Abstract
Purpose
To evaluate the prognostic significance of the international European LeukemiaNet (ELN) guidelines for reporting genetic alterations in acute myeloid leukemia (AML).
Patients and Methods
We analyzed 1,550 adults with primary AML, treated on Cancer and Leukemia Group B first-line trials, who had pretreatment cytogenetics and, for cytogenetically normal patients, mutational status of NPM1, CEBPA, and FLT3 available. We compared complete remission (CR) rates, disease-free survival (DFS), and overall survival (OS) among patients classified into the four ELN genetic groups (favorable, intermediate-I, intermediate-II, adverse) separately for 818 younger (age < 60 years) and 732 older (age ≥ 60 years) patients.
Results
The percentages of younger versus older patients in the favorable (41% v 20%; P < .001), intermediate-II (19% v 30%; P < .001), and adverse (22% v 31%; P < .001) genetic groups differed. The favorable group had the best and the adverse group the worst CR rates, DFS, and OS in both age groups. Both intermediate groups had significantly worse outcomes than the favorable but better than the adverse group. Intermediate-I and intermediate-II groups in older patients had similar outcomes, whereas the intermediate-II group in younger patients had better OS but not better CR rates or DFS than the intermediate-I group. The prognostic significance of ELN classification was confirmed by multivariable analyses. For each ELN group, older patients had worse outcomes than younger patients.
Conclusion
The ELN classification clearly separates the genetic groups by outcome, supporting its use for risk stratification in clinical trials. Because they have different proportions of genetic alterations and outcomes, younger and older patients should be reported separately when using the ELN classification.
INTRODUCTION
Identification of patients with acute myeloid leukemia (AML) who would likely respond to current therapies and those who are less likely to do well and are potential candidates for more aggressive treatment is of major clinical importance. Acquired cytogenetic and molecular alterations at diagnosis are among the most important independent factors used to stratify patients with AML into prognostic categories.1–4 Although the existing cytogenetic risk classifications of AML agree that patients with core-binding factor AML (CBF-AML) with t(8;21) (q22;q22) or inv(16)(p13.1q22)/t(16;16)(p13.1;q22) should be classified in the favorable-risk, those with cytogenetically normal AML (CN-AML) in the intermediate-risk, and those with a complex karyotype in the adverse-risk categories, these classifications differ in the way patients with many remaining recurrent cytogenetic abnormalities are classified.5–11 Moreover, none of these classifications include the results of molecular analyses, convincingly shown to provide important prognostic information, especially for patients with CN-AML.3,4
Therefore, in 2010, an international expert panel, working on behalf of the European LeukemiaNet (ELN), proposed a standardized system for reporting cytogenetic and selected molecular abnormalities in studies correlating genetic findings with treatment outcome in AML to facilitate meaningful comparisons among studies.12 This system stems from the 2008 revision of the WHO classification of myeloid neoplasms and acute leukemia13 and is based on published data on the prognostic significance of cytogenetic5–10,14–16 and molecular3,17–32 alterations. The novel aspect of the ELN classification is that it divides patients with CN-AML into genetic groups according to molecular alterations recognized in the WHO classification, namely NPM1, CEBPA, and FLT3 mutations. To the best of our knowledge, the ability of the four ELN genetic groups (hereafter referred to as “ELN groups”) favorable, intermediate-I, intermediate-II, and adverse (Table 1) to predict treatment outcome has not been tested in large cohorts of similarly treated patients, except for a recent study33 comprising patients with primary (de novo) and secondary AML. If the prognostic utility of ELN classification could be convincingly demonstrated, its use would become essential for risk stratification of patients with AML in prospective clinical trials. Hence, we have applied the ELN classification to a relatively large cohort of 1,550 adult patients with AML to assess its usefulness for the prognostic classification of both younger (age < 60 years) and older (age ≥ 60 years) patients. To avoid the confounding effects of AML type (primary v secondary) and different postremission therapies (chemotherapy v allogeneic stem-cell transplantation [SCT]), we included only patients with primary AML enrolled onto Cancer and Leukemia Group B (CALGB) first-line treatment trials who did not undergo allogeneic SCT in first complete remission (CR) per protocol. As recommended by ELN, we also analyzed outcomes of patients belonging to genetic subsets within each ELN group to gain further insights into the ELN classification.
Table 1.
European LeukemiaNet Standardized Reporting System for Correlation of Cytogenetic and Molecular Genetic Data in AML With Clinical Data12
| Genetic Group | Subsets |
|---|---|
| Favorable | t(8;21)(q22;q22); RUNX1-RUNX1T1 |
| inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB-MYH11 | |
| Mutated NPM1 without FLT3-ITD (normal karyotype) | |
| Mutated CEBPA (normal karyotype) | |
| Intermediate-I | Mutated NPM1 and FLT3-ITD (normal karyotype) |
| Wild-type NPM1 and FLT3-ITD (normal karyotype) | |
| Wild-type NPM1 without FLT3-ITD (normal karyotype) | |
| Intermediate-II | t(9;11)(p22;q23); MLLT3-MLL |
| Cytogenetic abnormalities not classified as favorable or adverse | |
| Adverse | inv(3)(q21q26.2) or t(3;3)(q21;q26.2); RPN1-EVI1 |
| t(6;9)(p23;q34); DEK-NUP214 | |
| t(v;11)(v;q23); MLL rearranged | |
| –5 or del(5q) | |
| –7 | |
| abnl(17p) | |
| Complex karyotype* |
Abbreviations: AML, acute myeloid leukemia; ITD, internal tandem duplication.
Complex karyotype is defined as three or more chromosome abnormalities in the absence of one of the WHO designated recurring translocations or inversions: t(8;21), inv(16) or t(16;16), t(15;17), t(9;11), t(v;11)(v;q23), t(6;9), inv(3) or t(3;3).
PATIENTS AND METHODS
Patients Studied
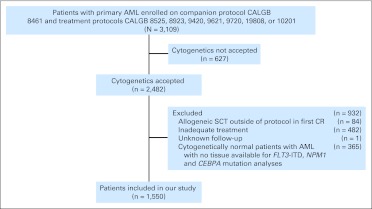
All patients were enrolled onto CALGB 8461, a prospective cytogenetics companion study,7 between 1985 and 2006. Only patients diagnosed with primary AML, defined by WHO criteria13 (except for acute promyelocytic leukemia), who had pretreatment cytogenetic results were eligible. All patients with CN-AML with material available were tested for an FLT3 internal tandem duplication (FLT3-ITD) and NPM1 and CEBPA mutations. The patients were enrolled onto CALGB first-line treatment protocols (Fig 1; Data Supplement).34–41 Per protocol, no patient received allogeneic SCT in first CR. The median follow-up time for living patients was 7.5 years (range, 0.6 to 19.1 years). All protocols were approved by the institutional review board of each participating institution, and written informed consent was obtained from all patients before enrollment in accordance with the Declaration of Helsinki.
Fig 1.
Overview of the study design. AML, acute myeloid leukemia; CALGB, Cancer and Leukemia Group B; CR, complete remission; ITD, internal tandem duplication; SCT, stem-cell transplantation.
Cytogenetic Studies
Cytogenetic analyses of bone marrow (BM) and/or blood were performed in institutional CALGB cytogenetics laboratories. Karyotypes were interpreted according to the International System for Human Cytogenetic Nomenclature,42 and results were confirmed by central review.43 The diagnosis of CN-AML was based on the analysis of ≥ 20 metaphases from BM subjected to short-term (24- to 48-hour) culture.
Analyses of FLT3-ITD and NPM1 and CEBPA Mutations
All patients with CN-AML were enrolled onto companion protocols CALGB 9665 (leukemia tissue bank) and CALGB 20202 (molecular studies in AML). Mononuclear cells were enriched through Ficoll-Hypaque gradient centrifugation and were cryopreserved. Genomic DNA and total RNA were extracted from BM or blood with ≥ 20% blasts, and FLT3-ITD20 and NPM122,25 and CEBPA29 mutations were analyzed centrally.
Statistical Analyses
The primary aim of our study was to assess differences in clinical outcome of patients with AML who were categorized into the ELN groups (Table 1). As recommended by ELN, we also tested for outcome differences among genetic subsets within each ELN group. Baseline characteristics were compared by using Fisher's exact test for categorical variables and Wilcoxon rank-sum and Kruskal-Wallis tests for continuous variables (Data Supplement). Clinical end points were defined according to published recommendations (Data Supplement).44 For time-to-event analyses, survival estimates were calculated by using the Kaplan-Meier method, and groups were compared by using the log-rank test. The Holm step-down procedure and Sidak adjustment, respectively, were used to adjust P values for multiple comparisons for CR and survival analyses concerning subsets within ELN groups.45 We constructed multivariable logistic regression models to analyze factors for CR achievement and multivariable Cox proportional hazards models for factors associated with survival end points (Data Supplement). All analyses were performed by the Alliance for Clinical Trials in Oncology Statistics and Data Center.
RESULTS
Pretreatment Characteristics and the Distribution of ELN Groups
The median age of all patients was 58 years (range, 17 to 86 years), and 55% were male. This patient population comprised 818 adults age younger than 60 years and 732 patients age 60 years or older. For baseline clinical features and outcomes of younger and older patients and pretreatment features of each ELN group in younger and older patients, see the Data Supplement.
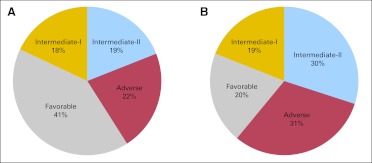
Among all patients, 31% were classified in the favorable, 18% in the intermediate-I, 24% in the intermediate-II, and 26% in the adverse group. However, the distribution of ELN groups among younger and older patients differed significantly (Fig 2). The proportion of younger patients classified in the favorable group was twice that in older patients (P < .001), whereas the proportion of younger intermediate-II (P < .001) and adverse (P < .001) groups was only about two thirds that of the respective ELN groups in older patients. The proportion of younger and older patients classified in the intermediate-I group was similar (P = .64).
Fig 2.
Distribution of the European LeukemiaNet genetic groups in younger (A) and older (B) adults with primary acute myeloid leukemia. The favorable group is more (P < .001) and the intermediate-II and adverse groups are less (P < .001) common among younger patients compared with older patients.
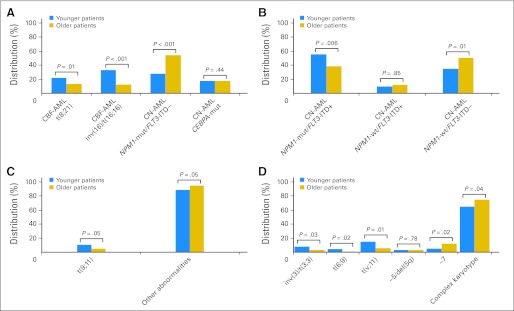
The proportion of genetic subsets within ELN groups also differed by age (Fig 3). Although more than half the younger patients in the favorable group (Fig 3A) had CBF-AML, only one fourth of older patients did (P < .001). Conversely, patients with CN-AML who had the NPM1 mutation without FLT3-ITD (hereafter designated NPM1-mut [mutation]/FLT3-ITD–) were twice as common among older as among younger patients (P < .001). In the intermediate-I group (Fig 3B), a larger proportion of younger patients had mutated NPM1 and FLT3-ITD (NPM1-mut/FLT3-ITD+; P = .006), and a smaller proportion had wild-type (wt) NPM1 without FLT3-ITD (NPM1-wt/FLT3-ITD–; P = .01) compared with older patients. In the intermediate-II group (Fig 3C), t(9;11)(p22;q23) was more common in younger patients, and the other abnormalities were more common in older patients. Most younger patients (65%) and older patients (75%) in the adverse group (Fig 3D) had a complex karyotype (P = .04), but among the remaining patients, balanced abnormalities (ie, inv(3)(q21q26.2)/t(3;3)(q21;q26.2) (P = .03); t(6;9)(p23;q34) (P = .02); t(v;11)(v;q23) (P = .01)) were found predominantly in younger patients and −7 (P = .02) was found in older patients. These data show that distributions of the ELN groups and subsets differ significantly between younger and older patients, underscoring the existence of important biologic differences between age groups and strongly supporting the need to assess the impact of ELN classification on outcome separately for younger and older adults.
Fig 3.
Distribution of the genetic subsets within European LeukemiaNet genetic groups in younger and older adults with primary acute myeloid leukemia (AML). (A) The favorable group consists of four genetic subsets. The first two subsets are patients with core-binding factor AML (CBF-AML) with either t(8;21) (ie, t(8;21)(q22;q22)/RUNX1-RUNX1T1) or inv(16)/t(16;16) (ie, inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB-MYH11). The second two subsets are patients with cytogenetically normal AML (CN-AML) with either NPM1-mut/FLT3-ITD– (ie, mutated NPM1 without FLT3-ITD [internal tandem duplication]) or CEBPA-mut (ie, mutated CEBPA). (B) The intermediate-I group consists of three genetic subsets of patients with CN-AML and either NPM1-mut/FLT3-ITD+ (ie, mutated NPM1 and FLT3-ITD) or NPM1-wt/FLT3-ITD+ (ie, wild-type NPM1 and FLT3-ITD) or NPM1-wt/FLT3-ITD– (ie, wild-type NPM1 without FLT3-ITD). (C) The intermediate-II group consists of two genetic subsets of patients with either t(9;11) (ie, t(9;11)(p22;q23)/MLLT3-MLL) or other abnormalities (ie, cytogenetic abnormalities not classified as favorable or adverse). (D) The adverse group consists of seven genetic subsets: (1) inv(3)/t(3;3) (ie, inv(3)(q21q26.2) or t(3;3)(q21;q26.2)/RPN1-EVI1), (2) t(6;9) (ie, t(6;9)(p23;q34)/DEK-NUP214), (3) t(v;11) (ie, t(v;11)(v;q23)/MLL rearranged), (4) −5/del(5q) (ie, monosomy of chromosome 5 or deletion of q), (5) −7 (ie, monosomy of chromosome ), (6) abnl(17p) (ie, abnormalities of the short arm or chromosome; no patient had this abnormality in our study), or () “complex karyotype” (ie, a complex karyotype containing three or more cytogenetic abnormalities).
Clinical Outcomes According to ELN Groups
Because of the aforementioned genetic differences and more intensive consolidation treatment received by younger patients, we performed outcome analyses separately for younger and older patients. CR rates differed among the ELN groups for both younger (P < .001) and older (P < .001) patients, with the highest rates observed in the favorable groups (96% and 83%, respectively) and the lowest rates observed in the adverse groups (50% and 39%, respectively; Table 2). Adjusted pairwise comparisons among the ELN groups yielded significant differences for all comparisons except for those between the intermediate-I and intermediate-II groups for which CR rates were not different in either younger (76% v 79%; P = .58) or in older (61% v 63%; P = .82) patients.
Table 2.
Treatment Outcomes of Younger (age < 60 years) and Older (age ≥ 60 years) Patients With Primary Acute Myeloid Leukemia According to European LeukemiaNet Genetic Groups
| Outcome End Point | Favorable |
Intermediate-I |
Intermediate-II |
Adverse |
P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | 95% CI | No. | % | 95% CI | No. | % | 95% CI | No. | % | 95% CI | ||
| Younger patients (n = 818) | (n = 339) | (n = 144) | (n = 156) | (n = 179) | |||||||||
| Complete remission rate | 324 | 96 | 109 | 76 | 123 | 79 | 90 | 50 | < .001a | ||||
| Disease-free survivalb | .001c | ||||||||||||
| Median, years | 5.5 | 0.8 | 1.2 | 0.6 | |||||||||
| Disease-free at 3 years | 55 | 49 to 60 | 23 | 16 to 31 | 34 | 26 to 45 | 10 | 5 to 17 | |||||
| Overall survivald | < .001e | ||||||||||||
| Median, years | 11.5 | 1.2 | 2.1 | 0.8 | |||||||||
| Alive at 3 years | 66 | 60 to 70 | 28 | 21 to 36 | 45 | 37 to 52 | 12 | 8 to 18 | |||||
| Older patients (n = 732) | (n = 145) | (n = 136) | (n = 222) | (n = 229) | |||||||||
| Complete remission rate | 120 | 83 | 83 | 61 | 139 | 63 | 89 | 39 | < .001a | ||||
| Disease-free survivalf | < .001f | ||||||||||||
| Median, years | 1.1 | 0.6 | 0.7 | 0.5 | |||||||||
| Disease-free at 3 years | 24 | 17 to 32 | 10 | 5 to 17 | 11 | 6 to 16 | 6 | 2 to 12 | |||||
| Overall survivalh | < .001a | ||||||||||||
| Median, years | 1.6 | 0.9 | 0.9 | 0.5 | |||||||||
| Alive at 3 years | 33 | 25 to 41 | 11 | 6 to 17 | 16 | 11 to 21 | 3 | 2 to 6 | |||||
The adjusted pairwise comparisons for favorable v intermediate-I, favorable v intermediate-II, favorable v adverse, intermediate-I v adverse, and intermediate-II v adverse were statistically significant, whereas there was no significant difference between the intermediate-I and intermediate-II groups.
The median follow-up time for younger patients who had not had an event was 7.9 years (range, 0.6-19.1 years).
The adjusted pairwise comparisons for favorable v intermediate-I, favorable v intermediate-II, favorable v adverse, and intermediate-II v adverse were statistically significant, whereas there was no significant difference between the intermediate-I and intermediate-II and between intermediate-I and adverse groups.
The median follow-up time for younger patients alive was 7.6 years (range, 0.6-19.1 years).
All adjusted pairwise comparisons were significant.
The median follow-up time for older patients who had not had an event was 5.9 years (range, 4.4-16.4 years).
The adjusted pairwise comparisons for favorable v intermediate-I, favorable v intermediate-II, and favorable v adverse were statistically significant. There were trends for longer disease-free survival of intermediate-I and intermediate-II groups when compared with the adverse group, whereas there was no significant difference between the intermediate-I and intermediate-II groups.
The median follow-up time for older patients alive was 6.1 years (range, 2.3-16.4 years).
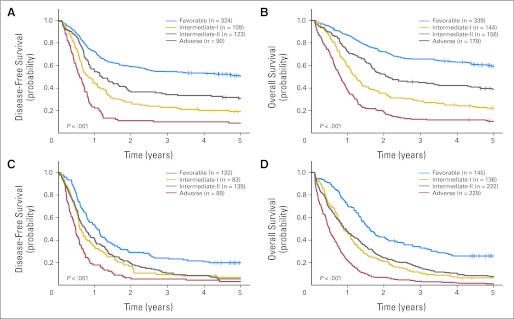
Similarly, disease-free survival (DFS; P = .001 and P < .001) and overall survival (OS; P < .001 and P < .001) differed across the ELN groups in younger and older patients. Patients in the favorable group had the longest and those in the adverse group the shortest DFS and OS, with patients classified in the intermediate-I and intermediate-II groups having DFS and OS significantly worse than those in the favorable group but significantly better than those in the adverse group (Table 2; Fig 4). Although OS of younger intermediate-II group patients was longer than that of younger intermediate-I group patients (3-year rates, 45% v 28%; P = .02; Table 2; Fig 4B), DFS (P = .33) in younger and DFS (P = .99) and OS (P = .99) in older patients in the intermediate-I and intermediate-II groups were similar.
Fig 4.
Outcome of patients with primary acute myeloid leukemia classified into the four European LeukemiaNet genetic groups according to the European LeukemiaNet recommendations. (A) Disease-free survival and (B) overall survival of patients younger than age 60 years; (C) disease-free survival and (D) overall survival of patients age 60 years or older.
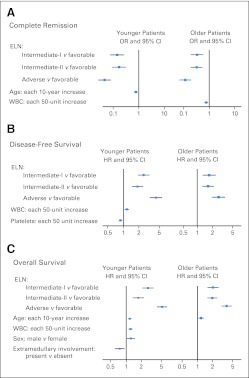
To determine whether ELN groups remain associated with outcome when controlling for established prognostic factors in AML, we performed multivariable analyses. For both age groups, CR rates, DFS, and OS remained better for patients in the favorable group and worse for patients in the adverse group compared with those in other ELN groups (P < .001 for all comparisons) after adjustment for other variables (Fig 5; Data Supplement).
Fig 5.
Forest plots summarizing multivariable analyses for (A) complete remission (CR), (B) disease-free survival (DFS), and (C) overall survival (OS) in younger and older patients. (A) Odds ratios (ORs) of less than 1 indicate lower CR rate for the first category listed for the categorical variables and the higher values of the continuous variables. (B and C) Hazard ratios (HRs) greater than 1 indicate higher risks and those less than 1 indicate lower risks of relapse or death (DFS) or death (OS) for the first category listed for the categorical variables and the higher values of the continuous variables. Variables considered for inclusion in the multivariable models had to have a univariable P value of less than .2 and were as follows for younger patients: for CR, European LeukemiaNet (ELN) groups, age (in 10-year increments), sex (male v female), and extramedullary involvement (present v absent); for DFS, ELN groups, WBC count (in 50-unit increments), platelets (in 50-unit increments), and extramedullary involvement; for OS, ELN groups, age, WBC count, sex, and extramedullary involvement; and for older patients: for CR, ELN groups, age, WBC count, and platelets; for DFS, ELN groups, platelets, sex, and extramedullary involvement; for OS, ELN groups, age, and sex. Only variables that were significant remained in the final models and are depicted in the forest plots.
In the multivariable modeling, we found that, compared with the favorable group, patients in the younger adverse group had almost 21 times lower odds of attaining CR, a 4.4 times increased risk of relapse or death, and a more than five times higher risk of death, whereas older adverse group patients had nine times lower odds of achieving CR, 2.6 times increased risk of relapse or death, and 3.7 times increased risk of death (Fig 5; Data Supplement).
Similarly, younger and older patients in the intermediate-I group had, respectively, approximately seven and three times lower odds of attaining CR, and these odds were also six and three times lower for the younger and older patients in the intermediate-II group than for those in the favorable group. Compared with the favorable group, the risk of relapse or death was increased 2.5-fold for younger and 1.7-fold for older patients in the intermediate-I group and almost two-fold for younger and 1.6-fold for older patients in the intermediate-II group. Likewise, the risk of death was increased 2.7-fold for younger and two-fold for older patients in the intermediate-I group and 1.7-fold for younger and almost two-fold for older patients in the intermediate-II group (Fig 5; Data Supplement).
Although direct comparisons between younger and older patients are limited by the fact that their treatment differed in intensity and the biology of the disease also differs, as exemplified by significant differences in the incidence of ELN groups (Fig 2) and subsets (Fig 3) between age groups, it is striking that for each ELN group, the outcome was worse for older patients (Fig 4). Consequently, DFS and OS of older patients in the favorable group were similar to those of younger patients in the intermediate-I group (Data Supplement), and DFS and OS of older patients in the intermediate-I and intermediate-II groups were similar to those of younger patients in the adverse group (Data Supplement).
Clinical Outcome of Patients Belonging to Genetic Subsets Within ELN Groups
The ELN guidelines recommend reporting response rates and outcome measures for specific subsets within each ELN group if sufficient numbers of patients are available.
ELN Favorable Group
Among younger patients, there was an overall difference in CR rates among the subsets (P = .04); although all CR rates were high, the highest were 99% and 98% for t(8;21) and inv(16) patients, respectively, and the lowest were 92% and 93% for patients with NPM1-mut/FLT3-ITD– and those with CEBPA mutations, respectively. In older patients, those with CBF-AML again had the highest CR rates (95% for t(8;21); 89% for inv(16)); the lowest CR rate, 69%, was observed in patients with CEBPA-mut (Data Supplement).
Although in younger patients DFS (P = .93) and OS (P = .30) were similar for all subsets, we found differences in OS (P = .02) but not DFS (P = .21) for older patients (Data Supplement). The overall difference in OS was likely affected by a shorter OS of older patients with CEBPA-mut compared with t(8;21) patients (adjusted P = .03; 3-year rates, 21% v 47%).
ELN Intermediate-I Group
Analysis of subsets within this group revealed differences with regard to CR rates (P = .02) and OS (P = .06; Data Supplement) only among older patients. Older NPM1-mut/FLT3-ITD+ patients had the highest CR rate (75%) and longest OS, and NPM1-wt/FLT3-ITD+ patients had the lowest CR rate (44%) and shortest OS (Data Supplement). All patients in the latter subset died within 26 months after diagnosis.
ELN Intermediate-II Group
This group contains two subsets, one comprising patients with t(9;11)(p22;q23) and the other including a heterogeneous set of patients harboring cytogenetic abnormalities not included in the favorable or adverse groups. A comparison of these subsets produced different results in younger and older patients (Data Supplement). In the former, despite similar CR rates (82% for t(9;11) v 78% for other abnormalities; P = 1.00), t(9;11) patients had a longer DFS (P = .04; 3-year rates, 57% v 31%) but not OS (P = .20) than those with other abnormalities. In contrast, older t(9;11) patients had a higher CR rate than those with other abnormalities (92% v 61%, P = .03), but their DFS was worse (P = .03; 3-year rates, 0% v 12%); OS was not significantly different (P = .24). The seemingly differing responses to treatment of subsets in younger and older patients are likely caused by cytogenetic heterogeneity of the other abnormalities subset in both age groups.
ELN Adverse Group
This group consists of several genetic subsets, of which the complex karyotype subset is the largest, constituting 65% of younger and 75% of older patients. In both age groups, the adverse group subsets differed with regard to CR rates (P < .001 for younger patients; P = .05 for older patients) and OS (P = .09 for younger patients; P = .10 for older patients). DFS also differed in older patients (P = .08), but too few younger patients achieved CR for analysis (Data Supplement).
Among younger patients, those with t(v;11)/MLL-rearranged (n = 26) had a high CR rate of 81%, higher than CR rates of patients with inv(3)/t(3;3) (20%; n = 15; adjusted P = .001), of those with a complex karyotype (48%; n = 117; adjusted P = .01), and of patients with −7 (33%; n = 9; adjusted P = .06). In older patients, who generally had lower CR rates, t(v;11)/MLL-rearranged patients (n = 14) also had the highest CR rate (57%), whereas those with inv(3)/t(3;3) (n = 7) again fared poorly (14%), as did patients with del(5q) (n = 8), none of whom attained CR. DFS and OS were short for all genetic subsets in both age groups (Data Supplement).
DISCUSSION
Our large study with prolonged follow-up demonstrates that application of the ELN reporting system to classify patients with AML allows a prognostic separation of the favorable and adverse groups from each other and from both intermediate groups. This has been achieved for all outcome end points analyzed and was shown to be independent from other prognostic factors by multivariable analyses. Moreover, the association of ELN groups with outcome was evident not only in younger adults, known to constitute a more prognostically heterogeneous patient population, but also in patients age 60 years or older, whose outcomes are generally worse,9,10,12 which makes discerning prognostically different subgroups in these patients more difficult. Our data also show that older patients, who received less intensive treatment, consistently had worse outcomes than younger patients classified in the same ELN group.
Although the ELN guidelines do not specify age of patients to be classified,12 a salient finding of our study is a demonstration that the distribution of all ELN groups except intermediate-I differs significantly between younger and older patients, as do distributions of several genetic subsets within each ELN group, including intermediate-I. These data are consistent with previous reports showing age-related differences in the distribution of particular cytogenetic abnormalities46,47 and strongly support the notion that the ELN classification should be applied to younger and older patients separately.
We observed a difference between younger and older patients concerning the intermediate-I and intermediate-II groups. Outcomes of older patients in these groups were virtually identical, whereas in younger patients, the intermediate-II group had a significantly longer survival than the intermediate-I group. Similar results (ie, no difference in outcome between the two intermediate groups in older patients and a better OS and relapse-free survival for the intermediate-II group in younger patients treated with chemotherapy) have been reported by Röllig et al,33 although only 80% of patients in their series had primary AML. However, these differences disappeared for patients undergoing allogeneic SCT,33 which underlines the significance of the treatment type for prognostic stratification.
The reasons for better outcome of younger, but not older, patients treated with chemotherapy in the intermediate-II as opposed to the intermediate-I group are unknown. The disparate results are likely related to marked cytogenetic heterogeneity of the other abnormalities subset in the intermediate-II group, which comprises numerous recurrent abnormalities. These outcome differences may also stem from a previously described phenomenon—that the prognostic significance of the same genetic alteration may vary in younger and older patients with AML.22,24,25 For instance, in younger patients, NPM1 mutations confer favorable prognosis mainly in the absence of FLT3-ITD,22,24 whereas in older patients, NPM1 mutations constitute a favorable prognostic factor independent from other molecular prognosticators.25 Likewise, younger patients with t(9;11) in our study had significantly longer DFS (P < .001) and OS (P < .001) than older patients with this translocation, and their DFS (P = .04) was longer than DFS of younger patients in the other abnormalities subset. In contrast, DFS of older t(9;11) patients was worse than DFS of patients with other abnormalities.
In summary, our large study of primary AML demonstrates a clear prognostic separation among the ELN genetic groups. This establishes clinical utility of the ELN classification and thus supports the mandatory application of this classification in future studies correlating genetic findings with clinical outcome and its use for risk-stratification of patients with AML in prospective clinical trials. However, to best capture the prognostic information provided by the ELN classification, younger and older patients should be considered separately because they differ with respect to the incidence of genetic alterations and outcome. We hope that addition of further genetic markers, such as those preliminarily tested in the context of the ELN classification (eg, TET2,48 ASXL1,49 RUNX150 mutations, FLT3-ITD allelic ratio33) and novel ones emerging from next-generation sequencing51,52 may, if their prognostic value is confirmed, refine the accuracy of patient risk stratification in clinical trials using the ELN classification.
Supplementary Material
Acknowledgment
We thank principal investigators and cytogeneticists from the Cancer and Leukemia Group B institutions participating in this study (see Appendix). We also thank Donna Bucci of the Cancer and Leukemia Group B Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services and Lisa J. Sterling, Christine Finks, and Colin G. Edwards, PhD, for data management.
Appendix
The following are Cancer and Leukemia Group B (CALGB) institutions, principal investigators, and cytogeneticists who participated in this study: Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, P. Nagesh Rao, Wendy L. Flejter, and Mark J. Pettenati (Grant No. CA03927); The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, Diane Minka, and Nyla A. Heerema (Grant No. CA77658); North Shore-Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R.K. Koduru (Grant No. CA35279); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (Grant No. CA02599); Dana-Farber Cancer Institute, Boston, MA: Harold J. Burstein, Ramana Tantravahi, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA32291); Duke University Medical Center, Durham, NC: Jeffrey Crawford, Sandra H. Bigner, Mazin B. Qumsiyeh, John Eyre, and Barbara K. Goodman (Grant No. CA47577); Washington University School of Medicine, St. Louis, MO: Nancy L. Bartlett, Michael S. Watson, Eric C. Crawford, Jaime Garcia-Heras, Peining Li, and Shashikant Kulkarni (Grant No. CA77440); University of Iowa Hospitals, Iowa City, IA: Daniel A. Vaena and Shivanand R. Patil (Grant No. CA47642); University of Chicago Medical Center, Chicago, IL: Hedy L. Kindler, Diane Roulston, Katrin M. Carlson, Yanming Zhang, and Michelle M. Le Beau (Grant No. CA41287); University of North Carolina, Chapel Hill, NC: Thomas C. Shea and Kathleen W. Rao (Grant No. CA47559); Dartmouth Medical School, Lebanon, NH: Konstantin Dragnev, Doris H. Wurster-Hill, and Thuluvancheri K. Mohandas (Grant No. CA04326); Rhode Island Hospital, Providence, RI: William Sikov, Teresita Padre-Mendoza, Jennifer A. Ahearn, Hon Fong L. Mark, Shelly L. Kerman, and Aurelia Meloni-Ehrig (Grant No. CA08025); Weill Medical College of Cornell University, New York, NY: John Leonard, Ram S. Verma, Prasad R.K. Koduru, Andrew J. Carroll, and Susan Mathew (Grant No. CA07968); Christiana Care Health Services, Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, Jeanne M. Meck, and Kathleen Richkind (Grant No. CA45418); Vermont Cancer Center, Burlington, VT: Steven M. Grunberg, Elizabeth F. Allen, and Mary Tang (Grant No. CA77406); Ft. Wayne Medical Oncology/Hematology, Ft. Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; University of Maryland Cancer Center, Baltimore, MD: Martin J. Edelman, Joseph R. Testa, Deana Hallman, Stuart Schwartz, Maimon M. Cohen, Judith Stamberg, and Yi Ning (Grant No. CA31983); University of Massachusetts Medical Center, Worcester, MA: William V. Walsh, Philip L. Townes, Vikram Jaswaney, Kathleen Richkind, Michael J. Mitchell, and Patricia Miron (Grant No. CA37135); University of California at San Diego: Barbara A. Parker, E. Robert Wassman Jr, Renée Bernstein, and Marie L. Dell'Aquila (Grant No. CA11789); Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA 12,449); Minneapolis Veterans Affairs Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (Grant No. CA47555); Western Pennsylvania Hospital, Pittsburgh, PA: John Lister and Gerard R. Diggans; University of Alabama at Birmingham: Robert Diasio and Andrew J. Carroll (Grant No. CA47545); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (Grant No. CA04457); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano, Larry Gordon, and Constance K. Stein (Grant No. CA21060); University of Illinois at Chicago: David J. Peace, Maureen M. McCorquodale, Kathleen E. Richkind, and Valerie Lindgren (Grant No. CA74811); Long Island Jewish Medical Center Community Clinical Oncology Program (CCOP), Lake Success, NY: Kanti R. Rai, Alan L. Shanske, and Prasad R. K. Koduru (Grant No. CA11028); Walter Reed National Military Medical Center, Washington, DC: Jeremy G. Perkins, Rawatmal B. Surana, Digamber S. Borgaonkar, and Karl S. Theil (Grant No. CA26806); Eastern Maine Medical Center, Bangor, ME: Thomas H. Openshaw and Laurent J. Beauregard (Grant No. CA35406); University of Minnesota, Minneapolis, MN: Bruce A. Peterson, Diane C. Arthur, and Betsy A. Hirsch (Grant No. CA16450); University of Puerto Rico School of Medicine, San Juan, PR: Eileen I. Pacheco, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton; University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Carl E. Freter, Judith H. Miles, Jeffrey R. Sawyer, Tim H. Huang, and Linda M. Pasztor (Grant No. CA12046); University of Nebraska Medical Center, Omaha, NE: Apar Ganti and Warren G. Sanger (Grant No. CA77298); Virginia Commonwealth University Minority-Based CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (Grant No. CA52784); University of California at San Francisco: Charles J. Ryan and Kathleen E. Richkind (Grant No. CA60138); Georgetown University Medical Center, Washington, DC: Minnetta C. Liu and Jeanne M. Meck (Grant No. CA77597); Medical University of South Carolina, Charleston, SC: Mark R. Green, Eduardo S. Cantu, G. Shashidhar Pai, and Daynna J. Wolff (Grant No. CA03927); Southern Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John Ellerton, Renée Bernstein, and Marie L. Dell'Aquila (Grant No. CA35421); McGill Department of Oncology, Montreal, Quebec, Canada: J.L. Hutchison and Jacqueline Emond (Grant No. CA31809); State University of New York Maimonides Medical Center, Brooklyn, NY: Sameer Rafla and Ram S. Verma (Grant No. CA25119); Columbia-Presbyterian Medical Center, New York, NY: Rose R. Ellison and Dorothy Warburton (Grant No. CA12011); Oklahoma University Health Sciences Center, Oklahoma City, OK: Howard Ozer and Shibo Li; Southeast Cancer Control Consortium, Winston-Salem, NC: James N. Atkins and Mark J. Pettenati; University of Cincinnati Medical Center, Cincinnati, OH: Orlando J. Martelo and Ashok K. Srivastava (Grant No. CA47515).
Footnotes
Supported in part by Grants No. CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657 from the National Cancer Institute, Bethesda, MD; The Coleman Leukemia Research Foundation; and the Deutsche Krebshilfe-Dr. Mildred Scheel Cancer Foundation (H.B.).
Presented in part at the 53rd Annual Meeting of the American Society of Hematology, San Diego, CA, December 10-13, 2011.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Joseph O. Moore, Novartis (C) Stock Ownership: None Honoraria: Joseph O. Moore, Novartis Research Funding: Joseph O. Moore, Novartis, Ariad Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Krzysztof Mrózek, Guido Marcucci, Deedra Nicolet, Kati S. Maharry, Clara D. Bloomfield
Financial support: Guido Marcucci, Clara D. Bloomfield
Administrative support: Richard A. Larson, Clara D. Bloomfield
Provision of study materials or patients: Guido Marcucci, Maria R. Baer, Jonathan E. Kolitz, Joseph O. Moore, Andrew J. Carroll, Richard M. Stone, Richard A. Larson
Collection and assembly of data: All authors
Data analysis and interpretation: Krzysztof Mrózek, Guido Marcucci, Deedra Nicolet, Kati S. Maharry, Heiko Becker, Susan P. Whitman, Klaus H. Metzeler, Jessica Kohlschmidt, Clara D. Bloomfield
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 2.Smith ML, Hills RK, Grimwade D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2011;25:39–51. doi: 10.1016/j.blre.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 4.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 6.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 7.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 8.Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): Analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 9.Farag SS, Archer KJ, Mrózek K, et al. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: Results from Cancer and Leukemia Group B 8461. Blood. 2006;108:63–73. doi: 10.1182/blood-2005-11-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fröhling S, Schlenk RF, Kayser S, et al. Cytogenetics and age are the main determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: Results from AMLSG trial AML HD98-B. Blood. 2006;108:3280–3288. doi: 10.1182/blood-2006-04-014324. [DOI] [PubMed] [Google Scholar]
- 11.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 12.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 13.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 14.Schlenk RF, Benner A, Krauter J, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: A survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2004;15:3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Marcucci G, Mrózek K, Ruppert AS, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 16.Mrózek K. Cytogenetic, molecular genetic, and clinical characteristics of acute myeloid leukemia with a complex karyotype. Semin Oncol. 2008;35:365–377. doi: 10.1053/j.seminoncol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 18.Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 19.Fröhling S, Schlenk RF, Breitruck J, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 20.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 21.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 22.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 23.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 24.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 25.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: A study from the Acute Leukemia French Association (ALFA) Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 27.Fröhling S, Schlenk RF, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 28.Bienz M, Ludwig M, Leibundgut EO, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 29.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CAJ, et al. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pabst T, Eyholzer M, Fos J, et al. Heterogeneity within AML with CEBPA mutations: Only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100:1343–1346. doi: 10.1038/sj.bjc.6604977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renneville A, Boissel N, Gachard N, et al. The favorable impact of CEBPA mutations in patients with acute myeloid leukemia is only observed in the absence of associated cytogenetic abnormalities and FLT3 internal duplication. Blood. 2009;113:5090–5093. doi: 10.1182/blood-2008-12-194704. [DOI] [PubMed] [Google Scholar]
- 33.Röllig C, Bornhäuser M, Thiede C, et al. Long-term prognosis of acute myeloid leukemia according to the new genetic risk classification of the European LeukemiaNet recommendations: Evaluation of the proposed reporting system. J Clin Oncol. 2011;29:2758–2765. doi: 10.1200/JCO.2010.32.8500. [DOI] [PubMed] [Google Scholar]
- 34.Mayer RJ, Davis RB, Schiffer CA, et al. Intensive postremission chemotherapy in adults with acute myeloid leukemia. N Engl J Med. 1994;331:896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 35.Stone RM, Berg DT, George SL, et al. Granulocyte-macrophage colony-stimulating factor after initial chemotherapy for elderly patients with primary acute myelogenous leukemia. N Engl J Med. 1995;332:1671–1677. doi: 10.1056/NEJM199506223322503. [DOI] [PubMed] [Google Scholar]
- 36.Lee EJ, George SL, Caligiuri M, et al. Parallel phase I studies of daunorubicin given with cytarabine and etoposide with or without the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age or older with acute myeloid leukemia: Results of Cancer and Leukemia Group B study 9420. J Clin Oncol. 1999;17:2831–2839. doi: 10.1200/JCO.1999.17.9.2831. [DOI] [PubMed] [Google Scholar]
- 37.Baer MR, George SL, Caligiuri MA, et al. Low-dose interleukin-2 immunotherapy does not improve outcome of patients age 60 years and older with acute myeloid leukemia in first complete remission: Cancer and Leukemia Group B study 9720. J Clin Oncol. 2008;26:4934–4939. doi: 10.1200/JCO.2008.17.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baer MR, George SL, Sanford BL, et al. Escalation of daunorubicin and addition of etoposide in the ADE regimen in acute myeloid leukemia patients aged 60 years and older: Cancer and Leukemia Group B study 9720. Leukemia. 2011;25:800–807. doi: 10.1038/leu.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcucci G, Moser B, Blum W, et al. A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a pro-apoptotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients > 60 years old. J Clin Oncol. 2007;25(suppl):360s. abstr 7012. [Google Scholar]
- 40.Kolitz JE, George SL, Dodge RK, et al. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B study 9621. J Clin Oncol. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 41.Kolitz JE, George SL, Marcucci G, et al. P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients younger than age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–1421. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitelman F, editor. Basel, Switzerland: Karger; 1995. ISCN (1995): An International System for Human Cytogenetic Nomenclature. [Google Scholar]
- 43.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 44.Cheson BD, Cassileth PA, Head DR, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–819. doi: 10.1200/JCO.1990.8.5.813. [DOI] [PubMed] [Google Scholar]
- 45.Westfall PH, Tobias RD, Rom D, et al. Cary, NC: SAS Institute; 1999. Multiple Comparisons and Multiple Tests Using the SAS System. [Google Scholar]
- 46.Moorman AV, Roman E, Willett EV, et al. Karyotype and age in acute myeloid leukemia: Are they linked? Cancer Genet Cytogenet. 2001;126:155–161. doi: 10.1016/s0165-4608(00)00414-3. [DOI] [PubMed] [Google Scholar]
- 47.Schoch C, Schnittger S, Kern W, et al. Acute myeloid leukemia with recurring chromosome abnormalities as defined by the WHO-classification: Incidence of subgroups, additional genetic abnormalities, FAB subtypes and age distribution in an unselected series of 1,897 patients with acute myeloid leukemia. Haematologica. 2003;88:351–352. [PubMed] [Google Scholar]
- 48.Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendler JH, Maharry K, Radmacher MD, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.