Abstract
Immunodepletion of abundant plasma proteins increases the depth of proteome penetration by mass spectrometry. However, the nature and extent of immunodepletion and the effect of off-target depletion on the quantitative comparison of the residual proteins have not been critically addressed. We performed mass spectrometry label-free quantitation to determine which proteins were immunodepleted and by how much. Two immunodepletion resins were compared: Qproteome (Qiagen) which removes albumin+immunoglobulins and Seppro IgY14+SuperMix (Sigma-Aldrich) which removes 14 target proteins plus a number of unidentified proteins. Plasma collected by P100 proteomic plasma collection tubes (BD) from 20 human subjects was individually immunodepleted to minimize potential variability, prior to pooling. The abundant proteins were quantified better when using only albumin+immunoglobulins removal (Qproteome) while lower abundance proteins were evaluated better using exhaustive immunodepletion (Seppro IgY14+SuperMix). The latter resin removed at least 155 proteins, 38% of the plasma proteome in protein number and 94% of plasma protein in mass. The depth of immunodepletion likely accounts for the effectiveness of this resin in revealing low abundance proteins. However, the more profound immunodepletion achieved with the IgY14+SuperMix may lead to false-positive fold-changes between comparison groups if the reproducibility and efficiency of the depletion of a given protein is not considered.
Keywords: immunodepletion, Seppro, IgY, Qproteome, iTRAQ, EMMOL normalization, off-target
INTRODUCTION
The large dynamic range of protein concentration in plasma, i.e., nine orders of magnitude1, has necessitated the removal of the higher abundance proteins to visualize the low abundance proteins. Immunodepletion of one or more high abundance proteins using antibody affinity immunoabsorption of specific antigens is one method used to accomplish this purpose. Initial application of immunodepletion to proteomics consisted of the depletion of serum albumin2. Subsequent methods, including the Qproteome (Qiagen) matrix, also remove immunoglobulins. Further generations of immunoaffinity columns, including the popular Multiple Affinity Removal System Columns (Agilent) and the ProteoPrep20 Immunodepletion Column (Sigma-Aldrich) remove several more proteins3–5.
To accomplish depletion of a greater number of proteins, the SuperMix resin was created by Sigma-Aldrich by immunization of rabbits with the protein fraction that did not bind to an IgY14 column. In principal, the medium abundance, immunogenic proteins, now enriched, in this fraction induced antibodies for the SuperMix column. However, the targets of the SuperMix resin have never been fully identified and the ability of the Seppro IgY14+SuperMix column (Sigma-Aldrich) to allow quantitation of low abundance proteins is unstudied.
When a given protein is targeted for antibody-mediated removal, protein-protein interactions can lead to the removal of multiple proteins. The terms albuminome and depletome have been proposed to describe the proteins that are bound to albumin or other proteins during immunodepletion 6–9. Estimations of the number of proteins that bind to albumin range from 24 to 67 depending on the methods used 8, 9. A recent report performed an IgY14 column and a SuperMix column in tandem, compared their depleted proteomes and concluded that numerous proteins were immunodepleted by taking the IgY14 exhaustive immunodepletion to the next higher level of exhaustive immunodepletion10. In contrast, our report quantified the immunodepletion efficiencies of individual plasma proteins by a single-stage pre-mixed IgY14+SuperMix column.
Success of immunodepletion approaches has often been measured as the detection of low abundance proteins. However, the nature and extent of immunodepletion and the effect of off-target depletion on the quantitative comparison of the residual proteins have not been critically addressed. We performed mass spectrometry label-free quantitation to determine which proteins were immunodepleted and by how much. Two popular immunodepletion resins were compared: Qproteome (Qiagen) which removes albumin and immunoglobulins and Seppro IgY14+SuperMix (Sigma-Aldrich) which removes 14 target proteins plus a number of unidentified proteins. We found that the IgY14+SuperMix column depleted a substantial portion of the plasma proteome including 155 proteins many of which were previously identified as disease biomarkers. The degree and variability of depletion of such biomarkers can be an issue when comparing their expression levels between groups. Thus exhaustive immunodepletion may lead to false positive or false negative results if both the variability and efficiency of the immunodepletion and the off-target removal of individual proteins are not addressed.
Finally, we combined the data from these two proteomes to yield a continuous quantitative picture of human plasma with 5 logs of dynamic range. The abundant proteins were favored when using only albumin+immunoglobulins removal while lower abundance proteins were evaluated better using exhaustive immunodepletion.
MATERIALS AND METHODS
Study Samples
Plasma samples were collected from 20 subjects using the BD™ P100 Blood Collection Tubes that contained protease inhibitors, according to manufacturer’s instructions. The subjects studied were Caucasian male ex-smokers enrolled in the NIH COPDGene® project, a large multi-center, genome-wide association study designed to elucidate the genetic basis for Chronic Obstructive Pulmonary Disease (COPD)11–18. The use of plasma from subjects in this study was incidental and resulted from a desire to use high quality samples in this analysis.
Study Design
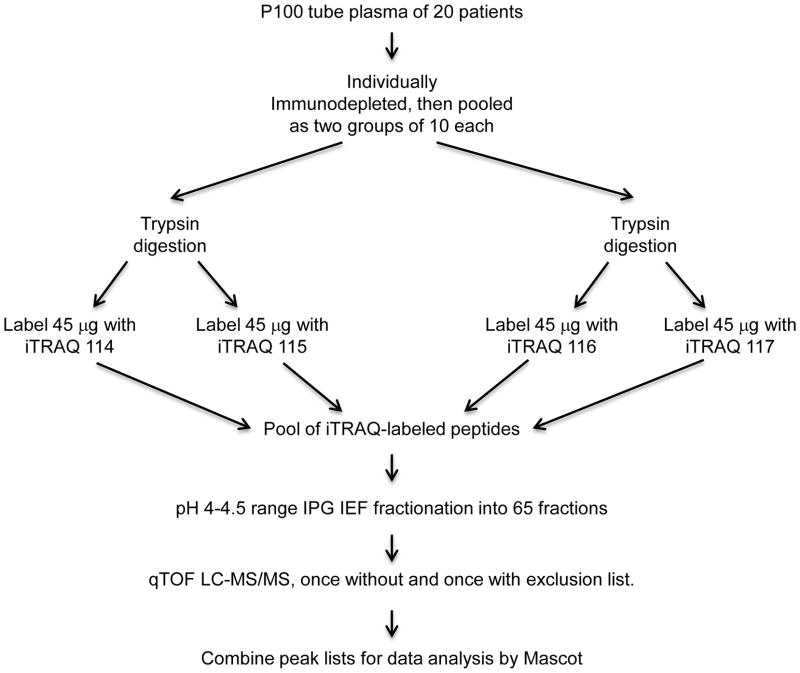
The study design for each of the two immunodepletion methods is shown in Figure 1. The 20 subjects were divided into two study groups of 10 subjects each i.e., Groups 1 and 2. Individual samples taken from disease and control groups were not mixed. To provide technical replicates for the study, iTRAQ 4-plex methodology was used since it allows four replicate experiments to be performed for each immunodepletion method and has the advantage of avoiding LC-MS/MS under-sampling.
Figure 1.
Work flow of an immunodepletion evaluation experiment. The process was done once for the Qproteome method and once for the Seppro IgY14+SuperMix method.
The Depletion of High-Abundance Proteins
Two methods of immunodepletion were used according to the manufacturer’s protocols i.e., the Qproteome spin column approach (Qproteome albumin/IgG Depletion Kit, Qiagen, Carson City, CA) 19 and the Seppro IgY14+SuperMix20 (Seppro Human IgY14 Human SuperMix LC5, catalog number SEP000-KT, Sigma-Aldrich Inc., St. Louis, MO). Researchers using immunodepletion have expressed concern over the possibility of run-to-run variations in the performance of immunodepletion chromatography. One way to prevent such potential variations from negatively impacting this study is to smooth the putative fluctuations by pooling the samples produced by ten chromatography runs. 100 microliter of plasma of each subject was individually immunodepleted by each method. Then aliquots of depleted plasma of 20 subjects were pooled as two groups of 10 each after quality analysis by SDS PAGE and quantitation.
iTRAQ Proteomics
The iTRAQ proteomic analysis procedures used in the current study have been described in detail previously21. From the Qproteome immunodepletion, 100 micrograms of the pooled immunodepleted plasma protein from Group 1 subjects was digested with trypsin and labeled with the iTRAQ 114 label and as a replicate by the iTRAQ 115 label. 100 micrograms of the pooled immunodepleted protein from Group 2 was labeled with the iTRAQ 116 label and as a replicate by the iTRAQ 117 label. These four samples were pooled into an iTRAQ peptide isoelectric focusing fractionation experiment 21–23 using the pH range 4.0–4.5 18 cm long region of a 24 cm IPG strip (pH 3.5–4.5 GE Healthcare) separating the peptides into 65 fractions as previously described in detail21. An identical approach was used for the Seppro IgY14+SuperMix immunodepletion flow through fraction. LC-MS/MS on an Applied Biosystems QSTAR XL qTOF mass spectrometer was performed on each IEF fraction as previously described in detail 24 including a second LC-MS/MS run for each sample using an exclusion list generated from the first LC-MS/MS run to obtain better peptide coverage. Peak lists were generated by the Mascot.dll script written by Matrix Science for the QSTAR instrument. Peptide charges +2 and +3, and monoisotopic. Enzyme was specified as trypsin. The peak lists were combined. Data processing of peptides based on 95% confidence for peptide assignment, with ion score >20 and at least one Bold Red by Mascot 2.2 software as described, allowing no trypsin missed cleavage and no methionine oxidation when calculating the emPAI score. Carbamidomethylation (Cys) was set as fixed modification. The precursor ion m/z tolerance was set at 150 ppm; the product ion m/z tolerance at 0.5 Da. SwissProt Database Release 2010_09 containing 519348 sequence entries was used. The false discovery rate was <3.5 % at the peptide level as determined by Mascot using a randomized SwissProt database. To avoid the possibility of missing identification of low abundance proteins, the database was searched again allowing one trypsin missed cleavage and variable methionine oxidation.
Both IEF (e.g.: OFFGEL of Agilent) and strong cation exchange HPLC (IEX) chromatography can produce the hyper-fractionation desired to minimize signal contamination in an iTRAQ quantitation experiment. IEF was used. Peptides focus at three tight pH ranges22. The narrow pH range of 4.0–4.5 contains about 1/3 of the total peptides and thus is another fractionation step. Having less peptide in the LC-MS/MS minimizes the amount of contamination of each targeted peptide peak being quantified.
Informatics
To calculate the immunodepletion efficiency of a protein, its concentration in the depleted plasma sample and in the initial plasma sample must be compared. Since our focus is the Seppro IgY14+Super Mix column, the Qproteome-depleted plasma was used as a surrogate for the initial plasma sample. This allowed us to assess the effect of the Seppro IgY14+Super Mix column on the lower abundance proteins that are difficult to detect in crude plasma.
Although the four samples in an iTRAQ 4-plex experiment were intended to be identical in total peptide quantity, we sought to correct for variations in protein load caused by protein assay, trypsin digestion, pipetting, and the labeling-chemistry efficiency in each of the iTRAQ channels. We thus used our recently-reported method of iTRAQ data normalization for hyper-fractionated samples called EMMOL21 (emPAI score × molecular weight normalization). emPAI is the exponentially modified Peptide Abundance Index. EMMOL uses mass spectrometry data to deduce the amounts of each protein in the four iTRAQ channels from the iTRAQ ratios. EMMOL normalization of the total protein quantity in each iTRAQ channel is guided by weighting towards the more abundant proteins where mass spectrometry measurements are more accurate for determining the relative values of total protein. In contrast, an iTRAQ normalization scheme that weighs all iTRAQ-labeled peptides equally regardless of protein abundance will suffer from the low accuracy of the identification and quantitation of low-abundance proteins. The peptides derived from low-abundance proteins are relatively low in intensity, producing iTRAQ reporter ions of low intensities. Moreover, fewer of the different peptides anticipated from the protein sequences will be observed by the mass spectrometer due to ion suppression by the higher abundance proteins and the threshold of sensitivity of the instrument.
Protein Quantitation
Key values included in Tables 1–4 are the iTRAQ reporter ratios, UniProt ID, Title, Gene Symbol, prot score, prot_mass, prot_matches, prot_cover, prot_pi, and emPAI.
Table 1.
Immunodepletion Efficiencies of the Targets of Qproteome and IgY14+SuperMix column
| Immuno-depletion method | Title | Gene Symbol | prot_score | prot_mass | prot_matches | prot_cover | emPAI | Average mg/mL in depleted plasma | % immunodepletion |
|---|---|---|---|---|---|---|---|---|---|
| Qproteome | Serum albumin | ALB | 4726 | 80107 | 236 | 39.2 | 2.71 | 1.17 | 97 |
| Seppro | Serum albumin | ALB | 3083 | 80107 | 137 | 38.4 | 2.24 | 0.09 | 99 |
| Qproteome | Ig gamma-1 chain C region | IGHG1 | 8609 | 40775 | 409 | 39.4 | 2.43 | 0.58 | 93 |
| Seppro | Ig gamma-1 chain C region | IGHG1 | 464 | 40775 | 28 | 10.3 | 0.42 | 0.01 | 99 |
| Qproteome | Ig alpha-1 chain C region | IGHA1 | 6711 | 40503 | 264 | 25.8 | 1.89 | 0.42 | 93 |
| Seppro | Ig alpha-1 chain C region | IGHA1 | 936 | 40503 | 43 | 22.4 | 1.03 | 0.02 | 96 |
| Qproteome | Ig kappa chain C region | IGKC | 6260 | 13070 | 168 | 69.8 | 3.84 | 0.29 | 93 |
| Seppro | Ig kappa chain C region | IGKC | 2458 | 13070 | 61 | 53.8 | 1.86 | 0.01 | 96 |
| Qproteome | Ig mu chain C region | IGHM | 5726 | 53274 | 209 | 40.7 | 2.38 | 0.72 | 93 |
| Seppro | Ig mu chain C region | IGHM | 4036 | 53274 | 196 | 44 | 2.87 | 0.08 | 87 |
| Qproteome | Immunoglobulin J chain | IGJ | 304 | 17049 | 8 | 7.5 | 0.5 | 0.05 | 93 |
| Seppro | Immunoglobulin J chain | IGJ | 174 | 19984 | 5 | 11.9 | 0.7 | 0.01 | 80 |
| Qproteome | Complement C3 | C3 | 29308 | 204997 | 1046 | 53.1 | 3.61 | 4.13 | |
| Seppro | Complement C3 | C3 | 1321 | 204997 | 51 | 13.1 | 0.31 | 0.03 | 99 |
| Qproteome | Serotransferrin | TF | 18294 | 87782 | 961 | 55.7 | 8.3 | 4.08 | |
| Seppro | Serotransferrin | TF | 1458 | 87782 | 64 | 27.9 | 1.1 | 0.05 | 98 |
| Qproteome | Haptoglobin-related protein | HPR | 7464 | 43675 | 334 | 30.7 | 3.4 | 0.82 | |
| Seppro | Haptoglobin-related protein | HPR | 875 | 43697 | 36 | 20.4 | 1.27 | 0.03 | 97 |
| Qproteome | Haptoglobin | HP | 10951 | 51048 | 620 | 47.3 | 6.74 | 1.90 | |
| Seppro | Haptoglobin | HP | 1523 | 51048 | 83 | 31.3 | 2.82 | 0.08 | 97 |
| Qproteome | Apolipoprotein A-I | APOA1 | 24599 | 34073 | 1192 | 76.4 | 58.75 | 11.12 | |
| Seppro | Apolipoprotein A-I | APOA1 | 11090 | 34073 | 532 | 77.5 | 34.37 | 0.62 | 95 |
| Qproteome | Apolipoprotein A-II | APOA2 | 3135 | 12867 | 334 | 74 | 17.91 | 1.29 | |
| Seppro | Apolipoprotein A-II | APOA2 | 508 | 12867 | 86 | 74 | 10.08 | 0.07 | 95 |
| Qproteome | Alpha-1-acid glycoprotein 1 | ORM1 | 3162 | 25886 | 150 | 48.3 | 4.19 | 0.61 | |
| Seppro | Glial fibrillary acidic protein | ORM1 | 4211 | 25886 | 214 | 46.8 | 3.52 | 0.05 | 92 |
| Qproteome | Alpha-1-acid glycoprotein 2 | ORM2 | 1418 | 25890 | 95 | 31.3 | 2.44 | 0.35 | |
| Seppro | Alpha-1-acid glycoprotein 2 | ORM2 | 2546 | 25890 | 156 | 29.9 | 2.44 | 0.03 | 91 |
| Qproteome | Alpha-2-macroglobulin | A2M | 19430 | 177570 | 838 | 46 | 3.2 | 3.18 | |
| Seppro | Alpha-2-macroglobulin | A2M | 36377 | 177584 | 1446 | 53.4 | 4.96 | 0.48 | 84 |
| Qproteome | Fibrinogen gamma chain | FGG | 4757 | 57150 | 302 | 33.6 | 3.55 | 1.12 | |
| Seppro | Fibrinogen gamma chain | FGG | 4348 | 57150 | 283 | 48.1 | 4.85 | 0.15 | 84 |
| Qproteome | Fibrinogen alpha chain | FGA | 12367 | 101996 | 656 | 36.5 | 3.01 | 1.69 | |
| Seppro | Fibrinogen alpha chain | FGA | 13296 | 101996 | 690 | 42.6 | 5.36 | 0.30 | 79 |
| Qproteome | Apolipoprotein B-100 | APOB | 13207 | 568255 | 471 | 25.2 | 0.91 | 2.90 | |
| Seppro | Apolipoprotein B-100 | APOB | 31350 | 568096 | 1259 | 48.3 | 3.12 | 0.97 | 66 |
Table 4.
Putative Concentrations of the Composite 412 Protein Plasma Proteome less Albumin and Immunoglobulins
| Immuno-depletion method | Title | Gene Symbol | prot_score | prot_mass | prot_matches | prot_cover | emPAI | Average mg/mL in depleted plasma |
|---|---|---|---|---|---|---|---|---|
| Qproteome | Centrosomal protein of 164 kDa | CEP164 | 94 | 179858 | 78 | 0.4 | 0.02 | 0.020 |
| Seppro | Attractin | ATRN | 803 | 172529 | 48 | 5.8 | 0.21 | 0.020 |
| Qproteome | Inner centromere protein | INCENP | 42 | 117004 | 16 | 2.7 | 0.03 | 0.020 |
| Qproteome | Tumor necrosis factor receptor superfamily member 8 | TNFRSF8 | 86 | 69388 | 10 | 1 | 0.05 | 0.019 |
| Seppro | Thrombospondin-1 | THBS1 | 774 | 141361 | 23 | 7.1 | 0.23 | 0.017 |
| Qproteome | Ras GTPase-activating protein SynGAP | SYNGAP1 | 33 | 158671 | 5 | 0.6 | 0.02 | 0.017 |
| Seppro | Multiple epidermal growth factor-like domains protein 8 | MEGF8 | 282 | 320217 | 10 | 3.2 | 0.07 | 0.012 |
| Seppro | Neural cell adhesion molecule L1-like protein | CHL1 | 246 | 147296 | 28 | 2.4 | 0.13 | 0.010 |
| Seppro | Phosphatidylcholine-sterol acyltransferase | LCAT | 288 | 51762 | 11 | 5 | 0.32 | 0.009 |
| Seppro | Pantetheinase | VNN1 | 295 | 60742 | 8 | 11.5 | 0.27 | 0.009 |
| Seppro | Intercellular adhesion molecule 1 | ICAM1 | 693 | 62046 | 17 | 8.8 | 0.26 | 0.009 |
| Seppro | GDH/6PGL endoplasmic bifunctional protein | H6PD | 118 | 93441 | 7 | 5.4 | 0.17 | 0.009 |
| Seppro | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 808 | 80161 | 23 | 9.4 | 0.2 | 0.009 |
| Seppro | Interleukin-1 receptor accessory protein | IL1RAP | 269 | 72170 | 12 | 5.8 | 0.22 | 0.008 |
| Seppro | Receptor-type tyrosine-protein phosphatase eta | PTPRJ | 421 | 156271 | 9 | 4.6 | 0.1 | 0.008 |
| Seppro | Nidogen-1 | NID1 | 71 | 143465 | 4 | 2.6 | 0.11 | 0.008 |
| Seppro | Neogenin | NEO1 | 147 | 170601 | 8 | 3.3 | 0.09 | 0.008 |
| Seppro | Laminin subunit alpha-2 | LAMA2 | 89 | 379925 | 22 | 2.1 | 0.04 | 0.008 |
| Seppro | Tenascin-X | TNXB | 306 | 488484 | 13 | 1.9 | 0.03 | 0.008 |
| Seppro | 1 Macrophage mannose receptor 1-like protein | MRC1L1 | 100 | 183170 | 4 | 3 | 0.08 | 0.008 |
| Seppro | Platelet basic protein | PPBP | 686 | 16188 | 22 | 18.8 | 0.91 | 0.008 |
| Seppro | Intercellular adhesion molecule 2 | ICAM2 | 345 | 32877 | 9 | 9.5 | 0.39 | 0.007 |
| Seppro | Cell surface glycoprotein MUC18 | MCAM | 108 | 77287 | 19 | 2.8 | 0.15 | 0.006 |
| Seppro | Hypoxia up-regulated protein 1 | HYOU1 | 392 | 122734 | 13 | 6 | 0.09 | 0.006 |
| Seppro | Vascular endothelial growth factor receptor 3 | FLT4 | 184 | 155572 | 7 | 4.3 | 0.07 | 0.006 |
| Seppro | Protein S100-A9 | S100A9 | 110 | 15020 | 4 | 24.6 | 0.59 | 0.005 |
| Seppro | Cystatin-C | CST3 | 379 | 17170 | 10 | 18.5 | 0.5 | 0.005 |
| Seppro | Coiled-coil domain-containing protein 126 | CCDC126 | 303 | 17244 | 8 | 8.6 | 0.5 | 0.005 |
| Seppro | Centromere protein F | CENPF | 36 | 417533 | 22 | 1 | 0.02 | 0.005 |
| Seppro | Metalloproteinase inhibitor 1 | TIMP1 | 281 | 25137 | 6 | 5.8 | 0.33 | 0.004 |
| Seppro | C-reactive protein | CRP | 289 | 27355 | 11 | 11.2 | 0.3 | 0.004 |
| Seppro | Multimerin-2 | MMRN2 | 195 | 110792 | 8 | 1.8 | 0.07 | 0.004 |
| Seppro | Ficolin-2 | FCN2 | 147 | 36885 | 8 | 6.7 | 0.21 | 0.004 |
| Seppro | Noelin | OLFM1 | 177 | 59540 | 8 | 4.7 | 0.13 | 0.004 |
| Seppro | L-selectin | SELL | 108 | 47532 | 9 | 7 | 0.16 | 0.004 |
| Seppro | Endoglin | ENG | 255 | 74730 | 10 | 5.8 | 0.1 | 0.004 |
| Seppro | Scavenger receptor cysteine-rich type 1protein M130 | CD163 | 246 | 136636 | 9 | 2.1 | 0.05 | 0.004 |
| Seppro | Plectin-1 | PLEC1 | 40 | 569055 | 21 | 1.1 | 0.01 | 0.003 |
| Seppro | Poly [ADP-ribose] polymerase 14 | PARP14 | 54 | 218539 | 12 | 0.8 | 0.02 | 0.002 |
| Seppro | Rapamycin-insensitive companion of mTOR | RICTOR | 63 | 208185 | 22 | 1.2 | 0.02 | 0.002 |
| Seppro | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform | PIK3CB | 74 | 135868 | 25 | 0.7 | 0.03 | 0.002 |
| Seppro | Nucleoporin NUP188 homolog | NUP188 | 37 | 209465 | 8 | 0.3 | 0.02 | 0.002 |
| Seppro | Pleckstrin homology domain-containing family A member 7 | PLEKHA7 | 38 | 135984 | 22 | 1.2 | 0.03 | 0.002 |
| Seppro | Enhancer of polycomb homolog 1 | EPC1 | 47 | 102045 | 69 | 1.8 | 0.04 | 0.002 |
| Seppro | Kinesin-like protein KIF21B | KIF21B | 104 | 199879 | 49 | 1 | 0.02 | 0.002 |
| Seppro | Atrophin-1 | ATN1 | 40 | 130632 | 28 | 1 | 0.03 | 0.002 |
| Seppro | MIF4G domain-containing protein | MIF4GD | 67 | 27334 | 18 | 2.7 | 0.14 | 0.002 |
| Seppro | Formin-2 | FMN2 | 36 | 194304 | 12 | 0.6 | 0.02 | 0.002 |
| Seppro | Probable ATP-dependent RNA helicase DDX41 | DDX41 | 44 | 77826 | 5 | 1.1 | 0.05 | 0.002 |
| Seppro | Uncharacterized protein C2orf77 | C2orf77 | 100 | 77323 | 22 | 1.1 | 0.05 | 0.002 |
| Seppro | Probable G-protein coupled receptor 25 | GPR25 | 46 | 39984 | 5 | 1.9 | 0.09 | 0.002 |
| Seppro | Tumor necrosis factor receptor superfamily member 10A | TNFRSF10A | 37 | 53964 | 12 | 1.5 | 0.07 | 0.002 |
| Seppro | Interleukin-12 receptor subunit beta-1 | IL12RB1 | 36 | 77456 | 12 | 0.9 | 0.05 | 0.002 |
| Seppro | Plexin domain-containing protein 2 | PLXDC2 | 153 | 62998 | 5 | 2.1 | 0.06 | 0.002 |
| Seppro | SPRY domain-containing protein 3 | SPRYD3 | 34 | 53346 | 6 | 1.4 | 0.07 | 0.002 |
| Seppro | HMG domain-containing protein 3 | HMGXB3 | 62 | 181488 | 9 | 0.5 | 0.02 | 0.002 |
| Seppro | Calcyclin-binding protein | CACYBP | 108 | 31207 | 43 | 5.7 | 0.12 | 0.002 |
| Seppro | UPF0505 protein C16orf62 | C16orf62 | 82 | 120515 | 4 | 2.1 | 0.03 | 0.002 |
| Seppro | Synaptotagmin-13 | SYT13 | 46 | 51806 | 15 | 2.6 | 0.07 | 0.002 |
| Seppro | Cholesteryl ester transfer protein | CETP | 169 | 59011 | 4 | 3 | 0.06 | 0.002 |
| Seppro | ATP-binding cassette sub-family B member 9 | ABCB9 | 232 | 88768 | 17 | 0.9 | 0.04 | 0.002 |
| Seppro | E3 ubiquitin-protein ligase TRIM37 | TRIM37 | 70 | 116552 | 6 | 1.7 | 0.03 | 0.002 |
| Seppro | 26S proteasome non-ATPase regulatory subunit 1 | PSMD1 | 38 | 117026 | 8 | 1 | 0.03 | 0.002 |
| Seppro | Sodium- and chloride-dependent transporter XTRP3 | SLC6A20 | 37 | 69839 | 7 | 1 | 0.05 | 0.002 |
| Seppro | Retina-specific copper amine oxidase | AOC2 | 38 | 86239 | 6 | 0.8 | 0.04 | 0.002 |
| Seppro | DNA ligase 1 | LIG1 | 31 | 112676 | 6 | 1.2 | 0.03 | 0.002 |
| Seppro | Leucine-rich repeat serine/threonine-protein kinase 2 | LRRK2 | 36 | 315074 | 4 | 0.4 | 0.01 | 0.002 |
| Seppro | Structural maintenance of chromosomes protein 3 | SMC3 | 35 | 160154 | 23 | 2.2 | 0.02 | 0.002 |
| Seppro | Cohesin subunit SA-1 | STAG1 | 54 | 156257 | 10 | 1 | 0.02 | 0.002 |
| Seppro | Centrosomal protein of 135 kDa | CEP135 | 34 | 149283 | 17 | 1 | 0.02 | 0.002 |
| Seppro | Voltage-dependent N-type calcium channel subunit alpha-1B | CACNA1B | 56 | 279251 | 22 | 0.6 | 0.01 | 0.001 |
| Seppro | Nuclear mitotic apparatus protein 1 | NUMA1 | 104 | 261102 | 21 | 0.7 | 0.01 | 0.001 |
EMMOL method of proteome normalization21 was used to calculate the protein concentration of each protein in each pooled plasma sample. EMMOL was demonstrated to be valid by using defined E. coli lysate samples over a 20 fold range of protein quantities and over three logs of dynamic range of protein abundance21.
EMMOL uses the relationship from equation 4 of Ishihama et al.25:
where Mr = molecular weight.
emPAI score is proportional to the fraction of observed peptides/theoretical peptides for a given protein (allowing no methionine oxidation and no missed trypsin cleavage). “emPAI × molecular weight” value is roughly proportional to the abundance of a protein.
Thus the EMMOL data processing workflow involves:
Calculate (emPAI × molecular weight) for each protein as its total in four iTRAQ channels
Normalize each protein to the sum of all proteins for the 4 iTRAQ channels, herein 180 μg protein (the total amount loaded into the 4 channels but any arbitrary total protein value such as the normal plasma protein concentration could have been used instead).
Use the iTRAQ ratios of each protein to calculate the μg of this protein in each iTRAQ channel
Sum the total protein of each iTRAQ channel
Normalize the four channels each to 45 μg of protein
Normalize each protein in each channel to the total protein concentration, determined by protein assay, for the immunodepleted plasma with respect to original plasma volume
Compare the corresponding concentrations from the two immunodepletion methods for each protein
Calculation of the Efficiencies of Immunodepletion of Individual Proteins in Two Immunodepletion Experiments
To calculate the immunodepletion efficiencies for removing albumin and immunoglobulins from plasma by the Qproteome method, the values of serum albumin before immunodepletion was stipulated as 44 mg/mL and immunoglobulins were 24 mg/mL. These are representative values from clinical test reports. After immunodepletion, the Qproteome depleted plasma had a protein concentration of 62.4 mg/mL when equated to the original plasma volume. The IgY14+SuperMix depleted plasma had a protein concentration of 7.4 mg/mL when equated to the original plasma volume. These values were used to normalize the measurements of individual protein quantities obtained from EMMOL calculations. The averaged results of the four iTRAQ channels are shown under the header “Average mg/mL in depleted plasma” in Tables 1–4. The ratio of this value for each protein from the IgY14+SuperMix column immunodepletion compared with the value from the Qproteome immunodepletion produces the values under the header “% immunodepletion”.
Construction of a Plasma Proteome from Two Immunodepletion Experiments
A subset of proteins was each quantified in both immunodepletion approaches. However, a given protein may be more abundant in the depleted proteome of one immunodepletion method than the other, resulting in better peptide statistics for identification and better iTRAQ quantitation accuracy. The better quality of these two results where available for each protein was used to construct a composite continuous proteome for further analysis. Moreover, the comparison of the proteomes of the two immunodepletion methods (Tables 1–4) was used to identify the medium abundance proteins and the non-targeted proteins removed by the Seppro IgY14+SuperMix column.
RESULTS AND DISCUSSION
A comparison of the proteins in the pooled plasma samples immunodepleted by two procedures is shown in Tables 1–4. Supporting Information for Publication is the full display of all the tables that are partially presented herein plus the original Mascot output data of the Qproteome proteome and the Seppro IgY14+SuperMix proteome. Although the intended quantity of protein in each label reaction was 45 micrograms, EMMOL calculations suggested that the values of the total peptides for each iTRAQ channel during LC-MS/MS varied by as much as 20%. For the Qproteome experiments, iTRAQ 114, 115, 116, and 117 channels were 40.1, 41.7, 47.6, and 50.6 micrograms, respectively. For the Seppro IgY14+SuperMix experiments, iTRAQ 114, 115, 116, and 117 channels were 39.9, 43.9, 46.6, and 49.9 micrograms, respectively. A 20% bias in an iTRAQ channel would tend to complicate accurate determination of small fold-changes among proteins that differ in expression in two proteomes and confound proteomics pathway analysis. Hence, normalization based on the mass spectrometry measurements of the high abundance proteins tends to restore precision for the iTRAQ comparison.
The resulting proteome contained 412 proteins with quantitation, of which 120 were present in both immunodepletion samples
IgY14 Depletion
The Qproteome and the Seppro IgY14+SuperMix methods removed most of the albumin and the immunoglobulins according to SDS PAGE (data not shown) and the latter matrix removed many other proteins not visualized in SDS PAGE. The IgY14 was designed to remove human serum albumin, IgG, fibrinogen, transferrin, IgA, IgM, haptoglobin, alpha2-macroglubulin, alpha1-acid glycoprotein, alpha1-antitrypsin, apo A-I HDL, apo A-II, complement C3, and apo B. Table 1 illustrates their immunodepletion efficiencies.
SuperMix Depletion
A reasonable indication that a protein has been immunodepleted by the SuperMix resin is that its level has decreased by about 80–90 % after passing through the SuperMix resin. This arbitrary cutoff range seemed suitable for this dataset and revealed that the resin removed most of another 76 proteins (Table 2), including: complement factor B, antithrombin-III, inter-alpha-trypsin inhibitor heavy chain H1, H2, and H4; ceruloplasmin, complement C4-A, vitronectin, hemoglobin subunit alpha, beta and delta, plasma protease C1 inhibitor, prothrombin, angiotensinogen, vitamin D-binding protein, histidine-rich glycoprotein, and alpha-1B-glycoprotein. In all, most of the 20 top-abundance plasma proteins were depleted, but not eliminated, by IgY14+SuperMix column.
Table 2.
Immunodepletion Efficiencies of the 76 Putative Targets of Seppro SuperMix Resin
| Immuno-depletion method | Title | Gene Symbol | prot_score | prot_mass | prot_matches | prot_cover | emPAI | Average mg/mL in depleted plasma | % immunodepletion |
|---|---|---|---|---|---|---|---|---|---|
| Qproteome | Complement factor B | CFB | 2265 | 94629 | 110 | 21.6 | 1.32 | 0.698 | |
| Seppro | Complement factor B | CFB | 102 | 94629 | 2 | 1.8 | 0.04 | 0.002 | 100 |
| Qproteome | Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 2298 | 109541 | 95 | 24.5 | 0.94 | 0.576 | |
| Seppro | Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 51 | 109573 | 3 | 1.8 | 0.07 | 0.004 | 100 |
| Qproteome | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 1671 | 116337 | 55 | 15 | 0.55 | 0.357 | |
| Seppro | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 198 | 116364 | 5 | 2.3 | 0.03 | 0.002 | 100 |
| Qproteome | Complement C4-A | C4A | 4364 | 205487 | 181 | 24.2 | 1.03 | 1.178 | |
| Seppro | Complement C4-A | C4A | 168 | 205487 | 8 | 2.5 | 0.07 | 0.008 | 100 |
| Qproteome | Antithrombin-III | SERPINC1 | 5520 | 58501 | 167 | 46.3 | 2.43 | 0.794 | |
| Seppro | Antithrombin-III | SERPINC1 | 160 | 58501 | 4 | 5 | 0.13 | 0.004 | 100 |
| Qproteome | Ceruloplasmin | CP | 3498 | 132638 | 124 | 25.1 | 0.93 | 0.687 | |
| Seppro | Ceruloplasmin | CP | 74 | 132638 | 2 | 2.1 | 0.06 | 0.004 | 99 |
| Qproteome | Vitronectin | VTN | 1661 | 58096 | 47 | 20.7 | 0.86 | 0.281 | |
| Seppro | Vitronectin | VTN | 210 | 58096 | 4 | 3.1 | 0.06 | 0.002 | 99 |
| Qproteome | Hemoglobin subunit alpha | HBA1 | 945 | 17034 | 37 | 35.2 | 2.43 | 0.231 | |
| Seppro | Hemoglobin subunit alpha | HBA1 | 55 | 17034 | 2 | 8.5 | 0.23 | 0.002 | 99 |
| Qproteome | Plasma protease C1 inhibitor | SERPING1 | 2382 | 59671 | 105 | 23.8 | 1.33 | 0.444 | |
| Seppro | Plasma protease C1 inhibitor | SERPING1 | 124 | 59671 | 3 | 5.4 | 0.13 | 0.004 | 99 |
| Qproteome | Vitamin D-binding protein | GC | 1757 | 61010 | 83 | 23.2 | 1.03 | 0.351 | |
| Seppro | Vitamin D-binding protein | GC | 112 | 61010 | 3 | 6.3 | 0.19 | 0.006 | 99 |
| Qproteome | Prothrombin | F2 | 1017 | 75798 | 28 | 12.5 | 0.4 | 0.169 | |
| Seppro | Prothrombin | F2 | 49 | 75798 | 2 | 1.6 | 0.05 | 0.002 | 99 |
| Qproteome | Angiotensinogen | AGT | 977 | 56576 | 42 | 13.2 | 0.47 | 0.149 | |
| Seppro | Angiotensinogen | AGT | 278 | 56576 | 9 | 2.7 | 0.07 | 0.002 | 99 |
| Qproteome | Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 2049 | 108699 | 55 | 13.4 | 0.4 | 0.242 | |
| Seppro | Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 179 | 108699 | 5 | 4.2 | 0.07 | 0.004 | 99 |
| Qproteome | Hemoglobin subunit beta | HBB | 2335 | 17832 | 71 | 67.3 | 7.62 | 0.761 | |
| Seppro | Hemoglobin subunit beta | HBB | 629 | 17832 | 15 | 44.9 | 1.66 | 0.016 | 98 |
| Qproteome | Histidine-rich glycoprotein | HRG | 945 | 63825 | 36 | 17.9 | 0.57 | 0.203 | |
| Seppro | Histidine-rich glycoprotein | HRG | 60 | 63825 | 4 | 5 | 0.12 | 0.004 | 98 |
| Qproteome | Alpha-1B-glycoprotein | A1BG | 249 | 56538 | 24 | 9.5 | 0.21 | 0.066 | |
| Seppro | Alpha-1B-glycoprotein | A1BG | 38 | 56538 | 3 | 2 | 0.07 | 0.002 | 98 |
| Qproteome | Hemoglobin subunit delta | HBD | 1079 | 17889 | 42 | 50.3 | 2.94 | 0.294 | |
| Seppro | Hemoglobin subunit delta | HBD | 87 | 17889 | 8 | 32 | 0.8 | 0.008 | 98 |
| Qproteome | Afamin | AFM | 1139 | 78456 | 40 | 20.7 | 0.59 | 0.259 | |
| Seppro | Afamin | AFM | 775 | 78456 | 31 | 8.2 | 0.2 | 0.008 | 97 |
| Qproteome | Complement component C7 | C7 | 200 | 103711 | 5 | 3 | 0.07 | 0.039 | |
| Seppro | Complement component C7 | C7 | 219 | 103711 | 5 | 1.8 | 0.04 | 0.002 | 96 |
| Qproteome | Alpha-1-antitrypsin | SERPINA1 | 16719 | 51922 | 799 | 64.6 | 8.86 | 2.550 | |
| Seppro | Alpha-1-antitrypsin | SERPINA1 | 4261 | 51922 | 220 | 53.1 | 4.66 | 0.129 | 96 |
| Qproteome | C4b-binding protein alpha chain | C4BPA | 770 | 74086 | 23 | 8.9 | 0.41 | 0.169 | |
| Seppro | C4b-binding protein alpha chain | C4BPA | 395 | 74086 | 9 | 3.9 | 0.16 | 0.006 | 96 |
| Qproteome | C4b-binding protein beta chain | C4BPB | 77 | 32046 | 4 | 4 | 0.25 | 0.045 | |
| Seppro | C4b-binding protein beta chain | C4BPB | 51 | 32046 | 2 | 4 | 0.12 | 0.002 | 96 |
| Qproteome | Dermcidin | DCD | 56 | 12976 | 2 | 10 | 0.3 | 0.022 | |
| Seppro | Dermcidin | DCD | 77 | 12976 | 2 | 10 | 0.3 | 0.002 | 95 |
| Qproteome | Kinesin-like protein KIF21B | KIF21B | 77 | 199879 | 30 | 1 | 0.04 | 0.045 | |
| Seppro | Kinesin-like protein KIF21B | KIF21B | 104 | 199879 | 49 | 1 | 0.02 | 0.002 | 95 |
| Qproteome | Apolipoprotein C-III | APOC3 | 1489 | 11854 | 56 | 55.6 | 6.52 | 0.437 | |
| Seppro | Apolipoprotein C-III | APOC3 | 1199 | 11854 | 49 | 53.5 | 3.23 | 0.021 | 95 |
| Qproteome | Complement C1q subcomponent subunit B | C1QB | 238 | 28976 | 7 | 5.5 | 0.28 | 0.045 | |
| Seppro | Complement C1q subcomponent subunit B | C1QB | 276 | 29238 | 10 | 5.5 | 0.13 | 0.002 | 95 |
| Qproteome | Heparin cofactor 2 | SERPIND1 | 917 | 62105 | 29 | 18.4 | 0.5 | 0.174 | |
| Seppro | Heparin cofactor 2 | SERPIND1 | 292 | 62105 | 10 | 9.6 | 0.26 | 0.009 | 94 |
| Qproteome | Ficolin-2 | FCN2 | 152 | 36885 | 7 | 9.6 | 0.34 | 0.070 | |
| Seppro | Ficolin-2 | FCN2 | 147 | 36885 | 8 | 6.7 | 0.21 | 0.004 | 94 |
| Qproteome | Interleukin-12 receptor beta-1 chain | IL12RB1 | 34 | 77456 | 6 | 2.3 | 0.05 | 0.022 | |
| Seppro | Interleukin-12 receptor subunit beta-1 | IL12RB1 | 36 | 77456 | 12 | 0.9 | 0.05 | 0.002 | 94 |
| Qproteome | Serum amyloid A-4 protein | SAA4 | 355 | 16007 | 12 | 26.2 | 0.92 | 0.082 | |
| Seppro | Serum amyloid A-4 protein | SAA4 | 250 | 16004 | 7 | 8.5 | 0.55 | 0.005 | 94 |
| Qproteome | Phospholipid transfer protein | PLTP | 182 | 57526 | 5 | 8.1 | 0.29 | 0.093 | |
| Seppro | Phospholipid transfer protein | PLTP | 381 | 57526 | 17 | 5.3 | 0.21 | 0.007 | 94 |
| Qproteome | Complement factor I | CFI | 306 | 74412 | 12 | 9.3 | 0.21 | 0.088 | |
| Seppro | Complement factor I | CFI | 154 | 74412 | 14 | 4.6 | 0.16 | 0.006 | 93 |
| Qproteome | Tripartite motif-containing protein 37 | TRIM37 | 46 | 116552 | 2 | 0.7 | 0.03 | 0.020 | |
| Seppro | E3 ubiquitin-protein ligase TRIM37 | TRIM37 | 70 | 116552 | 6 | 1.7 | 0.03 | 0.002 | 93 |
| Qproteome | Complement component C9 | C9 | 759 | 70091 | 29 | 8.4 | 0.36 | 0.140 | |
| Seppro | Complement component C9 | C9 | 337 | 70091 | 16 | 8.9 | 0.29 | 0.011 | 93 |
| Qproteome | R3H domain-containing protein 1 | R3HDM1 | 63 | 129519 | 2 | 0.6 | 0.03 | 0.022 | |
| Seppro | R3H domain-containing protein 1 | R3HDM1 | 54 | 129519 | 2 | 0.6 | 0.03 | 0.002 | 93 |
| Qproteome | Complement C1s subcomponent | C1S | 794 | 83650 | 29 | 14.2 | 0.54 | 0.251 | |
| Seppro | Complement C1s subcomponent | C1S | 766 | 83650 | 39 | 13.7 | 0.48 | 0.022 | 91 |
| Qproteome | Apolipoprotein C-I | APOC1 | 627 | 10767 | 46 | 32.5 | 3.85 | 0.232 | |
| Seppro | Apolipoprotein C-I | APOC1 | 373 | 10767 | 23 | 32.5 | 3.85 | 0.022 | 91 |
| Qproteome | Alpha-1-antichymotrypsin | SERPINA3 | 4424 | 51682 | 158 | 38.3 | 1.65 | 0.475 | |
| Seppro | Alpha-1-antichymotrypsin | SERPINA3 | 2975 | 51682 | 114 | 40.4 | 2.27 | 0.062 | 90 |
| Qproteome | Apolipoprotein C-II | APOC2 | 247 | 12285 | 13 | 45.5 | 2.05 | 0.142 | |
| Seppro | Apolipoprotein C-II | APOC2 | 313 | 12285 | 12 | 57.4 | 2.05 | 0.014 | 90 |
| Qproteome | UPF0505 protein C16orf62 | C16orf62 | 32 | 120515 | 3 | 2.1 | 0.03 | 0.020 | |
| Seppro | UPF0505 protein C16orf62 | C16orf62 | 82 | 120515 | 4 | 2.1 | 0.03 | 0.002 | 90 |
| Qproteome | Obscurin | OBSCN | 217 | 928336 | 71 | 0.3 | 0.01 | 0.051 | |
| Seppro | Obscurin | OBSCN | 87 | 928336 | 34 | 0.6 | 0.01 | 0.005 | 90 |
| Qproteome | Apolipoprotein D | APOD | 147 | 23276 | 9 | 10.1 | 0.35 | 0.045 | |
| Seppro | Apolipoprotein D | APOD | 128 | 23276 | 5 | 10.1 | 0.35 | 0.004 | 90 |
| Qproteome | Complement C1q subcomponent subunit C | C1QC | 155 | 27859 | 3 | 7.3 | 0.14 | 0.022 | |
| Seppro | Complement C1q subcomponent subunit C | C1QC | 174 | 27859 | 3 | 7.3 | 0.14 | 0.002 | 90 |
| Qproteome | A disintegrin and metalloproteinase with thrombospondin motifs 12 | ADAMTS12 | 37 | 196369 | 8 | 0.9 | 0.02 | 0.022 | |
| Seppro | A disintegrin and metalloproteinase with thrombospondin motifs 12 | ADAMTS12 | 33 | 196355 | 6 | 0.9 | 0.02 | 0.002 | 89 |
| Qproteome | Phosphatidylinositol-4,5- bisphosphate 3-kinase catalytic subunit beta isoform | PIK3CB | 61 | 135868 | 26 | 1.2 | 0.03 | 0.023 | |
| Seppro | Phosphatidylinositol-4,5- bisphosphate 3-kinase catalytic subunit beta isoform | PIK3CB | 74 | 135868 | 25 | 0.7 | 0.03 | 0.002 | 89 |
| Qproteome | Uncharacterized protein C2orf77 | C2orf77 | 47 | 77323 | 3 | 1.1 | 0.05 | 0.022 | |
| Seppro | Uncharacterized protein C2orf77 | C2orf77 | 100 | 77323 | 22 | 1.1 | 0.05 | 0.002 | 89 |
| Qproteome | Vacuolar fusion protein MON1homolog A | MON1A | 52 | 64884 | 8 | 1.4 | 0.06 | 0.022 | |
| Seppro | Vacuolar fusion protein MON1homolog A | MON1A | 62 | 64884 | 9 | 1.4 | 0.06 | 0.002 | 89 |
| Qproteome | T-complex protein 1 subunit zeta-2 | CCT6B | 127 | 65072 | 61 | 1.5 | 0.06 | 0.021 | |
| Seppro | T-complex protein 1 subunit zeta-2 | CCT6B | 42 | 65072 | 23 | 2.8 | 0.06 | 0.002 | 88 |
| Qproteome | ATP-binding cassette sub-family B member 9 | ABCB9 | 126 | 88768 | 7 | 0.9 | 0.04 | 0.020 | |
| Seppro | ATP-binding cassette sub-family B member 9 | ABCB9 | 232 | 88768 | 17 | 0.9 | 0.04 | 0.002 | 88 |
| Qproteome | Keratin, type I cytoskeletal 9 | KRT9 | 221 | 66211 | 13 | 7.7 | 0.24 | 0.087 | |
| Seppro | Keratin, type I cytoskeletal 9 | KRT9 | 138 | 66146 | 13 | 5.8 | 0.24 | 0.008 | 88 |
| Qproteome | Apolipoprotein L1 | APOL1 | 573 | 47751 | 16 | 9.5 | 0.46 | 0.123 | |
| Seppro | Apolipoprotein L1 | APOL1 | 517 | 47751 | 25 | 11.6 | 0.57 | 0.015 | 87 |
| Qproteome | Protein AMBP | AMBP | 934 | 42624 | 36 | 25 | 0.52 | 0.125 | |
| Seppro | Protein AMBP | AMBP | 1308 | 42624 | 37 | 25 | 0.66 | 0.015 | 87 |
| Qproteome | Fibronectin | FN1 | 1774 | 277418 | 49 | 7.4 | 0.17 | 0.261 | |
| Seppro | Fibronectin | FN1 | 1648 | 277437 | 44 | 6.7 | 0.19 | 0.029 | 86 |
| Qproteome | Hyaluronan-binding protein 2 | HABP2 | 95 | 70504 | 2 | 1.6 | 0.11 | 0.043 | |
| Seppro | Hyaluronan-binding protein 2 | HABP2 | 349 | 70504 | 12 | 3.8 | 0.17 | 0.006 | 86 |
| Qproteome | Complement-activating component of Ra-reactive factor | MASP1 | 161 | 86325 | 8 | 2.7 | 0.09 | 0.043 | |
| Seppro | Mannan-binding lectin serine protease 1 | MASP1 | 485 | 86325 | 20 | 2.7 | 0.13 | 0.006 | 86 |
| Qproteome | Isocitrate dehydrogenase [NAD]subunit alpha, mitochondrial | IDH3A | 131 | 43625 | 12 | 2.7 | 0.09 | 0.022 | |
| Seppro | Isocitrate dehydrogenase [NAD]subunit alpha, mitochondrial | IDH3A | 52 | 43625 | 3 | 2.7 | 0.18 | 0.004 | 84 |
| Qproteome | Keratin, type I cytoskeletal 10 | KRT10 | 728 | 63161 | 21 | 14.4 | 0.41 | 0.144 | |
| Seppro | Keratin, type I cytoskeletal 10 | KRT10 | 840 | 62478 | 29 | 17 | 0.68 | 0.023 | 84 |
| Qproteome | Apolipoprotein E | APOE | 1982 | 38263 | 45 | 41.3 | 2.71 | 0.579 | |
| Seppro | Apolipoprotein E | APOE | 2852 | 38263 | 100 | 46.1 | 5.5 | 0.113 | 83 |
| Qproteome | Gelsolin | GSN | 695 | 92672 | 16 | 11.8 | 0.22 | 0.114 | |
| Seppro | Gelsolin | GSN | 808 | 92672 | 24 | 13.9 | 0.37 | 0.019 | 83 |
| Qproteome | CD5 antigen-like | CD5L | 88 | 42052 | 2 | 4 | 0.09 | 0.022 | |
| Seppro | CD5 antigen-like | CD5L | 110 | 42052 | 4 | 6.1 | 0.19 | 0.004 | 82 |
| Qproteome | Keratin, type II cytoskeletal 2epidermal | KRT2 | 359 | 71298 | 12 | 7.5 | 0.23 | 0.092 | |
| Seppro | Keratin, type II cytoskeletal 2epidermal | KRT2 | 362 | 70866 | 18 | 11.7 | 0.43 | 0.016 | 80 |
| Qproteome | Hypoxia up-regulated protein 1 | HYOU1 | 99 | 122734 | 2 | 1.9 | 0.03 | 0.021 | |
| Seppro | Hypoxia up-regulated protein 1 | HYOU1 | 392 | 122734 | 13 | 6 | 0.09 | 0.006 | 80 |
| Qproteome | Corticosteroid-binding globulin | SERPINA6 | 259 | 47877 | 9 | 7.2 | 0.25 | 0.067 | |
| Seppro | Corticosteroid-binding globulin | SERPINA6 | 1344 | 47877 | 42 | 12.8 | 0.57 | 0.015 | 79 |
| Qproteome | Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | 262 | 108718 | 7 | 4 | 0.14 | 0.085 | |
| Seppro | Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | 513 | 108718 | 16 | 12 | 0.31 | 0.018 | 79 |
| Prostaglandin-H2 D-isomerase | PTGDS | 139 | 22829 | 3 | 8.9 | 0.17 | 0.022 | ||
| Seppro | Prostaglandin-H2 D-isomerase | PTGDS | 248 | 22829 | 15 | 12.6 | 0.36 | 0.004 | 79 |
| Qproteome | L-lactate dehydrogenase B chain | LDHB | 55 | 40791 | 2 | 7.5 | 0.19 | 0.043 | |
| Seppro | L-lactate dehydrogenase B chain | LDHB | 445 | 40791 | 11 | 12.3 | 0.42 | 0.009 | 79 |
| Qproteome | Keratin, type II cytoskeletal 1 | KRT1 | 380 | 70184 | 23 | 13.5 | 0.51 | 0.197 | |
| Seppro | Keratin, type II cytoskeletal 1 | KRT1 | 846 | 70349 | 40 | 19.9 | 1.16 | 0.043 | 78 |
| Qproteome | Selenoprotein P | SEPP1 | 163 | 48016 | 8 | 8.1 | 0.25 | 0.067 | |
| Seppro | Selenoprotein P | SEPP1 | 536 | 48016 | 25 | 16 | 0.57 | 0.015 | 77 |
| Qproteome | Cystatin-C | CST3 | 125 | 17170 | 2 | 11 | 0.23 | 0.022 | |
| Seppro | Cystatin-C | CST3 | 379 | 17170 | 10 | 18.5 | 0.5 | 0.005 | 77 |
| Qproteome | Transthyretin | TTR | 2397 | 17288 | 141 | 57.1 | 4.03 | 0.390 | |
| Seppro | Transthyretin | TTR | 2970 | 17288 | 191 | 73.5 | 8.21 | 0.078 | 76 |
| Qproteome | Cartilage oligomeric matrix protein | COMP | 131 | 88457 | 2 | 2.6 | 0.04 | 0.020 | |
| Seppro | Cartilage oligomeric matrix protein | COMP | 482 | 88457 | 9 | 5.8 | 0.13 | 0.006 | 76 |
| Qproteome | Coagulation factor XIII A chain | F13A1 | 195 | 89348 | 8 | 6.1 | 0.13 | 0.065 | |
| Seppro | Coagulation factor XIII A chain | F13A1 | 697 | 89348 | 22 | 10.5 | 0.28 | 0.014 | 76 |
| Qproteome | Plasma serine protease inhibitor | SERPINA5 | 90 | 49389 | 4 | 5.9 | 0.16 | 0.044 | |
| Seppro | Plasma serine protease inhibitor | SERPINA5 | 221 | 49389 | 10 | 11.1 | 0.34 | 0.009 | 75 |
| Qproteome | Monocyte differentiation antigen CD14 | CD14 | 162 | 42119 | 4 | 7.2 | 0.19 | 0.045 | |
| Seppro | Monocyte differentiation antigen CD14 | CD14 | 581 | 42119 | 21 | 14.7 | 0.53 | 0.012 | 75 |
| Qproteome | Coagulation factor V | F5 | 249 | 271099 | 9 | 2.1 | 0.06 | 0.091 | |
| Seppro | Coagulation factor V | F5 | 809 | 271099 | 33 | 5.5 | 0.16 | 0.023 | 74 |
| Qproteome | Serum paraoxonase/arylesterase 1 | PON1 | 1071 | 42921 | 25 | 18 | 0.29 | 0.070 | |
| Seppro | Serum paraoxonase/arylesterase 1 | PON1 | 1409 | 42921 | 59 | 27 | 0.95 | 0.022 | 74 |
Off-target Protein Removal
Many abundant proteins in serum double as carrier proteins with high affinity binding but this phenomenon has only been studied for a few abundant proteins. The carrier role of serum albumin for small molecules26 and proteins 9 is well known. Another example is alpha 2 macroglobulin, the most abundant globulin in the blood that is not an immunoglobulin, is an effective protease inhibitor, blocks fibrinolysis and is a known carrier of prostate-specific antigen PSA27, 28. However, it also binds to numerous growth factors and cytokines, such as platelet-derived growth factor, basic fibroblast growth factor, TGF-β, insulin, and IL-1β. Driven by affinity removal by the high-abundance carrier proteins, these off-target depleted proteins shown in Table 3 were not seen in the highly enriched Seppro IgY14+SuperMix fraction in spite of their relatively ease of identification, with many peptides each, in the Qproteome fraction. Table 3 illustrates 65 proteins of this class. Mass spectrometry under-sampling is unlikely to explain their absence in the Seppro IgY14+SuperMix fraction; and we designate these as off-target removals. Other explanations are plausible, including the possibility that some of these proteins were exceptionally immunogenic in production of SuperMix matrix. In all, about 155 plasma proteins, 38% of the plasma proteome in protein number in this report and 94% of plasma protein in mass, were removed from the plasma by the Seppro IgY14+SuperMix resin.
Table 3.
65 Proteins Putatively Absorbed by Protein-protein Interactions on Seppro IgY14+SuperMix Column
| Immuno-depletion method | Title | Gene Symbol | prot_score | prot_mass | prot_matches | prot_cover | emPAI | Average mg/mL in depleted plasma |
|---|---|---|---|---|---|---|---|---|
| Qproteome | Hemopexin | HPX | 3086 | 55555 | 219 | 32.3 | 2.02 | 0.628 |
| Qproteome | Phosphatidylinositol-4-phosphate 3-kinase C2domain-containing gamma polypeptide | PIK3C2G | 106 | 182657 | 102 | 1.3 | 0.04 | 0.041 |
| Qproteome | Centrosomal protein of 164 kDa | CEP164 | 94 | 179858 | 78 | 0.4 | 0.02 | 0.020 |
| Qproteome | Kinesin-like protein KIF16B | KIF16B | 41 | 169204 | 77 | 1.5 | 0.02 | 0.019 |
| Qproteome | Alpha-2-HS-glycoprotein | AHSG | 557 | 42548 | 76 | 19.1 | 0.97 | 0.232 |
| Qproteome | Dynein heavy chain 17, axonemal | DNAH17 | 214 | 563266 | 74 | 0.2 | 0.01 | 0.031 |
| Qproteome | Complement factor H | CFH | 834 | 155353 | 57 | 11.2 | 0.26 | 0.225 |
| Qproteome | Elongation factor Ts, mitochondrial | TSFM | 143 | 39169 | 52 | 2.2 | 0.1 | 0.022 |
| Qproteome | Protein FAM171B | FAM171B | 61 | 100849 | 49 | 1.4 | 0.04 | 0.022 |
| Qproteome | Kininogen-1 | KNG1 | 1267 | 80489 | 48 | 20.3 | 0.72 | 0.326 |
| Qproteome | WD repeat-containing protein 19 | WDR19 | 47 | 166391 | 38 | 0.4 | 0.02 | 0.018 |
| Qproteome | Plasminogen | PLG | 521 | 100452 | 34 | 5.1 | 0.2 | 0.112 |
| Qproteome | Beta-2-glycoprotein 1 | APOH | 282 | 44051 | 27 | 7.8 | 0.28 | 0.069 |
| Qproteome | Serum amyloid P-component | APCS | 780 | 27358 | 22 | 14.3 | 0.68 | 0.104 |
| Qproteome | Transmembrane protein 201 | TMEM201 | 42 | 76326 | 20 | 1.2 | 0.05 | 0.021 |
| Qproteome | Complement C5 | C5 | 504 | 207189 | 16 | 3.7 | 0.11 | 0.127 |
| Qproteome | Complement component C6 | C6 | 663 | 118022 | 16 | 6.7 | 0.13 | 0.085 |
| Qproteome | Dynein heavy chain 8, axonemal | DNAH8 | 59 | 565682 | 16 | 0.3 | 0.01 | 0.032 |
| Qproteome | Negative elongation factor B | COBRA1 | 87 | 71759 | 16 | 2.2 | 0.05 | 0.020 |
| Qproteome | Inner centromere protein | INCENP | 42 | 117004 | 16 | 2.7 | 0.03 | 0.020 |
| Qproteome | Formin-like protein 1 | FMNL1 | 32 | 132291 | 12 | 0.6 | 0.03 | 0.023 |
| Qproteome | Vacuolar protein-sorting-associated protein 36 | VPS36 | 40 | 48397 | 12 | 1.6 | 0.08 | 0.022 |
| Qproteome | Actin, cytoplasmic 1 | ACTB | 474 | 44934 | 11 | 15.5 | 0.49 | 0.125 |
| Qproteome | Pericentrin | PCNT | 48 | 411049 | 11 | 0.5 | 0.01 | 0.023 |
| Qproteome | Complement component C8 beta chain | C8B | 462 | 73758 | 10 | 7.1 | 0.22 | 0.090 |
| Qproteome | Tumor necrosis factor receptor superfamily member 8 | TNFRSF8 | 86 | 69388 | 10 | 1 | 0.05 | 0.019 |
| Qproteome | Staphylococcal nuclease domain-containing protein 1 | SND1 | 45 | 111120 | 9 | 2.3 | 0.07 | 0.044 |
| Qproteome | SET domain-containing protein 3 | SETD3 | 37 | 73898 | 9 | 0.8 | 0.05 | 0.021 |
| Qproteome | CAP-Gly domain-containing linker protein 1 | CLIP1 | 29 | 183824 | 9 | 1 | 0.02 | 0.021 |
| Qproteome | Retinol-binding protein 4 | RBP4 | 349 | 25067 | 8 | 19.4 | 0.53 | 0.075 |
| Qproteome | Alpha-2-antiplasmin | SERPINF2 | 396 | 57755 | 8 | 6.5 | 0.21 | 0.068 |
| Qproteome | Heparanase | HPSE | 46 | 67043 | 8 | 2.4 | 0.06 | 0.023 |
| Qproteome | Actin-binding LIM protein 1 | ABLIM1 | 32 | 97150 | 7 | 2.6 | 0.08 | 0.043 |
| Qproteome | Trichoplein keratin filament-binding protein | TCHP | 69 | 67056 | 7 | 2.8 | 0.06 | 0.023 |
| Qproteome | Uncharacterized protein FLJ44048 | - | 59 | 205325 | 7 | 1.1 | 0.02 | 0.023 |
| Qproteome | ADAMTS-like protein 1 | ADAMTSL1 | 29 | 64754 | 7 | 0.4 | 0.06 | 0.022 |
| Qproteome | P protein | OCA2 | 46 | 96527 | 7 | 0.7 | 0.04 | 0.022 |
| Qproteome | Kappa-actin | FKSG30 | 65 | 45645 | 6 | 5.5 | 0.17 | 0.044 |
| Qproteome | Probable E3 ubiquitin-protein ligase HECTD3 | HECTD3 | 35 | 104187 | 6 | 1.4 | 0.04 | 0.023 |
| Qproteome | Cyclin-dependent kinase-like 1 | CDKL1 | 39 | 46856 | 6 | 2 | 0.08 | 0.020 |
| Qproteome | Golgi resident protein GCP60 | ACBD3 | 40 | 65308 | 6 | 1.3 | 0.06 | 0.020 |
| Qproteome | ANKRD26-like family C member 1A | A26C1A | 161 | 135563 | 5 | 2.6 | 0.08 | 0.061 |
| Qproteome | Plasma kallikrein | KLKB1 | 97 | 79485 | 5 | 1.7 | 0.05 | 0.022 |
| Qproteome | Zinc finger protein 37A | ZNF37A | 47 | 74724 | 5 | 1.4 | 0.05 | 0.021 |
| Qproteome | 26S proteasome non-ATPase regulatory subunit 5 | PSMD5 | 40 | 59730 | 5 | 1.4 | 0.06 | 0.020 |
| Qproteome | Ras GTPase-activating protein SynGAP | SYNGAP1 | 33 | 158671 | 5 | 0.6 | 0.02 | 0.017 |
| Qproteome | N-acetylmuramoyl-L-alanine amidase | PGLYRP2 | 161 | 64621 | 4 | 8 | 0.18 | 0.065 |
| Qproteome | MAP7 domain-containing protein 1 | MAP7D1 | 42 | 102617 | 4 | 1.5 | 0.04 | 0.022 |
| Qproteome | Ladybird homeobox corepressor 1-like protein | CORL2 | 44 | 111363 | 4 | 0.8 | 0.03 | 0.019 |
| Qproteome | Eukaryotic translation initiation factor 3 subunit J | EIF3J | 29 | 34779 | 4 | 3.1 | 0.11 | 0.021 |
| Qproteome | Syntaxin-1B | STX1B | 41 | 37343 | 4 | 2.1 | 0.1 | 0.021 |
| Qproteome | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | OGT | 47 | 125886 | 4 | 0.6 | 0.03 | 0.021 |
| Qproteome | Envoplakin | EVPL | 38 | 252963 | 4 | 0.4 | 0.01 | 0.014 |
| Qproteome | Complement factor H-related protein 1 | CFHR1 | 115 | 41216 | 3 | 6.7 | 0.19 | 0.043 |
| Qproteome | DENN domain-containing protein 4C | DENND4C | 50 | 204270 | 3 | 0.7 | 0.02 | 0.023 |
| Qproteome | Disks large homolog 5 | DLG5 | 36 | 233891 | 3 | 0.7 | 0.02 | 0.026 |
| Qproteome | Putative myosin-XVB | MYO15B | 47 | 176166 | 3 | 0.5 | 0.02 | 0.020 |
| Qproteome | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-1 | PLCG1 | 42 | 160269 | 3 | 0.5 | 0.02 | 0.018 |
| Qproteome | Lebercilin-like protein | LCA5L | 49 | 88817 | 2 | 1.3 | 0.04 | 0.020 |
| Qproteome | FACT complex subunit SPT16 | SUPT16H | 28 | 135972 | 2 | 0.7 | 0.03 | 0.023 |
| Qproteome | Complement component C8 gamma chain | C8G | 120 | 23299 | 2 | 7.4 | 0.16 | 0.021 |
| Qproteome | Complement component C8 alpha chain | C8A | 75 | 71587 | 2 | 1.4 | 0.05 | 0.020 |
| Qproteome | Structural maintenance of chromosomes protein 1B | SMC1B | 32 | 165103 | 2 | 0.7 | 0.02 | 0.018 |
| Qproteome | Platelet-activating factor acetylhydrolase | PLA2G7 | 44 | 54825 | 2 | 2.3 | 0.07 | 0.021 |
| Qproteome | Tubulin polyglutamylase TTLL5 | TTLL5 | 41 | 155257 | 2 | 0.5 | 0.02 | 0.017 |
Proteins made Visible by Immunodepletion by IgY14+SuperMix
276 proteins were visible only in the Seppro IgY14+SuperMix sample (Supporting Information for Publication and partially in Table 4). Most of these proteins are of low abundance in the plasma.
Continuous Plasma Proteome is best Constructed from Two Immunodepletion Approaches
In general, proteins depleted by the Seppro IgY14+SuperMix column were analyzed with better peptide coverage in the Qproteome method. The opposite is true for proteins enriched by the IgY14+SuperMix column. Using EMMOL normalization to calculate the approximate concentrations of each protein in the plasma and the availability of two immunodepletion results, we combined the two proteomes to form a continuous protein concentration range of 5 logs. For each protein that can be quantified in the proteome of each immunodepletion protocol, the value with higher confidence level, because of higher peptide coverage (emPAI), was selected. A portion of this proteome is presented in Table 4 and in whole in Supporting Information for Publication.
Why is Assessment of the On-target and Off-target Removed Proteins Important to Blood Biomarker Studies?
Immunodepletion can be conveniently performed once on a pool of plasma of cases and then once on a pool of the plasma of the controls. That approach is expedient but variation in the efficiencies of the immunodepletion process from run to run between cases and controls can increase the potential for false discovery. The performance of immunodepletion may vary in different laboratories depending on the composition of the immunodepletion matrix used, the chosen ratio of plasma volume to immunodepletion column capacity, and the age of the column. Our comparison reported the overall immunodepletion efficiency of the combined resin for each protein in the proteome. Thus a useful database that indicates which proteins are at risk for false positives is provided. Based on our quantitation of the immunodepletion efficiency of each individual protein, we hypothesize that those proteins which have the highest percentage immunodepletion are at the highest risk of being identified as false-positives in differential proteomic studies. For example, 98% removal of a protein in cases versus 96% removal in controls may suggest one fold change incorrectly in the residual proteins when the two groups were equal in the original plasma. However, the design of our study does not allow us to identify individual at-risk proteins as false positives or false negatives. Some of the removed off-target proteins can be relatively low abundance proteins. Thus variation in their detection or quantification in comparator groups may cause them to be misinterpreted as disease biomarkers. On the other hand, we noticed that proteins that have been depleted by less than 50% in the Seppro IgY14+SuperMix immunodepletion method still report protein fold-changes comparable to the Qproteome method that did not deplete those proteins.
In summary, the current study illustrates that caution needs to be exercised in experiments involving immunodepletion. Nonetheless, after proper accounting for off-target protein loss, immunodepletion proteomics by two methods can provide accurate comparison of both high-abundance and low-abundance proteins. One method should account for the highly-abundant proteins and a separate method should more exhaustively deplete the less-abundant proteins.
Supplementary Material
Acknowledgments
We thank Brian Searle for critical reading of this manuscript and Dr. Karen Kawarta, Sigma-Aldrich, for performing the immunodepletion procedures.
This work was partially supported by the National Institutes of Health (NIH) grant RC2 HL101713, the National Cancer Institute, Work Assignment #16 of N01-CN-43309, the Driskill Foundation, Institutional Core Grant P30CA06927, Tobacco Settlement Funds from the Commonwealth of Pennsylvania, the Pew Charitable Trust, and the Kresge Foundation.
The abbreviations used are
- COPD
Chronic Obstructive Pulmonary Disease
- iTRAQ
Isobaric Tag for Relative and Absolute Quantitation
- IEF
Isoelectric Focusing
- IPG
Immobilized Polyacrylamide Gel
- EMMOL
emPAI-Molecular Weight method of proteome normalization
Footnotes
Conflicts of interests
Dr. Dian Er Chen who performed the immunodepletion steps was employed by Sigma-Aldrich that sells the IgY14+SuperMix column. Other authors declare no competing interest and no support by any company in connection with this work or future related work.
This article contains Supporting Information for Publication.
References
- 1.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3(4):311–26. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Burgess-Cassler A, Johansen JJ, Kendrick N. Immunodepletion of albumin from human serum samples. Clinica chimica acta; international journal of clinical chemistry. 1989;183(3):359–65. doi: 10.1016/0009-8981(89)90371-9. [DOI] [PubMed] [Google Scholar]
- 3.Beer LA, Tang HY, Sriswasdi S, Barnhart KT, Speicher DW. Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation. Journal of proteome research. 2011;10(3):1126–38. doi: 10.1021/pr1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maccarrone G, Milfay D, Birg I, Rosenhagen M, Holsboer F, Grimm R, Bailey J, Zolotarjova N, Turck CW. Mining the human cerebrospinal fluid proteome by immunodepletion and shotgun mass spectrometry. Electrophoresis. 2004;25(14):2402–12. doi: 10.1002/elps.200305909. [DOI] [PubMed] [Google Scholar]
- 5.Shen Z, Want EJ, Chen W, Keating W, Nussbaumer W, Moore R, Gentle TM, Siuzdak G. Sepsis plasma protein profiling with immunodepletion, three-dimensional liquid chromatography tandem mass spectrometry, and spectrum counting. Journal of proteome research. 2006;5(11):3154–60. doi: 10.1021/pr060327k. [DOI] [PubMed] [Google Scholar]
- 6.Koutroukides TA, Guest PC, Leweke FM, Bailey DM, Rahmoune H, Bahn S, Martins-de-Souza D. Characterization of the human serum depletome by label-free shotgun proteomics. Journal of separation science. 2011;34(13):1621–6. doi: 10.1002/jssc.201100060. [DOI] [PubMed] [Google Scholar]
- 7.Smith MP, Wood SL, Zougman A, Ho JT, Peng J, Jackson D, Cairns DA, Lewington AJ, Selby PJ, Banks RE. A systematic analysis of the effects of increasing degrees of serum immunodepletion in terms of depth of coverage and other key aspects in top-down and bottom-up proteomic analyses. Proteomics. 2011;11(11):2222–35. doi: 10.1002/pmic.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scumaci D, Gaspari M, Saccomanno M, Argiro G, Quaresima B, Faniello CM, Ricci P, Costanzo F, Cuda G. Assessment of an ad hoc procedure for isolation and characterization of human albuminome. Analytical biochemistry. 2011;418(1):161–3. doi: 10.1016/j.ab.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 9.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clinical applications. 2007;1(1):73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juhasz P, Lynch M, Sethuraman M, Campbell J, Hines W, Paniagua M, Song L, Kulkarni M, Adourian A, Guo Y, Li X, Martin S, Gordon N. Semi-targeted plasma proteomics discovery workflow utilizing two-stage protein depletion and off-line LC-MALDI MS/MS. Journal of proteome research. 2011;10(1):34–45. doi: 10.1021/pr100659e. [DOI] [PubMed] [Google Scholar]
- 11.Foreman MG, Zhang L, Murphy J, Hansel NN, Make B, Hokanson JE, Washko G, Regan EA, Crapo JD, Silverman EK, DeMeo DL. Early-onset chronic obstructive pulmonary disease is associated with female sex, maternal factors, and African American race in the COPDGene Study. American journal of respiratory and critical care medicine. 2011;184(4):414–20. doi: 10.1164/rccm.201011-1928OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han MK, Kazerooni EA, Lynch DA, Liu LX, Murray S, Curtis JL, Criner GJ, Kim V, Bowler RP, Hanania NA, Anzueto AR, Make BJ, Hokanson JE, Crapo JD, Silverman EK, Martinez FJ, Washko GR. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261(1):274–82. doi: 10.1148/radiol.11110173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DK, Jacobson FL, Washko GR, Casaburi R, Make BJ, Crapo JD, Silverman EK, Hersh CP. Clinical and radiographic correlates of hypoxemia and oxygen therapy in the COPDGene study. Respiratory medicine. 2011;105(8):1211–21. doi: 10.1016/j.rmed.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim V, Han MK, Vance GB, Make BJ, Newell JD, Hokanson JE, Hersh CP, Stinson D, Silverman EK, Criner GJ. The chronic bronchitic phenotype of COPD: an analysis of the COPDGene Study. Chest. 2011;140(3):626–33. doi: 10.1378/chest.10-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambod M, Porszasz J, Make BJ, Crapo JD, Casaburi R. Six-minute walk distance predictors, including CT scan measures, in the COPDGene cohort. Chest. 2012;141(4):867–75. doi: 10.1378/chest.11-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan ES, Hokanson JE, Murphy JR, Regan EA, Make BJ, Lynch DA, Crapo JD, Silverman EK. Clinical and radiographic predictors of GOLD-unclassified smokers in the COPDGene study. American journal of respiratory and critical care medicine. 2011;184(1):57–63. doi: 10.1164/rccm.201101-0021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washko GR, Lynch DA, Matsuoka S, Ross JC, Umeoka S, Diaz A, Sciurba FC, Hunninghake GM, San Jose Estepar R, Silverman EK, Rosas IO, Hatabu H. Identification of early interstitial lung disease in smokers from the COPDGene Study. Academic radiology. 2010;17(1):48–53. doi: 10.1016/j.acra.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geoui T, Urlaub H, Plessmann U, Porschewski P. Extraction of proteins from formalin-fixed, paraffin-embedded tissue using the Qproteome extraction technique and preparation of tryptic peptides for liquid chromatography/mass spectrometry analysis. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 10. Chapter 10. 2010. pp. 27pp. 1–12. [DOI] [PubMed] [Google Scholar]
- 20.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Molecular & cellular proteomics: MCP. 2008;7(10):1963–73. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim PD, Patel BB, Yeung AT. Isobaric Labeling and Data Normalization without Requiring Protein Quantitation. Journal of biomolecular techniques: JBT. 2012;23(1):11–23. doi: 10.7171/jbt.12-2301-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengqvist J, Uhlen K, Lehtio J. iTRAQ compatibility of peptide immobilized pH gradient isoelectric focusing. Proteomics. 2007;7(11):1746–52. doi: 10.1002/pmic.200600782. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson H, Lengqvist J, Hedlund J, Uhlen K, Orre LM, Bjellqvist B, Persson B, Lehtio J, Jakobsson PJ. Quantitative membrane proteomics applying narrow range peptide isoelectric focusing for studies of small cell lung cancer resistance mechanisms. Proteomics. 2008;8(15):3008–18. doi: 10.1002/pmic.200800174. [DOI] [PubMed] [Google Scholar]
- 24.Ke E, Patel BB, Liu T, Li XM, Haluszka O, Hoffman JP, Ehya H, Young NA, Watson JC, Weinberg DS, Nguyen MT, Cohen SJ, Meropol NJ, Litwin S, Tokar JL, Yeung AT. Proteomic analyses of pancreatic cyst fluids. Pancreas. 2009;38(2):e33–42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–72. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Human serum albumin: from bench to bedside. Molecular aspects of medicine. 2012;33(3):209–90. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Lin VK, Wang SY, Boetticher NC, Vazquez DV, Saboorian H, McConnell JD, Roehrborn CG. Alpha(2) macroglobulin, a PSA binding protein, is expressed in human prostate stroma. The Prostate. 2005;63(3):299–308. doi: 10.1002/pros.20183. [DOI] [PubMed] [Google Scholar]
- 28.Zhang WM, Finne P, Leinonen J, Salo J, Stenman UH. Determination of prostate-specific antigen complexed to alpha(2)-macroglobulin in serum increases the specificity of free to total PSA for prostate cancer. Urology. 2000;56(2):267–72. doi: 10.1016/s0090-4295(00)00609-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.