Abstract
80α is a temperate, double-stranded DNA bacteriophage of Staphylococcus aureus that can act as a “helper” for the mobilization of S. aureus pathogenicity islands (SaPIs), including SaPI1. When SaPI1 is mobilized by 80α, the SaPI genomes are packaged into capsids that are composed of phage proteins, but that are smaller than those normally formed by the phage. This size determination is dependent on SaPI1 proteins CpmA and CpmB. Here, we show that co-expression of the 80α capsid and scaffolding proteins in S. aureus, but not in E. coli, leads to the formation of procapsid-related structures, suggesting that a host co-factor is required for assembly. The capsid and scaffolding proteins also undergo normal N-terminal processing upon expression in S. aureus, implicating a host protease. We also find that SaPI1 proteins CpmA and CpmB promote the formation of small capsids upon co-expression with 80α capsid and scaffolding proteins in S. aureus.
Keywords: molecular piracy, virus assembly, size determination, procapsid, electron microscopy, scaffolding protein, ribosomal protein L27, co-expression, Staphylococcus aureus pathogenicity island 1
Introduction
Staphylococcus aureus is a Gram-positive bacterium that is often a harmless inhabitant of human skin and mucosal epithelia, but is also associated with serious systemic and local infections (Gordon and Lowy, 2008). The emergence of virulent strains of S. aureus that are also resistant to a range of antibiotics has become a significant public health problem (Sakoulas and Moellering, 2008; Otto, 2010). Antibiotic resistance and virulence factor genes are frequently carried on mobile genetic elements, including bacteriophages, plasmids and pathogenicity islands (Novick, 2003; Brussow et al., 2004; Los et al., 2010). Chromosomal and plasmid-encoded virulence and resistance genes can also be transferred by bacteriophages through the process of generalized transduction (Dyer et al., 1985; Novick et al., 1986). A novel type of high frequency transduction has been demonstrated for members of the SaPI family of S. aureus pathogenicity islands. SaPIs are a family of 14–27 kb mobile genetic elements that are integrated into the host genome and may carry genes for a variety of superantigen toxins, such as the toxic shock syndrome toxin (TSST-1), and other virulence and resistance factors (Novick, 2003; Novick et al., 2010). SaPIs are normally integrated stably into the host genome, but become mobilized by infection with certain “helper” bacteriophages or by the induction of endogenous prophages through the SOS response(Maiques et al., 2006).
Phage 80α is a temperate, double-stranded (ds) DNA bacteriophage of the Siphoviridae family that is nearly identical to staphylococcal typing phage 53 (Christie et al., 2010). 80α is capable of generalized transduction and can also act as a helper for the mobilization of several different SaPIs, including SaPI1, SaPI2, SaPIbov1 and SaPIbov2 (Christie et al., 2010; Novick et al., 2010). The 80α virion consists of a 63-nm-diameter icosahedrally symmetric capsid with T=7 architecture, attached to a 190-nm-long flexuous, non-contractile tail that is capped with an elaborate baseplate decorated with six tail fibers (Spilman et al., 2011). As in other bacteriophages, the 80α capsid is assembled as an empty precursor, the procapsid. 80α procapsids are composed of 415 copies of the major capsid protein, the gene product (gp) of open reading frame (orf) 47, 100–200 copies of a scaffolding protein (gp46), a dodecameric portal protein (gp42), and ≈20 copies of gp44, a minor capsid protein of unknown function (Tallent et al., 2007; Poliakov et al., 2008). The gp47 capsid protein has the HK97-like fold found in all tailed dsDNA bacteriophages analyzed to date (Johnson, 2010). The capsid and scaffolding proteins both undergo N-terminal processing, which removes 14 and 13 amino acids and yields proteins 47* and 46*, respectively (Poliakov et al., 2008). Headfuls of phage DNA are packaged into the procapsids through the portal vertex in an ATP-dependent process that requires the small (gp40, TerS) and large (gp41, TerL) terminase subunits. DNA packaging is accompanied by capsid expansion and structural reorganization of the shell. This expansion is mediated by a conformational switch at a Pro residue that introduces a kink in the middle of the α3 spine helix of gp47 (Spilman et al., 2011). The organization of the 80α structural gene cluster is conserved among a number of S. aureus phages, and is virtually identical to those of S. aureus strain Newman prophages φNM1 and φNM2 (Bae et al., 2006; Dearborn and Dokland, 2012), and φETA2, carrier of the gene for exfoliative toxin A.
When SaPI1 is mobilized by 80α, the 80α protein Sri (gp22) binds to and inactivates the SaPI1 repressor, Stl. This leads to expression of SaPI1 gene products, excision from the host genome and replication of the SaPI1 genome (Tormo-Mas et al., 2010). SaPI1 genomes are then packaged into phage-like transducing particles composed of structural proteins supplied by the helper phage (Tallent et al., 2007; Poliakov et al., 2008). This process requires the SaPI1-encoded TerS subunit, which specifically recognizes the SaPI1 DNA (Ubeda et al., 2009). However, the capsids produced in the presence of SaPI1 are smaller than the normal 80α capsids and have T=4 symmetry, commensurate with the smaller size of the SaPI1 genome (Ruzin et al., 2001; Dearborn et al., 2011). We recently demonstrated that SaPI1-encoded proteins CpmA (gp7) and CpmB (gp6) are required for small capsid formation and that dimers of CpmB bind to gp47 on the inside of SaPI1 procapsids (Dearborn et al., 2011; Damle et al., 2012).
Here, we show that 80α procapsids can be made by co-expression of just the capsid and scaffolding proteins. This assembly occurs in S. aureus, but not in E. coli, implicating host co-factors in the assembly process. We also show that SaPI1 proteins CpmA and CpmB are required and sufficient to switch the capsid assembly program to small capsids in the co-expression system. Consistent with its role as a scaffolding protein, CpmB can to some extent substitute for the 80α scaffolding protein during capsid assembly. Cleavage of the capsid and scaffolding proteins appear to involve a host protease, rather than one encoded by the phage. The cleaved sequence is very similar to the N-terminus of S. aureus ribosomal protein L27, suggesting that both are cleaved by the same protease. The involvement of host factors in both processing and assembly of 80α capsids is a novel aspect of this system.
Results
Expression of gp46 and gp47 in E. coli
The gene (orf47) encoding the 80α major capsid protein (gp47) was cloned alone and together with orf46, encoding the gp46 scaffolding protein, into the E. coli expression vector pET21a, yielding plasmids pPD1 and pPD2, respectively (Table 1). Upon expression and lysis, cellular debris and insoluble material were collected by centrifugation at 17,000g for 30 min, yielding pellet 1 (P1). The supernatant (S1) was further centrifuged at 160,000g for 1 hr to pellet any capsid-related structures (pellet P2). Soluble (unassembled) proteins remain in supernatant 2 (S2).
Table 1. Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
|
| ||
| pET21a | E. coli T7 expression vector | Novagen |
| pMAD | Allelic exchange vector | (Arnaud et al., 2004) |
| pPD1 | pET21a derivative; 80α gp47 | This work |
| pPD2 | pET21a derivative; 80α gp46, gp47 | (Poliakov et al., 2008) |
| pPD8 | pMAD derivative; 80α Δorf46 and flanking region | This work |
| pJRC102 | pET21a derivative; 80α gp46*, gp47* | This work |
| pJRC110 | pET21a derivative; 80α gp47, gp46, gp42, gp44 | This work |
| pJRC130 | pET21a derivative, 80α gp47 + SaPI1 gp6, gp7 | This work |
| pJRC131 | pET21a derivative; 80α gp46, gp47 + SaPI1 gp6, gp7 | This work |
| pG164 | S. aureus T7 expression plasmid | (D'Elia et al., 2006) |
| pPD18 | pG164 derivative; 80α gp47, native RBS§ | This work |
| pPD20 | pG164 derivative; 80α gp47 | This work |
| pPD21 | pG164 derivative; 80α gp46, gp47 | This work |
| pEW4 | pG164 derivative; 80α 47* + 46* | This work |
| pPD22 | pG164 derivative; 80α gp42 − gp47 | This work |
| pPD44 | pG164 derivative; 80α gp47 + SaPI1 CpmB | This work |
| pPD45 | pG164 derivative; 80α gp47 + SaPI1 CpmA | This work |
| pPD46 | pG164 derivative; 80α gp47 + SaPI1 CpmA, CpmB | This work |
| pPD51 | pG164 derivative; 80α gp46, gp47 + SaPI1 CpmA, CpmB | This work |
Unless otherwise noted, all pG164-derived plasmids use the vector RBS for the first gene and native RBS for subsequent genes.
Upon expression of gp47 alone from pPD1, >99% of the gp47 protein accumulated in the low-speed P1 pellet (Fig. 1A) where it appeared to form large sheet-like structures (Fig. 1B). A small amount of protein formed what appeared to be small (24–40 nm) capsid-like particles in the P2 high-speed pellet (Fig. 1C). Co-expression of gp47 and gp46 from pPD2 yielded similar results: gp47 accumulated in P1, whereas gp46 remained soluble (Fig. 1D), demonstrating that the two proteins did not form stable interactions and that gp46 did not promote capsid assembly. Co-expression of gp47 and gp46 together with gp42 (portal) and the minor capsid protein gp44 from the plasmid pJRC110 also did not yield capsids, and most gp47 aggregated in P1 (data not shown), showing that the failure to form capsids was not only due to the absence of these proteins.
Figure 1.
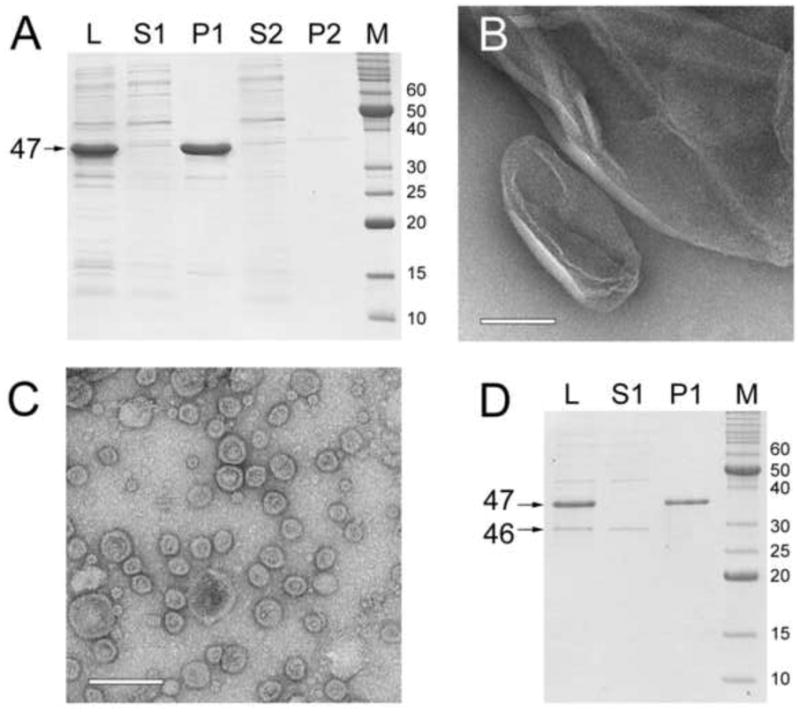
Expression of 80α proteins in E. coli. (A) Coomassie-stained SDS-PAGE of proteins expressed from plasmid pPD1 (orf47 only). L, total lysate; S1, low-speed supernatant; P1, low-speed pellet; S2, high-speed supernatant; and P2, high-speed pellet; M, marker, molecular weights (kDa) of bands indicated. The gp47 band is indicated. The volumes of the different fractions were kept equivalent for direct comparison of protein amounts. (B) Electron micrograph of material found in the P1 low speed pellet from the pPD1 expression. (C) Electron micrograph of particles found in a 50× concentrated P2 pellet from the pPD1 expression. Scale bars equal 100 nm. (D) SDS-PAGE of pPD2 (orf47+orf46) expression; lanes and markers as in (A). Bands corresponding to gp47 and gp46 are indicated.
Expression of gp46 and gp47 in S. aureus
Based on these results, we surmised that the failure to assemble was due to the lack of host-specific co-factors, such as chaperones, that are found in S. aureus, but not in E. coli. Consequently, we cloned orf47 alone, orf47 and orf46 together, and the entire orf42–orf47 capsid gene cluster as a unit into the S. aureus expression vector pG164 (D'Elia et al., 2006)(Table 1). In most constructs, the first gene in the plasmid used the optimized ribosome binding site (RBS) found in the vector, whereas downstream genes carried their native RBS and intergenic sequences from the phage genome. The resulting plasmids, pPD18 (orf47 alone with native RBS), pPD20 (orf47 alone, vector RBS), pPD21 (orf46–orf47) and pPD22 (orf42–orf47) were introduced by electroporation into S. aureus SA178RI (D'Elia et al., 2006), which carries an IPTG-inducible copy of the T7 RNA polymerase gene, yielding strains ST66, ST70, ST71 and ST72, respectively (Table 2). After expression, the cells were lysed, and capsid-related structures were harvested by centrifugation, as described above for the E. coli expression studies.
Table 2. S. aureus strains used in this study.
| Strains | Description | Reference |
|---|---|---|
|
| ||
| RN10616 | RN4220 (80α | (Ubeda et al., 2009) |
| RN10628 | RN4220 (80α) SaPI1 tst∷tetM | (Ubeda et al., 2009) |
| SA178RI | RN4220 derivative expressing T7 RNA polymerase | (D'Elia et al., 2006) |
| ST51 | RN4220 (80α Δorf46) SaPI1 tst∷tetM | This work |
| ST66 | SA178RI [pPD18] | This work |
| ST70 | SA178RI [pPD20] | This work |
| ST71 | SA178RI [pPD21] | This work |
| ST72 | SA178RI [pPD22] | This work |
| ST91 | RN4220 (80α Δorf46) | This work |
| ST112 | SA178RI [pPD44] | This work |
| ST113 | SA178RI [pPD45] | This work |
| ST114 | SA178RI [pPD46] | This work |
| ST118 | SA178RI [pPD51] | This work |
| ST188 | SA178RI [pEW4] | This work |
Expression of orf47 alone (strain ST70) resulted in the accumulation of limited amounts of gp47 in the P2 high-speed pellet (Fig. 2), but no obvious capsid-related structures were observed by EM (Fig. 3A). The same result was obtained for strain ST66, in which orf47 is expressed using the native phage RBS (Fig. 2). Upon co-expression of gp47 and gp46 in strain ST71, however, large amounts of gp47 were found in the P2 pellet (Fig. 2). Gp46 levels, were lower, albeit visible on the gel (Fig. 2). Several additional bands, especially at higher MW, were also seen in all pellets, apparently corresponding to bacterial proteins. The ST71 P2 pellet contained numerous capsid-related structures (Fig. 3B). Many of these capsids were aberrant, unclosed shells known as “monsters” (including the “monster truck” seen in Fig. 3B), and angular, double-layered shells, but a subset of the particles had the characteristic appearance of 80α procapsids (Poliakov et al., 2008) and contained an inner core, consistent with the presence of scaffolding protein.
Figure 2.
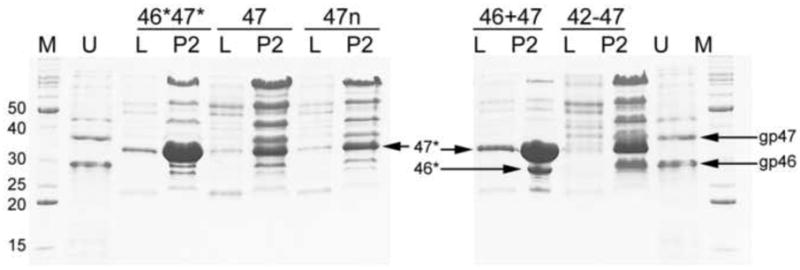
Expression of 80α proteins in S. aureus, separated by Coomassie-stained SDS-PAGE. M, marker (molecular weights in kDa indicated); U, uncleaved gp46 and gp47 from E. coli expression; 46*47*, expression of 46* and 47* from strain ST188; 47, gp47 only (ST70); 47n, gp47 only, with native RBS (ST66); 46+47, gp46 and gp47 (ST71); 42-47, expression of the entire cluster from gp42 to gp47 (ST72). Lanes L are bacterial lysate; lanes P2 are from a 25× concentrated, capsid-containing high-speed pellet. The positions of the bands corresponding to the uncleaved gp46 and gp47 and cleaved 46* and 47* are indicated. Several bands at higher MW correspond to bacterial proteins that co-sediment in the P2 pellet.
Figure 3.
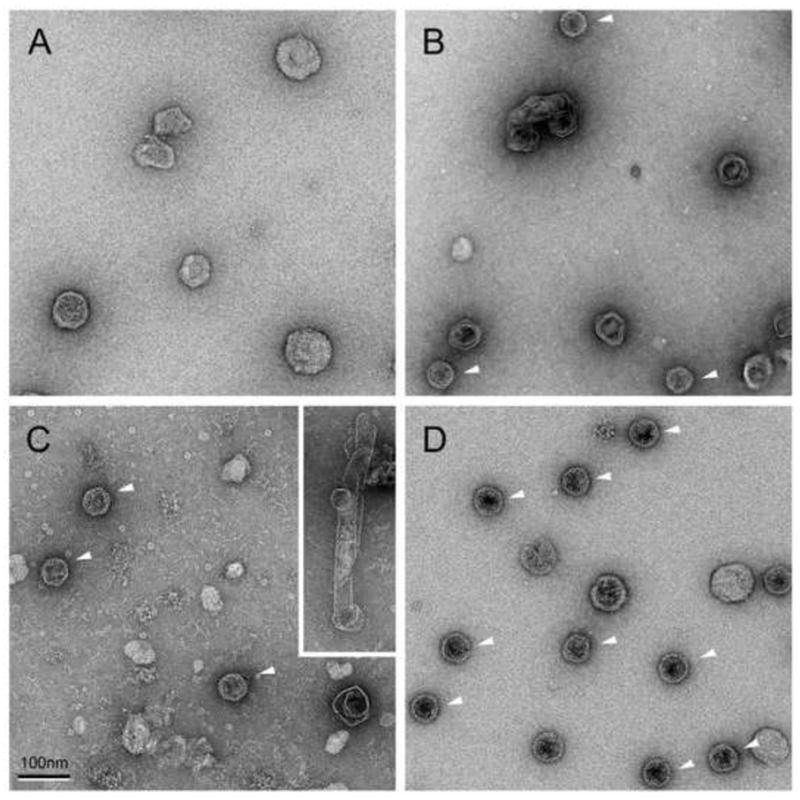
EM of particles formed upon expression of 80α proteins in S. aureus. (A) gp47 alone (strain ST70), P2 pellet after sucrose gradient purification; (B) gp46 + gp47 (ST71), P2 pellet; (C) 46* + 47* (ST188), P2 pellet; inset, example of a polyhead-like tubular structure; (D) gp42−gp47 (ST72), P2 pellet after sucrose gradient purification. The white arrowheads point to well-formed procapsids. The scale bar equals 100 nm.
Strain ST72 expresses the whole orf42–orf47 gene cluster, including portal protein (gp42), gp44, gp46 and gp47 (Tables 1 and 2). All genes except orf42 had their native ribosome binding sites and other 5′ sequences. The amount of gp47 accumulating upon induction of expression in this strain was not as high as in ST71, but greater than the amount seen with orf47 alone (ST70) (Fig. 2). Expression from this plasmid also yielded many capsid-related structures, including a highly homogeneous population of 80α procapsids when separated on a 10-40% sucrose gradient (Fig. 3D).
Cleavage of gp46 and gp47
When gp46 and gp47 were co-expressed in S. aureus, both proteins underwent proteolytic processing to their cleaved form, as did gp47 expressed alone (Fig. 2). Mass spectrometry confirmed that the cleavage was identical to that of 46* and 47* found in native 80α procapsids and virions (Poliakov et al., 2008). An intrinsic protease activity of gp46 or gp47 seems unlikely, since the proteins were not cleaved when expressed in E. coli, and they have no recognizable protease domains. Therefore, the maturational cleavage of gp46 and gp47 in 80α appears to be due to a S. aureus-specific host protease not found in E. coli.
The absence of cleavage was not in itself the reason for the failure to assemble correctly in E. coli, since expression from plasmid pJRC102 of truncated forms of gp46 and gp47 equivalent to 46* and 47* resulted in aggregation of 47* in P1 while 46* remained soluble (not shown). This is similar to what was observed upon expression of the full-length proteins. However, expression of the same truncated proteins (46* and 47*) in S. aureus, from plasmid pEW4, led to the formation of some well-formed procapsids, although 46* levels appeared to be low (Fig. 2) and the predominant assembly product was tubular “polyheads” (Fig. 3C).
Deletion of orf46
To better understand the role of the gp46 scaffolding protein in protein expression and capsid formation, we generated the mutant strain ST91, in which orf46 had been deleted in-frame from the 80α prophage in RN10616 (Table 2). This mutant was induced with mitomycin C and purified by PEG precipitation and CsCl gradient centrifugation, according to standard procedures for phage inductions. As expected, there was no band corresponding to DNA-filled capsids. A band higher up in the gradient contained numerous tails and portals as well as other structures most likely of bacterial origin, but no capsid-related structures and little gp47 protein (Fig. 4A, B). Since tail proteins are encoded by genes downstream of orf47, the low level of gp47 was not due to polar effects of the orf46 deletion.
Figure 4.
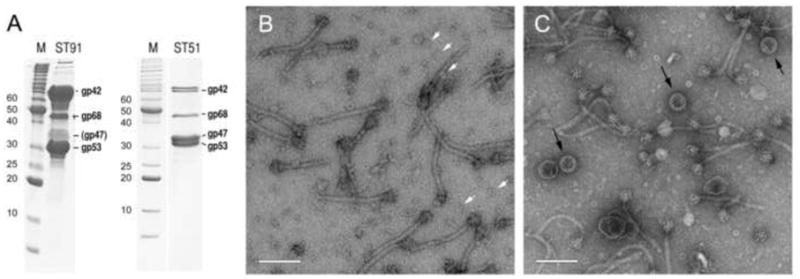
Capsid formation in the orf46 deletion strains. (A) SDS-PAGE of the CsCl-gradient purified fractions of ST91 (80α Δorf46) and ST51 (80α Δorf46 + SaPI1). M, marker, with molecular weights (kDa) shown. Bands corresponding to major structural proteins are identified according to Poliakov et al. (2008) (B) EM of ST91. The white arrows point to a few examples of portals. (C) EM of ST51. SaPI1 procapsid-like shells are indicated by black arrows. Scale bars equal 100 nm.
When this same deletion was made in RN10628, an 80α lysogen containing SaPI1 (yielding strain ST51), the resulting CsCl band contained substantial amounts of gp47 protein and numerous capsid-like structures in addition to tails and portals (Fig. 4A, C). Some of these structures had the appearance of well-formed procapsids. However, these procapsids were only 40 nm in diameter, rather than the 51-nm size of regular 80α procapsids. This size difference is presumably due to SaPI1-encoded proteins CpmA and CpmB, which were previously implicated in capsid size determination (Poliakov et al., 2008; Dearborn et al., 2011; Damle et al., 2012; Dearborn and Dokland, 2012). No transducing activity was found in the lysate, however, suggesting that gp46 serves additional functions that cannot be complemented by CpmB.
Expression of gp46 and gp47 with SaPI1 proteins CpmA and CpmB in S. aureus
To see if CpmA and CpmB could induce small capsid formation in the co-expression system, cpmA and cpmB were cloned either individually or in tandem together with 80α orf46 and/or orf47 in the S. aureus expression vector pG164, yielding plasmids pPD44 (gp47+CpmB), pPD45 (gp47+CpmA), pPD46 (gp47+CpmA+CpmB) and pPD51 (gp46+gp47+CpmA+CpmB), resulting in S aureus strains ST112, ST113, ST114 and ST118, respectively (Tables 1 and 2).
As shown above, gp47 does not assemble efficiently in the absence of gp46 (Figs. 3A and 4). However, co-expression of gp47 with SaPI1 CpmB (strain ST112) led to the formation of numerous 80α-sized procapsids containing 47* as well as aberrant, thin-shelled capsids (Fig. 5A) that formed a broad band on sucrose gradients (Fig. 6A). There was also considerable accumulation of 47* at the bottom of the gradient, presumably due to aggregation (Fig. 6A). Although little CpmB could be discerned on the gel (Fig. 6A), this suggests that CpmB can to some extent substitute for gp46 as a scaffold. However, the lack of small, SaPI1-sized capsids indicates that CpmB alone is insufficient for size determination, which is consistent with experiments utilizing deletions and insertions of cpmA and cpmB (Damle et al., 2012). In contrast, co-expression of gp47 and CpmA (ST113) yielded no capsid-related structures (not shown). Indeed, even when CpmA and CpmB were both co-expressed with gp47 (ST114), no procapsids were observed (Fig. 5B) and protein production was poor (Fig. 6B). Apparently, CpmA inhibits the ability of CpmB to act as an alternative scaffold for gp47. Co-expression of both gp46 and gp47 with CpmA and CpmB (ST118), however, yielded large numbers of capsid-related structures containing 46* and 47* that formed a band on sucrose gradients (Fig. 6C). A faint band probably corresponding to CpmB was also visible, but no band corresponding to CpmA, which superimposes on 46* and is present in procapsids in only small amounts (Poliakov et al., 2008), could be discerned. The particles included numerous 80α-sized (large) procapsids with a scaffolding core, small, SaPI1-sized procapsids with a less defined core, and thin-walled, expanded shells of various sizes (Fig. 5C, D).
Figure 5.
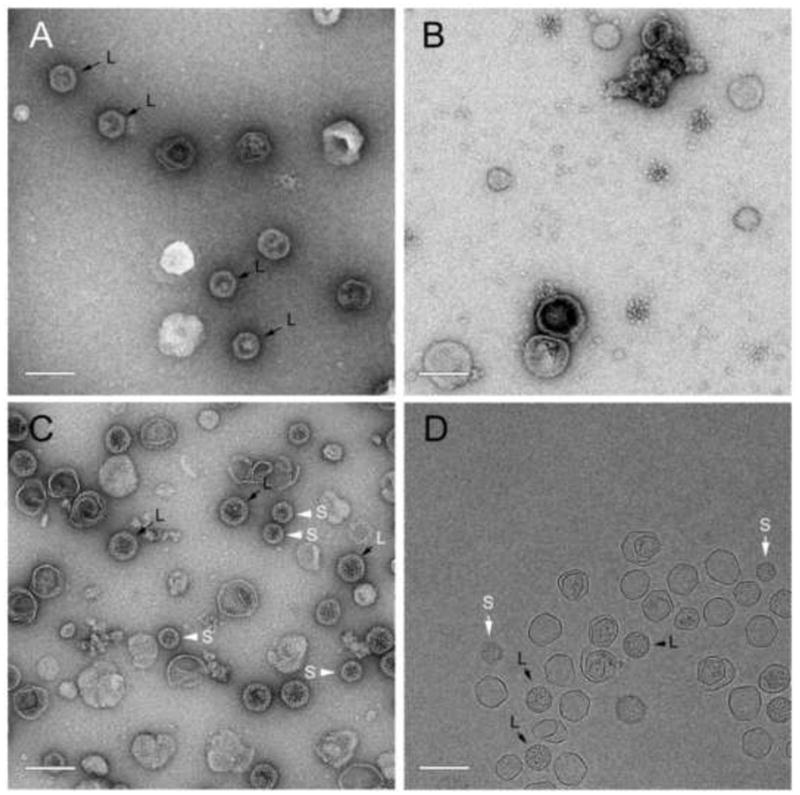
EM of particles formed upon co-expression of 80α and SaPI1 proteins in S. aureus. (A) 80α gp47 + SaPI1 CpmB (strain ST112); (B) gp47 + SaPI1 CpmA and CpmB (ST114); (C) gp46+gp47+CpmA+CpmB (ST118). (D) Cryo-EM of ST 118. (B) is a P2 pellet; others are sucrose-purified fractions. Examples of small and large procapsids are indicated by S (white arrows) and L (black arrows), respectively. Scale bars equal 100nm.
Figure 6.
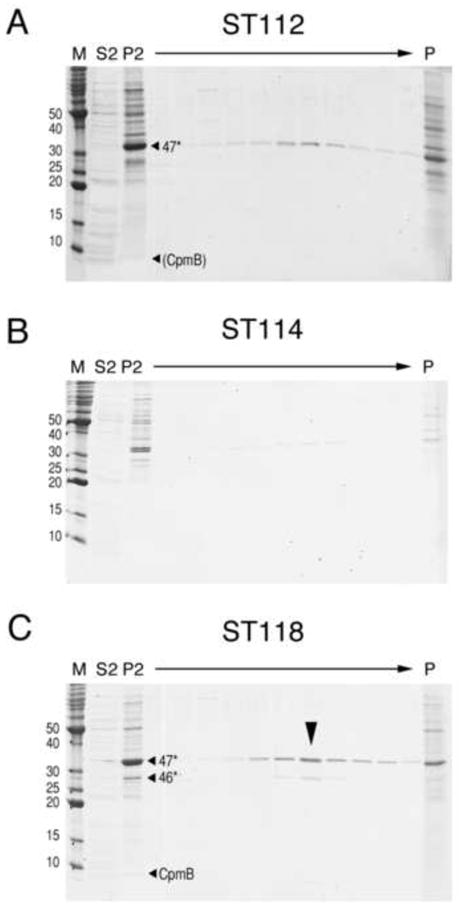
SDS-PAGE of 10–40% sucrose gradients of material produced by co-expression of (A) 80α gp47 + SaPI1 CpmB (strain ST112); (B) gp47 + SaPI1 CpmA and CpmB (ST114); (C) gp46+gp47+CpmA+CpmB (ST118). Lanes M, marker, MW (kDa) indicated; S2, high-speed supernatant; P2, high-speed pellet. The arrow indicates the direction of the gradient. Lane P is the gradient pellet fraction. Bands corresponding to the known positions of 47*, 46* and CpmB are indicated, where appropriate. The procapsid fraction is indicated by the big arrowhead in (C).
In E. coli, in contrast, expression of gp47 together with CpmA and CpmB, with or without gp46 (pJRC130 and pJRC131) did not yield any capsid-related structures. In each case, gp47 was insoluble and located in P1, whereas gp46, CpmA and CpmB remained in the S2 supernatant (data not shown).
Discussion
Protein cleavage
Cleavage of structural proteins such as that of 80α proteins gp46 and gp47 (Poliakov et al., 2008) is a common occurrence in bacteriophages, including P2 (Rishovd and Lindqvist, 1992; Chang et al., 2008), T4 (Laemmli, 1970) and HK97 (Conway et al., 1995), and is generally considered to occur after procapsid assembly. Most phages that undergo cleavage encode a prohead protease between the portal and scaffolding/capsid genes, though in some cases two or more of the genes encoding these activities are overlapping or fused (Effantin et al., 2010). An example of such a fusion is found in bacteriophage P2, where gpO serves a dual role as both scaffolding protein and protease (Chang et al., 2008; Chang et al., 2009). 80α does not have a protease gene in this position, nor does there appear to be a protease domain associated with the scaffolding protein. Gene 44, which occupies this location in the 80α genome, is equivalent to gene 7 of SPP1 (Vinga et al., 2006), and appears to be involved in DNA stability within the capsid (Dearborn et al., 2011). Here, we have shown that gp46 and gp47 are processed normally when expressed in S. aureus, in the absence of any other phage proteins (Fig. 2), implicating a host protease in this process. The cleavage does not appear to be autoproteolytic, and no cleavage of either protein occurs in E. coli. While this could be because cleavage is dependent on correct assembly, the fact that pre-cleaved gp46 (46*) and gp47 (47*) could assemble into procapsids in S. aureus may suggest that cleavage occurs prior to assembly in this system. Although an involvement of host proteases in bacteriophage assembly is not unprecedented (Conlin et al., 1992), this is the first demonstration of the involvement of a host protease in cleavage of the major capsid protein.
A search for a host protein with a potential cleavage site similar to gp46 and gp47 identified a homologous sequence at the N-terminus of S. aureus ribosomal protein L27, the product of the rpmA gene (Fig. 7). This sequence is conserved in a number of Gram-positive organisms, including Bacillus, Listeria, Clostridium and Streptococcus and represents an N-terminal extension that is not found in Gram-negative organisms such as E. coli. It is not clear whether L27 does get cleaved, but if so, the same protease is likely responsible for the cleavage of gp46 and gp47 as well. L27 has an unusual structure with a long N-terminal extension that reaches into the peptidyl transfer site of the ribosome (Wang et al., 2004; Trobro and Aqvist, 2008), and is a target for several antibiotics (Sonenberg et al., 1973; Tejedor and Ballesta, 1986; Colca et al., 2003). It is not unlikely that the N-terminal extension in S. aureus L27 affects ribosome function and that its cleavage would be an essential process. The presumptive protease might then be a potential target for novel antibiotics that would specifically target Gram-positive bacteria.
Figure 7.
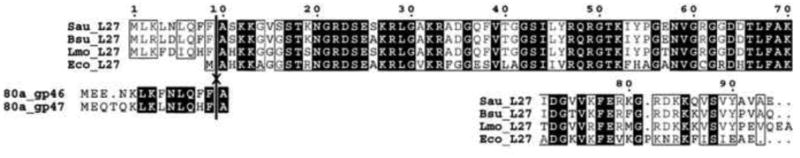
Sequence alignment of ribosomal protein L27 from S. aureus (Sau), Bacillus subtilis (Bsu), Listeria monocytogenes (Lmo) and E. coli (Eco). The alignment was generated with CLUSTALW (Thompson et al., 2002) and ESPript 2.2 (http://espript.ibcp.fr). The sequences of the N-terminal peptides of gp46 and gp47 from 80α are aligned with L27 below, showing the conserved sequence of the cleaved peptide. The cleavage site known from gp46 and gp47 (Poliakov et al., 2008) is indicated by the vertical line through all the sequences.
Capsid assembly
Gp47 did not accumulate at high levels upon expression in S. aureus (Fig. 2), nor upon induction of an 80α Δorf46 lysogen (Fig. 4). This could be due to protein instability, rapid degradation or regulation at the translational level. The effect was seen whether the vector RBS or the native RBS was used. Co-expression of gp46 and gp47, however, led to high level production of gp47 and the formation of numerous capsid-like structures, many of which had the appearance of normal 80α procapsids (Fig. 3B-D). This suggests that gp46 may act as a chaperone that protects gp47 against degradation or downregulation, e.g. through rapid incorporation into procapsid shells. It is unclear why ST72, which includes the whole ORF 42–47 gene cluster, also produces relatively low levels of gp47 (Fig. 2). In this case, however, the proportion of well-formed capsids was higher, perhaps due to the presence of minor capsid proteins gp42 and gp44, or because the ratio of gp46 to gp47 was closer to that seen in a normal 80α infection (Tallent et al., 2007; Poliakov et al., 2008). The relatively high proportion of aberrant capsids formed in most co-expressions compared to normal phage production could be due to the lack of other required phage-encoded factors and feedback mechanisms, the absence of phage genomic templates, and the local concentrations of SaPI1 gene products during assembly. Additionally, the available cellular machinery and host factors likely differ between the IPTG-based co-expression system and the mitomycin C-activated SOS response in lysogen strains.
In E. coli, gp47 alone is expressed at high levels (Fig. 1), but does not form capsids even in the presence of gp46. The failure to assemble in E. coli was quite surprising, since phage capsid assembly typically works well in heterologous systems. For example, the structural proteins from Bacillus phages φ29 and SPP1 both produce well-formed capsids upon expression in E. coli (Guo et al., 1991; Droge et al., 2000). Presumably, E. coli lacks a chaperone found in S. aureus and perhaps other Gram-positive bacteria that is required for the gp47 folding, assembly or interaction with gp46. This failure cannot be simply attributed to lack of proper cleavage of capsid precursors, since expression of 46* and 47* also failed to assemble properly. The requirement for host chaperones is another unusual aspect of the 80α system.
SaPI1-mediated size determination
In the bacteriophage P2/P4 system, only one P4-encoded protein, Sid, is sufficient to promote small capsid formation, even in the absence of the P2 scaffolding protein, gpO (Marvik et al., 1994; Wang et al., 2006; Dearborn et al., 2012). We previously showed that SaPI1 proteins CpmA and CpmB are both required and sufficient for inducing small capsid formation with 80α (Damle et al., 2012). Both proteins were present in SaPI1 procapsids(Poliakov et al., 2008), and deletion of either cpmA or cpmB from a SaPI1-containing 80α lysogen abolished or greatly reduced small capsid formation(Dearborn et al., 2011; Damle et al., 2012). Dimers of CpmB appear to bridge gp47 capsomers on the inside surface of the procapsid (Dearborn et al., 2011).
Here, we have characterized the effect of CpmA and CpmB on capsid assembly in a minimal co-expression system, which confirms the requirement for both proteins in SaPI1 capsid size determination (Fig. 5C, D). Moreover, CpmB, but not CpmA, could rescue gp47 accumulation in the absence of gp46, both during co-expression (Fig. 5A) and in an 80α Δorf46 lysogen (Fig. 4). These observations support a role for CpmB and gp46 as alternative assembly chaperones for gp47. Indeed, structural analysis and sequence considerations previously indicated that the two proteins likely bind to the same site on the inside of the gp47 shell (Dearborn et al., 2011). Our hypothesis for CpmA is that it is required for reorganizing the scaffolding core made of gp46 that would otherwise prevent the formation of small capsids, and/or to remove gp46 in order to allow access to gp47 by CpmB.
Materials and Methods
Strains and vectors
The strains and plasmids used in this study are listed in Tables 1 and 2. E. coli expression plasmids are derivatives of pET21a (Novagen). Plasmids were introduced initially into E. coli strain DH5α (Invitrogen) and expression carried out in BL21 (DE3) (Invitrogen). All S. aureus strains are derivatives of the phage-cured, restriction-defective strain RN4220 (Kreiswirth et al., 1983). Expression in S. aureus was carried out using vector pG164, an E. coli-S. aureus shuttle vector carrying a T7 late promoter into which a lac operator, a multiple cloning site, an optimized gram-positive ribosome binding site, and a constitutively expressed copy of the lac repressor gene were introduced (D'Elia et al., 2006). The expression strain, SA178RI, carries a lac operator-regulated T7 RNA polymerase expression cassette (D'Elia et al., 2006).
Fragments of phage 80α and SaPI1 were amplified using primers carrying appropriate restriction sites for ligation into the plasmid vectors; primers for each construct are listed in Table 3. Plasmid pEW4 was constructed using the In-Fusion PCR cloning system (Clontech) and primers carried 15 bp extensions homologous to the ends of the assembled fragments and the linearized vector. All inserts were verified by DNA sequencing. All genes expressed in E. coli using an optimized E. coli ribosome binding site (RBS) either existing in the vector or introduced in the primers. For expression in S. aureus, plasmid plasmid pPD18 uses the native phage RBS for expression of orf47; all other pG164-derived plasmids use the vector RBS for expression of the first gene in the construct and the native RBS for downstream genes. Plasmids pPD44, pPD45 and pPD46 were derived from pPD20 into which SaPI1 cpmA, cpmB or both were inserted downstream of the phage capsid gene. pPD51 was similarly derived from pPD21.
Table 3. Oligonucleotides used in this study.
| Plasmid | Oligonucleotide | Sequence (5′-3′)* |
|---|---|---|
| pPD1 | SMT34 | GACTCATATGGAACAAACACAAAAATTAAAAT |
| SMT35 | GACTGGATCCTTATTAAACTTCTCCTGGAACT | |
|
| ||
| pPD8 | PKD-9 | GAGGATCCGGTCGAAAACAAGGACTTTAGCGATAGAG |
| PKD-10 | ATGCCTCCGTTAATTTTTAATAATTCTATTTTCTTCCATGAGATATACCTCCATTTATAGTCTGTC | |
| PKD-11 | ATGGAAGAAAATAGAATTATTAAAAATTAACGGAGGCATTTAAATGGAACAAAC | |
| PKD-12 | GAGGATCCCAATGATTTCGGGCATGTTACCACTCC | |
|
| ||
| pPD18 | pKD57 | AACTGCAGAACGGAGGCATTTAAATGGAAC |
| pKD58 | AACTCGAGAAGTCAGGCGCGCCAATTGTTTATTAAACTTCTCCTGGAACT | |
|
| ||
| pPD20 | PKD-78 | CACAGGATCCATGGAACAAACACAAAAATTAAAATTAAATTTGC |
| PKD-58 | AACTCGAGAAGTCAGGCGCGCCAATTGTTTATTAAACTTCTCCTGGAACT | |
|
| ||
| pPD21 | PKD-79 | CACAGGATCCATGGAAGAAAATAAACTTAAGTTTAATTTGCAA |
| PKD-58 | AACTCGAGAAGTCAGGCGCGCCAATTGTTTATTAAACTTCTCCTGGAACT | |
|
| ||
| pPD22 | PKD-66 | TTTTGGATCCATGTTAAAAGTAAACGAATTTGAAACAGATACA |
| PKD-67 | AAAACTGCAGTAATTGTTTATTAAACTTCTCCTGGAACTG | |
|
| ||
| pPD41 | PKD-104 | CACAGGATCCATGGCAGACCAATCAGATGATCCG |
| PKD-58 | AACTCGAGAAGTCAGGCGCGCCAATTGTTTATTAAACTTCTCCTGGAACT | |
|
| ||
| pPD44 | PKD-86 | GGCAAAGGCGCGCCAAGAAAGGGTAATTAAATGGAAACAAAATACGA |
| PKD-87 | GGCAACTCGAGTTGCTATTTAATAATCCTGTTTTGCTTAGCTAAATTT | |
|
| ||
| pPD45 | PKD-88 | GGCAAAGGCGCGCCAAGGGGATAAAAATGAAAACTGAATCGTAC |
| PKD-89 | GGCAACTCGAGTTAACGTTTTAAAAACAACTTGTTATTGTGTTCG | |
|
| ||
| pPD46, pPD51 | PKD-88 | GGCAAAGGCGCGCCAAGGGGATAAAAATGAAAACTGAATCGTAC |
| PKD-87 | GGCAACTCGAGTTGCTATTTAATAATCCTGTTTTGCTTAGCTAAATTT | |
Restriction sites are underlined
In-frame deletions of orf46 were made by allelic exchange in the 80α lysogenic strain RN10616 and its SaPI1-containing derivative RN10628 using a derivative of pMAD (Arnaud et al., 2004), as described previously (Poliakov et al., 2008), yielding strains ST91 and ST51, respectively. The 80α lysogens were grown in CY+GL broth at 32°C and induced by mitomycin C as previously described (Novick, 1991). The particles were purified by PEG precipitation, chloroform extraction and banding on CsCl gradients as previously described (Spilman et al., 2011).
Expression in E. coli
Cells harboring E. coli expression plasmids were grown in LB with 100 μg/ml ampicillin at 37°C and expression was induced at OD600=0.6 by the addition of 0.5 mM IPTG. Cultures were collected by centrifugation, resuspended in 100 mM Tris pH 7.4, 200 mM NaCl, 0.5% deoxycholate, 1% Triton X-100 and 1 mM PMSF, frozen overnight at −20°C, thawed, lysed in a high pressure cell disruptor (Avestin, Ottawa, ON), and clarified by centrifugation at 17,000g for 30 min, yielding pellet P1 and supernatant S1. The S1 supernatant was pelleted at 160,000g for 1 hr, yielding supernatant S2 and pellet P2, which was resuspended in 50 mM Tris pH 8.0, 100 mM NaCl and 10 mM MgCl2. Samples were analyzed by SDS-PAGE and by electron microscopy.
Expression in S. aureus
S. aureus cells were grown in CY+GL medium (Novick, 1991) with 15 μg/ml chloramphenicol to a density of OD600 = 0.5, induced with 0.5 mM IPTG and grown for 4 hours at 32°C. The cells were by harvested by centrifugation and resuspended in 100 mM Tris pH 8.0 with 500 mM NaCl and frozen overnight at −20°C. Cells were lysed by the addition of 50 μg/ml lysostaphin (Sigma) or by multiple passages through a high-pressure cell disruptor (Avestin, Ottawa, Ontario). The lysate was processed the same way as the E. coli expression samples, except that S1 was pelleted at 250,000g for 30 min and resuspended in 100 mM Tris pH 7.4, 500 mM NaCl and 1 mM DTT. This was layered on a 10–40% sucrose gradient and separated by centrifugation at 110,000g for 2 hr. The gradients were manually fractionated from the top and each fraction was analyzed by SDS-PAGE. Fractions containing capsid-related proteins were analyzed by electron microscopy.
Electron microscopy
Samples for electron microscopy were placed on glow-discharged 400 mesh carbon-coated grids (Electron Microscopy Sciences, Hatfield, PA), washed with 20 mM Tris pH 7.8, 50 mM NaCl, 2 mM CaCl2, 1 mM MgSO4 and negatively stained with 1% uranyl acetate. The grids were observed in an FEI Tecnai F20 microscope operated at 200kV and images were collected on a Gatan Ultrascan 4000 CCD camera at a typical magnification of 65,500×. For cryo-EM, the sample was applied to C-flat holey film (Electron Microscopy Sciences), flash frozen, transferred to a Gatan 626 cryo-specimen holder and observed in an FEI Tecnai F20 electron microscope operated at 200 kV at a magnification of 81,220× (Dokland and Ng, 2006; Spilman et al., 2011).
Acknowledgments
We are grateful to Dr. Eric Monroe for assistance with mass spectrometry, and to Todd Black (Merck & Co.) for providing pG164 and SA178RI. This work was supported by National Institutes of Health grants R01 AI083255 to T.D. and R56 AI081837 to G.E.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Molec Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology. 2008;370:352–361. doi: 10.1016/j.virol.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Spilman MS, Rodenburg CM, Dokland T. Functional domains of the bacteriophage P2 scaffolding protein: identification of residues involved in assembly and protease activity. Virology. 2009;384:144–150. doi: 10.1016/j.virol.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80 alpha - implications for the specificity of SaPI mobilization. Virology. 2010;407:381–390. doi: 10.1016/j.virol.2010.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colca J, Mcdonald W, Waldon D, Thomasco L, Gadwood R, Lund E, Cavey G, Mathews W, Adams L, Cecil E, Pearson J, Bock J, Mott J, Shinabarger D, Xiong L, Mankin A. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J Biol Chem. 2003;278:21972–21979. doi: 10.1074/jbc.M302109200. [DOI] [PubMed] [Google Scholar]
- Conlin C, Vimr E, Miller C. Oligopeptidase A is required for normal phage P22 development. J Bacteriol. 1992;174:5869–5880. doi: 10.1128/jb.174.18.5869-5880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- D'Elia MA, Pereira M, Chung Y, Zhao W, Chau A, Kenney T, Sulavik M, Black T, Brown E. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol. 2006;188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle PK, W EA, Spilman MS, Dearborn AD, Ram G, Novick RP, Dokland T, Christie GE. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology. 2012 doi: 10.1016/j.virol.2012.05.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn AD, Dokland T. Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages. Bacteriophage. 2012;2 doi: 10.4161/bact.20632. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn AD, Spilman MS, Damle PK, Chang JR, Monroe EB, Saad JS, Christie GE, Dokland T. The Staphylococcus aureus pathogenicity island protein gp6 functions as an internal scaffold during capsid size determination. J Mol Biol. 2011;412:710–722. doi: 10.1016/j.jmb.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn A, Laurinmaki P, Chandramouli P, Rodenburg C, Wang S, Butcher S, Dokland T. Structure and size determination of bacteriophage P2 and P4 procapsids: Function of size responsiveness mutations. J Struct Biol. 2012;178:215–224. doi: 10.1016/j.jsb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Ng ML. In: Transmission electron microscopy of biological specimens. Dokland T, Hutmacher DW, Ng ML, Schantz JT, editors. World Scientific Press; Singapore: 2006. pp. 153–208. [Google Scholar]
- Droge A, Santos MA, Stiege AC, Alonso JC, Lurz R, Trautner TA, Tavares P. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J Mol Biol. 2000;296:117–132. doi: 10.1006/jmbi.1999.3450. [DOI] [PubMed] [Google Scholar]
- Dyer DW, Rock MI, Lee CY, Iandolo JJ. Generation of transducing particles in Staphylococcus aureus. J Bacteriol. 1985;161:91–95. doi: 10.1128/jb.161.1.91-95.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Figueroa-Bossi N, Schoehn G, Bossi L, Conway J. The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology. 2010;402:355–365. doi: 10.1016/j.virol.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of phage phi29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE. Virus particle maturation: insights into elegantly programmed nanomachines. Curr Opin Struct Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B, Lofdahl S, Betley M, O'Reilly M, Schlievert P, Bergdoll M, Novick R. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Los M, Kuzio J, Mcconnell MR, Kropinski AM, Wegrzyn G, Christie GE. In: Lysogenic conversion in bacteria of importance to the food industry. Sabour MP, Griffiths M, editors. ASM Press; Washington, D.C.: 2010. pp. 157–198. [Google Scholar]
- Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbe J, Penades JR. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvik OJ, Sharma P, Dokland T, Lindqvist BH. Bacteriophage P2 and P4 assembly: alternative scaffolding proteins regulate capsid size. Virology. 1994;200:702–714. doi: 10.1006/viro.1994.1234. [DOI] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in Staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Edelman I, Lofdahl S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J Mol Biol. 1986;192:209–220. doi: 10.1016/0022-2836(86)90360-8. [DOI] [PubMed] [Google Scholar]
- Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- Poliakov A, Chang JR, Spilman MS, Damle PK, Christie GE, Mobley JA, Dokland T. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J Mol Biol. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishovd S, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: protein processing and capsid size determination. Virology. 1992;187:548–554. doi: 10.1016/0042-6822(92)90457-z. [DOI] [PubMed] [Google Scholar]
- Ruzin A, Lindsay JA, Novick RP. Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- Sakoulas G, Moellering RJ. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46(5):S360–7. doi: 10.1086/533592. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Wilchek M, Zamir A. Mapping of Escherichia coli ribosomal components involved in peptidyl transferase activity. Proc Natl Acad Sci U S A. 1973;70:1423–1426. doi: 10.1073/pnas.70.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman MS, Dearborn AD, Chang JR, Damle PK, Christie GE, Dokland T. A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80alpha capsids. J Mol Biol. 2011;405:863–876. doi: 10.1016/j.jmb.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallent SM, Langston TB, Moran RG, Christie GE. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J Bacteriol. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor F, Ballesta J. Reaction of some macrolide antibiotics with the ribosome. Labeling of the binding site components. Biochemistry. 1986;25:7725–7731. doi: 10.1021/bi00371a066. [DOI] [PubMed] [Google Scholar]
- Thompson J, Gibson T, Higgins D. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2 doi: 10.1002/0471250953.bi0203s00. Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Tormo-Mas MA, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa I, Barbe J, Novick RP, Christie GE, Penades JR. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. Role of ribosomal protein L27 in peptidyl transfer. Biochemistry. 2008;47:4898–4906. doi: 10.1021/bi8001874. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Olivarez NP, Barry P, Wang H, Kong X, Matthews A, Tallent SM, Christie GE, Novick RP. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol Microbiol. 2009;72:98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga I, Droge A, Stiege A, Lurz R, Santos M, Daugelavicius R, Tavares P. The minor capsid protein gp7 of bacteriophage SPP1 is required for efficient infection of Bacillus subtilis. Mol Microbiol. 2006;61:1609–1621. doi: 10.1111/j.1365-2958.2006.05327.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Takemoto C, Murayama K, Sakai H, Tatsuguchi A, Terada T, Shirouzu M, Kuramitsu S, Yokoyama S. Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8. Protein Sci. 2004;13:2806–2810. doi: 10.1110/ps.04864904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]


