Abstract
We are reporting that the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G promotes the expression of hyper-inflammation in response to M. tuberculosis infection, inducing extensive tissue damage and severe TB disease. Carriers of this two-locus genotype have a 13-fold higher chance of developing severe disease and 6.5-fold higher chance of developing permanent lesions, and a 3.864-fold higher chance of delayed response to first-line standardized treatment than carriers of any other relevant combination of genotypes at those two loci. Thus, these persons have an increased likelihood of poor health-related quality of life and of transmitting M. tuberculosis to other members of the community. In addition, through the analysis of human lung tissues, serum/plasma, and in vitro experiments, including in vitro infections of THP-1 cells with M. tuberculosis and micro-array analysis, we determined that this hyper-inflammation state is potentially driven by the MCP-1/MMP-1/PAR-1 pathway. Hence, we are providing markers for the identification of TB cases that may develop severe pulmonary disease and delayed response to treatment, and are providing the basis for development of novel host-targeted clinical interventions to ameliorate the severity of pulmonary TB.
Keywords: Tuberculosis, genetics, disease severity, MCP-1, MMP-1
INTRODUCTION
Knowledge of genetic predictors of disease severity may be useful as markers for detection of TB patients at risk of developing a severe TB disease. Revealing the mechanisms of severe disease in these patients may provide molecular targets for new clinical interventions. It is well established that functional genetic polymorphisms in specific molecules of the immune/inflammatory response increase the likelihood of progression from M. tuberculosis infection to active pulmonary TB (1–7). Active TB may evolve into severe lung disease, or even to extra-pulmonary disease if left untreated (1–7). We propose that expression of severe pulmonary TB disease is genetically controlled.
Human host-encoded susceptibility to expression of severe pulmonary TB may involve: (i) the spread of infection beyond a critical number of cells because of a failing immune response against this challenge (4–7); and (ii) an excessive inflammatory response (hyper-inflammatory response), resulting in extensive inflammation-caused tissue damage (3). These mechanisms are not necessarily mutually exclusive. We previously reported that a functional single nucleotide polymorphism (SNP) located in position -2518 of the enhancing promoter region of the macrophage chemoattractant protein (MCP)-1 gene (the genotype GG) is associated with the expression of active TB disease in humans (1). Our ex vivo and in vitro studies indicated that increased MCP-1 production in response to infection is driving the expression of disease in carriers of that susceptibility genotype (1, 2). MCP-1 is a potent chemoattractant of macrophages that, in excess, down-regulates IL-12p40 production (1, 8–10), and up-regulates matrix metalloproteinase (MMP)-1 production by these cells in response to M. tuberculosis antigens, in vivo and in vitro (2). IL-12p40 is a subunit of two cytokines that are essential in the development of cellular immunity against TB, IL-12 and IL-23 (7). On the other hand, human MMP-1 is a potent collagenase, and collagen is an essential component of the granulomatous matrix over which cells (such as macrophages, T-cells, etc.) become organized to form granulomas and also a very important component of the lung matrix (2, 11). Thus, MMP-1 may be involved in liquefaction of mature granulomas. MMP-1 may also oppose to granuloma formation and maturation. MCP-1, MMP-1 and MMP-9 may contribute to non-granulomatous damage of lung tissue in patients affected by TB. Given that granulomas may serve to enclose infected cells and free bacteria in a confined environment, we and other investigators hypothesize that excessive MMP-1 levels may promote the spread of infection and inflammation (2, 12–17).
In search for genetic modifiers of the -2518 MCP-1 genotype GG, we reported that Hispanics with Amerindian ancestry carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G are at a significant increased chance of progression from M. tuberculosis infection to active TB disease (2). Of note, the -2518 MCP-1 allele G and the -1607 MMP-1 allele 2G creates a transcription activator-like effector (TALE) binding site for PREP1/PBX2 transcription factor complexes (18, 19), and increases the rate of transcription of the MCP-1 gene, which is active in TB (1, 2, 20). Likewise, the -1607 MMP-1 allele 2G, which consists of the insertion of a guanine at position -1607 (the allele 2G), creates an Ets-1 transcription factor binding site (21, 22), and enhances expression of the gene in normal and tumor cells, and in active TB (21, 22).
Whether the two-locus susceptibility genotype -2518 MCP-1 GG and -1607 MMP-1 2G/2G may also influence the expression of a more severe disease is unknown. Thus, in this study we address this question. We also conducted ex vivo and in vitro experiments to unveil potential mechanisms of disease severity in carriers of this two-locus susceptibility genotype. Data from this study could guide the development of new therapeutic approaches to overcome host-encoded deleterious hyper-inflammatory response to M. tuberculosis infection and expression of severe pulmonary TB. Here, we are also demonstrating that excessive MMP-1 may potentiate the MCP-1 and M. tuberculosis-induced inflammatory response through activation of protease activated receptor (PAR)-1, and thereby increases the likelihood of developing a severe TB disease.
RESULTS
Demographic and clinical features of TB cases studied
We recruited de novo 224 patients with TB that met the enrollment criteria and were tightly followed through the directly observed therapy program (DOT), ens uring adherence to therapy. We did not see differences in the proportion of males and females, nor in the mean age between groups (Table 1). None of the patients self-reported consumption of alcohol or tobacco. All of them were of low socioeconomic status and Peruvian Mestizo to the third generation (an admixture of mainly Amerindian and Spaniards). All of the patients were recruited in similar geographical areas, where they had lived for more than 2 years, making it more likely that they were infected by the same bacterial strains circulating in the region. They were all BCG-vaccinated, and had a disease confined to the lungs of no more than 3 months of evolution. We did not see differences in duration of disease (the length of time that passed since the first symptom was noticed by the patient to the time of sampling) (Table 1). To test for differences in the admixture of TB cases stratified according to genotypes, we genotyped 42 genomic controls and compared the allele frequencies across strata using a chi2 test. We used these chi2 values to calculate a λ value of admixture as we did before (1, 2). The λ value of admixture was 0.95 (Table S1). As this was close to 1, we concluded that there was not a significant difference in the level of admixture between TB cases in each stratum.
Table 1.
TB cases – characteristics
| two-locus genotypes | Group 1: A/- 1G/- | Group 2: A/- 2G/2G | Group 3: GG 1G/- | Group 4: GG 2G/2G | Statistics |
|---|---|---|---|---|---|
| TB cases per genotype n (%) (total n = 224) | 51 (23%) | 59 (26%) | 43 (19%) | 71 (32%) | n.s.* |
| Mean age ± SD | 33.9 ± 10.8 | 31.63 ± 10.9 | 28.95 ± 9.3 | 29.7 ± 10.06 | n.s.* |
| Women n (%) | 23 (45%) | 27 (46% | 20 (47%) | 31 (44%) | n.s.* |
| Duration of disease (days) | 63 ± 20 | 64 ± 20 | 65 ± 22 | 64 ± 24 | n.s.* |
| BMI ≤ the median n (%) Median = 21.21 | 19 (37.3) | 26 (44.06) | 22 (51.2) | 45 (63.4) | p = 0.026** |
| Severity score > median n (%) Median = 44 | 14 (27.5) | 20 (33.9) | 8 (18.6) | 59 (83) | p < 0.001** |
Group 1 = genotypes AA 1G/1G, AA 1G/2G, AG 1G/1G, AG 1G/2G
Group 2 = genotypes AA 2G/2G, AG 2G/2G
Group 3 = genotypes GG 1G/1G, GG 1G/2G
Group 4 = genotypes GG 2G/2G
SD = standard deviation of the mean
n = number of cases
BMI = Body mass index
n.s. = not significant
One-way ANOVA was used to test for significant differences
Chi-squared test was used to test for significant differences
The two-locus genotype -2518 MCP-1 GG and -1607 MMP-1 2G/2G is associated with expression of severe TB disease
To assess the severity of disease, we modified previously published severity scores developed to capture clinical information available in low-resource countries (23–26; Table 2). An expert physician classified the patients into two categories: severe or non-severe disease, according to the clinical characteristics exposed in Table 2. Following this analysis, we proceeded to assign scores and classified the cases according to whether their scores were above the median (severe disease) or were ≤ the median values obtained (non-severe disease). The median score was 44. We obtained a significant agreement between the clinical diagnosis of severe disease and the classification according to the severity score (κ coefficient = 0.83). Disagreements were observed in 20 cases with scores ≤ 44. Of note, the partial scores obtained from the assessment of clinical signs and symptoms and that obtained from the assessment of posteroanterior full-size chest X-rays were consistent, since a significant direct (positive) correlation was observed between them (Spearman’s Rho = 0.83; p = 0.00001).
Table 2.
Severity score
| I. Clinical signs and symptoms | Score if present |
|---|---|
|
| |
| Cough | 10 |
|
| |
| Dyspnoea | 10 |
|
| |
| Night sweats | 10 |
|
| |
| Haemoptysis | |
| At least one episode | 10 |
| More than one episode | 20 |
|
| |
| Chest pain | 10 |
|
| |
| Decreased body mass index (BMI) in adults | |
| BMI < 25 to 18.5 | 2 |
| BMI < 18.5 to 17 | 4 |
| BMI < 17 to 16 | 6 |
| BMI < 16 | 10 |
|
| |
| Low BMI for age percentile in children | |
| BMI < 50th but > 25th percentile for age | 2 |
| BMI in the 25th to no more than 10th percentile | 4 |
| BMI in the 10th to more than 5th percentile | 6 |
| BMI below the 5th percentile | 10 |
|
| |
| Anemia | 10 |
|
| |
| Fever | 10 |
|
| |
| Signs at lung auscultation | 10 |
|
| |
| Extra-thoracic involvement | 20 |
|
| |
| II. Chest-X-ray assessment | Score if present |
|
| |
| Extension of the lesion (each lung is assessed independently) | |
| No more than one third of the lung field affected | 10 |
| One third to no more than two third of the lung field affected | 20 |
| Two third or more of the lung field affected | 30 |
|
| |
| Type of lesion (each lung is assessed independently) | |
| Infiltration patterns | 10 |
| Pleural effusion | 10 |
| Cavitary lesion | 20 |
| Caseating pneumonia | 20 |
| Miliary tuberculosis | 20 |
| Lung fibrosis | 20 |
| Intra-thoracic lymphadenopathy | 10 |
There was a greater proportion of cases with severity scores higher than the median among those TB cases carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G, than in TB cases carrying any other relevant combination of genotypes at those two loci (Table 1). There was a significant difference in BMI values across stratum (Table 1). The proportion of TB cases with BMI < median was higher among carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G (Table 1).
We run a logistic regression analysis to assess whether the main or joint effects of the -2518 MCP-1 genotype GG and the -1607 MMP-1 genotype 2G/2G would predict the severity of disease, adjusting for age and gender (Table 3). We used the severity score as a surrogate marker of disease severity, as explained in materials and methods. We observed that the two-locus genotype -2518 MCP-1 genotype GG -1607 MMP-1 genotype 2G/2G was significantly associated with expression of a severe TB disease, with strong point estimates of association (Table 3). None of the other two-locus genotypes, or age and gender contributed significantly to the expression of severe TB disease in this sample (Table 3). The observed model did not differ from that predicted (Pearson goodness-of-fit test, Table 3). Using a second analytical approach, we corroborated our findings and observed significant differences in the mean of severity scores between groups (Table 4). The carriers of the two-locus -2518 MCP-1 GG -1607 MMP-1 2G/2G had the highest score (Table 4). Six TB cases carrying the two-locus -2518 MCP-1 GG -1607 MMP-1 2G/2G underwent surgery to remove damaged lung tissue versus none in the groups of carriers of any other combination of genotypes at these two loci.
Table 3.
Contribution of age, gender and -2518 MCP-1 -1607 MMP-1 two-locus genotypes in TB disease severity
| Number of observations = 224 LR chi-square (5) = 68.62 Probability > chi2 = 0.00001 Log likelihood = −119.87 Pseudo R2 = 0.223 | |||||
|---|---|---|---|---|---|
| Dependent Variables | OR | S.E. | Z | Prob > |Z| | 95% C.I. |
| Age | 0.998 | 0.0154 | − 0.10 | 0.921 | 0.969 – 1.029 |
| Gender (female) | 0.667 | 0.215 | − 1.26 | 0.209 | 0.355 – 1.254 |
| Group 2: A/- 2G/2G | 1.358 | 0.571 | 0.73 | 0.467 | 0.595 – 3.095 |
| Group 3: GG 1G/- | 0.6 | 0.306 | − 1.00 | 0.317 | 0.222 – 1.63 |
| Group 4: GG 2G/2G | 13.13 | 5.941 | 5.69 | 0.0001 | 5.41 – 31.88 |
| Pearson Goodness-of-fit: | Chi2 = 151.44; p = 0.13 | ||||
Genotype A/- 1G/- = genotypes AA 1G/1G or AA 1G/2G, AG 1G/1G or AG 1G/2G
Genotype A/- 2G/2G = genotypes AA 2G/2G, AG 2G/2G
Genotype GG 1G/- = genotypes GG 1G/1G, GG 1G/2G
Group 1 was used as reference
OR = Odds Ratio; SE = standard error; Z = Z score; 95% C.I. = 95% confidence interval
Table 4.
Mean score value associations with the -2518 MCP-1 and -1607 MMP-1 two-locus genotypes
| Two-locus genotypes | Cases (n) | Score mean ± SD | Rank sum | Bonferroni LSD |
|---|---|---|---|---|
| Group 1: A/- 1G/- | 51 | 39.45 ± 11.97 | 4047 | G1 versus G2 p = n.s. G1 versus G3 p = n.s. G1 versus G4 p < 0.0001 |
| Group 2: A/- 2G/2G | 59 | 43.96 ± 12.1 | 6004.5 | G2 versus G3 p = n.s. G2 versus G4 p < 0.0001 |
| Group 3: GG 1G/- | 43 | 39.81 ± 15.21 | 3439 | G3 versus G4 p < 0.0001 |
| Group 4: GG 2G/2G | 71 | 61.58 ± 17.3 | 11709.5 | |
| Kruskal-Wallis test | Chi-squared with ties = 72.689, d.f. = 3, p = 0.001 | |||
Genotype A/- 1G/- = genotypes AA 1G/1G or AA 1G/2G, AG 1G/1G or AG 1G/2G
Genotype A/- 2G/2G = genotypes AA 2G/2G, AG 2G/2G
Genotype GG 1G/- = genotypes GG 1G/1G, GG 1G/2G
n = number of cases; n.s. = not significant
SD = standard deviation of the mean
LSD = least significant difference
G1 = group 1; G2 = group 2; G3 = group 3, G4 = group 4
d.f. = degrees of freedom
Given all of this evidence, we concluded that adult TB cases carrying the two-locus genotype -2518 MCP-1 GG and -1607 MMP-1 2G/2G express a more severe TB disease than carriers of any other relevant combination of genotypes at these two loci.
TB patients carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G expressed a delayed response to standardized treatment and extensive fibrosis surrounding cavities and bronchiectasis at the end of the treatment
We asked whether disease severity, which reflects excessive tissue damage, will influence the response to the standardized TB treatment. Most of our TB cases observed a sputum smear test conversion from positive to negative at the second month of treatment. In contrast, we observed that 29.6% of TB patients carrying the two-locus genotype -2518 MCP-1 GG and -1607 MMP-1 2G/2G experienced a significant delayed response to treatment (Table 5). The test for homogeneity of the odds and the score Test for Trend in Odds indicate the presence of a dose-response relationship, were carrying the genotypes -2518 MCP-1 GG and the -1607 MMP-1 2G/2G conferred the highest odds of a sputum test conversion from positive to negative at more than 3 months (mean ± SD = 3.7 ± 0.6), a delayed response to treatment according to the WHO (Table 5). At the end of the treatment, a greater proportion of TB cases, all carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G, had more than 2/3 of lung parenchyma compromised by fibrotic lesions than carriers of any other genotypes, which was reflected by the presence of Dyspnoea (difficulty breathing). Thirty three out of 71 TB cases carrying the susceptibility two-locus genotype (83%) versus 18 out of 153 TB cases (37.8%) in carriers of any other combination of genotypes expressed those permanent lesions (OR = 6.5, 95% CI = 3.14–13.6; chi-squared= 33.23, p < 0.00001). Lung fibrosis was mainly observed surrounding cavities and bronchiectasis.
Table 5.
Associations of the -2518 MCP-1 and -1607 MMP-1 two-locus genotypes and sputum conversion at more than 3 months
| Two-locus genotypes | Cases n (%) | OR | Prob > chi2 | 95% C.I. |
|---|---|---|---|---|
| Group 1: A/- 1G/- | 5 (9.8) | 1 | - | - |
| Group 2: A/- 2G/2G | 9 (15.3%) | 1.4 | 0.57 | 0.44 – 4.5 |
| Group 3: GG 1G/- | 6 (13.9%) | 1.492 | 0.54 | 0.41 – 5.33 |
| Group 4: GG 2G/2G | 21 (29.6%) | 3.864 | 0.009 | 1.3 – 11.5 |
| Test of homogeneity (equal odds): Score test for trend of odds: |
Chi2 = 10.41, d.f. = 3; prob > chi2 = 0.0154 Chi2 = 8.61, d.f. = 1; prob > chi2 = 0.0034 |
|||
Genotype A/- 1G/- = genotypes AA 1G/1G or AA 1G/2G, AG 1G/1G or AG 1G/2G
Genotype A/- 2G/2G = genotypes AA 2G/2G, AG 2G/2G
Genotype GG 1G/- = genotypes GG 1G/1G, GG 1G/2G
n = number of cases showing sputum conversion > 3 months
OR = Odds Ratio; 95% C.I. = 95% confidence interval, d.f. = degrees of freedom
Carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G express the highest levels of MMP-9
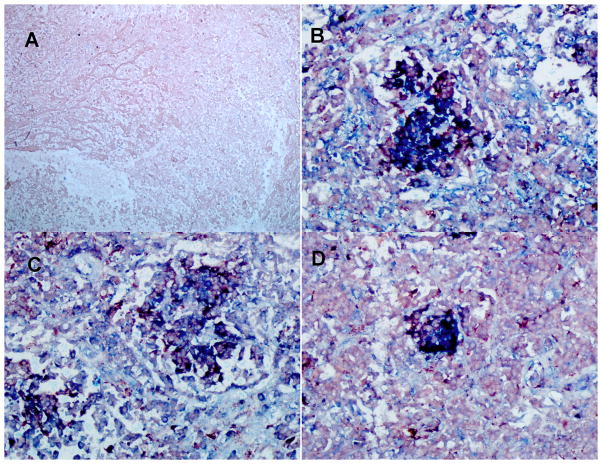
Using immunohistochemistry (IHC), we assessed the levels of MCP-1, MMP-1, and MMP-9 expression by alveolar macrophages in lung tissue from those six TB cases that required surgery to remove damaged tissue. MMP-9 was tested because, in the Zebra fish model of tuberculosis, MMP-9 was shown to drive the recruitment of monocytes to the granulomatous inflammatory foci, where these cells were exposed and infected with M. marinum, resulting in a significant increase in the burden of infected cells (27). Because of the extensive lung damage observed, we focused our analysis on granulomas located in areas of lung parenchyma surrounding extensive necrotic areas. We observed several macrophages, and other cells (i.e. lung epithelial cells) expressing copious amounts of MCP-1, MMP-1, and MMP-9 in these tissues (Fig. 1). The disease tissue slices showed multiple dark purple cells, which could be interpreted as macrophages (red for CD68) expressing MCP-1, MMP-1, MMP-9 (blue for each factor). We could not assess whether those levels observed in lung tissues were higher in carriers of the -2518 MCP-1 GG -1607 MMP-1 2G/2G than in carriers of any other combination of genotypes at these two loci because of the lack of tissues from the latter groups. Hence, to further determine whether genotypes will correlate with phenotypes in these TB cases we measured the serum levels of MCP-1 and plasma levels of MMP-1 and MMP-9 (Fig. 2).
Figure 1. TB patients carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G express MCP-1, MMP-1, and MMP9.
We used double IHC analysis of MCP-1 or MMP-1 or MMP-9 and CD68 in paraffin-embedded diseased lung from six TB patients who underwent surgery to remove damaged tissue. We are showing representative data. A: negative control (irrelevant antibodies isotype control); B: MCP-1 (blue) and CD68 (red); C: MMP-1 (blue) and CD68 (light red); D: MMP-9 (blue) and CD68 (light red). Images were acquired at 200X total magnification. IHC analysis shows granulomas with CD68-positive cells (macrophages) expressing copious amounts of MCP-1, MMP-1, and MMP-9.
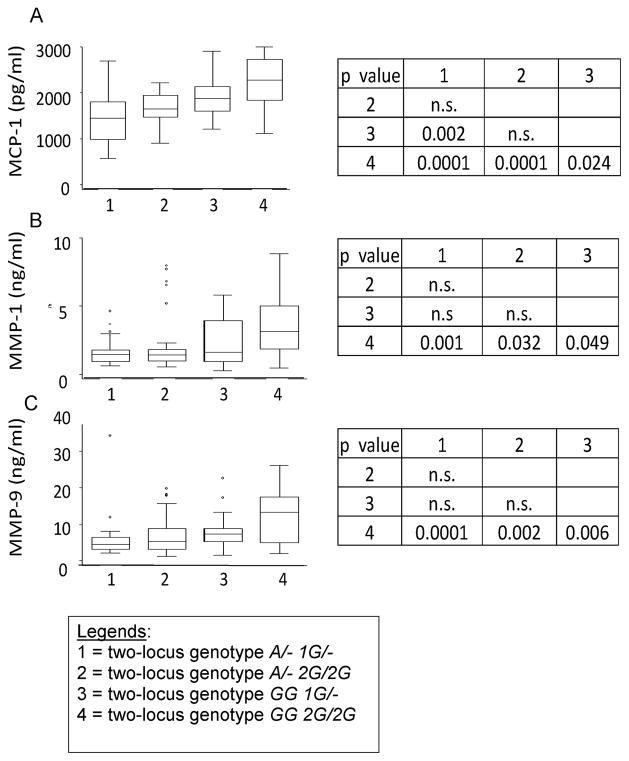
Figure 2. TB patients carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G have the highest serum levels of MCP-1 and the highest plasma levels of MMP-1 and MMP9.
Values are shown as medians (horizontal lines), the 25th and 75th percentiles (boxes), and ranges (whiskers). Legends in the x-axis mean: 1 = two-locus genotype A/- 1G/-; 2 = two-locus genotype A/- 2G/2G; 3 = two-locus genotype GG 1G/-; 4 = two-locus genotype GG 2G/2G). Section A: Distribution of serum MCP-1 values for tuberculosis patients stratified according to the relevant two-locus genotypes. We observed a significant difference in the serum levels of MCP-1 across genotypes (ANOVA F = 16.31; p = 0.0001). Section B: Distribution of plasma MMP-1 values for tuberculosis patients stratified according to the relevant two-locus genotypes. We observed a significant difference in the plasma levels of MMP-1 across genotypes (ANOVA F = 5.76; p = 0.001). Section C: Distribution of plasma MMP-9 values for tuberculosis patients stratified according to the relevant two-locus genotypes. We observed a significant difference in the plasma levels of MMP-9 across genotypes (ANOVA F = 8.25; p = 0.0001). Comparison of means and standard deviations by genotypes are shown at the right of the whiskers and box figures. The p-values are based on the Bonferroni least significant difference test.
Thirty randomly selected cases per each of the four relevant -2518 MCP-1 -1607 MMP-1 two-locus genotypes, and 20 healthy controls (only as reference) were assessed. MMP-9 was significantly over expressed in TB patients carrying the two-locus genotype -2518 MCP-1 GG and -1607 MMP-1 2G/2G, as compared to the levels of MMP-9 in carriers of any other combination of genotypes at these two loci (Fig. 2). A less significant increase in MCP-1 and MMP-1 was also observed in carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G, when compared to the levels of these factors in carriers of other combinations of genotypes at these two loci (Fig. 2). Controls had only normal levels of MMP-9 (24.760 ± 13.050 ng/ml) and MMP-1 (6.12 ± 2.76 ng/ml) in plasma, and MCP-1 (563 ± 328 pg/ml) in serum. Furthermore, we obtained significant positive correlations between MCP-1 levels (Spearman’s Rho = 0.91; p < 0.00001) or MMP-1 levels (Spearman’s Rho = 0.93; p < 0.00001) or MMP-9 levels (Spearman’s Rho = 0.9; p < 0.00001) and the scores in TB cases carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. All together, this indicates that carriers of the susceptibility genotype have the higher severity scores and express the higher levels of MCP-1, MMP-1 and MMP-9.
In vitro studies revealed underlying mechanisms of increased MMP-9
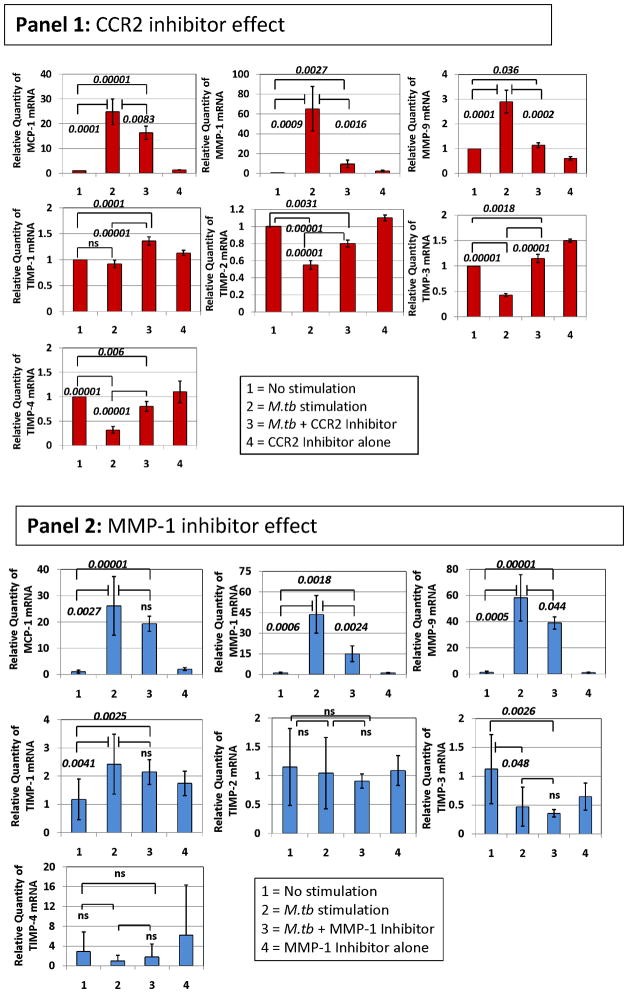
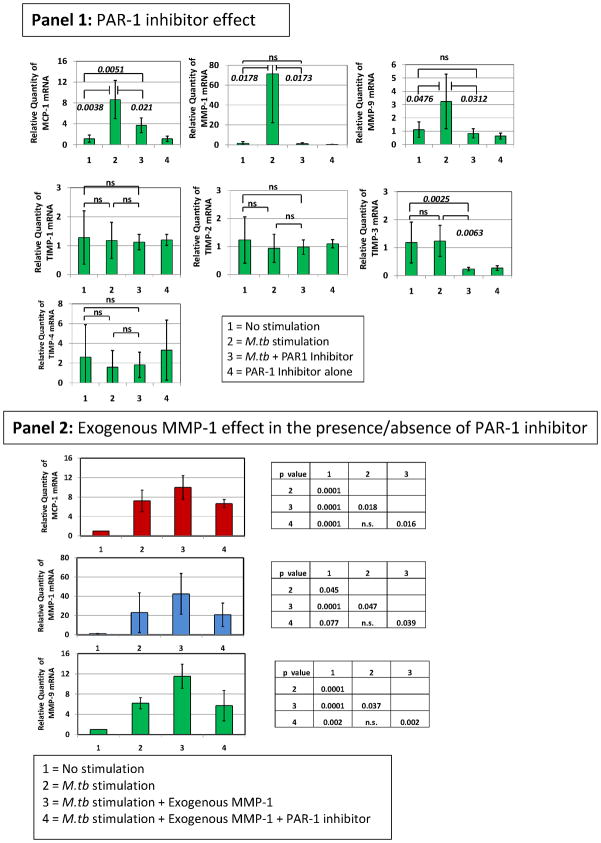
Given that we observed the highest plasma levels of MMP-9 in TB patients carrying the two-locus genotype – 2518 MCP-1 GG -1607 MMP-1 2G/2G, we reasoned that this might result from more complex preceding molecular events, where MCP-1 and MMP-1 were involved. We used in vitro exposure of human THP-1 monocytic cells to sonicated H37Rv M. tuberculosis to model the exposure of massively recruited monocytes to the lung inflammatory foci during TB-induced inflammatory response (28–31). We observed that exposure of THP-1 cells to sonicated H37Rv M. tuberculosis for 24 hrs caused a significant increase in the expression of MCP-1, MMP-1, and MMP-9 (Fig. 3). In contrast, this stimulus inhibited the expression of tissue inhibitor of metalloproteinase (TIMP)-1, -2, -3, and 4 (Fig. 3). To test the notion that MCP-1 increases MMP-9 production, we activated THP-1 cells with sonicated H37Rv M. tuberculosis and inhibited CCR2 with the compound RS504393 (Fig. 3, Panel 1). MCP-1 effects are likely inhibited by this compound since CCR2 is the only receptor for MCP-1 (32). MMP-9 mRNA expression in response to the sonicated H37Rv M. tuberculosis stimuli was significantly decreased by this CCR2 inhibiting compound (Fig. 3, Panel 1). MCP-1 and MMP-1 mRNA were also significantly down-regulated by this CCR2 inhibitor (Fig. 3, Panel 1). Although the expression of natural inhibitors of MMPs (TIMP-1, -2, -3, and -4) was poor, as compared to that of MMP-1 and MMP-9, RS504393 appeared to slightly counteract the inhibitory effect of the sonicated H37Rv M. tuberculosis exposure on TIMPs expression (Fig. 3, Panel 1). We next investigated whether inhibition of MMP-1 would provide similar results. Using the 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide (at 1 μM concentration) to inhibit MMP-1 functions we could also down-regulate MMP-9 (Fig. 3, Panel 2). Although MCP-1 was not significantly down-regulated, MMP-1 expression was significantly inhibited by addition of the 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide to cultures of THP-1 cells activated with sonicated H37Rv M. tuberculosis (Fig. 3, Panel 2). TIMP expression was not significantly modulated by this MMP-1 inhibitor (Fig. 3, Panel 2).
Figure 3. CCR2 RS504393 and MMP-1 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide inhibitors modulate the expression of MCP-1, MMP-1, and MMP-9 and, to a lesser extent the expression of TIMPs inTHP-1 monocytic cells stimulated by sonicated H37Rv M. tuberculosis.
We measured the relative changes in MCP-1, MMP-1, MMP-9, and TIMP gene expression by real-time PCR. Data are presented as the fold change in gene expression normalized to the endogenous reference gene PDHB and relative to untreated controls. Panel 1, the effect of 10 μM concentration of CCR2 RS504393 inhibiting compound was assessed following 24 hr in vitro stimulation of THP-1 cells with 5 μg/ml sonicated H37Rv M. tuberculosis. Cultures proceeded in 500 μl serum-free RPMI. CCR2 RS504393 inhibiting compound was diluted with DMSO and dispensed in 5 μl volume to produce a final culture concentration of 0.01% DMSO. We also added 5 μl of DMSO to control cultures. The results presented are from six independent experiments showing the mean and standard deviations. We consistently observed significant differences in the mean values across variables (Kruskal-Wallis p < 0.01) when testing MCP-1, MMP-1 and MMP-9. We show corrected p-values obtained from student t-tests. Notably, levels of specific mRNAs from non-stimulated cells were not significantly different than those obtained from cultures that proceeded with the CCR2 inhibitor alone. Panel 2, the effect of 1 μM concentration of MMP-1 inhibitor 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide was assessed following 24 hr in vitro stimulation of THP-1 cells with 5 μg/ml sonicated H37Rv M. tuberculosis. Cultures proceeded in 500 μl serum-free RPMI. 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide was diluted in incomplete RPMI. The results presented are from six independent experiments showing the mean and standard deviations from the mean. We consistently observed significant differences in the mean values across variables (Kruskal-Wallis p < 0.01) when testing MCP-1, MMP-1 and MMP-9. We show corrected p-values obtained from student t-tests. Of note, levels of specific mRNAs from non-stimulated cells were not significantly different from those obtained from cultures that proceeded with the 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide inhibitor alone. The inhibitors’ concentrations were selected from dose response-experiments.
MMP-1 may activate PAR-1 expression on monocytes/macrophages in tuberculosis to induce MCP-1, MMP-1, and MMP-9 expression
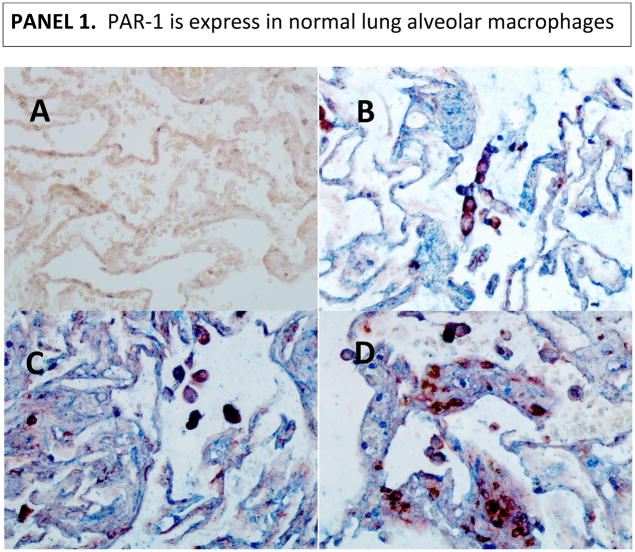
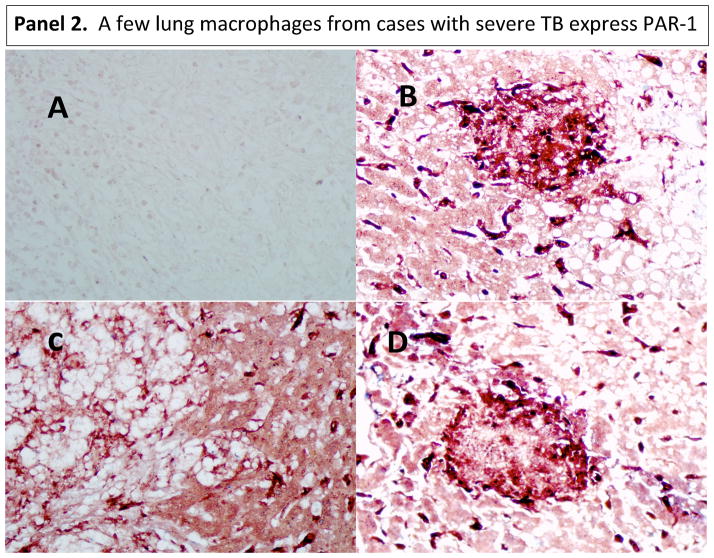
Given the fact that MMP-1 can activate protease-activated receptor (PAR)-1 (33), we asked whether this pathway might operate in tuberculosis. Alveolar macrophages are the first line of defense in the lungs and the initiators of peripheral monocyte recruitment into the lungs’ inflammatory foci (28–31). Alveolar macrophages express CD14 and CD68 markers (34–36). IHC analysis of PAR-1 expression revealed that human alveolar macrophages in normal lungs constitutively express PAR-1. Of note, there were some macrophages in the interstitium of normal tissues (the tissue and space around the air sacs of the lung); we reasoned that this might be a displacement artifact produced while processing the samples (Fig. 4, panel 1). Few macrophages in lung tissues from individuals with severe TB disease do express PAR-1 (Fig. 4, panel 2). Indeed, some dark purple cells could be observed which could be interpreted as macrophages (red for CD68) expressing PAR-1 (blue). Images taken at lower magnification of tissues presented in figure 1, panel 2, corroborates this statement (Fig. S1).
Figure 4. PAR-1 is expressed in alveolar macrophages from normal lungs, while only few macrophages expressed PAR-1 in diseased lung tissues from severe TB cases.
We used double IHC analysis of PAR-1 and CD68 in paraffin-embedded normal (Panel 1) and diseased lung (Panel 2). Sections B, C, and D show three different normal persons (Panel 1). One representative image from three of the six TB cases that underwent surgery to remove damaged tissue is shown in Panel 2. In each panel, Section A shows the negative control (irrelevant antibodies isotype control). Sections B, C, and D show PAR-1 (blue) and CD68 (dark red) macrophages. Images were acquired at 200X total magnification.
PAR-1 was also expressed by epithelial cells in normal and disease lungs (Fig. 4). Low PAR-1 expression in diseased tissues might result from the enzymatic activity of several proteases in the lung milieu of these severe cases that may digest several proteins, including PAR-1. In conjunction with this, PAR-1 may have been down-regulated in response to activation by thrombin or/and MMP-1. Of note, the same anti-PAR-1 antibody was used to stain normal lung tissue, and both experiments were done simultaneously. Thus, lack of specificity of the antibody does not explain our results.
MMP-1 may activate PAR-1 in THP-1 cell exposed to M. tuberculosis sonicate
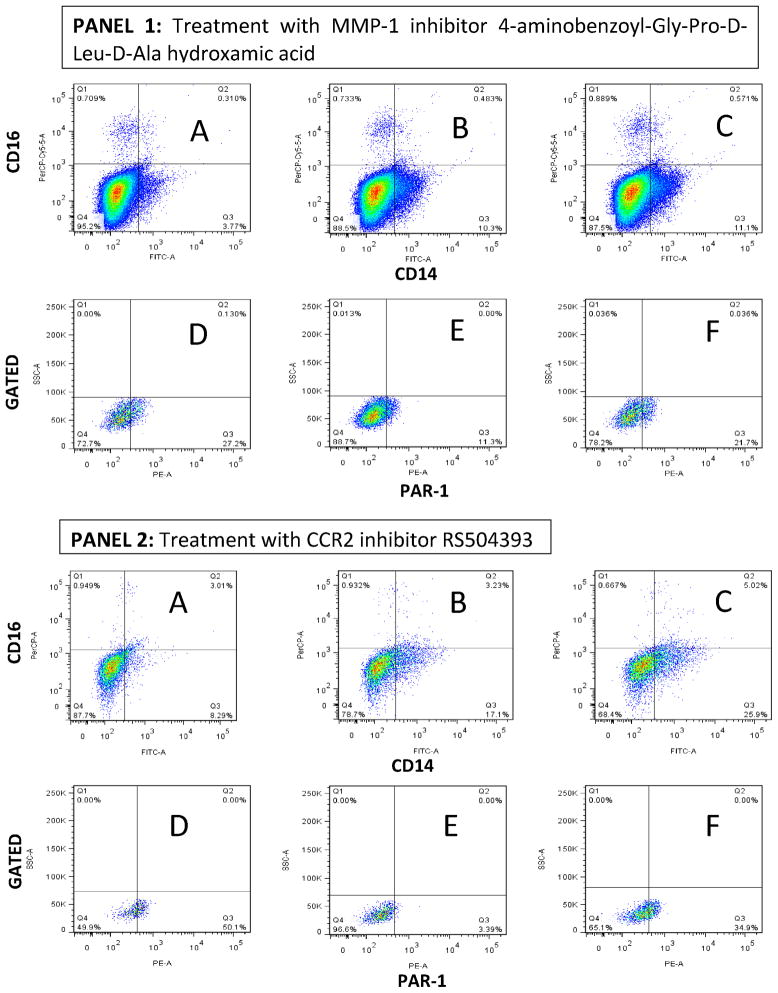
To test the whether MMP-1 activates PAR-1 in THP-1 cells exposed to sonicated M. tuberculosis H37Rv, we took advantage of the fact that PAR-1 is a G-coupled protein receptor and that as such, PAR-1 undergoes recycling upon activation by thrombin or MMP-1 (33). We focused our analysis of PAR-1 expression in THP-1 cells expressing a phenotype close to that of macrophages. We observed by FACS analysis, that a low proportion of quiescent THP-1 cells expressed CD14, and that a very low proportion of these cells expressed CD16 (Fig. 5, Panels 1 and 2, Section A). CD14-positive/CD16-negative THP-1 cells also expressed CD68 (data not shown). Exposure of THP-1 cells to sonicated H37Rv M. tuberculosis mainly induced their differentiation into CD14-positive/CD16-negative cells (Fig. 5, Panels 1 and 2, Section B) (34–36). Interestingly, none of the compounds or peptides used perturbed the differentiation of THP-1 cells into CD14-positive/CD16-negative cells (Fig. 5, Panels 1 and 2, Section C). We next gated into CD14-positive/CD16-negative THP-1 cells to determine whether PAR-1 expression is regulated by exposure of these cells to sonicated H37Rv M. tuberculosis ± MMP-1 inhibitor 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide (Fig. 5, Panel 1, Sections D, E, and F). We observed that 27% of the gated CD14-positive/CD16-negative quiescent THP-1 cells expressed PAR-1 (Fig. 5, Panel 1, Section D). Exposure of THP-1 cells to sonicated H37Rv M. tuberculosis significantly down-regulated the expression of PAR-1 to 11.3% (D versus E: OR = 2.96, 95% CI = 2.6–3.3, p < 0.00001; Fig. 5, Panel 1). The addition of MMP-1 inhibitor to M. tuberculosis stimulated THP-1 cells preserved the expression of PAR-1 in approximately 22% of cells (F versus E: OR = 2.2, 95% CI = 1.95–2.4, p < 0.00001; Fig. 5, Panel 1).
Figure 5. Down-regulation of PAR-1 expression by exposure of THP-1 cells to sonicated H37Rv M. tuberculosis and counter effect of MMP-1 and CCR-2 inhibitor.
We used three-color FACS analysis for these experiments. Exposure of quiescent THP-1 cells to sonicated H37Rv M. tuberculosis for 24 hr induced the differentiation of these cells into CD14-positive/CD16-negative cells. In Panels 1 and 2, Section A shows a low proportion of CD14-positive/CD16-negative cells in quiescent THP-1 cells; Section B shows an increment in the proportion of CD14-positive/CD16-negative THP-1 cells in response to sonicated H37RV M. tuberculosis exposure.; Section C shows minimal (non-significant) variation in this response to sonicated H37Rv M. tuberculosis exposure in the presence of the MMP-1 inhibitor (Panel 1) or CCR2 inhibitor (Panel 2). In Sections D, E, and F of Panel 1, we show that the presence of MMP-1 inhibitor 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide prevents the down-regulation of PAR-1 expression by THP-1 cells exposed to sonicated H37Rv M. tuberculosis exposure. In Sections D, E, and F of Panel 2, we show that the presence of CCR2 inhibitor RS504393 also prevents the down-regulation of PAR-1 expression by THP-1 cells exposed to sonicated H37Rv M. tuberculosis exposure. We acquired 100,000 events for experiments in Panel 1, while we acquired 10,000 events for experiments in Panel 2, according to the number of live cells gathered in each experiment. Of note, the MMP-1 inhibitor was dissolved in incomplete RPMI while the CCR2 inhibitor was dissolved in DMSO to obtain a DMSO final culture concentration of 0.01%. Three experiments were done for the assessment of each inhibitor.
We then asked whether the CCR2 inhibitor RS504393 would provide similar results since MCP-1 activates CCR2 and through this it potentiates the expression of MMP-1 by M. tuberculosis-stimulated THP-1 cells (Fig. 3, Panel 1). Approximately 50% of the gated CD14-positive/CD16-negative quiescent THP-1 cells expressed PAR-1 (Fig. 5, Panel 2, Section D). Once again, exposure of THP-1 cells to sonicated H37Rv M. tuberculosis significantly down-regulated (to 3.39%) the expression of PAR-1 (D versus E: OR = 28.6, 95% CI = 20.2–41.1, p < 0.00001; Fig. 5). The addition of CCR2 inhibitor to M. tuberculosis-stimulated THP-1 cells preserved the expression of PAR-1 in approximately 35% of the cells (F versus E: OR = 15.24, 95% CI = 11.1–21.5; p < 0.00001; Fig. 5, Panel 2). Notably, THP-1 cells cultured under any of the aforementioned conditions did not express thrombin (data not shown). Hence, we considered that this results added evidence that MMP-1 could be activating PAR-1 in this in vitro system and inducing recycling of this receptor. Since we partially recovered PAR-1 cell membrane expression by treating M. tuberculosis-antigen-activated THP-1 cells with a MMP-1 or a CCR2 inhibitor, we do not exclude the possibility that other macrophage proteases might be acting on PAR-1 as well.
Treatment of H37Rv M. tuberculosis-infected THP-1 cells with PAR-1 inhibitor SCH79797 down-regulates MMP-1 induced MCP-1, MMP-1 and MMP-9
We first used the SCH79797 PAR-1 inhibitor to test whether PAR-1 inhibition regulates the expression of MCP-1, MMP-1, MMP-9, and the TIMPs by M. tuberculosis-stimulated THP-1 cells. We observed that PAR-1 inhibitor SCH79797, at nanomolar concentrations (50 nM), potently down-regulated MCP-1, MMP-1, and MMP-9 mRNA expression by THP-1 cells stimulated with sonicated H37Rv M. tuberculosis (Fig. 6, Panel 1). This inhibitor did not significantly regulate the expression of the TIMPs (Fig. 6, Panel 1). To confirm that MMP-1 activates PAR-1, we added exogenous human purified MMP-1 (hMMP-1) to M. tuberculosis-stimulated THP-1 cells and observed that exogenous MMP-1 increased MCP-1, MMP-1, and MMP-9 expression to levels that were higher than those observed in cultures of THP-1 cells stimulated with sonicated H37Rv M. tuberculosis alone (Fig. 6, Panel 2). Addition of PAR-1 inhibitor to THP-1 cells stimulated with sonicated H37Rv M. tuberculosis in the presence of hMMP-1 significantly decreased the expression of MCP-1, MMP-1, and MMP-9 (Fig. 6, Panel 1). Thus, we confirmed that MMP-1 activation of PAR-1 regulates the expression of MCP-1, MMP-1, and MMP-9 by M. tuberculosis-stimulated THP-1 cells.
Figure 6. PAR-1 inhibitor SCH79797 regulates the expression of MCP-1, MMP-1, and MMP-9 in THP-1 monocytic cells stimulated by sonicated H37Rv M. tuberculosis and THP-1 cells stimulated by sonicated H37Rv M. tuberculosis in the presence of exogenous purified human MMP-1.
Panel 1, we measured the relative changes in MCP-1, MMP-1, MMP-9, and TIMP gene expression by real-time PCR. Data are presented as the fold change in gene expression normalized to the endogenous reference gene PDHB and relative to untreated controls. The effect of 50 nM concentration of SCH79797 PAR-1 inhibiting compound was assessed following 24 hr in vitro stimulation of THP-1 cells with 5 μg/ml sonicated H37Rv M. tuberculosis. Cultures proceeded in 500 μl incomplete RPMI. PAR-1 SCH79797 inhibiting compound was diluted with DMSO and dispensed in 5 μl volume to produce a final culture concentration of 0.01% DMSO. We also added 5 μl of DMSO to control cultures. The results presented are from six independent experiments showing the mean and standard deviations. We consistently observed significant differences in the mean values across variables (Kruskal-Wallis p < 0.001) when testing MCP-1, MMP-1 and MMP-9. We show corrected p-values obtained from student t-tests. Of note, levels of specific mRNAs from non-stimulated cells were not significantly different from those obtained from cultures that proceeded with the PAR-1 inhibitor alone. Panel 2, the effect of 1 nM exogenous human purified MMP-1 on THP-1 cells exposed to 5 μg/ml sonicated H37Rv M. tuberculosis was assessed. We also assessed the effect of 1 nM exogenous human purified MMP-1 on THP-1 cells exposed to 5 μg/ml sonicated H37Rv M. tuberculosis in the presence or absence of 50 nM concentration of SCH79797 PAR-1 inhibiting compound. We measured the relative changes in MCP-1, MMP-1, and MMP-9 gene expression by real-time PCR as explained above. Cultures proceeded in 500 μl of incomplete RPMI. The results presented are from three independent experiments showing the mean and standard deviations. We consistently observed significant differences in the mean values across variables (Kruskal-Wallis p < 0.001) when testing MCP-1, MMP-1 and MMP-9. We show corrected p-values obtained from student t-tests. The inhibitor’s concentration was selected from dose response-experiments.
Based on the results described above, we concluded that PAR-1 activation by MMP-1 emerged as a key event in M. tuberculosis-induced and MCP-1-potentiated inflammatory response in vitro. Thus, we further tested the effect of SCH79797 PAR-1 inhibitor on the expression of MCP-1, MMP-1, MMP-9 and additional 31,000 annotated genes in M. tuberculosis H37Rv-infected THP-1 cells, using an whole genome expression microarray.
We first selected those genes regulated by H37Rv M. tuberculosis infection. To do so, we compared the gene expression profiles of infected cells and non-infected cells and selected those genes significantly regulated, according to our stringent parameters (Table 6). We then followed the effect of treatment on the expression of those genes by comparison of infected cells treated with SCH79797 (50 nM) versus infected cells untreated. Among those genes significantly regulated by PAR-1 inhibitor treatment were our primary target genes MCP-1 (CCL2), MMP-1, and MMP-9 (Table 6). In addition, IL-7 receptor (IL7R), Toll-like receptor 8 (TLR8), and SPP-1 genes were also modulated by this treatment (Table 6). Thus, we confirmed the down-regulation of MCP-1, MMP-1 and MMP-9 by PAR-1 inhibitor in M. tuberculosis infected cells. Interestingly, PAR-1 treatment did not significantly modify the expression TNFα in response to infection (Table 6), which is required to mount a protective immune response against M. tuberculosis (37). In addition, CD14 gene expression was up-modulated by M. tuberculosis infection, in concordance with the notion that infection causes maturation of THP-1 (Table 6) (38). PAR-1 treatment of THP-1 cells did not modify significantly this trend (Table 6). We further confirmed the down-regulation of MCP-1, MMP-1 and MMP-9 by PAR-1 inhibitor testing the concentrations of functional proteins in supernatants of THP-1 cells infected with M. tuberculosis H37Rv (Table 7).
Table 6.
Differentially express genes in H37Rv M. tuberculosis infected THP-1 cells and its regulation by PAR-1 inhibitor SCH79797
| TargetID | CYTOBAND | ProbeID | NINT AVG_Sig | NINT ARRAY_SD | YINT AVG_Sig | YINT ARRAY_SD | YINT Diff. Score | YINT Diff. P-val | Log-ratios (YINT/NINT) | YIYT AVG_Sig | YIYT ARRAY_SD | YIYT Diff. Score | YIYT Diff. P-val | Log-ratios (YIYT/YINT) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBI3 | 19p13.3 | 2810767 | 16.7 | 9.268 | 910.1 | 311.641 | 31.915 | 0.00064 | 5.8 | 511.5 | 257.594 | 0 | 0.99998 | −0.8 |
| SPP1 | 4q22.1 | 3840154 | 22.1 | 21.323 | 615.2 | 235.702 | 20.707 | 0.0085 | 4.8 | 112.2 | 102.994 | −8.322 | 0.14717 | −2.5 |
| IL4I1 | 19q13.33 | 540670 | 22.2 | 1.29 | 606.9 | 146.803 | 75.194 | 0 | 4.8 | 344.7 | 206.619 | 0 | 0.99998 | −0.8 |
| MMP1 | 11q22.2 | 3360224 | 4.2 | 2.889 | 112.9 | 26.33 | 333.614 | 0 | 4.8 | 35.3 | 10.905 | −15.437 | 0.02859 | −1.7 |
| IL1B | 2q13 | 840685 | 29.5 | 9.647 | 681 | 106.828 | 333.614 | 0 | 4.5 | 401 | 146.768 | −2.186 | 0.60445 | −0.8 |
| CCL3 | 17q12 | 6590682 | 13.2 | 4.007 | 217.2 | 27.128 | 333.614 | 0 | 4.0 | 173.5 | 83.115 | 0 | 0.99998 | −0.3 |
| CXCL10 | 4q21.1 | 6270553 | 14.3 | 4.081 | 206.9 | 55.44 | 51.663 | 0.00001 | 3.8 | 219.4 | 92.483 | 0 | 0.99998 | 0.1 |
| CCL3L1 | 17q12 | 610524 | 7.6 | 8.675 | 106.2 | 1.037 | 69.425 | 0 | 3.8 | 88.9 | 35.98 | 0 | 0.99998 | −0.3 |
| CCL2 | 17q12 | 1030333 | 83.6 | 41.342 | 1064.4 | 209.641 | 83.693 | 0 | 3.7 | 290.2 | 135.327 | −34.545 | 0.00035 | −1.9 |
| ADAMDEC1 | 8p21.2 | 1690192 | 17.7 | 5.507 | 180.5 | 49.54 | 43.13 | 0.00005 | 3.4 | 50.8 | 44.977 | −7.746 | 0.16803 | −1.8 |
| ADAMDEC1 | 8p21.2 | 2470184 | 45.6 | 28.138 | 457 | 160.361 | 21.198 | 0.00759 | 3.3 | 113.9 | 76.87 | −7.985 | 0.15903 | −2.0 |
| CNTNAP1 | 17q21.31 | 4570358 | 6.7 | 7.157 | 64.5 | 10.33 | 33.995 | 0.0004 | 3.3 | 74.2 | 11.383 | 0 | 0.99998 | 0.2 |
| IL8 | 4q13.3 | 1980309 | 108.5 | 28.184 | 1007.9 | 353.371 | 21.558 | 0.00699 | 3.2 | 134.5 | 78.161 | −6.288 | 0.23506 | −2.9 |
| CCL3L1 | 17q12 | 4250053 | 26.2 | 2.748 | 217.3 | 42.472 | 90.901 | 0 | 3.1 | 166.9 | 69.44 | 0 | 0.99998 | −0.4 |
| SLAMF8 | 1q23.2 | 6040577 | 28.3 | 16.329 | 207.3 | 71.669 | 18.141 | 0.01534 | 2.9 | 42.3 | 37.615 | −9.425 | 0.11416 | −2.3 |
| CD14 | 5q31.3 | 7000369 | 27.7 | 15.637 | 197.5 | 58.132 | 28.835 | 0.00131 | 2.8 | 131.4 | 27.984 | 0 | 0.99998 | −0.6 |
| IL7R | 5p13.2 | 3830349 | 13.1 | 10.911 | 78 | 23.206 | 15.948 | 0.02542 | 2.6 | 10.6 | 14.198 | −14.125 | 0.03868 | −2.9 |
| SGK | 6q23.2 | 4390450 | 71.2 | 11.95 | 418 | 88.396 | 70.069 | 0 | 2.6 | 293.2 | 101.02 | 0 | 0.99998 | −0.5 |
| CCL3L3 | 17q12 | 2810010 | 79.2 | 10.877 | 460.5 | 26.218 | 108.993 | 0 | 2.5 | 391 | 123.518 | 0 | 0.99998 | −0.2 |
| SGK1 | 6q23.2 | 1410209 | 50.9 | 8.127 | 271.9 | 53.795 | 74.496 | 0 | 2.4 | 195.2 | 77.904 | 0 | 0.99998 | −0.5 |
| SGK1 | 6q23.2 | 2030324 | 51.9 | 1.156 | 246.2 | 53.282 | 56.057 | 0 | 2.3 | 173.7 | 60.625 | 0 | 0.99998 | −0.5 |
| IL8 | 4q13.3 | 1570553 | 66.3 | 3.606 | 310.1 | 46.316 | 86.246 | 0 | 2.2 | 134.5 | 78.161 | −6.288 | 0.23506 | −1.2 |
| BCL3 | 19q13.31 | 6330725 | 43.6 | 13.98 | 202.9 | 3.745 | 69.425 | 0 | 2.2 | 256.8 | 93.862 | 0 | 0.99998 | 0.3 |
| NFKB2 | 10q24.32 | 1710504 | 22.1 | 8.518 | 101.9 | 7.329 | 42.738 | 0.00005 | 2.2 | 123.9 | 32.161 | 0 | 0.99998 | 0.3 |
| MMP9 | 20q13.12 | 3180528 | 72.3 | 17.171 | 324.8 | 73.222 | 48.888 | 0.00001 | 2.2 | 41.4 | 17.344 | −58.228 | 0 | −3.0 |
| IFI44L | 1p31.1 | 3870338 | 33.9 | 15.679 | 150.7 | 41.158 | 22.758 | 0.0053 | 2.2 | 193.5 | 60.91 | 0 | 0.99998 | 0.4 |
| TNF | 6p21.33 | 2640301 | 202.2 | 71.993 | 869.5 | 212.143 | 35.423 | 0.00029 | 2.1 | 731.9 | 321.682 | 0 | 0.99998 | −0.2 |
| TLR8 | Xp22.2 | 6480360 | 28 | 11.682 | 117.2 | 25.951 | 33.995 | 0.0004 | 2.1 | 31.4 | 18.932 | −18.749 | 0.01334 | −1.9 |
| SPP1 | 4q22.1 | 3460070 | 100.5 | 8.21 | 416.7 | 89.322 | 53.789 | 0 | 2.1 | 53.7 | 19.68 | −64.456 | 0 | −3.0 |
| RGL1 | 1q25.3 | 3180039 | 55.5 | 14.147 | 228.3 | 73.208 | 15.048 | 0.03128 | 2.0 | 64.9 | 18.321 | 0 | 0.99998 | −1.8 |
| CD83 | 6p23 | 6620026 | 44.6 | 12.391 | 179.4 | 23.918 | 58.588 | 0 | 2.0 | 185.7 | 42.845 | 0 | 0.99998 | 0.5 |
| PLA2G4C | 19q13.32 | 1990672 | 192.8 | 30.83 | 766.4 | 102.941 | 88.184 | 0 | 2.0 | 661.2 | 289.94 | 0 | 0.99998 | −0.2 |
| NFE2L3 | 7p15.2 | 5820035 | 100.8 | 25.404 | 395.5 | 104.806 | 27.147 | 0.00193 | 2.0 | 268.6 | 114.723 | 0 | 0.99998 | −0.6 |
| TNFAIP3 | 6q23.3 | 3360681 | 99.1 | 25.272 | 384.1 | 13.89 | 72.06 | 0 | 2.0 | 348.5 | 128.42 | 0 | 0.99998 | −0.1 |
| SOD2 | 6q25.3 | 3420373 | 124.4 | 25.642 | 469.7 | 153.48 | 12.912 | 0.05115 | 1.9 | 297.2 | 186.744 | 0 | 0.99998 | −0.7 |
| PMP22 | 17p12 | 7560138 | 18.3 | 8.967 | 65.8 | 12.688 | 15.303 | 0.02949 | 1.9 | 101.5 | 25.474 | 0 | 0.99998 | 0.7 |
| FEZ1 | 11q24.2 | 3290458 | 387.3 | 24.881 | 1388.1 | 121.045 | 83.665 | 0 | 1.8 | 1156.9 | 230.953 | 0 | 0.99998 | −0.3 |
| NFKB2 | 10q24.32 | 3310615 | 24.4 | 8.507 | 85.9 | 14.941 | 25.951 | 0.00254 | 1.8 | 112.6 | 18.106 | 0 | 0.99998 | 0.4 |
| EPSTI1 | 13q14.11-q14.11 | 5700725 | 98.9 | 16.449 | 346.5 | 66.314 | 58.14 | 0 | 1.8 | 513.5 | 156.037 | 0 | 0.99998 | 0.6 |
| GPR84 | 12q13.13 | 2070646 | 57.6 | 8.674 | 200.7 | 34.292 | 56.221 | 0 | 1.8 | 89.9 | 60.458 | −3.008 | 0.50028 | −1.2 |
| RELB | 19q13.32 | 730440 | 50.5 | 7.492 | 173.3 | 23.16 | 51.233 | 0.00001 | 1.8 | 181.2 | 56.865 | 0 | 0.99998 | 0.1 |
| MSC | 8q13.3 | 7510377 | 25.6 | 6.291 | 85.7 | 5.224 | 25.491 | 0.00282 | 1.7 | 73.5 | 9.918 | 0 | 0.99998 | −0.2 |
| FTH1 | 11q12.3 | 5220240 | 501.6 | 130.423 | 1673.9 | 487.744 | 15.825 | 0.02615 | 1.7 | 2232.1 | 993.713 | 0 | 0.99998 | 0.4 |
| NCF1 | 7q11.23 | 3940438 | 320.9 | 93.979 | 1051.9 | 295.417 | 16.749 | 0.02114 | 1.7 | 1328.8 | 323.479 | 0 | 0.99998 | 0.3 |
| NCF1C | 7q11.23 | 3180681 | 345.9 | 74.534 | 1114.1 | 163.744 | 67.61 | 0 | 1.7 | 1384.7 | 303.348 | 0 | 0.99998 | 0.3 |
| FEZ1 | 11q24.2 | 360427 | 54.9 | 17.449 | 176.8 | 19.607 | 40.193 | 0.0001 | 1.7 | 163.7 | 46.344 | 0 | 0.99998 | −0.1 |
| OAS2 | 12q24.13 | 240722 | 28 | 5.225 | 88.6 | 19.942 | 21.198 | 0.00759 | 1.7 | 116.4 | 24.927 | 0 | 0.99998 | 0.4 |
| GBP2 | 1p22.2 | 1940162 | 80.4 | 25.95 | 238.9 | 45.217 | 35.423 | 0.00029 | 1.6 | 268.4 | 55.122 | 0 | 0.99998 | 0.2 |
| SOD2 | 6q25.3 | 3890326 | 535 | 89.897 | 1585.9 | 213.363 | 65.492 | 0 | 1.6 | 1130.1 | 383.698 | 0 | 0.99998 | −0.5 |
| ECGF1 | 22q13.33 | 7650128 | 64.9 | 22.1 | 191.2 | 33.538 | 33.18 | 0.00048 | 1.6 | 91.3 | 31.354 | −9.698 | 0.1072 | −1.1 |
| MMP25 | 16p13.3 | 6020630 | 33.7 | 6.297 | 97.3 | 3.732 | 23.134 | 0.00486 | 1.5 | 53.5 | 8.542 | −4.147 | 0.38489 | −0.9 |
| OAS3 | 12q24.13 | 990768 | 80 | 19.678 | 230.2 | 43.56 | 38.675 | 0.00014 | 1.5 | 366.1 | 91.638 | 1.296 | 0.74201 | 0.7 |
| MX1 | 21q22.3 | 1690066 | 890.6 | 376.587 | 2562.3 | 589.164 | 17.622 | 0.01729 | 1.5 | 3602 | 1081.058 | 0 | 0.99998 | 0.5 |
| ZC3H12A | 1p34.3 | 2490017 | 57.7 | 14.209 | 165.5 | 16.804 | 35.386 | 0.00029 | 1.5 | 186.3 | 25.133 | 0 | 0.99998 | 0.2 |
| IRF7 | 11p15.5 | 1470382 | 52.1 | 17.993 | 149.1 | 33.814 | 18.858 | 0.01301 | 1.5 | 156.6 | 29.869 | 0 | 0.99998 | 0.1 |
| CD83 | 6p23 | 5050162 | 94.2 | 18.895 | 267.1 | 22.24 | 46.019 | 0.00003 | 1.5 | 267.3 | 58.124 | 0 | 0.99998 | 0.0 |
Data from those genes with scanning detection p-values ≤ 0.05, differentially express p-values ≤ 0.05 after FDR correction (Diff. p-val.), and Log-ratios ≥ 1.5 (regardless of the orientation) are shown. The differential scores (Diff. score) are a function of the p-values (Illumina Inc.). Log-ratios are the logarithm in base 2 of the average signal (AVG. sig.) ratios. We also show the array standard deviation of the average signals (Array_SD). NINT = Not infected not treated; YINT = Yes infected not treated; YIYT = Yes infected yes treated. We show highlighted in red the results from our primary targets. In black bold we show genes with scanning detection p-values ≤ 0.05, differentially express p-values ≤ 0.05 before FDR correction, and Log-ratios ≥ 1.5 (regardless of the orientation). All cultures proceed and were processed according to the protocols described in material and methods, including washing steps.
Table 7.
PAR-1 inhibitor treatment of infected THP-1 cells down-regulates MCP-1, MMP-1 and MMP-9 protein secretion, but does not modify M. tuberculosis intra-cellular proliferation
| Culture condition/measurement | M. tuberculosis- infected, untreated | M. tuberculosis- infected, treated | Two-sample t-test |
|---|---|---|---|
| MCP-1 (pg/ml ± SD) | 136 ± 39.7 | 0 | p = 0.004 |
| MMP-1 (pg/ml ± SD) | 744.4 ± 103.4 | 0 | p < 0.00001 |
| MMP-9 (pg/ml ± SD) | 951 ± 23 | 0 | p < 0.0001 |
| CFU/ml (103) | 9 ± 2.65 | 8.3 ± 3.1 | ns |
Soluble factors were determined in supernatants using ELISA test for MCP-1 (BD Biosciences) and functional specific-peptide-cleavage ELISA tests for MMP-1 and MMP-9 (R&D). Cell pellets lysed in Triton X-100 were used to recover intracellular bacteria and processed as indicated in material and methods to obtain the colony forming units (CFU) counts. SD = standard deviation; ns = not significant. The values for uninfected untreated negative control THP-1 cells were cero for MCP-1, MMP-1, and MMP-9.
We also analyzed those genes that were not regulated by infection per se, but were significantly regulated by PAR-1 treatment in infected cells. Of note, CCR2 (the receptor of MCP-1) and TIMP3 were differentially and significantly down-regulated by PAR-1 treatment of those cells (Table S2). We did not observed significant differences in the CFU in infected THP-1 cells treated with PAR-1 inhibitor or untreated (Table 7). In our model, PAR-1 plays a significant role in the amplification of the inflammatory response to M. tuberculosis infection (Fig. S2).
DISCUSSION
Our studies in human populations combined with our ex vivo analysis of tissues from TB cases, and our in vitro model using the human THP-1 monocytic cell line, led us to conclude that the -2518 MCP-1 genotype GG and the -1607 MMP-1 genotype 2G/2G are key genetic loci associated with the expression of severe TB disease. The presence of both genotypes increases by 13-fold the chance of developing severe TB disease, by 3.864-fold the chance of a delayed sputum conversion from positive to negative, and by 6.5-fold the chance of developing extensive areas of lung fibrosis. Moreover, the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G operates with high penetrance in this population since 83% of the carriers expressed severe disease. Neither the -2518 MCP-1 genotype GG nor the -1607 MMP-1 genotype 2G/2G alone were sufficient to produce a severe disease. Interestingly, a higher proportion of TB patients carrying the susceptibility two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G have had BMI values at first visit lesser than the median BMI value of the entire sample. Interestingly, weight loss is per se a key marker of severity of TB disease in animal models (39). All together, our observations have important implications when the interest is to detect, as early as possible, those persons who are at increased chance of developing a severe TB disease, may remain contagious for a longer period than any other TB case, and will develop a poor health related-quality of life. We must remember that these TB cases were tightly followed up which may have decreased the chance of abandonment of treatment and progression to a potentially lethal illness. This may also have implications for the treatment time required to attain sputum smear test conversion in TB cases infected with multi-drug resistant (MDR)-M. tuberculosis strains and the increased mortality observed among them.
Six TB cases in carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G expressing severe disease (score > 44) underwent surgery to remove damaged lung tissue versus none in the groups of carriers of any other combination of genotypes at these two loci. The IHC analysis of these lung tissues revealed macrophages expressing copious amounts of MCP-1/CCL2, MMP-1, and MMP-9. Cell of other lineages (CD68-negative) were also present in the lesions and expressing these factors, indicating a possible crosstalk between macrophages and perhaps, lung epithelial cells. Our IHC analysis is in agreement with results from a genome-wide comparison of gene expression in human granulomatous lesions (40). Access to their database allowed us to rank the genes according to the magnitude of the ratios and observed that MCP-1/CCL2, MMP-1 and MMP-9 were also among the most significantly up-regulated genes in human granulomatous lesions (40).
We tested serum and plasma samples and observed the highest serum levels of MCP-1, and the highest plasma levels of MMP-1 and MMP-9 in carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G as compared to levels of these molecules in carriers of any other relevant combination of genotypes at those loci. It was unexpected to see the highest levels of MMP-9 in carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. We thought that preceding molecular events involving increased production of MCP-1 and MMP-1 levels might explain why these TB cases expressed the highest levels of MMP-9. We first tested whether endogenous MCP-1 and MMP-1 were involved in the induction of MMP-9 using CCR2 inhibiting compound RS504393 and MMP-1 inhibiting 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide. We clearly established in vitro that the MCP-1/MMP-1 loop drives the expression of MMP-9 to its highest levels in THP-1 cells activated by sonicated H37Rv M. tuberculosis. Moreover, addition of exogenous human (h)MMP-1 to THP-1 cells activated by sonicated H37Rv M. tuberculosis potentiated significantly the production of MMP-9. Hence, these in vitro data may explain why we observed the highest levels of MMP-9 in carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. Given these observations, we concluded that it is very likely that the MCP-1/MMP-1/MMP-9 pathway is driving an intense inflammatory response to infection in lung tissues of patients carrying that susceptibility two-locus genotype. Notable, MMP-9 not only helps in recruiting macrophages to the inflammatory foci (27), but can also degrade collagen byproducts of MMP-1-digested fibrilar collagen (41).
Since it is known that MMP-1 cleaves the PAR-1 N-terminal exodomain to generate a SFLLRN-containing motif that activates the PAR-1 signaling pathway (33), we investigated whether this pathway may operate in TB. We observed by IHC that alveolar macrophages in normal lungs constitutively express PAR-1 while in few macrophages express PAR-1 in TB cases suffering from severe lung disease. Nevertheless, we concluded that lung macrophages express PAR-1 in healthy and to a lesser extend in disease lung parenchyma. Our FACS analysis experiments suggested that MMP-1 activates PAR-1 in monocytic cells exposed to M. tuberculosis antigens. We first established that as a result of exposure to M. tuberculosis antigens, PAR-1 expressed in CD14-positive/CD16-negative THP-1 cells was significantly down-regulated, which may indicate that an specific ligand was activating, and subsequently inducing the recycle of this receptor. Addition of MMP-1 inhibitor 4-Aminobenzoyl-Gly-Pro-D-Leu-D-Ala hydroxamic peptide to the cultures significantly neutralized M. tuberculosis antigen-induced down-regulation of PAR-1 expression in these cells, indicating that MMP-1 was one of the molecules activating PAR-1. Furthermore, in agreement with our observation that MCP-1 potentiates MMP-1 secretion by THP-1 cells exposed to sonicated H37Rv M. tuberculosis, we also observed that the addition of CCR2 inhibitor RS504393 prevents the activation, and consequently, the down-regulation of PAR-1 expression by CD14-positive/CD16-negative THP-1 cells. Since THP-1 cells did not express thrombin under those conditions, we concluded that MMP-1 might be one of the proteases activating PAR-1, and thereby inducing the recycling of activated PAR-1 in our in vitro system.
We also conducted studies to directly determine the possible role of PAR-1 activation by MMP-1 in TB, adding exogenous human MMP-1 to THP-1 cells activated with sonicated M. tuberculosis H37Rv. These experiments corroborated a link between the MCP-1/MMP-1 up-regulation of MMP-9 with activation of PAR-1 in monocytes/macrophages. Addition of PAR-1 inhibitor SCH79797 at nanomolar concentration to THP-1 cells stimulated with sonicated M. tuberculosis H37Rv or THP-1 cells infected in vitro with M. tuberculosis H37Rv significantly down-regulated MCP-1, MMP-1, and MMP-9 expression, while the TIMPs remained poorly expressed. Likewise, addition of this PAR-1 inhibitor, at nanomolar concentration, to cultures of M. tuberculosis-stimulated THP-1 cells that proceeded in the presence of exogenous hMMP-1 resulted in inhibition of MCP-1, MMP-1, and MMP-9 expression. Interestingly, this PAR-1 inhibitor down-regulated the expression of CCR2 in infected cells, which may contribute to decrease the effect of excessive MCP-1 production.
The similarities between the results from the reanalysis of data from a genome-wide comparison of gene expression in human granulomatous lesions mentioned above (40), and the data we have obtained from M. tuberculosis infections of THP-1 cells, provides physiological relevance to our in vitro observations. It will be interesting to study the effects of the compound or peptide inhibitors we have tested, alone or in combinations, on primary human CD14+ CD16− and CD14+ CD16+ macrophages. Likewise, since we did not attain an efficient transient transformation of a significant proportion of THP-1 cells with specific siRNA, it will be important to generate, for future experiments, stable THP-1 transfectants expressing specific siRNAs.
It is extremely interesting that a functional SNP in the promoter region of leukotriene A4 hydrolase (LTA4H), the genotype TT, drives increased expression of the LTA4H gene and decreased survival of TB patients affected with meningitis (3). The significance of this finding is that LTA4H is a key enzyme in the generation of leukotriene B4 (LTB4), and that an excess of LTB4 results in excessive production of TNFα and subsequent macrophage necrosis (3). Since the PAR-1 and LTB4 receptors BLT1 and BLT2 are G-protein-coupled receptors (GPCR) expressed in macrophages and it is known that crosstalk exist between GPCRs (3, 42), it will be interesting to investigate whether CCR2 or PAR-1 and LTB4 receptors crosstalk, and if they do crosstalk, whether they crosstalk in an agonistic or antagonistic fashion. Notably, we do not rule out that the -2518 MCP-1 genotype GG or the -1607 MMP-1 genotype 2G/2G may drive, alone or in interaction with other genes, the expression of severe TB disease in other populations. In fact, there is evidence that the -2518 MCP-1 genotype GG influences the expression of meningitis in Chinese TB patients of pediatric age (43).
We know at least that PAR-1 inhibition do not increase M. tuberculosis proliferation in THP-1 cells, as we assessed this experimentally. However, more in-depth studies are needed to determine whether disease modification by inhibitors of the MCP-1/MMP-1/PAR-1 pathway may serve as an adjuvant of current chemotherapy to avoid the expression of a severe disease and permanent lung damage in human carriers of the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. Targeting the MCP-1/MMP-1/PAR-1 pathway to decrease active tissue damage may also help to shorten the time needed to achieve sputum sterilization by chemotherapy in these patients.
All together, our results uncovered a new concept that the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G increases the chance of developing severe pulmonary TB. We are also reporting that MMP-1 can operate a positive feed-back induction of MCP-1 and MMP-9 through PAR-1 activation, and that inhibition of PAR-1 activation down-regulates the MCP-1/MMP-1/MMP-9 pathway in monocytic cells exposed to sonicated H37Rv or infected with H37Rv M. tuberculosis strain. That in fact TNFα up-regulation in response to infection is not modified in THP-1 cells by treatment with SCH79797 PAR-1 inhibitor is encouraging since this is a factor critically involve in the development of cellular immune response against M. tuberculosis infection (37). Studies in animal models are needed to confirm in vivo these molecular mechanisms and to confirm that the PAR-1 inhibitor SCH79797 constitutes a promising candidate for the treatment of MCP-1/MMP-1 mediated tissue damage in TB. Thus, our finding that MMP-1, through PAR-1 activation, is driving a hyper-inflammatory in response to infection highlights key molecular events for the development of new clinical interventions to attenuate tissue damage.
PATIENTS, MATERIALS AND METHODS
Ethics Statement
All subjects provided informed consent, under protocols approved by the Institutional Review Boards of The Methodist Hospital Research Institute (TMHRI) in Houston (Texas, USA), and the Peruvian Ministry of Health.
Recruitment Base
People from Peru are of Hispanic ancestry whose genetic composition consists mainly of an admixture of Amerindians and Spaniards. People with this genetic composition are the main ethnic group in most Latin-American countries and of Hispanics living in the United States (US). Peru is a moderate-to-high tuberculosis-burdened country (44). The TB Incidence in 2010 was 118 per 100,000 person-years and the prevalence of TB was 187 per 100,000 person-years (44). The existence of government-run, community-based programs designed to detect new TB cases in Peru facilitate identification of well-characterized study subjects (45). We recruited our cases using a sentinel surveillance network comprised of primary (community) health centers under the jurisdiction of two National Hospitals (serving a total population of 10 million individuals) in Lima Province, the Hospital Hipolito Unanue and the Hospital Maria Auxiliadora. All TB cases were residents of communities sharing similar TB incidence, socioeconomic status and life styles, and very likely similar M. tuberculosis strains. Most of the individuals suffering from TB in Peru are of low socioeconomic status (44, 45).
Inclusion and exclusion criteria
We conducted a case-case study of homogeneous populations of Peruvian Mestizo ancestry. From July 2009 to July 2011, we recruited 224 TB cases meeting our inclusion/exclusion criteria. TB cases were unrelated to the third generation as determined by a questionnaire, aged > 18 to ≤ 50 years old. They were of Peruvian ethnicity to the third generation, increasing the likelihood that this population is ethnically homogeneous. Patients were not HIV-seropositive, and/or did not have co-morbidities affecting immunity, including autoimmunity, cancer, organ transplantation, primary immunodeficiency, were not on therapy with immunosuppressive drugs (such as corticosteroids), and did not suffer from endocrine disorders, or chronic cardiopulmonary, hepatic, or renal disease. Patients did not suffer from alcoholism and were not illegal drug users. Body mass index (BMI) was used as an indicator of malnourishment in adults (46, 47). The BMI was determined based on weight and height measured by a nurse, as we did before (1, 2). We selected TB patients meeting the following criteria: 1) Completeness of demographic and clinical data; 2) all TB cases were new with a disease of no more than 3 months of duration. We excluded cases of recurrent or re-infected TB. Recruitment of new cases avoids confounding of data by unmeasured (unknown) factors that increase the chance of recurrent or re-infected TB; 3) Patients who developed TB disease within a period of two years following the most recent close exposure to an index case; 4) TB patients identified at the local (community) TB control center to avoid selection bias of choosing a disproportionate number of TB cases with a severe disease presentation that needed specialized clinical interventions at secondary or tertiary hospitals. Physicians at the local TB control center identified all cases; 5) Patients with positive adherence to treatment. Patients had: (i) signs and symptoms of active TB; (ii) a full-size posteroanterior chest X-ray suggestive of active pulmonary TB; (iii) disease confirmed by at least one of three positive sputum smear test; (iv) monthly sputum smear tests to follow up sputum smear test conversion from positive to negative. New TB patients in Peru received a four-drug standardized treatment consisting of 300 mg of Rifampicin + 300 mg of Isoniazid + 1500 mg of Pyrazinamide + 1200 mg of Ethambutol daily for 2 months, followed by 300 mg of Rifampicin + 300 mg of Isoniazid twice a week for 2 months (45).
The severity score
We defined disease duration as the length of time since the first symptom was noticed by the patient to the time of sampling. The total score is the sum of a score based on the assessment of clinical signs and symptoms plus a score based on the assessment of posteroanterior full-size chest X-rays (total from the independent assessment of the two lungs; Table 2). BMI in adults was assessed according to the World Health Organization (WHO) chart. Anemia was assessed by levels of hemoglobin according to gender and laboratory standards. Fever was assessed by axillary temperature ≥ 37.0 °C measured by an electronic thermometer in a closed axillary fold. Any abnormal sign at lung auscultation provided by a physician was used to assign a score to this item. The extent of lung lesions and type of lesions were evaluated for each lung independently. Clinical presentations (e.g. miliary and meningitis TB) were also scored when present.
Sample size calculation of the case-case study
Based on preliminary data from the analysis of 108 TB cases, a logistic regression of a binary response variable (Y) on an independent variable with 4 levels, adjusted for an additional 2 independent variables with a sample size of 206 observations will achieve 90% power at a 0.01 significance level to detect a change in probability (Y=1) from the baseline value of 0.05 to 0.24 in the comparison of the strata of interest with other stratum. This change corresponds to an odds ratio of 6 (48).
Blood Samples and SNP Analysis
Genomic DNA was isolated from blood cell pellets using DNA extraction kits (Qiagen, Valencia, CA). We genotyped the -2518A>G SNP in MCP-1 (rs1024611), the -1607_1608insG variant in MMP-1 (rs1799750), and 42 genomic control SNPs (2). The rs1024611 and rs1799750 SNPs were genotyped in duplicate using the tetra-primer ARMS PCR technique (2). We did not find discrepancies in the typing results for MCP-1 and MMP-1. We will make primer sequences available and procedures available upon request. To confirm our results for the MCP-1 and MMP-1 gene analysis, we sequenced 10 randomly selected samples. The 42 SNPs used as genomic controls in Peruvians were genotyped using an Illumina platform (2). We obtained λ values of admixture as we did before (1, 2). Given that Africans are the founders of all human races and that the -2518 MCP-1 allele A and the -1607 MMP-1 allele 1G are the most frequent in this population (2), we used them as reference alleles.
We randomly selected 30 TB cases from each stratum by genotypes and 20 normal subjects to assess the levels of MCP-1 in serum, and MMP-1 and MMP-9 in plasma. We used serum to assess MCP-1 levels because we figured up that the data obtained from serum was more reproducible compared to that of plasma. Serum was tested at a 1:4 dilution and plasma for MMP-1 in 1:4 and for MMP-9 in 1:8 dilutions. MCP-1 levels were measured by ELISA (BD Biosciences). MMP-1 and MMP-9 were measured using Fluorokine E, Human Active MMP-1 or Fluorokine E, and Human Active MMP-9 kits (R&D Systems). These Fluorokine assays measured the amount of cleaved collagen peptide by monitoring fluorescence. These assays are functional assays since the antibodies capture only free MMP-1 and MMP-9. The levels of MCP-1, MMP-1, and MMP-9 were obtained using a Multi-mode Microplate Reader Synergy 2 (BioTek Instruments Inc., Winooski, VT).
IHC analysis of lung tissues
For IHC, lung tissue samples from the six TB cases that underwent surgery were fixed in paraformaldehyde and embedded in paraffin. Lung samples from normal individuals were provided by the Department of Pathology and Genomic Medicine at the Methodist Hospital. For IHC, we used heat-induced epitope retrieval in citrate buffer (Thermo/Fisher Scientific Inc., Waltham, MA), rabbit anti-human CCL2/MCP-1 polyclonal antibody LS-C112304 (LifeSpan Bioscience, Inc), rabbit anti-human MMP-1/Collagenase-1 Ab-6 (Thermo/Fisher Scientific, Inc), rabbit anti-human MMP-9 ab13458 (Millipore), rabbit anti-human PAR-1 ab13398 (abcam), mouse anti-human CD68 KP1 (abcam), and the MultiVision Polymer Detection System/anti-mouse-HRP (red) + anti-rabbit-AP (blue) detection system (Lab Vision Products, Thermo Scientific). We immunostained 5-μm thick sections in duplicate for each case. Negative controls were sections incubated with irrelevant normal Rabbit IgG (KPL Inc., Gaithersburg, ME) or irrelevant normal mouse IgG antibody (R&D Systems). We used 1X Automation buffer pH 7.5 (Biomeda Inc., Albuquerque, NM) in all wash steps. SuperMount permanent aqueous mounting media (BioGenex, San Ramon, CA) was used for mounting immunostained tissue sections. Immunostained tissue sections were assessed at 200X and 100X total magnification and 0.8 numerical aperture of the objective lenses. Image acquisition was performed using a computerized analysis system comprising: a BX41 microscope with a U-TVIX-2 and a U-CMAD3 tube and adapter attached for on-screen viewing, a C3040 4.1 megapixel digital camera, and Magnafire-SP software (Olympus America Inc.).
Culture Conditions and RT-PCR Assay
For the assessment of MCP-1, MMP-1, MMP-9, TIMP-1, -2, -3, and -4 expression, we used the 7500 Fast Real-Time PCR System and assay-on-demand primers for MCP-1 (ID Hs00234140), MMP-1 (ID Hs00233958), MMP-9 (ID Hs00234579), TIMP-1 (ID Hs00171558), TIMP-2 (ID Hs01091319), TIMP-3 (ID Hs00227214), TIMP-4 (ID Hs00162784), and the house-keeping gene PDHB (Pyruvate dehydrogenase beta, ID Hs00168650) (Applied Biosystems). To calculate the relative quantity (RQ) of mRNA expression, we used the 2 −ΔΔCT method implemented in the software (Applied Biosystems). The data are presented as the fold change in gene expression normalized to the endogenous reference gene PDHB and relative to the untreated controls (RQ values). THP-1 cells (1×106 cells/ml) were stimulated with the indicated amounts of H37Rv M. tuberculosis lysate obtained after sonication (sonicated H37Rv M. tuberculosis) as described previously (Ganachari et al. 2010), or the indicated amounts of CCR2 RS504393 (Tocris), MMP-1 4-Aminobenzoyl-Gly-ProD-Leu-D-Ala hydroxamic peptide (SIGMA), PAR-1 SCH79797 (Tocris) inhibitors, and human purified MMP-1 (hMMP-1) activated with p-aminophenylmercuric acetate (EMD Chemicals). THP-1 cells were cultured for 24 hr in incomplete RPMI as indicated in the figure legends. Cells were then harvested and total RNA was extracted using TRIzol (Invitrogen). Complementary DNA (cDNA) was obtained from 3 μg of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Approximately 100 ng cDNA was used to determine the expression levels of the housekeeping gene PDHB and MCP-1, MMP-1, MMP-9, and TIMPs.
Three-color FACS analysis
THP-1 cells cultured as described above were harvested and washed twice in cold incomplete RPMI, then re-suspended in 100 μl of cold incomplete RPMI containing 10 μg/ml of human IgG (SIGMA), and incubated on ice for 15 min. Cells were washed once in PBS + 10% BSA (SIGMA) and re-suspended at 5 × 105 cells/100 μl of this buffer. FITC-anti-human CD14 mouse monoclonal antibody 61D3 (Santa Cruz Biotechnology, Inc), PerCP-Cy5.5 anti-human CD16 mouse monoclonal antibody DJ130c (Santa Cruz Biotechnology, Inc), and APC anti-human F2R/PAR-1 mouse monoclonal antibody ATAP2 or appropriately labeled isotype controls were added to the cells and incubated for 30 min on ice. Cells were then washed thoroughly and re-suspended in cold PBS. Acquisition was done using the BD LSR II instrument and software. We acquired 10,000 to 100,000 events as indicated in the figure legends. The analysis of data was done using the FlowJo 7.6.4 software. CD14-positive/CD16-negative cells were gated for the analysis of F2R/PAR-1 expression.
Infections of THP-1 cells and gene expression microarrays
We used the M. tuberculosis H37Rv (ATCC) grown in Middlebrook 7H9 broth supplemented with ADC enrichment (Difco Laboratories), 2% glucose, and 1% glycerol. Cultures proceeded at 37°C and slow constant shaking to mid-log growth phase (7 to 10 days). The Bacteria were quantified using McFarland equivalence turbidity standards (Thermo Scientific). Fresh cultures of 106 to 108 bacilli/ml bacteria were immediately used for infections. To determine M. tuberculosis CFU cell pellets were obtained by centrifugation at 1,500 rpm and the cell pellets were lysed in 0.1% Triton X-100 (Sigma) before preparation of plating dilutions. We counted colony forming units (CFU)/ml by serial titration of cultures 1:10, 1:100, and 1:1,000 in 7H9 plated on 7H11 agar plates, and left in culture for 3 weeks at 37°C.
THP-1 cells were infected with M. tuberculosis bacilli at a multiplicity of infection (MOI) of 5 bacteria per cell in complete RPMI (10% FBS) without antibiotics. We used this culture media throughout the experiments, unless indicated. THP-1 cells and M. tuberculosis bacilli were incubated for 6 hours at 37°C in 5% CO2, then washed 3 times in incomplete RPMI and plated at 1 million cells/0.5 ml of culture media in 48-well plates. We then added the PAR-1 inhibitor SCH79797 (Tocris) at a final culture concentration of 50 nM. The PAR-1 inhibitor was previously diluted in DMSO at a 5 μM working concentration to dispense a 5 μl volume per well and to produce the desired concentration of PAR-1 inhibitor and a final culture concentration of 0.01% DMSO. Control cultures of infected cells contained 0.01% DMSO alone. Then the cultures proceed for additional 18 hours when we proceed to harvest the THP-1 cells for isolation of RNA or for CFU determinations, and to harvest the culture supernatants for ELISA tests. We did not induce differentiation of THP-1 cells previous to infection since exposure of THP-1 cells to M. tuberculosis (or M. tuberculosis-sonicate) induces differentiation of these cells into macrophage-like adherent cells.
Total RNA from these samples was extracted using Trizol followed by total RNA purification using Qiagen Kit as per manufacturer’s instructions. We assessed the quality of RNA using Agilent RNA 6000 Nano Kit. Gene expression profiling was performed using the Illumina HumanHT-12v4 BeadChip (Illumina Inc.). Two hundred nanograms of total RNA were labeled using a TotalPrepRNA Amplification Kit (Applied Biosystems) to obtain biotinylated-cRNA. We quantified and ascertained the fragment size using the Agilent RNA 6000 Nano Kit. We loaded the beadchip’s arrays with 750 ng of biotinylated-cRNA and left they hybridized overnight. We then washed and scanned the BeadChip arrays according to the manufacturer’s instructions. The chips were scanned using the Illumina BeadArray Reader.
Statistical analysis
Statistical analysis was done with Intercoiled STATA10 software (Stata Corporation). Chi2 test or student t-test was used to assess differences between categorical or continuous variables, respectively. The severity of disease was first established by expert clinicians in the field. Each TB case had a clinical diagnosis of disease severity as: non-severe or severe disease. Once the severity scores were obtained, we classified the cases according to whether they had severity score values above or ≤ the median. We used the Kappa statistic and a modified Rietveld and van Houd scale (49), to evaluate agreement between: (i) an expert clinician assessment of the cases, and (ii) the severity of disease classification obtained by another clinician using our clinical charts and table 2 (severity scores). According to this scale, the interpretation of Kappa coefficients is as follows: <0 = Less than chance agreement, 0.01–0.2 = Slight agreement, 0.21–0.40 = Fair agreement, 0.41–0.60 = Moderate agreement, 0.61–0.8 = Substantial agreement, 0.81–0.99 = Almost perfect agreement, 1 = perfect agreement (49). We treated the severity scores as a dependent binary random variable [0 = non-severe score (score ≤ 44); 1 = severe disease (score > 44); Table 3], and assessed the contribution on severity of disease of each locus and the two-locus genotypes, adjusting for age and gender using logistic regression analysis. We performed Pearson Goodness-of-Fit x2 tests to determine whether the observed models differed from the predicted. We used the non-parametric Kruskal-Wallis test, followed by the Bonferroni least significant difference (LSD) test for multiple comparisons to analyze differences in the mean severity score stratified according to genotypes. Spearman’s test was used to evaluate correlations. We used multiple 2 × 2 tables, with genotype arranged in an ordinal scale, and Chi2 Mantel-Haenszel statistics to test for genotypes associations with delayed (> 3 months) sputum smear test conversion primarily to obtain valid estimates for association for each stratum, and to assess dose effects and homogeneity of the odd ratios (OR) across strata. To test for homogeneity of OR, we used the Breslow and Day’s test provided in the SATA10 output. One-way ANOVA followed by a Bonferroni LSD test was used to evaluate results from the assessment of serum/plasma levels of MCP-1, MMP-1, and MMP9, and to evaluate results from in vitro experiments designed to test contributions of MCP-1/CCR2, MMP-1, and PAR-1 pathways in models of M. tuberculosis-induced expression of inflammatory factors by THP-1 cells.
We subtracted the background to the raw data obtained from the Illumina platform and proceed to normalized data using quantile normalization (50). We excluded probes showing detection p-values less than 0.05. All the statistical analyses of the expression data were conducted using the GenomeStudio package (Illumina). The p-values were obtained using false discovery rate (FDR) for multiple testing adjustments (51). Given that the scores obtained for each gene were a function of the p-values, we calculated the log2 of the average-signal’s ratios (log-ratios) to assess the magnitude and orientation (up- or down-regulation) of the gene regulation. We declare significant change in expression when FDR-adjusted p-values were ≤ 0.05 and the log2-ratios ≥ 1.5, regardless of the orientation.
Supplementary Material
We used double IHC analysis of PAR-1(blue) and CD68 (red) in paraffin-embedded diseased lung. Representative images from three of the six TB cases that underwent surgery to remove damaged tissue are shown. In section A we show the negative control (irrelevant antibodies isotype control). Images were acquired at 100X total magnification.
A model of MCP-1/MMP-1/PAR-1 pathway-caused tissue damage in TB cases carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. PHASE I: After exposure to M. tuberculosis, ganuloma macrophages express MCP-1 and MMP-1. Persons carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G produce excessive amounts of these factors. PHASE II: MCP-1 activates CCR2 receptors and potentiates M. tuberculosis-induced MMP-1 production. MMP-1 targets fibrilar collagen in the granulomatous matrix, resulting in matrix degradation. Persons carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G experience matrix degradation, an event that, in normal individuals, is not as intense and is part of the normal healing process. PHASE III: The excess of MMP-1 intensifies the granulomatous matrix degradation and activates the PAR-1 receptors. PHASE IV: Activation of PAR-1 increases MCP-1, MMP-1, and induces secretion of MMP-9. MMP-9 may contribute to MCP-1-mediated monocyte recruitment and further degradation of the granulomatous matrix. PAR-1 activation is a key event in the amplification of the inflammatory process (a hyper-inflammatory state) and in the induction of extensive tissue damage.
Acknowledgments
We are grateful to all patients and healthy donors for their kind cooperation. We are grateful to Ms. Maritza L. Flores, study coordinator, for the superb work done, and Dr. Federico Monzon-Bordonaba and Mr. Raul Gonzales for providing normal lung samples. We are grateful with Mr. Philip Randall for his superb editorial assistance. We are grateful to Dr. Edmond J. Yunis in the Department of Pathology of Harvard Medical School and Dr. Mauro Ferrari at The Methodist Hospital Research Institute for their superb leadership. We are grateful to the Peruvian Ministry of Health and the Peruvian National Institute of Health, both of which allowed appropriate access to the patients included in this study. Funding: We are grateful to the National Institutes of Health (NIH) and The Methodist Hospital Research Institute (TMHRI), both of which provided funding for this study. This work was supported by NIH grant R01 HL084347-06.
Footnotes
Table S1. Admixture λ value ; Table S2. Proportions of cases with severity scores above the median according to the two-locus genotypes -2518 MCP-1 and -1607 MMP-1; Table S2. Additional genes modulated by PAR-1 treatment of H37Rv M. tuberculosis infected THP-1 cells; Figure S1: Images of lung tissues from severely affected TB cases at low magnification; Figure S2. A model of MCP-1/MMP-1/PAR-1 pathway-caused tissue damage in TB cases carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G.
CONFLICT OF INTEREST
The authors have no conflicting financial interests. DNA samples are available through an MTA.
References
- 1.Flores-Villanueva PO, Ruiz-Morales JA, Song CH-H, Flores LM, Jo E-K, Montaño M, et al. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. J Exp Med. 2005;12:1649–1658. doi: 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganachari MM, Ruiz-Morales JA, Gomez de la Torre Pretell JC, Dinh J, Granados J, Flores-Villanueva PO. Joint effect of MCP-1 genotype GG and MMP-1 genotype 2G/2G increases the likelihood of developing pulmonary tuberculosis in BCG-vaccinated individuals. PLoS ONE. 2010;5:e8881. doi: 10.1371/journal.pone.0008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tobin DM, Roca FJ, Sungwhan FO, McFarland R, Vickery TV, Ray JP, et al. Host genotype-specific therapies can optimize the inflammatory response to Mycobacterial infections. Cell. 2012;148:434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, Levin M. A mutation in the interferon-γ-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 5.Ottenhoff TH, Kumararatne D, Casanova JL. Novel human immunodeficiencies reveal the essential role of type-I cytokines in immunity to intracellular bacteria. Immunol Today. 1998;19:491–494. doi: 10.1016/s0167-5699(98)01321-8. [DOI] [PubMed] [Google Scholar]
- 6.Dorman SE, Holland SM. Interferon-gamma and interleukin-12 pathway defects and human disease. Cytokine Growth Factor Rev. 2000;11:321–333. doi: 10.1016/s1359-6101(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 7.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 8.Braun MC, Lahey E, Kelsall BL. Selective suppression of IL-12 production by chemoattractants. J Immunol. 2000;164:3009–3017. doi: 10.4049/jimmunol.164.6.3009. [DOI] [PubMed] [Google Scholar]
- 9.Omata N, Yasutomi M, Yamada A, Iwasaki H, Mayumi M, Ohshima Y. Monocyte chemoattractant protein-1 selectively inhibits acquisition of CD40 ligand-dependent IL-12-producing capacity of monocyte-derived dendritic cells and modulates Th1 immune response. J Immunol. 2002;169:4861–4866. doi: 10.4049/jimmunol.169.9.4861. [DOI] [PubMed] [Google Scholar]
- 10.Rutledge BJ, Rayburn H, Rosenberg R, North RJ, Glaude RP, Corless CL, Rollins BJ. High level monocyte chemoattractant protein-1 expression in transgenic mice increases their susceptibility to intracellular pathogens. J Immunol. 1995;155:4838–4843. [PubMed] [Google Scholar]
- 11.Bradley K, McConnell-Breui S, Crystal RG. Collagen in the human lung - Quantitation of rates of synthesis and partial characterization. J Clin Invest. 1975;55:543–550. doi: 10.1172/JCI107961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JC, Wysocki A, Tchou-Wong KM, Moskowitz N, Zhang Y, Rom WN. Effect of Mycobacterium tuberculosis and its components on macrophages and the release of matrix metalloproteinases. Thorax. 1996;51:306–311. doi: 10.1136/thx.51.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedland JS, Shaw TC, Price NM, Dayer JM. Differential regulation of MMP-1/9 and TMP-1 secretion in human monocytes cells in response to Mycobacterium tuberculosis. Matrix Biol. 2002;21:103–110. doi: 10.1016/s0945-053x(01)00175-5. [DOI] [PubMed] [Google Scholar]
- 14.Hoheisel GU, Sack U, Hui DS, Lai F, Chan KS, Choi CH, et al. Immunochemical localization of matrix metalloproteinases (MMP) and tissue inhibitors of metalloproteinases (TIMP) in tuberculosis pleuritis. Pneumologie. 2004;58:305–308. doi: 10.1055/s-2004-818452. [DOI] [PubMed] [Google Scholar]
- 15.Hoheisel GU, Sack U, Hui DS, Huse K, Chan KS, Chan KK, et al. Occurrence of matrix metalloproteinases and tissue inhibitors of metalloproteinases in tuberculosis pleuritis. Tuberculosis. 2001;81:203–209. doi: 10.1054/tube.2000.0276. [DOI] [PubMed] [Google Scholar]
- 16.Iglesias D, Alegre J, Aleman C, Ruiz E, Soriano T, Armadans LI, et al. Metalloproteinases and tissue inhibitors of metalloproteinases in exudative pleural effusions. Eur Respir J. 2005;25:104–109. doi: 10.1183/09031936.04.00010504. [DOI] [PubMed] [Google Scholar]
- 17.Elkington P, Shiomi T, Breen R, Nuttall RK, Ugarte-Gil CA, Walker NF, et al. MMP-1 drives immunopathology in human and transgenic mice. J Clin Invest. 2011;121(5):1827–1833. doi: 10.1172/JCI45666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright EK, Page SH, Barber SA, Clements JE. Prep1/Pbx2 complexes regulate CCL2 expression through the -2578 guanine polymorphism. Genes Immun. 2008;9:419–430. doi: 10.1038/gene.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page SH, Wright EK, Gama L, Clements JE. Regulation of CCL2 expression by upstream TALE homeodomain protein-binding site that synergizes with the site created by the A-2578G SNP. Plos One. 2011;6:e22052. doi: 10.1371/journal.pone.0022052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rovin BH, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun. 1999;259:344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 21.Wyatt CA, Coon CI, Gibson JJ, Brinckerhoff CE. Potential for the 2G single nucleotide polymorphism in the promoter of matrix metalloproteinase to enhance gene expression in normal stromal cells. Cancer Res. 2002;62:7200–7202. [PubMed] [Google Scholar]
- 22.Reutter JL, Mitchell TI, Buttice G, Meyers J, Gusella JF, Ozelius LJ, Brinkckerhoff CE. A single nucleotide polymorphism in the matrix metalloproteinase promoter creates an Ets binding site and augments transcription. Cancer Res. 1998;58:5321–5325. [PubMed] [Google Scholar]
- 23.Wejse C, Gustafson P, Nielsen J, Gomes VF, Aaby P, Andersen PL, Sodemann M. Signs and symptoms from tuberculosis patients in a low-resource setting have predictive value and may be used to assess clinical course. Scand J Infect Dis. 2008;40:111–120. doi: 10.1080/00365540701558698. [DOI] [PubMed] [Google Scholar]
- 24.Raaby L, Bendix-Struve M, Nielsen J, Wejse C. Inter-observer variation of the Bandim TB-score. Scand J Infect Dis. 2009;41:220–223. doi: 10.1080/00365540802684070. [DOI] [PubMed] [Google Scholar]
- 25.Harrison AC. Guidelines for tuberculosis control in New Zealand. Publishers: New Zealand Ministry of Health; 2003. Clinical investigation and assessment of tuberculosis. [Google Scholar]
- 26.Gie R. Diagnostic atlas of intrathoracic tuberculosis in children – A guide for low income countries. Publishers: International Union Against Tuberculosis and Lung Disease; 2003. [Google Scholar]
- 27.Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculosis infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott HM, Flynn JL. Mycobacterium tuberculosis in chemokine receptor 2-deficient mice: influence of dose on disease progression. Infect Immun. 2002;70:5946–5954. doi: 10.1128/IAI.70.11.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters W, Cyster JG, Mack M, Schlondorff D, Wolf AJ, Ernst AJ, Charo IF. CCR2-dependent trafficking of F4/80dim macrophages and CD11cdim/intermediate dendritic cells is crucial for T cell recruitment to lungs infected with Mycobacterium tuberculosis. J Immunol. 2004;172:7647–7653. doi: 10.4049/jimmunol.172.12.7647. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt E, Meuret G, Stix L. Monocyte recruitment in tuberculosis and sarcoidosis. Br J Haematol. 1977;35:11–17. doi: 10.1111/j.1365-2141.1977.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 31.Basaraba RJ. Cell-mediated immune responses in tuberculosis. Ann Rev Immunol. 2008;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charo IF, Myers SJ, Herman A, Franci C, Connolly AJ, Coughlin SR. Molecular and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc Natl Acad USA. 1994;91:2752–2756. doi: 10.1073/pnas.91.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boire S, Covic L, Agarvai A, Jacques S, Sherifi S, Kuliopulos A. PAR1 is a matrix metalloproteinase-1 receptor that promotes invasion and tumorogenesis of breast cancer cells. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Pforte A, Schiessler A, Gais P, Beer B, Ehlers M, Schutt C, Ziegler-Heitbrock JW. Expression of CD14 correlates with lung function impairment in pulmonary sarcoidosis. Chest. 1994;105:349–354. doi: 10.1378/chest.105.2.349. [DOI] [PubMed] [Google Scholar]
- 35.Umino T, Skols CM, Pirruccello SJ, Spurzem JR, Rennard SI. Two-colour flow-cytometric analysis of pulmonary alveolar macrophages from smokers. Eur Respir J. 1999;13:894–899. doi: 10.1034/j.1399-3003.1999.13d33.x. [DOI] [PubMed] [Google Scholar]
- 36.Saito N, Pulford KA, Breton-Gorious J, Masse JM, Mason DY, Cramer EM. Ultraestructural localization of the CD68 macrophage-associated antigen in human neutrophils and monocytes. Am J Pathol. 1991;139:1053–1059. [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn JL, Goldsten MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 38.Skold M, Behar SM. Tuberculosis triggers a tissue-dependent program of differentiation and acquisition of effector functions by circulating monocytes. J Immunol. 2008;181(9):6349–6360. doi: 10.4049/jimmunol.181.9.6349. [DOI] [PubMed] [Google Scholar]
- 39.Lavebratt C, Apt AS, Nokonenko BV, Schalling M, Shurr E. Severity of tuberculosis in mice is linked to distal chromosome 3 and proximal chromosome 9. J Infect Dis. 1999;180(1):150–155. doi: 10.1086/314843. [DOI] [PubMed] [Google Scholar]
- 40.Kim M-J, Wainwright HC, Locketz M, Bekker L-G, Walther GB, Dittrich C, et al. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMB0 Mol Med. 2010;2:258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elkington PT, Ugarte-Gil CA, Friendland JS. Matrix metalloproteinases in tuberculosis. Eur Respir J. 2011;38:456–464. doi: 10.1183/09031936.00015411. [DOI] [PubMed] [Google Scholar]
- 42.Werry TD, Wilkinson GF, Willars GB. Mechanisms of cross-talk between G-protein-coupled receptors resulting in enhanced release of intracellular Ca2+ Biochem J. 2003;374:281–296. doi: 10.1042/BJ20030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng WX, Mokrousov I, Wang BB, Nelson N, Jiao WW, Wang J, et al. Tag SNP polymorphism of CCL2 and its role in clinical tuberculosis in Han Chinese pediatric population. PLoS One. 2011;6:e14652. doi: 10.1371/journal.pone.0014652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.http://www.who.int
- 45.http://www.minsa.gob.pe
- 46.WHO Working Group. Use and interpretation of anthropometric indicators of nutritional status. Bull World Health Organ. 1986;64:929. [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey BE, Ferro-Luzzi A. Use of body mass index of adults in assessing individual and community nutritional status. Bull World Health Organ. 1995;3:673. [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh FY, Bloch DA, Larsen MD. A simple method of sample size calculation for linear and logistic regression. Stat Med. 1998;17:1623–1624. doi: 10.1002/(sici)1097-0258(19980730)17:14<1623::aid-sim871>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 49.Viera AJ, Garrett JM. Understanding interobserver agreement: The Kappa Statistic. Fam Med. 2005;37:360–363. [PubMed] [Google Scholar]
- 50.Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We used double IHC analysis of PAR-1(blue) and CD68 (red) in paraffin-embedded diseased lung. Representative images from three of the six TB cases that underwent surgery to remove damaged tissue are shown. In section A we show the negative control (irrelevant antibodies isotype control). Images were acquired at 100X total magnification.
A model of MCP-1/MMP-1/PAR-1 pathway-caused tissue damage in TB cases carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G. PHASE I: After exposure to M. tuberculosis, ganuloma macrophages express MCP-1 and MMP-1. Persons carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G produce excessive amounts of these factors. PHASE II: MCP-1 activates CCR2 receptors and potentiates M. tuberculosis-induced MMP-1 production. MMP-1 targets fibrilar collagen in the granulomatous matrix, resulting in matrix degradation. Persons carrying the two-locus genotype -2518 MCP-1 GG -1607 MMP-1 2G/2G experience matrix degradation, an event that, in normal individuals, is not as intense and is part of the normal healing process. PHASE III: The excess of MMP-1 intensifies the granulomatous matrix degradation and activates the PAR-1 receptors. PHASE IV: Activation of PAR-1 increases MCP-1, MMP-1, and induces secretion of MMP-9. MMP-9 may contribute to MCP-1-mediated monocyte recruitment and further degradation of the granulomatous matrix. PAR-1 activation is a key event in the amplification of the inflammatory process (a hyper-inflammatory state) and in the induction of extensive tissue damage.