriophage biology will consider participating in a similar activity.
One recent effort to enlist community expertise to improve annotation of bacteriophage genomes is the PhAnToMe project (http://phantome.org). In 2011, PhAnToMe gathered experts in bacteriophage biology at BioSphere 2. The focus of the workshop was to improve tools for automated annotation based on RAST annotation pipeline (Aziz et al., 2008) and the SEED functional classification of gene products into subsystems (Overbeek et al., 2005). More than 80 subsystems for phage-specific biology were added to SEED through PhAnToMe, improving bacteriophage gene prediction, prophage identification, and viral metagenomics capabilities of these pipelines. Although PhAnTome periodically updates automated annotations for more than 839 bacteriophages, no stable mechanism was established to facilitate ongoing literature-based manual annotation of reference proteins such as those needed to seed SwissProt or Protein Clusters entries.
One possible approach to sustain community annotation of Protein Clusters is illustrated by the Community Assessment of Community Annotation with Ontologies (CACAO), organized by the PortEco project to couple annotation to undergraduate education. CACAO uses the teaching of critical reading of the scientific literature to have students participate in functional annotation based on the GO (Ashburner et al., 2000). Teams of students compete to make annotations of any protein in UniProt via the GONUTS (http://gowiki.tamu.edu) website (Renfro et al., 2012). Annotation is done in two week “innings”. Annotations are entered during the first week. During the second week, teams can take points from one another by identifying and correcting problematic annotations. The competitive nature of CACAO has stimulated students to submit thousands of literature-based annotations per semester. Annotations are judged by experts before being submitted back to the GO consortium for further review prior to inclusion in data resources.
Applying CACAO more broadly to viral genome annotation will require not only recruiting CACAO mentors with virology expertise, but as with the PhAnToMe efforts with the SEED, improvements in GO are needed to better cover phage biology. We are currently working with the virus group of the GO consortium to incorporate ontology terms from the ACLAME MEGO system (Leplae, Lima-Mendez, and Toussaint, 2010) into GO, and to cross-reference GO and SEED functional categories. Improvements to the ontologies to better reflect the relevant biology is another area where community involvement is needed. In addition to capturing literature-based annotation with existing GO terms, CACAO provides an opportunity to identify areas where GO needs improvement through new terms or reorganization of its structure.
The path forward
The emerging genome annotation community includes not only public databases like those at NCBI and UniProtKB/Swiss-Prot, but also large sequencing centers, independent databases like ACLAME, community efforts like PhAnToMe, as well as individual researchers. Given this landscape, we are now challenged to leverage contributions among these stakeholders and capture them in a single aggregate space. Though this proposition will certainly require unprecedented coordination, it also offers the promise of an adaptable, system-wide approach capable of keeping pace with the dynamic demands of microbial virus genome annotation.
We propose a hybrid model of genome annotation wherein expert curators from public databases collaborate with independent and community curation groups to best exploit the unique strengths of each group. This collaboration will require coordination of both pursuit and practice, and nomenclature and feature annotation conventions will be necessary to facilitate consistent contributions from all groups. To orchestrate this cabal, we propose the formation of a microbial virus genome annotation working group - PhaGeAn - which will help develop standards for virus genome annotation, prepare “gold standard” records, and coordinate the distribution of annotation from independent databases and community efforts.
Our hybrid model relies on the use of Protein Clusters to aggregate and disseminate annotation data. This resource supports a unique combination of data aggregation, visualization, and propagation features which should provide the flexibility necessary to coordinate data entry from a myriad of sources and allow review of complete data sets prior to dissemination (Klimke et al., 2009). Annotation data will be directly captured from NCBI, SwisProt, and other resources, parsing well annotated “gold standard” records, or uploading simple tables containing protein accession numbers, protein names, and other annotations. This resource should provide a single, public depository where any uploaded data can be freely accessed and used in annotation pipelines or other tools.
Conclusion
With the horizon filled with viral genome sequencing projects, the coming years portend a struggle to accurately annotate a building torrent of sequence data. To transform this sequence stream into usable, well annotated genome data sets, previously disjoined stakeholders - public databases, sequencing centers, and community groups - must articulate common annotation goals and leverage their combined knowledge and individual strengths. Such collaborative efforts are critical to the coverage, consistency, and community participation necessary for a sustainable virus genome annotation model. Despite some very real challenges engaging the broader scientific community and coordinating annotation efforts among disparate groups, in an era of restrained research budgets, this is the only path forward.
The article includes highlights from the NCBI Genome Annotation Workshop and offers an overview of current microbial virus genome annotation efforts at NCBI, SwissProt, and within the greater scientific community. Each of these efforts has short comings, and it is difficult to imagine that the current, disparate approach can mitigate the growing need for better microbial virus genome annotation. This reality leads us to argue for a collaborative effort between stakeholders, and we present a blueprint for a hybrid annotation model within the manuscript.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. Work by PLM was supported by SIB Swiss Institute of Bioinformatics. Work by JH was supported by NIGMS U24GM088849 and NIH R01 GM089636.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JRB wrote the first drafts of the “”Introduction,” the sections on “Virus genome annotation efforts at NCBI,” “Protein Clusters,” “The path forward,” and the “Conclusions and perspectives.” PL wrote the first draft of the section on “UniProt-KB/SwissProt.” JCH wrote the section “Community annotation.” All authors revised all sections and read and approved the final draft.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
J. Rodney Brister, Email: philippe.lemercier@isb-sib.ch.
James C. Hu, Email: jimhu@tamu.edu.
Bibliography
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Li D, Muller R, Chetty R, Ploetz L, Singh S, Wensel A, Huala E. Assessment of community-submitted ontology annotations from a novel database-journal partnership. Database (Oxford) 2012;(0):bas030. doi: 10.1093/database/bas030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33(Web Server issue):W451–4. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- Brister JR, Bao Y, Kuiken C, Lefkowitz EJ, Le Mercier P, Leplae R, Madupu R, Scheuermann RH, Schobel S, Seto D, Shrivastava S, Sterk P, Zeng Q, Klimke W, Tatusova T. Towards Viral Genome Annotation Standards, Report from the 2010 NCBI Annotation Workshop. Viruses. 2010;2(10):2258–68. doi: 10.3390/v2102258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt SM, Grumbling GB, Field HI, Marygold SJ, Brown NH, Millburn GH. Directly e-mailing authors of newly published papers encourages community curation. Database (Oxford) 2012:bas024. doi: 10.1093/database/bas024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TU. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(Database issue):D71–5. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AMQ International Committee on Taxonomy of Viruses; International Union of Microbiological Societies. Virology Division. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; Amsterdam: 2012. [Google Scholar]
- Karsch-Mizrachi I, Nakamura Y, Cochrane G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40(Database issue):D33–7. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009;37(Database issue):D216–23. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke W, O’Donovan C, White O, Brister JR, Clark K, Fedorov B, Mizrachi I, Pruitt KD, Tatusova T. Solving the Problem: Genome Annotation Standards before the Data Deluge. Stand Genomic Sci. 2011;5(1):168–93. doi: 10.4056/sigs.2084864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Lima-Mendez G, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38(Database issue):D57–61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39(Database issue):D52–7. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Brown G, Tatusova T, Maglott DR, MJaOJ . The NCBI Handbook [Internet] National Center for Biotechnology Information (US); Bethesda (MD): 2002. The Reference Sequence (RefSeq) Database. [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37(Database issue):D32–6. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33 (Database issue):D501–4. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfro DP, McIntosh BK, Venkatraman A, Siegele DA, Hu JC. GONUTS: the Gene Ontology Normal Usage Tracking System. Nucleic Acids Res. 2012;40(Database issue):D1262–9. doi: 10.1093/nar/gkr907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, Huala E. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36(Database issue):D1009–14. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]