Abstract
ENOD40, an early nodulin gene, is expressed following inoculation with Rhizobium meliloti or by adding R. meliloti-produced nodulation (Nod) factors or the plant hormone cytokinin to uninoculated roots. We isolated two MsENOD40 clones, designated MsENOD40–1 and MsENOD40–2, with distinct promoters from an alfalfa (Medicago sativa cv Chief) genomic library. The promoters were fused to the reporter gene uidA (gus), and the constructs were introduced into alfalfa. We observed that the MsENOD40–1 construct was expressed almost exclusively under symbiotic conditions. The MsENOD40–2 construct was transcribed under both symbiotic and nonsymbiotic conditions and in nonnodular and nodular tissues. Both MsENOD40 promoter-gus constructs were similarly expressed as nodules developed, and both were expressed in roots treated with 6-benzylaminopurine or purified Nod factor. However, no blue color was detected in nodule-like structures induced by the auxin transport inhibitor N-1-(naphthyl)phthalamic acid on roots of plants containing the MsENOD40–1 promoter construct, whereas pseudonodules from plants containing the MsENOD40–2 promoter construct stained blue. A 616-bp region at the distal 5′ end of the promoter is important for proper spatial expression of MsENOD40 in nodules and also for Nod-factor and cytokinin-induced expression.
The symbiotic interaction between leguminous plants and rhizobia results in the development on plant roots of a novel organ, the nodule, in which rhizobia provide fixed nitrogen to the host in exchange for carbon and protection. To establish this mutualistic relationship, the host plant and rhizobia must continually exchange molecular signals. Rhizobial nodulation (nod) gene expression is induced in response to different flavonoids that are excreted by plant roots or seeds (for reviews, see McKhann and Hirsch, 1994; Long, 1996). In turn, nod gene products synthesize and secrete nodulation signal molecules called nodulation factors (Nod factors), which have been identified as modified lipo-chitooligosaccharide molecules (Lerouge et al., 1990; Truchet et al., 1991; for reviews, see van Rhijn and Vanderleyden, 1995; Dénarié et al., 1996; Long, 1996). Nod factor is considered a primary morphogenetic signal for nodulation because it triggers on a compatible host the earliest stages of nodule development, including root hair deformation and curling, as well as cortical cell divisions (Lerouge et al., 1990; Spaink et al., 1991; Truchet et al., 1991). Furthermore, Nod factor elicits the expression of several early nodulin (ENOD) genes, such as ENOD12 and ENOD40 (Horvath et al., 1993; Vijn et al., 1993, 1995a; Bauer et al., 1994; Crespi et al., 1994; Journet et al., 1994; Hirsch et al., 1997). Nodulins are plant-encoded proteins that are expressed during nodule development. ENOD40 is one of the earliest nodulins to be expressed upon Rhizobium inoculation, and has been cloned from a number of legumes (Kouchi and Hata, 1993; Yang et al., 1993; Asad et al., 1994; Crespi et al., 1994; Matvienko et al., 1994; Vijn et al., 1995b; Papadopoulou et al., 1996) as well as one nonlegume, tobacco (van de Sande et al., 1996). In alfalfa (Medicago sativa) ENOD40 transcripts have been detected in dividing cells or cells that are competent to divide (Asad et al., 1994; Crespi et al., 1994).
The effects of Nod factor on legume plants can be phenocopied by modulating the balance of endogenous plant hormones. For example, applying 2,3,5-triiodobenzoic acid or NPA, both of which presumably block auxin polar transport in the root, elicits the formation of nodule-like structures (pseudonodules) on alfalfa, Afghanistan pea roots, or sweetclover sym mutants (Hirsch et al., 1989; Scheres et al., 1992; Wu et al., 1996). Transcripts for the early nodulin genes, ENOD2, ENOD8, and ENOD12, were detected in these pseudonodules. Similarly, when alfalfa roots were inoculated with R. meliloti nodulation mutants carrying the Agrobacterium tumefaciens tzs gene (encoding DMA transferase, an enzyme which is associated with trans-zeatin biosynthesis; [Beaty et al., 1986]), small, uninfected nodules developed. MsENOD2 (Cooper and Long, 1994) and MsENOD40 (Hirsch et al., 1997) transcripts were detected in these nodules. Likewise, cytokinin application triggered ENOD2 expression in Sesbania rostrata roots (Dehio and deBruijn, 1992) and ENOD2 and ENOD12A expression in alfalfa roots (Bauer et al., 1996). Furthermore, exogenous cytokinin can bypass the block of a nonnodulating alfalfa mutant, MN1008, such that both MsENOD2 and MsENOD40 were expressed in the mutant roots, although at a reduced level when compared with the wild-type alfalfa level (Hirsch et al., 1997). These results suggest that a change in endogenous hormone level and/or sensitivities may be the consequence of Nod factor perception in a responsive host plant (Hirsch and Fang, 1994).
How the change of hormone (presumably auxin/cytokinin) balance is brought about remains unknown. However, the fact that some early nodulin genes are hormone responsive may lead to the identification of specific transcription factors or cis-acting elements that are involved in regulating these genes upon Nod factor application. Thus, studying the promoter elements of MsENOD40 should help us elucidate the cis-acting region(s) responsible for its expression, as well as those necessary for induction by Nod factors and/or cytokinin. In this way we can achieve a better understanding of the signal transduction pathway in the nitrogen-fixing symbiosis, as well as plant hormone (especially cytokinin) regulation of plant genes.
In this report we cloned the promoter regions from two independent genomic clones (1a and 6c) of MsENOD40 isolated from an alfalfa genomic library. The two discrete promoters were individually fused to a reporter gene uidA (gus), and introduced into alfalfa via Agrobacterium tumefaciens-mediated transformation. We followed the spatial and temporal expression patterns of the two ENOD40 genes under both nonsymbiotic and symbiotic conditions. The expression patterns of both promoter constructs were also examined in NPA- or R. meliloti exopolysaccharide mutant-induced nodules, both of which are bacteria free. To define the cis-acting region of the MsENOD40 promoter required for its induction by Nod factor or cytokinin, a series of nested truncated promoters of the MsENOD40–1 promoter was constructed and introduced into alfalfa plants. The activities of these truncated promoters were studied in the transgenic plants.
MATERIALS AND METHODS
Isolation of MsENOD40 Genomic Clones and Sequencing Analysis
One million plaques from an alfalfa (Medicago sativa cv Chief) genomic library (Clontech, Palo Alto, CA) were screened with a 32P-radiolabeled MsENOD40 cDNA (Asad et al., 1994). Five individual plaques, 1a, 2b, 4B, 6c, and 9B′, that hybridized to the probe were isolated and the phage DNAs were purified. Two clones, 1a and 6c, were selected, and the putative promoter regions located within the HindIII fragments were subcloned into the pBluescript II KS vector (Stratagene) to generate 1a1 (approximately 2.8 kb) and 6c1 (approximately 2.6 kb), respectively. Fragment 1a1 was then designated as the MsENOD40-1 promoter and 6c1 was designated as the MsENOD40-2 promoter. A set of nested deletions from either the 5′ or 3′ end of both clones was generated using the method described by Henikoff (1984).
Both DNA strands were sequenced using the double-stranded dideoxy chain-termination method according to the manufacturer's protocol (Sequenase kit version 2, United States Biochemical). Sequence analyses were performed on a VAX/VMS computer using the University of Wisconsin Genetics Computer Group software package (Madison).
Southern Analysis
Phage DNAs were isolated according to the method described by Kernodle et al. (1993). DNA was digested with different enzymes and blotted for Southern analysis (Asad et al., 1994).
Construction of 5′-Truncated MsENOD40–1 Promoters and Other Plasmids
The HindIII-BclI fragments of 1a1 and 6c1 were individually ligated into the HindIII-BamHI site of the binary vector pBI101.3 (Clontech) to create pBI1a1–1 and pBI6c1–1, respectively. The BclI site is about 35 bp downstream of the putative TATA box of the MsENOD40 gene (Asad et al., 1994). A SpeI-BclI (approximately 2.3 kb) and a ClaI-BclI (approximately 1.6 kb) fragment from 1a1 were cloned into pBI101.3 to produce pBI1a1–2 and pBI1a1–4, respectively. The EcoRV-BclI (231 bp) fragment was used to create pBI1a1–5, which is considered to be the minimal promoter in this paper. The SpeI-ClaI fragment of 1a1 was fused at the 5′ end of pBI1a1–5 to produce pBI1a1–7, a composite promoter for MsENOD40–1, consisting of the minimal promoter and the 616-bp SpeI-ClaI fragment (see Fig. 2). All pBI plasmids were electroporated into Agrobacterium tumefaciens strain LBA4404 and then transformed into cv Regen using the alfalfa transformation and regeneration procedure described in Hirsch et al. (1995).
Figure 2.
The diagram shows the restriction map of the constructs used for alfalfa transformation. The vector pBI101.3 is a promoterless construct and is not shown here. The black boxes represent the reporter gene uidA (gus). The unshaded boxes are the regions shared by both promoters. The hatched box is the upstream region in the MsENOD40–1 promoter that is only 40% similar to the lined region in the MsENOD40–2 promoter. Hind, HindIII; Spe, SpeI; Cla, ClaI; Eco, EcoRV; Bcl, BclI; Acc, AccI; and Bam, BamHI.
Transgenic Plant Propagation and Treatments
All of the transgenic plants used for the experiments were primary transformants that were propagated vegetatively. Healthy stems were cut and rooted in a vermiculite/perlite mixture soaked with one-quarter-strength complete Hoagland plant medium. After 12 to 14 d the rooted plants were transferred into sterile 50-mL Falcon tubes containing nitrogen-free Jensen's medium and placed in a growth chamber (Conviron, Permbina, ND). The tubes were wrapped with aluminum foil to exclude light from the roots. After 6 to 7 d the plants were transferred to fresh Jensen's medium without any additives (control) or inoculated with Rhizobium meliloti strain 1021, or treated with 10−8 m PNF (NodRmIV[C16:1, S]) or 10−6 m BAP. These concentrations were determined previously to give an optimal response (Hirsch et al., 1997). Roots were harvested 4 d after treatment for GUS histochemical staining or for total protein extraction. To obtain nodules at different stages, the rooted plants were either spot inoculated (see Asad et al., 1994) or flood inoculated with R. meliloti strain 1021 (wild type) or strain 7094 (exoB::Tn5), or were treated with 10−5 m NPA. Nodules or other tissues (e.g. roots) were harvested approximately 2 to 5 weeks postinoculation.
GUS Histochemical and Colorimetric Assay
We used the method developed by Jefferson (1987) for assaying GUS activity histochemically. Plant tissues were placed in the fixing buffer (Jefferson, 1987) for 45 min, rinsed three times with 50 mm phosphate buffer, and incubated with 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid substrate at 37°C from 1 h to overnight. After staining, the tissues were rinsed in phosphate buffer and kept in 50% ethanol. Some tissues were further cleared using a 20% bleach solution under a vacuum for 15 to 30 min. The blue-stained tissues were either used directly for observation after washing in distilled water or embedded in paraffin via the TBA series (McKhann and Hirsch, 1993). If paraffin embedded, the tissues were sectioned at 15 to 25 μm and mounted onto slides for observation and photography using an Axiophot microscope (Zeiss).
To assay the enzymatic activity of GUS protein, total protein was extracted from roots 4 d following the various treatments (see above). Twenty nanograms of protein was used for the GUS assays in microtiter plates using N-nitrophenyl glucuronide as a substrate. The assay was adapted from the method described by Breyne et al. (1993). The plates were read in a Titertek plate reader (Eflab for Flow Laboratories, Finland) at a wavelength of 405 nm after a 6-h incubation at 37°C. Ten independent transgenic plants generated from each construct were tested, and each plant was assayed three times with three individual sets of plant cuttings per treatment. The colorimetric GUS assay was repeated, and the readings were averaged and plotted using Microsoft Excel and Cricket Graph programs.
RESULTS
There Are at Least Two Different MsENOD40 Genes in Alfalfa
After three rounds of screening 1,000,000 plaques, five independent phage clones were isolated from an alfalfa genomic library using the full-length MsENOD40 cDNA as a probe. Two phage clones were found to be identical to each other; this was confirmed later by sequence analysis (data not shown). The putative promoter region, which was located within the HindIII fragment in each phage clone, varied in size (data not shown). We knew that the HindIII fragment contained the potential promoter region based on our previous finding that there is a HindIII site at the very 5′ end of the MsENOD40 cDNA sequence (Asad et al., 1994). This was verified by sequencing analysis (Fig. 1). Genomic Southern analysis had suggested that one or two MsENOD40 genes were present in the alfalfa genome (Asad et al., 1994). Preliminary analysis showed that the putative promoters in phages 1a and 6c were the largest fragments, and differed in their restriction patterns (data not shown). On this basis, phages 1a and 6c were selected for further characterization.
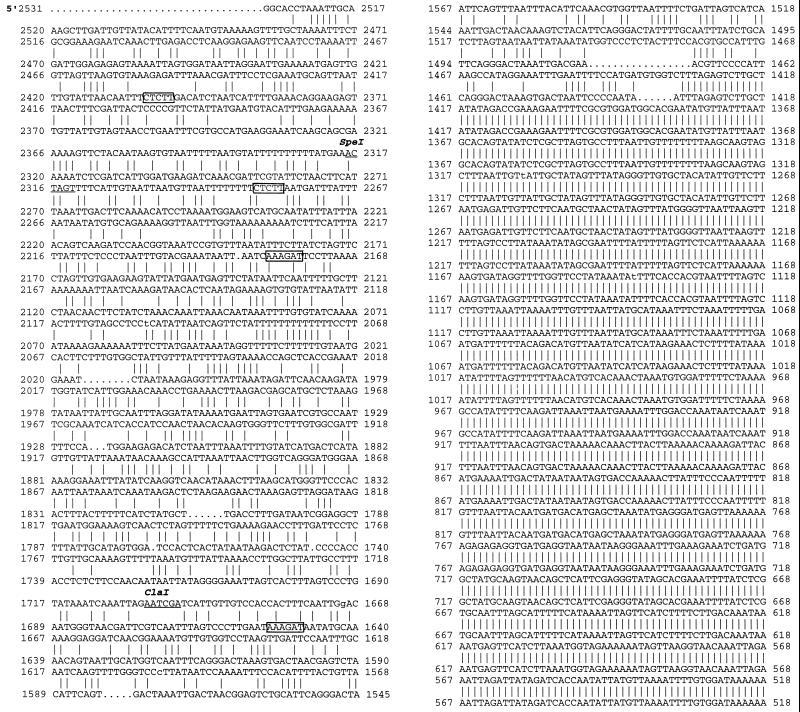
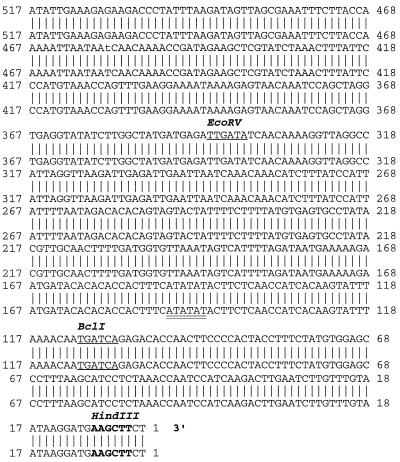
Figure 1.
Comparison of the promoter sequences of MsENOD40–1 (top sequence) and MsENOD40–2 (bottom sequence). The lines between the two sequences represent the identical nucleotides between them. Sequence analyses were performed on a VAX/VMS computer using program GAP in the University of Wisconsin Genetics Computer Group software package. Note that the first 1431 bp (1–1431) of the 3′ end were identical between the two promoters, and they are at the proximal end of the promoters (the unshaded regions in Fig. 2). The remaining 5′ distal sequences are only 40% similar to each other. Approximately 200 bp of sequence of the 5′ end of MsENOD40–1 is not shown here, but the promoter sequence of MsENOD40–2 is complete. The bold letters indicate the HindIII restriction site. The single-underlined letters are the BclI site; and the double-underlined letters indicate the putative TATA box in the promoter. The boxed regions are the putative “nodule-specific motifs.”
Sequencing analysis indicated that the 2.8-kb (1a1) HindIII fragment in clone 1a and the 2.6-kb (6c1) HindIII fragment in clone 6c most likely contain the promoters for the MsENOD40 genes. These HindIII fragments also included the first 75-bp sequence found at the 5′ end of the MsENOD40 cDNA, including the TATA box. Sequence comparison revealed that the proximal sequences of these two promoters, which span approximately 1.4 kb, were identical. However, the upstream sequences shared only 40% similarity (Fig. 1). The different restriction sites in these regions also supported this conclusion (Figs. 1 and 2). This finding suggested that these two clones (1a and 6c) were likely to represent promoters of two different MsENOD40 genes in alfalfa (Asad et al., 1994). Thus, the designation MsENOD40–1 was assigned to clone 1a1, and MsENOD40–2 was used for clone 6c1.
The Nonsymbiotic Expression of the MsENOD40 Promoters in Transgenic Alfalfa
To investigate the expression patterns of the two different MsENOD40 genes, each promoter was fused to the reporter gene uidA (gus) to produce plasmids pBI1a1–1 and pBI6c1–1, respectively (Fig. 2). The putative transcriptional start site in the promoter was determined by primer-extension experiments (data not shown); thus, these constructs were transcriptional fusions. Both constructs were subsequently introduced into cv Regen via A. tumefaciens-mediated transformation to generate transgenic plants (Hirsch et al., 1995). More than 30 individual transgenic plants generated for each construct were examined by the histochemical assay, and approximately 70% of them expressed the gus gene in nodules at locations determined previously by in situ hybridization analysis (Asad et al., 1994). On this basis, it seemed unlikely that sequences other than those located within the MsENOD40 promoter were affecting the spatial patterns of gene expression. Twenty of the 30 independent transgenic lines from each construct were selected for more detailed studies. Using Southern analysis we found that most lines contained multiple inserts of the T-DNA. However, the level of gus gene expression did not correlate with the copy number of the inserts (data not shown).
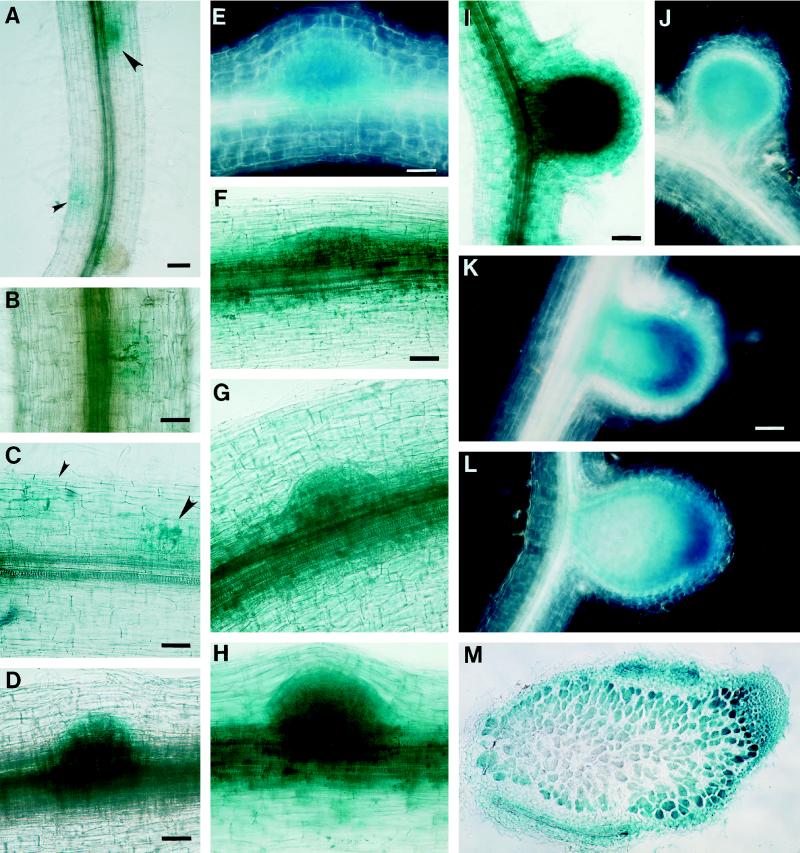
By performing histochemical staining for GUS protein (Jefferson, 1987) in various plant organs, the expression patterns of the two MsENOD40 genes were shown to differ under nonsymbiotic conditions. During the process of somatic embryogenesis, we could not detect any gus expression in callus cells or somatic embryos for either construct. In mature transgenic plants the MsENOD40–1 promoter was usually not active in uninoculated roots. For example, 16 of 20 plants showed no blue staining in roots, whereas the remaining 4 plants expressed the gus gene in the vascular tissues of the root and the stem (data not shown). Of the 20 plants, 9 of them (45%) showed the blue staining indicative of GUS protein in emerging lateral root primordia. When the lateral root emerged from the parent root, however, the blue color was no longer apparent (data not shown).
In contrast, 17 of 21 (81%) of the transgenic plants containing the MsENOD40–2 promoter-gus construct expressed gus in the root stele and in the stem procambium/phloem region even in the absence of R. meliloti (Fig. 3, A and B). Furthermore, gus expression was detected at the root tip (sometimes in the root cap) in 40% of the plants examined (Fig. 3C). About 80% of the plants studied also expressed gus in developing lateral roots (Fig. 3, D–F). The blue stain indicating GUS protein was seen in the central cells of the lateral root primordium (Fig. 3D). Upon lateral root elongation and emergence from the parent root, gus expression became restricted to the central vasculature at the proximal end of the lateral root, at the point where it was attached to the parent root; however, it remained high in the root tip (Fig. 3, E and F).
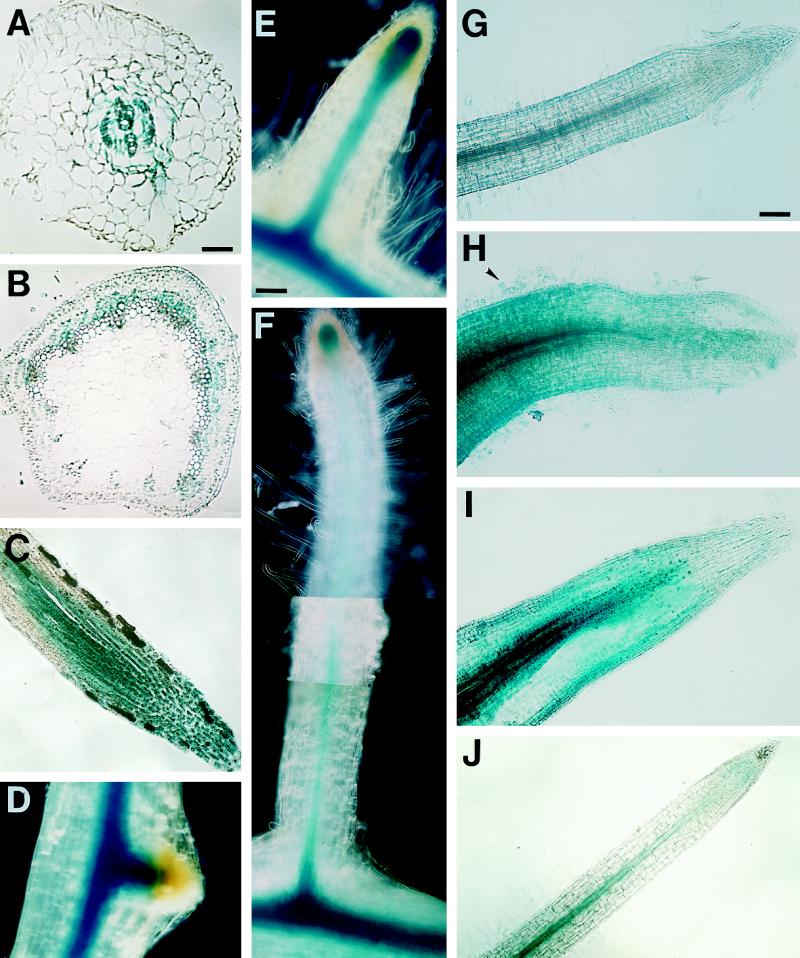
Figure 3.
The localization of GUS protein in the transgenic alfalfa plants in the absence of R. meliloti. A, Transverse section of a root of plant line A27 (MsENOD40–2). Blue color was detected in the stele, in the pericycle, endodermis, and inner cortex; bar = 55 μm. B, Cross-section of a stem of plant line A17 (MsENOD40–2). Blue color is localized to the stem procambium and the protophloem; same magnification as in C. C, Longitudinal section of a root tip of plant line A17 (MsENOD40–2). Note that the blue staining is present even in the root cap; bar = 110 μm. D, Lateral root primordium on transgenic plant line A27 (MsENOD40–2) same magnification as in C. E and F, Elongated lateral roots on the same plant shown in D; bar = 110 μm. G, Root of plant line a18 (MsENOD40–1) without any treatment; bar = 110 μm. H, Root of plant line a18 4 d after treatment with 10−6 m BAP. I, Root of plant line a18 4 d after treatment with 10−8 m PNF. J, Root of plant line A27 (MsENOD40–2) without any treatment. H through J are the same magnification as in G. K, A 10−6 m BAP-treated root of line A27 after 4 d; bar = 110 μm. L, Root of line A27 treated with 10−8 m PNF for 4 d. M, Root of line A23 treated with 10−6 m 2,4-D for 4 d; no blue color is present. L and M are the same magnification as in K. N, Nodule primordium formed on plant line a18 (MsENOD40–1) after BAP treatment for 4 d; bar = 55 μm. O, Cross-section of a primordium in N that was induced by BAP. P, Primordium formed on plant line A27 (MsENOD40–2) after BAP treatment for 4 d. Same magnification as N. Q, Nodule primordium formed on plant line A27 (MsENOD40–2) following 4 d of PNF treatment; bar = 55 μm. Arrows indicate starch grains and the arrowhead points to a root hair that is out of the plane of section.
Despite the differences between the two promoters, their combined expression patterns in transgenic alfalfa correlated with our previous findings of MsENOD40 transcript localization using in situ hybridization (Asad et al., 1994). Two new locations were detected: the root tip and the node where the leaf is attached (data not shown). Only the MsENOD40–2 promoter was active in the root tip and occasionally in the root cap of some transgenic plants (Fig. 3C). The MsENOD40–1 promoter construct was occasionally expressed in the node but not in other parts of the stem (data not shown).
The Expression of the MsENOD40 Promoters during Wild-Type Nodule Development
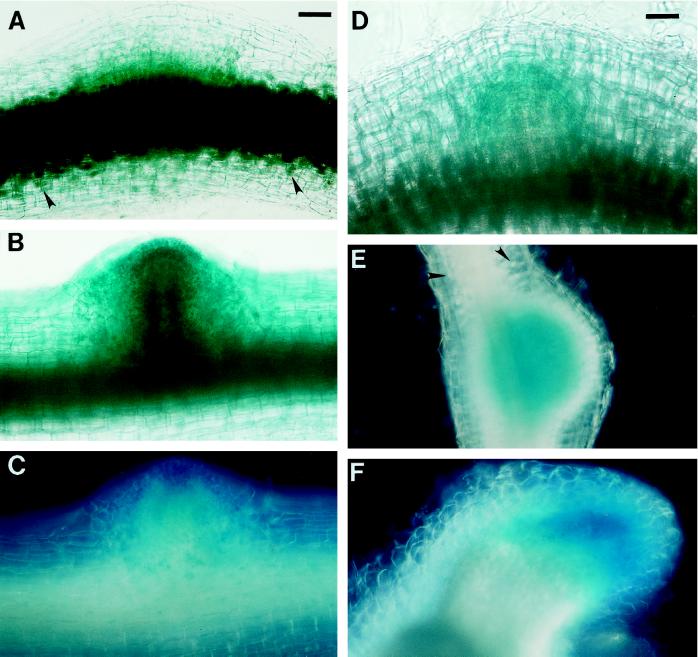
We then followed gus gene expression driven by the MsENOD40 promoter constructs in the plants inoculated with wild-type R. meliloti strain 1021. The two promoters were found to respond to R. meliloti inoculation to different extents. For the transgenic plants bearing the MsENOD40–1 promoter-gus fusion construct, less than 50% of them expressed gus in the stele upon inoculation (Fig. 4, A–E), whereas nearly 90% of the transgenic alfalfa plants containing the MsENOD40–2 promoter responded in this way (Fig. 4, F–I). However, both constructs demonstrated similar expression patterns as the nodule developed. Before root cortical cell divisions were apparent, gus expression was detected in the root outer cortex, as well as in the epidermal cells (small arrowhead in Fig. 4C). When the inner cortical cells were activated to divide, the blue color indicating GUS protein was found in the dividing inner cortical cells, as well as in the pericycle (large arrowhead in Fig. 4, A and C; Fig. 4F). Blue color was subsequently detected in all cells of the nodule primordium (Fig. 4, D, E, G, and H).
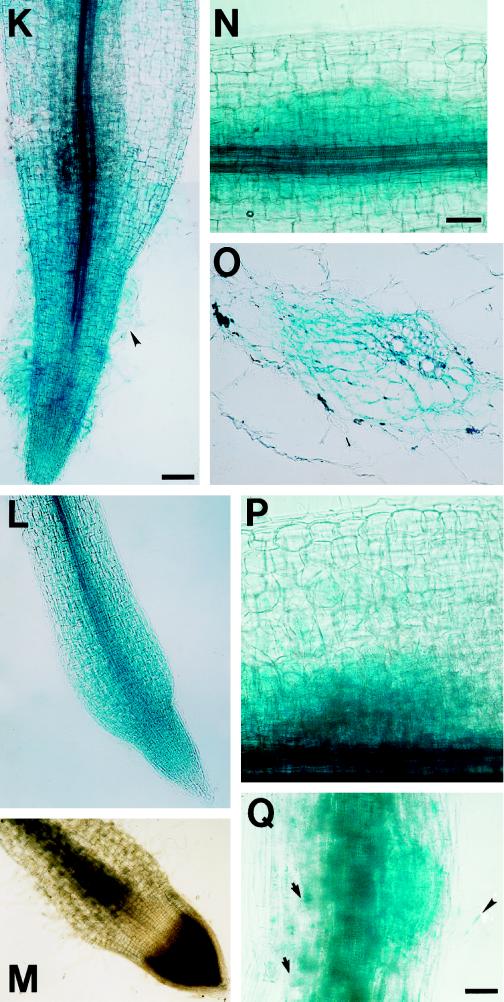
Figure 4.
The localization of GUS protein during wild-type nodule development on roots of plants containing MsENOD40–1 and MsENOD40–2 promoter-gus constructs. The roots illustrated here were flood inoculated, but each developmental stage is comparable to that found by spot inoculation. Plants shown in A through E and J through M contained the MsENOD40–1 promoter construct. Plants shown in F through I contained the MsENOD40–2 promoter construct. A, Two areas showing the blue color on a root of plant line a18. The area indicated by the small arrowhead shows the blue color mainly in the outer cortical cells and the epidermis. The area indicated by the large arrowhead shows the blue color in the dividing inner cortical cells, including the pericycle; bar = 114 μm. B, An enlargement of the area indicated by the large arrowhead in A; bar = 57 μm. C, Two adjacent areas show the blue color on a root of plant line a33 at two stages similar to A; bar = 55 μm. D, A small nodule primordium on a root of plant line a18 (cell divisions in the inner cortical cells). The blue staining of GUS protein is displayed mainly in the inner cortical cells and its derivatives as well as in the root pericycle; bar = 110 μm. E, A well-developed nodule primordium on a root of plant line a33. Same magnification as in D. F through I, Nodule primordia/nodules at different developmental stages for MsENOD40–2 transgenic plants. The blue staining for GUS protein is present in the cortex, as well as in the root stele. F was from plant line A4, whereas G through I were from plant line A27. F, Bar = 55 μm. G is the same magnification as B. H is the same magnification as F. I, Bar = 110 μm. J through L, Nodules at different stages (postprimordium) on plant line a18 or a33. J is the same magnification as I. K, Bar = 110 μm. M, Longitudinal section of a mature nodule on plant line b5–2 (containing construct pBI1a1–2). L and M are the same magnification as K. Note that the blue color is present in the nodule meristem, in peripheral infected cells, and in cells associated with the nodule vascular bundles.
Following initiation of a nodule meristem and the further development of the nodule, gus expression became localized to the nodule meristem (including the prefixing zone), to cells on the periphery of the central region, and to cells associated with the nodule vascular bundles (Fig. 4, I–L). This pattern persisted in the mature nodule (Fig. 4M). For plants bearing the MsENOD40–2 promoter construct, gus expression was much stronger. The time required to detect the appearance of the blue color was 1 to 6 h, depending on the plant line, compared with 6 to 24 h for the MsENOD40–1 construct. In addition, blue staining indicating GUS protein was detected in the root cortex following R. meliloti inoculation for the MsENOD40–2 construct (Fig. 4, F–I). The localization of the reporter gene driven by the MsENOD40 promoters in nodules formed on the transgenic plants is consistent with the in situ hybridization results found earlier (Asad et al., 1994).
For the transgenic plants containing the vector control pBI101.3, no gus expression was detected in any tissue or organ, even after incubation in the solution with the 5-bromo-4-chloro-3-indolyl-β-glucuronic acid substrate for 2 to 3 d (more than 30 plants examined). Thus, gus expression directed by the MsENOD40 promoters in the transgenic alfalfa plants was specific.
The Expression Pattern of the MsENOD40 Promoters in Bacteria-Free Nodules
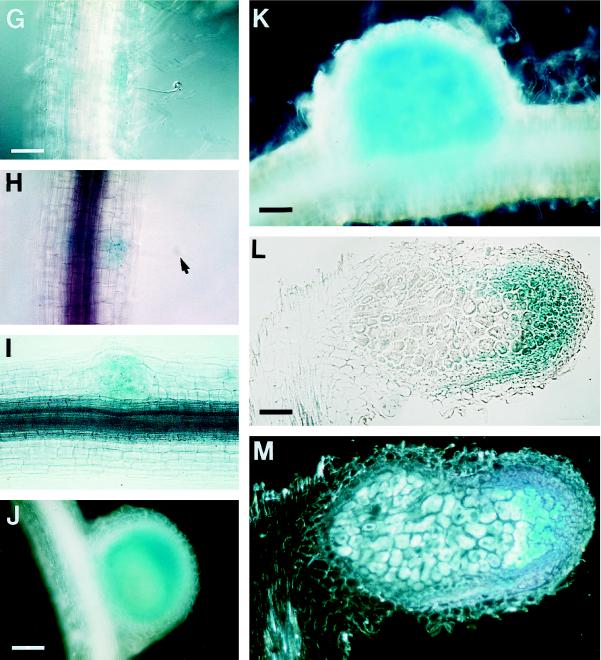
Earlier we found using northern-blot analysis that MsENOD40 is expressed in ineffective nodules elicited by NPA and R. meliloti exo mutants (Asad et al., 1994). To examine whether both promoter constructs are expressed in these bacteria-free nodules, 16 individual plants that contained either the MsENOD40–1- or the MsENOD40–2-gus construct were tested. The NPA-induced nodule-like structures gave the largest difference with respect to the expression patterns of the two promoter constructs. No gus expression was detected in the NPA-induced pseudonodules that were formed on the plants carrying the MsENOD40–1 promoter construct, unless these plants expressed the gus gene in the root stele constitutively (20% of the plants). In comparison, the majority (80%) of the plants containing the MsENOD40–2 promoter expressed gus in NPA-induced nodules. Following treatment with 10−5 m (Continued on p. 61.) NPA, gus expression was detected in the dividing cortical cells and pericycle (Fig. 5A). In an older NPA-induced pseudonodule, GUS activity was detected in the area around the central vascular bundle, as well as in the peripheral area consisting of pericycle and cortical cell derivatives (Fig. 5, B and C).
Figure 5.
GUS localization during the development of ineffective nodules (A through F) and during nodule formation on plants containing the composite promoter in pBI1a1–7 (see Fig. 2; plants with the composite promoter are represented by the letter H). A through C represent different stages of nodules induced by 10−5 m NPA on plants containing the MsENOD40–2 promoter construct (seven different lines were examined). The blue color is not only found in the nodule primordium, but also in the root cortex. A, Very young nodule primordium on plant line A27. Arrowheads indicate starch grains. B, NPA-induced nodule on plant line A32. C, Dark-field picture of B. D through F show the different stages of R. meliloti exo (strain 7094) mutant-induced nodule development (eight different lines were examined). D, Nodule primordium on plant line a18 (MsENOD40–1); bar = 55 μm. E, Well-formed nodule primordium on plant line a18 (MsENOD40–1). Arrowheads indicate starch grains. F, Mature exo nodule on plant line A27. The blue color is mainly located in the apical and peripheral area of the nodule. A through C and E through F are the same magnification; bar = 110 μm. G, An infection thread formed in a curled root hair on plant line H2. The picture was taken with Nomarski optics; bar = 92 μm. H, Bright-field picture of G, showing the blue color in the cortical cells where the infection thread has penetrated. The arrow indicates the curled root hair. I, Very young nodule primordium on plant line H35. Same magnification as A. J, Nodule primordium on plant line H2; bar = 55 μm. K, Young nodule on plant line H15; bar = 460 μm. L, Section of a mature nodule formed on plant line H25; bar = 110 μm. M, Dark-field picture of L.
In contrast, when roots of the transgenic plants containing either of the promoter-gus constructs were inoculated with R. meliloti strain 7094 (exoB::Tn5), the blue staining for gus protein was detected in the uninfected nodules that developed on the roots. Both promoters, including the composite MsENOD40–1 promoter (pBI1a1–7, Fig. 2), expressed the gus gene in a pattern similar to that of wild-type nodules, i.e. in dividing cortical cells (Fig. 5D), in the nodule primordium (Fig. 5E), and in the peripheral region that is comparable to the meristem of the wild-type nodule (Yang et al., 1992) in mature, bacteria-free nodules (Fig. 5F). In addition, we observed amyloplast deposition in roots (arrowheads in Fig. 5, A and E) following treatment with NPA or inoculation with R. meliloti strain 7094.
The Expression Patterns of the Two Different MsENOD40 Promoter Constructs in Cytokinin- or Nod Factor-Treated Roots
Earlier we found using northern-blot analysis that MsENOD40 was induced by BAP application (van Rhijn et al., 1997). Treatment with 2,4-D or other plant hormones did not elicit MsENOD40 expression (Hirsch et al., 1997; Fig. 3M). The expression patterns of the two MsENOD40 promoter constructs were observed after treating the transgenic plant roots with BAP or Nod factor (six independent plant lines for each construct were examined). For both constructs, Nod factor or cytokinin triggered similar responses. In roots without any treatment (Jensen's medium alone), GUS protein was not detected in roots of the MsENOD40–1 transgenic plants (Fig. 3G) or was seen in the root stele and root tip of the MsENOD40–2 transgenic plants (Fig. 3J). In roots treated with BAP or PNF for 4 d, blue staining indicating GUS protein was present in the root cortex and epidermal cells for both promoter constructs (Fig. 3, H, I, K, and L). In some cases, gus expression was also found in root hairs (arrowheads in Fig. 3, H and K). gus expression in the root cortex and epidermis was seen mainly in the root-elongation zone, where the root expanded laterally following BAP or PNF treatment (Fig. 3, H, I, K, and L). This change in morphology and induction of gus expression took place as early as 2 d after treatment (data not shown). In the root treated with either BAP or PNF, small nodule primordia resulting from inner cortical cell divisions developed; blue staining indicated that GUS protein was present in all the cells of the nodule primordium (Fig. 3, N–Q). Starch grain accumulation in BAP- or PNF-treated roots was also observed (arrows in Fig. 3Q).
A 616-bp Region in the MsENOD40–1 Promoter Is Essential and Sufficient for MsENOD40–1 Expression in Nodules
To define the cis-acting region in the MsENOD40 promoter that is important for its activity, a series of 5′-truncated promoters of MsENOD40–1 was constructed, fused to uidA (Fig. 2), and subsequently transformed into alfalfa. The MsENOD40–1 promoter was chosen for this study because its expression levels in roots and nodules correlated well with our previous study (Asad et al., 1994), and also because it was expressed almost exclusively under symbiotic conditions in contrast to the MsENOD40–2 promoter construct (this report). By performing the GUS histochemical assay on 30 independent lines, the promoter in pBI1a1–4 (see Fig. 2) was found to be completely inactive in any tissues or organs of the transgenic plants, including nodules (data not shown). This promoter contained the region common to both promoters. If the promoter contained the upstream region up to SpeI site (pBI1a1–2; Fig. 2), the same gus expression patterns as the full-length MsENOD40–1 promoter were detected in 30 independent lines (data not shown). The smallest construct from the MsENOD40–1 promoter (231 bp, pBI1a1–5 in Fig. 2) behaved exactly the same as the promoterless vector pBI101.3 (30 independent lines examined for each construct). These results suggest that the region between SpeI and ClaI site was required for the MsENOD40–1 promoter activity.
To test this hypothesis, a composite promoter was created by directly linking the SpeI-ClaI fragment (616 bp) to the minimal promoter in pBI1a1–5 (pBI1a1–7, see Fig. 2). This construct was introduced into alfalfa plants to produce transgenic lines. In 40% of these transgenic plants gus expression was detected in the inner and outer root cortical cells at early stages of R. meliloti infection, but was absent or below detection levels in the root stele (including the pericycle) (see Fig. 5, G and H). When the nodule primordium was formed, gus expression was found within the cells of the primordium (Fig. 5, I and K). In the nodule gus expression was detected at locations similar to those of the full-length MsENOD40–1 promoter, e.g. in the nodule meristem and in the cells associated with the nodule vascular bundle (Fig. 5, J, L, and M).
Compared with the full-length promoter, only about 40% (versus >70%) of the transgenic plants examined (11 out of 25 plants) demonstrated the expected expression patterns in nodules. Like the full-length promoter construct, the composite promoter construct was active in nodules induced by R. meliloti exo mutants, but not by NPA (data not shown). In addition, the composite promoter in pBI1a1–7 was exclusively expressed in nodules. It completely lost its activity under nonsymbiotic conditions, even in young lateral root primordia (data not shown).
The Region Responsible for Inducing the MsENOD40–1 Expression by Nod Factors and Cytokinin Is Overlapping
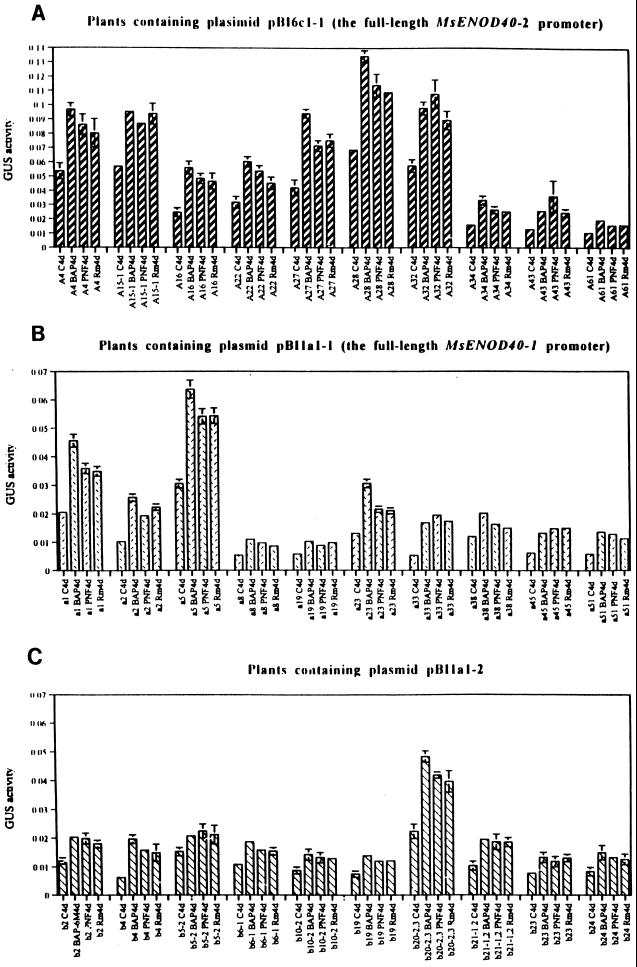
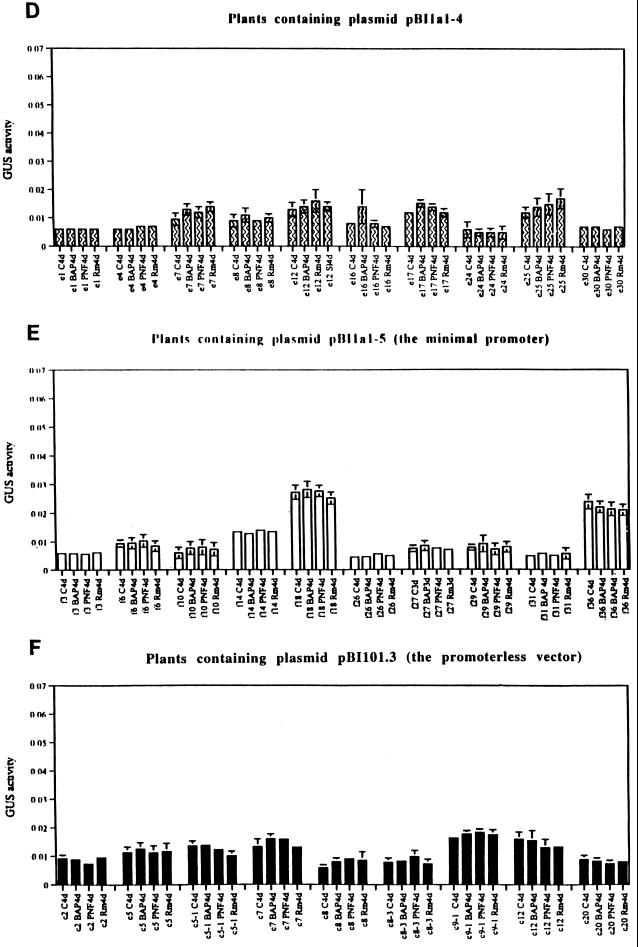
Having the truncated promoters of the MsENOD40–1, we were able to study the cis-acting region required for BAP and/or PNF induction. Ten independent transgenic alfalfa plants from each transformation with each construct, including that of the vector control, were tested. Rooted plants generated from the stem cuttings were treated with wild-type R. meliloti strain 1021, 10−6 m BAP, or 10−8 m PNF. Total protein was extracted from roots 4 d following each treatment, and GUS activity was assayed with a microtiter plate assay using N-nitrophenyl glucuronide as a colorimetric substrate (see Methods). Both promoters (MsENOD40–1 and MsENOD40–2) were enhanced 2- to 3-fold on average in roots by exogenous BAP, as well as by R. meliloti inoculation or addition of PNF (Fig. 6). Furthermore, the average basal level of the MsENOD40–2 promoter activity was measured at approximately 3-fold higher than that of the MsENOD40–1 promoter (compare the different scales in Fig. 6, A and B). When the MsENOD40–1 promoter was deleted from the 5′ end to the SpeI site (pBI1a1–2 in Fig. 2), it acted like the full-length promoter (Fig. 6C). However, for the transgenic plants containing the construct pBI1a1–4, gus expression could no longer be induced by BAP, by PNF, or by R. meliloti inoculation (Fig. 6D). The same result was obtained for the smallest promoter (pBI1a1–5) and the vector control (pBI101.3) (Fig. 6, E and F). These results suggest the 616-bp region in the MsENOD40–1 promoter is involved in regulating expression induced by either Nod factors or cytokinin.
Figure 6.
The results of GUS colorimetric assays in the transgenic alfalfa roots. A, Plants with construct pBI6c1–1. B, Plants with construct pBI1a1–1. C, Plants with construct pBI1a1–2. D, Plants with construct pBI1a1–4. E, Plants with construct pBI1a1–5. F, Plants with vector pBI101.3. GUS activity is expressed as per milligram per hour per milliliter. Plants containing the same construct are represented with the same letter, e.g. letter “A” for construct pBI6c1–1, “a” for pBI1a1–1, “b” for pBI1a1–2, “c” for pBI101.3, “e” for pBI1a1–4, and “f” for pBI1a1–5. The number following the letter represents an individual plant line; a different number represents a different transgenic plant. Plant roots were harvested after a 4-d treatment with the following: C, Jensen's medium only; 10−6 m BAP; 10−8 m PNF; Rm, wild-type R. meliloti strain 1021. The line above and below the average values represent the sd. If absent, the sds were too small to be displayed.
DISCUSSION
Here we have extended our investigations on ENOD40 gene expression in alfalfa to studying promoter-gus fusions in transgenic plants. Five independent genomic clones were isolated from an alfalfa genomic library using the MsENOD40 cDNA as a probe, and two of them were found to be identical. The promoter sequences from two distinct clones, 1a and 6c, were cloned for detailed characterizations. Sequence analyses revealed that the two promoters differed at their 5′ distal end (40% similarity), but were identical at the proximal region (1431 bp long) (Fig. 1). Based on these results and our previous Southern analysis (Asad et al., 1994), these two clones (1a and 6c) are proposed to represent two distinct ENOD40 genes in the alfalfa genome. We have designated them MsENOD40–1 and MsENOD40–2, respectively.
More than one ENOD40 gene has also been isolated from other legumes, such as soybean and French bean (Kouchi and Hata, 1993; Papadopoulou et al., 1996). A soybean GmENOD40–2 promoter-gus fusion has been introduced into a heterologous plant, vetch, and found to be expressed only in the pericycle of the nodule vascular bundle and at the region of the root where the nodule was attached (Roussis et al., 1995). However, the spatial and temporal expression patterns of the second soybean ENOD40 gene in a transgenic plant have not yet been investigated. We fused the promoters of the two MsENOD40 genes individually to gus, and subsequently transformed these constructs into alfalfa plants. By doing so, we were able to investigate the expression patterns of these two genes in a homologous transgenic plant system with or without R. meliloti inoculation.
During the process of alfalfa transformation and regeneration, no gus expression was detected in the callus cells or somatic embryos using the gus histochemical assay. When using northern analysis, we found that MsENOD40 was not expressed in alfalfa cell cultures, even after cytokinin treatment (data not shown). Therefore, we conclude that the MsENOD40 gene is probably not expressed during somatic embryogenesis or in cell cultures.
Under nonsymbiotic conditions MsENOD40–1 was usually not expressed in the uninoculated root of the transgenic alfalfa plants, but was activated when lateral roots or nodule primordia are initiated. In contrast, the expression of MsENOD40–2 is likely to be constitutive, because the promoter construct was expressed in the vascular tissues of the root and the stem even in the absence of R. meliloti (see Fig. 3). In addition, the expression of the MsENOD40–2 promoter construct was also detected at the root tip and throughout lateral root development (Fig. 3). The expression pattern of MsENOD40–2 in lateral roots is very similar to that found for PvENOD40 in bean (Papadopoulou et al., 1996), suggesting that one of the PvENOD40 genes may be a MsENOD40–2 homolog.
Using in situ-hybridization analysis (Asad et al., 1994), we detected the combined expression patterns of MsENOD40–1 and MsENOD40-2. Transcripts were localized in the stem procambium, in the root stele, in the margins of the young leaf primordia, and in emerging lateral roots (see Table I). We found two new sites of expression by using the GUS histochemical assay. Both promoter constructs were expressed in the stem node at the point of petiole attachment in some transgenic plants, a site which had not been examined previously using in situ hybridizations. Moreover, MsENOD40–2 was expressed in the root tip, including the root cap in some transgenic plants. Comparing our previous findings (Asad et al., 1994) and the results from studying the transgenic plants, the expression of the two MsENOD40 genes in alfalfa appears to be differentially regulated under nonsymbiotic conditions. Transcripts detected in the stem and in the lateral roots by the in situ method are likely to be derived from the expression of MsENOD40–2.
Table I.
Summary of the expression patterns of the two MsENOD40 genes in transgenic plants
| Location | In Situ Results | pBI6c1-1 | pBI1a1-1 | pBI1a1-2 | pBI1a1-4 | pBI1a1-5 | pBI1a1-7 | pBI101.3 |
|---|---|---|---|---|---|---|---|---|
| Root stele (uninoculated) | +/− | +(80%) | +(20%) | +(20%) | − | − | − | − |
| Stem procambium | + | +(80%) | +(20%) | +(20%) | − | − | − | − |
| Lateral root primordium | + | +(90%) | +(45%) | +(45%) | − | − | − | − |
| Root tip | +/− | +(∼50%) | − | − | − | − | − | − |
| Nodule primordium | + | + | + | + | − | − | + | − |
| Nodule meristem and vascular bundles | + | + | + | + | − | − | + | − |
| NPA-induced nodules | + | +(80%) | +(10%) | +(10%) | − | − | − | − |
| Rhizobium exo− mutant-induced nodules | + | + | + | + | − | − | + | − |
Plasmids represent the different promoter constructs shown in Figure 3. The percentage is the number of plants that demonstrated the indicated GUS localization over the total number of the plants examined. −, Not expressed; +, expressed.
When transgenic plants were inoculated with wild-type R. meliloti strain 1021, both MsENOD40–1 and MsENOD40–2 constructs were expressed in a similar fashion throughout all stages of nodule development. Both promoter constructs were activated in the cortical cells/pericycle in the preinfection and infection stages. They were subsequently expressed in the nodule primordium, and later in the nodule meristem and peripheral cells around the central tissues, as well as in the nodule vascular bundles of the mature nodule (see Fig. 4). Although MsENOD40–2 expression appears to be stronger than MsENOD40–1 (3-fold higher), we cannot distinguish between the two promoters in terms of GUS localization as the nodule develops. Also, because GUS localization correlates with our previous findings obtained by in situ hybridization (Asad et al., 1994), we could not determine whether only one or both genes are expressed during the alfalfa-R. meliloti interaction.
On the other hand, even though the two promoter constructs behaved similarly during nodule development, they are regulated differently in the NPA-induced pseudonodules. Only the MsENOD40–2 promoter construct was expressed in the nodule-like structures elicited by NPA treatment (Fig. 5, A–C). In contrast, both promoter constructs were expressed in nodules induced by R. meliloti exo mutants (Fig. 5, D–F). Part of the explanation for the differential expression may be the fact that NPA-elicited nodules morphologically and physiologically resemble lateral roots more closely than nodules, whereas R. meliloti exo mutant-induced nodules follow the wild-type nodule-developmental pathway (Yang et al., 1992; Asad et al., 1994). Thus, the differential expression patterns of the two MsENOD40s may reflect two distinct pathways for these different types of ineffective nodules. MsENOD40–1 is not usually active in the root stele or in the lateral root, and consequently it is not expressed in NPA-induced nodules. In contrast, the MsENOD40–2 gene is constitutively active in the main root or lateral roots; thus, it is expressed in NPA-elicited nodules (Fig. 5, A–C). Both genes show similar expression patterns during the formation of wild-type nodules, and consequently both genes are expressed in R. meliloti exo mutant-induced nodules (Fig. 5, D–F). These results suggest that these two MsENOD40 genes have different but complementary roles in the development of different organs.
We have found that MsENOD40 gene expression was enhanced in roots treated with either cytokinin or PNFs (Hirsch et al., 1997; van Rhijn et al., 1997). Here we confirm this observation by testing gus gene expression in roots of transgenic plants carrying either of the MsENOD40 promoters. The expression of MsENOD40–1 as well as MsENOD40–2 was induced by BAP or Nod factor treatment, and the extent of induction is similar for both genes (2- to 3-fold), although the basal level of MsENOD40–2 expression is generally higher (Fig. 6). In addition, both genes were also expressed in a similar pattern in roots following treatment with cytokinin or Nod factor for 4 d (see Fig. 3, G–Q). If both promoter constructs were induced by cytokinin or Nod factors in alfalfa, we would expect that the expression levels of MsENOD40 expression would increase at least 4- to 6-fold. However, our previous northern analysis showed that MsENOD40 was enhanced only approximately 2-fold in roots after cytokinin or Nod factor treatment. Two possible explanations are suggested. One is that only one MsENOD40 gene is induced by cytokinin or Nod factors in alfalfa. The second is that both genes are involved in cytokinin or Nod factor induction, but they are coordinately regulated so the overall level stays the same.
The differential expression patterns of MsENOD40–1 and MsENOD40–2 may reflect their different sensitivities to hormone changes or other signals. This is based on several observations. Under nonsymbiotic conditions 80% of plants (compared with 20% of the MsENOD40–1 transgenic plants) containing the MsENOD40–2-gus construct expressed gus in the root stele as well as in lateral roots. Following inoculation with R. meliloti strain 1021, MsENOD40–2 was expressed not only in nodules, but also in the root cortex; this latter location was not observed for the MsENOD40–1 construct. MsENOD40–2 expression was also detected in the root cortex after addition of either NPA or R. meliloti exo mutants. This difference between the two genes may be dependent on the 5′ upstream regions, which are only 40% similar between the two promoters.
To define the cis-acting region involved in cytokinin or Nod factor induction, a series of 5′ deletions of the MsENOD40–1 promoter was constructed and introduced into plants to study their activities. By assaying GUS enzymatic activities in roots after various treatments, a 616-bp SpeI-ClaI region in the MsENOD40–1 promoter was found to be absolutely required for its expression. This region plus the minimal promoter (the composite promoter) is sufficient to drive reporter gene expression at similar locations in the nodule as the full-length promoter (see Fig. 5). Furthermore, this region is also involved in the induction of MsENOD40–1 gene by cytokinin or by Nod factors. However, this region does not contain any known consensus sequences needed for hormonal regulation, such as auxin-responsive elements, ethylene-responsive elements, G-boxes, etc., confirming our finding that MsENOD40 is not induced by other plant hormones (Hirsch et al., 1997).
Similar to the GmENOD40-2 promoter, the two MsENOD40 promoters also contain two conserved sequence motifs, AAAGAT and CTCTT (see boxed regions in Fig. 1), that have been identified in several nodulin gene promoters (Sandal et al., 1987; Bak Ramlov et al., 1993; Miao and Verma, 1993; Roussis et al., 1995). Using deletion analysis and site-specific mutagenesis, these sequences have been found to be required for nodule-specific expression for certain genes, such as lbc3 and nodulin-26 (Stougaard et al., 1990; Bak Ramlov et al., 1993; Miao and Verma, 1993; Szcyzglowski et al., 1994). Whether these motifs determine or are responsible for nodule-specific expression of MsENOD40 will be investigated in future work.
Our studies do not clarify whether the ENOD40 gene product functions as a small peptide (van de Sande et al., 1996) or as a riboregulator (Crespi et al., 1994). However, it is very likely that the induction of expression of ENOD40 by cytokinin or other endogenous molecules upon R. meliloti inoculation could serve as an amplification mechanism thereby triggering a localized hormone imbalance, a state that initiates cell divisions in the root cortex. We should caution that we have analyzed the expression patterns of transgenes in transgenic plants in this study. Most of our results corresponded very well with our previous in situ results with few exceptions. These may reflect the difference between detecting the transcripts from the endogenous genes versus detecting the reporter gene, i.e. gus, from the transgenes introduced.
ACKNOWLEDGMENTS
This paper was written in partial fulfillment of the Ph.D. thesis of Y.F. to the Department of Molecular, Cell and Developmental Biology, University of California-Los Angeles (UCLA). We would like to acknowledge the following UCLA undergraduate students for their help in the experiments: Yousun Kim, Chrystene Ngyuen, Richard Na, John Chen, and James Shih. We are grateful to Weigang Yang for taking care of the transgenic plants in the greenhouse. We thank S.R. Long's lab (Stanford University, CA) for their generous gift of PNF and B. Bonavida's lab (UCLA) for letting us use the plate reader for the GUS colorimetric assay. We thank Margaret Kowalczyk at UCLA for preparing the color plates.
Abbreviations:
- BAP
6-benzylaminopurine
- NPA
N-1-(naphthyl)phthalamic acid
- PNF
purified Nod factor
Footnotes
LITERATURE CITED
- Asad S, Fang Y, Wycoff KL, Hirsch AM. Isolation and characterization of cDNA and genomic clones of MsENOD40: transcripts are detected in meristematic cells of alfalfa. Protoplasma. 1994;183:10–23. [Google Scholar]
- Bak Ramlov K, Bech Laursen N, Stougaard J, Marcker KA. Site-directed mutagenesis of the organ-specific element in the soybean leghaemobloin lbc3 gene promoter. Plant J. 1993;4:577–580. doi: 10.1046/j.1365-313x.1993.04030577.x. [DOI] [PubMed] [Google Scholar]
- Bauer P, Crespi MD, Szecsi J, Allison LA, Schultze M, Ratet P, Kondorosi E. Alfalfa Enod12 genes are differentially regulated during nodule development by Nod factors and Rhizobium invasion. Plant Physiol. 1994;105:585–592. doi: 10.1104/pp.105.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Ratet P, Crespi MD, Schultze M, Kondorosi A. Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and Msenod12A expression in alfalfa roots. Plant J. 1996;10:91–105. [Google Scholar]
- Beaty JS, Powell GK, Licia L, Rigier DA, MacDonald EMS, Hommes NG, Morris RO. Tzs, a nopaline Ti plasmid gene from Agrobacterium tumefaciens associated with trans-zeatin biosynthesis. Mol Gen Genet. 1986;203:274–280. [Google Scholar]
- Breyne P, De Loose M, Dedonder A, van Montagu M, Depicker A. Quantitative kinetic analysis of β-glucuronidase activities using a computer-directed microtiter plate reader. Plant Mol Biol. 1993;11:21–31. [Google Scholar]
- Cooper JB, Long SR. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell. 1994;6:215–225. doi: 10.1105/tpc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Hurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. ENOD40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C, deBruijn FJ. The early nodulin gene SrEnod2 from Sesbania rostrata is inducible by cytokinin. Plant J. 1992;2:117–128. doi: 10.1046/j.1365-313x.1992.t01-51-00999.x. [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Bhuvaneswari TV, Torrey JG, Bisseling T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc Natl Acad Sci USA. 1989;86:1244–1248. doi: 10.1073/pnas.86.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Brill LM, Lim PO, Scambray J, van Rhijn P. Steps toward defining the role of lectins in nodule development in legumes. Symbiosis. 1995;19:155–173. [Google Scholar]
- Hirsch AM, Fang Y. Plant hormones and nodulation: what's the connection? Plant Mol Biol. 1994;26:5–9. doi: 10.1007/BF00039514. [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Fang Y, Asad S, Kapulnik Y. The role of phytohormones in plant-microbe symbioses. Plant Soil. 1997;194:171–184. [Google Scholar]
- Horvath B, Heidstra R, Lados M, Moerman M, Spaink HP, Promé J-C, van Kammen A, Bisseling T. Lipooligosaccharides of Rhizobium induce infection-related early nodulin gene expression in pea root hairs. Plant J. 1993;4:727–733. doi: 10.1046/j.1365-313x.1993.04040727.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the gus gene-fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Journet EP, Pichon M, Dedieu A, de Billy F, Truchet G, Barker DG. Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J. 1994;6:241–249. doi: 10.1046/j.1365-313x.1994.6020241.x. [DOI] [PubMed] [Google Scholar]
- Kernodle SP, Cannon RE, Scandalios JG. Rapid and simple phage DNA isolation. Biotechniques. 1993;14:360–361. [PubMed] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roché P, Faucher C, Maillet F, Truchet G, Promé J-C, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M, van de Sande K, Yang WC, van Kammen A, Bisseling T, Franssen H. Comparison of soybean and pea ENOD40 cDNA clones representing genes expressed during both early and late stages of nodule development. Plant Mol Biol. 1994;26:487–493. doi: 10.1007/BF00039559. [DOI] [PubMed] [Google Scholar]
- McKhann HI, Hirsch AM. In situ localization of specific mRNAs in plant tissues. In: Glick BR, Thompson JE, editors. Methods in Plant Molecular Biology and Biotechnology. Boca Raton, FL: CRC Press; 1993. pp. 179–203. [Google Scholar]
- McKhann HI, Hirsch AM. Does Rhizobium avoid the host response? In: Dangl J, editor. Bacterial Pathogenesis in Plants and Animals. Current Topics in Microbiology and Immunology. Berlin: Springer-Verlag; 1994. pp. 132–162. [DOI] [PubMed] [Google Scholar]
- Miao GH, Verma DPS. Soybean nodulin-26 gene encoding a channel protein is expressed only in the infected cells of nodules and is regulated differently in roots of homologous and heterologous plants. Plant Cell. 1993;5:781–784. doi: 10.1105/tpc.5.7.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K, Roussis A, Katinakis P. Phaseolus ENOD40 is involved in symbiotic and non-symbiotic organogenetic processes: expression during nodule and lateral root development. Plant Mol Biol. 1996;30:403–417. doi: 10.1007/BF00049320. [DOI] [PubMed] [Google Scholar]
- Roussis A, van de Sande K, Papadopoulos K, Drenth J, Bisseling T, Franssen H, Katinakis P. Characterization of the soybean gene GmENOD40-2. J Exp Bot. 1995;46:719–724. [Google Scholar]
- Sandal NN, Bojsen K, Marcker KA. A small family of nodule specific genes from soybean. Nucleic Acids Res. 1987;15:1507–1519. doi: 10.1093/nar/15.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, McKhann HI, Zalensky A, Löbler M, Bisseling T, Hirsch AM. The PsENOD12 gene is expressed at two different sites in Afghanistan pea pseudonodules induced by auxin transport inhibitors. Plant Physiol. 1992;100:1649–1655. doi: 10.1104/pp.100.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP, Sheeley DM, van Brussel AAN, Glushka J, York WS, Tak T, Geiger O, Kennedy EP, Reinhold VN, Lugtenberg BJJ. A novel highly saturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991;354:125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- Stougaard J, Jorgensen JE, Christensen T, Kuhle A, Marcker KA. Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghaemoglobin lbc3 and N23 gene promoters. Mol Gen Genet. 1990;220:353–360. doi: 10.1007/BF00391738. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Szabados L, Fujimoto SY, Silver D, de Bruijn FJ. Site-specific mutagenesis of the nodule-infected cell expression (NICE) element and the AT-rich element ATRE-BS2* of the Sesbania rostrata leghaemoglobin glb3 promoter. Plant Cell. 1994;6:317–332. doi: 10.1105/tpc.6.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G, Roche P, Lerouge P, Vasse J, Camut S, de Billy F, Promé J-C, Dénarié J. Sulphated lipooligosaccharide signals from Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature. 1991;351:670–673. [Google Scholar]
- van de Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A, and others (1996) Modification of phytohormone response by a peptide encoded by ENOD40 of legumes and a nonlegume. Science 273: 370–373 [DOI] [PubMed]
- van Rhijn P, Fang Y, Galili S, Shaul O, Atzmon N, Wininger S, Eshed Y, Lum M, Li Y, To V and others. Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc Natl Acad Sci USA. 1997;94:5467–5472. doi: 10.1073/pnas.94.10.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijn I, das Neves L, van Kammen A, Franssen H, Bisseling T. Nod factors and nodulation in plants. Science. 1993;260:1764–1765. doi: 10.1126/science.8511583. [DOI] [PubMed] [Google Scholar]
- Vijn I, Matinez-Abarca F, Yang WC, das Neves L, van Brussel A, van Kammen A, Bisseling T. Early nodulin gene expression during Nod factor-induced processes in Vicia sativa. Plant J. 1995a;8:111–119. doi: 10.1046/j.1365-313x.1995.08010111.x. [DOI] [PubMed] [Google Scholar]
- Vijn I, Yang WC, Pallisgard N, Jensen EO, van Kammen A, Bisseling T. VsENOD5, VsENOD12 and VsENOD40 expression during Rhizobium-induced nodule formation on Vicia sativa roots. Plant Mol Biol. 1995b;28:1111–1119. doi: 10.1007/BF00032671. [DOI] [PubMed] [Google Scholar]
- Wu C, Dickstein R, Cary AJ, Norris JH. The auxin transport inhibitor N-(1-naphthyl)phthalamic acid elicits pseudonodules on nonnodulating mutants of white sweetclover. Plant Physiol. 1996;110:501–510. doi: 10.1104/pp.110.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Signer ER, Hirsch AM. Nodule initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol. 1992;98:143–151. doi: 10.1104/pp.98.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Katinakis P, Hendriks P, Smolders A, de Veries F, Spee J, van Kammen A, Bisseling T, Franssen H. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993;3:573–585. doi: 10.1046/j.1365-313x.1993.03040573.x. [DOI] [PubMed] [Google Scholar]