Abstract
Hearing and balance use hair cells in the inner ear to transform mechanical stimuli into electrical signals1. Mechanical force from sound waves or head movements is conveyed to hair-cell transduction channels by tip links2,3, fine filaments formed by two atypical cadherins: protocadherin-15 and cadherin-234,5. These two proteins are products of deafness genes6–10 and feature long extracellular domains that interact tip-to-tip5,11 in a Ca2+-dependent manner. However, the molecular architecture of the complex is unknown. Here we combine crystallography, molecular dynamics simulations, and binding experiments to characterize the cadherin-23 and protocadherin-15 bond. We find a unique cadherin interaction mechanism, with the two most N-terminal cadherin repeats (EC1+2) of each protein interacting to form an overlapped, antiparallel heterodimer. Simulations predict that this tip-link bond is mechanically strong enough to resist forces in hair cells. In addition, the complex becomes unstable upon Ca2+ removal due to increased flexure of Ca2+-free cadherin repeats. Finally, we use structures and biochemical measurements to understand molecular mechanisms by which deafness mutations disrupt tip-link function. Overall, our results shed light on the molecular mechanics of hair-cell sensory transduction and on new interaction mechanisms for cadherins, a large protein family implicated in tissue and organ morphogenesis12,13, neural connectivity14, and cancer15.
Hair cell mechanotransduction happens within each bundle of stereocilia (Fig. 1a), which is deflected by mechanical stimulation1. Deflection results in tension applied to tip links, protein filaments linking the tip of each stereocilium to its tallest neighbour2,3. The tip links, acting in series with an elastic “gating spring,” pull open transduction channels1. Recently, protocadherin-15 and cadherin-23, which feature exceptionally long extracellular domains with 11 and 27 extracellular cadherin (EC) repeats (Fig. 1b), were shown to form the tip link4,5. To elucidate the tip-link heterophilic molecular bond between protocadherin-15 and cadherin-23, we determined the crystallographic structure of their interacting N-termini (Fig. 1c; results summary in Supplementary Fig. 1). Size exclusion chromatography (SEC) of co-refolded protein fragments comprising the EC1+2 repeats of protocadherin-15 and EC1+2 of cadherin-23 (referred to here as pcdh-15 and cdh-23, respectively) showed a monodisperse peak with the two protein fragments interacting in solution (Supplementary Fig. 2a). The complex crystallized in two packing arrangements and two independent models were fully refined (S1a–S1b and S2, respectively; Supplementary Table 1 and Supplementary Fig. 3).
Figure 1.
Structure of tip-link protocadherin-15 bound to cadherin-23. a, Hair-cell stereocilia bundle. A tip-link filament extends from the tip of each stereocilium to the side of its tallest neighbour. b, The tip link formed by a protocadherin-15 parallel dimer interacting tip-to-tip with a cadherin-23 parallel dimer5. These proteins feature 11 and 27 extracellular cadherin (EC) repeats, respectively. Inset shows possible arrangement at the junction. c, Ribbon diagram of protocadherin-15 EC1+2 (pcdh-15; purple) bound to cadherin-23 EC1+2 (cdh-23; blue) with Ca2+ ions as green spheres. Arrowheads indicate pcdh-15’s RGGPP loop and cdh-23’s 310 helix. Residues R113, C11, and C99 of pcdh-15 are shown in stick representation. d, Detail of disulfide bond C11-C99 and isoform-dependent residues D4-Y8 at the pcdh-15 N-terminus. e, Detail of Ca2+-binding sites 1, 2, and 3 at the pcdh-15 linker. Protein backbone and sidechains are in cartoon and stick representations, respectively. f, Surface representation of pcdh-15 (purple and pink) and cdh-23 (blue and cyan) as in (c). g, Pcdh-15 and cdh-23 interaction surfaces exposed with interfacing residues labeled.
The structures show that pcdh-15 and cdh-23 form an overlapping and antiparallel heterodimer (pcdh-15+cdh-23; Figs 1c, f and g). The interaction resembles an “extended handshake” and involves repeats EC1 and EC2 from both proteins. The overall fold of pcdh-15 and cdh-23 matched the well-known Greek-key motif of classical cadherins (Supplementary Fig. 4). As expected, three Ca2+ ions are found in a canonical arrangement (sites 1, 2, and 3) at the linker region between repeats EC1 and EC2 of each protein (Fig. 1c,e). However, several novel structural features within pcdh-15 and cdh-23 enable the handshake interaction.
Pcdh-15 has an elongated N-terminus clamped by an intramolecular disulfide bond (Fig. 1d), which is followed by a conserved RXGPP motif that forms a rigid and bulky loop (Supplementary Fig. 5a&b). This RXGPP loop, within strand A of protocadherin-15 EC1, is tucked against the narrow wrist of the adjacent cdh-23’s linker region (Fig. 1c). Similarly, cdh-23 has an elongated N-terminus, stabilized at the tip by Ca2+-binding site 016,17, which is followed by a bulky 310 helix within strand A that sits at the narrow wrist of the adjacent pcdh-15’s linker (Fig. 1c). Thus, the pcdh-15+cdh-23 interface exploits unique structural protrusions within strand A of each EC1 repeat, which in turn are stabilized by a disulfide bond and a Ca2+-binding site and lead to two main areas of interaction described below.
The pcdh-15+cdh-23 heterophilic interface differs from the strand-exchanged or X-dimer homophilic interfaces of classical cadherins18–20. Furthermore, this interface is not directly mediated by Ca2+ as previously speculated16,17. However, several factors indicate that this is a robust interface. The buried surface area is ~1,000 Å2 per protomer (see Supplementary Tables 1&2), similar to that of classical cadherin interfaces (850 Å2 and 1,270 Å2 for type I and type II, respectively). The interface is amphiphilic (Supplementary Fig. 6); all its residues are highly conserved in mouse, human, and chicken homologues and none are predicted to be glycosylated (Fig. 1g and Supplementary Fig. 7). Finally, the same interface was observed in two different crystal lattices, so it is unlikely to represent unphysiological crystal packing interactions.
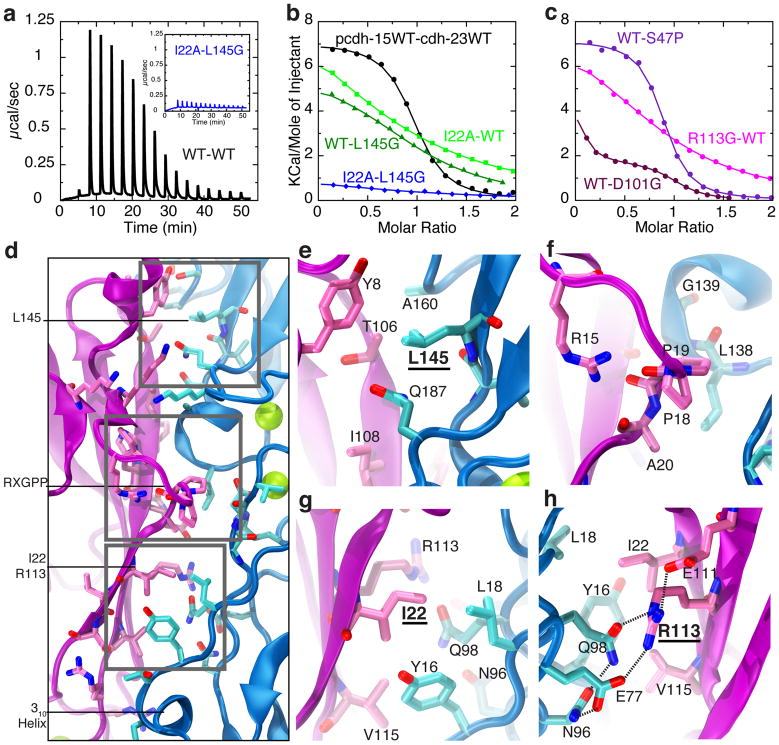
To further validate the pcdh-15+cdh-23 interface we used isothermal titration calorimetry (ITC) and site-directed mutagenesis. The stoichiometry of the wild-type complex was determined to be N = 0.88 ± 0.1, consistent with the one-to-one crystallographic arrangement (Fig. 1). The measured dissociation constant was KD = 2.9 ± 0.4 μM (T = 10 °C, ΔH = 7084 ± 233 cal/mol, ΔS = 50.4 ± 1.1 cal/mol/deg, two trials; Fig. 2a,b and Supplementary Discussion). The tip link is thought to be a heterotetramer of parallel protocadherin-15 and cadherin-23 dimers5 (Fig. 1b). However, our biochemical and crystallographic data do not show homophilic binding of cdh-23 or pcdh-15, suggesting that parallel dimerization is mediated by repeats other than EC1+2. If so, the binding affinity for the heterotetramer is expected to be significantly higher21.
Figure 2.
Pcdh-15+cdh-23 complex formation probed using isothermal titration calorimetry (ITC) and site-directed mutagenesis. a, Raw power vs. time data for pcdh-15 (111 μM) titrated with cdh-23 (1.1 mM) at 10°C (black, WT-WT). Inset shows raw data (blue) for pcdh-15I22A (114 μM) titrated with cdh-23L145G (1.2 mM). b–c, Change in molar enthalpy for pcdh-15 titrated with cdh-23 (black); pcdh-15I22A with cdh-23 (light green); pcdh-15 with cdh-23L145G (dark green); pcdh-15I22A with cdh-23L145G (blue); pcdh-15 with cdh-23S47P (violet); pcdh-15R113G with cdh-23 (magenta); and pcdh-15 with cdh-23D101G (indigo; concentrations in Supplementary Fig. 8). Sigmoidal isothermals were observed only for pcdh-15+cdh-23 and pcdh-15+cdh-23S47P. d–h, Details of pcdh-15+cdh-23 interface, highlighting residue L145 (e), the RXGPP loop (f), and residues I22 (g) and R113 (h). Panel (h) is a 180°-rotated version of panel (g). Protein backbone and interfacing residues (as identified by PISA) are in purple/pink for pcdh-15 and blue/cyan for cdh-23.
The “extended handshake” features two main areas of interaction. The first one is located at and above pcdh-15’s RXGPP loop and centers on Y8, P19 and I108 in pcdh-15, and L145 and Q187 in cdh-23 (Figs 1g, 2d–f). The second, located between the RXGPP loop and cdh-23’s 310 helix, involves I22, R113 and V115 in pcdh-15 along with Y16 and Q98 in cdh-23 (Figs. 1g &2d,g). To test the two interaction areas we introduced mutations predicted to disrupt them: I22A in pcdh-15 (pcdh-15I22A) and L145G in cdh-23 (cdh-23L145G; Fig. 2d,e,g). SEC confirmed proper folding and structural integrity of the mutant proteins. ITC experiments, testing binding with either one or both mutant partners, showed decreased affinity for each single-mutant complex (pcdh-15I22A+cdh-23, KD > 100μM; pcdh-15+cdh-23L145G, KD > 30 μM), and complete lack of interaction for the double-mutant complex (pcdh-15I22A+cdh-23L145G; Fig. 2a,b,d,e,g and Supplementary Figs 2, 8&9). Likewise, SEC of pcdh-15+cadherin-23 EC1 repeats alone did not show complex formation (Supplementary Fig. 2b). Taken together, these results show that the interface observed in the crystals is consistent with the interface observed in solution.
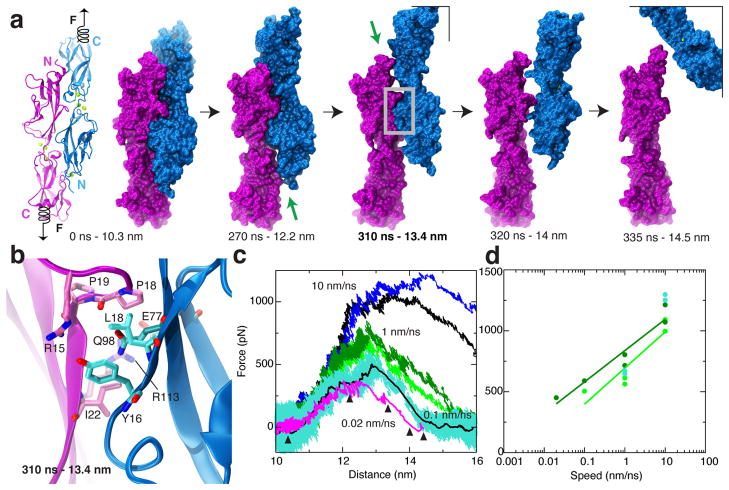
Does the interface have the properties expected for a tip-link bond? Tip links are regularly subjected to (and must withstand) forces ranging from 10 to 100 pN, both in vivo and in physiological experiments. While SEC and ITC experiments provide a characterization of the bond in thermodynamic equilibrium, they do not probe its response to mechanical force. To determine whether the pcdh-15+cdh-23 interface is mechanically strong we used steered molecular dynamics (SMD) simulations (Methods and Supplementary Table 3). Force was applied to the C-terminus of each protomer to induce complex dissociation (Fig. 3a). In all SMD simulations of pcdh-15+cdh-23 with Ca2+, unbinding was observed without unfolding of repeats. Partial rupture of the binding interface at contacts formed by residues pcdh-15T106 – cdh-23L145 and pcdh-15R84 – cdh-23N96 was followed by sliding of the 310 helix in strand A of cadherin-23 EC1 over the pcdh-15 RXGPP loop, and simultaneous rupture of a salt bridge between pcdh-15R113 and cdh-23E77 (Fig. 3b and Supplementary Discussion, Figs 10, 11, and Movies I&II). Simulations performed using different stretching speeds, initial conditions, and thermodynamic ensembles revealed a similar scenario with at least one force peak of > 400 pN associated with complex unbinding (Fig. 3c,d and Supplementary Figs 10&11).
Figure 3.
Mechanical strength of the pcdh-15+cdh-23 complex probed by steered molecular dynamics (SMD) simulations. a, Snapshots of pcdh-15 (purple) and cdh-23 (blue) unbinding during simulation SNA7 (Supplementary Table 3). The complex is shown in both cartoon and surface representations at the beginning, and in surface representation at indicated time points. Force was applied to the C-termini of both protomers (Supplementary Movies I&II). Green arrows point to broken interfaces. b, Region of gray box in panel (a), showing interacting residues during unbinding. c, Force applied to one C-terminus versus distance between C-termini ends of pcdh-15 and cdh-23. Different traces correspond to independent simulations performed at stretching speeds of 10 (blue and black), 1 (light and dark green), 0.1 (cyan, 1-ns running average shown in black), and 0.02 nm/ns (magenta, 1-ns running average). Snapshots in (a) are indicated by arrowheads. d, Maximum force-peak values vs. stretching speed for unbinding simulations of pcdh-15+cdh-23 started after a 1-ns or 1-μs equilibration (light green, SN2 to SN6; dark green, SNA2 to SNA7; cyan, SN10 to SN13). Simulations SN2-SN6 and SNA2-SNA7 used the S1b structure and SN10-SN13 used S1a; unbinding forces for all three sets were equivalent.
Unbinding forces followed the well-known dependence on stretching speed22, with less force required when the stretch was slower. The slowest speed used in our simulations matched the measured velocity of the basilar membrane induced by loud sound23 as well as speeds of mechanical stimulators used in ex-vivo electrophysiological experiments24 (see Supplementary Discussion). In all our simulations the pcdh-15+cdh-23 interface was stronger than that of the classical C-cadherin interface pulled under identical conditions (Supplementary Fig. 12). Furthermore, the predicted force required to unbind parallel complexes was almost double that required to unbind a single pcdh-15+cdh-23 complex (Supplementary Figs 12b,c), which may correspond to the actual force that heterotetrameric tip links can withstand in vivo before rupture due to large mechanical stimuli, e.g., loud sound.
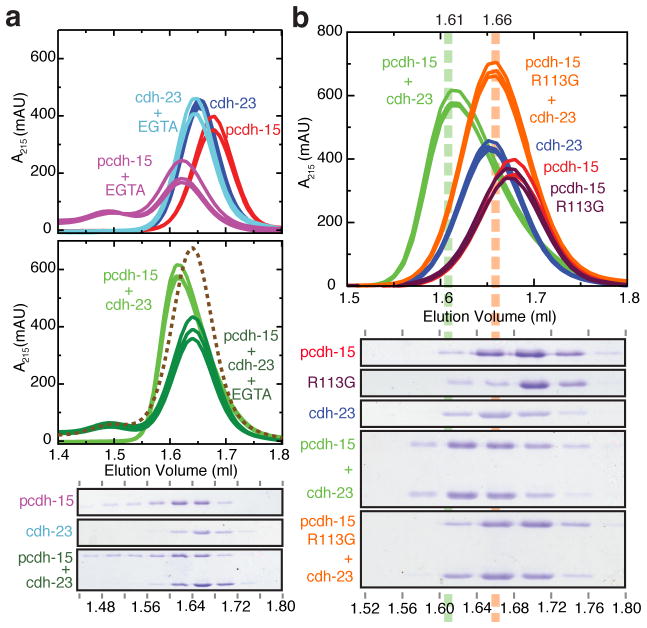
The integrity of tip links in hair cells is Ca2+-dependent3, yet Ca2+ does not directly participate in the binding interface. We therefore determined whether the pcdh-15+cdh-23 complex is disrupted by Ca2+ removal using SEC experiments in the presence of EGTA (Fig. 4a). The cdh-23 fragment by itself did not show changes in its elution volume, while pcdh-15 in the presence of EGTA showed a shifted elution trace. Importantly, the pcdh-15+cdh-23 complex was disrupted by addition of EGTA: its elution trace did not match that in the presence of Ca2+, but rather corresponded to the summation of the individual components’ traces without the shift in elution volume that indicates interaction. The complex used for crystallization is thus Ca2+-dependent at equilibrium, in line with what is known about the full-length protocadherin-15 and cadherin-23 proteins and the tip link3,5. To understand the basis of the Ca2+ dependence, we performed microsecond-long MD simulations of the Ca2+-free complex. These suggest a molecular mechanism: removal of Ca2+ is predicted to cause dissociation indirectly, through entropic stress (see Supplementary Discussion, Supplementary Figs 13 & 14, and Movies III, IV & V), as observed for other cadherins25.
Figure 4.
Pcdh-15+cdh-23 complex formation, its Ca2+-dependence, and the role of the deafness mutation R113G, probed using analytical size exclusion chromatography (SEC). Individual traces represent independent experiments. a, top, SEC traces for pcdh-15 and cdh-23 with Ca2+ (red and blue) or with 5 mM EGTA (purple and cyan). A shift upon Ca2+ removal by EGTA was observed for pcdh-15 (purple vs. red curves). middle, SEC traces for pcdh-15+cdh-23 in the presence of Ca2+ (light green) or 5 mM EGTA (dark green). The summation of a purple and a cyan curve from above is shown as a dashed line. The EGTA-treated complex behaved as the sum of its EGTA-treated components, indicating Ca2+-dependent complex formation. bottom, Coomassie-stained SDS-PAGE of eluted fractions from EGTA-treated proteins. b, top, SEC traces for mutant pcdh-15R113G alone (maroon), and mixed with cdh-23 (orange). Wild-type proteins from (a) are shown for comparison. bottom, Coomassie-stained SDS-PAGE of eluted fractions aligned to chromatogram. A reproducible shift in elution volume was observed for the wild-type (green) but not for the mutant mixture (pcdh-15R113G+cdh-23; orange). The shifted peak (1.61 ml) contained both proteins (1.56 to 1.64 ml fractions).
Over 40 missense mutations associated with deafness in humans or mice target the extracellular domains of protocadherin-15 and cadherin-23. Most modify Ca2+-binding residues26. Three human mutations causing inherited deafness (pcdh-15D157G27, cdh-23D101G28, and pcdh-15R113G27) and one mouse mutation that accelerates progressive hearing loss (cdh-23S47P29) are located within the crystallized pcdh-15+cdh-23 complex (Supplementary Figs 1c & 15). Our data provide a structural context to interpret their effect on tip link function. We also constructed all four mutants to test formation of heterophilic complexes in vitro with ITC and SEC, finding that they each affect the pcdh-15+cdh-23 complex in different ways. Cdh-23D101G, cdh-23S47P, and pcdh-15R113G refolded well as assessed by SEC, whereas pcdh-15D157G did not and its analysis was not possible. The D101G and S47P cdh-23 fragments crystallized in complex with pcdh-15 and show only minor changes in the interface and in binding (Fig. 2c, Supplementary Discussion and Supplementary Fig. 16). On the other hand, pcdh-15R113G showed impaired binding to cdh-23.
Residue pcdh-15R113 is of particular interest because it is at the interface between pcdh-15 and cdh-23 (Fig. 2h). Mutation R113G, causing human non-syndromic deafness DFNB2327, eliminates the long arginine sidechain that flanks the hydrophobic core of this interface and disrupts its integrated hydrogen-bond network. SEC of either co-refolded or independently refolded proteins showed no evidence of pcdh-15R113G+cdh-23 complex formation (Fig. 4b and Supplementary Fig. 2). Furthermore, ITC experiments show impaired binding and indicate an estimated KD at least an order of magnitude larger than that measured for wild-type pcdh-15+cdh-23 (>20 μM, Fig. 2c and Supplementary Fig. 8). In other work, R113G impaired binding of full-length protocadherin-15 and cadherin-23 in vitro5, as well as binding of protein fragments to hair-cell tip links ex vivo11. Together, these observations help validate the interface observed in our crystal structure and indicate that this mutation causes deafness by directly interfering with binding between cadherin-23 and protocadherin-15. Residual interactions detected in ITC experiments may explain why vestibular function is not affected in human subjects carrying this mutation27.
In summary (Supplementary Fig. 1), the pcdh-15+cdh-23 structure provides the first view of a heterophilic cadherin complex, revealing a novel “extended handshake” interface that simulations predict to be mechanically stronger than required to resist forces produced by moderate sound. The structure helps explain the Ca2+ sensitivity of the tip link, and the etiology of certain inherited deafnesses. For other cadherins, both the existence of heterophilic cadherin bonds and the possibility of interdigitation have been debated20. While protocadherin-15 and cadherin-23 are rather specialized members of the cadherin family, the overlapping heterophilic complex formed by these molecules suggests structural determinants that could also favor this type of interactions in related members of the cadherin family, such as the fat3 and fat4 cadherins that control neuronal morphology and morphogenesis30 (Supplementary Discussion).
Online Methods
Cloning, expression, and purification of protocadherin-15 and cadherin-23 repeats
Clones of mouse cadherin-23 repeats EC1 and EC1+2 were previously described16. Numbering corresponds to mouse cadherin-23 and protocadherin-15 without their signal sequences. Mouse protocadherin-15 EC1+2 comprising residues Q1 to D233 (Q27 to D259 in NP_001136218.1) was subcloned into the NdeI and XhoI sites of the vector pET21a (C-terminal His-tag). The signal sequence was replaced by a methionine at position 0. The R113G, D157G, and I22A mutations in pcdh-15, as well as the L145G and S47P mutations in cdh-23 were generated using the QuikChange Lightning mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing. Pcdh-15, pcdh-15R113G, pcdh-15D157G, pcdh-15I22A, cdh-23, cdh-23D101G, cdh-23S47P, and cadherin-23 EC1 were expressed independently in BL21CodonPlus(DE3)-RIPL (Stratagene) cultured in LB and induced at OD600=0.6 with 100 μM IPTG at room temperature for ~16 hrs. Cells were lysed by sonication in denaturing buffer (20 mM HEPES at pH 7.5, 6 M guanidine hydrochloride [GuHCl], 10 mM CaCl2, 20 mM imidazole at pH 7.0). The cleared lysates were loaded onto Ni-sepharose (GE Healthcare) and eluted with denaturing buffer supplemented with 500 mM imidazole. Wild-type and mutant Pcdh-15 protein fragments were reduced by adding 1 mM DTT and incubating at 37°C for ~30 min. Purified and denatured samples were mixed (pcdh-15+cdh-23, pcdh-15R113G+cdh-23, pcdh-15D157G+cdh-23, pcdh-15+cdh-23D101G, pcdh-15+cdh-23S47P, pcdh-15+cadherin-23 EC1, pcdh-15R113G+cadherin-23 EC1, and pcdh-15I22A+cadherin-23 EC1) and co-refolded in six steps at 4°C using MWCO 2000 membranes (protocol adapted from ref 31). First, the mixture was dialyzed for 24 hrs against D buffer (20 mM Tris HCl pH 8.0, 10 mM CaCl2) with 6 M GuHCl, followed by two 24-hr dialyses against D buffer with 3 and 2 M GuHCl, respectively. The last three steps consisted of 12-hr dialyses against D buffer with decreasing GuHCl concentration (1, 0.5, and 0 M) plus 400 mM L-Arg and 375 μM GSSG. Refolded protein used for crystallization was further purified in two consecutive size-exclusion chromatography (SEC) experiments with relevant fractions on a Superdex75 column (GE Healthcare) in 20 mM TrisHCl pH 8.0, 200 mM KCl, with 10 and 1 mM CaCl2, respectively. Predicted and apparent molecular weights (SDS-PAGE) for pcdh-15 and cdh-23 fragments were 27.5/37 kDa and 23.8/25 kDa, respectively. Identity of pcdh-15 was confirmed through N-terminal sequencing.
Crystallization, data collection, and structure determination
Crystals were grown by vapor diffusion at 4°C by mixing equal volumes of protein (5–10 mg/ml) and reservoir solution of (0.1 M MES pH 6.5, 8% w/v PEG 8000) for S1a, (0.1 M MES pH 6.5, 15% w/v PEG 550 MME) for S1b, (0.1 M HEPES pH 7.5, 10% w/v PEG 8000) for S2, (0.1M MES pH 6.5, 8% w/v PEG 20000) for S3, and (0.1M MES pH 6.5, 12% PEG 4000) for S4. All crystals were cryoprotected in reservoir solution plus 25% glycerol and cryo-cooled in N2. X-ray diffraction data were collected as indicated in Supplementary Tables 1&2 and processed with HKL200032. Structures were determined by molecular replacement using the cdh-23 structure (pdb code 2wcp) and a pcdh-15 homology model (based on the same cdh-23 structure) as search models with Phaser33. Model building was done using COOT34 and restrained TLS refinement using REFMAC535. The final models include residues M0 to E207 (cdh-23) and M0 to H237 (pcdh-15) for S1a, M0 to E207 (cdh-23) and Q1 to H236 (pcdh-15) for S1b, M0 to E207 (cdh-23 A and B) and Q1/Y2 to H237 (pcdh-15 C and D) for S2, M0 to D205 (cdh-23D101G and cdh-23S47P) and M0 to H236/H237 (pcdh-15) for S3 and S4. Data collection and refinement statistics are provided in Supplementary Tables 1&2. Coordinates have been deposited in the Protein Data Bank with entry codes 4apx (S1a), 4axw (S1b), 4aq8 (S2), 4aqa (S3), and 4aqe (S4).
Isothermal titration calorimetry
ITC experiments were carried out using a MicroCal ITC200 calorimeter and designed following guidelines from Thomson and Ladbury36 and the manufacturer’s manual. Wild type, I22A and R113G pcdh-15 fragments were co-refolded with cadherin-23 EC1 as described above, and subsequent SEC was performed on a Superdex75 column with 20 mM TrisHCl pH 8.0, 300 mM NaCl, and 1 mM CaCl2. Fractions with pure pcdh-15, pcdh-15I22A, and pcdh-15R113G were collected, concentrated (to 100 to 150 μM) and placed in the calorimeter’s sample cell. Similarly, wild type, L145G, D101G, and S47P cdh-23 fragments were refolded as previously described16, concentrated (to 1 to 2 mM), buffer-matched through SEC, and then used as titrants. Protein concentrations were measured using samples’ absorbances at 280 nm and theoretical extinction coefficients. All experiments were performed at 10°C, as no signal was detected and the protein tended to be unstable at room temperature. In a typical experiment, an initial 0.5-μl injection (disregarded in analyses) was followed by fifteen 2.44-μl injections of titrant (Δt = 3 min). Experiments and controls with each combination of fragments were repeated at least once (Supplementary Fig. 8). Fittings and binding constants were obtained using the MicroCal Software, a model with one set of sites, and blank-subtracted data. Fitting parameters are reported for saturating sigmoidal curves as the average±s.d. of two independent experiments. Attempts to obtain saturating sigmoidal curves and precise KDs for mutant proteins were hampered by aggregation of protein samples at the high concentration required (>2 mM).
Analytical size exclusion chromatography
SEC of co-refolded proteins (pcdh-15+cdh-23, pcdh-15R113G+cdh-23, pcdh-15D157G+cdh-23, pcdh-15+cdh-23D101G, pcdh-15+cdh-23S47P, pcdh-15+cadherin-23 EC1, and pcdh-15R113G+cadherin-23 EC1) was performed on a Superdex75 16/60 column with 20 mM TrisHCl pH 8.0, 300 mM NaCl and 1 mM CaCl2 (Supplementary Fig. 2). Fractions with pure cdh-23 (excess from pcdh-15+cdh-23), pcdh-15 (from pcdh-15+cadherin-23 EC1), and pcdh-15R113G (from pcdh-15R113G+cadherin-23 EC1) were collected, concentrated (~0.5 mg/ml), and used for subsequent analytical SEC (Fig 4a,b) on a Superdex200 PC3.2/3.0 column equilibrated with the same buffer (plus 5 mM EGTA when indicated). Experiments were performed at 4°C using a 10 μl loop and a 50 μl/min flow rate on an AKTAmicro system equipped with a fused silica capillary tubing for collection of 40 μl fractions. EGTA was added to individual or pre-mixed concentrated samples followed by a 1 hr mild shaking prior to SEC.
Simulated systems
The psfgen, solvate, and autoionize VMD37 plug-ins were used to build all systems (Supplementary Table 3) as previously reported16. Most of the pcdh-15+cdh-23 complex simulations used structure S1b (which we determined first) except simulations SN9 to SN13 (which used higher resolution structure S1a; Supplementary Tables 1 and 3). Structures with non-native N- and C-terminal tails were modified back to native sequences. Systems without bound Ca2+ were prepared by replacing Ca2+ atoms with K+.
Molecular dynamics simulations using NAMD
MD simulations were performed using NAMD 2.738, the CHARMM27 force field for proteins with CMAP correction39,40, and the TIP3P model for water. Simulation parameters were as in ref 16, except for simulations SNA7 and SNC6, in which a multiple-time-step scheme was used with electrostatic interactions evaluated every other time step. Parameters for Ca2+ were from ref 41. Each system was energy-minimized, then equilibrated in the constant number, pressure, and temperature ensemble (NpT), and the resulting state used to perform subsequent Anton or SMD simulations42. All simulations used T=310 K. Coordinates of all atoms were saved for analysis every picosecond. Constant velocity stretching simulations used the SMD method and NAMD Tcl Forces interface16,43.
Molecular dynamics simulations using Anton
Anton is a massively parallel special-purpose machine for molecular dynamics simulations44. Systems pre-equilibrated in NAMD (1.1 ns, T=310 K) were converted to the Anton-compatible Maestro format using the convertNAMDtoMaestro.py script provided by NRBSC/PSC. Anton and NAMD simulations used the same force field. Hydrogen atoms were constrained with SHAKE. Restraints were applied to Cα atoms of residues 121, 173, and 205 of cdh-23 to avoid rotation of the complex and contact between periodic images. A multiple-time-step scheme was used with interactions evaluated every 2.5 fs, except for non-bonded interactions computed every other time step. A set of cutoff radii and parameters for evaluation of electrostatic forces was automatically generated for each simulated system using the guess_anton_config script. Simulations were performed in either the constant number, volume, and temperature ensemble (NVT) using the Nose-Hoover thermostat, or the constant number, pressure, and temperature ensemble (NpT) using the Berendsen thermostat/barostat. Center of mass motion was removed. Coordinates of all atoms were saved for analysis every 50 (NpT) or 200 (NVT) picoseconds. Anton simulations are restricted in size (<120,000 atoms) and cannot incorporate SMD-like forces, hence the use of complementary NAMD simulations.
Analysis tools
The Protein Interfaces, Surfaces and Assemblies (PISA) server was used to analyze complex interfaces45 and identify residues shown in Figs 1g and 2d. The VMD “measure SASA” command was used to determine interface area throughout simulations with a probe radius of 1.4 Å. Interface area was defined as the difference in total solvent-accessible surface areas for each isolated protomer and for the complex divided by two. Glycosylation sites were predicted using OGPET46, NetNGlyc47 and NetOGlyc48. Intradomain RMSD and interdomain flexing were analyzed using DynDom49. Regression fits to data points of maximum force peaks versus stretching speeds were performed using a logarithmic expression of the form y = a + b log x. Plots and curve fits were prepared using xmgrace. Molecular images in this paper were created with the molecular graphics program VMD37, except for Supplementary Fig. 5 which used PyMOL (Schrödinger, LLC).
Supplementary Material
Acknowledgments
We thank Bruce Derfler for assistance with mutagenesis, Dr. Victoria D’Souza and her laboratory for advice on calorimetry, and the Corey and Gaudet laboratories for helpful discussions. Full-length cDNAs of cdh23 and pcdh15 used as template for some of our constructs were kindly provided by Dr. U. Müller (TSRI) and Dr. T. B. Friedman (NIH). This work was supported by NIH (R01 DC02281 to D.P.C.; RC2GM093307 to NRBSC/PSC) and NSF through TeraGrid/XSEDE (TRAC MCB080015). Simulations were performed at the NCSA-Abe, NICS-Kraken, TACC-Ranger, and PSC-Anton supercomputers. Use of APS beamlines was supported by NIH award RR-15301 and DOE contract No. DE-AC02-06CH11357. Use of ALS beamline 4.2.2 was supported by DOE contract No. DE-AC02-05CH11231. M.S. was a HHMI Fellow of the Helen Hay Whitney Foundation and D.P.C. is an HHMI Investigator.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
All authors participated in all parts of this study. MS did all experiments and simulations.
Author Information
Atomic coordinates and structure factors for the reported crystals structures have been deposited with the Protein Data Bank under accession codes 4apx (S1a), 4axw (S1b), 4aq8 (S2), 4aqa (S3), and 4aqe (S4). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to D.P.C. (dcorey@hms.harvard.edu) or R.G. (gaudet@mcb.harvard.edu).
References
- 1.Gillespie PG, Müller U. Mechanotransduction by hair cells: Models, molecules, and mechanisms. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickles JO, Comis SD, Osborne MP. Cross-links between stereocilia in the guinea pig organ of corti, and their possible relation to sensory transduction. Hear Res. 1984;15:103–112. doi: 10.1016/0378-5955(84)90041-8. [DOI] [PubMed] [Google Scholar]
- 3.Assad JA, Shepherd G, Corey DP. Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron. 1991;7:985–994. doi: 10.1016/0896-6273(91)90343-x. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed ZM, et al. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J of Neurosci. 2006;26:7022–7034. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 6.Alagramam KN, et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed ZM, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolz H, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 9.Bork JM, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–27. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Palma, et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 11.Lelli A, Kazmierczak P, Kawashima Y, Muller U, Holt JR. Development and regeneration of sensory transduction in auditory hair cells requires functional interaction between cadherin-23 and protocadherin-15. J Neurosci. 2010;30:11259–11269. doi: 10.1523/JNEUROSCI.1949-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 13.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 15.Rouget-Quermalet, et al. Protocadherin 15 (PCDH15): a new secreted isoform and potential marker for NK/T cell lymphomas. Oncogene. 2006;25:2807–2811. doi: 10.1038/sj.onc.1209301. [DOI] [PubMed] [Google Scholar]
- 16.Sotomayor M, Weihofen WA, Gaudet RG, Corey DP. Structural determinants of cadherin-23 function in hearing and deafness. Neuron. 2010;66:85–100. doi: 10.1016/j.neuron.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elledge HM, et al. Structure of te N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members. Proc Natl Acad Sci USA. 2010;107:10708–100712. doi: 10.1073/pnas.1006284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. The International Journal of Biochemistry and Cell Biology. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- 21.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 22.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robles L, Ruggero MA. Mechanics of Mammalian Cochlea. Physiological Reviews. 2001;81:1305–1352. doi: 10.1152/physrev.2001.81.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stauffer EA, Holt JR. Sensory Transduction and Adaptation in Inner and Outer Hair Cells of the Mouse Auditory System. J Neurophysiol. 2007;98:3360–3369. doi: 10.1152/jn.00914.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häussinger D, et al. Calcium-dependent homoassociation of E-cadherin by NMR-spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–839. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 26.de Brouwer APM, et al. Mutations in the calcium-binding motifs of cdh23 and the 35delg mutation in gjb2 cause hearing loss in one family. Hum Genet. 2003;112:156–163. doi: 10.1007/s00439-002-0833-0. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed ZM, et al. Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum Genet. 2008;124:215–223. doi: 10.1007/s00439-008-0543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Astuto LM, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with usher syndrome and nonsyndromic deafness. Am J Hum Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han F, et al. A new mouse mutant of the Cdh23 gene with early-onset hearing loss facilitates evaluation of otoprotection drugs. Pharmacogenomics J. 2012;12:30–44. doi: 10.1038/tpj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deans MR, et al. Control of neuronal morphology by the atypical cadherin fat3. Neuron. 2011;71:820–832. doi: 10.1016/j.neuron.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsumoto K, et al. Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent–application to a human single-chain Fv fragment. J Immmuno Meth. 1998;219:119–129. doi: 10.1016/s0022-1759(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Thomson JA, Ladbury JE. Isothermal Titration Calorimetry: a Tutorial. In: Ladbury John E, Doyle Michael L., editors. Biocalorimetry 2: Applications of Calorimetry in the Biological Sciences. John Wiley & Sons Ltd; 2004. pp. 37–58. [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell A, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 41.Marchand S, Roux B. Molecular dynamics study of calbindin d9k in the apo and singly and doubly calcium-loaded states. Proteins: Struct, Func, Gen. 1998;33:265–284. [PubMed] [Google Scholar]
- 42.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 43.Sotomayor M, Schulten K. The allosteric role of the Ca++ switch in adhesion and elasticity of C-cadherin. Biophys J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–799. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 46.http://ogpet.utep.edu/OGPET/
- 47.http://www.cbs.dtu.dk/services/NetNGlyc/
- 48.http://www.cbs.dtu.dk/services/NetOGlyc/
- 49.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lyzozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.